Abstract
Background
Exact comprehension of the prevalence of SARS-CoV-2 infection is essential for the preventive measures. In the clinical settings, however, patients infected with SARS-CoV-2 may not be fully detected by PCR. In the long-term prevalence study, cut-off of IgG assay may not be appropriate due to waning IgG titer.
Methods
24 PCR-negative subjects suspected of COVID-19 were categorized into cohorts termed “presumed COVID-19 positive” and “presumed COVID-19 negative” by chest CT images. IgG against nucleocapsid protein of SARS-CoV-2 (IgG (N)) and IgG against receptor biding domain of SARS-CoV-2 (IgG (RBD)) were measured in sera of the subjects and the concordance with the cohort categorization was assessed by receiver operating characteristics (ROC) analyses.
Results
Area under the curves (AUC's) by the ROC analyses with the 24 subjects were 0.982 with IgG (N) and 0.854 with IgG (RBD). Even when we excluded the subjects whose initial PCR was performed after five days from symptom onset, the AUC's were 0.967 with IgG (N) and 0.800 with IgG (RBD). The ROC analysis indicated 0.2 S/C as the optimum cut-off forIgG (N).
Conclusion
Both IgG (N) and IgG (RBD) titers were significantly elevated in subjects whose PCR never showed positive but suggestive of SARS-CoV-2 infection, which indicated the necessity of serological tests in complementing the shortcomings of PCR. For a long-term prevalence study, a cut-off lower than the one used in the ongoing infection phase (e.g. 0.2 S/C vs. 1.4 S/C) was indicated to be more appropriate for IgG (N).
Keywords: COVID-19, SARS-CoV-2, PCR, IgG, Serological test, Prevalence
Introduction
COVID-19, a disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 and has caused a worldwide pandemic.1 As of September 2021, number of confirmed cases and that of deaths exceeded 217 million and 4.5 million, respectively,2 the mortality rate of which is estimated to be 2.1%. The underlying conditions leading to cause of death are mostly due to pneumonia or acute respiratory distress syndrome.3 The vaccination that started in late 2020 in several countries is expected to be a powerful measure to overcome this pandemic and over 4.8 billion shots were administered globally as of August 2021.2 The aggregated number of new cases per day, however, is still at the high level so that it is still a tremendous threat to the public health of the world.
It is important to comprehend the exact degree of prevalence for several reasons; to capture the number of cases with on-going SARS-CoV-2 infection is essential to the prevention of the viral transmission to the others including healthcare workers, to have data on people with a history of the infection is necessary to trace the mode of the infection, to have data on percentage of the population whose immune system encountered with the viral antigens would provide important information for the prediction of future spread and for the planning of vaccination strategy.
Since the emergence of COVID-19, PCR has been used as the gold standard for the detection of SARS-CoV-2 due to its sensitivity and specificity. It is reported, however, that there was a limitation to the detection of SARS-CoV-2 by PCR due to several factors including varying viral load by the sample sources such as nasopharyngeal mucus, oropharyngeal saliva, sputum or bronchial fluid and the rapidly declining viral load after the onset of the disease.1 In fact, Kanji et al. reported that false negative rate was 9.3% when discordant repeat PCR test results were examined.4 Mallet et al. also reported that the percentage of virus detection from nasopharyngeal swabs in subjects infected with SARS-CoV-2 was 89% when collected within four days post symptom onset and it dropped to 54% when collected after 10–14 days post symptom onset.5 Cao et al. reported that negative PCR results with nasopharyngeal swabs in suspected COVID-19 patients could lead to increased risk of infection clusters,6 therefore it is critical to devise a way to complement the shortcomings of PCR.
Combination test with chest computed tomography (CT) and serological tests in addition to PCR would be one of the feasible ways to complement the shortcomings of PCR in diagnosing COVID-19. As Guan et al. reported, abnormalities on chest CT were detected in 86.2% of COVID-19 patients.7 Wen et al. reported that the positive and negative predictive value of chest CT for COVID-19 are estimated to be 92% and 42%, respectively, in a population with high pretest probability for the disease.8 Bryan et al. reported that the positivity of an IgG assay against SARS-CoV-2 nucleocapsid protein (abbreviated as “IgG (N)”) was >84% at day 7 after symptom onset and reached 100% at day 17 after symptom onset.9
In addition to the diagnosis of COVID-19 in clinical settings, IgG test is considered useful for the surveillance of the prevalence of SARS-CoV-2 infection. However, there have not been many discussions on the appropriate cut-off for the long-term surveillance taking the factor of waning IgG titer into consideration, so the prevalence may have been underestimated.
