Abstract
Survival data is being analysed here under the middle censoring scheme, using specifically quantile function modelling under competing risks. The use of middle censoring scheme has been shown to be very appropriate under the COVID-19 pandemic scenario. Cause-specific quantile inference under middle censoring is employed. Such quantile inferences are obtained through cumulative incidence function based on cause-specific proportional hazards model. The baseline lifetime is assumed to follow a very general parametric model namely the Weibull distribution, and is independent of the censoring mechanism. We obtain estimates of the unknown parameters and cause specific quantile functions under classical as well as a Bayesian set-up. A Monte Carlo simulation study assesses the relative performance of the different estimators. Finally, a real life data analysis is given for illustration of the proposed methods.
Keywords: Middle censoring, COVID-19, Competing risks, Quantile function, Weibull distribution, Bayes estimation
1. Introduction
After the Coronavirus Infectious Disease (COVID-19) first appeared in Wuhan, China in December 2019, the World Health Organization (WHO) declared it a global pandemic on March 11, 2020 due to its global spread [1]. COVID-19 is transmitted mainly during coughing and sneezing; fecal-oral transmission is also reported in a few cases. According to the worldometer website (https://www.worldometers.info/coronavirus/), as of 10 June 2021, there were more than 175 million cases and around 3.78 million deaths globally. COVID-19 has now been reported on every continent. The first case of COVID-19 in India is reported in the state of Kerala on 30 January 2020, after a medical student returned back from China. COVID-19 has occurred at an unprecedented period, and the lockdown measures enacted have influenced human life with serious economic and social development concerns. In comparison to high-income nations, low- and middle-income nations with less developed health systems appear to have greater obstacles and remain vulnerable in regulating and controlling COVID-19 [2].
The COVID-19 outbreak has changed the health system’s responsibilities, and it now finds itself not only overloaded, but also with limited capacity to provide services that it had previously extended to communities. COVID-19 patients are clogging up hospitals and health facilities, making it difficult for other symptomatic patients with acute or chronic illnesses to receive standard care [3]. When the entire health system is focused on fighting against the COVID-19 pandemic, medical (clinical trials) and surgical emergencies (including road accidents) are neglected. COVID-19’s disruption of health services is especially problematic for people with noncommunicable diseases (NCDs) who require regular care [4]. NCDs includes mostly cardiovascular diseases, cancers, diabetes and chronic respiratory diseases. NCDs are responsible for over 70% of all deaths, with nearly 80% of these deaths occurring in low and middle-income countries. In addition, NCDs constitute approximately 80% of all years lived with disability globally [4].
Aside from the aforementioned effects of COVID-19 on the health-care system, elective care cancellations and lack of transportation due to imposed lock-downs, insufficient staff, and hospital closures are the most common causes of health care service disruptions. In this scenario, medical professionals require more precise information about the patients’ follow-up. However, in the current COVID-19 pandemic situation, follow-up data consists of a set of exact event times as well as interval event times, and it is important to include such exact and interval event times in any analysis to obtain an accurate estimate of the patients’ survival after surgery. This is exactly the context and type of data where the idea of “middle-censoring scheme” [5] is most appropriate and which we investigate.
Censoring is a key feature of survival analysis in statistics, and it is commonly used when the exact lifetimes of individuals are not known except for a few in the study. [5] introduced a modern concept of censoring scheme known as middle censoring, which has received considerable attention in the statistical literature. In this censoring scheme the exact lifetimes of some individuals are observed while for others, it becomes unobservable as they fall in some random censoring intervals. This censoring scheme arises prominently in time to event analysis in clinical trial studies. Moreover, middle censoring also occurs when the mechanism where the observations are being taken, is closed for a period, due to an external emergency such as the outbreak of disease, war or a strike.
In the current scenario of COVID-19 pandemic, the need for the use and application of middle censoring scheme is highly relevant. For example, in a breast cancer clinical trial centre, patients are registered with node-negative breast cancer. Some of the patients may receive the tamoxifen-alone arm and remaining patients received the combination of radiation and tamoxifen arm. Investigators may be interested to observe events such as local relapse, auxiliary relapse, remote relapse, second malignancy of any kind, and death. After the surgery patients are discharged and they are instructed that they have to come to the hospital for routine check-up and follow-up. But in the pandemic situation, many countries declared a nationwide complete lock-down so that many patients fail to get routine check-ups, and the investigators may also lose the follow-up on these patients. In the interim, some of the patients may experience an event of interest. In this situation the exact time of the event occurrence cannot be observed for some patients except for noting the interval for the event-time, and learning on later inspection that the actual event has occurred during this interval. Thus, middle censoring is highly relevant with immediate application in real life.
To define middle censoring, let and be the lifetimes and random censoring intervals respectively of the individuals who are under observation. Intervals are independent and identically distributed (i.i.d.) with some unknown bivariate distribution and they are independent of . Under the notion of the middle censoring, lifetime becomes observable if with , otherwise unobservable. Middle censored competing risks data with exponential distribution was studied by [6] and [7]. See also the references cited therein.
A single long-term survivor may have a major impact on mean life in survival study, particularly in the case of heavy tailed models, which are widely used for lifetime data. This type of situation is commonly encountered when the subjects can experience types of mutually exclusive competing risks of death/event. For example, in a cancer clinical trial, the primary risk of concern may be a full or partial reaction to therapy, with death as a competing risk, and death may be attributed due to various risks such as cardiac arrest, corona virus infection etc. Similarly, in liver transplantation an individual can experience one of the three possible outcomes such as death, transplantation and withdrawal from the waiting list.
In modelling of survival data with competing risks, two basic quantities such as cumulative incidence function (CIF) and cause specific hazard function (CSHF) get considerable attention in the statistical literature, see [8]. The CIF represents the cumulative probability of failure due to cause up to a certain time point , conditional on vector of covariates , which is described as follows
| (1) |
where is the time to failure, is the cause of failure. The CSHF simply gives the instantaneous failure rate from the cause at time among the individuals who survives up to . Mathematically, CSHF is defined as
| (2) |
Although, in general the distribution function reaches as , in the presence of competing risks, the asymptote of the CIF is less than , implying that the proportion of the due to cause increases up to some time point, and then plateaus. Therefore, obtaining the mean survival time does not make much sense because it will always be infinite. Hence, in such a situation, quantile-based estimates, which are finite and may be identifiable from observed data are generally found to be more precise and robust. These measures can be used for summarizing a CIF curve.
