Graphical abstract
Keywords: COVID-19, Machine learning, Datasets, X-Ray, Imaging, Review, Bias, Confounding
MSC: 62H35, 68T45, 92C55, 92C60
Abstract
Computer-aided-diagnosis and stratification of COVID-19 based on chest X-ray suffers from weak bias assessment and limited quality-control. Undetected bias induced by inappropriate use of datasets, and improper consideration of confounders prevents the translation of prediction models into clinical practice. By adopting established tools for model evaluation to the task of evaluating datasets, this study provides a systematic appraisal of publicly available COVID-19 chest X-ray datasets, determining their potential use and evaluating potential sources of bias. Only 9 out of more than a hundred identified datasets met at least the criteria for proper assessment of risk of bias and could be analysed in detail. Remarkably most of the datasets utilised in 201 papers published in peer-reviewed journals, are not among these 9 datasets, thus leading to models with high risk of bias. This raises concerns about the suitability of such models for clinical use. This systematic review highlights the limited description of datasets employed for modelling and aids researchers to select the most suitable datasets for their task.
1. Introduction
Since the end of 2019, the novel coronavirus SARS-CoV-2 gained worldwide attention and eventually developed to the global COVID-19 pandemic in early 2020 (Sohrabi et al., 2020). Reliable diagnosis and stratification play a vital role in the management of cases and the allocation of potentially limited resources, like intensive care unit (ICU) beds. Hence, there is an urgent necessity to create trustworthy tools for diagnosis and prognosis of the disease. While most of the people with COVID-19 infection do not develop pneumonia (Cleverley et al., 2020), the early identification of COVID-19 induced pneumonia cases is essential.
To this end, imaging modalities such as planar X-ray, computed tomography (CT) and sometimes ultrasound are employed. When assessing the different pros and cons of the imaging modalities, the global scale of the COVID-19 pandemic and the need for imaging also in countries with less developed healthcare systems impose additional constraints to consider (cf. Greenspan et al., 2020).
Ultrasound is a very interesting non-invasive complementary modality described to assess lung damage. Some COVID-19 ultrasound datasets have been released recently (Born, Wiedemann, Cossio, Buhre, Brändle, Leidermann, Aujayeb, Moor, Rieck, Borgwardt, 2021, Born, Wiedemann, Cossio, Buhre, Brändle, Leidermann, Aujayeb, 2021, Almeida, Bilbao, Ruby, Rominger, López-De-Ipiña, Dahl, ElKaffas, Sanabria, 2020). However, ultrasound needs medical specialist expertise to be carried out and the amount of publicly available data is still limited. Therefore, CT and Chest X-ray (CXR) might be considered the primary modalities in COVID-19 imaging.
CT yields the highest diagnostic sensitivity, however, it is very complex to apply in an intensive care unit setting, and expected to be frequently inaccessible for patients in less developed health care systems. In contrast, CXR yields lower diagnostic sensitivity but is a widely available, fast, minimal-invasive, and relatively cheap tool to diagnose and monitor COVID-19 induced pneumonia (Aljondi and Alghamdi, 2020). CXR is easy to apply, being applicable even in anaesthetised patients receiving intensive care treatment using portable scanners and expected to be much more widely available in less developed healthcare systems than any other imaging method. Additionally, the nature of the data - individual 2D images - is resulting in much easier data management compared to the image stacks of CT or the (arbitrary angle) sequences of ultrasound. As a result, very large public datasets are available for CXR and the present work prioritised their analysis.
1.1. Motivation
Machine learning, and in particular deep learning methods, promise to assist medical staff in coherent diagnosis and interpretation of images (Choy, Khalilzadeh, Michalski, Do, Samir, Pianykh, Geis, Pandharipande, Brink, Dreyer, 2018, McBee, Awan, Colucci, Ghobadi, Kadom, Kansagra, Tridandapani, Auffermann, 2018). A remarkable amount of machine learning models has been proposed in a very short amount of time to tackle the problems of COVID-19 diagnosis, quantification and stratification from X-Ray imaging (Shoeibi, Khodatars, Alizadehsani, Ghassemi, Jafari, Moridian, Khadem, Sadeghi, Hussain, Zare, et al., Islam, Karray, Alhajj, Zeng, et al., Ilyas, Rehman, Naït-Ali).
However, there is a growing awareness in the community that the presence of different sources of bias significantly decreases the overall generalisation ability of the models, leading to overestimated model performance reported in internal validation compared to evaluation on independent test data (Soneson, Gerster, Delorenzi, 2014, Cohen, Hashir, Brooks, Bertrand, Zech, Badgeley, Liu, Costa, Titano, Oermann, 2018, Maguolo, Nanni). In addition, numerous journal editorials are calling for better development, evaluation and reporting practices of machine learning models aimed for clinical application (Mateen, Liley, Denniston, Holmes, Vollmer, 2020, Nagendran, Chen, Lovejoy, Gordon, Komorowski, Harvey, Topol, Ioannidis, Collins, Maruthappu, 2020, Campbell, Lee, Abrmoff, Keane, Ting, Lum, Chiang, 2020, Health, 2020, O’Reilly-Shah, Gentry, Walters, Zivot, Anderson, Tighe, 2020, Health, 2019, Stevens, Mortazavi, Deo, Curtis, Kao, 2020). Underneath, there are growing concerns about ethics and the risk of harmful outcomes of using AI in medical applications (Campolo, Sanfilippo, Whittaker, Crawford, 2018, Geis, Brady, Wu, Spencer, Ranschaert, Jaremko, Langer, Borondy Kitts, Birch, Shields, van den Hoven van Genderen, Kotter, Wawira Gichoya, Cook, Morgan, Tang, Safdar, Kohli, 2019, Brady, Neri, 2020).
To avoid or at least be able to detect potential bias, it is important that datasets and models are well documented. Some aspects of dataset building, such as criteria for subject inclusion and exclusion, recruitment method, patterns in missing data, and many more, may influence model accuracy and introduce bias in prediction models. Among the most common sources of bias are unknown confounders and selection bias (Steyerberg, 2009, Griffith, Morris, Tudball, Herbert, Mancano, Pike, Sharp, Sterne, Palmer, Davey Smith, Tilling, Zuccolo, Davies, Hemani, 2020, Greenland, Robins, Pearl, 1999, Heckman, 1979). In both cases, the presence of a spurious association between predictors and outcomes might be learned by the model, leading to undetected overfitting resulting in models not capable of generalizing and eventually failing transportation to clinical application (see Fig. 1 and Wolff et al. (2019)).
Fig. 1.
Importance of identified meta-information during model development. (a) Given a dataset with unidentified composition of the dataset population, there is a high risk of bias, i.e. a model is systematically prejudiced to faulty assumptions. (b) For example in an extreme case almost all of the control cases form a special sub-population of young age. With knowledge on the dataset age composition one is at least aware that any model developed with this dataset has a high risk of being biased by age (Model 1) or can even choose a model mitigating the age influence (Model 2). (c) Biased models are very likely to lead to impaired performance in the target population hampering generalizability.
1.2. Tools for model evaluation and information extraction
In the last years, several guidelines and checklists to evaluate prediction model quality, risk of bias and transparency, depending on model characteristics, have been proposed. An essential requirement to apply such tools is access to detailed dataset documentation.
Model evaluation
PROBAST (Prediction model Risk Of Bias Assessment Tool, Wolff et al. (2019) was developed to evaluate the risk of bias and the applicability to the intended population and setting of diagnostic and prognostic prediction model studies. TRIPOD (Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis, Moons et al. (2015) aims to improve reporting and understanding of prediction model studies. A third example of a quality assessment tool, in this case specifically designed for machine learning and artificial intelligence research, is given by a set of 20 critical questions proposed in Vollmer et al. (2020), to account for transparency, reproducibility, ethics, and effectiveness (TREE). These tools require answering specific questions about participant selection criteria and setting, its numbers, information about predictors and outcomes, and whether all of these choices were appropriate for the model intended use.
Information extraction
The CHARMS checklist (Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies, Moons et al. (2014) was designed as a data extraction tool for systematic review of prediction modelling studies, including machine learning models. It is not specifically designed to evaluate datasets, however a large part of its items are devoted to extract information from data used in the studies. The BIAS checklist (Biomedical Image Analysis Challenges, Maier-Hein et al., 2020) is intended to improve the transparency of reporting biomedical image analysis challenges regardless of application field, image modality or task category. The main aim of the initiative is to standardise and facilitate the review process to raise the interpretability and the reproducibility of the results of biomedical challenges by better reporting, as a potential solution to fully exploit their potential and maximise their capacity to move forward in the field (Maier-Hein et al., 2018). Up to our knowledge, there are no other tools, protocols or statements, exclusively designed for dataset evaluation.
1.3. State-of-the-Art
In a recent review and critical appraisal of prediction models for diagnosis and prognosis of COVID-19 (Wynants et al., 2020), all evaluated models were rated at high risk of bias. The authors concluded that they “do not recommend any of these reported prediction models for use in current practice”. For the subset of diagnostic models based on medical imaging two main causes for high risk of bias were identified: 1) a lack of information to assess selection bias (such as how controls were selected or which subset of patients underwent imaging); and 2) a lack of clear reporting of image annotation procedures and quality control measures. Similar conclusions were obtained in another publication specifically addressing machine learning models using chest X-ray and CT images (Roberts et al., 2021). They found a high or unclear risk of bias in all studies and that the reported results were extremely optimistic, mainly due to limitation in the datasets or combination of datasets used.
1.4. Aim & contribution
Given the previously described disappointing state-of-the-art, we hypothesise that the current main obstacle towards building clinically applicable machine learning models for COVID-19 is not the machine learning techniques per se, but instead access to reliable training data that, on the one hand captures the problem complexity, but on the other does not induce undetected bias to the models.
Therefore, there is a need to raise awareness of such problems and to aid modellers to efficiently find the right dataset for their particular problem, supporting the creation of robust models. Consequently, this paper focuses on data instead of models, giving an overview of current publicly available COVID-19 chest X-ray datasets and identifying strengths and limitations, including the most evident potential sources of bias.
We systematically evaluated the quality of COVID-19 chest X-ray datasets and their utility for training prediction models using an adapted version of the CHARMS tool and an adapted version of BIAS. Dataset quality is measured by the amount and the detail in the description of the dataset variables and of the dataset building process. Model designers need this information to evaluate the risk of bias and the generalizability, for example, using tools such as PROBAST, TRIPOD or TREE. The utility is determined by the data structure (e.g. cross-sectional vs longitudinal), the available potential outcomes (e.g. image annotations or survival information), and the amount of dataset information provided (e.g. sample size and information on missing data).