In this study, we used two types of IgG assay, IgG (N) and IgG (RBD), and investigated whether these IgG tests could detect SARS-CoV-2 infection in subjects suspected of the infection but who never showed PCR positive. We also sought cut-offs for these IgG assays appropriate for long-term surveillance so that those with previous infection are properly detected.
Methods
Study subjects
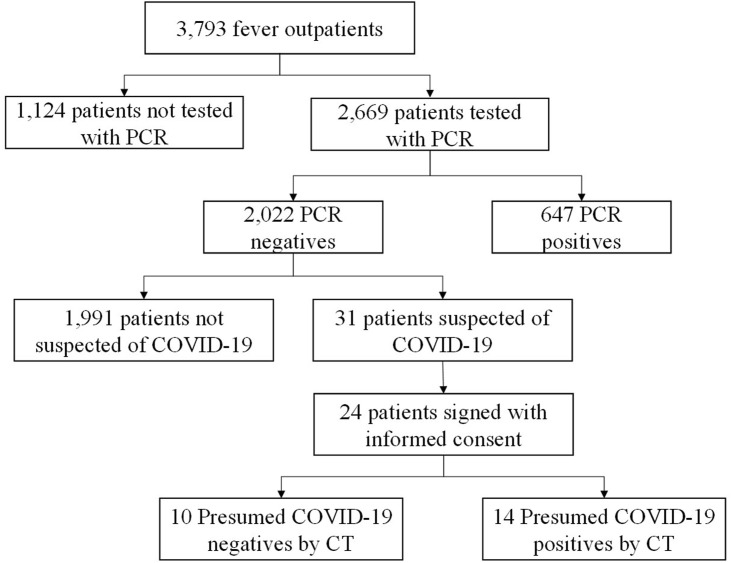
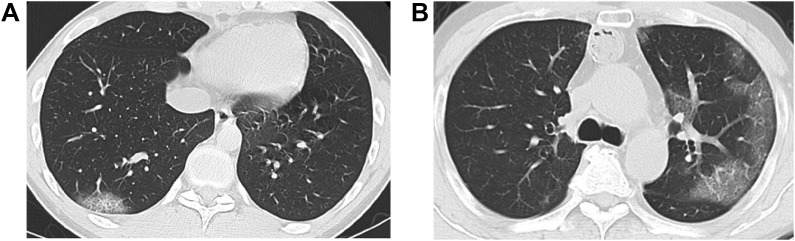
This study was conducted at Tokyo Shinagawa Hospital. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics Committee at Tokyo Shinagawa Hospital (approval no. 20-A-26) prior to the start of the study. We summarized the inclusion criteria in Fig. 1 . To complement this criteria, we selected 31 subjects from 2,022 PCR-negative subjects by symptoms suggestive of COVID-19 and/or history of close contact with infected people. Among the 31 subjects, 24 subjects who agreed on the participation to the study orally and in the written form, were included. Two pulmonologists with more than ten years of experience categorized the 24 subjects into cohorts of presumed COVID-19 negative and positive by pathological abnormalities shown in chest CT images (e.g. peripheral distribution of ground-glass opacities or consolidation, presence of a crazy-paving pattern or intralobular lines) referring to Radiological Society of North America Expert Consensus Statement on Reporting Chest CT Findings Related to COVID-19.10 In case of the discordance between the two pulmonologists, a third pulmonologist decided the categorization. Typical CT imaging features for COVID-19 are shown in Fig. 2 A and B.
Figure 1.
Flow chart of subject inclusion criteria.
Figure 2.
Typical CT imaging features for COVID-19. Unenhanced, thin-section axial images of two subjects presumed of COVID-19 postive who showed rounded and peripheral GGO with superimposed interlobular septal thickening and visible intralobular lines (“crazy-paving”). A) 27-year-old male (subject ID #17). B) 51-year-old male (subject ID #21). GGO; ground-glass opacity.