A good discussion of the various theoretical aspects of the quantile function may be found in [9]. Let be a right-continuous distribution function of a random variable . The quantile function of , say is defined by
| (3) |
The quantile function has a number of unique features that a distribution function does not have. For example, the sum of the quantiles, product of the positive quantiles, and the monotonic transformation of the quantile functions are also quantile functions. These properties make the quantile function a preferred alternative to the distribution function in statistical modelling. The quantile function is gaining popularity as a comprehensive tool for statistical analysis of lifetime data. Quantiles are frequently used in medical research to summarize the survival function. For example, the median survival time has long been used to assess the survival curve. In survival studies, quantile regression has gained increasing interest as a viable alternative to methods using distribution functions. For more details on quantile function one may refer to [10] and the references therein.
Modelling of competing risks survival data using the quantile function has been studied by several researchers. [11], [12] discuss non-parametric and regression model approach of quantile function with competing risks. [10] propose a quantile based test for comparing the equality of CIFs. [13] considered parametric and non-parametric inferences for cumulative incidence quantiles without covariates. [14] proposed covariate adjusted quantiles inferences through cause specific proportional hazards (PH) model of the CIF. [15] considered the parametric modelling of the cause specific quantiles with covariates through Weibull PH model and direct semi-parametric improper Gompertz model. Recently, [16] has reviewed various aspects of quantile regression analysis of survival data with competing risks and without competing risks under randomly censored and left truncation mechanism based on the semiparametric approach.
The primary goal of this paper is to obtain quantile based inference for middle censored competing risks survival data based on the parametric regression modelling of CIF. We define the CIF through Weibull PH model. Under this set-up, we obtain both maximum likelihood and Bayes estimates under reasonable priors. The Bayes estimates are obtained using two different loss functions, namely the squared error loss function and LINEX loss function. As one may expect, explicit form of the posterior densities turn out to be intricate, and so we adopt the Markov Chain Mote Carlo (MCMC) simulation algorithms for generating the posterior samples.
The rest of the article is organized as follows. In Section 2 we define the parametric cause-specific quantile functions based on Weibull PH model. In Section 3 we obtain the maximum likelihood estimates under middle censoring scheme for cause specific quantile functions. Bayes estimates based on squared error and LINEX loss functions are provided in Section 4. Section 5 presents a Monte Carlo simulation study to compare the relative performance of the proposed methods. A real data application of the proposed approach is given in Section 6. Finally Section 7 concludes with some remarks.
2. Parametric cause specific quantile functions
Regression models in survival studies may be developed via Cox’s Proportional Hazards (PH) [17] model, in which the effect of the covariates is multiplicative on some baseline hazard function. For parametric regression modelling of survival time one could use some well known distribution for the baseline function. More details can be found in [18] and references therein. The CSHF in terms of the well known Cox’s PH model turns out to be of the following form:
| (4) |
where is the baseline CSHF and is the vector of regression coefficients of cause . The CIF can be formulated in terms of all the CSHFs as follows
| (5) |
The overall survival function is obtained in terms of cumulative CSHFs as , where . In this article, we consider is corresponding to a Weibull distribution i.e. . The CIF under the cause specific PH assumption (4) is then given by
| (6) |
where is the vector of parameters and . From (6) it can be seen that the closed form expression for exists if the shape parameter is common for all causes i.e. . Under this assumption, it takes the form
| (7) |
The corresponding cause specific probability density function is obtained by differentiating , and we get
| (8) |
Following general notation for quantile function as in (3), the cause specific quantile/ sub-quantile function is defined as
| (9) |
The estimate of cause specific quantile is obtained as . Note that, if is continuous and strictly increasing function, then is the unique value of such that . Therefore, cause specific quantile function of from Eq. (7), yields
| (10) |
We used Weibull distribution for parametric modelling of cause specific quantile functions under middle censoring scheme because it is a very broad and flexible distribution for lifetime data analysis. A detailed discussion on parametric modelling of middle censored lifetime data with covariates can be found in [19]. We now proceed to obtain estimates of the unknown parameters of the Weibull distribution, the regression coefficients, and cause specific quantile functions, under both classical and Bayesian approaches in the following Sections 3, 4 respectively.
3. Maximum likelihood estimation
The maximum likelihood estimation (MLE) is widely used among the statistical inference methods because of its desirable properties such as consistency, asymptotic efficiency, and invariance. In the middle censoring scenario, we assume that the lifetime is middle censored by random censoring interval which having a bivariate cumulative distribution function . Moreover, we assumed that are i.i.d. pairs of bivariate random observations, where left end point and length of the censoring interval are independently follow exponential distributions i.e. and . For individuals, let lifetime ’s, and censoring interval ’s are independent, given the covariate . The observed lifetime for the th individual is given by
where is a censoring indicator. In this study it is assumed that when , then the causes of failure can be observed on later inspection. We assume that are i.i.d. observations of corresponding to individuals under study. For the observed data , the likelihood function is then given by
| (11) |
where is the indicator function for th cause. It is also assumed that and do not depend on . Without loss of generality we assume that first are the uncensored and remaining are censored observations respectively. Let and are the number of the observed events of type with respect to uncensored and censored individuals respectively with where .
The likelihood based on Eqs. (7), (8) can be written as
| (12) |
The log-likelihood function is given by
| (13) |
The MLEs of unknown parameters are obtained by maximizing the log-likelihood (13). The system of equations with respect to each of the parameters is given in Appendix. Since these equations are not in an explicit form, analytical solutions are not possible. So, we utilize iterative methods such as Newton–Raphson or other techniques to solve the system of equations. We used the optim function in R software for obtaining MLEs of the unknown parameters. By using the invariance property of MLEs we obtain the MLEs of cause specific quantile functions. Suppose that the is the MLE of then the estimator of is given by .
