Abstract
Background
Based on reports on elevated cholesterol levels in cancer cells, strategies to lower cholesterol synthesis have been suggested as an antitumour strategy. However, cholesterol depletion has also been shown to induce tumour-promoting actions in tumour-associated macrophages (TAMs).
Methods
We performed lipidomic and transcriptomic analyses of human lung cancer material. To assess whether the TAM phenotype is shaped by secreted factors produced by tumour cells, primary human monocyte-derived macrophages were polarized towards a TAM-like phenotype using tumour cell-conditioned medium.
Findings
Lipidomic analysis of lung adenocarcinoma (n=29) and adjacent non-tumour tissues (n=22) revealed a significant accumulation of free cholesterol and cholesteryl esters within the tumour tissue. In contrast, cholesterol levels were reduced in TAMs isolated from lung adenocarcinoma tissues when compared with alveolar macrophages (AMs) obtained from adjacent non-tumour tissues. Bulk-RNA-Seq revealed that genes involved in cholesterol biosynthesis and metabolism were downregulated in TAMs, while cholesterol efflux transporters were upregulated. In vitro polarized TAM-like macrophages showed an attenuated lipogenic gene expression signature and exhibited lower cholesterol levels compared with non-polarized macrophages. A genome-wide comparison by bulk RNA-Seq confirmed a high similarity of ex vivo TAMs and in vitro TAM-like macrophages. Modulation of intracellular cholesterol levels by either starving, cholesterol depletion, or efflux transporter inhibition indicated that cholesterol distinctly shapes macrophage gene expression.
Interpretation
Our data show an opposite dysregulation of cholesterol homeostasis in tumour tissue vs. TAMs. Polarization of in vitro differentiated macrophages by tumour cell-conditioned medium recapitulates key features of ex vivo TAMs.
Keywords: Non-small cell lung cancer, innate immune response, ABCA1, ABCG1, ATR-101
Research in context.
Evidence before this study
Lung cancer is the leading cause of cancer-related mortality worldwide. Its predominant type, non-small cell lung cancer (NSCLC), accounts for approximately 85% of all lung cancers. Cholesterol plays a vital role for tumour growth since cancer cell proliferation heavily relies on its availability. Thus, strategies to either lower cholesterol synthesis or inhibit cholesterol uptake havex been suggested as potential antitumour therapies, but clinical data do not support improved survival. Immune cells present at the tumour site, including macrophages, have received increasing attention over the last years, and the phenotype of macrophages has been suggested to influence both tumour progression and survival. The cholesterol content of macrophages affects their phenotype. However, cholesterol homeostasis in macrophages from human lung cancer tissue had not been studied so far.
Added value of this study
We show that macrophages within tumours were depleted of cholesterol, although the lung tumours themselves were cholesterol-rich. In line with this finding, tumour-associated macrophages showed a decreased expression of genes involved in either cholesterol biosynthesis or uptake, whereas cholesterol efflux transporters were upregulated. These effects were also observed when polarizing macrophages differentiated from human blood monocytes with tumour cell-conditioned medium, implicating that this in vitro model recapitulates central attributes of macrophages that were directly obtained from tumour material. Furthermore, our data suggest that modulation of cholesterol levels in tumour-associated macrophages by efflux inhibition may help to reprogram these cells in order to inhibit their tumour-promoting properties.
Implications of all the available evidence
Our work identifies cholesterol depletion as a key feature of tumour-associated macrophages and urges new efforts to elucidate the interplay between lipid metabolism and the phenotype of macrophages within tumours. As described in this study, TAM-like macrophages recapitulate much of the in vivo polarization of tumour-promoting macrophages and can serve as a model for mechanistic and therapeutic approaches.
Alt-text: Unlabelled box
1. Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide. Its predominant type, non-small cell lung cancer (NSCLC), accounts for approximately 85% of all lung cancers and can be further classified into several subtypes based on histological characteristics. The most common subtypes are adenocarcinoma (40%), squamous-cell carcinoma (30%), and large-cell carcinoma (15%) [1,2].
Cancer research used to focus on the cancer cell itself. In recent years, however, the tumour microenvironment (TME) and its essential function in supporting malignancy have received increasing attention, especially in the context of immunotherapy. Among the diverse cell types of the TME, macrophages are the most abundant non–tumour cell type in most cancer types. These tumour-associated macrophages (TAMs) are discussed as a promising target for novel therapeutic approaches [3,4]. In NSCLC, a correlation between TAM density or phenotype and clinical outcomes has been described [3,5,6]. Cell culture models mimicking human TAMs to test therapeutic strategies are lacking, though.
Macrophages maintain tissue homeostasis by engulfing apoptotic or senescent cells, debris, and foreign material, orchestrate the immune response to pathogens by generating and resolving the inflammatory reaction, contribute to tissue development and repair, and have distinct metabolic functions. This broad spectrum of macrophage functions is only possible due to their high plasticity, which allows them to adopt diverse phenotypes in response to equally diverse microenvironmental conditions. Therefore, macrophages harbour a broad transcriptional repertoire with their extremes termed M1 or M2 macrophages. In an inflammatory environment, microbial stimuli, such as lipopolysaccharide (LPS), either alone or in concert with Th1-related cytokines like interferon-γ (IFNγ), induce classically activated M1 macrophages. After an acute inflammatory phase, macrophages undergo an M1 to M2 phenotype switch caused by exogenous and endogenous stimuli, gradually acquiring an anti-inflammatory phenotype and initiating a resolution phase associated with the inhibition of inflammation, scavenging of debris, angiogenesis, and tissue repair. IL4 and IL10 are regarded as typical inducers of an M2 phenotype [7,8].
In cancer, a high prevalence of TAMs with an M2-like polarization within the tumour stroma correlates with a poor prognosis. M2-like TAMs exert several tumour-promoting functions, including stimulation of angiogenesis, matrix remodelling, promotion of cancer cell proliferation, invasion, extravasation and metastasis, and immunosuppression [[9], [10], [11], [12]]. In the early stages of NSCLC, the density of tumour-preventing M1-TAMs in tumour islets is associated with extended survival [[13], [14], [15]].
Cholesterol plays a vital role within the tumour since cancer cell proliferation heavily relies on its availability [16]. Thus, strategies to either lower cholesterol synthesis or inhibit cholesterol uptake have been suggested as potential antitumour therapies [16,17]. While some observational evidence suggested positive effects upon respective interventions for some tumour entities, randomized control trials could not support survival benefits for lung cancer [18]. Why cholesterol reduction was not effective in the treatment of lung cancer remains elusive.
One might speculate that such approaches support a tumour-promoting macrophage phenotype: A recent study reported that ovarian cancer cells induced membrane-cholesterol efflux and depletion of lipid rafts from murine macrophages. Increased cholesterol efflux promoted IL4-mediated reprogramming, whereas IFNγ-induced gene expression was inhibited [19]. Moreover, depletion of the cholesterol efflux transporter ATP-binding cassette transporter G1 (ABCG1) inhibited tumour growth through the accumulation of cholesterol within macrophages in a mouse model of bladder cancer [20].
Cholesterol homeostasis in primary human TAMs has not been studied so far. Thus, the aim of the current study was to test the hypothesis that cholesterol levels are inversely regulated in cancer tissue and TAMs by lipidomic and transcriptomic analyses of human lung cancer material.
2. Methods
2.1. Materials
Cell culture medium RPMI 1640, FCS, penicillin, streptomycin, glutamine, trypsin-EDTA, and Accutase® were purchased from Sigma-Aldrich (Darmstadt, Germany). TE buffer and water molecular biology grade for RNA analysis were from AppliChem (Darmstadt, Germany). ATR-101 (#SML2802) and Methyl-β-cyclodextrin (#C4555) were from Sigma-Aldrich. Other chemicals were purchased from either Sigma-Aldrich (Darmstadt, Germany), Carl Roth (Karlsruhe, Germany), or VWR (Radnor, USA) unless stated otherwise.
2.2. Human lung tumour and normal lung tissue
Human lung tumour tissue and the autogenic non-tumour tissue was obtained from patients undergoing potentially curative resections (Völklingen Heart Center, Germany). The use of human material was reviewed and approved by the local ethics committee (State Medical Board of Registration, Saarland, Germany; permission no. 213/06). The informed consent of all participating subjects was obtained. The age, sex, and tumour stage of all donors are listed in Supplemental Table 1.
2.3. Lipidomic analysis of lung tumour and normal lung tissues
For lipidomic analyses, 22 normal lung tissues and 29 tumour tissues from adenocarcinoma patients undergoing lung resection were analyzed (for patient data, see supplemental table 1). 25 – 30 mg tissue were placed into a 1:1 mixture of water and methanol (20 µl/mg tissue) in 2 ml tubes filled with 2.8 mm ceramic beads (Precellys Ceramic Kit 2.8 mm, #10479394, Peqlab Biotechnologie, Erlangen, Germany). The samples were homogenized using a Precellys 24 (Peqlab) with the following program: 4 × 30 s at 6,000 rpm with 45 s breaks in between.
Lipid extraction was performed according to the method described by Bligh and Dyer [21] in the presence of non-naturally occurring lipid species as internal standards. The following lipid species were added as internal standards: PC 14:0/14:0, PC 22:0/22:0, PE 14:0/14:0, PE 20:0/20:0 (di-phytanoyl), PS 14:0/14:0, PS 20:0/20:0 (di-phytanoyl), PI 17:0/17:0, LPC 13:0, LPC 19:0, LPE 13:0, Cer d18:1/14:0, Cer d18:1/17:0, D7-FC, CE 17:0, CE 22:0, TG 17:0/17:0/17:0, TG 19:0/19:0/19:0, DG 14:0/14:0 and DG 20:0/20:0. The batch contained murine liver and brain pools as well as cell homogenates as internal quality control samples. Solvent and internal blanks were used to evaluate carry-over and background signals.
Tissue homogenates containing a wet weight of 2 mg, corresponding to 40 µl of the homogenate, were extracted. The chloroform phase was recovered by a pipetting robot (Tecan Genesis RSP 150, Tecan, Männedorf, Switzerland) and vacuum dried. The residues were dissolved in either 7.5 mM ammonium acetate in methanol/chloroform (3:1, v/v) (for low mass resolution tandem mass spectrometry) or chloroform/methanol/2-propanol (1:2:4 v/v/v) with 7.5 mM ammonium formate (for high resolution mass spectrometry).
The analysis of lipids was performed by direct flow injection analysis (FIA) using a triple quadrupole mass spectrometer (FIA-MS/MS; QQQ triple quadrupole; Micromass Quattro Ultima, Waters, Elstree, UK) and a hybrid quadrupole-Orbitrap mass spectrometer (FIA-FTMS; high mass resolution; QExactive Orbitrap, ThermoFisher Scientific, Waltham, MA, USA).
FIA-MS/MS (QQQ) was performed in positive ion mode using the analytical setup and strategy described previously [22,23]. A fragment ion of m/z 184 was used for phosphatidylcholine (PC), sphingomyelin (SM), [22] and lysophosphatidylcholine (LPC) [24]. The following neutral losses were applied: Phosphatidylethanolamine (PE) 141, phosphatidylserine (PS) 185, phosphatidylglycerol (PG) 189 and phosphatidylinositol (PI) 277 [25]. PE-based plasmalogens (PE P) were analyzed according to the principles described by Zemski-Berry [26]. Sphingosine-based ceramides (Cer) and hexosylceramides (HexCer) were analyzed using a fragment ion of m/z 264 [27]. Quantification for QQQ was achieved by calibration lines generated by addition of naturally occurring lipid species to pooled lung homogenate. Calibration lines were generated for the following naturally occurring species: PC 34:1, 36:2, 38:4, 40:0 and PC O-16:0/20:4; SM d18:1/16:0, 18:1, 18:0; LPC 16:0, 18:1, 18:0; PE 34:1, 36:2, 38:4, 40:6, and PE P-16:0/20:4; PS 34:1, 36:2, 38:4, 40:6; Cer d18:1/16:0, 18:0, 20:0, 24:1, 24:0; FC, CE 16:0,18:2,18:1,18:0.