Clinical and laboratory tests
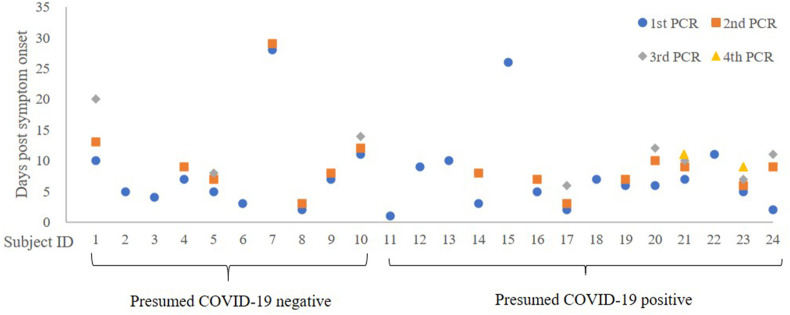
All subjects underwent routine biochemical and hematological blood tests for the assessment of health status at the time of the first visit to the hospital. The biochemistry and hematology tests were performed on TBA-c16000 (Canon Medical Systems, Otawara-city, Tochigi, Japan) and XN-3000 (Sysmex, Kobe-city, Hyogo, Japan), respectively. Real-time RT-PCR test for SARS-CoV-2 was performed with TaqMan™ Fast Virus 1-Step Master Mix which targets N1 and N2 regions of SARS-CoV-2 on QuantStudio™ 5 Real-Time PCR System (Thermo Fisher SCIENTIFIC, Waltham, MA, USA). Sensitivity and specificity of the PCR were both 100% by a report from National Institute of Infectious Diseases in Japan.11 The PCR result was defined as positive when cycle threshold (Ct) was equal to or less than 40. Nasopharyngeal swabs for the PCR were collected by nurses who had ample experience of routine swab collections from fever outpatients and hospitalized patients. Time points of the swab collections are shown in Fig. 3 . Chest CT was performed on Revolution Maxima (GE Healthcare, IL, USA) around the same time as the first PCR sample was collected. Titer of IgG against SARS-CoV-2 nucleocapsid protein (abbreviated as “IgG (N)”) was measured by an ARCHITECT SARS-CoV-2 IgG assay on Architect i2000 CS5100 (Abbott Laboratories, Abbott Park, IL, USA) and that of IgG against SARS-CoV-2 receptor binding domain (abbreviated as “IgG (RBD)”) was measured by an ARCHITECT SARS-CoV-2 IgG II Quant assay on Architect i2000 CS5100 (Abbott Laboratories, Abbott Park, IL, USA). According to the package insert of the IgG (N) assay, cut-off index is 1.4 S/C and CV% at a mean index of 0.04 S/C of 50 negative controls is 5.9%. According to the package insert of the IgG (RBD) assay, cut-off index is 50.0 AU/mL and lowest concentration at which CV% is within 20% is 7.8 AU/mL.
Figure 3.
Days of PCR sample collection from symptom onset. Subject #1–10: presumed COVID-19 negative. Subject #11–24: presumed COVID-19 positive. Blue circle: 1st PCR sample collection. Orange square: 2nd PCR sample collection. Grey diamond: 3rd PCR sample collection. Yellow triangle: 4th PCR sample collection.
Statistical analysis
We used JMP 15.1.0 (SAS Institute Inc., Cary, NC, USA) for statistical analyses including Wilcoxon signed-rank test and receiver operating characteristic (ROC) analysis. P-value of <0.01 was considered significant.
Results
Characteristics of the study subjects
The characteristics of the two cohorts, presume COVID-19 negative and positive, are presented in Table 1 . The p-values shown on the right were obtained by Wilcoxon signed-rank test between the two cohorts. Briefly, none of the parameters were significantly different except for age, albumin, blood urea nitrogen (BUN), C-reactive protein (CRP), IgG (N) and IgG (RBD). In the cohort of presumed COVID-19 positive, the age, BUN and CRP were lower than that of presumed COVID-19 negative while albumin, IgG (N) and IgG (RBD) were higher.
Table 1.
Clinical data of the study subjects.