4. Bayes estimation
Bayesian inference is distinctive in that it incorporates prior information with the observed data. For obtaining the Bayes estimates of unknown parameters and cause specific quantile , first we need to define the suitable priors of unknown parameters and appropriate loss functions. It is known that there is limited information about the unknown parameters except that and . If all the parameters are unknown then it is difficult to obtain the joint conjugate prior for the parameters. We therefore assumed informative prior by choosing independent gamma priors for and and normal priors for as follows
| (14) |
where , , and are the hyper parameters. The hyperparameters are assumed to be known and are chosen in such a way that reflects the degree of belief about the unknown parameters. The joint prior distribution of and from (14) up to the proportionality is given by
| (15) |
The joint posterior density of and is obtained as follows
| (16) |
where is the likelihood function based on observed data as given in Eq. (12) and is the joint prior density (15). The denominator part of Eq. (16) involve multiple integrals and it is difficult to obtain the posterior densities of random variables and in explicit form. Thus the analytical evaluation of posterior samples is impossible. Therefore, in this situation MCMC method can be used to approximate the integrals [20]. Popularly used MCMC algorithms are Gibbs sampling algorithm [21] and Metropolis–Hastings (M–H) algorithm [22]. Since, marginal posterior densities of random variables and are not obtained in closed form, and so we employ the M–H algorithm.
In this article we consider two different types of loss functions, namely the commonly used squared error (symmetric) loss function and the LINEX (asymmetric) loss function for the purpose of comprehensive comparison of Bayes estimates. Squared error loss function (SELF) is defined as for a parameter . Then the Bayes estimate for parameter and under SELF can be obtained as the posterior mean and calculated as
where are the MCMC posterior random samples drawn from the marginal posterior distribution of random variables and , and is the number of iteration used in burn-in period.
Note that SELF is a symmetric loss function but it is not useful for the situations when under/over estimation is more costly than the over/under estimation and it is considered the equal weight for both under and over estimation. For example, over estimation of survival function and failure rate function is usually much more serious than under estimation. To overcome this difficulty we also consider as an alternative the LINEX loss function (LLF) which is an asymmetric loss function given by . Under LLF the Bayes estimates of parameter and can be obtained as follows
where is the hyper parameter of the LLF and magnitude of reflect the degree of asymmetry. For the LLF is quite asymmetric about 0 with overestimation being more serious than underestimation. The opposite is true with . If is close to zero then estimates under LLF are approximately equal to estimates obtained under SELF.
5. A Monte Carlo comparison of the estimates
We conducted a Monte Carlo simulation study to observe the finite sample behaviour of the MLE and Bayes estimators of the unknown parameters and cause specific quantile functions. We generate the random samples through inverse transformation for four different sample sizes i.e. , and . For each sample sizes, we simulated sets of data. In this scenario, we computed average estimates (AVE) and mean square error (MSE) for and . Besides that we obtained the average length (AVL) along with coverage probability (CP) of the asymptotic confidence interval (ACI) of the MLE and Bayes credible interval (BCI) of the Bayes estimates to compare the precision of the estimates. We also consider two different censoring percentage viz., mild (approximately, 10%), and heavy (approximately, 30%) for observing the impact of censoring. The censoring effect is explained as follows: if lock-down is extended during the COVID-19 pandemic, the percentage of censored observations will increase. As a result, COVID-19 cases are on the rise, posing a threat to the health-care system. We refer these censoring percentages as censoring scheme 1 (CS-1) and censoring scheme 2 (CS-2) respectively. The results of simulation study based on CS-1 and CS-2 are available in Table 1, Table 2 respectively.
Table 1.
AVE and MSE values under CS-1 when , and .