The Fourier Transform Mass Spectrometry (FIA-FTMS) setup was previously described [28]. Triacylglycerol (TG), diacylglycerol (DG), and cholesteryl ester (CE) were recorded in positive ion mode FTMS in m/z range 500 – 1,000 for 1 min and a target resolution of 140,000 (at 200 m/z). CE was corrected for their species-specific response [29]. PC and PC ether (PC O) were measured in negative ion mode in m/z range 520 - 960. Multiplexed acquisition (MSX) was applied for the [M+NH4]+ of free cholesterol (FC) and the internal standard (D7-FC). Data processing details were described in Höring et al. [28] using the ALEX software [30], which includes peak assignment and intensity picking. The extracted data were exported to Microsoft Excel 2016 and further processed by self-programmed Macros.
FIA-FTMS quantification was performed by multiplication of the spiked IS amount with the analyte-to-IS ratio.
Lipid species were annotated according to the proposal for shorthand notation of lipid structures that are derived from mass spectrometry.[31] For QQQ, glycerophospholipid species annotation was based on the assumption of even-numbered carbon chains only. SM species annotation is based on the assumption that a sphingoid base with two hydroxyl groups is present.
For PE, PE P, PS, PI, PG, LPC, LPE, SM, Cer, and HexCer, QQQ data were analyzed. For PC, PC O, DG, TG, FC, and CE, FTMS data were used. FTMS permits differentiation of isobaric PC/PC O and direct analysis of FC [28].
Data for all lipids were used as input for hierarchical clustering and PCA plots. These analyses were performed with functions of the R package. Hierarchical clustering was performed with the hclust function. The heat map was drawn using the heatmap.2 function and PCA was done using the prcomp function. Data are available in the Mendeley repository (doi:10.17632/d57xrfymgm.1).
2.4. Human tumour-associated macrophages (TAMs) and alveolar macrophages (AMs)
TAMs and AMs were isolated from human lung tumour tissue or the autogenic non-tumour lung tissue obtained from patients undergoing lung resection as previously described [32]. In brief, TAMs were isolated after the tumour tissue had been enzymatically digested using the human tumour dissociation kit (#130-095-929, Miltenyi Biotec, Bergisch Gladbach, Germany) and the gentleMACS Octo Dissociator (Miltenyi Biotec) according to the manufacturer's instructions. Cells were washed, resuspended in RPMI 1640 medium without supplements, and incubated for 30 minutes in a T175 flask. Adherent cells were thoroughly washed with PBS to remove non-adherent cells and further cultivated in AM/TAM medium (RPMI 1640, 5% FCS, 100 U/ml penicillin G, 100 µg/ml streptomycin, 2 mM glutamine). TAMs were detached with accutase and seeded at a density of 5 × 105 cells per well into a 12-well plate.
AM isolation was performed according to a previously described method [33,34] with minor modifications. After visible bronchi were removed, the lung tissue was chopped and washed with 100–200 ml PBS. The washing buffer was collected, and AMs were obtained by centrifugation. The remaining erythrocytes were lysed by quickly resuspending the pellet in autoclaved water, immediately followed by washing with PBS. The cell pellet was mock-digested and seeded as described for TAMs. AM and TAM purity was > 95%, as assessed by CD68 staining [32].
2.5. Human monocyte-derived macrophages (MDMs)
Buffy coats were obtained from healthy adult blood donors (Blood Donation Center, Klinikum Saarbrücken, Germany). The use of human material for the isolation of primary cells was approved by the local ethics committee (permission no. 173/18).
Monocytes were isolated and polarized as previously described [35,36]. In brief, peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats by density gradient centrifugation using Lymphocyte Separation Medium 1077 (#C-44010, PromoCell, Heidelberg, Germany) and LeucoSEP tubes (#227290, Greiner Bio-One, Kremsmünster, Austria). Monocytes were purified by magnetic cell sorting using anti-CD14 microbeads (#130-050-201, Miltenyi Biotec) and LS Columns (#130-042-401, Miltenyi Biotec) according to the manufacturer's instructions, except that only 10% of the recommended bead amount was used [35]. Monocyte purity was > 95% as assessed by CD14 expression (data not shown).
Monocytes were seeded into a 12-well plate at a density of 5 × 105 cells/well or into a 24-well plate at a density of 2.5 × 105 cells/well, and differentiated into macrophages in MDM medium, i.e., standard growth medium (RPMI 1640 with 10% FCS, 100 U/ml penicillin G, 100 mg/ml streptomycin, 2 mM glutamine) supplemented with 20 ng/ml macrophage-colony stimulating factor (M-CSF, Miltenyi Biotec, #130-096-492) for six days. After differentiation, MDMs were polarized for another 24 hours. To this end, MDM medium was supplemented with 20 ng/ml IFNγ (Miltenyi Biotec, #130-096-484) and 100 ng/ml LPS (Ultrapure LPS from Escherichia coli K12 #tlrl-peklps, Invivogen, San Diego, CA, USA) for M1 polarization; either 20 ng/ml IL4 (Miltenyi Biotec, #130-093-921) or IL10 (Miltenyi Biotec, #130-093-948) for M2 polarization; or left without further supplementation for M0 macrophages. TAM-like macrophages were generated by cultivation in tumour cell-conditioned medium (TCM) supplemented with 20 ng/ml M-CSF. TCM was generated by seeding 0.5-1 × 106 A549 lung adenocarcinoma cells into a T75 culture flask and grown until confluency for three days. Subsequently, the supernatant was discarded, and standard growth medium was added to the cells. After 48 hours, the medium was sterile filtered (0.22 µm) and used immediately for macrophage polarization.
MDM Samples from one female (donor 1) and two male donors (donor 2 and 3) were used for RNA sequencing. For ATR-101 treatment, cell viability was assessed by MTT assay as previously described [36] to ensure that only non-toxic concentrations were used (data not shown).
2.6. RNA isolation
Total RNA from TAMs/AMs and polarized MDMs for RNA-Seq was extracted using the Direct-zol™ RNA MiniPrep Kit (#2050, Zymo Research, Irvine, CA, USA) according to the manufacturer's instructions. Prior to RNA isolation, cells were lysed with QIAzol Lysis Reagent (#79306, Qiagen, Hilden, Germany). Residual genomic DNA was removed by treatment with Ambion DNase I (#AM2222, ThermoFisher Scientific, Waltham, MA, USA).
For RT-qPCR, the High Pure RNA Isolation Kit (#11828665001, Roche Diagnostics International, Rotkreuz, Switzerland) was used according to the supplier's recommendations.
2.7. RNA sequencing
For transcriptome analysis of TAMs/AMs and MDM subsets, high throughput sequencing of cDNA using next-generation sequencing (NGS) was performed.
Libraries were prepared from 250 ng total RNA for MDM and 500 ng for TAM/AM samples with an RNA integrity (RIN) > 9, according to Agilent2100 Bioanalyzer and Agilent RNA 6000 Pico Kit (#5067-1513, Agilent Technologies, Santa Clara, CA, USA). Poly(A) enrichment was performed on the input total RNA using the NEBNext® Poly(A) mRNA Magnetic Isolation Module (#E7490, New England Biolabs, Ipswich, USA) according to the manufacturer's instructions. The cDNA library preparation was conducted with the NEBNext® UltraTM Directional RNA Library Prep Kit for Illumina® (#E7420, New England Biolabs) as recommended by the supplier. In brief, first- and second-strand cDNA synthesis were performed, followed by adapter ligation and PCR amplification of the final library (10 PCR cycles for AM/TAM samples and 12 cycles for MDM samples). PCR cleanup was performed using Agencourt AM-Pure® XP beads (# A63881, Beckmann Coulter, Krefeld, Germany). RNA libraries were sequenced for 2 × 55 bp on an Illumina HiSeq2500 Sequencer using a V3 paired-end flow cell.
Raw reads were subjected to quality control (QC) through FastQC v 0.11.2. Library adaptor removal and trimming were done using Fastx (v 0.0.13, http://hannonlab.cshl.edu/fastx_toolkit/).
Gene expression was quantified using the Salmon software v 0.8.2 [37] using gencode gene annotation (v 26). For the computation of PCA plots, gene expression values were normalized using the RUV approach [38]. All analyses were conducted in the R programming language. Genes were screened for the enrichment of transcription factor binding using the iRegulon software (Cytoscape plugin v 1.3). Processed data are available in the Mendeley repository (doi:10.17632/c3ntj95zgg.1), and raw data were deposited in the Gene Expression Omnibus (GEO) database (GEO datasets GSE162669 and GSE162698).
Gene ontology (GO) terms matching to differentially expressed genes were determined by the "protein annotation through evolutionary relationship" (PANTHER) Gene Ontology (GO) term classification system (v 14.0) [39], or ShinyGO (v 0.61) [40].
Gene expression clustering was done with hierarchical clustering in R. Cluster-specific lists of genes, associated transcription factors, and GO terms are available in the Mendeley repository (doi:10.17632/c3ntj95zgg.1).
The Seurat R-package (version 4.0.1)[41] was used for the analysis of single-cell data. UMI counts of single-cell data from the work of Lavin et al. [42] were obtained from GEO (GSE97168). Default parameters were used for the clustering of the single-cell data, and the top 2,000 most variable genes were analyzed. Elbow plot analysis suggested the use of the first 14 PCA components to capture expression variability as part of the UMAP-based clustering.
Expression of the known macrophage marker genes CD68, CSF1R, MAFB, MARCO, and MSR1 was used to identify macrophages in the resulting clustering. Differential gene expression between tumour (n=6) and lung (n=4) annotated cells was determined by the MAST statistical approach [43] with Benjamini-Hochberg correction. A threshold of FDR ≤ 0.05 was used to determine significantly differentially expressed genes.
2.8. Quantitative RT-PCR (qPCR)
Quaitative RT-PCR was performed as described previously [35,36,44]. RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (#4368813, Thermo Fisher Scientific, Waltham, MA, USA), in the presence of RNase inhibitor (RNaseOUT™, #10777019, Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer's instructions. qPCR was performed using the 5xHotFirePol EvaGreen qPCR Mix and a total volume of 20 µl. Primer sequences are given in supplementary table 2. The CFX96 touch™ Real-Time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) was used to quantify gene expression. Data were analyzed by absolute quantification using a standard curve of the PCR product cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA). ACTB served as a housekeeping gene.
2.9. Cholesterol quantification in macrophage samples
The quantification of cholesterol was performed using the Amplex Red® Cholesterol Assay Kit (#A12216, ThermoFisher Scientific, Waltham, MA, USA) as recommended by the supplier with minor modifications. In brief, cells were lysed by the addition of 400 µl Amplex Red® Cholesterol Assay Kit Reaction Buffer per 1 × 106 cells, followed by two freeze-thawing cycles (-80°C / room temperature). 50 µl of each sample were used for cholesterol quantification according to the manufacturer's instructions. Fluorescence was measured with a GloMax® Discover microplate reader (excitation: 520 nm, emission 580-640 nm; Promega, Madison, WI, USA). Background measurements were performed under the same conditions except that the reaction mix did not contain cholesterol oxidase. The cholesterol concentration was calculated with background-subtracted values and either expressed as µg cholesterol / 106 cells or ng cholesterol / µg cellular protein. Protein concentrations were determined by using the Pierce™ BCA™ Protein-Assay (#23225, ThermoFisher Scientific).
