Abstract
An inexpensive virus detection scheme with high sensitivity and specificity is desirable for broad applications such as the COVID-19 virus. In this article, we introduce the localized surface plasmon resonance (LSPR) principle on the aggregation of antigen-coated gold nanoparticles (GNPs) to detect SARS-CoV-2 Nucleocapsid (N) proteins. Experiments show this technique can produce results observable by the naked eye in 5 min with a LOD (Limits of Detection) of 150 ng/ml for the N proteins. A comprehensive numerical model of the LSPR effect on the aggregation of GNPs has been developed to identify the key parameters in the reaction processes. The color-changing behaviors can be readily utilized to detect the existence of the virus while the quantitative concentration value is characterized with the assistance of an optical spectrometer. A parameter defined as the ratio of the light absorption intensity at the upper visible band region of 700 nm to the light absorption intensity at the peak optical absorption spectrum of the GNPs at 530 nm is found to have a linear relationship with respect to the N protein concentrations. As such, this scheme could be utilized as an inexpensive testing methodology for applications in POC (Point-of-Care) diagnostics to combat current and future virus-induced pandemics.
Keywords: SARS-CoV-2 detection, Plasmonic GNP, LSPR, Colorimetric, Point-of-Care, Biosensors
1. Introduction
The COVID-19 pandemic since 2020 has resulted in the losses of money and lives (Almaghaslah et al., 2020; Hu et al., 2020). One key area to combat the coronavirus is large-scale testing for both symptomatic and asymptomatic patients. However, the state-of-art tests are often conducted by trained workers in laboratories or hospitals which severely limit the testing capacity (Udugama et al., 2020). Specifically, body-temperature screening is not to detect patients without symptoms (Bielecki et al., 2020) and the nucleic acid test based on polymeric chain reaction (PCR) (Pfefferle et al., 2020; Udugama et al., 2020), requires sophisticated instruments and a long processing time (a few hours) (Smyrlaki et al., 2020). To address these issues, many new detection methods have been reported (Alafeef et al., 2020, Behrouzi and Lin, 2021; Broughton et al., 2020; Cheong et al., 2020; Iravani, 2020; Miripour et al., 2020; Moitra et al., 2020; Qiu et al., 2020; Seo et al., 2020; Wang et al., 2020), including the nucleic acid test assisted by CRISPR-Cas12 (Broughton et al., 2020), GFET-based biosensors (Seo et al., 2020), plasmonic thermocycling RNA detections (Cheong et al., 2020), and functionalized terahertz plasmonic metasensors (Ahmadivand et al., 2021). However, these aforementioned approaches all require sophisticated instruments and/or expensive bio-agents.
A point-of-care (POC) detection method that is easy to operate, inexpensive, and rapid to provide high specificity results would be desirable. Previously, schemes based on antigens have shown promising results for patients with severe symptoms, such as the Abbott BinaxNOW kit. This tool does not require any filtration steps, but the sensitivity and specificity are often inferior to that of the PCR tests. Previously, optical detection schemes based on the amplification of weak sensing signals have been shown to achieve low limit-of-detection (LOD) in various biosensors (Ahmadivand et al., 2020; Draz and Shafiee, 2018; Iarossi et al., 2018; Kim et al., 2017, 2010; Liu et al., 2016, 2015; Moitra et al., 2020; Nie et al., 2014; Valentini et al., 2013). Here, we propose and demonstrate the application of localized surface plasmon resonance of gold nanoparticles in the visible light range for the detection of the SARC-CoV-2 Nucleocapsid (N) proteins.
Electron clouds can oscillate at a certain frequency at the surface of metals, which is so called the surface plasmon (SP) and the resonant frequency depends on the refractive index of the metal and surrounding materials (Zhang et al., 2012). When SPs become localized around nanoparticles, it is known as the localized surface plasmons (LSPs) and the oscillation frequency can be modulated based on the shape, size and interparticle distance of metallic nanoparticles. Under electromagnetic (EM) waves, LSPs could be resonated at a specific frequency to amplify the electric field at near-fields which can lead to the higher optical absorptivity of material (Mayer and Hafner, 2011; Petryayeva and Krull, 2011). As such, the color change of solutions upon the aggregation of nanoparticles (Iarossi et al., 2018) have been utilized in many applications and GNPs are often chosen for better biocompatibility (Shukla et al., 2005). Several previous papers have modeled the LSPs effect on GNPs and most of them have focused on applications for surface enhanced Raman spectroscopy (SERS) (Zong et al., 2018). To analyze the effect of aggregation on the optical responses of GNPs (Gomes et al., 2020; Link et al., 1999; Myroshnychenko et al., 2008; Shabaninezhad and Ramakrishna, 2019; Wang et al., 2021; Yang and Hallinan, 2016), we analyze this phenomenon numerically for GNPs in a variety of aggregate sizes and validate simulation results with experimental tests.