Thus, this paper provides a more in-depth description of datasets than previous works (Shuja, Alanazi, Alasmary, Alashaikh, 2020, Sohan, Shoeibi, Khodatars, Alizadehsani, Ghassemi, Jafari, Moridian, Khadem, Sadeghi, Hussain, Zare, et al., Islam, Karray, Alhajj, Zeng, et al., Ilyas, Rehman, Naït-Ali), that have focused on surveying papers describing methods, and on identifying the datasets used to train these methods, without assessing the quality or utility of the datasets.
1.5. Paper structure
In Section 2 the methodology used to systematic search for datasets and evaluate their potential for clinical prediction models is given. Next, in Section 3, the datasets selected for review are analysed in detail on their provided information content to assess the risk of bias. Moreover, in order to put this review in the context of the current model development scenario, an analysis on dataset usage in published papers was conducted and a general overview and discussion of the most commonly used datasets is given. In Section 4 the insights from this analysis are discussed. Finally, some recommendations for researchers aiming at clinical prediction model building are given in Section 5.
2. Methods
In this section, the search strategy for datasets is explained, the eligibility criteria are discussed, and subsequently the application of information extraction tools is described.
2.1. Dataset search, eligibility & selection
To identify publicly available COVID-19 X-ray datasets which are sufficiently documented to asses their risk of bias, two search strategies were combined: a direct search for datasets using a specialised dataset search engine, and an indirect search for datasets by screening in published papers for dataset references. To this end the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses, (Moher et al., 2015) statement was adapted to the given special situation, where the final objects of interest are datasets instead of studies (see Fig. 2 ). Therefore, two separate PRISMA flows from identification to eligibility, for the direct and indirect case, were introduced.
Fig. 2.
Adapted PRISMA workflow for the analysis of COVID-19 X-ray datasets. Boxes in blue indicate papers from the additional indirect search for papers using datasets, from which datasets (yellow/green) were extracted. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The identification step of the direct approach consisted of querying the google research dataset search 1 with “covid” & “x-ray”. For the indirect approach PubMed, medRxiv and arXiv were queried for any combination of {“covid”, “covid-19”}, {“x-ray”} and {“dataset”, “data set”, “machine learning”, “deep learning”}. The search was restricted to the time interval between 1st of January 2020 to 31st of March 2021.
Next, in the PRISMA screening step, all papers obtained in the indirect approach were excluded if they were not concerned with COVID-19, the main text was not written in English or they did not contain any reference to a chest X-ray dataset. From the remaining papers all references to X-ray datasets were extracted and assigned as an annotation to the paper record. Datasets with no clear origin were tagged as not identified and those that were not openly available (at least by simple registration) were marked as private. All other datasets were labelled with an identifier and, if not yet previously encountered, added to a running list of identified publicly available datasets. In the end this procedure yielded the set of all unique dataset references encountered.
Datasets from the direct search approach were excluded if they were not (publicly) available or did not contain any chest X-ray data. The remaining datasets were de-duplicated to yield a second set of unique chest X-ray datasets.
In the eligibility step of both the direct and indirect approach the extracted datasets were investigated in further detail and excluded if they did not qualify for an in-depth risk of bias analysis of COVID-19 X-ray datasets. First of all, this excludes all datasets which do not contain any real COVID-19 related cases, excluding non-COVID and synthetic datasets.
Furthermore, to be able to address questions on the risk of bias, a sufficient documentation of the dataset origin and population composition has to be available (see Section 1.2). Any dataset, from which one cannot reconstruct the collection procedure of the samples must be deemed of high risk of bias per se. Thus, all datasets containing no further information except the images themselves (no info), or solely the origin of their collection (no meta) were excluded. Additionally, to be able to coherently address questions on patient selection/enrolment criteria and outcome definition, all datasets that were not collected under the same protocol but are merely a collection of case reports (case collection) were also excluded.
Finally all datasets which are copies, modifications or aggregations (remix) of other datasets were excluded. They carry at least the same risk of bias as their underlying primary source datasets, and thus their minimal risk can be derived from the analysis of primary datasets included in the analysis. Additionally, merged dataset carry a special risk of being confounded by dataset identity (DeGrave, Janizek, Lee, 2020, Garcia Santa Cruz, Sölter, Bossa, Husch, 2020) and thus have to be deemed at high risk of bias if not treated accordingly (see also Section 3.3.3).
The full code and annotation data (including a list with the discarded datasets) to reproduce the extraction process is made publicly available2 .
2.2. Dataset information extraction
Due to the lack of specific tools to evaluate datasets for their suitably to train reliable models, two different information extraction tools, the CHARMS checklist and the BIAS tool (see Section 1.2), were adapted and employed to conduct an in-deep analysis of the selected sets.
2.2.1. CHARMS
The following domains from the CHAMRS checklist were used for dataset evaluation: data source, participant description, outcomes, predictors, sample size and missing data. The domains about model (development, performance and evaluation), results and interpretation were omitted. The detailed definition of each item can be found in the Supplementary material 1, Table S1.
The information on Participant description, including recruitment method, inclusion and exclusion criteria, is needed to determine the applicability and generalizability of the model, whether the study population is representative of the target population, and to discard the presence of selection mechanisms that can introduce bias. Information about received treatments could be relevant if they affect the outcome of prognostic models.
The Outcomes to be predicted will depend on the purpose of the intended model, i.e. whether it is a prognostic or a diagnostic model. Radiological findings, lesion segmentations and differential diagnosis could be suitable outcomes for diagnostic models, when they are measured close in time to the image acquisition. When there are multiple images from the same subject acquired at different time-points (longitudinal data), a prognostic model could be trained, where features from the later image are predicted from the earlier one. Time to death or discharge, or whether ICU, supplementary oxygen or other life support treatments were needed, could also be used in prognostic models.
Outcomes based on image findings could sometimes be determined a posteriori (e.g. with a post-hoc annotation by a radiologist), however a precise definition of them is essential to describe model applicability. It is also important to specify whether outcomes were obtained blinded to predictors and/or the other way round, because this affects the causal model assumption and the strategies to mitigate bias (see Castro et al., 2020). Furthermore, model performance could be overestimated in the absence of blinding, in particular when the outcome require subjective interpretation, as is the case for radiological diagnosis (Moons et al., 2014).
The main predictor considered here is the X-ray lung image, however other measurements could be incorporated (additionally) into prediction models. Details of the image acquisition protocol and acquisition device description are important as they could be a significant source of confounding when merging images from different sources. In addition, the model performance could be reduced if applied to images acquired using a different setting than in the training set.
Finally, a large enough sample size and the amount and treatment of missing data are highly relevant to avoid overfitting and confounding, respectively (Moons et al., 2019).
Appropriate sample size will depend on several aspects of the model development process, such as the number of predictors, its preprocessing and the magnitude of the effect to be predicted. For example, a model using as predictors the volume of a lung lesion and a few other prespecified scalar biomarkers may need much less training data than a neural network system using the whole image, because the latter model includes several orders of magnitude more of learnable parameters.
2.2.2. BIAS
Despite not been a dataset review tool, the BIAS checklist provides specific questions about dataset origin, purpose, distribution and intended use which are considered highly valuable for our in-depth dataset analysis.
The adapted version of BIAS checklist employed in this work is given in (Supplementary material 1, Table S2). For further reading refer to the Appendix A: BIAS Reporting Guideline of Maier-Hein et al. (2020). Similarly to CHARMS, only items related to datasets were kept.
2.3. Analysis of the dataset use frequency
To analyse the dataset usage pattern in peer-reviewed publications, the result of the dataset screening was further evaluated for the subset of PubMed papers. In the screening procedure the records of these papers got annotated with their dataset reference (see Section 2.1). Additionally, the date of the article was extracted from the <MedlineCitation><Article><ArticleDate> meta data tag.3 When this date was not speficied the <MedlineCitation><Article><Journal><JournalIssue><PubDate> was used instead.
3. Results
This section is structured as follows: First, the results of the adapted PRISMA search are presented and then the extracted eligible datasets are analysed in detail. Afterwards, an analysis of dataset usage frequency is presented and the most popular datasets not eligible for the detailed analysis are briefly described, with special emphasis on their risk of bias.
3.1. Dataset search & selection
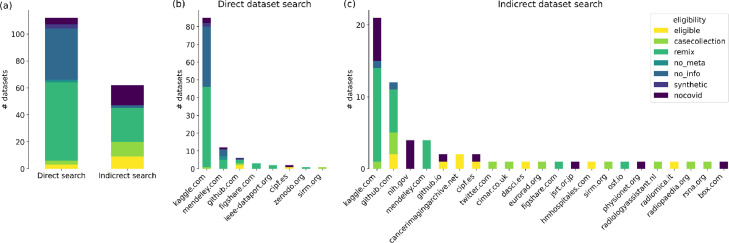
The dataset extraction process consisted of two complementary approaches: A direct search for datasets using the google research dataset search and an indirect search of dataset references in published peer-reviewed papers on PubMed and in pre-prints on medRxiv and arXiv. The direct search revealed 112 unique COVID-19 related X-ray datasets, from which only three were eligible for a risk of bias analysis (Fig. 3 a). Most of them were deemed non-eligible for being either remixes of other datasets or providing no meta-information. A further breakdown of dataset origin by top-level domain (Fig. 3b) reveals that the overwhelming majority of these poorly documented or remixed datasets is hosted on kaggle.com.
Fig. 3.
Identified datasets and their eligibility classification (a) While the direct search identified more datasets overall, the indirect search for referenced datasets in papers yielded more eligible datasets. (b) Most of the papers found in the direct search were sourced from www.kaggle.com and either contained no accompanying information at all or were remixes of other datasets. (c) Likewise, the most common origin of dataset references in papers is kaggle.com, but many eligible datasets have been found on dedicated medical imaging websites.
Conversely, the indirect search identified only about half of the potentially relevant datasets (64), but in turn, nine of them (including all three from the direct search) met the criteria for further analysis (Fig. 3a).
Additional to the original primary release of the data, the indirect search revealed two re-annotations, i.e. expert post-hoc annotations of previously released datasets. Notably, almost none of the datasets in the papers were among the worst with no documentation at all (see “noinfo” label in Fig. 3). Nevertheless, many paper utilise remix datasets sourced from kaggle.com and github.com (Fig. 3c).