| Unit | Presumed COVID-19 (−) | Presumed COVID-19 (+) | p-value | |
|---|---|---|---|---|
| Subject number | N | 10 | 14 | |
| Male | n (%) | 5 (50.0%) | 7 (50.0%) | 1.000 |
| Age | Years | 69.0 (63.3–72.5) | 42.5 (37.0–51.8) | 0.001 |
| BMI | kg/m2 | 23.4 (21.9–24.8) | 21.4 (20.0–24.7) | 0.396 |
| ADL | scale of 1–10 | 9.0 (8.0–9.8) | 10.0 (8.0–10.0) | 0.516 |
| Headache | n (%) | 4 (44.4%) | 7 (50.0%) | 0.660 |
| Fever | n (%) | 8 (88.9%) | 12 (85.7%) | 0.799 |
| Cough | n (%) | 4 (44.4%) | 9 (64.3%) | 0.515 |
| Sore throat | n (%) | 6 (66.7%) | 2 (14.3%) | 0.024 |
| Fatigue | n (%) | 6 (66.7%) | 10 (71.4%) | 0.970 |
| Dyspnea | n (%) | 1 (11.1%) | 7 (50.0%) | 0.049 |
| Anosmia | n (%) | 1 (11.1%) | 5 (35.7%) | 0.172 |
| Body temperature | °C | 37.1 (36.7–37.8) | 36.7 (36.3–37.4) | 0.379 |
| SpO2 | % | 96.5 (93.5–97.8) | 96.5 (95.0–98.0) | 0.697 |
| SBP | mmHg | 143.5 (140.5–152.8) | 118.5 (111.5–136.5) | 0.028 |
| DBP | mmHg | 79.0 (74.8–87.8) | 73.0 (69.3–93.0) | 0.500 |
| HR | BPM | 89.0 (71.5–112.3) | 86.0 (77.0–93.0) | 0.576 |
| Initial PCR | days from onset | 6.0 (4.3–9.3) | 6.0 (3.5–8.5) | 0.768 |
| IgG sample collection | days from onset | 151.0 (44.3–273.0) | 285.0 (217.5–302.3) | 0.033 |
| WBC | /uL | 7750.0 (6850.0–8975.0) | 6000.0 (4300.0–6775.0) | 0.015 |
| Neutrophil | % | 78.1 (75.8–82.5) | 66.8 (58.7–79.5) | 0.069 |
| Lymphocyte | % | 13.5 (7.4–16.0) | 20.7 (13.6–33.6) | 0.026 |
| Neutrophil/Lymphocyte Ratio | 5.8 (4.7–11.7) | 3.2 (1.8–6.2) | 0.024 | |
| Monocyte | % | 5.6 (5.2–6.7) | 5.0 (3.7–7.1) | 0.578 |
| Hemoglobin | g/dL | 12.8 (12.4–14.1) | 14.0 (13.5–14.3) | 0.207 |
| Platelet | 104/uL | 26.1 (17.4–31.6) | 23.8 (20.7–30.2) | 0.977 |
| Total protein | g/dL | 6.9 (6.5–7.4) | 7.3 (7.1–7.6) | 0.206 |
| Albumin | g/dL | 3.5 (3.3–3.6) | 4.3 (3.9–4.4) | 0.007 |
| LDH | U/L | 224.5 (177.5–296.5) | 221.5 (175.3–252.8) | 0.725 |
| AST | U/L | 24.0 (19.8–30.5) | 29.5 (20.0–35.5) | 0.428 |
| ALT | U/L | 23.0 (11.5–27.8) | 24.0 (18.0–41.8) | 0.333 |
| γGTP | U/L | 31.5 (15.8–59.3) | 31.5 (25.5–64.8) | 0.501 |
| BUN | mg/dL | 19.7 (14.3–22.2) | 11.0 (8.0–13.1) | 0.001 |
| Creatinine | mg/dL | 0.9 (0.7–1.2) | 0.7 (0.6–0.9) | 0.043 |
| Creatine Kinase | U/L | 94.0 (57.5–147.0) | 73.0 (48.5–97.5) | 0.482 |
| CRP | mg/dL | 7.4 (4.1–12.0) | 0.8 (0.4–1.9) | 0.002 |
| D-dimer | μg/mL | 1.2 (0.4–1.6) | 0.2 (0.2–0.5) | 0.052 |
| IgG (N) | S/C | 0.03 (0.02–0.04) | 0.60 (0.26–1.55) | <0.001 |
| IgG (RBD) | AU/mL | 4.2 (2.3–6.1) | 238.4 (8.2–332.4) | 0.004 |
BMI: body mass index, ADL: activities of daily living, SpO2: percutaneous oxygen saturation, SBP: systolic blood pressure, DBP: diastolic blood pressure, HR: heart rate, WBC: white blood cell count, LDH: lactate dehydrogenase, AST: aspartate aminotransferase, ALT: alanine aminotransferase, γGTP: gamma-glutamyl transpeptidase, BUN: blood urea nitrogen, CRP: c-reactive protein, IgG (N): IgG against nucleocapsid protein of SARS-CoV-2, IgG (RBD): IgG against receptor binding domain of SARS-CoV-2.
Time points at which PCR samples were collected
Days from symptom onset when nasopharyngeal swabs were collected for PCR are shown by each subject in Fig. 3. All the PCR samples shown here resulted negative.