When interacting with analytes, nanoparticles could conjugate with receptors to make aggregates and the spaces between nanoparticles could vary significantly to cause changes in the optical intensity spectrum (Fig. 1 ) (Ghosh and Pal, 2007). By conjugating GNPs with antibodies specific to the SARC-CoV-2 antigens, GNPs can make aggregate in the presence of antigens (Iarossi et al., 2018; Mayer and Hafner, 2011; Petryayeva and Krull, 2011). Aggregation of GNPs causes the optical absorption spectrum shifts towards larger wavelengths (red-shift), and the solution turns to blue-spectrum color as a result (Ghosh and Pal, 2007; Iarossi et al., 2018; Mayer and Hafner, 2011; Petryayeva and Krull, 2011). This behavior could be utilized as a simple colorimetric method to detect viruses by the naked eye. This reaction comes from the accumulation of antigens around GNPs which brings individual GNPs together, couples their plasmons and changes the refractive index of the surrounding environment, consequently this process causes the red-shift of the optical intensity spectrum (Mayer and Hafner, 2011). Previously, Iarossi et al. (Iarossi et al., 2018) have shown the possibility of using this method to detect human IgG with a key drawback of limited working range as the wavelength shift has reverted back to the analyte-free case at high protein concentrations. Here, a new parameter is utilized to address this problem. The ratio of the light absorption intensity of the solution containing GNPs at the upper visible band region of 700 nm to that at the peak optical absorption spectrum at 530 nm is found to have a linear relationship with respect to the N protein concentrations.
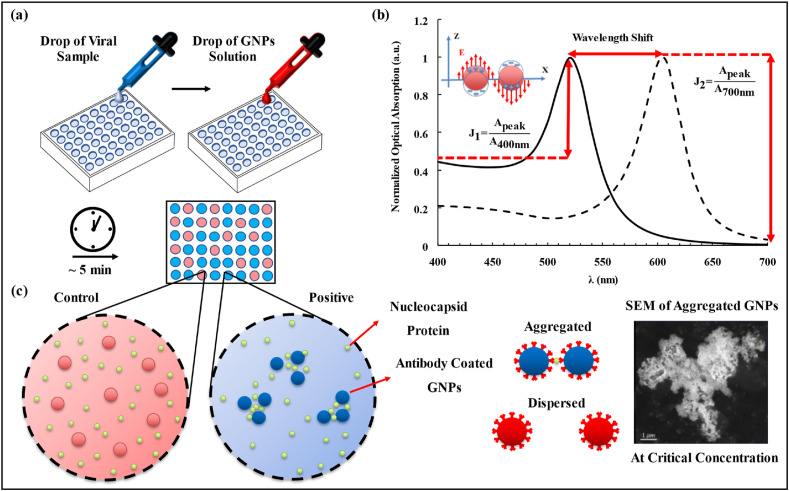
Fig. 1.