3.2. Availability of metadata for risk of bias assessment
The selected datasets contain different degree of additional information. On the lower end of the spectrum are ACTUALMED, HM HOSPITALES and ML HANNOVER which include only a short description and mainly provide additional data in tabular form. All other datasets are described in more detail in an accompanying paper. Notably, three of them deal exclusively with the dataset itself (RICORD, COVID-19-AR, BIMCV-COVID19), whereas all other papers are a combination of dataset and modeling description. Based on the provided information, in the following sections the datasets are analysed in detail using the dataset-adapted CHARMS and BIAS tools.
3.2.1. CHARMS analysis
In the CHARMS analysis all primary datasets are evaluated for their information content with respect to sample size, participant data, outcome and predictor information. Additionally, the two re-annotation datasets (CARING and AR-OPC) are analysed with respect to outcome information only, as they share the participant data and predictor information with their primary datasets. Analysis of the adapted CHARMS items is depicted in Tables 2 and 3 . Additionally, an extended version of the answers can be found in Supplementary Material 2.
Table 1.
Key definitions.
| Target population: Set of people with certain common characteristics (disease, age, localisation) for whom the model is aimed to be applied. |
| Predictors: Independent variables or inputs of the prediction system. It is assumed that they are always available at the time of prediction. |
| Outcome: The dependent variable or output of the prediction system. |
| Bias: Systematic error that leads to distorted estimates of the models predictive performance. |
| Selection bias: A.k.a. collider bias, happens when some samples are more likely to be selected than others, making the sample not representative of the population. |
| Generalizability: Capacity of a model to correctly predict unseen data from the same population as the training sample. Can be determined with internal validation. |
| Transportability: Measure of the extent to which a predictive model performs well across different populations. Can be determined with external validation. |
| Confounder: Variable that has an influence in both, the predictor and the outcome. The presence of uncontrolled confounders leads to spurious associations hampering generalizability and transportability. |
Table 2.
CHARMS analysis - Part I (Datasets 1–5).
| Domain | Items Datasets: | BRIXIA a | ML HANNOVER b | COVID-19-AR c | HM HOSPITALES d | ACTUALMED e | |||
|---|---|---|---|---|---|---|---|---|---|
| Participant data | Eligibility recruitment | Yes | Unclear | Yes | Yes | No | |||
| Participant description | Age, sex, location, study setting | Sex | Age, sex, race, location, weight, height, comorbidities | Age, sex | No | ||||
| Treatment info | No | No | No | Yes | No | ||||
| Study dates | Yes | No | No | Yes | No | ||||
| Country | Italy | Germany | USA | Spain | Unknown | ||||
| Outcome information | Candidate outcomes | radiological severity | clinical outcome | clinical outcome | radiological diagnosis | AR-OPCf radiological diagnosis | clinical outcome | radiological diagnosis | |
| Train | Test | ||||||||
| Labels | 4 severity level at 6 locations | ICU, survival | ICU, survival | flowing text | segmented opacity | ICU, survival, discharge | normal, covid uncertain | ||
| # of labelled images | 4553 | 150 | - | - | 256 | 221 | - | 238 | |
| Method definition | Yes | Yes | - | - | No | Yes | - | No | |
| Outcome timing | Yes | Yes | Yes | No | Yes | Yes | Yes | - | |
| Diagnosis / Prognosis | Both | Both | Prog. | Prog. | Both | Both | Prog. | Diag. | |
| Predictor information | Candidate predictors | DICOM header | view, modality, laboratory data, vital signs | DICOM header, comorbidities | DICOM header, medications, laboratory data, vital signs | view | |||
| Method definition | Yes | No | Yes | Yes | No | ||||
| Measurement timing | Yes | Yes | Yes | Yes | - | ||||
| Sample size | # subjects / images | 2351/4703 | 71/243 | 105/256 | 2307/2310 | 215/238 | |||
Brixia score COVID-19 dataset (Signoroni et al., 2020) (https://brixia.github.io/).
COVID-19 Image Repository (https://github.com/ml-workgroup/covid-19-image-repository).
COVID-19-AR,(Desai et al., 2020), (https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=70226443).
Covid Data Save Lives (https://www.hmhospitales.com/coronavirus/covid-data-save-lives/english-version).
AR-OPC, external annotation (Tang et al., 2020) (https://github.com/haimingt/opacity_segmentation_covid_chest_X_ray).
Table 3.
CHARMS analysis - Part II (Datasets 6–9).
| Domain | Items Datasets: | BIMCV-COVID19 a | COVIDGR b | RICORD c | AIforCOVID d | ||||
|---|---|---|---|---|---|---|---|---|---|
| Participant data | Eligibility recruitment | Yes | Unclear | Yes | Yes | ||||
| Participant description | Age and sex | No | Age, sex, testing method | Sex, age, symptoms, comorbidities | |||||
| Treatment info | No | No | No | Yes | |||||
| Study dates | Yes | No | No | No | |||||
| Country | Spain | Spain | Turkey, USA, Canada, Brazil | Italy | |||||
| Outcome information | Candidate outcomes | radiological diagnosis | diagnostic test | CARINGe radiological diagnosis | diagostic test | radiological severity | radiological diagnosis | radiological severity | clinical outcome |
| Labels | radiological findings (336) | PCR, IgG, IgM f | radiogical findings (22) + BBoxes | PCR | normal, mild, moderate, severe | typical, uncertain atypical, negative | mild, moderate, severe | mild, severe, death | |
| Labelled images | 23k | – | 1749 | – | 426 | 998 | 998 | – | |
| Method definition | Yes | Yes | Yes | Yes | Yes | Yes | Yes | ||
| Outcome timing | – | – | – | No | - | Yes | No | ||
| Diagnosis/Prognosis | Both | Diag. | Both | Diag. | Both | Both | Prog. | ||
| Predictor information | Candidate predictors | DICOM header | DICOM header | DICOM header, comorbidities, medications, laboratory data, vital signs. | |||||
| Method definition | – | - | - | Yes | |||||
| Measurement timing | Yes | - | Yes | No | |||||
| Sample size | # subjects / images | 4706/16840(COVID+) 3238/6540 (COVID-) | 426/426 (COVID+) 426/426 (Control -) | 361/998 | 983/983 | ||||
BIMCV-COVID19, Valencian Region Medical ImageBank (Vayá et al., 2020)) (https://bimcv.cipf.es/bimcv-projects/bimcv-covid19/).
RICORD (Tsai et al., 2021)(https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=80969742).
AIforCOVID (Soda et al., 2020),(https://aiforcovid.radiomica.it).
CARING, external annotation (Mittal et al., 2021) (https://osf.io/b35xu/).
Immunoglobulin G (IgG) and Immunoglobulin M(IgM).
Participants. In general, there was limited information about participants (see Tables 2 and 3, Domain: Participant data). Some datasets (6/9) provided a clear, albeit very brief, description of the eligibility and recruitment method (See Description column of “Eligibility/recruitment” rows in Supplementary Material 2). Most of the datasets included participant data such as sex (7/9) and age (6/9), but only one of them provided other relevant information, such as height, weight and race, and another one included clinical symptoms. Only two included information about comorbidities, which might be particular relevant for prognostic models for COVID-19, given the strong evidence of interactions between comorbidities and worse outcome (Yang et al., 2020). In general, the participant description is too scarce to asses if the training population is representative of a potential target population, hampering the applicability of the models. It is also difficult to determine if there are hidden selection mechanisms, including inclusion and exclusion criteria, that could be a source of strong confounding and limit generalizability and transportability of developed models. Only two datasets provide information about treatment, which could be another serious limitation for prognostic models, in particular taking into account that this is a new disease and different experimental treatments might have been applied in each country or hospital unit.
Candidate Outcomes. Across and within datasets a variety of potential outcome variables are available (see Tables 2 and 3, Domain: Outcome information). They can be useful for diagnostic or prognostic models, depending on whether the variable can be assessed at the time of image acquisition or whether one has to wait some time for the variable to change. Diagnostic outcomes might be either obtained by means of a diagnostic test independent of the image (e.g. a Polymerase chain reaction test, PCR) or derived from the image through direct image interpretation by a trained doctor. Such radiological annotations by doctors range from simple labels on the presence of COVID-19 (ACTUALMED, RICORD), over quantitatively rated severity of COVID-19 (RICORD, BRIXIA, COVIDGR) to detailed labelling of particular radiolgical findings, like consolidations or ground glass patterns (BIMCV). All radiological labels can either be globally attributed to the image or localised at certain parts of the image. Such localisation range from roughly defined areas according to anatomical landmarks (BRIXIA), over annotated bounding boxes (BBoxes) in the image (CARING) to pixelwise segmentation (AR-OPC).
Typical prognostic variables are clinical outcomes like ICU admission and survival (ML HANNOVER, COVID-19-AR and HM HOSPITALES). But also quantitative diagnostic outcomes, like severity, might be used for prognostic models if they are available longitudinally.
In general, a lack of image derived annotations is not a critical issue because these could be obtained post-hoc by independent radiologists. This is, for example, the case for the COVID-AR and BIMCV-COVID19 datasets, for which independent post-hoc annotations are provided in AR-OPC and CARING, respectively. However, if researchers are going to use image annotations provided with the dataset, a precise definition and method description is needed. In particular, definitions and methods for image annotations are completely missing for the ACUTALMED dataset.
Candidate predictors. The number of predictors, in addition to X-ray scans and demographic variables, vary widely between the datasets (see Table 2 & 3, Domain: Predictor information). Two datasets have no additional potential predictors apart from image, view and demographics.
The DICOM (Digital Imaging and Communications in Medicine standard) header is included as candidate predictor because it may contain potentially useful information (Mustra et al., 2008). Most datasets (6/9) include images in DICOM format. The other 3 are ML HANNOVER, which used NIfTI (Neuroimaging Informatics Technology Initiative) format for privacy reasons, and COVIDGR and ACTUALMED, that provide only post-processed.jpg and.png, respectively.
Except for the evident cases, it was difficult to assess whether the predictors are blinded to the outcomes, and whether the outcomes are blinded to the predictors. For example, when the outcomes are some kind of radiological annotation, the former are clearly not blinded to the latter. On the other hand, when the outcome is the patient survival, the predictor (i.e. the image) is blinded to the outcome. Other combinations are more subtle and a careful protocol description is needed to determine blinding. For example, we can not guaranty that the date of ICU admission is not affected by the X-ray image which, after all, is a diagnosis tool that help to make such decisions. Therefore, blinding is not reported for being either trivial or difficult to assess in the studied datasets.
