Abstract
The Saccharomyces cerevisiae DNA repair gene PHR1 encodes a photolyase that catalyzes the light-dependent repair of pyrimidine dimers. PHR1 expression is induced at the level of transcription by a variety of DNA-damaging agents. The primary regulator of the PHR1 damage response is a 39-bp sequence called URSPHR1 which is the binding site for a protein(s) that constitutes the damage-responsive repressor PRP. In this communication, we report the identification of two proteins, Rph1p and Gis1p, that regulate PHR1 expression through URSPHR1. Both proteins contain two putative zinc fingers that are identical throughout the DNA binding region, and deletion of both RPH1 and GIS1 is required to fully derepress PHR1 in the absence of damage. Derepression of PHR1 increases the rate and extent of photoreactivation in vivo, demonstrating that the damage response of PHR1 enhances cellular repair capacity. In vitro footprinting and binding competition studies indicate that the sequence AG4 (C4T) within URSPHR1 is the binding site for Rph1p and Gis1p and suggests that at least one additional DNA binding component is present in the PRP complex.
In the yeast Saccharomyces cerevisiae, more than 20 different genes are induced in response to UV radiation and a variety of chemical agents that damage DNA (1, 16). Induction is the final step in a series of events that includes damage recognition, signal transduction, and modification of transcription factors regulating expression of damage-responsive genes. Damage recognition and/or early steps in signal transduction are carried out by proteins encoded by RAD9, RAD17, RAD24, and MEC3, while MEC1, RAD53, and DUN1 encode downstream protein kinases that are required for most transcriptional induction (reviewed in reference 48). In contrast to the components of the signaling pathway, little is known about the transcription factors that act as downstream effectors of the pathway.
To date, two transcriptional regulators targeted by the MEC1/RAD53 pathway have been identified: Swi6p and Crt1p (also known as Rfx1p). Swi6p is the regulatory subunit for the G1-specific transcription factors MBF and SBF. In response to methyl methanesulfonate (MMS)-generated damage, Swi6p is phosphorylated and represses transcription of the cyclin genes CLN1 and CLN2, thereby contributing to delay of G1 progression (41). Crt1p represses transcription of the RNR2, RNR3, and RNR4 genes by binding to X boxes found in the 5′ flanking regions of these genes. Hyperphosphorylation of Crt1p in response to DNA damage or replication stress leads to dissociation of Crt1p from the X boxes and derepression (20). Genes containing X boxes or binding sites for MBF or SBF make up only a small subset of the known damage-inducible genes in yeast. Thus, additional damage-responsive regulators remain to be identified. Of particular interest are regulators of genes encoding DNA repair enzymes.
PHR1 encodes the apoenzyme for the DNA repair enzyme photolyase (31). Transcription of the gene is induced in response to a large number of different DNA-damaging agents, as well as by passage through the diauxic shift (38, 44). Three promoter elements control basal-level and induced expression of PHR1 (35). An upstream activation sequence, UASPHR1, is required for both basal-level and induced expression and is the promoter element responsible for induction at the diauxic shift (44). The damage response is regulated primarily through an upstream repressing sequence, URSPHR1, which consists of a 39-bp region containing a 22-bp palindrome (35, 39). Mutations within the palindrome reduce or abolish repression, as does deletion of the entire 39-bp region, while transfer of URSPHR1 into the context of a heterologous promoter both represses expression and confers a low level of damage inducibility (35, 39). Crude extracts from nonirradiated cells contain a protein(s), called PRP, that binds to this region, while extracts from irradiated cells do not (39). Efficient derepression requires a third promoter element called an upstream essential sequence which consists of three related elements (35). In this communication, we describe the isolation and initial characterization of two damage-responsive transcriptional regulators, RPH1 and GIS1, that control PHR1 expression by binding to URSPHR1.
MATERIALS AND METHODS
Plasmids.
Standard recombinant DNA techniques (25) were used to construct the plasmids described here. The structures of all plasmids were confirmed by restriction analysis and in many cases by DNA sequence analysis across crucial regions.
pGBS116 is a 2μm-based PHR1-lacZ reporter plasmid described previously (35, 38). pGBS408 is a derivative of pBM1499 (15) in which the EcoRI fragment containing UASGAL was replaced with a 53-bp oligonucleotide containing URSPHR1 and several flanking nucleotides (−32 to −83 of the PHR1 promoter [35]), thus placing HIS3 expression under the control of URSPHR1. The URS fragment was generated by PCR using oligonucleotides EcoRI-URStop (GAAGCAGTCGAATTCAACCTTAAGG) and EcoRI-URSbot (TGTTCTGTGAATTCAATTGTAAAGAGG) as primers and pGBS116 as the template. (Oligonucleotide sequences are given only when they differ from the wild-type sequence, in which case alterations are indicated in italics. Numbering is relative to the first ATG in a given open reading frame [ORF]. A prime indicates a sequence on the noncoding strand.) pGBS116 was also used as the template in a PCR to produce pGBS759 and pGBS723. In pGBS759, the AG4 sequence in the RPH1 binding site in pGBS116 was mutated to TC3G by using oligonucleotides mURS-TC3 (TCGCTTTTACTGGCGCCACTTTTCTTCCTCGTTTTTCGAGGAAGCAGTCAAATTAAACCTTACTTTGTGAAAGTATGCTTACTT) and BglIIbot (PHR1 66′→34′). pGBS723 is a derivative of the CEN-ARS lacZ reporter plasmid pRW95-3 (49). It was constructed by using primers Bam-URStop (CGGGATCCACCTTAAGGGGTGAAAGTATGC) and Bam-URSbot (CGGGATCCTGTAAAGAGGAATAAGTGTCAA) to generate a 65-bp fragment containing URSPHR1 which was inserted into BglII-digested pRW95-3. pLG669Z contains the CYC1 promoter fused to lacZ and has been described previously (18).