| Method | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 25 | MLE | AVE | 1.6307 | 0.5108 | 0.1225 | 0.4188 | 0.1013 | 0.7059 | 0.9202 |
| MSE | 0.9298 | 0.0978 | 1.2951 | 0.0937 | 1.4748 | 0.4114 | 1.0638 | ||
| ACI | AVL | 1.0253 | 0.3506 | 1.1851 | 0.3420 | 1.3856 | 0.7198 | 1.0804 | |
| CP | 0.9440 | 0.9260 | 0.9140 | 0.9280 | 0.9460 | 0.8520 | 0.8500 | ||
| Bself | AVE | 1.5974 | 0.5043 | 0.1049 | 0.4081 | 0.0844 | 0.6919 | 0.9138 | |
| MSE | 0.2932 | 0.0235 | 0.9010 | 0.0234 | 0.9447 | 0.1435 | 0.3554 | ||
| Bllf-1 | AVE | 1.5726 | 0.5014 | 0.0478 | 0.4053 | 0.0090 | 0.6787 | 0.8802 | |
| MSE | 0.2381 | 0.0228 | 0.9365 | 0.0226 | 1.0459 | 0.1222 | 0.2542 | ||
| Bllf-2 | AVE | 1.6234 | 0.5073 | 0.1623 | 0.4109 | 0.1595 | 0.7061 | 0.9681 | |
| MSE | 0.3647 | 0.0243 | 0.9426 | 0.0244 | 0.9751 | 0.1718 | 0.6732 | ||
| BCI | AVL | 0.7165 | 0.2448 | 1.0647 | 0.2386 | 1.2184 | 0.5206 | 0.8390 | |
| CP | 0.9800 | 0.9920 | 0.9220 | 0.9840 | 0.9580 | 0.9820 | 0.9880 | ||
| 50 | MLE | AVE | 1.5521 | 0.5111 | 0.1021 | 0.4053 | 0.1005 | 0.6511 | 0.8754 |
| MSE | 0.3745 | 0.0414 | 0.4913 | 0.0375 | 0.6337 | 0.1387 | 0.3688 | ||
| ACI | AVL | 0.6837 | 0.2479 | 0.7701 | 0.2405 | 0.9115 | 0.5137 | 0.7499 | |
| CP | 0.9480 | 0.9560 | 0.9300 | 0.9460 | 0.9520 | 0.9280 | 0.9120 | ||
| Bself | AVE | 1.5523 | 0.5070 | 0.0930 | 0.4021 | 0.0896 | 0.6580 | 0.8877 | |
| MSE | 0.1922 | 0.0183 | 0.4244 | 0.0171 | 0.5244 | 0.0794 | 0.2319 | ||
| Bllf-1 | AVE | 1.5371 | 0.5051 | 0.0663 | 0.4003 | 0.0531 | 0.6505 | 0.8683 | |
| MSE | 0.1716 | 0.0178 | 0.4369 | 0.0169 | 0.5451 | 0.0738 | 0.1904 | ||
| Bllf-2 | AVE | 1.5678 | 0.5090 | 0.1200 | 0.4040 | 0.1260 | 0.6658 | 0.9121 | |
| MSE | 0.2183 | 0.0188 | 0.4279 | 0.0174 | 0.5324 | 0.0866 | 0.3174 | ||
| BCI | AVL | 0.5585 | 0.1998 | 0.7339 | 0.1930 | 0.8547 | 0.3927 | 0.6321 | |
| CP | 0.9660 | 0.9860 | 0.9340 | 0.9840 | 0.9580 | 0.9740 | 0.9760 | ||
| 100 | MLE | AVE | 1.5448 | 0.5016 | 0.1005 | 0.4076 | 0.1056 | 0.6587 | 0.8522 |
| MSE | 0.1623 | 0.0193 | 0.2048 | 0.0186 | 0.2593 | 0.0763 | 0.1556 | ||
| ACI | AVL | 0.4787 | 0.1711 | 0.5344 | 0.1665 | 0.6266 | 0.3646 | 0.4962 | |
| CP | 0.9660 | 0.9660 | 0.9420 | 0.9460 | 0.9580 | 0.9360 | 0.9380 | ||
| Bself | AVE | 1.5445 | 0.5006 | 0.0952 | 0.4044 | 0.0988 | 0.6610 | 0.8642 | |
| MSE | 0.1169 | 0.0127 | 0.1935 | 0.0122 | 0.2400 | 0.0570 | 0.1284 | ||
| Bllf-1 | AVE | 1.5357 | 0.4994 | 0.0820 | 0.4034 | 0.0807 | 0.6566 | 0.8546 | |
| MSE | 0.1074 | 0.0127 | 0.1961 | 0.0120 | 0.2429 | 0.0539 | 0.1166 | ||
| Bllf-2 | AVE | 1.5534 | 0.5017 | 0.1085 | 0.4055 | 0.1168 | 0.6654 | 0.8745 | |
| MSE | 0.1281 | 0.0128 | 0.1947 | 0.0124 | 0.2440 | 0.0606 | 0.1442 | ||
| BCI | AVL | 0.4254 | 0.1521 | 0.5184 | 0.1476 | 0.6027 | 0.2995 | 0.4440 | |
| CP | 0.9640 | 0.9740 | 0.9380 | 0.9640 | 0.9620 | 0.9640 | 0.9620 | ||
| 200 | MLE | AVE | 1.5122 | 0.5028 | 0.0986 | 0.4014 | 0.1045 | 0.6384 | 0.8415 |
| MSE | 0.0706 | 0.0096 | 0.0945 | 0.0091 | 0.1184 | 0.0323 | 0.0708 | ||
| ACI | AVL | 0.3303 | 0.1222 | 0.3718 | 0.1183 | 0.4404 | 0.2668 | 0.3620 | |
| CP | 0.9540 | 0.9320 | 0.9360 | 0.9500 | 0.9560 | 0.9680 | 0.9600 | ||
| Bself | AVE | 1.5136 | 0.5020 | 0.0953 | 0.4001 | 0.1003 | 0.6410 | 0.8487 | |
| MSE | 0.0588 | 0.0077 | 0.0930 | 0.0073 | 0.1152 | 0.0273 | 0.0644 | ||
| Bllf-1 | AVE | 1.5090 | 0.5014 | 0.0888 | 0.3995 | 0.0912 | 0.6387 | 0.8436 | |
| MSE | 0.0570 | 0.0077 | 0.0941 | 0.0073 | 0.1159 | 0.0268 | 0.0612 | ||
| Bllf-2 | AVE | 1.5183 | 0.5027 | 0.1019 | 0.4007 | 0.1094 | 0.6433 | 0.8538 | |
| MSE | 0.0610 | 0.0078 | 0.0927 | 0.0073 | 0.1163 | 0.0280 | 0.0684 | ||
| BCI | AVL | 0.3080 | 0.1139 | 0.3647 | 0.1102 | 0.4298 | 0.2166 | 0.3210 | |
| CP | 0.9600 | 0.9480 | 0.9380 | 0.9640 | 0.9500 | 0.9560 | 0.9560 | ||
Bself, Bllf-1, Bllf-2 denotes the Bayes estimates under SELF, llf-1 and llf-2 respectively.
Table 2.
AVE and MSE values under CS-2 when , and .
| Method | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 25 | MLE | AVE | 1.6454 | 0.5128 | 0.1261 | 0.4214 | 0.1085 | 0.7084 | 0.9279 |
| MSE | 1.0921 | 0.1025 | 1.3280 | 0.0989 | 1.5060 | 0.4187 | 1.2594 | ||
| ACI | AVL | 1.0844 | 0.3596 | 1.2237 | 0.3495 | 1.4154 | 0.7492 | 1.1557 | |
| CP | 0.9420 | 0.9320 | 0.9240 | 0.9320 | 0.9380 | 0.8580 | 0.8640 | ||
| Bself | AVE | 1.6021 | 0.5051 | 0.1071 | 0.4090 | 0.0887 | 0.6930 | 0.9153 | |
| MSE | 0.3056 | 0.0229 | 0.9158 | 0.0236 | 0.9752 | 0.1424 | 0.3608 | ||
| Bllf-1 | AVE | 1.5759 | 0.5021 | 0.0468 | 0.4061 | 0.0108 | 0.6794 | 0.8811 | |
| MSE | 0.2452 | 0.0223 | 0.9546 | 0.0228 | 1.0670 | 0.1205 | 0.2569 | ||