Sample size. The sample size is critical for clinical prediction models based on medical images, because of the high risk of overfitting due to the high dimensionality of the input.
Lack of large enough sample sizes is a common issue in all medical applications, but COVID-19 data is especially scarce. The sample size of the reviewed datasets ranges from nearly 8k subjects and more 23k images (BIMCV-COVID19) to only 71 participants and 243 images (ML HANNOVER). Two datasets include 2.3k subjects, another two less than 1.000 and the other two 361 and 105 subjects.
3.2.2. BIAS analysis
Comprehensive and standardised reporting of datasets is key to address questions of generalizability and transportability in models. The coverage of essential reporting elements is evaluated using the adapted BIAS tool (see Table 4 ). Additionally, an extended version of the answers can be found in Supplementary Material 3. While some questions have a full or very high coverage, such as the description of a principal objective (8/9), general image information (9/9) official website or associated platform (9/9) or regional origin of the data (8/9), others have poor coverage. Information on potential limitations is available for only 3 of the 9 datasets. Information on whether the population distribution of the dataset matches with the general or expected population is even more scarce, available in only 2 datasets. In general, the coverage for potential error sources is very low, only 3 datasets acknowledge potential error sources and none of them has a proper quantification of such errors.
Table 4.
BIAS analysis of the nine selected datasets: Using an adapted version of BIAS tools the quality of dataset description was evaluated.
| BRIXIA | ML HANNOVER | COVID-19-AR | HM HOSPITALES | ACTUALMED | BIMCV-COVID 19 | COVIDGR | RICORD | AIforCOVID | Coverage (%) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Principal objective | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | 88 |
| Team description | Yes | No | Yes | No | No | Yes | Yes | Yes | Yes | 66 |
| Oficial website or platform | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 100 |
| Open or on request | On request | Open | Open | On request | Open | On request | Open | Open | On request | 100 (55) |
| Include potential limitations | Yes | No | No | No | No | No | No | Yes | Yes | 33 |
| Ethical approval | Yes | No | No | No | No | Yes | Yes | Yes | Yes | 55 |
| Usage agreement specification | Yes | Yes | Yes | Yes | No | Yes | No | Yes | Yes | 77 |
| General image information | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 100 |
| General patient information | Yes | Yes | Yes | Yes | No | Yes | No | Yes | Yes | 77 |
| Acquisition device | Yes | No | Yes | Yes | No | Yes | No | Yes | Yes | 66 |
| Acquisition protocol | Yes | No | Yes | Yes | No | Yes | No | Yes | Yes | 66 |
| Post-procesing tecniques | Yes | No | Yes | Yes | No | Yes | Yes | Yes | Yes | 77 |
| Regional origin of data | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | 88 |
| Whole or training/test division | Whole * | Whole | Whole | Whole | Whole | Whole | Whole | Whole | Whole | 100 (Whole) |
| Different population distribution explanation | No | No | Yes | No | No | No | No | No | Yes | 22 |
| Description of annotation methods | Yes | No | Yes | No | No | Yes | Some | Yes | Yes | 66(55) |
| If human annotations, description | Yes | – | – | – | – | Yes | No | Yes | – | 75% When apply |
| If multiple annotations merge, description | Yes | – | – | – | – | – | – | Yes | – | 100% When apply |
| Indication of potential error sources | Yes | No | No | No | No | Yes | No | No | Yes | 33 |
| Quantification of potential error sources | Some | No | No | No | No | Some | No | No | Some | 33(0) |
*The dataset was realised as a whole, but the annotation method for training and test differ, hence in other parts of the paper training and test subsets are analysed separately.
Since the scope of the present work focused on publicly available datasets, we categorise their openness as ‘On request’, when some registration is needed, and ‘Open’, when the access to the dataset is straightforward. The access to specific information about the image, such as acquisition device (6/9), acquisition protocol (6/9) and post-processing techniques (7/9), has a medium-high coverage. Importantly, datasets sharing images in DICOM format were more likely to provide these information, as it could partially be derived from the DICOM headers.
All datasets were released “as a whole” in contrast to situations where datasets are distributed split into train and test subsets, and the access to the test set labels may be limited (as it is typical for challenges). However, the BRIXIA dataset constitutes a special case, using two different methods to generate “train” and “test” labels, i.e. all data have label annotations (whole), but a “test” subset has labels generated with another method. The property description of the annotation methods is partially covered, with the datasets ML HANNOVER, HM HOSPITALES and ACTUALMED lacking such description. Importantly, these datasets also lack a corresponding paper describing the dataset.
3.3. Usage of COVID-19 datasets in peer-reviewed papers
Within the dataset extraction procedure 201 peer-reviewed papers that reference COVID-19 X-ray datasets have been identified on PubMed. A temporal breakdown on publication date shows an steady increase in publication numbers per month since the first paper appeared in March 2020 (Fig. 4 ).
Fig. 4.
Usage pattern of datasets employed in peer-reviewed papers (PubMed). Top-Panel: Number of COVID-19 X-Ray imaging papers per month and total number of referenced datasets therein. Left-Panel: Percentage of all screened papers (201) that reference at least one dataset of an exclusion category (hollow bars) and the percentage of those that reference a specific dataset (full bars). All datasets with less than 5 mentions are sub-summed in the “other” category except all eligible ones (in bold). Central-Panel: Temporal evolution of the dataset reference count by month.
Since all these paper passed the quality control mechanism of peer revision, it is interesting to see how the dataset usage in these papers relates to our analysis on the datasets’ risk of bias. Intriguingly, all the datasets meeting inclusion criteria for analysis in the previous section are rarely employed, with only the least well documented dataset, ACTUALMED, having more than 2 references (Fig. 4). This might be partly explained by the fact that all these dataset got mentioned for their first time not before November 2020 and thus it is taking time for the community to recognise and incorporate them.
Thus, most papers published so far relied on one or more datasets excluded from the analysis. To better understand the implication of this usage pattern, a short overview on the most frequently used datasets (more than 4 references) and their risk of bias is presented in the following, categorised according to their exclusion criterion.
3.3.1. Non-COVID-19 datasets
60% of all papers employ datasets built before the COVID-19 pandemic. Either for pre-training or, more commonly, to enrich training data with non-COVID-19 controls. A short summary of these datasets is given in the supplementary material (Supplementary Material 1, Table S3) and more comprehensive overviews are provided in the recent reviews of Sogancioglu et al. (2021) and Garcia Santa Cruz et al. (2020). In general, these datasets are much larger and better curated than those for COVID-19. For example, there exists a quite exemplary datasheet for the CheXpert dataset (Garbin et al., 2021).
But even if the datasets themselves are at lower risk of bias, each of them reflects a certain sub-population. Thus, combining them with COVID-19 datasets always introduces the risk of confounding by dataset peculiarities. For example, the most frequently used non-COVID dataset, Guangzhou pediatric (Kermany et al., 2018), is composed of paediatric patients (aged 1–5 years) in Guangzhou, China. It is commonly used in combination with COVID-19 datasets representing an adult population outside of China. Models trained on such combinations are at high risk of being confounded by age, for example.
3.3.2. Case collections
70% of all papers utilise a case collection dataset. These datasets consists of cases published on various websites to facilitate knowledge transfer between radiologists. Usually, these are websites of radiological associations like sirm.org and radiopedia.org, but sometimes also more peculiar sites like a radiologist feed on twitter.com 4 .
It is worth emphasising that case collections are originally made public with educational reasons in mind, and not for training prediction models. Thus, they provide no clear protocol of subject enrolment and are not representative of any defined population, but are a selection of cases deemed interesting. Thus, for any model trained and/or validated on them it is unclear how this model performs on a population in an actual clinical setting. Several initiatives like Cohen/IEEE8023 (Cohen et al., 2020b), kaggle/andrewmd 5 and agchung/Figure 1 6 collect case collections and provide them in a structured dataset format (Fig. 5 ). Among those, the Cohen/IEEE8023 dataset is by far the most popular COVID-19 dataset and is referenced in 68% of all papers. It was one of the first available datasets (already in April 2020), it is easily accessible through cloning a Github repository, and it provides a well maintained meta-data table. However, none of the meta-data variables can account for the aforementioned potential inherent selection bias of case collections, due to their selection for educational purposes of fellow radiologists.
Fig. 5.
Overview on the relationships of popular COVID-19 case collections, remixes and non-COVID datasets. Grey boxes represent datasets which can be downloaded as one entity. Green boxes indicate primary COVID-19 datasets meeting the inclusion criteria for review. Transparent boxes indicate data sources not directly available as downloadable datasets. Diamond shaped symbols indicate that the attached source dataset is (partially) included in an aggregated dataset. Dotted lines describe the case when images of a source dataset are included in an aggregated dataset but labels have been re-annotated. The colouring of the circle in the upper right corner of each datasets indicates the approximate proportion of COVID-19 patients (yellow) and control subjects (gray). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3.3. Remix datasets
Remix datasets are referenced in about one third of the papers. We use the term Remix to refer to aggregations of datasets being redistributed as a new dataset. The aggregated and then redistributed source datasets are often primary datasets, but can also be other remixes or even case collections, forming a potentially very complex aggregation hierarchy (see Fig. 5).
The most frequent examples of theses kind are the kaggle/tawsifurrahman (Chowdhury et al., 2020) and the COVIDx dataset (Linda Wang and Wong, 2020) which combine different sources of COVID-19 and non-COVID cases (Fig. 5).
Whereas at first glance it seems convenient to obtain such datasets as compilations instead of separate parts, the aggregation obfuscates the individual story of each dataset and the associated risk of bias as well. In the case of the kaggle/tawsifurrahman dataset the most obvious risk is induced again (cf. Section 3.3.1) by mixing in a pediatric population from the Guangzhou pediatric dataset as control cases.
But the most general risk to all such compilations is confounding by dataset identity. This effect was for example clearly demonstrated for the COVIDx dataset by means of an external test set in Robinson et al. (2021) and more general for different COVID-19 related dataset merges in Ahmed, Goldgof, Paul, Goldgof, Hall, 2021, Ahmed, Hall, Goldgof, Goldgof, Paul.