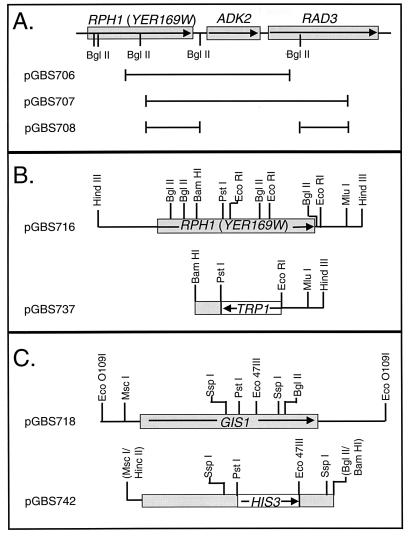
pGBS706 and pGBS707 (Fig. 1A) are plasmids recovered from the yeast genomic library screen described below and contain GAL4AD-RPH1 translational fusions. pGBS708 (Fig. 1A) is a derivative of pGBS707 from which a 2.2-kbp BglII fragment of yeast genomic DNA was removed. A size-selected yeast genomic DNA library containing HindIII restriction fragments from strain GBS76 (38) inserted into pBlueScript SK(+) was screened by colony hybridization (3) for clones containing RPH1. Plasmid pGBS716 (Fig. 1B) was isolated in this screen and contains the entire RPH1 ORF and approximately 1,500 bp of 5′ and 3′ flanking sequences. pGBS737 (Fig. 1B) contains TRP1 flanked by 557 bp of RPH1 coding sequence and 473 bp of RPH1 3′ flanking sequence and was used for targeted disruption of RPH1. pGBS712 contains the 3.8-kbp HindIII fragment from pGBS716 cloned into the HindIII site of pRS415 (42). PCR amplification using primers 096Ecotop (GIS1 −604→−585) and 096Ecobot (GIS1 4078′→4059′) and GBS76 (38) genomic DNA yielded a 4.5-kbp GIS1-containing fragment which was cloned into the EcoO109I site in pBlueScript SK(+), generating pGBS718 (Fig. 1C). pGBS718ΔCT contains a 2.4-kbp MscI-BglII fragment from pGBS718 inserted into the BamHI and HincII sites of pBlueScript SK(+). Subsequently, a 186-bp PstI-Eco47III fragment was deleted from this construct and a 975-bp Eco47III-PstI fragment containing HIS3 from pJJ217 (22) was inserted, yielding pGBS742 (Fig. 1C). pDB81 (a kind gift from Hans Ronne) contains the entire GIS1 gene, including promoter sequences. A GIS1-containing MluI-SacI fragment from pDB81 was inserted into unique SmaI and SacI sites in pRS415, yielding pGBS207.
FIG. 1.
Restriction maps of yeast chromosomal inserts in selected plasmids used in cloning and disruption of YER169w (RPH1) and YDR096w (GIS1). Arrows indicate the direction of transcription of genes indicated by boxes. (A) Sketch of the region of chromosome V carrying YER169w and adjacent genes which were included in the GAL4AD-YER169w fusions that activated the URSPHR1 reporter constructs. The chromosomal DNA carried by plasmids pGBS706, pGBS707, and pGBS708 are indicated by the black lines beneath the map. (B) Restriction map of the yeast chromosomal DNA fragments carried by pGBS716 and by the derivative plasmid pGBS737 which was used to disrupt YER169w. (C) Restriction map of the 4.6-kbp chromosomal DNA fragment carried by pGBS718 and of the gene disruption in plasmid pGBS742. Among the SspI and Eco47III sites in the fragments, only those sites used in subcloning and directed homologous recombination are shown. Restriction sites in parentheses were lost during subcloning.
Plasmids expressing glutathione S-transferase (GST)-Rph1p fusion proteins were constructed in pGEX18 (30). pGBS727 contains a 0.9-kbp EcoRI-BglII fragment from RPH1 (Fig. 1B) subcloned into pBlueScript SK(+). pGBS731, which expresses the C-terminal third of Rph1p fused to GST (Rph1p-CT), was constructed by inserting a 0.9-kbp EcoRI-XbaI fragment from pGBS727 into pGEX18. pGST169w contains the entire RPH1 ORF fused to GST. The plasmid was constructed in two steps. The first 340 bp of the coding sequence of RPH1 were amplified in a PCR using PFU polymerase, primers GBT169-Bam CGGGATCCCGATGACGAAACTAATC) and GBT169-BglII (GAAGATCTTCCGGAGGCACATAGTCC), and pGBS716 as the template. After digestion with BamHI and BglII, the PCR product was subcloned into BamHI-digested pGBS716. The resulting plasmid, pGBS733, contains RPH1 flanked by a unique BamHI site 6 nucleotides 5′ to the first ATG and a SalI site immediately 3′ to the yeast genomic insert. In the second step, this BamHI-SalI fragment was ligated to pGEX18 digested with the same enzymes.
pGBS763 carries a portion of RAD2 and was constructed by insertion of a 1.9-kbp EcoO109I-SacI fragment from pNF2005 (28) into pBlueScript SK(+). A 2.0-kbp fragment containing the LEU2 gene from pJJ283 (22), flanked by a filled-in HindIII site and a BamHI site, was ligated into BglII-EcoRV-digested pGBS763, yielding the RAD2 knockout plasmid pGBS764.
Strains.
The parental S. cerevisiae strains used in this study are listed in Table 1 and were constructed and propagated by using standard techniques. RE1006 was transformed with PvuII-digested pGBS408, thereby targeting insertion of the URSPHR1-HIS3 reporter gene to LYS2. Ura+ transformants were subsequently subjected to selection on 5-fluoroorotic acid, and stable Ura− derivatives were tested by Southern analysis to confirm integration of the reporter at LYS2 and loss of URA3. The resulting strain, GBS157, was transformed with the lacZ reporter plasmid pGBS723, generating GBS1659. Strain GBS1391 carries a marked disruption of RPH1 and was constructed by transforming YPH499 with a 1.8-kbp BamHI-MluI fragment from pGBS737 (Fig. 1B). Replacement of RPH1 was confirmed by PCR of DNA from Trp+ transformants using primer KO169-5′ (RPH1 174→193) in combination with KOTRP-5′ (TRP1 305′→285′) or KO169-out (RPH1 2966′→2948′). A marked disruption of GIS1 was constructed by transforming YPH500 with a 1.5-kbp SspI fragment from pGBS742 (Fig. 1C), yielding strain GBS1396. Gene replacement was verified in His+ transformants by PCR using primers KO096-5′ (RPH1 1033→1050) and KO096-3′ (RPH1 2234′→2215′) or KOHIS-5′ (HIS3 611→628). GBS1406 is a diploid strain obtained by mating GBS1391 and GBS1396. Strains GBS1734, GBS1736, and GBS1738 are haploid meiotic segregants of GBS1406. Strains GBS1867, GBS1869, GBS1872, and GBS1875 contain marked deletions of rad2 and were constructed by transforming YPH499, GBS1734, GBS1736, and GBS1738, respectively, with a 3.5-kbp EcoO109I-SacI fragment from pGBS764, selecting for Leu+ transformants. Replacement of rad2 was confirmed by PCR using primers KO-rad2-5′ (RAD2 5→14) and KO-rad2-3′ (RAD2 1889′→1872′). All other strains are derivatives of these and were constructed by transformation with various plasmids as indicated in the figure legends.
TABLE 1.