| Bllf-2 | AVE | 1.6294 | 0.5081 | 0.1680 | 0.4119 | 0.1664 | 0.7078 | 0.9710 | |
| MSE | 0.3845 | 0.0238 | 0.9659 | 0.0247 | 1.0249 | 0.1720 | 0.6903 | ||
| BCI | AVL | 0.7354 | 0.2480 | 1.0936 | 0.2410 | 1.2399 | 0.5282 | 0.8487 | |
| CP | 0.9860 | 0.9960 | 0.9340 | 0.9860 | 0.9580 | 0.9900 | 0.9880 | ||
| 50 | MLE | AVE | 1.5580 | 0.5118 | 0.1037 | 0.4061 | 0.1017 | 0.6528 | 0.8771 |
| MSE | 0.4252 | 0.0426 | 0.5142 | 0.0393 | 0.6595 | 0.1459 | 0.3879 | ||
| ACI | AVL | 0.7162 | 0.2540 | 0.7926 | 0.2449 | 0.9316 | 0.5349 | 0.7800 | |
| CP | 0.9440 | 0.9640 | 0.9320 | 0.9460 | 0.9460 | 0.9240 | 0.9080 | ||
| Bself | AVE | 1.5565 | 0.5074 | 0.0943 | 0.4027 | 0.0910 | 0.6596 | 0.8890 | |
| MSE | 0.2059 | 0.0182 | 0.4442 | 0.0174 | 0.5474 | 0.0815 | 0.2424 | ||
| Bllf-1 | AVE | 1.5403 | 0.5054 | 0.0659 | 0.4009 | 0.0531 | 0.6518 | 0.8692 | |
| MSE | 0.1817 | 0.0177 | 0.4559 | 0.0172 | 0.5677 | 0.0754 | 0.1991 | ||
| Bllf-2 | AVE | 1.5731 | 0.5095 | 0.1227 | 0.4046 | 0.1288 | 0.6678 | 0.9147 | |
| MSE | 0.2366 | 0.0187 | 0.4503 | 0.0178 | 0.5585 | 0.0892 | 0.3369 | ||
| BCI | AVL | 0.5759 | 0.2030 | 0.7548 | 0.1954 | 0.8699 | 0.4010 | 0.6401 | |
| CP | 0.9720 | 0.9840 | 0.9300 | 0.9800 | 0.9560 | 0.9720 | 0.9600 | ||
| 100 | MLE | AVE | 1.5475 | 0.5022 | 0.1026 | 0.4080 | 0.1081 | 0.6596 | 0.8531 |
| MSE | 0.1742 | 0.0205 | 0.2130 | 0.0189 | 0.2726 | 0.0808 | 0.1564 | ||
| ACI | AVL | 0.4993 | 0.1752 | 0.5480 | 0.1695 | 0.6384 | 0.3790 | 0.5155 | |
| CP | 0.9680 | 0.9580 | 0.9500 | 0.9560 | 0.9560 | 0.9340 | 0.9340 | ||
| Bself | AVE | 1.5473 | 0.5011 | 0.0968 | 0.4048 | 0.1018 | 0.6620 | 0.8655 | |
| MSE | 0.1228 | 0.0133 | 0.2015 | 0.0122 | 0.2500 | 0.0594 | 0.1289 | ||
| Bllf-1 | AVE | 1.5378 | 0.4999 | 0.0828 | 0.4037 | 0.0830 | 0.6574 | 0.8556 | |
| MSE | 0.1120 | 0.0132 | 0.2038 | 0.0120 | 0.2518 | 0.0560 | 0.1164 | ||
| Bllf-2 | AVE | 1.5569 | 0.5022 | 0.1109 | 0.4059 | 0.1205 | 0.6667 | 0.8762 | |
| MSE | 0.1357 | 0.0134 | 0.2033 | 0.0124 | 0.2557 | 0.0633 | 0.1456 | ||
| BCI | AVL | 0.4401 | 0.1546 | 0.5334 | 0.1497 | 0.6152 | 0.3066 | 0.4521 | |
| CP | 0.9680 | 0.9720 | 0.9480 | 0.9660 | 0.9600 | 0.9640 | 0.9540 | ||
| 200 | MLE | AVE | 1.5139 | 0.5025 | 0.0985 | 0.4013 | 0.1044 | 0.6396 | 0.8429 |
| MSE | 0.0748 | 0.0102 | 0.1028 | 0.0095 | 0.1257 | 0.0344 | 0.0720 | ||
| ACI | AVL | 0.3437 | 0.1249 | 0.3811 | 0.1201 | 0.4484 | 0.2774 | 0.3762 | |
| CP | 0.9560 | 0.9420 | 0.9360 | 0.9600 | 0.9600 | 0.9760 | 0.9660 | ||
| Bself | AVE | 1.5159 | 0.5018 | 0.0954 | 0.4000 | 0.1006 | 0.6424 | 0.8502 | |
| MSE | 0.0619 | 0.0081 | 0.1005 | 0.0075 | 0.1215 | 0.0290 | 0.0655 | ||
| Bllf-1 | AVE | 1.5109 | 0.5012 | 0.0886 | 0.3994 | 0.0912 | 0.6400 | 0.8450 | |
| MSE | 0.0597 | 0.0080 | 0.1017 | 0.0075 | 0.1222 | 0.0284 | 0.0621 | ||
| Bllf-2 | AVE | 1.5209 | 0.5025 | 0.1023 | 0.4007 | 0.1100 | 0.6449 | 0.8555 | |
| MSE | 0.0647 | 0.0081 | 0.1002 | 0.0075 | 0.1226 | 0.0298 | 0.0697 | ||
| BCI | AVL | 0.3201 | 0.1160 | 0.3738 | 0.1118 | 0.4368 | 0.2224 | 0.3268 | |
| CP | 0.9620 | 0.9520 | 0.9340 | 0.9640 | 0.9560 | 0.9620 | 0.9680 | ||
Bself, Bllf-1, Bllf-2 denotes the Bayes estimates under SELF, llf-1 and llf-2 respectively.
In the simulation study, we consider two causes of failure for simplicity i.e. and one single covariate . The covariate is generated from . The survival time is generated by using the steps given in [23]. Without loss of generality the true parameter values arbitrary taken as and . For obtaining the endpoints of middle censoring the random variable and are generated from independent exponential distributions with fixed means and respectively. For CS-1 and CS-2 we choose two pairs of values for () as and which gives the average censoring proportion approximately 10% (mild) and 30% (heavy) respectively. Based on different sample sizes and CSs, the MLE and Bayes estimates of and are calculated. The estimates of are obtained at and covariate for both the causes and denoted as and with true values and .
The Bayes estimates are obtained based on assumed priors. The hyperparameters of gamma priors under informative prior are calculated using the likelihood estimates of and based on 1000 iteration of sample size 25. Now, we compute the mean and variance of and and compare with the mean and variance of gamma priors. Subsequently, we get the hyper parameters values as and . For regression parameters and we assumed as informative priors. The hyper parameter of LLF is fixed at , and it is known as llf-1 and llf-2.