Concordance of presumed COVID-19 diagnosis with IgG titer by ROC analyses
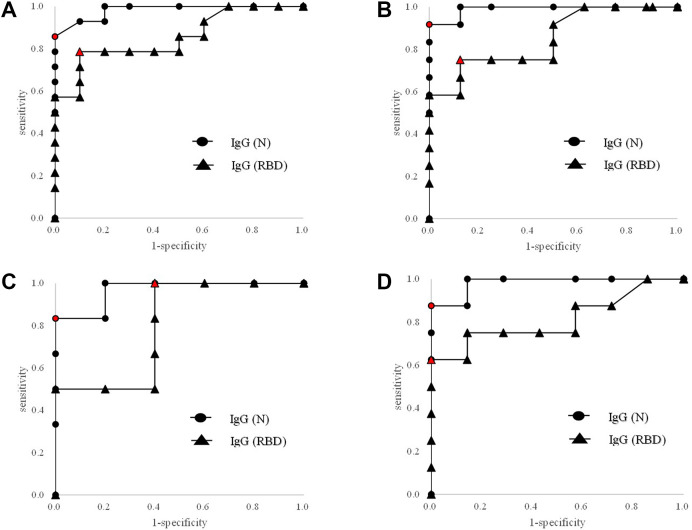
We performed ROC analyses for the concordance between the presumed COVID-19 categorization with IgG (N) titers or with IgG (RBD) titers as shown in Fig. 4 A–D and Table 2 . With the 24 subjects, area under the curves (AUC's) were 0.982 with IgG (N) and 0.854 with IgG (RBD). With subjects with PCR confirmed within ten days from symptom onset, AUC's were 0.990 with IgG (N) and 0.849 with IG (RBD). With subjects with PCR confirmed within five days from symptom onset, AUC's were 0.967 with IgG (N) and 0.800 with IG (RBD). With subjects with repeated PCR tests, AUC's were 0.982 with IgG (N) and 0.813 with IG (RBD). The optimum cut-offs with IgG (N) indicated by the ROC analyses were in the range of 0.20–0.26 S/C while those with IgG (RBD) were in the range of 2.9–262.9 AU/mL.
Figure 4.
ROC analyses for presumed COVID-19 with IgG (N) or IgG (RBD). Circle: IgG (N). Triangle: IgG (RBD). The circle or triangle in red signifies the point that gave the optimum cut-off. A) Total subjects. B) Subjects with PCR confirmed within ten days from symptom onset. C) Subjects with PCR confirmed within five days from symptom onset. D) Subjects with more than one PCR tests.
Table 2.
Comparison of AUC's cut-offs indicated by ROC analyses among subject groups: total subjects, subjects with PCR confirmed within ten days from symptom onset, subjects with PCR confirmed within five days from symptom onset and subjects with more than one PCR tests. AUC's by IgG (N) or IgG (RBD) were shown side-by-side in each group.
| Subject number |
IgG (N) |
IgG (RBD) |
||||
|---|---|---|---|---|---|---|
| Presumed COVID-19 (−) | Presumed COVID-19 (+) | AUC | Cut-off (S/C) | AUC | Cut-off (AU/mL) | |
| Total subject | 10 | 14 | 0.982 | 0.20 | 0.854 | 7.2 |
| PCR confirmed within 10 days from onset | 8 | 12 | 0.990 | 0.20 | 0.849 | 11.3 |
| PCR confirmed within 5 days from onset | 5 | 6 | 0.967 | 0.20 | 0.800 | 2.9 |
| Subjects with repeated PCR tests | 7 | 8 | 0.982 | 0.26 | 0.813 | 262.9 |
AUC: area under the curve.
Positive rates by different cut-offs
Positive rates of IgG (N) in the cohort of presumed COVID-19 positive was 28.6% with the cut-off 1.4 S/C and was 85.7% with the cut-off 0.2 S/C indicated by the ROC analysis with the total cohort above. Positive rates of IgG (RBD) in the cohort of presumed COVID-19 positive was 57.1% with the cut-off 50 AU/mL and was 78.6% with the cut-off 7.2 AU/mL indicated by the ROC analysis with the total subjects above.