2.10. Flow cytometry
Flow cytometric analyses were performed as previously described [36]. Briefly, MDMs were differentiated for five to six days and polarized as described above. 2.5-5 × 105 cells were resuspended in FACSwash (PBS, 2.5% FCS, 0.1% sodium azide), blocked in human Fc Block solution (#564220, BD Biosciences, San Jose, CA, USA) for 15 minutes, and stained for 30 minutes on ice with anti-CD14-APC (#555399, BD Biosciences), anti-CD163-PE-CF594 (#562670, BD Biosciences), anti-CD80-BB515 (#565008, BD Biosciences) or anti-CD86-BB515 (#564544, BD Biosciences), and anti-HLA-DR-PerCP-Cy5.5 (#560652, BD Biosciences) antibodies. After washing, stained cells were resuspended in 1% paraformaldehyde in PBS prior to flow cytometric analysis on a BD LSRFortessa. Data were analyzed using BD FACSDiva software (BD Biosciences). Median fluorescence intensities of singlet cells were used to quantify surface marker expression.
2.11. Statistics
Data are shown as means ± SEM (bar graphs) or box plots with 25th/75thpercentile boxes, geometric medians (line), means (square), and SD as whiskers unless stated otherwise. Sample sizes for major readout parameters are listed in Table 1. An extended table detailing sample sizes and statistical tests is given in the Supplement (Supplemental Table 3). The Shapiro-Wilks test was used to determine whether data were distributed normally. Two groups were compared by Student's t-test (normal distribution) or Mann-Whitney U test (no normal distribution). For multiple groups, p-values were determined by ANOVA with post-hoc Bonferroni correction for normally distributed data or Mann-Whitney U test with Bonferroni correction for not normally distributed data. Outliers were identified using the Grubbs' test. The Wald test with Benjamini-Hochberg correction was used to identify DEGs within RNA-Seq datasets. The OriginPro 2020b software (OriginLab, Northampton, MA, USA) was used for statistical analyses and illustrations unless stated otherwise.
Table 1.
Sample sizes for major readouts.
| Lipidomics | Macrophages: RNA-Seq, cholesterol content | |
|---|---|---|
| Lung | 22 | RNA-Seq: 3, cholesterol: 4 (AM) |
| Tumour | 29 | RNA-Seq: 3, cholesterol: 4 (TAM) |
| M0 macrophages | 3 | |
| M1 macrophages | 3 | |
| M2(IL10) macrophages | 3 | |
| M2(IL4) macrophages | 3 | |
| TAM-like macrophages | 3 |
2.12. Role of the funding source
The study was supported by the Deutsche Forschungsgemeinschaft (DFG, #KI702) and the Landesforschungsförderungsprogramm Saarland (LFFP, #17/08). The funders did not participate in study design, data collection, data analyses, interpretation, or writing.
3. Results
3.1. Increased cholesteryl ester levels in human lung adenocarcinoma vs. normal lung tissue
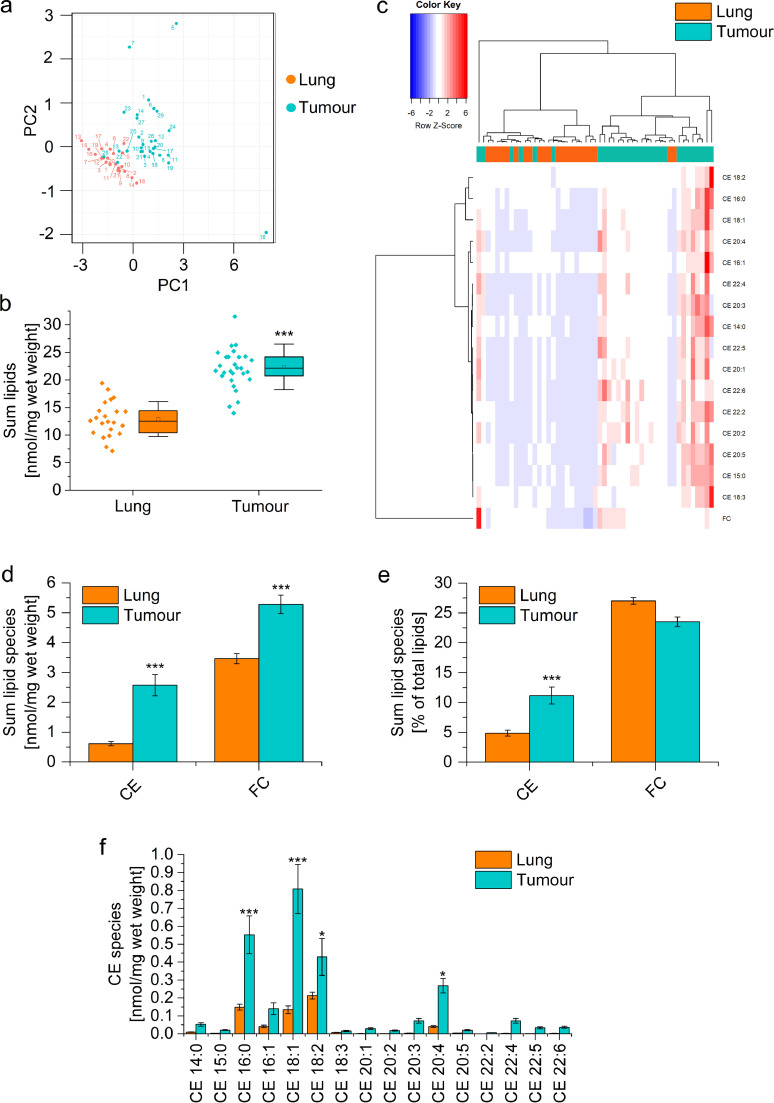
The lipid content of 29 lung adenocarcinoma tissues was compared with 22 samples from non-tumour adjacent tissue by lipidomic analysis. Principal component analysis (PCA) showed a clear distinction between normal and tumour tissue based on the abundance of lipid species (Fig. 1a). The total lipid concentration was distinctly higher within tumour tissues (Fig. 1b), with significant increases in cholesteryl esters (CE) and free cholesterol (FC) (Fig. 1c). Hierarchical clustering by Ward's method revealed two distinct clusters of lung and tumour samples based on CE and FC content (Fig. 1d).
Fig. 1.
Lipid content of human lung adenocarcinoma vs. adjacent non-tumour tissue. Lipid concentrations were determined in 22 normal and 29 tumour tissues by mass spectrometry. a: Principal component analysis. Sample numbers are annotated. b: Accumulated lipid concentrations of all detected lipids. c: Concentration of free cholesterol (FC) and accumulated concentration of cholesteryl esters (CE). d: Hierarchical clustering of sterol lipid concentrations using Ward's linkage and Euclidean distances. Normal tissues are highlighted as orange and tumour samples as green. e: CE and FC concentrations were normalized against total lipid content. f: Concentration of CE species. *p<0.05, ***p<0.001 vs. normal lung (B: Student's t-test, D-F: ANOVA with Bonferroni's post hoc test).
When CE concentrations were either expressed as a percentage of total lipids or as a percentage of total CE, the data showed that not only the lipid amount but also the lipid composition was altered within tumour tissue (Fig. 1e, Supplemental Figures 2a and b). CE 18:1 and 20:4 were increased in tumour vs. normal lung tissue when expressed either way, suggesting that these CE species are the most characteristic sterol lipids in lung adenocarcinomas (Fig. 1f, Supplemental Figure 2a and b). This assumption was supported by comparison to a previously published data set (Supplemental Figure 2c and d) [45].
In addition, we observed a higher overall concentration of phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), and triacylglycerol (TG) within the tumour tissues (Supplemental Figure 3a). However, only PI was also increased percentage-wise (Supplemental Figure 3b), which was also reflected on the level of individual PI species (Supplemental Figure 3c). In contrast, tumour samples contained lower levels of typical surfactant components, such as PC 30:0, PC 32:0, and the phosphatidylglycerols PG 32:0, PG 34:1, and PG 36:1, than normal lung tissues (Supplemental Figure 3c).
No correlation between the lipid composition within the tissue samples and the available patient data, i.e., age, gender, and tumour stage, was observed, which may be due to the limited size of our data set (data not shown).
3.2. Cholesterol depletion in ex vivo TAMs isolated from lung adenocarcinoma tissue
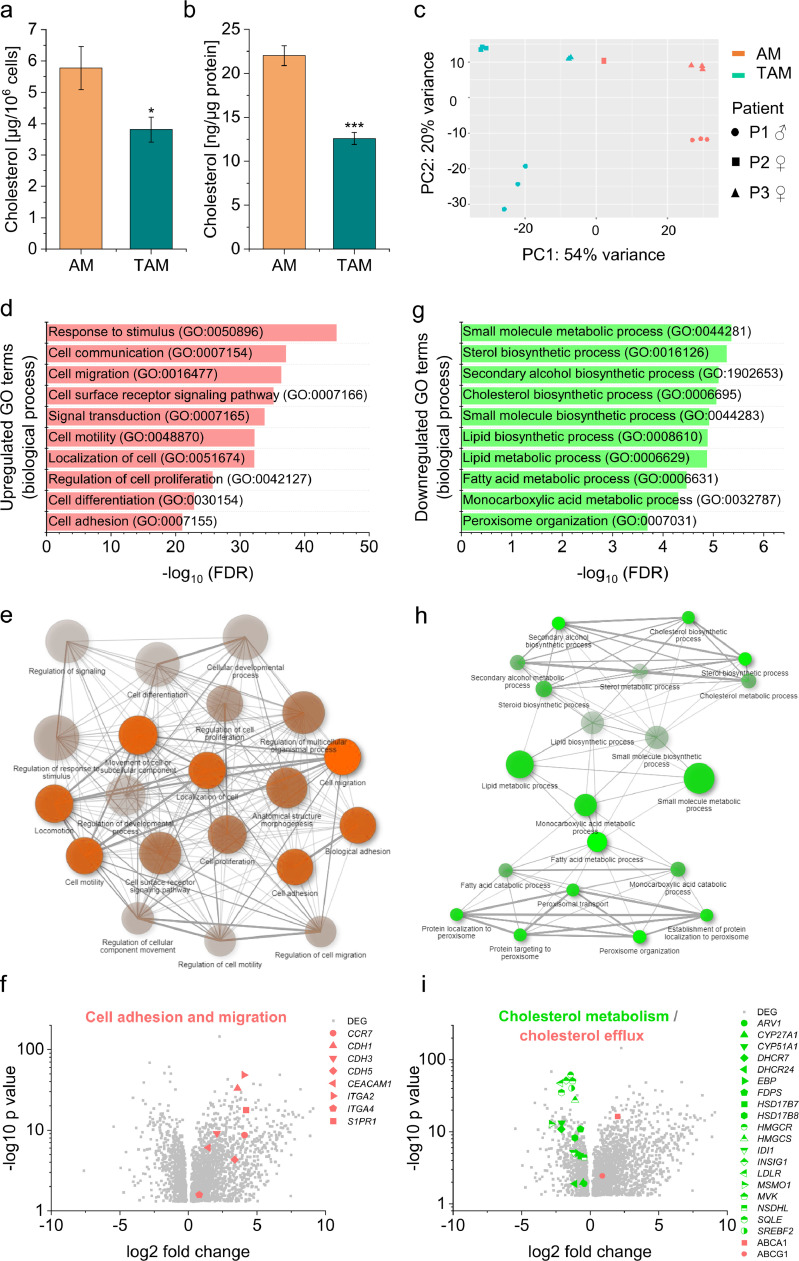
The cholesterol content of macrophages has been suggested to shape their phenotype [19,20]. Thus, we examined whether the cholesterol content was altered in TAMs isolated from lung adenocarcinoma tissue when compared with AMs obtained from adjacent normal lung tissue. Despite the high abundance of cholesteryl esters and free cholesterol in lung tumour tissue, TAMs showed lower cholesterol concentrations, both after normalization to cell count (Fig. 2a) or total protein content (Fig. 2b).