Parental S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| RE1006 | MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-52 | M. Johnston |
| GBS157 | MATa can1-100 his3-11,15 lys2::URSPHR1-HIS3 leu2-3,112 trp1-1 ura3-52 | This work |
| GBS1391 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801amber rph1Δ::TRP1 trp1-Δ63 ura3-52 | This work |
| GBS1396 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801amber gis1Δ::HIS3 trp1-Δ63 ura3-52 | This work |
| GBS1406 | MATa/MATα ade2-101/ade2-101 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 lys2-801amber/lys2-801amber rph1Δ::TRP1/RPH1 gis1Δ::HIS3/GIS1 trp1-Δ63/trp1-Δ63 ura3-52/ura3-52 | This work |
| GBS1734 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801amber rph1Δ::TRP1 trp1-Δ63 ura3-52 | This work |
| GBS1736 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801amber gis1Δ::HIS3 trp1-Δ63 ura3-52 | This work |
| GBS1738 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801amber rph1Δ::TRP1 gis1Δ::HIS3 trp1-Δ63 ura3-52 | This work |
| GBS1867 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801amber rad2Δ::LEU2 trpl-Δ63 ura3-52 | This work |
| GBS1869 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801amber rad2Δ::LEU2 rphΔ1::TrP1 trp1-Δ63 ura3-52 | This work |
| GBS1873 | MATa ade2-101 gis1Δ::HIS3 his3-Δ200 leu2-Δ1 lys2-801amber rad2Δ::LEU2 trp1-Δ63 ura3-52 | This work |
| GBS1875 | MATa ade2-101 gis1Δ::HIS3 his3-Δ200 leu2-Δ1 lys2-801amber rad2Δ::LEU2 rphΔ1:: TRP1 trp1-Δ63 ura3-52 | This work |
| YPH499 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801amber trp1-Δ63 ura3-52 | 42 |
| YPH500 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801amber trp1-Δ63 ura3-52 | 42 |
Library screening.
The GAL4AD fusion yeast genomic library constructed by Paetkau and coworkers (29) was screened for genes encoding proteins that bind to URSPHR1. This library consists of high-copy-number LEU2 plasmids carrying the Gal4 transcriptional activation domain fused to random yeast genomic DNA fragments. We used three libraries covering all three possible reading frames to transform GBS1659 and screened Leu+ His+ transformants for increased β-galactosidase activity by using a nonlethal colony assay (13). Plasmids from positive clones were recovered in Escherichia coli DH5α and used to transform naive GBS1659 to confirm the Leu+ His+ phenotype and increased β-galactosidase production.
Expression and purification of GST fusion proteins.
E. coli BL21 was used for the expression of GST fusion proteins. Cells were grown in Luria broth to an A595 of 0.5, at which point isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM and growth was continued for 2 h at 27°C. Cells were lysed, and the proteins were purified by glutathione affinity chromatography as described by the manufacturer (Pharmacia). Both the fusion protein containing only the Rph1p C-terminal region (Rph1p-CT) and the fusion protein containing full-length Rph1p (Rph1) were proteolyzed to a significant extent. Based upon the intensity of bands in sodium dodecyl sulfate-polyacrylamide gels stained with Coomassie blue, we estimate that approximately 30% of the protein in the Rph1p-CT preparations was of the expected length while approximately 10% of the protein from the Rph1p preparation was full length.
EMSAs and footprinting.
Radiolabeled substrate was prepared by hybridization of oligonucleotides URStop (PHR1 −85→−40) and URSbot (PHR1 −40′→−85′) followed by end filling using Klenow fragment and [α-32P]dATP using conditions previously described (44). Unlabeled competitors were prepared by hybridization of oligonucleotide pairs AG4TG (PHR1 −85→−65 and PHR1 −65′→−85′) or various derivatives (see Fig. 6). The buffer for Rph1 binding assays contained 4 mM Tris HCl (pH 8.0), 4 mM MgCl2, 40 mM NaCl, 10 μM ZnCl2, 10% glycerol, bovine serum albumin at 100 μg/ml, 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, aprotinin at 10 μg/ml, soybean trypsin inhibitor at 10 μg/ml, and leupeptin at 4 μg/ml. Rph1p or Rph1p-CT was incubated on ice for 20 min with the various oligonucleotides at the concentrations indicated in the figure legends. Free and bound DNAs were separated by electrophoresis through 6% polyacrylamide gels in 1× Tris-borate-EDTA and quantitated by PhosphorImager analysis or an Ambis Radioanalytic System as previously described (44).
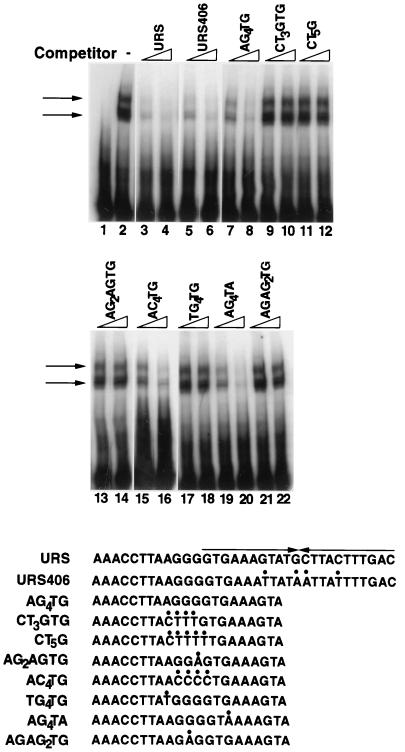
FIG. 6.
Binding competition assays to determine the sequences required for Rph1p binding. Radiolabeled URS oligonucleotide was incubated with Rph1p-CT as described in the legend to Fig. 4, either in the absence or in the presence of the indicated competing unlabeled double-stranded oligonucleotides, and the bound and free portions of the substrate were separated by electrophoresis and autoradiographed. Two concentrations are shown for each competitor, 1 μM (lanes 3, 5, 7, 9, 11, 13, 15, 17, 19, and 21) and 4 μM (lanes 4, 6, 8, 10, 12, 14, 16, 18, 20, and 22). Lane 1 contained the substrate only, and lane 2 contained the substrate and Rph1p-CT without a competitor. Arrows indicate the bound substrate. The sequences of the competitors are shown below the autoradiograms. Sites changed relative to the wild-type sequence are indicated by dots above the changed bases. The 12 bp 3′ to URSPHR1 are not shown for oligonucleotides URS and URS406; however, they are identical.