Next, as we discussed in Section 4 that marginal posterior densities of unknown parameters are not in a closed form, so we utilized the MCMC procedure for generating the random samples from marginal posteriors. For this purpose we used the BUGS software via R2OpenBUGS package in R software [24]. We generate, Markov chains for each parameter and the first samples were used in burn-in period. Furthermore, for minimizing the effect of the autocorrelation every second equally spaced outcome is considered i.e. thin=2. By the visualization of the convergence diagnostics plots it is realized that chains are converging nicely. Therefore, the last 6000 MCMC samples are used to obtained the Bayes estimates of and based on SELF and LLF.
From Table 1 it is clear that for fixed censoring proportion as the sample size increases, MSEs decreases for MLE and Bayes estimates, which verifies the consistency property of all the estimators. As expected, for small sample size the Bayes estimates under both the loss functions are better than MLE in terms of MSE, AVL and CP. It is also noticed that the CPs for ACIs of the cause specific quantile functions for sample size 25 are little bit away from the nominal level (95%). Similarly, for and we can say that Bayes estimates of baseline parameters and cause specific quantiles under both the loss functions are quite better except some values in sample size 200. The Bayes estimates for llf-1 are smaller as compared to llf-2. From Table 1, Table 2 it is observed that for fixed sample sizes the MSE and AVL of all the estimates is increases as the censoring percentage increases except for some values. Therefore, it indicates that as the censored observations are increased this will leads to the less efficient estimates. This implies that if the spread of corona virus is not under control in a reasonable period of time, then it will have a significant impact on the human health-care system. Overall it is observed that the CPs maintain the nominal level (95%) of all the estimates for both the censoring schemes.
6. An illustrative application
In this section, for illustrative purposes a real life application is considered. We have taken real data from a Mayo Clinic study on primary biliary cirrhosis (PBC) of the liver conducted between 1974 to 1984. This data set is available in survival package of R software. During this ten-year period, 312 patients were randomly assigned to receive D-penicillamine or placebo treatment from a total of 424 patients. The remaining 112 patients did not take part in the clinical trial but agreed to have their basic measurements taken and to be followed for survival. Six of those patients were lost to follow-up shortly after diagnosis, so these patients were removed from the study. 161 patients died at the end of the study, 25 patients received a liver transplant, and 232 patients were lost to follow-up. As a result, for two competing outcome variables, liver transplant and death, the competing risks model becomes more reasonable. All the survival times are measured in days. For more information on this PBC data, one may refer to [25] and application of competing risk on PBC data is available in [26].
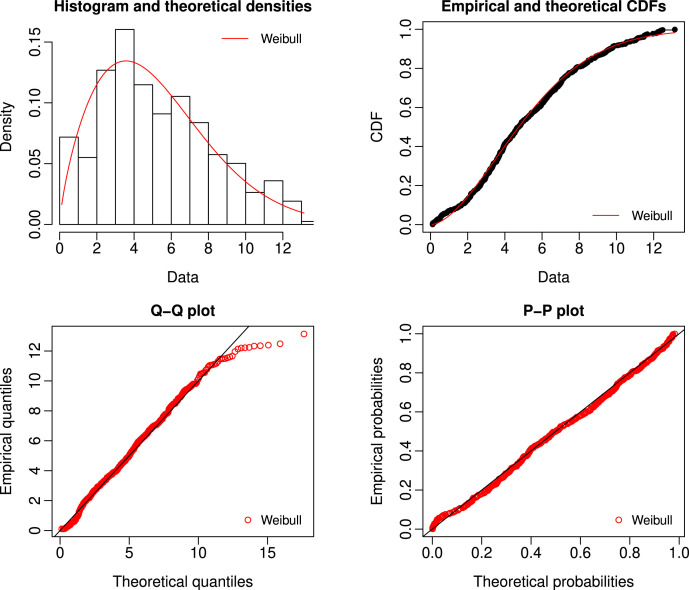
In order to compute the survival time in years, it divided by 365, which yielded a median survival time of 4.74 years. First, we check the goodness of fit of the Weibull distribution using fitdistrplus package in R software with assumption that the data is complete. The Kolmogorov–Smirnov distance between the empirical distribution function and the fitted Weibull distribution function is 0.0331 and the corresponding -value is 0.7378. Therefore, Weibull model appears to be reasonable and cannot be rejected. We also consider the graphical method of goodness of fit to check the appropriateness of the model as given in Fig. 1 and it indicates that the model fits well to the data.
Fig. 1.
Graphical fitting of the Weibull distribution.
As we have discussed in Section 1 the middle censoring may arise due to COVID-19 pandemic. But this PBC data set does not have middle censored observations and currently we do not have any middle censored follow-up data. However, once the COVID-19 pandemic is over, it is possible that middle censored follow-up data will be available. Therefore, we created an artificial data set using middle censoring, whose left end point is equal to the observed time and the right end point is equal to the , where is the width of the interval, which is generated from an exponential distribution with a mean value of 10. Then, all the censored individuals in the original data set are considered as the middle censored observations. The competing outcome variables for middle censored observation are randomly assigned from transplant and death.
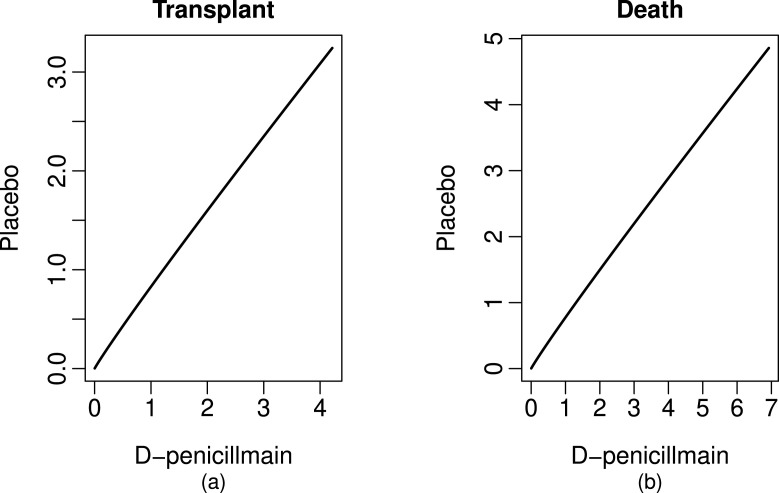
For this new data set which consists observed exact lifetimes and censored intervals, we check the PH assumption of the model (4) by considering treatment as a covariate. To examine the PH assumption for transplant and death we utilize the graphical method known as Andersen plot [27]. The covariate treatment is discrete and take two values 1 and 2 for D-penicillmain and placebo respectively. We also assume that 106 patients who do not participate in the trial they received the D-penicillmain treatment. Thus data are divided into two strata, corresponding to D-penicillmain and placebo individuals. Suppose that be the estimate of baseline cumulative CSHF for th cause in rth stratum, for transplant and death, and for D-penicillmain and placebo respectively. We plot versus for transplant and death. If the proportionality assumptions holds then these plots should be a straight line passing through origin which is verifying by Fig. 2.