Fig. 2.
Cholesterol content and gene expression in macrophages from lung adenocarcinoma (TAMs) and adjacent non-tumour tissue (AMs). a, b: Cholesterol concentrations were measured in AM and TAM lysates and either normalized to cell count (a) or protein content (b) (n=4 in duplicate or triplicate) *p<0.05. ***p<0.001 vs. AMs (Student's t-test). c: Principal component analysis plot of RNA-Seq results from AMs (red) and TAMs (green) from three different patients (P1 – P3) prepared in technical triplicates. d-i: Analysis of genes differentially expressed in TAM vs. AMs. d-f: Genes upregulated in TAMs vs. AMs. g-h: Genes downregulated in TAMs vs. AMs. d, g: Top 10 gene ontology (GO) biological processes according to the PANTHER classification system. e, h: The relationship between enriched pathways was visualized with ShinyGO. Two pathways (nodes) are connected if they share 20% or more of the genes. More intensely coloured nodes contain more significantly enriched gene sets. Bigger nodes represent larger gene sets. Thicker edges represent more overlapped genes. f, i: Differentially expressed genes (DEGs) in TAMs vs. AMs are shown as volcano plots. Log2 fold change is plotted against –log10 p-value. Upregulated genes are highlighted in red, downregulated genes in green. H: Genes involved in cell adhesion and migration are highlighted. CCR7: CC chemokine receptor 7; CDH1/3/5: cadherin 1/3/5; CEACAM1: Carcinoembryonic antigen-related cell adhesion molecule 1; ITGA2/4: integrin alpha 2/4; S1PR1: sphingosine-1-phosphate receptor 1. I: Genes involved in cholesterol biosynthesis, metabolism, uptake, and efflux are highlighted. ARV1: acyl-CoA acyltransferase-related enzyme 2 required for viability; CYP27A1/51A1: cytochrome P450 family 27/51 subfamily A member 1; DHCR7/24: dehydrocholesterol reductase 7/24; EBP: emopamil binding protein; FDPS: farnesyl diphosphate synthase; HSD17B7/8: 17-beta hydroxysteroid dehydrogenase 7/8; HMGCR/S: HMG-CoA reductase/synthase; IDI1: isopentenyl-diphosphate delta isomerase 1; INSIG1: insulin-induced gene 1; LDLR: low-density lipoprotein receptor; MSMO1: methylsterol monooxygenase 1; MVK: mevalonate kinase; NSDHL: NAD(P)H steroid dehydrogenase-like; SQLE: squalene epoxidase; SREBF2: sterol regulatory element-binding protein 2; ABCA1: ATP-binding cassette transporter A1; ABCG1: ATP-binding cassette transporter G1.
TAMs and AMs were further analyzed by bulk mRNA sequencing. Cells were isolated from tumour tissue and autogenic lung tissue from one male and two female adenocarcinoma patients at comparable age and cancer stage. mRNA was prepared from both macrophage types of each individual in technical triplicates, and each replicate was sequenced separately.
PCA revealed a prominent discrimination between not only AMs and TAMs (right and left side of the plot, Fig. 2c) but also between all three patients. The matching technical replicates clustered, indicating that the sample and library preparation gave reproducible results.
A total of 4,812 genes were differentially expressed in TAMs compared with AMs, with 3,018 upregulated and 1,794 downregulated genes. We then compared our bulk RNA-Seq data to publicly available scRNA-Seq data from non-lymphocytic immune cells sorted from human early-stage lung adenocarcinoma lesions and adjacent non-involved lung tissue (GSE97168). This approach revealed a clear enrichment of differentially expressed genes (Supplemental Figure 4), which substantiates the validity of our data.
Genes that were upregulated in TAMs play a role in signal processing, cell adhesion, and cell motility according to GO term analysis (Fig. 2d). Network analysis indicated that the top 20 enriched pathways were highly interconnected (Fig. 2e). Specific examples of upregulated genes involved in cell migration and adhesion, e.g., C-C chemokine receptor type 7 (CCR7), cadherins 1-3 (CDH1-3), and sphingosine-1-phosphate receptor 1 (S1PR1), are highlighted within in the volcano plot in Fig. 2f. Furthermore, the expression of genes promoting extracellular matrix degradation, angiogenesis, and chemotaxis was also substantially enhanced in TAMs (Supplemental Figure 5).
Many downregulated genes were associated with biological processes involving lipid metabolism or biosynthesis (Fig. 2g and h). In accordance, transcription factor analyses revealed a downregulation of the central lipogenic transcription factors SREBF1 and 2 (Table 2). Downregulated genes encoding for enzymes required for cholesterol biosynthesis included, e.g., HMG-CoA reductase (HMGCR), mevalonate kinase (MVK), and 7-dehydrocholesterol reductase (DHCR7). Moreover, the LDL receptor (LDLR) was downregulated while the cholesterol efflux transporters ATP-binding cassette transporter A1 and G1 (ABCA1, ABCG1) were upregulated, suggesting that TAMs favour cholesterol export over import (Fig. 2i).
Table 2.
Transcription factors regulating DEGs in TAMs vs. AMs as predicted by iRegulon
| Upregulated in TAMs |
Downregulated in TAMs |
||
|---|---|---|---|
| TF | No. of targets | TF | No. of targets |
| GATA2 | 969 | MYB | 699 |
| HNF18 | 852 | SPI1 | 544 |
| CEBPG | 589 | KLF6 | 504 |
| ZNF503 | 473 | SOX4 | 183 |
| NAP1L1 | 370 | GABPA | 121 |
| FOXM1 | 314 | SREBF1 | 85 |
| TEAD4 | 239 | TAF1A | 82 |
| BATF | 202 | SRF | 74 |
| TCF12 | 135 | SREBF2 | 34 |
| E2F4 | 118 | ||
3.3. Characterization of an in vitro TAM model
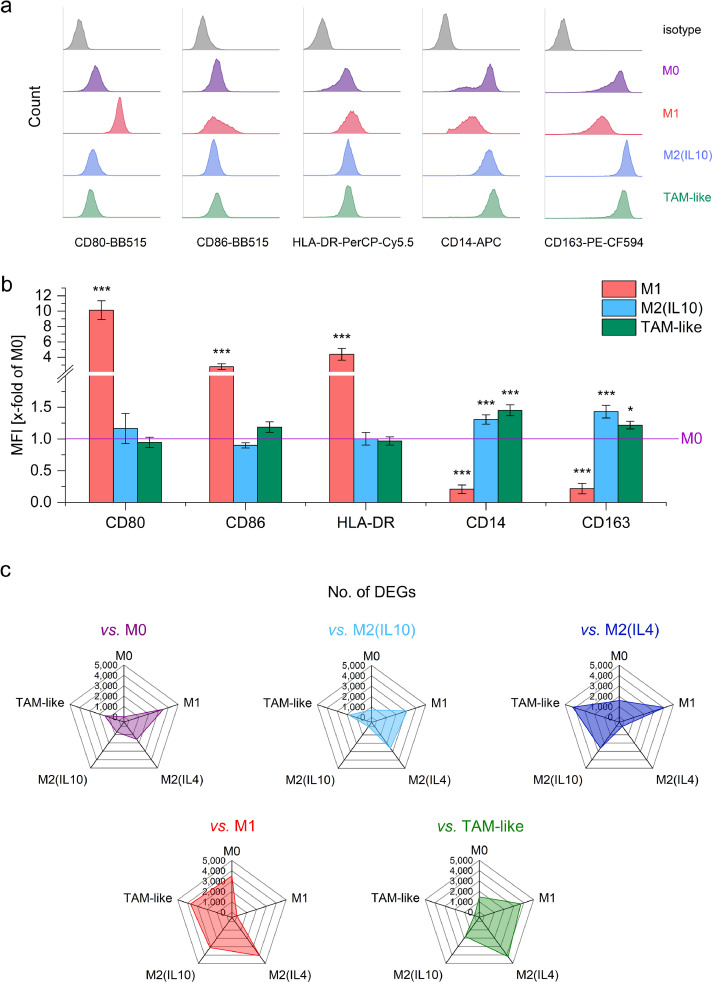
In order to assess whether differences in the transcriptomic profile of TAMs vs. AMs are caused by factors secreted by tumour cells, primary in vitro differentiated human MDMs were polarized towards a TAM-like phenotype by cultivating them in medium conditioned by the lung adenocarcinoma cell line A549. In addition, MDMs were polarized with either LPS/IFNγ towards an M1 phenotype or with either IL10 or IL4 towards an M2 phenotype as established models of macrophage polarization [46,47].
Flow cytometric analysis suggested that TAM-like macrophages resemble M2 macrophages (Fig. 3 a and b). Further characterization was performed by bulk RNA-Seq of M0, M1, M2(IL4), M2(IL10), and TAM-like macrophages. MDMs from three different donors were polarized and analyzed as biological replicates. M1, as well as M2(IL4) macrophages, expressed a high number of genes that were differentially expressed when compared with TAM-like cells, indicating that they are highly different from not only TAM-like but also from all the other macrophage types (Fig. 3c). The lower number of DEGs in TAM-like vs. M0 or M2(IL10) macrophages suggested that TAM-like cells are more similar to these two types.
Fig. 3.
In vitro polarized macrophages. a, b: Human MDMs were polarized with LPS/IFNγ (M1), IL10 (M2(IL10)), or A549-conditioned supernatant for 24 hours, and surface marker expression was quantified by flow cytometry. a: Representative histograms. b: Median fluorescence intensities (MFIs) were expressed as x-fold of unpolarized M0 cells. *p<0.05, ***p<0.001 vs. M0 (ANOVA with Bonferroni's post hoc test; CD80, CD86: n=4 individual donors in duplicates; HLA-DR, CD14, CD163: n=7 donors in duplicates. c: Human monocyte-derived macrophages were polarized with LPS/IFNγ (M1), IL10 (M2(IL10)), IL4 (M2(IL4), or A549-conditioned supernatant for 24 h and bulk RNA-Seq was performed (n=3 individual donors). The radar graphs show the number of differentially expressed genes in each group vs. all other polarization schemes.