32P-labeled substrates for footprinting were prepared by using kinase-treated oligonucleotide UEStop (PHR1 −155→−134) or PHR10 (PHR1 10′→−10′) as the primer in a PCR (25) in which pGBS116 was the template. Copper phenanthroline (OP-Cu) footprinting was performed as previously described (39). For DNase I footprinting, 2 ng of probe was incubated with various concentrations of Rph1p-CT or Rph1p at concentrations sufficient to produce 60 to 80% bound substrate as judged by electrophoretic mobility shift assay (EMSA). The binding buffer used was the same as that described above, except that 0.5 μg of poly(dA-dT) was included. Following a 20-min incubation on ice, 1 U of DNase I (Promega) and 1 μl of 50 mM CaCl2 were added, the reaction was allowed to proceed at room temperature for 45 s to 2 min, and then 20 μl of stop solution (1% sodium dodecyl sulfate, 200 mM NaCl, 20 mM EDTA, 40 μg of tRNA per ml) was added. The products were purified by phenol extraction and ethanol precipitation and displayed on 8% polyacrylamide–7 M urea gels (39).
In vivo expression and UV survival studies.
β-galactosidase assays were performed as previously described (44). Cells were grown in liquid YPAD or synthetic complete medium lacking appropriate components to maintain plasmid selection (40), and 1-ml samples were harvested at an A600 of 0.1 to 0.5. The damage response was assessed by using MMS (2.3 mM final concentration) or UV irradiation. MMS was added to cultures at an A600 of 0.1 to 0.2, and cells were incubated at 30°C for 3 h prior to harvesting. To correct for variations in reporter plasmid copy number, DNA was extracted from control cultures (2), digested with EcoRI, and subjected to Southern analysis (25). Probe for plasmid-borne lacZ was synthesized in a PCR using pGBS116 as the template and oligonucleotides lac-top (lacZ 571→592) and lac-bot (lacZ 2700′→2681′). Probe for the single-copy chromosomal gene ACT1 was obtained by PCR of YPH499 genomic DNA using the primers act-top (ACT1 405→428) and act-bot (ACT1 1414′→1393′). Probes were labeled with either [32P]dATP (random primer method [25]) or horseradish peroxidase (ECL; Amersham life Science). Band intensity was determined by using a Molecular Dynamics Storm 860 PhosphorImager and ImagQuant software.
For UV survival and photoreactivation experiments, cultures were harvested in early log phase (A600 of <0.3), washed with and suspended in phosphate-buffered saline, and irradiated at 254 nm as previously described (36). Following irradiation, aliquots of cells were transferred to culture tubes on a tissue culture roller drum placed 9 in. from a bank of two 15-W Cool White fluorescent lamps. Cells were sampled at various times, diluted, and plated on YPAD, and surviving colonies were counted after 3 days of growth at 30°C in the dark.
RESULTS
Identification of YER169w and GIS1 as putative regulators acting through URSPHR1.
We utilized the one-hybrid method to identify putative PRP-encoding genes. URSPHR1 was inserted into the promoter regions of two reporter genes, HIS3 and lacZ, in the reporter strain GBS1659. Because both reporter genes are devoid of upstream activation sequences, GBS1659 is a histidine auxotroph and produces extremely low levels of β-galactosidase regardless of whether URSPHR1 is present. In principle, expression of a gene encoding the DNA binding domain of PRP fused to the transcriptional activation domain of GAL4 should confer high-level expression of the reporter genes. We transformed GBS1659 with a series of GAL4 fusion yeast genomic libraries carried on the 2μm LEU2 plasmids pDP4, pDP7, and pDP12 (29). Approximately two million Leu+ transformants from each library were tested for histidine prototrophy, and a total of 85 His+ Trp+ Leu+ clones were obtained. In a secondary screening for increased β-galactosidase activity using a colony color assay (13), four of these clones (URS39, URS48, URS67, and URS72) consistently produced dark blue colonies on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) indicator plates. Plasmids carrying the GAL4 fusion genes were rescued from these clones, and the DNA was sequenced at the 5′ and 3′ fusion sites. Each plasmid carried GAL4 fused in frame to sequences from the carboxy-terminal half of yeast ORF YER169w (17), followed by an intact copy of ADK2 and variable amino-terminal portions of RAD3 (Fig. 1). Plasmids from transformants URS48 and URS67 were identical to one another and were designated pGBS706; similarly, plasmids from URS39 and URS72 were identical and were designated pGBS707. To confirm that the Gal4-Yer169w fusion protein was responsible for enhanced expression from the reporter genes, a 2.2-kbp BglII fragment containing the entire ADK2 gene and the RAD3 promoter and translational start site was removed from pGBS707. The resulting plasmid (pGBS708, Fig. 1) conferred histidine prototrophy and high-level β-galactosidase expression on naive GBS1659, whereas the vector alone had no effect on expression (data not shown).
YER169w is a 2,388-bp ORF with an unknown function that was identified in the course of the S. cerevisiae genome sequencing project (9). It encodes a highly basic 90-kDa protein containing, near the carboxy terminus, a classical C2H2 zinc finger followed by a C2HC zinc finger (6) (Fig. 2). Deletion of the zinc fingers abolishes transcriptional activation by the Gal4-Yer169w fusion protein in vivo (data not shown), suggesting that the Zn fingers are required for binding to URSPHR1. ORF YER169w has been renamed RPH1 (regulator of PHR1). Comparison of the predicted amino acid sequence of RPH1 to all other yeast ORFs revealed striking homology to the protein encoded by GIS1 (9). GIS1 has been previously isolated as an overexpression suppressor of gig1-2 (5), a mutation in the SRB8 gene encoding a subunit of the cyclin C-dependent protein kinase complex (4). As is shown in Fig. 2, the two proteins are 92.7% identical over the 55-amino-acid region comprising the zinc fingers of the two proteins, 100% identical in the regions of the zinc fingers thought to interact with DNA, and 34.7% identical overall. In addition to the zinc finger region, scattered regions of homology are found throughout the molecules. Two particularly interesting regions near the amino terminus also show 30 to 40% identity with human retinoblastoma binding protein 2 (14), human cDNA XE169 (50), the mouse jumonji-encoded protein (45), and the product of ZK593.4, a gene with an unknown function identified during the Caenorhabditis elegans genome sequencing project (8). While the function of this region is not known, its conservation across phylogenetic lines suggests it is an important structural or functional motif.
FIG. 2.
Alignment of the Rph1p and Gis1p proteins. The predicted amino acid sequences of the proteins were aligned by using the program WU-BLAST 2.0 (30a). Open boxes indicate the regions of homology to RBP2, while filled boxes indicate the region containing the two zinc finger motifs (6). The amino acids within the zinc fingers thought to be involved in DNA binding are overlined. Asterisks indicate identical amino acids.
RPH1 and GIS1 are required for repression of PHR1.