Days from symptom onset and IgG titers
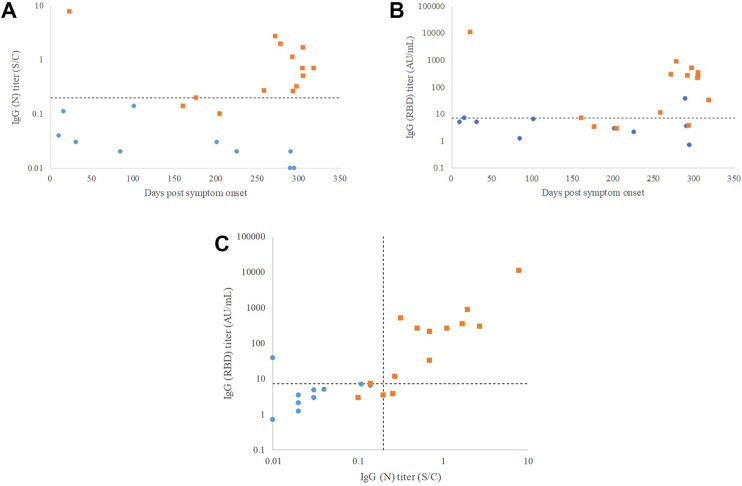
Plots to show the relationship between the days from the onset when the samples for the serological tests were collected (X-axis) and the titers of IgG (N) or IgG (RBD) (Y-axis) and are shown in Fig. 5 A and B, respectively. The days from the onset to the sample collection spanned from 23 to 318 days. Correlation between IgG (N) and IgG (RBD) titers is shown in Fig. 5C. The correlation coefficient (R) was 0.929 with the p-value of <0.001.
Figure 5.
IgG titers against days of sample collection from symptom onset and correlation between IgG (N) and IgG (RBD) titers. Blue circle: presumed COVID-19 negative. Orange square: presumed COVID-19 positive. A) IgG (N). B) IgG (RBD). C) Correlation between IgG (N) and IgG (RBD) titers. Dotted lines indicate the cut-offs derived from the ROC analyses; 0.2 S/C and 7.2 AU/mL for IgG (N) and IgG (RBD), respectively. ROC; receiver operating characteristic.
Discussion
In this study, we looked at IgG but not IgM because elevation of IgG titer from symptom onset was as fast as that of IgM13 and because IgG titer was sustained longer than IgM titer. We confirmed here that both IgG (N) and IgG (RBD) titers were significantly higher in the cohort of presumed COVID-19 positive than that of presumed negative (Table 1) and good concordance between the presumed categorization with IgG (N) or IgG (RBD) was seen by the ROC analyses as shown by the AUC's with the total subjects being 0.982 with IgG (N) and 0.854 with IgG (RBD) (Fig. 4A). The medians of days from symptom onset for the first PCR sample collections were five and six days in the cohorts of presumed COVID-19 negative and positive, respectively, but the days of the initial sample collection exceeded 10 days in two subjects (subject ID #7 and #10) in the presumed negative cohort and two subjects (subject ID #15 and #17) in the presumed positive cohort (Fig. 3). Because the sensitivity of PCR declines after ten days from symptom onset as mentioned earlier, we performed additional ROC analyses in conditions in which the influence of late sampling was minimized by excluding the subjects with the initial PCR performed after ten days (Fig. 4B) or five days (Fig. 4C) from symptom onset. We also performed an ROC analysis by excluding subjects with single PCR to minimize the influence of potential fluctuation of PCR testing (Fig. 4D). In these subgroups, the AUC's were retained at or above 0.967 with IgG (N) and 0.800 with IgG (RBD). These results indicate that the influence of the analytical fluctuation by single PCR test was minimal and that the PCR negativity could be seen even in subjects tested promptly from symptom onset. These results are consistent with the report by Cao et al. in which the existence of people infected with SARS-CoV-2 despite the absence of PCR positivity was confirmed by serological tests.6
If not a small number of patients infected with SARS-CoV-2 but not detected by PCR are there, consideration of transmissibility would be of critical importance for the prevention of potential infection. One of the reasons why SARS-CoV-2 infection was not reflected to PCR positivity could be unequal distribution of the viral loads by the part of the body. Wölfel et al. reported that RNA copies of SARS-CoV-2 in most of the nasopharyngeal swab were lower in number than those in sputum or stool samples,12 so that there may be a case in which the viral load in nasopharyngeal swab is below the detection limit while the virus is present elsewhere in the body. As shown by a report by Fajnzylber et al. in which the viral load was associated with the severity of the disease,13 another reason could be that the disease status of the subjects in this study may not have been severe to the extent to yield the viral loads detectable in the nasopharyngeal swabs. Woloshin et al. estimated chance of SARS-CoV-2 infection when a person was given a negative PCR result.14 They reported that post-test probability of infection was a function of pre-test probability and sensitivity of PCR, and estimated the post-test probability to be more than 50% if the pre-test probability of a subject was more than 90% and the subject was given a negative result by PCR of 95% sensitivity. With the sensitivity of the PCR we used in this study (100%), post-test probability of transmission by a person with SARS-CoV-2 infection could be more than 50% according to this estimation. On the other hand, Kawasuji et al. reported the transmissibility was dependent on the viral load in nasopharyngeal swab,15 so that the post-test probability could be lower than 50%. As pointed out by Jones et al., however, SARS-CoV-2 shed in feces and urine may have a potential role in person-to-person transmission,16 so that we may have to take these routes into consideration in addition to nasopharyngeal route. The cases of SARS-CoV-2 transmission from patients with negative PCR swab tests to others reported by Cao et al.,6 shows a necessity to be cautious about the potential transmission, but under the circumstances with the factors of uncertainty mentioned above, the exact number of the probability remains to be seen.