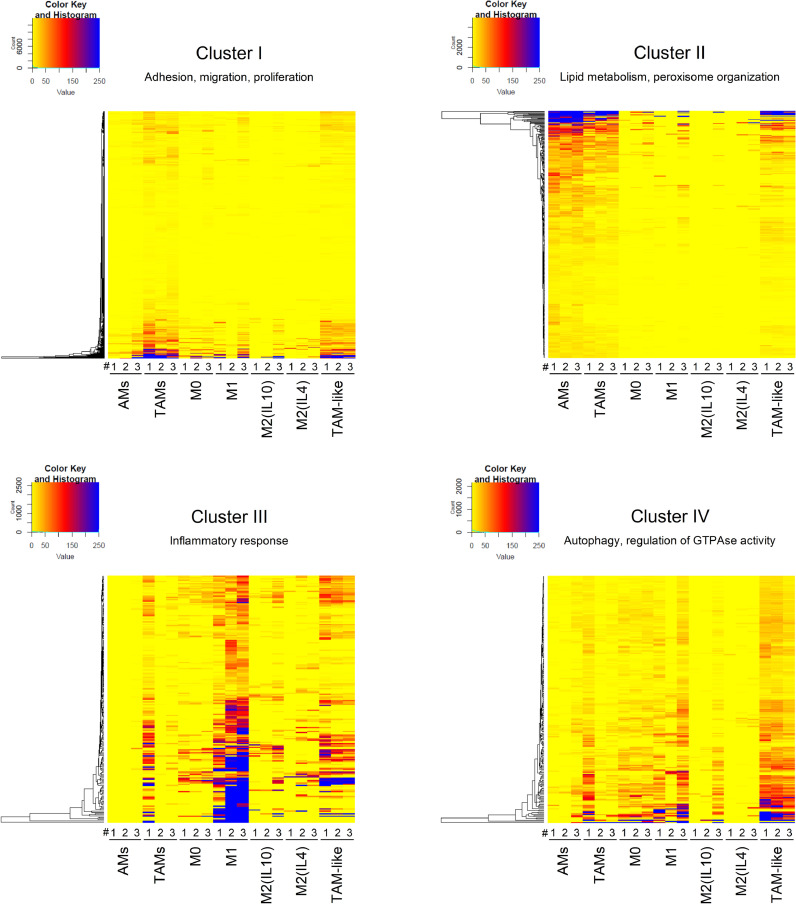
To compare ex vivo TAMs with in vitro TAM-like macrophages, TAM vs. AM DEGs were clustered based on their expression pattern. Among the resulting eight clusters, clusters I-IV contained the highest number of genes, i.e., 88% of all DEGs. GO term analysis indicated that the 1,806 genes in cluster I were associated with cell migration, adhesion, and proliferation. Cluster II contained 741 genes involved in lipid metabolism and peroxisome organization, whereas cluster III (433 genes) was linked to inflammation and immune response. The 403 genes within cluster IV were mostly related to autophagy and the regulation of GTPase activity (Fig. 4). The heat maps in Fig. 4 show the gene expression signatures within clusters I-IV in AM/TAM and in in vitro polarized macrophages. TAM and TAM-like macrophages displayed similar signatures within clusters I and II, i.e., the clusters containing the highest numbers of genes. Similar to TAMs, TAM-like macrophages exhibited a mixed M1-/M2-like signature (cluster III). Cluster IV genes were induced in TAM-like macrophages, whereas the expression pattern was not consistent in ex vivo TAMs. Gene expression data from clusters V-VIII did not reveal additional cell-type-specific characteristics, in part due to the limited number of genes contained within these clusters (Supplemental Figure 6). In summary, bulk-RNA-Seq suggested a high similarity of ex vivo TAMs and in vitro TAM-like macrophages.
Fig. 4.
Comparison of gene expression signatures of TAMs/AMs and in vitro polarized macrophages. Genes differentially expressed in TAMs vs. AMs were clustered according to their expression pattern, and gene expression in in vitro polarized cells and ex vivo macrophages is visualized as one heatmap per cluster. Clusters were linked to distinct functions based on the predominant GO terms associated with genes contained within each cluster. TAMs/AMs: mean values of technical replicates for each donor are shown. MDMs: Individual values per donor are shown.
3.4. Cholesterol depletion in in vitro generated TAM-like macrophages
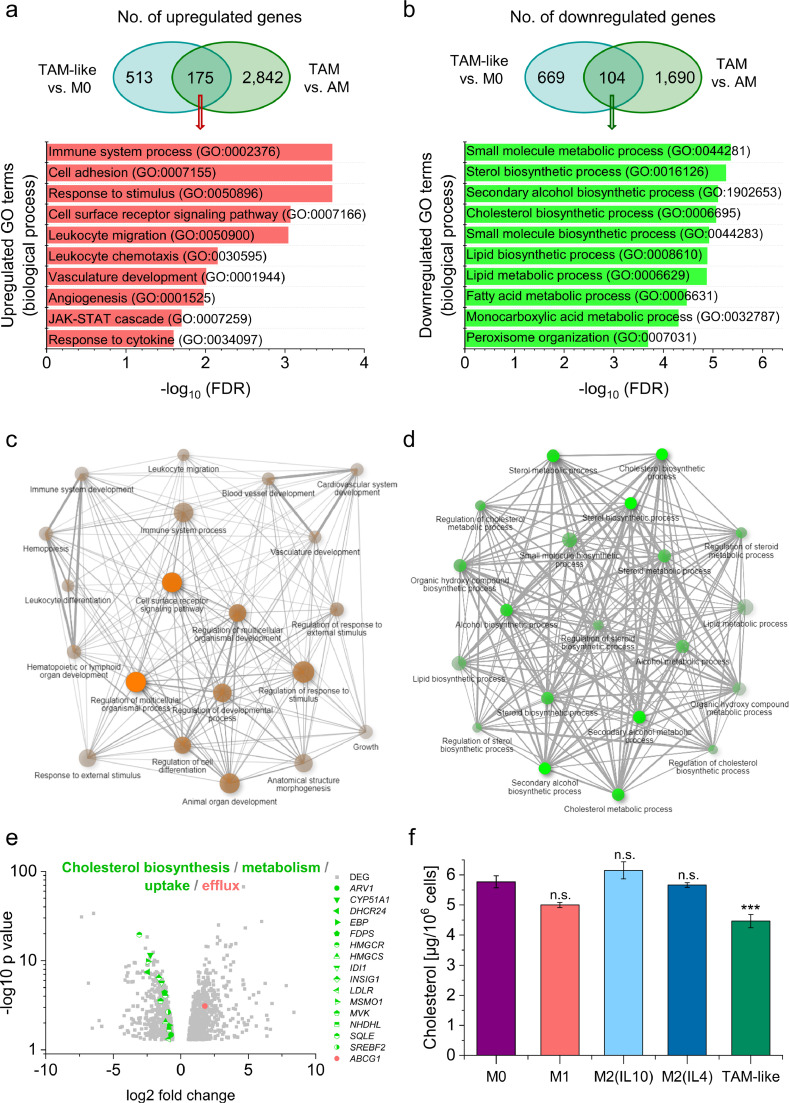
Next, we evaluated the function of genes that were similarly regulated by the addition of A549 supernatant and the TME by intersection analysis (Fig. 5a-d). The 175 genes that were upregulated in both TAM-like vs. M0 and TAM vs. AM are, e.g., involved in cell adhesion, migration, chemotaxis, and angiogenesis (Fig. 5a and c). Primary TAMs and TAM-like cells shared 104 downregulated genes, which were mainly associated with cholesterol biosynthesis and metabolism (Fig. 5b), and the respective enriched GO terms were highly interconnected (Fig. 5d). Fig. 5e highlights the differential expression of specific genes involved in cholesterol homeostasis in TAM-like vs. M0 macrophages. As seen in primary TAMs (Fig. 2g), downregulated genes involved HMGCR, MVK, SREBF2, and LDLR, whereas the cholesterol efflux transporter ABCG1 was upregulated. In contrast, of the 16 genes highlighted within Fig. 5e, only four genes were regulated similarly in M1 vs. M0 (ARV1, DHCR24, EBP, INSIG1), two in M2(IL4) vs. M0 (HMGCS, LDLR), and not a single one in M2(IL10) vs. M0, suggesting that only the TAM-like phenotype recapitulates the changes in cholesterol homeostasis found in ex vivo TAMs. This assumption was supported by the finding that TAM-like cells, but not M1, M2(IL10), or M2(IL4) macrophages, exhibited a lowered cholesterol content when compared with M0 cells (Fig. 5f). These results indicate that TME- and A549-derived factors alter cholesterol homeostasis in TAMs and TAM-like macrophages in a similar manner.
Fig. 5.
TAM-like macrophages resemble ex vivo TAMs. A-D: Genes induced (a, c) or downregulated (b, d) by treatment of MDMs with A549-conditioned supernatant (TAM-like vs. M0) that were similarly regulated in TAMs vs. AMs were subjected to GO term analysis. a, b: Significantly enriched GO terms according to the PANTHER classification system. c, d: The relationship between enriched pathways was visualized with ShinyGO. Two pathways (nodes) are connected if they share 20% or more of the genes. More intensely coloured nodes contain more significantly enriched gene sets, and bigger nodes represent larger gene sets. Thicker edges represent more overlapped genes. e: Differentially expressed genes (DEGs) involved in cholesterol biosynthesis, metabolism, uptake, and efflux in TAM-like vs. M0 macrophages are shown as a volcano plot. Log2 fold change is plotted against –log10 p-value. Upregulated genes are highlighted in red, downregulated genes in green. ARV1: acyl-CoA acyltransferase-related enzyme 2 required for viability; CYP51A1: cytochrome P450 family 51 subfamily A member 1; DHCR24: dehydrocholesterol reductase 24; EBP: emopamil binding protein; FDPS: farnesyl diphosphate synthase; HMGCR/S: HMG-CoA reductase/synthase; IDI1: isopentenyl-diphosphate delta isomerase 1; INSIG1: insulin-induced gene 1; LDLR: low density lipoprotein receptor; MSMO1: methylsterol monooxygenase 1; MVK: mevalonate kinase; NSDHL: NAD(P)H steroid dehydrogenase-like; SQLE: squalene epoxidase; SREBF2: sterol regulatory element-binding protein 2; ABCG1: ATP-binding cassette transporter G1. f: The cholesterol content was measured in lysates of differentially polarized MDMs and normalized to cell count (n=3, triplicates). ***p<0.001 (ANOVA with Bonferroni's post hoc test).
Since the A549-conditioned medium showed lower levels of cholesterol in comparison to fresh medium (Supplemental Figure 7a), we hypothesized that deprivation of cholesterol and other nutrients might contribute to the phenotypic changes in TAM-like cells. Thus, we compared TAM-like macrophages with cells kept in starving medium containing only 1% instead of 10% FCS for 24 hours. Starving led to a decline of intracellular cholesterol levels when normalized to initial cell count (Supplemental Figure 7b). This effect was not seen when normalizing cholesterol concentrations to total protein content, presumably due to a reduction of total protein levels under starving conditions (Supplemental Figure 7c).