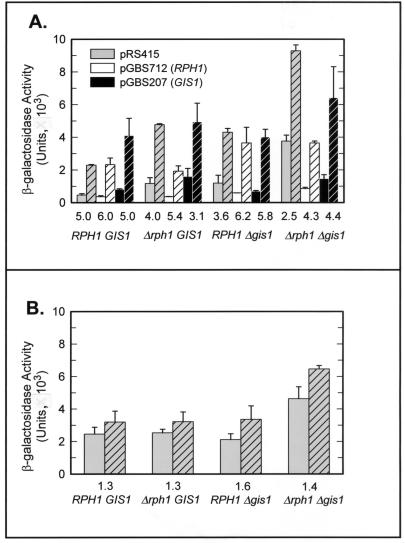
We constructed targeted disruptions of RPH1 and GIS1 to assess the effect of loss of function on cell growth and viability and on PHR1 expression. Disruption of either RPH1 or GIS1 in haploid strains of either mating type had no discernible effect on the viability of log-phase cells grown in YPAD at 30°C (data not shown), indicating that neither RPH1 nor GIS1 is an essential gene under these conditions. This was confirmed by tetrad analysis of sporulated GBS1406, a diploid strain in which a single copy of each gene was disrupted; all four expected classes of segregants were recovered, and there was no consistent difference in viability on YPAD of any segregant class (data not shown). The effect of RPH1 and GIS1 disruption on PHR1 expression was assessed by using pGBS116, which contains the intact PHR1 promoter, including URSPHR1, fused to lacZ. As can be seen in Fig. 3A, strains containing a disruption of either RPH1 or GIS1 displayed a modest increase in basal-level expression, as well as a decrease in the induction ratio (defined as the ratio of damage-induced expression to basal-level expression), following treatment with the DNA-damaging agent MMS. Simultaneous disruption of both RPH1 and GIS1 had a synergistic effect, producing a sixfold increase in basal-level expression and a 50% decrease in the induction ratio. Both the increase in basal-level expression and the decrease in the induction ratio upon deletion of either or both genes are consistent with the encoded proteins acting as damage-responsive negative regulators of PHR1. The synergistic effect observed when both genes are disrupted suggests that the proteins are redundant with respect to PHR1 repression. It is somewhat surprising, then, that while multiple copies of RPH1 complement a deletion of GIS1, multiple copies of GIS1 do not complement an RPH1 deletion (Fig. 3). It is unlikely that this reflects a unique requirement for RPH1 in PHR1 expression or GIS1 function, since GIS1 alone partially restores repression in a Δrph1 Δgis1 mutant (Fig. 3A). At present, we believe that the failure of multiple copies of GIS1 to complement an RPH1 deletion may be due to differences in the expression levels of the two genes or in the strength of repression conferred by the two proteins. RPH1 mRNA is approximately threefold more abundant in undamaged S. cerevisiae cells than is GIS1 mRNA (19). In these experiments, extra copies of RPH1 and GIS1 are expressed from their own promoters and are carried on centromeric plasmids that average one to two copies per haploid genome (46). Thus, in all likelihood, GIS1 was overexpressed only two- to threefold, a level that is apparently insufficient to fully repress PHR1.
FIG. 3.
Effects of deletion of RPH1 and GIS1 on basal-level expression and damage induction of PHR1. Strains YPH499, GBS1734, GBS1736, and GBS1738 were transformed with a PHR1-lacZ reporter plasmid and with pRS415, pGBS712 (RPH1), or pGBS207 (GIS1), and the effect on expression was assessed with (cross-hatched bars) or without (open bars) MMS treatment. The chromosomal genotypes are indicated below the ordinate, and the induction ratio following MMS treatment is indicated immediately above the chromosomal genotype. Error bars show the standard deviations from three or four independent determinations. (A) Effects on expression from a reporter plasmid (pGBS116) that contains the intact PHR1 promoter. (B) Effect on expression of a pGBS116 derivative (pGBS759) in which the AG4 sequence has been mutated.
Rph1p binds to URSPHR1 in vitro.
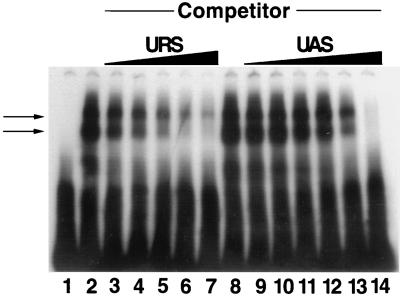
While the simplest interpretation of the in vivo data is that RPH1 and GIS1 encode DNA-binding proteins that recognize sequences within URSPHR1, secondary or indirect effects cannot be ruled out by these studies. Therefore, we expressed the protein encoded by RPH1 in E. coli and tested whether the purified protein binds specifically and with high affinity to URSPHR1. EMSAs shown in Fig. 4 demonstrate that this is indeed the case. Rph1p bound to an oligonucleotide containing URSPHR1 (Fig. 4, lanes 2 and 8). Sequence-specific binding was confirmed by competition studies in which a homologous oligonucleotide competed much more efficiently for binding of Rph1p than did a heterologous oligonucleotide (Fig. 4). Thus far, excessive proteolysis and insolubility have made it impossible to perform similar binding experiments with purified Gis1p.
FIG. 4.
EMSA testing the affinity and binding specificity of Rph1p-CT for URSPHR1. 32P-labeled URS oligonucleotide (20 nM), either without (lane 1) or incubated with Rphp-CT (100 nM; lanes 2 to 14), was electrophoresed as described in Materials and Methods. In lanes 3 to 7 and 9 to 14, the indicated unlabeled competitor oligonucleotide was present during the incubation. Competitor concentrations (lanes): 3 and 9, 200 nM; 4 and 10, 400 nM; 5 and 11, 1 μM; 6 and 12, 2 μM; 7 and 13, 4 μM; 14, 8 μM. Arrows indicate the major Rph1p-URS complexes which appear as a doublet. We believe this is due to partial proteolysis of Rph1p (see Materials and Methods).