Another evident risk are case duplications caused by the intertwined aggregation process illustrated in Fig. 5. This can lead to duplicate cases in train and test data as demonstrated in Garcia Santa Cruz et al. (2020) and thus severely biased evaluation scores (data leakage). This risk is even increased by further dataset merging of modellers when no attention is paid to the dataset aggregation hierarchy. For example the kaggle/bachrr 7 dataset is occasionally mixed with the Cohen/IEEE8023 dataset (Albahli, Albattah, 2020, Ismael, Şengür, 2020) despite providing the same collection of cases (Fig. 5). In another example, Gomes et al. (2020) use the Cohen/IEEE8023 dataset for training and ML HANNOVER for validation, despite ML HANNOVER being a subset of the former (see Fig. 5).
3.3.4. Private datasets
Private datasets are employed in 17% of the investigated papers. These datasets are typically sourced from a single hospital or a regional hospital association and thus they represent a very specific population. Some models are purely based on such a private dataset (e.g. Shamout, Shen, Wu, Kaku, Park, Makino, Jastrzȩbski, Witowski, Wang, Zhang, et al., 2021, Castiglioni, Ippolito, Interlenghi, Monti, Salvatore, Schiaffino, Polidori, Gandola, Messa, Sardanelli, 2021, Xia, Chen, Ren, Zhao, Wang, Jin, Zhou, Wang, Yan, Zhang, et al., 2021). But many authors utilise small private dataset together with larger public datasets. An exemplary case is the use of private datasets as external test data, therewith assessing the transportability of models trained on (merged) public datasets to the private test data population and thus to the underlying hospital setting (Park, Kim, Oh, Seo, Lee, Kim, Moon, Lim, Ye, Kim, Park, Oh, Seo, Lee, Kim, Moon, Lim, Ye, Elgendi, Nasir, Tang, Smith, Grenier, Batte, Spieler, Leslie, Menon, Fletcher, et al., 2021, Robinson, Trivedi, Blazes, Ortiz, Desbiens, Gupta, Dodhia, Bhatraju, Liles, Lee, et al., 2021).
4. Discussion
This work highlights a widespread problem in prediction models for medical image analysis. While the problem of overfitting is commonly acknowledged when dealing with a small number of images, other sources of bias such as confounders and selection bias are not as frequently considered. This is evident by the careless use of datasets, where critical questions about the population, such as recruitment procedures, inclusion and exclusion criteria, or outcome measurement procedures, are not addressed. Some authors (Cohen et al., 2020b) have already acknowledged that many datasets do not represent the real-world distribution of cases, that the presence of selection bias is highly probable (particularly on case study collections), and therefore that clinical claims must take into account these limitations.
However, the first step to tackle these issues is to have a good description of datasets. Only then strategies to reduce the bias can be implemented and model limitations, including the range of applicability, can be acknowledged.
Unknown confounders and collider bias are not as problematic in prediction models as they are in causal inference (Griffith, Morris, Tudball, Herbert, Mancano, Pike, Sharp, Sterne, Palmer, Davey Smith, Tilling, Zuccolo, Davies, Hemani, 2020, Wynants, Van Calster, Collins, Riley, Heinze, Schuit, Bonten, Dahly, Damen, Debray, de Jong, De Vos, Dhiman, Haller, Harhay, Henckaerts, Heus, Kammer, Kreuzberger, Lohmann, Luijken, Ma, Martin, McLernon, Andaur Navarro, Reitsma, Sergeant, Shi, Skoetz, Smits, Snell, Sperrin, Spijker, Steyerberg, Takada, Tzoulaki, van Kuijk, van Bussel, van der Horst, van Royen, Verbakel, Wallisch, Wilkinson, Wolff, Hooft, Moons, van Smeden, 2020). However, model generalizability is compromised and its prediction power can only be maintained when training and target population remain similar and are subject to the same sampling mechanism. Even in this particular case, specifying the optimal target population cannot be done without knowing the training population characteristics.
Recently, there have been some efforts to address the general problem of bias in AI, in particular regarding the use of human data. In Mitchell et al. (2019), for example, authors encourage transparent model reporting and propose a framework to describe many aspects of model building, including dataset description. Other tools in progress, such as datasheets for datasets, propose systematic documentation of datasets (Gebru et al., 2018).
General considerations about clinical prediction models (Steyerberg, 2009) are as relevant in complex AI models as in simple linear regression, however they are much more difficult to address in the former case. To this end, several protocols for AI model development are being developed (Collins, Moons, 2019, Liu, Faes, Calvert, Denniston, 2019, Faes, Liu, Wagner, Fu, Balaskas, Sim, Bachmann, Keane, Denniston, 2020, Stevens, Mortazavi, Deo, Curtis, Kao, 2020).
4.1. Bias in medical imaging models
Medical imaging based machine learning models, in particular Convolutional Neural Networks (CNNs), are known not only to learn underlying diagnostic features, but also to exploit confounding image information. For example, it was shown that the acquisition site, regarding both the hospital and the specific department within a hospital, can be predicted with very high accuracy () (Zech et al., 2018). Furthermore, CNN model were able to identify the source dataset with a high accuracy () solely from the image border region i.e. image parts containing no pathology information at all (Maguolo and Nanni, 2020). If disease prevalence is associated with the acquisition site, as it is often the case, this can induce a strong confounder. Thus, in any aggregated dataset composed from originally separated sub-datasets for COVID-19 and control cases, the dataset identity is fully confounded with the group label. Therefore, it is difficult to isolate the disease effect from dataset effect, making the desired learning almost impossible and posing a high risk of overestimating prediction performance. Indeed it has been observed that, by training on different COVID-19 and non-COVID-19 dataset combinations, the “deep model specialises not in recognising COVID features, but in learning the common features [of the specific datasets]” (Tartaglione et al., 2020), i.e. “these models likely made diagnoses based on confounding factors such as [...] image processing artifacts, rather than medically relevant information” (Ahmed et al., 2021b).
Besides features of the acquisition site, the demographic characteristics of the populations can also yield a strong confounder. An example are remix datasets that merged adult COVID-19 subjects with non-COVID-19 controls from the Guang-zhou pediatric dataset (from age 1–5 years old), posing the very high risk that models will associate anatomical features of age with the diagnosis.
However, difficulties are not limited to aggregated datasets, but also single-source datasets are not free of potential confounders and other sources of bias. The classical example is a different imaging protocol depending on the patient’s health status. For example, the PA prone protocol (posterior-anterior, standing in front of the detector) is the preferred imaging setup for lung X-ray in general. However, if the patient is bed-bound, as its common in severely ill COVID-19 cases, the clinical staff is forced to carry out AP supine imaging (anterior-posterior, while laying on the back), using a portable scanner. As a result, a naive machine learning system may associate PA imaging with better outcome by fitting to features induced by the confounders.
Another confounding factor might be the presence of medical devices like ventilation equipment or Electrocardiogram (ECG) cables, which allow a model to associate images with patient treatment instead of disease status. For example, for the NIH ChestXray dataset, a critical evaluation has shown that “in the pneumothorax class, [...] 80% of the positive cases have chest drains. In these examples, there were often no other features of pneumothorax” (Oakden-Rayner, 2020). Datasets that provide additional annotations on the presence of medical devices (e.g. BIMCV-COVID19 facilitate a risk analysis on this confounding effect and also enable mitigation strategies in training.
In general, one has to distinguish between labels that have been annotated by taking only the image itself into account and labels that have been generated by a different source, i.e. from another diagnostic method like CT or PCR. Unfortunately, radiological reports done in clinical routine are a mixture of both. Radiologists are often aware of the patients clinical context (and thus “not blinded to the predictors”). This extra information is reflected in the reports, especially because they are done to communicate information between different doctors. For example, it has been shown for the NIH ChestXray dataset that, in a substantial fraction of images, the associated finding extracted from the reports can not be confirmed by a post-hoc assessment of the images alone (Oakden-Rayner, 2020).
Bias arises more easily when the intended application of the prediction model is not clearly defined. For example, if the model objective is to find radiological manifestation of the disease in the images, that are not necessarily apparent to the radiologist naked eye, the labels should be generated by the best possible diagnostic test that does not rely on imaging information from the same modality. For instance, a perfectly valid goal could be to determine whether a feature observed in CT, but not visible in XR, could still be detected by an ML model. In contrast, if the goal is to reproduce radiological findings (for example, to save radiologist time) the label should be radiological annotations assessed by an independent clinician that has no information except for the image (i.e. is “blinded to the predictors”). Otherwise, the risk of bias increases significantly and the generalisation ability is compromised because we can not understand where the key information is coming from, what the model is learning, and what the possible sources of bias are. In this sense, it is worth noting that RICORD, BRIXIA test and the COVIDGR datasets do provide such annotations solely derived from the images. Furthermore, as such image-based annotation is independent of the collection process itself, such annotations could be independently created post-hoc like in the case of BIMCV-COVID19 and COVIDGR with AR-OPC and CARING, respectively.
All in all, datasets with an inherently high risk of inducing bias might still be useful for training models if applied appropriately. First, in contrast to classical statistical or standard machine learning methods, deep learning models are highly complex systems that may include several building steps and include auxiliary training tasks.
Quality standards for datasets used to (pre-)train these building blocks may not necessarily be as high as the ones for evaluating the final model. Some of these datasets that are deemed “close to useless” for training a serious medical diagnostic tool may be perfectly appropriate for pre-training and auxiliary tasks.
In this case, it is important that model performance metrics are reported also for an appropriate external test dataset, which can reveal any model bias especially concerning the transportability to the intended application domain. Ideally, such an external test dataset should be in the public domain to enable reliable benchmarking.
Report of external validation results is also important if mitigation strategies for known sources of bias are applied. Such strategies are for example re-balancing or re-weighting of outcome prevalence for each of the key demographic variables (Jiang, Nachum, Amini, Soleimany, Schwarting, Bhatia, Rus, 2019). Another mitigation strategy, special to deep learning, is the adversarial training of models to explicitly ignore confounding variables (Zhao et al., 2020). This approach is a conceptual extension of adversarial domain adaption (Ganin et al., 2016) which has been shown to mitigate some of the dataset identity confounding in models trained on the COVIDx dataset (Robinson et al., 2021). Nevertheless, for all mitigation strategies to work, at least the confounders have to be documented. Furthermore, there is, to the best knowledge of the authors, no mitigation strategy in the extreme case of complete confounding, e.g. in the previously mentioned examples of dataset mixtures of only adult COVID-19 samples combined with only paediatric control cases.