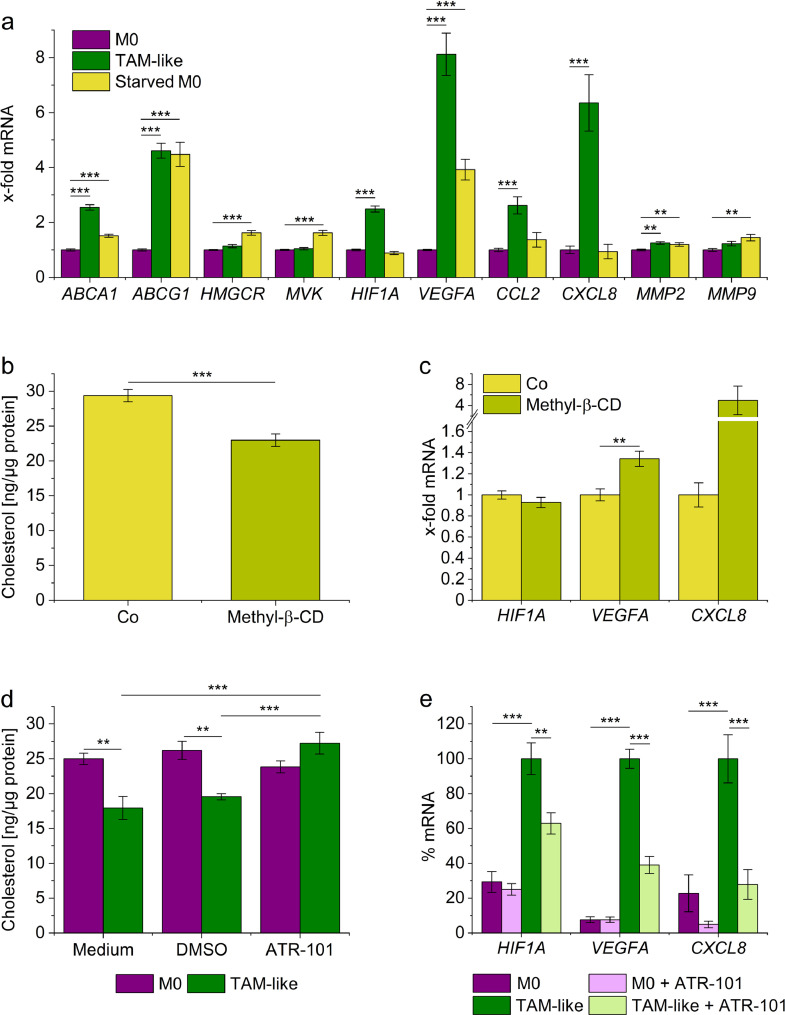
We then quantified a small set of TAM signature genes by qPCR. Whereas the expression levels of ABCA1 and ABCG1 were increased by starving, genes associated with cholesterol biosynthesis, i.e., HMGCR and MVK, were also induced, suggesting differential regulation of cholesterol homeostasis in TAM-like and starved macrophages. Although starving induced some characteristic TAM-like marker genes (VEGFA, MMP2, MMP9), others (HIF1A, CCL2, CXCL8) remained unaffected (Fig. 6a). Moreover, starving did not result in the typical changes in surface marker expression previously seen in TAM-like cells, indicating that starving alone is not sufficient to induce a full TAM-like phenotype (Supplemental Figure 8a).
Fig. 6.
Modulation of the cholesterol content affects the expression of TAM-specific genes in macrophages. a: Human MDMs were cultured in medium containing 10% FCS (normal medium, M0) or 1% FCS (starving medium, starved M0) for 24 hours. mRNA expression levels were quantified by qPCR, normalized to ACTB, and expressed as x-fold of M0 (n=4, triplicates). b, c: MDMs were incubated with PBS (Co) or Methyl-β-cyclodextrin (Methyl-β-CD, 5 mM in PBS) for one hour, followed by incubation in media containing 1% FCS for 24 hours. b: Cellular cholesterol content (n=4, triplicates). c: mRNA expression was measured by qPCR using ACTB as a housekeeping gene (n=3, triplicates). d: MDMs were cultured in normal medium (M0) or A549 tumour cell-conditioned medium (TAM-like) and either left otherwise untreated (Medium), co-treated with the solvent control DMSO (0.25%), or ATR-101 (25 µM) for 24 hours. Cholesterol levels were normalized against the total cellular protein (n=4, triplicates). e: MDMs were cultured in normal medium (M0) or A549 tumour cell-conditioned medium (TAM-like) and either co-treated with the solvent control DMSO (0.25%) or ATR-101 (25 µM) for 24 hours. mRNA expression was measured by qPCR, normalized against ACTB, and expressed as a percentage of DMSO-treated TAM-like macrophages (n=3, duplicates). **p<0.01, ***p<0.001 (a, c-e: ANOVA with Bonferroni's post hoc test; b: t-test).
The potential causal relationship between cholesterol content and macrophage phenotype was further assessed after depleting cholesterol with methyl-β-cyclodextrin, a method widely used in the literature [19,48]. A substantial cholesterol depletion that lasted for at least 24 hours was only possible when keeping the cells in starving medium, implicating that cholesterol levels might be easily replenished by cholesterol present in full medium (Fig. 6b, Supplemental Figure 7d and e). Thus, the observed effects should be considered as additive to starving. We found indeed that VEGFA expression can be further increased by cholesterol depletion and observed a similar tendency for CXCL8 (Fig. 6c). However, neither HIF1A expression levels nor surface marker expression patterns (Supplemental Figure 8b) were similar to TAM-like cells, supporting the assumption that the overall effect of A549-conditioned medium represents a result of an interplay of nutrient deprivation and multiple secreted factors.
Next, we used ATR-101, an inhibitor of the cholesterol efflux transporters ABCA1 and ABCG1 [49], to elucidate the role of these transporters for the polarization of macrophages by A549-conditioned medium. ATR-101 effectively inhibited cholesterol depletion in TAM-like macrophages (Fig. 6d). Importantly, ATR-101 treatment also partially reversed the effects of tumour-cell conditioned medium on the expression of HIF1A, VEGFA, and CXCL8 in TAM-like cells, whereas M0 macrophages remained unaffected by the treatment.
In summary, the modulation of cellular cholesterol levels by tumour-cell conditioned media contributes to the TAM-like-specific gene expression pattern, which may be targeted using inhibitors of cholesterol efflux transporters.
4. Discussion
An altered lipid metabolism represents a common feature of cancer cells. Cancer cells adjust their metabolic programs to meet abnormal energy demands caused by enhanced proliferation and survival. Changes in lipid metabolism have been reported in lung cancer, with varying phenotypes across subtypes and tumour stages [50].
Only a few lipidomic analyses of primary human lung cancer material have been performed to date. Marien et al. showed that phospholipid profiles were highly altered in non-small cell lung cancer vs. normal lung tissues [51]. Whereas most sphingomyelins and phosphatidylserine species were decreased, the abundance of specific phosphatidylinositols, as well as long phosphatidylethanolamines and phosphatidylcholine, was increased within tumour tissue [51]. We also found that sphingomyelin species, e.g., SM 36:1, 40:1, 42:1, and 42:2, were reduced, whereas phosphatidylinositol species, such as PI 38:3 and PI 40:4, were elevated within tumours. Interestingly, these phospholipids represent a source of lipid second messengers that can activate the phosphatidylinositol 3‐kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway. Increased activation of the PI3K/Akt/mTOR pathway has been linked to both tumorigenesis and the progression of cancer, including non-small cell lung cancer [52].
Another study on lipidomic profiles of tissues from non-small cell lung cancer and corresponding control tissues included not only the major phospholipid classes but also neutral lipids, sphingolipids, and glycerophospholipids [45]. In line with our observations, this study not only showed that tumour tissues exhibited higher levels of free cholesterol and cholesteryl esters but also identified CE 18:1 and 20:4 as CE species characteristic for non-small cell lung cancer. Furthermore, the main surfactant components phosphatidylcholine (e.g., PC 30:0, PC 32:0) and phosphatidylglycerol (e.g., PG 32:0, PG 34:1, and PG 36:1) were less abundant in tumour tissue, which was confirmed by our study. As expected because of the small sample size, we did not observe any correlation of the lipid profile with tumour stage, age, or gender. Of note, additional patient characteristics were not available, and potential confounding can therefore not be excluded.
Interestingly, Eggers et al. [45] also reported that an increased concentration of free cholesterol and cholesteryl esters correlated with the fraction of necrotic areas within the tumour. Necrotic areas are formed in tumours when the cellular requirement for oxygen exceeds its supply by the poorly organized tumour vasculature. When TAMs enter these areas, hypoxia-inducible factors (HIFs), particularly HIF1α, and various HIF target genes, such as vascular endothelial growth factor A, are upregulated [53,54]. In our study, TAMs from human lung tumours showed a transcriptomic profile that suggested pro-angiogenic functions along with an increased migratory capacity when compared with AMs from normal tissue.
Reports on the metabolic phenotype of TAMs are highly diverse, which may be due to the dynamic adaptation of TAMs in response to metabolic changes accompanying tumour progression [54]. For example, human TAMs from colon carcinoma patients, and M2-like TAMs, in particular, exhibit lower glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and succinate dehydrogenase (SDH) activities than normal macrophages, suggesting that TAMs reduce glycolysis rates as an adaption to the nutrient-deprived TME [55]. However, TAMs can switch from oxidative phosphorylation to glycolysis in response to cancer cell-derived signals. For example, lactate as a by-product of aerobic glycolysis in thyroid carcinoma cells caused increased glycolysis rates, along with an M1-like polarization, in TAMs [56]. Due to their poor glycolytic activity, M2-like TAMs show an elevated consumption of fatty acids as an energy source, at least in several murine models [54]. Thus, inhibition of fatty acid metabolism in TAMs has been suggested as a strategy to enhance the antitumour effect of cancer therapies [57]. Our study does not support these conclusions. Indeed, we found that genes associated with fatty acid metabolism were downregulated in TAMs, which may point towards species-specific effects. This assumption is further supported by a report showing that fatty acid oxidation is dispensable for IL4-mediated polarization in human, but not in murine macrophages [58].
Despite the diversity of TAM phenotypes across the literature, we found a substantial overlap of DEGs identified in our data and a previous study employing scRNA-Seq to decipher the innate immune landscape in early-stage lung adenocarcinoma [42]. Of note, the total number of genes detected by bulk RNA-Seq was much higher and thus revealed a larger number of differentially expressed genes that could not be identified previously. However, we cannot exclude potential confounding due to the limited sample size and the fact that additional patient data were not available.
Two other studies that performed RNA-Seq with FACS-sorted macrophages from squamous cell carcinoma patients compared non-tumour macrophages (i.e., a mixture of alveolar macrophages, interstitial macrophages, and monocyte-derived macrophages) with TAMs. The study by Sarode et al. [59] heavily focused on the Wnt signalling pathway, and only a GO term analysis was given in the paper. We also found that GO terms associated with Wnt signalling were enriched in TAMs vs. AMs, but these GO terms were not the most prominent ones. These differences might be related to cancer-type specific effects.
The study by Zheng et al. [60] gave no details about the DEGs between non-tumour and tumour macrophages, except for a brief comparison of non-tumour macrophages with different TAM subtypes, which identified M1-TAM and M2-TAM markers. In our hands, the proposed M1-TAM markers UBXN4 and ACTR6 were not overexpressed in TAMs, whereas the M2-TAM marker MFSD12 was (1.5-fold ± 0.1, p=0.005). However, we did not observe a clear M1/M2 phenotype within the TAM population. Thus, it remains unclear whether this gene can serve as an M2-TAM marker across distinct lung cancer subtypes.
Cancer cells can scavenge cholesterol from the tumour microenvironment through upregulation of apolipoproteins and their receptors, which may result in cholesterol depletion in other cells within the tumour, such as TAMs [16]. The major finding of our study is that cholesterol, although highly enriched in tumour tissue, is depleted in TAMs. This effect correlated with an upregulation of the cholesterol efflux transporters ABCA1 and ABCG1, which are known to show additive activity in promoting macrophage reverse cholesterol transport [61]. A recent study reported increased activity of ABCA1 in TAMs in a mouse model of ovarian cancer, leading to an enhanced efflux of membrane cholesterol. Subsequently, the loss of cholesterol-rich membrane micro-domains caused an amplification of IL4 receptor activity, whereas IFNγ signalling was impaired. Thus, loss of cholesterol supported the M2-like tumour-promoting TAM phenotype [19]. A prominent role of cholesterol transporters in macrophage polarization had previously been suggested by a report showing that ABCA1 knockout macrophages were more prone to M1 polarizing signals, whereas the response to M2 signals was attenuated [62]. In accordance, ABCA1 was reported to inhibit TLR signalling through changes in membrane lipid organization that disrupt the recruitment of TLR adapter molecules [63]. Furthermore, ABCG1 knockout in myeloid cells also led to a phenotypic shift of TAMs from an M2-like to an M1-like tumouricidal phenotype in murine models of bladder cancer and melanoma. The mechanisms by which the expression and activity of ABC transporters are upregulated in TAMs remain to be elucidated.