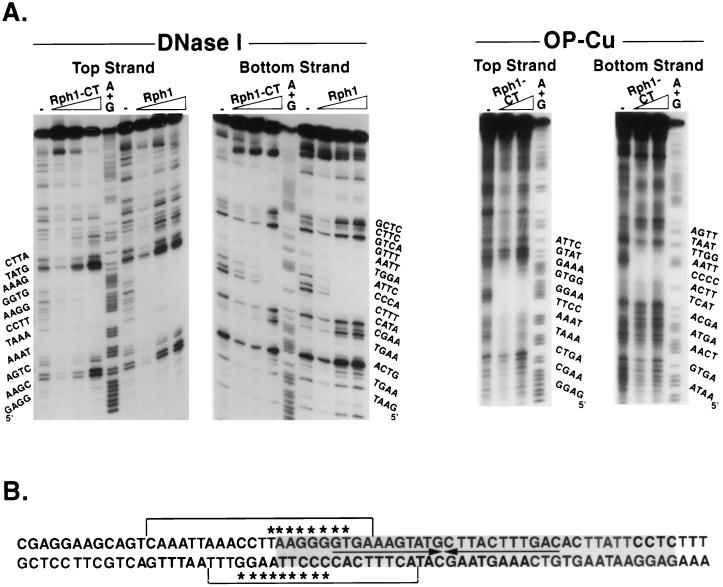
DNase I footprinting was used to determine the region within URSPHR1 that is bound by Rph1p. The 39-bp region footprinted by PRP contains a 22-bp palindrome, as well as flanking sequences (39). Surprisingly, Rph1p protected only the 5′ portion of the URS from attack by DNase I (Fig. 5). It should be noted that full-length Rph1p and Rph1p-CT, which contains only the C-terminal one-third of Rph1p, including the zinc fingers, yielded identical DNase I footprints (Fig. 5A), thereby validating the use of Rph1p-CT for DNA binding and footprinting experiments. DNase I overestimates the region of DNA in intimate contact with binding proteins, and therefore a more accurate estimation of the DNA binding site was obtained by using OP-Cu as a footprinting agent. Rph1p protected an 8-bp region, TAAGGGGT, from attack on the top strand and a 10-bp region, CCCCTTAAGG, on the bottom strand (Fig. 5B). The protected region partially overlaps the 39-bp region protected by partially purified PRP (39). A likely explanation for the smaller footprint compared to PRP is that the latter is composed of proteins in addition to Rph1p and/or Gis1p. This is supported by previous work demonstrating that changing the four central GC base pairs within the URSPHR1 palindrome to AT base pairs abolishes repression of PHR1 in vivo (35). However, currently we cannot rule out effects of proteolysis on the extent of the footprint (see Materials and Methods). That the Rph1p footprint extends outside of the previously footprinted region may be due to the relatively weak OP-Cu cleavage at the boundary regions or may reflect conformational differences between Rph1p in isolation versus Rph1p in a multisubunit complex.
FIG. 5.
Footprinting of Rph1p on the PHR1 transcriptional regulatory region. Oligonucleotides containing the PHR1 transcriptional regulatory region and labeled at the 5′ end on either the top or bottom strand were exposed to DNase I or OP-Cu in the absence or presence of increasing concentrations of Rph1p-CT or Rph1p as described in Materials and Methods. (A) Autoradiograms of the partial digestion products separated on denaturing acrylamide gels. The sequence of the oligonucleotide in the region of the footprint is shown to the left of each autoradiogram. Lanes: −, no protein added; A + G, products of a Maxam-Gilbert reaction which cleaves at A’s and G’s. (B) Sequence within and surrounding the region footprinted by Prp (gray area) and the region protected by Rph1p and Rph1p-CT from attack by DNase I (brackets above and below the sequence) and by OP-Cu (asterisks above and below the sequence).
Delineation of Rph1p binding specificity.
To further define the binding specificity of Rph1p, we compared the ability of oligonucleotides containing mutations within URSPHR1 to compete with the wild-type sequence for binding of Rph1p in vitro. As can be seen in Fig. 6, oligonucleotides containing either a deletion or a point mutation outside of the AG4 sequence were still able to compete effectively for binding of Rph1p (oligonucleotides AG4TG, URS406, and AG4TA). In contrast, oligonucleotides containing mutations within the AG4 sequence reduced competition to undetectable levels (oligonucleotides CT3GTG, CT5G, AG2AGTG, TG4TG, and AGAG2TG). The one exception to this pattern was the oligonucleotide AC4TG, in which the AG4 sequence was switched to the bottom strand while retaining the same polarity. We conclude that the AG4 sequence is both necessary and sufficient for binding by Rph1p in vitro.
To determine whether AG4 is the sequence through which Rph1p and Gis1p act in vivo, we constructed pGBS759, which contains a PHR1-lacZ fusion in which the AG4 sequence in URSPHR1 was mutated to TC3G, and assayed expression of the reporter gene in various genetic backgrounds (Fig. 3B). This mutation reduced induction in response to MMS by 70 to 75% in strains with intact RPH1 and GIS1 genes and rendered expression of the reporter gene almost completely insensitive to loss of either or both genes. Together, these data strongly argue that Rph1p and Gis1p regulate the damage response of PHR1 by binding to the AG4 sequence in URSPHR1.
Derepression of PHR1 enhances UV survival.
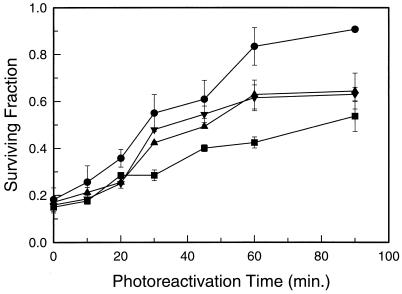
To determine whether derepression of PHR1 results in enhanced repair capacity, we tested the survival of wild-type, Δrph1, Δgis1, and Δrph1 Δgis1 strains following UV irradiation, with or without subsequent photoreactivation. Strains bearing deletions of rad2 were used because the effect of photoreactivation on survival is often difficult to see in cells with an intact nucleotide excision repair pathway. As can be seen in Fig. 7, deletion of rph1, gis1, or both genes enhanced both the rate and extent of light-dependent repair and the relative enhancement of survival mirrored the enhanced PHR1 expression seen in these strains. It should be noted that under these experimental conditions, both the rate and extent of the light-dependent increase in survival are decreased by the presence of 6-4 photoproducts which are lethal lesions that are not repaired by the Phr1 photolyase (7, 32). Thus, the survival data underestimate the extent of PHR1 derepression.
FIG. 7.
Effect of derepression of PHR1 on the UV survival of Δrad2 strains. Log-phase cells were exposed to 4.5 J of 254-nm radiation per m2 and then to photoreactivating light as described in Materials and Methods. Samples were taken at the indicated times and plated for survival determination. The data points are averages from three independent experiments, and the error bars indicate the standard deviation. Symbols: ■, GBS1867 (RPH1 GIS1); ▴, GBS1869 (Δrph1 GIS1); ▾, GBS1873 (RPH1 Δgis1); ●, GBS1875 (Δrph1 Δgis1).