Although IgG (N) and IgG (RBD) showed the correlation coefficient of 0.929 (Fig. 5C), the AUC's with IgG (RBD) were lower and less reproducible than AUC's with IgG (N) as shown in Table 2. We speculate this is due, at least partly, to the difference in the pattern of overlap between presumed negative and positive cohorts with IgG (N) and that with IgG (RBD) as shown in Fig. 5A and B. In a report by Burbelo et al., antibody response to the nucleocapsid protein was faster and showed less individual variability than that to the spike protein.17 Elslande et al. reported, on the other hand, that half-life of IgG (RBD) titer was more than two-fold than that of IgG (N) titer.18 Therefore, the kinetic differences could be involved in the different performances. Moreover, influence of cross reactivities with other corona virus strains could also influence the performances. Fraley et al. detected IgG antibody responses to SARS-CoV-2 nucleocapsid and S1 proteins in individuals recruited prior to the COVID-19 outbreak.19 The immunogenicity and duration of these cross-reactivities could be different between IgG (N) and IgG (RBD).
As mentioned earlier, Bryan et al. reported that the positive rate of IgG (N) reached 100% at day 17 after symptom onset.9 In the same report, the titer of IgG (N) was above the cut-off 1.4 S/C until 28 days after the onset of the symptom. According to report by Maine et al., the median titer of IgG (N) reached its peak at 6–7 weeks after symptom onset and it stayed above the cut-off 1.4 S/C up until five months after symptom onset,20 so this cut-off seems to be valid during this period. Elslande et al. reported, on the other hand, that the titer of IgG (N) started to decline after about a month, continuously declined and, as the result, seropositivity above the cut-off 1.4 S/C of severe patients after 180–240 days from positive PCR was 69.0% and that of mild patients was 33.3%.21 Considering this, we may need to adopt more appropriate cut-off for the assessment of long-term history of infection. Narasimhan et al. evaluated IgG (N) titer in 16 ex-COVID-19 patients who had recovered from COVID-19 three to 11 months earlier and showed that the titer was within the range of 0.2–1.4 S/C,22 which is consistent with the cut-offs indicated by the ROC analyses with IgG (N) shown in Table 2. Therefore, a cut-off lower than 1.4 S/C, such as 0.2 S/C, may be more suited to the assessment of long-term history of SARS-CoV-2 infection. This could also mean that the prevalence of SARS-CoV-2 infection when assessed by IgG (N) with the cut-off 1.4 S/C, could underestimate the actual prevalence of the population with the history of infection. In fact, the positive rate of IgG (N) in the cohort of presumed COVID-19 positive with the cut-off 1.4 S/C was 28.6% while that with the cut-off 0.2 S/C was 85.7%.
Conclusions
Both IgG (N) and IgG (RBD) titers were significantly elevated in subjects whose PCR never showed positive but suggestive of SARS-CoV-2 infection, which indicated the usefulness of the serological tests in complementing the shortcomings of PCR. The diagnostic performance of IgG (N) was more consistent than that of IgG (RBD). For a long-term prevalence study, a cut-off lower than the one indicated in the package insert (e.g. 0.2 S/C vs. 1.4 S/C with IgG (N)) was indicated to be more appropriate.
Acknowledgments
The authors are grateful to those at Tokyo Shinagawa Hospital who were involved in the promotion of this study including those who performed clinical testing, devotedly treated the COVID-19 patients, and those who promoted the enrollment of the study subjects, notably Ms. Kiyomi Kabasawa who strenuously promoted the data entry. This study was partly funded by Abbott Japan LLC.