Apart from shaping the macrophage phenotype, cholesterol efflux in TAMs has been speculated to support tumour growth by feeding cholesterol to cancer cells, thereby reducing their energetic costs [19,64]. This mechanism would require an increase of cholesterol biosynthesis in macrophages, which was indeed detected in the aforementioned murine model of ovarian cancer [19]. In our hands, however, genes involved in cholesterol biosynthesis were downregulated in TAMs vs. AMs, suggesting that the proposed feeding mechanism does not apply.
It has not yet been clearly defined which signals mediate cholesterol depletion in TAMs. Hyaluronic acid, an extracellular matrix component that is highly abundant in the TME, has been suggested to promote the reduction of membrane cholesterol in murine TAMs. This assumption was supported by the finding that cancer cell-conditioned medium did not affect cholesterol levels in macrophages when pretreated with hyaluronidase, and hyaluronic acid itself was able to reduce the cholesterol content [19]. Again, the mechanisms may differ between mice and men: while Goossens et al. [19] observed an increased activity of cholesterol efflux transporters, Abca1 or Abcg1 mRNA levels remained unchanged. In contrast, our data indicate that ABCA1 and ABCG1 transcripts are highly induced in human TAMs.
In order to evaluate whether reduced cholesterol levels and differences in the transcriptomic profile of TAMs vs. AMs were caused by factors secreted by tumour cells, we polarized primary human MDMs towards a TAM-like phenotype by cultivating them in tumour cell-conditioned medium. The cholesterol content of the conditioned medium was decreased, most likely due to its uptake by A549 cells. Therefore, A549-conditioned medium may mimic a cholesterol-depleted tumour-microenvironment. The treatment with tumour cell-conditioned medium replicated changes in macrophage gene expression and cholesterol content seen in ex vivo TAMs, e.g., upregulation of ABCG1. TAM-like macrophages also exhibited lower cholesterol levels than all other MDM phenotypes. Therefore, this cell model might represent a highly suitable model to gain mechanistic insights into TAM actions and, more importantly, to test potential therapeutic interventions. The inhibition of cholesterol efflux by ABCA1/ABCG1 inhibitors, such as ATR-101, might represent a promising tool to reprogram TAMs, as indicated by our observation that ATR-101 inhibits the induction of genes involved in angiogenesis and chemotaxis in TAM-like cells. ATR-101 has been considered to be a candidate for the treatment of adrenocortical cancer, mainly due to its ability to inhibit acyl-coenzyme A:cholesterol O-acyltransferase 1 (ACAT1), an enzyme that catalyzes the esterification of intracellular FC. In this context, the accumulation of FC within tumour cells was suggested to result in tumour cell apoptosis [65]. So far, it is unclear whether ATR-101 may have additional beneficial effects based on its influence on TAMs via ABCA1/ABCG1 inhibition. Further studies are required to determine whether ATR-101 also influences the TAM-phenotype in vivo.
In summary, our work identifies altered cholesterol homeostasis as a key feature of TAMs and urges new efforts to elucidate the interplay between lipid metabolism and the TAM phenotype.
5. Contributors
JH wrote the manuscript and prepared the figures. AD prepared tissue samples for lipidomic analysis, which was performed by MH and GL. MH, GL, JH, AS, and VH analyzed lipidomics data. JH quantified cholesterol in macrophage samples. AD prepared samples for RNA sequencing under the supervision of MS. Sequencing was performed by GG and JW. Sequencing data were analyzed by MHS, NW, AD, and JH. HSS, CD, SA, BD, and JH acquired and analyzed flow cytometry data. qPCR data were acquired and analyzed by HSS. HH provided the lung tissue and patient data. All authors participated in study design and data interpretation. AKK initiated and directed the study and revised the manuscript. JH and AKK verified all underlying data. All authors have read and approved the final version of the manuscript.
Data sharing statement
Processed sequencing data are available in the Mendeley repository (doi:10.17632/c3ntj95zgg.1), and raw data were deposited in the Gene Expression Omnibus (GEO) database (GEO datasets GSE162669 and GSE162698). Lipidomics data are available in the Mendeley repository (doi:10.17632/d57xrfymgm.1).
Funding
Deutsche Forschungsgemeinschaft (DFG), Landesforschungsförderungsprogramm Saarland (LFPP)
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
The authors thank Theo Ranßweiler and Daniela Oster for excellent technical assistance. The study was financially supported by the Deutsche Forschungsgemeinschaft (DFG, #KI702) and the Landesforschungsförderungsprogramm Saarland (LFFP, #17/08).
We acknowledge support by the Deutsche Forschungsgemeinschaft (DFG) and Saarland University within the funding programme Open Access Publishing.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103578.
Appendix. Supplementary materials
References
- 1.Rivas-Fuentes S., Salgado-Aguayo A., Pertuz Bell.oso S., Gorocica R.osete P., Alvarado-Vasquez N., Aquino-Jarquin G. Role of chemokines in non-small cell lung cancer: angiogenesis and inflammation. J Cancer. 2015;6:938–952. doi: 10.7150/jca.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad A., Gadgeel S.M. Lung cancer and personalized medicine: Novel therapies and clinical management. Preface. Adv Exp Med Biol. 2016;890:v–vi. [PubMed] [Google Scholar]
- 3.Conway E.M., Pikor L.A., Kung S.H., Hamilton M.J., Lam S., Lam W.L., Bennewith K.L. Macrophages, inflammation, and lung cancer. Am J Respir Crit Care Med. 2016;193:116–130. doi: 10.1164/rccm.201508-1545CI. [DOI] [PubMed] [Google Scholar]
- 4.Mills C.D., Lenz L.L., Harris R.A. A breakthrough: macrophage-directed cancer immunotherapy. Cancer Res. 2016;76:513–516. doi: 10.1158/0008-5472.CAN-15-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quatromoni J.G., Eruslanov E. Tumor-associated macrophages: function, phenotype, and link to prognosis in human lung cancer. Am J Transl Res. 2012;4:376–389. [PMC free article] [PubMed] [Google Scholar]
- 6.Takeya M., Komohara Y. Role of tumor-associated macrophages in human malignancies: friend or foe? Pathol Int. 2016;66:491–505. doi: 10.1111/pin.12440. [DOI] [PubMed] [Google Scholar]
- 7.Wang N., Liang H., Zen K. Molecular mechanisms that influence the macrophage M1-M2 polarization balance. Front Immunol. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sica A., Erreni M., Allavena P., Porta C. Macrophage polarization in pathology. Cell Mol Life Sci. 2015;72:4111–4126. doi: 10.1007/s00018-015-1995-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohtaki Y., Ishii G., Nagai K., Ashimine S., Kuwata T., Hishida T., Nishimura M., Yoshida J., Takeyoshi I., Ochiai A. Stromal macrophage expressing CD204 is associated with tumor aggressiveness in lung adenocarcinoma. J Thorac Oncol. 2010;5:1507–1515. doi: 10.1097/JTO.0b013e3181eba692. [DOI] [PubMed] [Google Scholar]
- 10.Zhang B., Yao G., Zhang Y., Gao J., Yang B., Rao Z., Gao J. M2-polarized tumor-associated macrophages are associated with poor prognoses resulting from accelerated lymphangiogenesis in lung adenocarcinoma. Clinics (Sao Paulo) 2011;66:1879–1886. doi: 10.1590/S1807-59322011001100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D., Nair V.S., Xu Y., Khuong A., Hoang C.D., Diehn M., West R.B., Plevritis S.K., Alizadeh A.A. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan A., Hsiao Y.J., Chen H.Y., Chen H.W., Ho C.C., Chen Y.Y., Liu Y.C., Hong T.H., Yu S.L., Chen J.J., Yang P.C. Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci Rep. 2015;5:14273. doi: 10.1038/srep14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeni E., Mazzetti L., Miotto D., Lo Cascio N., Maestrelli P., Querzoli P., Pedriali M., De Rosa E., Fabbri L.M., Mapp C.E., Boschetto P. Macrophage expression of interleukin-10 is a prognostic factor in nonsmall cell lung cancer. Eur Respir J. 2007;30:627–632. doi: 10.1183/09031936.00129306. [DOI] [PubMed] [Google Scholar]
- 14.Ohri C.M., Shikotra A., Green R.H., Waller D.A., Bradding P. Macrophages within NSCLC tumour islets are predominantly of a cytotoxic m1 phenotype associated with extended survival. Eur Respir J. 2009;33:118–126. doi: 10.1183/09031936.00065708. [DOI] [PubMed] [Google Scholar]
- 15.Ma J., Liu L., Che G., Yu N., Dai F., You Z. The M1 form of tumor-associated macrophages in non-small cell lung cancer is positively associated with survival time. BMC Cancer. 2010;10:112. doi: 10.1186/1471-2407-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guillaumond F., Bidaut G., Ouaissi M., Servais S., Gouirand V., Olivares O., Lac S., Borge L., Roques J., Gayet O., Pinault M., Guimaraes C., Nigri J., Loncle C., Lavaut M.N., Garcia S., Tailleux A., Staels B., Calvo E., Tomasini R., Iovanna J.L., Vasseur S. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2015;112:2473–2478. doi: 10.1073/pnas.1421601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding X., Zhang W., Li S., Yang H. The role of cholesterol metabolism in cancer. Am J Cancer Res. 2019;9:219–227. [PMC free article] [PubMed] [Google Scholar]
- 18.Xia D.K., Hu Z.G., Tian Y.F., Zeng F.J. Statin use and prognosis of lung cancer: a systematic review and meta-analysis of observational studies and randomized controlled trials. Drug Des Devel Ther. 2019;13:405–422. doi: 10.2147/DDDT.S187690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goossens P., Rodriguez-Vita J., Etzerodt A., Masse M., Rastoin O., Gouirand V., Ulas T., Papantonopoulou O., Van Eck M., Auphan-Anezin N., Bebien M., Verthuy C., Vu Manh T.P., Turner M., Dalod M., Schultze J.L., Lawrence T. Membrane cholesterol efflux drives tumor-associated macrophage reprogramming and tumor progression. Cell Metab. 2019;29:1376–1389. doi: 10.1016/j.cmet.2019.02.016. e1374. [DOI] [PubMed] [Google Scholar]
- 20.Sag D., Cekic C., Wu R., Linden J., Hedrick C.C. The cholesterol transporter ABCG1 links cholesterol homeostasis and tumour immunity. Nat Commun. 2015;6:6354. doi: 10.1038/ncomms7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 22.Liebisch G., Lieser B., Rathenberg J., Drobnik W., Schmitz G. High-throughput quantification of phosphatidylcholine and sphingomyelin by electrospray ionization tandem mass spectrometry coupled with isotope correction algorithm. Biochim Biophys Acta. 2004;1686:108–117. doi: 10.1016/j.bbalip.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Liebisch G., Binder M., Schifferer R., Langmann T., Schulz B., Schmitz G. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS) Biochim Biophys Acta. 2006;1761:121–128. doi: 10.1016/j.bbalip.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Liebisch G., Drobnik W., Lieser B., Schmitz G. High-throughput quantification of lysophosphatidylcholine by electrospray ionization tandem mass spectrometry. Clin Chem. 2002;48:2217–2224. [PubMed] [Google Scholar]