DISCUSSION
In this work, we have identified the proteins encoded by RPH1 and GIS1 as DNA damage-responsive repressors of PHR1 transcription and have demonstrated that derepression of PHR1 enhances light-dependent repair of UV-induced DNA damage. Rph1p recognizes a single AG4 sequence found in previously defined URSPHR1, and Rph1p binding to this site requires the two zinc fingers near the carboxy terminus of the protein. The key residues for sequence-specific binding by zinc fingers are at positions −1, 2, 3, and 6 relative to the beginning of the finger helix (reviewed in reference 21). These residues, and indeed all amino acids in the helical domain of the fingers, are identical in Rph1p and Gis1p, strongly suggesting that these two proteins recognize identical sequences. Additionally, altering the AG4 sequence in URSPHR1 eliminates Rph1p binding in vitro, derepresses PHR1 expression, and almost entirely eliminates the effects of deletion of RPH1 and GIS1 in vivo. Together with the observation that both RPH1 and GIS1 must be deleted to fully derepress PHR1 expression, the data indicate that RPH1 and GIS1 are functionally redundant with respect to PHR1 repression.
Several pairs of transcription factors that recognize identical sequences have been identified in yeast; however, functional redundancy of the type seen for RPH1 and GIS1 is unusual. The repressors Mig1p and Mig2p regulate SUC2 expression, but unlike Rph1p and Gis1p, Mig1p alone is sufficient to confer complete repression and Mig2p activity is only seen in strains lacking Mig1p (24). Ace2p and Swi5p activate the CTS1 and HO promoters, respectively, and can substitute for one another only when present in high copy number or in specific genetic backgrounds (11, 12). Perhaps the closest parallel to the functional redundancy of RPH1 and GIS1 is the situation observed with Msn2p and Msn4p, two activators of the multistress response in yeast that bind to the STRE (stress response element) (26, 37). While deletion of MSN2 reduces expression from an STRE-driven reporter gene by 80% (37), deletion of both genes is required to observe the full repertoire of phenotypes associated with loss of the multistress response (17, 26). A further similarity among Msn2p, Msn4p, Rph1p, and Gis1p is that each of these proteins binds specifically to the sequence AG4 (26, 37, and this work). Preliminary results indicate that deletion of RPH1 and GIS1 derepresses basal-level expression from an STRE-driven reporter gene (33). This suggests either that there is cross talk between the multistress response and the RPH1/GIS1 DNA damage response pathway or that deletion of RPH1 and GIS1 produces a signal that activates the stress response pathway.
An important question that remains to be addressed is whether RPH1 and GIS1 regulate DNA damage-responsive genes in addition to PHR1. The AG4 sequence recognized by these proteins occurs much too often in the yeast genome for a search based simply on this sequence to be meaningful. Nevertheless, it is probably significant that one or more AG4 sequences are found within 500 bp of the translational start site of half of the 28 known damage-inducible DNA repair and metabolism genes of yeast (PHR1, RAD5, RAD6, RAD7, RAD16, RAD27, RAD51, RAD54, DUN1, REV3, RFX1 [CRT1], RNR2, RNR3, and RNR4 [1, 16, 20, 23, 27, 43, 47]), while less than 20% of noninducible repair genes contain this sequence. Since most of these damage-responsive genes are not induced by heat shock, it is unlikely that the AG4 sequence is targeted by Msn2p and Msn4p in these promoters. The availability of MSN2, MSN4, RPH1, and GIS1 deletion mutants makes it possible to test directly whether RPH1 and GIS1 control a damage response regulon and whether MSN2 and MSN4 contribute to this response.
Repression by RPH1 and GIS1 differs in at least two respects from that mediated by CRT1, a homolog of the mammalian RFX family of DNA binding proteins and the only other characterized regulator of damage-inducible DNA repair genes in yeast (20). Despite the fact that the canonical RFX-X box contains the AG4 sequence recognized by Rph1p and Gis1p, none of the Crt1p binding sites thus far identified contain the AG4 sequence. In addition, repression by Crt1p requires the corepressors Ssn6p and Tup1p. In contrast, repression by RPH1 and GIS1 is TUP1 -independent (10). Another striking difference is that derepression of CRT1-regulated genes requires both the RAD53 and DUN1 protein kinases (20) while derepression of PHR1 requires RAD53 but not DUN1 (34). Thus, it appears not only that there are multiple damage-responsive transcriptional regulators but also that the signal transduction pathway differs to some extent, depending upon the target. This conclusion is consistent with studies by Kiser and Weinert (23) that suggested that at least four transcriptional pathways are activated by the damage response in yeast.
URSPHR1 was originally identified by OP-Cu footprinting as a 39-bp region that is bound by a protein or proteins present in partially purified extracts from nonirradiated cells and absent from extracts from UV-irradiated cells (39). The binding site for Rph1p identified in the current studies lies at the extreme 5′ end of URSPHR1 and includes only 2 bp of a 22-bp palindrome which we have previously shown to be required for repression of PHR1 (35). Taken together, these results indicate that an additional protein(s) is bound to URSPHR1 in vivo. This may explain the residual damage response of PHR1 when both RPH1 and GIS1 are deleted (Fig. 3). In addition, the fact that mutations in the palindrome abolish repression (35) indicates that the protein functions synergistically with Rph1p and Gis1p to repress transcription of PHR1. Experiments are in progress to identify additional components of the repressor complex and to determine the mechanisms that govern loss of DNA binding in response to damage.
ACKNOWLEDGMENTS
We gratefully acknowledge Hans Ronne for providing plasmids containing functional copies of GISI and for sharing information prior to publication. We thank Errol Friedberg, Louise Prakash, and Aziz Sancar for providing plasmids.
This work was supported by grant GM35123 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Aboussekhra A, Vialard J E, Morrison D E, de la Torre-Ruiz M A, Cernakova L, Fabre F, Lowndes N F. A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J. 1996;15:3912–3922. [PMC free article] [PubMed] [Google Scholar]
- 2.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 3.Adzuma K, Ogawa T, Ogawa H. Primary structure of the RAD52 gene in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2735–2744. doi: 10.1128/mcb.4.12.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balciunas, D., and H. Ronne. 1999. Personal communication.
- 5.Balciunas D, Ronne H. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res. 1995;23:4421–4425. doi: 10.1093/nar/23.21.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohm S, Frishman D, Mewes H W. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 1997;25:2464–2469. doi: 10.1093/nar/25.12.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brash D E, Franklin W A, Sancar G B, Sancar A, Haseltine W A. Escherichia coli DNA photolyase reverses cyclobutane pyrimidine dimers but pyrimidine-pyrimidone (6-4) photoproducts. J Biol Chem. 1985;260:11438–11441. , No. 21. [PubMed] [Google Scholar]
- 8.C. elegans Genome Project. November 1998, posting date. C. elegans genome database website. [Online.] http://www.sanger.ac.uk/Projects/C_elegans. [15 September 1999, last date accessed.]