Fig. 2.
Andersen plots for transplant and death.
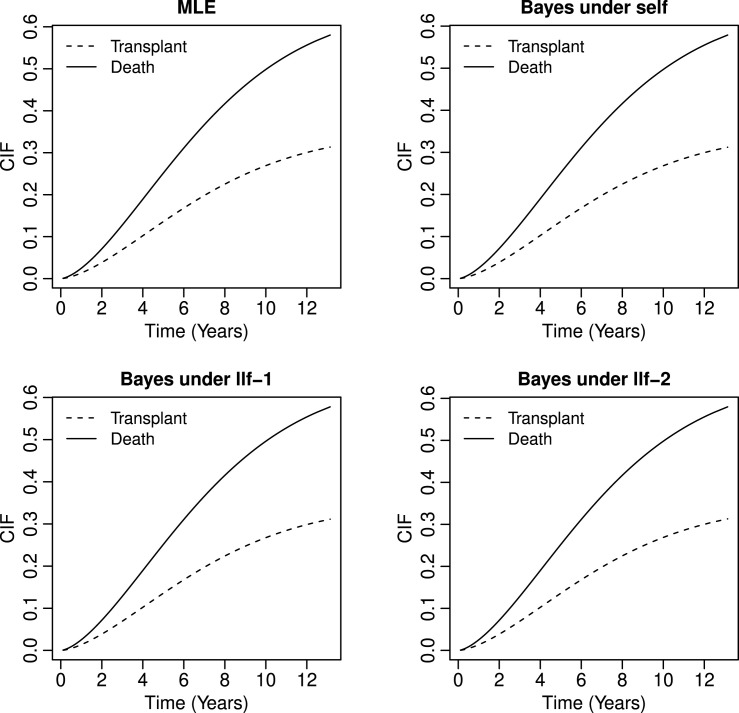
We then apply the proposed methods of estimation to obtain the estimates of unknown parameters and cause specific quantile functions. These are presented in Table 3, Table 4 under proposed methods of estimation. The Bayes estimates are obtained under SELF and LLF loss functions based on non-informative priors because we have no past information about the unknown parameters. For non-informative priors we assume that and where and are said to follow normal distribution with mean zero and large variance. From Table 3 it is observe that the MLE and Bayes estimates of the unknown parameters are very close. We also estimate the baseline CIFs based on Eq. (7) for transplant and death under MLE and Bayes estimates. Plots of the baseline CIFs are presented in Fig. 3 in which solid line represents the estimates of the CIF due to death and dotted line represents the estimates of the CIF due to transplant. From Fig. 3 it is observed that the estimates of CIFs have smaller value for transplant as compared to death.
Table 3.
Point estimates of unknown parameters.
| Method | |||||
|---|---|---|---|---|---|
| MLE | 1.5755 | 0.0654 | 0.0272 | 0.0966 | 0.2079 |
| MLE S.E. | 0.0692 | 0.0061 | 0.1829 | 0.0067 | 0.1351 |
| Bayes self | 1.5700 | 0.0650 | 0.0300 | 0.0964 | 0.2105 |
| Bayes llf-1 | 1.5665 | 0.0650 | 0.0048 | 0.0964 | 0.1969 |
| Bayes llf-2 | 1.5735 | 0.0650 | 0.0555 | 0.0965 | 0.2242 |
| Bayes S.E. | 0.0682 | 0.0061 | 0.1839 | 0.0067 | 0.1350 |
Table 4.
Parametric cause specific quantile estimates for transplant and death.
| Method | Placebo |
D-penicillamine |
||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||
| MLE | 3.935 | 7.069 | 2.529 | 4.173 | 3.929 | 7.306 | 2.209 | 3.630 |
| Bayes self | 3.976 | 7.225 | 2.531 | 4.186 | 3.955 | 7.447 | 2.204 | 3.627 |
| Bayes llf-1 | 3.845 | 6.703 | 2.494 | 4.104 | 3.866 | 7.001 | 2.182 | 3.587 |
| Bayes llf-2 | 4.132 | 12.60 | 2.570 | 4.275 | 4.055 | 9.162 | 2.225 | 3.668 |
Fig. 3.
CIF plots for transplant and death.
Table 4 shows the parametric estimates of quantile functions for time to transplant and death under both the treatment groups. Under placebo treatment, the 10% quantile estimates for time to transplant are approximately years, and 20% quantile estimates are approximately years based on MLE and Bayes estimates, except for Bayes estimate under llf-2 for 20% quantile. However, under placebo treatment, 10% and 20% quantile estimate for death are approximately 2.5 and years respectively. Similarly, under D-penicillamine treatment, 10% and 20% quantile estimate for transplant are approximately and years (except for Bayes estimate under llf-2 for 20% quantile) and for death are approximately 2.2 and 3.6 years respectively. Quantile event times gives the information about the stay of the patients in waiting queue, this implies that the 10% and 20% patients will receive the transplant soon after and years respectively and 10% and 20% patients died soon after 2.2 and 3.6 years respectively. This shows that the waiting time of the patients to receive the transplant is roughly two times larger as compared to the death under both the treatment groups. It is observed that the quantile estimates under both the treatment groups have minor differences. This indicates that the effect of treatment is not significantly different on transplant and death.
7. Concluding remarks
This article considers parametric cause specific quantile inference of the CIF under middle censoring scheme. In this study we have discussed the use of middle censoring scheme in the context of the current COVID-19 pandemic. This research could provide a useful statistical framework for medical practitioners to obtain precise survival analysis for patients who were lost to follow-up due to this pandemic. We believe this to be first such attempt to model the quantile event times of the middle censored data under competing risks. The regression model was developed based on Cox’s PH model by assuming a very flexible Weibull distribution for the baseline hazard function. Also, we provide the estimates of unknown parameters and cause specific quantiles under both the classical and Bayesian set-up. The simulation study shows that the Bayes estimates perform well based on informative priors under squared error loss function in terms of MSE as compared to MLE. However, all the estimates exhibit the consistency property, as also the identifiability and appropriate convergence of the proposed model. Overall, the proposed model performed well in simulation studies. In a real data analysis on Primary Biliary Cirrhosis of the liver, goodness of fit criteria verify that the Weibull model fits well to the data. Also the covariate treatment maintains the assumed model PH assumption. Other semi-parametric regression models, such as additive hazard regression model and proportional odds model, may also be appropriate in this context and will be explored elsewhere.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank the Editor-in-Chief and anonymous reviewers for a careful reading of the manuscript and several helpful suggestions that helped to improve the quality of the paper.