- 25.Matyash V., Liebisch G., Kurzchalia T.V., Shevchenko A., Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zemski-Berry K.A., Murphy R.C. Electrospray ionization tandem mass spectrometry of glycerophosphoethanolamine plasmalogen phospholipids. J Am Soc Mass Spectrom. 2004;15:1499–1508. doi: 10.1016/j.jasms.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Liebisch G., Drobnik W., Reil M., Trumbach B., Arnecke R., Olgemoller B., Roscher A., Schmitz G. Quantitative measurement of different ceramide species from crude cellular extracts by electrospray ionization tandem mass spectrometry (ESI-MS/MS) J Lipid Res. 1999;40:1539–1546. [PubMed] [Google Scholar]
- 28.Höring M., Ejsing C.S., Krautbauer S., Ertl V.M., Burkhardt R., Liebisch G. Accurate quantification of lipid species affected by isobaric overlap in fourier-transform mass spectrometry. J Lipid Res. 2021;62 doi: 10.1016/j.jlr.2021.100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Höring M., Ejsing C.S., Hermansson M., Liebisch G. Quantification of cholesterol and cholesteryl ester by direct flow injection high-resolution fourier transform mass spectrometry utilizing species-specific response factors. Anal Chem. 2019;91:3459–3466. doi: 10.1021/acs.analchem.8b05013. [DOI] [PubMed] [Google Scholar]
- 30.Husen P., Tarasov K., Katafiasz M., Sokol E., Vogt J., Baumgart J., Nitsch R., Ekroos K., Ejsing C.S. Analysis of lipid experiments (ALEX): a software framework for analysis of high-resolution shotgun lipidomics data. PLoS One. 2013;8:e79736. doi: 10.1371/journal.pone.0079736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liebisch G., Vizcaino J.A., Kofeler H., Trotzmuller M., Griffiths W.J., Schmitz G., Spener F., Wakelam M.J. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res. 2013;54:1523–1530. doi: 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoppstädter J., Seif M., Dembek A., Cavelius C., Huwer H., Kraegeloh A., Kiemer A.K. M2 polarization enhances silica nanoparticle uptake by macrophages. Front Pharmacol. 2015;6:55. doi: 10.3389/fphar.2015.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoppstädter J., Diesel B., Eifler L.K., Schmid T., Brüne B., Kiemer A.K. Glucocorticoid-induced leucine zipper is downregulated in human alveolar macrophages upon toll-like receptor activation. Eur J Immunol. 2012;42:1282–1293. doi: 10.1002/eji.201142081. [DOI] [PubMed] [Google Scholar]
- 34.Hoppstädter J., Diesel B., Zarbock R., Breinig T., Monz D., Koch M., Meyerhans A., Gortner L., Lehr C.M., Huwer H., Kiemer A.K. Differential cell reaction upon toll-like receptor 4 and 9 activation in human alveolar and lung interstitial macrophages. Respir Res. 2010;11:124. doi: 10.1186/1465-9921-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dembek A., Laggai S., Kessler S.M., Czepukojc B., Simon Y., Kiemer A.K., Hoppstädter J. Hepatic interleukin-6 production is maintained during endotoxin tolerance and facilitates lipid accumulation. Immunobiology. 2017;222:786–796. doi: 10.1016/j.imbio.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Dahlem C., Siow W.X., Lopatniuk M., Tse W.K.F., Kessler S.M., Kirsch S.H., Hoppstadter J., Vollmar A.M., Muller R., Luzhetskyy A., Bartel K., Kiemer A.K. Thioholgamide a, a new anti-proliferative antitumor agent, modulates macrophage polarization and metabolism. Cancers (Basel) 2020;12 doi: 10.3390/cancers12051288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Risso D., Ngai J., Speed T.P., Dudoit S. Normalization of RNA-Seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. Panther version 14: more genomes, a new panther go-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge S.X., Jung D., Shinygo Ya.o R. A graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavin Y., Kobayashi S., Leader A., Amir E.D., Elefant N., Bigenwald C., Remark R., Sweeney R., Becker C.D., Levine J.H., Meinhof K., Chow A., Kim-Shulze S., Wolf A., Medaglia C., Li H., Rytlewski J.A., Emerson R.O., Solovyov A., Greenbaum B.D., Sanders C., Vignali M., Beasley M.B., Flores R., Gnjatic S., Pe'er D., Rahman A., Amit I., Merad M. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750–765. doi: 10.1016/j.cell.2017.04.014. e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., Linsley P.S., Gottardo R. Mast: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valbuena Per.ez J.V., Linnenberger R., Dembek A., Bruscoli S., Riccardi C., Schulz M.H., Meyer M.R., Kiemer A.K., Hoppstädter J. Altered glucocorticoid metabolism represents a feature of macroph-aging. Aging Cell. 2020;19:e13156. doi: 10.1111/acel.13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eggers L.F., Müller J., Marella C., Scholz V., Watz H., Kugler C., Rabe K.F., Goldmann T., Schwudke D. Lipidomes of lung cancer and tumour-free lung tissues reveal distinct molecular signatures for cancer differentiation, age, inflammation, and pulmonary emphysema. Sci Rep. 2017;7:11087. doi: 10.1038/s41598-017-11339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sica A., Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shapouri-Moghaddam A., Mohammadian S., Vazini H., Taghadosi M., Esmaeili S.A., Mardani F., Seifi B., Mohammadi A., Afshari J.T., Sahebkar A. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 48.Mahammad S., Parmryd I. Cholesterol depletion using methyl-beta-cyclodextrin. Methods Mol Biol. 2015;1232:91–102. doi: 10.1007/978-1-4939-1752-5_8. [DOI] [PubMed] [Google Scholar]
- 49.Burns V.E., Kerppola T.K. ATR-101 inhibits cholesterol efflux and cortisol secretion by ATP-binding cassette transporters, causing cytotoxic cholesterol accumulation in adrenocortical carcinoma cells. Br J Pharmacol. 2017;174:3315–3332. doi: 10.1111/bph.13951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L., Zhu B., Zeng Y., Shen H., Zhang J., Wang X. Clinical lipidomics in understanding of lung cancer: opportunity and challenge. Cancer Lett. 2020;470:75–83. doi: 10.1016/j.canlet.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 51.Marien E., Meister M., Muley T., Fieuws S., Bordel S., Derua R., Spraggins J., Van de Plas R., Dehairs J., Wouters J., Bagadi M., Dienemann H., Thomas M., Schnabel P.A., Caprioli R.M., Waelkens E., Swinnen J.V. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int J Cancer. 2015;137:1539–1548. doi: 10.1002/ijc.29517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan A.C. Targeting the PI3K/Akt/mTOR pathway in non-small cell lung cancer (NSCLC) Thorac Cancer. 2020;11:511–518. doi: 10.1111/1759-7714.13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porta C., Sica A., Riboldi E. Tumor-associated myeloid cells: New understandings on their metabolic regulation and their influence in cancer immunotherapy. FEBS J. 2018;285:717–733. doi: 10.1111/febs.14288. [DOI] [PubMed] [Google Scholar]
- 54.Vitale I., Manic G., Coussens L.M., Kroemer G., Galluzzi L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019;30:36–50. doi: 10.1016/j.cmet.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 55.Miller A., Nagy C., Knapp B., Laengle J., Ponweiser E., Groeger M., Starkl P., Bergmann M., Wagner O., Haschemi A. Exploring metabolic configurations of single cells within complex tissue microenvironments. Cell Metab. 2017;26:788–800. doi: 10.1016/j.cmet.2017.08.014. e786. [DOI] [PubMed] [Google Scholar]
- 56.Arts R.J., Plantinga T.S., Tuit S., Ulas T., Heinhuis B., Tesselaar M., Sloot Y., Adema G.J., Joosten L.A., Smit J.W., Netea M.G., Schultze J.L., Netea-Maier R.T. Transcriptional and metabolic reprogramming induce an inflammatory phenotype in non-medullary thyroid carcinoma-induced macrophages. Oncoimmunology. 2016;5 doi: 10.1080/2162402X.2016.1229725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hossain F., Al-Khami A.A., Wyczechowska D., Hernandez C., Zheng L., Reiss K., Valle L.D., Trillo-Tinoco J., Maj T., Zou W., Rodriguez P.C., Ochoa A.C. Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances cancer therapies. Cancer Immunol Res. 2015;3:1236–1247. doi: 10.1158/2326-6066.CIR-15-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Namgaladze D., Brüne B. Fatty acid oxidation is dispensable for human macrophage IL-4-induced polarization. Biochim Biophys Acta. 2014;1841:1329–1335. doi: 10.1016/j.bbalip.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 59.Sarode P., Zheng X., Giotopoulou G.A., Weigert A., Kuenne C., Gunther S., Friedrich A., Gattenlohner S., Stiewe T., Brune B., Grimminger F., Stathopoulos G.T., Pullamsetti S.S., Seeger W., Savai R. Reprogramming of tumor-associated macrophages by targeting beta-catenin/FOSL2/ARID5a signaling: A potential treatment of lung cancer. Sci Adv. 2020;6:eaaz6105. doi: 10.1126/sciadv.aaz6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng X., Weigert A., Reu S., Guenther S., Mansouri S., Bassaly B., Gattenlohner S., Grimminger F., Pullamsetti S., Seeger W., Winter H., Savai R. Spatial density and distribution of tumor-associated macrophages predict survival in non-small cell lung carcinoma. Cancer Res. 2020;80:4414–4425. doi: 10.1158/0008-5472.CAN-20-0069. [DOI] [PubMed] [Google Scholar]
- 61.Wang X., Collins H.L., Ranalletta M., Fuki I.V., Billheimer J.T., Rothblat G.H., Tall A.R., Rader D.J. Macrophage ABCA1 and ABCG1, but not SR-BI, promote macrophage reverse cholesterol transport in vivo. J Clin Invest. 2007;117:2216–2224. doi: 10.1172/JCI32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pradel L.C., Mitchell A.J., Zarubica A., Dufort L., Chasson L., Naquet P., Broccardo C., Chimini G. ATP-binding cassette transporter hallmarks tissue macrophages and modulates cytokine-triggered polarization programs. Eur J Immunol. 2009;39:2270–2280. doi: 10.1002/eji.200838867. [DOI] [PubMed] [Google Scholar]
- 63.Ito A., Hong C., Rong X., Zhu X., Tarling E.J., Hedde P.N., Gratton E., Parks J., Tontonoz P. LXRs link metabolism to inflammation through ABCA1-dependent regulation of membrane composition and TLR signaling. Elife. 2015;4:e08009. doi: 10.7554/eLife.08009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sica A., Bleve A., Garassino M.C. Membrane cholesterol regulates macrophage plasticity in cancer. Cell Metab. 2019;29:1238–1240. doi: 10.1016/j.cmet.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 65.LaPensee C.R., Mann J.E., Rainey W.E., Crudo V., Hunt S.W., Hammer G.D. ATR-101, a selective and potent inhibitor of acyl-coa acyltransferase 1, induces apoptosis in H295r adrenocortical cells and in the adrenal cortex of dogs. Endocrinology. 2016;157:1775–1788. doi: 10.1210/en.2015-2052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.