4.2. Advice for modellers
To avoid the risk of inducing bias by inappropriate use of datasets it is important that researchers follow transparent practices and adequate reporting guidelines. To assess whether the chosen datasets are appropriate for the intended use, it is advisable that researchers address the following:
-
•
Be very careful when relying on remix datasets. Merging subjects from different datasets should be done directly from primary data sources, ensuring that potential sources of bias can be transparently evaluated and are not hidden in the data aggregation hierarchy. Furthermore, accidental double inclusions are ruled out by this procedure. The selected images from each dataset should be listed (e.g. as supplementary material), including the respective subjects available information. The reasoning behind the inclusion of subjects from particular datasets and their specific characteristics should be explained in the context of the intended use of the model.
-
•
Ask oneself which population is represented by the datasets, i.e. which were the recruitment procedures, location and setting, the inclusion and exclusion criteria, and subjects demographics. They should also address how exactly the outcome was obtained and how is related to the disease and the application.
-
•
Explain how the model can be applied to a clinical setting, which is the benefit for the patient or how it would help medical personal to make the decision.
-
•
The strategies followed to evaluate and mitigate the potential biases should be explained.
4.3. Limitations of this review
We acknowledge several limitations of this study. While this work is focused on the extensive field of X-ray imaging for the aforementioned reasons, related modalities useful for the diagnosis of respiratory diseases using machine learning models, such as CT and ultrasound, are not covered. Additionally, the missing values of both, the predictors and the outcomes, are not analysed in detail, although they are reported in Supplementary Material 2. Finally, we have insisted that availability of detailed dataset documentation is important to reduce the risk of bias in models trained with such datasets. However, we haven’t addressed the conflicting goals of extensive dataset documentation for risk of bias assessment on one side, and patient privacy rights, including compliance with data protection regulations such as General Data Protection Regulation (GDPR), on the other.
Taking into account that medical data includes highly sensitive information, this opens an additional important field of problems to study.
5. Conclusion
This work presents a first attempt to systematically evaluate X-Ray imaging datasets in terms of their utility to train COVID-19 predictions models. We followed the PRISMA guidelines to systematically search for X-ray chest images databases of COVID-19 subjects, either screening papers reporting models where these images are used (indirect search) or directly searching for datasets using a dataset search engine. Inspired by the PROBAST, TRIPOD and TREE statements, this work aimed to answer whether the available COVID-19 X-ray datasets could be used to train or validate clinical prediction models with a low risk of bias. With this objective in mind, the CHARMS and BIAS checklists were adapted to extract the relevant information about participants, outcomes, predictors and sample size.
The information provided in almost all extracted datasets is too scarce to guarantee that a model can be built with a low risk of bias. For example, key questions regarding participant information and their appropriateness for a given application can not be answered. This finding is consistent with results presented in a systematic methodological review of Machine learning for COVID-19 prediction models using chest X-rays and CT scans (Roberts et al., 2021), where PROBAST assessment rated all X-ray-based models as having a high or unclear risk of bias in the Participant domain. Hence, claims about efficacy could be highly biased, and generalizability and transportability are uncertain. Applicability to clinical settings is therefore extremely risky and not recommended.
With time passing by, more and better documented datasets potentially less prone to induce model bias are becoming publicly available. New models should be developed and older models evaluated using these datasets. So far, very few prediction models used these promising datasets (see Fig. 4).
In general, datasets owners should make an effort to improve the documentation about the whole dataset building process to increase the dataset value and the quality of models trained on them. For example, there should be a clear statement of dataset intended use and explicit warning of common misuse cases. Label definition and generating procedure should be reported in detail, so that other researchers can verify the accuracy of label assignments and evaluate the utility and adequacy to the problem at hand. Finally, datasets should contain cohort characteristics and subject selection criteria information. This is important to evaluate the risk of selection bias and to check if the training and target population features similar characteristics.
This review should help modellers to efficiently choose the appropriate datasets for their modelling needs and to raise awareness of biases to look out for while training models. It is also encouraged that everyone validates models by reporting benchmark results on a very well curated publicly available external dataset, which is carefully selected to represent the real clinical use case as close as possible.
Although dataset quality is arguably the most important requirement for building a medical diagnostic system, other aspects of the model building process are also prone to biases. Following the TRIPOD reporting guideline, answering the critical questions of TREE, and assessing the risk of bias with the PROBAST tool (Parikh, Teeple, Navathe, 2019, Sounderajah, Ashrafian, Aggarwal, De Fauw, Denniston, Greaves, Karthikesalingam, King, Liu, Markar, McInnes, Panch, Pearson-Stuttard, Ting, Golub, Moher, Bossuyt, Darzi, 2020) could be a promising starting point. However, extensions of these established guidelines are required to be fully applicable to deep learning systems (Wynants et al., 2020). Efforts are already being done in this direction: extension of TRIPOD (TRIPOD-AI, Collins and Moons, 2019) and CONSORT-AI/SPIRIT-AI (Liu et al., 2019) are currently being developed, focused on model validation and clinical trials, respectively. Recent considerations for critically appraising ML studies are given in Faes et al. (2020), and reporting recommendations can be found in Stevens et al. (2020).
CRediT authorship contribution statement
Beatriz Garcia Santa Cruz: Conceptualization, Methodology, Validation, Visualization, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing. Matías Nicolás Bossa: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing. Jan Sölter: Conceptualization, Methodology, Validation, Software, Visualization, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing. Andreas Dominik Husch: Conceptualization, Methodology, Validation, Formal analysis, Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Luxembourg National Research Fund (FNR) COVID-19/2020-1/14702831/AICovIX/Husch. Beatriz Garcia Santa Cruz is supported by the FNR within the PARK-QC DTU (PRIDE17/12244779/PARK-QC). Andreas Husch was partially supported by the Fondation Cancer Luxembourg. The authors would like to thank Prof. Peter Gemmar, Trier University of Applied Sciences, Trier, Germany and Prof. Frank Hertel, Centre Hospitalier de Luxembourg, Luxembourg, for discussions.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.media.2021.102225.
Appendix A. Supplementary materials
Table S1: Adapted items from CHARMS tool. Table S2: Adapted items from BIAS tool. Table S3: Non-COVID datasets analysed in the paper.
Extended answers of CHARMS analysis.
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
Extended answers of BIAS analysis.
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
References
- Ahmed K.B., Goldgof G.M., Paul R., Goldgof D.B., Hall L.O. Discovery of a generalization gap of convolutional neural networks on covid-19 x-rays classification. IEEE Access. 2021 doi: 10.1109/ACCESS.2021.3079716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, K. B., Hall, L. O., Goldgof, D. B., Goldgof, G. M., Paul, R., 2021b. Deep learning models may spuriously classify covid-19 from x-ray images based on confounders. arXiv:2102.04300.
- Albahli S., Albattah W. Deep transfer learning for COVID-19 prediction: case study for limited data problems. Curr. Med. Imaging. 2020 doi: 10.2174/1573405616666201123120417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljondi R., Alghamdi S. Diagnostic value of imaging modalities for COVID-19: scoping review. J. Med. Internet Res. 2020;22:e19673. doi: 10.2196/19673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A., Bilbao A., Ruby L., Rominger M.B., López-De-Ipiña D., Dahl J., ElKaffas A., Sanabria S.J. 2020 IEEE International Ultrasonics Symposium (IUS) IEEE; 2020. Lung ultrasound for point-of-care COVID-19 pneumonia stratification: computer-aided diagnostics in a smartphone. first experiences classifying semiology from public datasets. [Google Scholar]; 1–4.
- Amini A., Soleimany A.P., Schwarting W., Bhatia S.N., Rus D. Proceedings of the 2019 AAAI/ACM Conference on AI, Ethics, and Society. Association for Computing Machinery; New York, NY, USA: 2019. Uncovering and Mitigating Algorithmic Bias through Learned Latent Structure; pp. 289–295. [Google Scholar]; 10.1145/3306618.3314243.
- Born J., Wiedemann N., Cossio M., Buhre C., Brändle G., Leidermann K., Aujayeb A. L2 accelerating COVID-19 differential diagnosis with explainable ultrasound image analysis: an AI tool. Thorax. 2021;76:A230–A231. doi: 10.1136/thorax-2020-BTSabstracts.404. [DOI] [Google Scholar]
- Born J., Wiedemann N., Cossio M., Buhre C., Brändle G., Leidermann K., Aujayeb A., Moor M., Rieck B., Borgwardt K. Accelerating detection of lung pathologies with explainable ultrasound image analysis. Appl. Sci. 2021;11:672. doi: 10.3390/app11020672. [DOI] [Google Scholar]
- Brady A.P., Neri E. Artificial intelligence in radiology-ethical considerations. Diagnostics. 2020;10:231. doi: 10.3390/diagnostics10040231. [DOI] [PMC free article] [PubMed] [Google Scholar]; https://pubmed.ncbi.nlm.nih.gov/32316503.
- Campbell J.P., Lee A.Y., Abrmoff M., Keane P.A., Ting D.S., Lum F., Chiang M.F. Reporting guidelines for artificial intelligence in medical research. Ophthalmology. 2020 doi: 10.1016/j.ophtha.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://www.sciencedirect.com/science/article/pii/S0161642020308812
- Campolo A., Sanfilippo M., Whittaker M., Crawford K. ai now 2017 symposium and workshop. AI Now Institute at New York University; 2018. AI Now 2017 report. [Google Scholar]; https://www.microsoft.com/en-us/research/publication/ai-now-2017-report/
- Castiglioni I., Ippolito D., Interlenghi M., Monti C.B., Salvatore C., Schiaffino S., Polidori A., Gandola D., Messa C., Sardanelli F. Machine learning applied on chest x-ray can aid in the diagnosis of covid-19: a first experience from lombardy, italy. Eur. Radiol. Exp. 2021;5:1–10. doi: 10.1186/s41747-020-00203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro D.C., Walker I., Glocker B. Causality matters in medical imaging. Nat. Commun. 2020;11:3673. doi: 10.1038/s41467-020-17478-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, M. E., Rahman, T., Khandakar, A., Mazhar, R., Kadir, M. A., Mahbub, Z. B., Islam, K. R., Khan, M. S., Iqbal, A., Al-Emadi, N., et al., 2020. Can AI help in screening viral and COVID-19 pneumonia?arXiv:2003.13145.