Appendix.
Details of the system of normal equations used in Section 3 are provided here after introducing some notations that help reduce the length of expressions viz.
Partial derivatives of the log-likelihood of Eq. (13) with respect to each of the parameters gives the following equations.
| (17) |
| (18) |
| (19) |
References
- 1.Dima A., Balaban D.V., Jurcut C., Berza I., Jurcut R., Jinga M. Healthcare. Vol. 9. Multidisciplinary Digital Publishing Institute; 2021. Perceptions of Romanian physicians on lockdowns for COVID-19 prevention; p. 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shrestha N., Shad M.Y., Ulvi O., Khan M.H., Karamehic-Muratovic A., Nguyen U.-S.D., Baghbanzadeh M., Wardrup R., Aghamohammadi N., Cervantes D., et al. The impact of COVID-19 on globalization. One Health. 2020 doi: 10.1016/j.onehlt.2020.100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khetrapal S., Bhatia R. Impact of COVID-19 pandemic on health system & sustainable development goal 3. Indian J. Med. Res. 2020;151(5):395. doi: 10.4103/ijmr.IJMR_1920_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Organization W.H., et al. World Health Organization; 2020. The impact of the COVID-19 pandemic on noncommunicable disease resources and services: results of a rapid assessment. [Google Scholar]
- 5.Jammalamadaka S., Mangalam V. Nonparametric estimation for middle-censored data. J. Nonparametr. Stat. 2003;15(2):253–265. [Google Scholar]
- 6.Ahmadi K., Rezaei M., Yousefzadeh F. Statistical analysis of middle censored competing risks data with exponential distribution. J. Stat. Comput. Simul. 2017;87(16):3082–3110. [Google Scholar]
- 7.Abuzaid A.H., El-Qumsan M.K.A., El-Habil A.M. On the robustness of right and middle censoring schemes in parametric survival models. Comm. Statist. Simulation Comput. 2017;46(3):1771–1780. [Google Scholar]
- 8.Kalbfleisch J.D., Prentice R.L. John Wiley & Sons, New York; 2002. The Statistical Analysis of Failure Time Data. [Google Scholar]
- 9.Parzen E. Nonparametric statistical data modeling. J. Amer. Statist. Assoc. 1979;74(365):105–121. [Google Scholar]
- 10.Sankaran P., Nair N.U., Sreedevi E. A quantile based test for comparing cumulative incidence functions of competing risks models. Statist. Probab. Lett. 2010;80(9):886–891. doi: 10.1016/j.spl.2010.01.023. [DOI] [Google Scholar]
- 11.Peng L., Fine J.P. Nonparametric quantile inference with competing–risks data. Biometrika. 2007;94(3):735–744. [Google Scholar]
- 12.Peng L., Fine J.P. Competing risks quantile regression. J. Amer. Statist. Assoc. 2009;104(488):1440–1453. [Google Scholar]
- 13.Lee M., Fine J.P. Inference for cumulative incidence quantiles via parametric and nonparametric approaches. Stat. Med. 2011;30(27):3221–3235. doi: 10.1002/sim.4349. [DOI] [PubMed] [Google Scholar]
- 14.Lee M., Han J. Covariate-adjusted quantile inference with competing risks. Comput. Statist. Data Anal. 2016;101:57–63. [Google Scholar]
- 15.Lee M. Parametric inference for quantile event times with adjustment for covariates on competing risks data. J. Appl. Stat. 2019;46(12):2128–2144. [Google Scholar]
- 16.Peng L. Quantile regression for survival data. Annu. Rev. Stat. Appl. 2021;8:413–437. doi: 10.1146/annurev-statistics-042720-020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox D.R. Regression models and life-tables. J. R. Stat. Soc. Ser. B Stat. Methodol. 1972;34(2):187–220. [Google Scholar]
- 18.Rehman H., Chandra N., Hosseini-Baharanchi F.S., Baghestani A.R., Pourhoseingholi M.A. Cause-specific hazard regression estimation for modified Weibull distribution under a class of non-informative priors. J. Appl. Stat. 2021:1–18. doi: 10.1080/02664763.2021.1882407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett N., Iyer S.K., Jammalamadaka S. Analysis of gamma and Weibull lifetime data under a general censoring scheme and in the presence of covariates. Comm. Statist. Theory Methods. 2017;46(5):2277–2289. [Google Scholar]
- 20.Robert C.P., Casella G. Springer; 2010. Introducing Monte Carlo Methods with R. [Google Scholar]
- 21.Geman S., Geman D. Stochastic relaxation, gibbs distributions, and the Bayesian restoration of images. IEEE Trans. Pattern Anal. Mach. Intell. 1984;PAMI-6(6):721–741. doi: 10.1109/tpami.1984.4767596. [DOI] [PubMed] [Google Scholar]
- 22.Hastings W.K. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970;57(1):97–109. [Google Scholar]
- 23.Beyersmann J., Allignol A., Schumacher M. Springer Science & Business Media; 2012. Competing Risks and Multistate Models with R. [Google Scholar]
- 24.Lunn D., Jackson C., Best N., Spiegelhalter D., Thomas A. Chapman and Hall/CRC; 2012. The BUGS book: A practical introduction to Bayesian analysis. [Google Scholar]
- 25.Therneau T.M., Grambsch P.M. Springer; 2000. Modeling Survival Data: Extending the Cox Model. [Google Scholar]
- 26.Lai X., Yau K.K., Liu L. Competing risk model with bivariate random effects for clustered survival data. Comput. Statist. Data Anal. 2017;112:215–223. [Google Scholar]
- 27.Andersen P.K. Testing goodness of fit of Cox’s regression and life model. Biometrics. 1982;38(1):67–77. [Google Scholar]