- 9.Cherry, J. M., C. Adler, C. Ball, S. Dwight, S. Chervitz, G. Juvik, T. Roe, S. Weng, and D. Botstein. 25 August 1999, posting date. Saccharomyces Genome Database. [Online.] http://genome-www.stanford.edu/Saccharomyces/. [15 September 1999, last date accessed.]
- 10.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 11.Dohrmann P R, Butler G, Tamai K, Dorland S, Greene J R, Thiele D J, Stillman D J. Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 1992;6:93–104. doi: 10.1101/gad.6.1.93. [DOI] [PubMed] [Google Scholar]
- 12.Dohrmann P R, Voth W P, Stillman D J. Role of negative regulation in promoter specificity of the homologous transcriptional activators Ace2p and Swi5p. Mol Cell Biol. 1996;16:1746–1758. doi: 10.1128/mcb.16.4.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duttweiler H M. A highly sensitive and non-lethal β-galactosidase plate assay for yeast. Trends Genet Sci. 1996;12:340–341. doi: 10.1016/s0168-9525(96)80008-4. [DOI] [PubMed] [Google Scholar]
- 14.Fattaey A R, Helin K, Dembski M S, Dyson N, Harlow E, Vuocolo A, Hanobik M G, Haskell K M, Oloff A, Defeo-Jones D, Jones R E. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene. 1993;8:3149–3156. [PubMed] [Google Scholar]
- 15.Flick J S, Johnston M. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4757–4769. doi: 10.1128/mcb.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedberg E C, Walker G C, Siede W, editors. DNA repair and mutagenesis, 595–631. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 17.Gorner W, Durchschlag E, Martinez-Pastor M T, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guarente L, Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuit of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 20.Huang M, Zhou Z, Elledge S. The DNA replication and damage checkpoint pathways induce transcription by induction of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 21.Jamieson A C, Wang H, Kim S H. A zinc finger directory for high-affinity DNA recognition. Proc Natl Acad Sci USA. 1996;93:12834–12839. doi: 10.1073/pnas.93.23.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones J S, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 23.Kiser G L, Weinert T A. Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol Biol Cell. 1996;7:703–718. doi: 10.1091/mbc.7.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutfiyya L L, Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol Cell Biol. 1996;16:4790–4797. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Martinez-Pastor M T, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald J P, Levine A S, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1998;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolet C M, Chenevert J M, Friedberg E C. The RAD2 gene of Saccharomyces cerevisiae: nucleotide sequence and transcript mapping. Gene. 1985;36:225–234. doi: 10.1016/0378-1119(85)90177-5. [DOI] [PubMed] [Google Scholar]
- 29.Paetkau D W, Riese J A, MacMorran W S, Woods R A, Gietz R D. Interaction of the yeast RAD7 and SIR3 proteins: implications for DNA repair and chromatin structure. Genes Dev. 1994;8:2035–2045. doi: 10.1101/gad.8.17.2035. [DOI] [PubMed] [Google Scholar]
- 30.Park C-H, Mu D, Reardon J T, Sancar A. The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J Biol Chem. 1995;270:4896–4902. doi: 10.1074/jbc.270.9.4896. [DOI] [PubMed] [Google Scholar]
- 30a.Saccharomyces cerevisiae WU-Blast 2 Website. 1998, copyright date. [Online.] Washington University. http://genome-www2.stanford.edu/cgi-bin/SGD/nph-blast2sgd. [15 September 1999, last date accessed.]
- 31.Sancar G B. Sequence of the Saccharomyces cerevisiae PHR1 gene and homology of the PHR1 photolyase to E. coli photolyase. Nucleic Acids Res. 1985;13:8231–8246. doi: 10.1093/nar/13.22.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sancar G B. DNA photolyases: physical properties, action mechanism, and roles in dark repair. Mutat Res. 1990;236:147–160. doi: 10.1016/0921-8777(90)90002-m. [DOI] [PubMed] [Google Scholar]
- 33.Sancar, G. B. 1999. Unpublished data.
- 34.Sancar, G. B. 1999. Unpublished data.
- 35.Sancar G B, Ferris R, Smith F W, Vandeberg B. Promoter elements of the PHR1 gene of Saccharomyces cerevisiae and their roles in the response to DNA damage. Nucleic Acids Res. 1995;21:4320–4328. doi: 10.1093/nar/23.21.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sancar G B, Smith F W. Interactions between yeast photolyase and nucleotide excision repair proteins in Saccharomyces cerevisiae and Escherichia coli. Mol Cell Biol. 1989;9:4767–4776. doi: 10.1128/mcb.9.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt A P, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sebastian J, Kraus B, Sancar G B. Expression of the yeast PHR1 gene is induced by DNA-damaging agents. Mol Cell Biol. 1990;10:4630–4637. doi: 10.1128/mcb.10.9.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sebastian J, Sancar G B. A damage-responsive DNA binding protein regulates transcription of the yeast DNA repair gene PHR1. Proc Natl Acad Sci USA. 1991;88:11251–11255. doi: 10.1073/pnas.88.24.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherman F. Getting started with yeast. Methods Enzymol. 1991;••:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 41.Sidorova J M, Breeden L L. Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev. 1997;11:3032–3045. doi: 10.1101/gad.11.22.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sikorski R S, Hieter P. A system for shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singhal R K, Hinkle D C, Lawrence C W. The REV3 gene of Saccharomyces cerevisiae is transcriptionally regulated more like a repair gene than one encoding a DNA polymerase. Mol Gen Genet. 1992;236:17–24. doi: 10.1007/BF00279638. [DOI] [PubMed] [Google Scholar]
- 44.Sweet D H, Jang Y K, Sancar G B. Role of UME6 in transcriptional regulation of a DNA repair gene in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6223–6235. doi: 10.1128/mcb.17.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- 46.Tschumper G, Carbon J. Copy number control by a yeast centromere. Gene. 1983;23:221–232. doi: 10.1016/0378-1119(83)90054-9. [DOI] [PubMed] [Google Scholar]
- 47.Vallen E A, Cross F R. Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol Cell Biol. 1995;15:4291–4302. doi: 10.1128/mcb.15.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinert T. DNA damage and checkpoint pathways: molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- 49.Wolf S S, Roder K, Schweizer M. Construction of a reporter plasmid that allows expression libraries to be exploited for the one-hybrid system. BioTechniques. 1996;20:568–574. doi: 10.2144/19962004568. [DOI] [PubMed] [Google Scholar]
- 50.Wu J, Ellison J, Salido E, Yen P, Mohandas T, Shapiro L J. Isolation and characterization of XE169, a novel human gene that escapes X-inactivation. Hum Mol Genet. 1994;1:153–160. doi: 10.1093/hmg/3.1.153. [DOI] [PubMed] [Google Scholar]