References
- 1.Hu B., Guo H., Zhou P., Shi Z. Characteristics of SARS- CoV-2 and COVID-19. Nat Rev Microbiol. 2020 doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elezkurtaj S., Greuel S., Ihlow J., Michaelis E.G., Bischoff P., Kunze C.A., et al. Causes of death and comorbidities in hospitalized patients with COVID-19. Sci Rep. 2021;11:4263. doi: 10.1038/s41598-021-82862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanji J.N., Zelyas N., MacDonald C., Pabbaraju K., Khan M.N., Prasad A., et al. False negative rate of COVID-19 PCR testing: a discordant testing analysis. Virol J. 2021;18:13. doi: 10.1186/s12985-021-01489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallett S., Allen A.J., Graziadio A., Taylor S.A., Sakai N.S., Green K., et al. At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data. BMC Med. 2020;18:346. doi: 10.1186/s12916-020-01810-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao G., Tang S., Yang D., Shi W., Wang X., Wang H., et al. The potential transmission of SARS-CoV-2 from patients with negative RT-PCR swab tests to others: two related clusters of COVID-19 outbreak. Jpn J Infect Dis. 2020;73:399–403. doi: 10.7883/yoken.JJID.2020.165. [DOI] [PubMed] [Google Scholar]
- 7.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wen Z., Chi Y., Zhang L., Liu H., Du K., Li Z., et al. Coronavirus disease 2019: initial detection on chest CT in a retrospective multicenter study of 103 Chinese patients. Radiology: Cardiothoracic Imaging. 2020;2(2) doi: 10.1148/ryct.2020200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryan A., Pepper G., Wener M.H., Fink S.L., Morishima C., Chaudhary A., et al. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence testing in Idaho. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.00941-20. e00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simpson S., Kay F.U., Abbara S., Bhalla S., Chung J.H., Chung M., et al. Radiological society of North America Expert Consensus statement on reporting chest CT Findings related to COVID-19. Endorsed by the society of thoracic radiology, the American college of radiology and RSNA. Radiology: Cardiothoracic Imaging. 2020;2(2) doi: 10.1148/ryct.2020200152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genetic tests of 2019-nCoV assessed by clinical specimens. National Institute of Infectious Diseases; 2020. https://www.niid.go.jp/niid/images/lab-manual/2019-nCoV-17-current.pdf [Google Scholar]
- 12.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange1 S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–473. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 13.Fajnzylber J., Regan J., Coxen K., Corry H., Wong C., Rosenthal A., et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection - challenges and implications. N Engl J Med. 2020;383:6. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 15.Kawasuji H., et al. Transmissibility of COVID-19 depends on the viral load around onset in adult and symptomatic patients. PLoS One. 2020;15(12) doi: 10.1371/journal.pone.0243597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones D.L., et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci Total Environ. 2020;749:41364. doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., et al. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus in patients with coronavirus disease 2019. J Infect Dis. 2020;222:206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elslande J.V., Gruwier L., Godderis L., Vermeersch P. Estimated half-life of SARS-CoV-2 anti-spike antibodies more than double the half-life of anti-nucleocapsid antibodies in healthcare workers. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraley, et al. Cross-reactive antibody immunity against SARS-CoV-2 in children and adults. Cell Mol Immunol. 2021;18:1826–1828. doi: 10.1038/s41423-021-00700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maine G.N., Lao K.M., Krishnan S.M., Afolayan-Oloye O., Fatemi S., Kumar S., et al. Longitudinal characterization of the IgM and IgG humoral response in symptomatic COVID-19 patients using the Abbott Architect. J Clin Virol. 2020;133:104663. doi: 10.1016/j.jcv.2020.104663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elslande J.V., Oyaert M., Ailliet S., Ranst M.V., Lorent N., Weygaerde Y.V., et al. Longitudinal follow-up of IgG anti-nucleocapsid antibodies in SARS-CoV-2 infected patients up to eight months after infection. J Clin Virol. 2021;136:104765. doi: 10.1016/j.jcv.2021.104765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narasimhan M., Mahimainathan L., Araj E., Clark A.E., Markantonis J., Green A., et al. Clinical evaluation of the Abbott Alinity SARS-CoV-2 spike-specific quantitative IgG and IgM assays in infected, recovered, and vaccinated groups. J Clin Microbiol. 2021;59(7):7. doi: 10.1128/JCM.00388-21. e00388-21. [DOI] [PMC free article] [PubMed] [Google Scholar]