- Choy G., Khalilzadeh O., Michalski M., Do S., Samir A.E., Pianykh O.S., Geis J.R., Pandharipande P.V., Brink J.A., Dreyer K.J. Current applications and future impact of machine learning in radiology. Radiology. 2018;288:318–328. doi: 10.1148/radiol.2018171820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleverley J., Piper J., Jones M.M. The role of chest radiography in confirming covid-19 pneumonia. BMJ. 2020;370 doi: 10.1136/bmj.m2426. [DOI] [PubMed] [Google Scholar]
- Cohen, J. P., Hashir, M., Brooks, R., Bertrand, H., 2020a. On the limits of cross-domain generalization in automated x-ray prediction. arXiv:2002.02497.
- Cohen, J. P., Morrison, P., Dao, L., Roth, K., Duong, T. Q., Ghassemi, M., 2020b. Covid-19 image data collection: prospective predictions are the future. arXiv:2006.11988.
- Collins G.S., Moons K.G.M. Reporting of artificial intelligence prediction models. Lancet. 2019;393:1577–1579. doi: 10.1016/S0140-6736(19)30037-6. [DOI] [PubMed] [Google Scholar]
- DeGrave A.J., Janizek J.D., Lee S.I. AI for radiographic COVID-19 detection selects shortcuts over signal. medRxiv. 2020 doi: 10.1101/2020.09.13.20193565. [DOI] [Google Scholar]
- Desai S., Baghal A., Wongsurawat T., Jenjaroenpun P., Powell T., Al-Shukri S., Gates K., Farmer P., Rutherford M., Blake G., et al. Chest imaging representing a COVID-19 positive rural US population. Sci. Data. 2020;7:1–6. doi: 10.1038/s41597-020-00741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgendi M., Nasir M.U., Tang Q., Smith D., Grenier J.P., Batte C., Spieler B., Leslie W.D., Menon C., Fletcher R.R., et al. The effectiveness of image augmentation in deep learning networks for detecting covid-19: a geometric transformation perspective. Front. Med. 2021;8 doi: 10.3389/fmed.2021.629134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faes L., Liu X., Wagner S.K., Fu D.J., Balaskas K., Sim D.A., Bachmann L.M., Keane P.A., Denniston A.K. A clinician’s guide to artificial intelligence: how to critically appraise machine learning studies. Transl. Vis. Sci. Technol. 2020;9 doi: 10.1167/tvst.9.2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]; 7–7.
- Ganin Y., Ustinova E., Ajakan H., Germain P., Larochelle H., Laviolette F., Marchand M., Lempitsky V. Domain-adversarial training of neural networks. J. Mach. Learn. Res. 2016;17 [Google Scholar]; 2096–2030.
- Garbin, C., Rajpurkar, P., Irvin, J., Lungren, M. P., Marques, O., 2021. Structured dataset documentation: a datasheet for chexpert. arXiv:2105.03020.
- Garcia Santa Cruz B., Sölter J., Bossa M.N., Husch A. On the composition and limitations of publicly available COVID-19 x-ray imaging datasets. BioRxiv. 2020 [Google Scholar]
- Gebru, T., Morgenstern, J., Vecchione, B., Vaughan, J. W., Wallach, H., Daumé, III, H., Crawford, K., 2018. Datasheets for datasets. arXiv:1803.09010.
- Geis J.R., Brady A.P., Wu C.C., Spencer J., Ranschaert E., Jaremko J.L., Langer S.G., Borondy Kitts A., Birch J., Shields W.F., van den Hoven van Genderen R., Kotter E., Wawira Gichoya J., Cook T.S., Morgan M.B., Tang A., Safdar N.M., Kohli M. Ethics of artificial intelligence in radiology: summary of the joint european and north american multisociety statement. Radiology. 2019;293:436–440. doi: 10.1148/radiol.2019191586. [DOI] [PubMed] [Google Scholar]; PMID: 31573399.
- Gomes, D. P., Horry, M. J., Ulhaq, A., Paul, M., Chakraborty, S., Saha, M., Debnath, T., Rahaman, D., 2020. Mavidh score: A corona severity scoring using interpretable chest x-ray pathology features. arXiv:2011.14983.
- Greenland S., Robins J.M., Pearl J. Confounding and collapsibility in causal inference. Stat. Sci. 1999;14:29–46. doi: 10.1214/ss/1009211805. [DOI] [Google Scholar]
- Greenspan H., Estépar R.S.J., Niessen W.J., Siegel E., Nielsen M. Position paper on COVID-19 imaging and AI: from the clinical needs and technological challenges to initial AI solutions at the lab and national level towards a new era for AI in healthcare. Med. Image Anal. 2020;66:101800. doi: 10.1016/j.media.2020.101800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith G.J., Morris T.T., Tudball M.J., Herbert A., Mancano G., Pike L., Sharp G.C., Sterne J., Palmer T.M., Davey Smith G., Tilling K., Zuccolo L., Davies N.M., Hemani G. Collider bias undermines our understanding of COVID-19 disease risk and severity. Nat. Commun. 2020;11:5749. doi: 10.1038/s41467-020-19478-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health T.L.D. Walking the tightrope of artificial intelligence guidelines in clinical practice. Lancet Digit. Health. 2019;1:e100. doi: 10.1016/S2589-7500(19)30063-9. [DOI] [PubMed] [Google Scholar]
- Health T.L.D. Guiding better design and reporting of AI-intervention trials. Lancet Digit. Health. 2020;2:e493. doi: 10.1016/S2589-7500(20)30223-5. [DOI] [PubMed] [Google Scholar]
- Heckman J.J. Sample selection bias as a specification error. Econometrica. 1979;47:153–161. [Google Scholar]; http://www.jstor.org/stable/1912352
- Ilyas, M., Rehman, H., Naït-Ali, A., 2020. Detection of covid-19 from chest x-ray images using artificial intelligence: an early review. arXiv:2004.05436.
- Islam, M., Karray, F., Alhajj, R., Zeng, J., et al., 2020. A review on deep learning techniques for the diagnosis of novel coronavirus (COVID-19). arXiv:2008.04815. [DOI] [PMC free article] [PubMed]
- Ismael A.M., Şengür A. The investigation of multiresolution approaches for chest x-ray image based COVID-19 detection. Health Inf. Sci. Syst. 2020;8:1–11. doi: 10.1007/s13755-020-00116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H., Nachum, O., 2019. Identifying and correcting label bias in machine learning. arXiv:1901.04966.
- Kermany D.S., Goldbaum M., Cai W., Valentim C.C., Liang H., Baxter S.L., McKeown A., Yang G., Wu X., Yan F., et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- Kim, G., Park, S., Oh, Y., Seo, J. B., Lee, S. M., Kim, J. H., Moon, S., Lim, J. K., Ye, J. C., 2021. Severity quantification and lesion localization of covid-19 on CXR using vision transformer. arXiv:2103.07062.
- Linda Wang, Z. Q. L., Wong, A., 2020. COVID-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv:2003.09871. [DOI] [PMC free article] [PubMed]
- Liu X., Faes L., Calvert M.J., Denniston A.K. Extension of the CONSORT and SPIRIT statements. Lancet. 2019;394:1225. doi: 10.1016/S0140-6736(19)31819-7. [DOI] [PubMed] [Google Scholar]
- Maguolo, G., Nanni, L., 2020. A critic evaluation of methods for covid-19 automatic detection from x-ray images. arXiv:2004.12823. [DOI] [PMC free article] [PubMed]
- Maier-Hein L., Eisenmann M., Reinke A., Onogur S., Stankovic M., Scholz P., Arbel T., Bogunovic H., Bradley A.P., Carass A., et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-018-07619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier-Hein L., Reinke A., Kozubek M., Martel A.L., Arbel T., Eisenmann M., Hanbury A., Jannin P., Müller H., Onogur S., et al. BIAS: transparent reporting of biomedical image analysis challenges. Med. Image Anal. 2020;66:101796. doi: 10.1016/j.media.2020.101796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateen B.A., Liley J., Denniston A.K., Holmes C.C., Vollmer S.J. Improving the quality of machine learning in health applications and clinical research. Nat. Mach. Intell. 2020;2:554–556. doi: 10.1038/s42256-020-00239-1. [DOI] [Google Scholar]
- McBee M.P., Awan O.A., Colucci A.T., Ghobadi C.W., Kadom N., Kansagra A.P., Tridandapani S., Auffermann W.F. Deep learning in radiology. Acad. Radiol. 2018;25:1472–1480. doi: 10.1016/j.acra.2018.02.018. [DOI] [PubMed] [Google Scholar]
- Mitchell M., Wu S., Zaldivar A., Barnes P., Vasserman L., Hutchinson B., Spitzer E., Raji I.D., Gebru T. Proceedings of the Conference on Fairness, Accountability, and Transparency. Association for Computing Machinery; New York, NY, USA: 2019. Model cards for model reporting; pp. 220–229. [Google Scholar]; 10.1145/3287560.3287596
- Mittal S., Venugopal V.K., Agarwal V.K., Malhotra M., Chatha J.S., Kapur S., Gupta A., Batra V., Majumdar P., Malhotra A., et al. A novel abnormality annotation database for covid-19 affected frontal lung x-rays. medRxiv. 2021 doi: 10.1371/journal.pone.0271931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Shamseer L., Clarke M., Ghersi D., Liberati A., Petticrew M., Shekelle P., Stewart L.A., et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-p) 2015 statement. Syst. Rev. 2015;4:1. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons K.G., Altman D.G., Reitsma J.B., Ioannidis J.P., Macaskill P., Steyerberg E.W., Vickers A.J., Ransohoff D.F., Collins G.S. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann. Intern. Med. 2015;162:W1–W73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- Moons K.G.M., de Groot J.A.H., Bouwmeester W., Vergouwe Y., Mallett S., Altman D.G., Reitsma J.B., Collins G.S. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med. 2014;11:1–12. doi: 10.1371/journal.pmed.1001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons K.G.M., Wolff R.F., Riley R.D., Whiting P.F., Westwood M., Collins G.S., Reitsma J.B., Kleijnen J., Mallett S. PROBAST: A tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann. Intern. Med. 2019;170:W1–W33. doi: 10.7326/M18-1377. [DOI] [PubMed] [Google Scholar]; PMID: 30596876.
- Mustra M., Delac K., Grgic M. 2008 50th International Symposium ELMAR. IEEE; 2008. Overview of the DICOM standard; pp. 39–44. [Google Scholar]
- Nagendran M., Chen Y., Lovejoy C.A., Gordon A.C., Komorowski M., Harvey H., Topol E.J., Ioannidis J.P.A., Collins G.S., Maruthappu M. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ. 2020;368 doi: 10.1136/bmj.m689. [DOI] [PMC free article] [PubMed] [Google Scholar]; https://www.bmj.com/content/368/bmj.m689.full.pdf
- Oakden-Rayner L. Exploring large-scale public medical image datasets. Acad. Radiol. 2020;27:106–112. doi: 10.1016/j.acra.2019.10.006. [DOI] [PubMed] [Google Scholar]
- O’Reilly-Shah V.N., Gentry K.R., Walters A.M., Zivot J., Anderson C.T., Tighe P.J. Bias and ethical considerations in machine learning and the automation of perioperative risk assessment. Br. J. Anaesth. 2020 doi: 10.1016/j.bja.2020.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://www.sciencedirect.com/science/article/pii/S0007091220306310.
- Parikh R.B., Teeple S., Navathe A.S. Addressing bias in artificial intelligence in health care. JAMA. 2019;322:2377–2378. doi: 10.1001/jama.2019.18058. [DOI] [PubMed] [Google Scholar]
- Park, S., Kim, G., Oh, Y., Seo, J. B., Lee, S. M., Kim, J. H., Moon, S., Lim, J. K., Ye, J. C., 2021. Vision transformer for covid-19 cxr diagnosis using chest x-ray feature corpus. arXiv:2103.07055. [DOI] [PMC free article] [PubMed]
- Roberts M., Driggs D., Thorpe M., Gilbey J., Yeung M., Ursprung S., Aviles-Rivero A.I., Etmann C., McCague C., Beer L., Weir-McCall J.R., Teng Z., Gkrania-Klotsas E., Ruggiero A., Korhonen A., Jefferson E., Ako E., Langs G., Gozaliasl G., Yang G., Prosch H., Preller J., Stanczuk J., Tang J., Hofmanninger J., Babar J., Sánchez L.E., Thillai M., Gonzalez P.M., Teare P., Zhu X., Patel M., Cafolla C., Azadbakht H., Jacob J., Lowe J., Zhang K., Bradley K., Wassin M., Holzer M., Ji K., Ortet M.D., Ai T., Walton N., Lio P., Stranks S., Shadbahr T., Lin W., Zha Y., Niu Z., Rudd J.H.F., Sala E., Schönlieb C.B., AIX-COVNET Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat. Mach. Intell. 2021;3:199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- Robinson C., Trivedi A., Blazes M., Ortiz A., Desbiens J., Gupta S., Dodhia R., Bhatraju P.K., Liles W.C., Lee A., et al. Deep learning models for COVID-19 chest x-ray classification: preventing shortcut learning using feature disentanglement. medRxiv. 2021 doi: 10.1371/journal.pone.0274098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamout F.E., Shen Y., Wu N., Kaku A., Park J., Makino T., Jastrzȩbski S., Witowski J., Wang D., Zhang B., et al. An artificial intelligence system for predicting the deterioration of covid-19 patients in the emergency department. NPJ Digit. Med. 2021;4:1–11. doi: 10.1038/s41746-021-00453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoeibi, A., Khodatars, M., Alizadehsani, R., Ghassemi, N., Jafari, M., Moridian, P., Khadem, A., Sadeghi, D., Hussain, S., Zare, A., et al., 2020. Automated detection and forecasting of COVID-19 using deep learning techniques: a review. arXiv:2007.10785.
- Shuja J., Alanazi E., Alasmary W., Alashaikh A. COVID-19 Open source data sets: a comprehensive survey. Applied Intelligence. 2020 doi: 10.1007/s10489-020-01862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoroni, A., Savardi, M., Benini, S., Adami, N., Leonardi, R., Gibellini, P., Vaccher, F., Ravanelli, M., Borghesi, A., Maroldi, R., Farina, D., 2020. End-to-end learning for semiquantitative rating of COVID-19 severity on chest x-rays. arXiv:2006.04603. [DOI] [PMC free article] [PubMed]
- Soda, P., D’Amico, N. C., Tessadori, J., Valbusa, G., Guarrasi, V., Bortolotto, C., Akbar, M. U., Sicilia, R., Cordelli, E., Fazzini, D., et al., 2020. AIforCOVID: predicting the clinical outcomes in patients with COVID-19 applying AI to chest-x-rays. an italian multicentre study. arXiv:2012.06531. [DOI] [PMC free article] [PubMed]
- Sogancioglu, E., Çallı, E., van Ginneken, B., van Leeuwen, K. G., Murphy, K., 2021. Deep learning for chest x-ray analysis: a survey. arXiv:2103.08700. [DOI] [PubMed]
- Sohan, M. F., 2020. So you need datasets for your COVID-19 detection research using machine learning? arXiv:2008.05906.
- Sohrabi C., Alsafi Z., O’Neill N., Khan M., Kerwan A., Al-Jabir A., Iosifidis C., Agha R. World health organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020 doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Gerster S., Delorenzi M. Batch effect confounding leads to strong bias in performance estimates obtained by cross-validation. PLoS One. 2014;9:e100335. doi: 10.1371/journal.pone.0100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sounderajah V., Ashrafian H., Aggarwal R., De Fauw J., Denniston A.K., Greaves F., Karthikesalingam A., King D., Liu X., Markar S.R., McInnes M.D.F., Panch T., Pearson-Stuttard J., Ting D.S.W., Golub R.M., Moher D., Bossuyt P.M., Darzi A. Developing specific reporting guidelines for diagnostic accuracy studies assessing AI interventions: the STARD-AI steering group. Nat. Med. 2020;26:807–808. doi: 10.1038/s41591-020-0941-1. [DOI] [PubMed] [Google Scholar]
- Stevens L.M., Mortazavi B.J., Deo R.C., Curtis L., Kao D.P. Recommendations for reporting machine learning analyses in clinical research. Circulation. 2020;0 doi: 10.1161/CIRCOUTCOMES.120.006556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyerberg E. Springer International Publishing; 2009. Clinical Prediction Models: a Practical Approach to Development, Validation, and Updating. [DOI] [Google Scholar]; 19
- Tabik S., Gómez-Rıos A., Martın-Rodrıguez J.L., Sevillano-Garcıa I., Rey-Area M., Charte D., Guirado E., Suárez J.L., Luengo J., Valero-González M., et al. COVIDGR dataset and COVID-SDNet methodology for predicting COVID-19 based on chest x-ray images. IEEE J. Biomed. Health Inform. 2020;24:3595–3605. doi: 10.1109/JBHI.2020.3037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Sun N., Li Y. Segmentation model of the opacity regions in the chest x-rays of the covid-19 patients in the us rural areas and the application to the disease severity. medRxiv. 2020 [Google Scholar]
- Tartaglione, E., Barbano, C. A., Berzovini, C., Calandri, M., Grangetto, M., 2020. Unveiling COVID-19 from chest x-ray with deep learning: a hurdles race with small data. arXiv:2004.05405. [DOI] [PMC free article] [PubMed]
- Tsai E.B., Simpson S., Lungren M., Hershman M., Roshkovan L., Colak E., Erickson B.J., Shih G., Stein A., Kalpathy-Cramer J., et al. The RSNA international COVID-19 open annotated radiology database (RICORD) Radiology. 2021:203957. doi: 10.1148/radiol.2021203957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayá, M. d. l. I., Saborit, J. M., Montell, J. A., Pertusa, A., Bustos, A., Cazorla, M., Galant, J., Barber, X., Orozco-Beltrán, D., Garcia, F., et al., 2020. BIMCV COVID-19+: a large annotated dataset of RX and CT images from COVID-19 patients. arXiv:2006.01174.
- Vollmer S., Mateen B.A., Bohner G., Király F.J., Ghani R., Jonsson P., Cumbers S., Jonas A., McAllister K.S.L., Myles P., Grainger D., Birse M., Branson R., Moons K.G.M., Collins G.S., Ioannidis J.P.A., Holmes C., Hemingway H. Machine learning and artificial intelligence research for patient benefit: 20 critical questions on transparency, replicability, ethics, and effectiveness. BMJ. 2020;368 doi: 10.1136/bmj.l6927. [DOI] [PMC free article] [PubMed] [Google Scholar]; https://www.bmj.com/content/368/bmj.l6927.full.pdf
- Wolff R.F., Moons K.G., Riley R.D., Whiting P.F., Westwood M., Collins G.S., Reitsma J.B., Kleijnen J., Mallett S. PROBAST: A tool to assess the risk of bias and applicability of prediction model studies. Ann. Intern. Med. 2019;170:51–58. doi: 10.7326/M18-1376. [DOI] [PubMed] [Google Scholar]
- Wynants L., Van Calster B., Collins G.S., Riley R.D., Heinze G., Schuit E., Bonten M.M.J., Dahly D.L., Damen J.A., Debray T.P.A., de Jong V.M.T., De Vos M., Dhiman P., Haller M.C., Harhay M.O., Henckaerts L., Heus P., Kammer M., Kreuzberger N., Lohmann A., Luijken K., Ma J., Martin G.P., McLernon D.J., Andaur Navarro C.L., Reitsma J.B., Sergeant J.C., Shi C., Skoetz N., Smits L.J.M., Snell K.I.E., Sperrin M., Spijker R., Steyerberg E.W., Takada T., Tzoulaki I., van Kuijk S.M.J., van Bussel B.C.T., van der Horst I.C.C., van Royen F.S., Verbakel J.Y., Wallisch C., Wilkinson J., Wolff R., Hooft L., Moons K.G.M., van Smeden M. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;369 doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Chen W., Ren H., Zhao J., Wang L., Jin R., Zhou J., Wang Q., Yan F., Zhang B., et al. A rapid screening classifier for diagnosing covid-19. Int. J. Biol. Sci. 2021;17:539. doi: 10.7150/ijbs.53982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q., Ji R., Wang H., Wang Y., Zhou Y. Prevalence of comorbidities and its effects in patients infected with SARS-cov-2: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech J.R., Badgeley M.A., Liu M., Costa A.B., Titano J.J., Oermann E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15:e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Adeli E., Pohl K.M. Training confounder-free deep learning models for medical applications. Nat. Commun. 2020;11:1–9. doi: 10.1038/s41467-020-19784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Adapted items from CHARMS tool. Table S2: Adapted items from BIAS tool. Table S3: Non-COVID datasets analysed in the paper.
Extended answers of CHARMS analysis.
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
Extended answers of BIAS analysis.
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/