The concept of the plasmonic GNPs for the SARC-CoV-2 N-protein detection. (a) A droplet of a viral sample solution and a droplet of the antibody coated GNPs solution are mixed. After 5 min, the color of the solution may remain as red (control) or change to blue as the positive identification of the virus. (b) Schematic of the LSPR effect over the 400–700 nm optical spectra for the GNPs. The ratio of the highest optical absorption intensity of the testing solution to the optical absorption intensity at 400 nm is defined as J1 and the ratio of the highest optical absorption intensity of the testing solution to the optical absorption intensity at 700 nm is defined as J2. (c) Antibody coated GNPs interact with antigens to make aggregates with a variety of sizes. The LSPR effects in aggregated GNPs result in the optical responses (color changes) of the solution. SEM image of the aggregated GNPs shows a very large aggregate (micrometer in size). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In this article, we present a naked eye-based colorimetric sensing scheme using the plasmonic reactions of antibody-coated GNPs to detect SARS-CoV-2 Nucleocapsid (N) proteins. Two parameters, J1 and J2, are defined as the ratio of the peak optical absorption to the optical absorption at 400 nm and at 700 nm of the testing solution, respectively, as illustrated in Fig. 1b. Here, N proteins have been chosen over the Spike (S) proteins since they have significantly lower variations over different SARS-CoV-2 mutants for our proof-of-concept demonstrations (Diao et al., 2021). The proposed method provides results in less than 5 min based on the LSPR effect of bio-conjugated GNPs in the aggregated state (see Fig. 1c). The key comparisons of various other biosensors and this work are summarized in Table S1.
2. Materials and methods
2.1. Materials
NHS (N-hydroxysuccinimide)-dried gold nanoparticles with 40 nm diameter (<15% variation in size) have been purchased from Nanocomposix. The SARS-CoV-2 Nucleocapsid polyclonal antibodies and Nucleocapsid-His recombinant proteins have been provided by Sino Biological. Millipore Amicon Ultra 0.5 ml centrifugal filters have been used for the antibody purification process. All reagents in the conjugation process have been purchased from Nanocomposix.
2.2. Antibody purification
The solution containing antibodies may have small amounts of salts and proteins such as BSA as a stabilizer, and amines such as Sodium Azide and Tris for the preservation purpose. These proteins must be removed before the conjugation step since they can interfere with NHS-esters in the process. We spilt the antibody solution inside a microcentrifuge tube containing the 10 kDa filter with 10 mM of potassium phosphate to reach 450 μl. Then, we centrifuged the solution at 13.8 k RCF (relative centrifugal field) for 5 min. After discarding supernatants, we added 350 μl of 10 mM potassium phosphate and repeated washing and centrifuging steps for 4 times. To collect the purified antibodies, we centrifuged one more time at 1 k RCF and added 10 mM of potassium phosphate to reach the 1 mg/ml concentration.
2.3. Covalently conjugation of GNPs
We added 1 ml of the 5 mM potassium phosphate, 0.5% 20 K MW PEG (Polyethylene glycol) to the NHS-dried gold nanoparticles to activate NHS-esters for antibody conjugations. Next, we mixed the GNPs solution with purified antibodies at the specific concentration and incubate the mixture for an hour. After the incubation, we added 5 μl of 5% (w/v) hydroxylamine to block any non-bonded NHS-esters and let them interact with each other for additional 10 min. To remove the excess antibodies inside the solution, we centrifuged covalently conjugated GNPs three times at 3.8 k RCF. Finally, after removing all non-boded antibodies, we added the required volume of 0.1X PBS, 0.5% BSA, 0.5% Tween 20, 0.05% Sodium Azide to conjugate GNPs with antibody at a 5 μg/ml concentration (see Fig. S16).
2.4. Colorimetric diagnostic assay
We prepared samples containing different concentrations of Nucleocapsid from 150 ng/ml to 900 ng/ml and mixed them with 5 μl of antibody-coated GNPs solution of 40 μl in each well and incubated them for 10 min. After the incubation, we measured the absorption spectra. We mixed 1 μl of sample with 1 μl PBS and compared its concentration with the sample of 2 μl with Direct Detect Spectrometer. We found out that the final concentration is about one sixth of the original value, such that the conversion scale of the dilution process was about one-third. For time evolution results, we measured the optical intensity spectra without the prior incubation step and waited 30 s after the addition of GNPs to have the uniform mixture. The shifts in the maximum absorption wavelength, J1, J2, and were calculated from measured optical absorption spectra. We subtracted the background absorptions and normalized the spectra with respect to their maximum absorption values. We observed the visible color change at different antigen concentrations and estimated the corresponding naked eye LOD. In the experiments, 6 different batches of antibody conjugated GNPs have been utilized and the experimental results have shown about 5% and 15% deviations away and around the relative maximum concentration point in the recorded UV–Visible spectrometry, including experimental errors.
2.5. Instruments
We used microcentrifuge Eppendorf 5415C for antibody purification and Molecular Devices Spectramax M5 plate reader with Falcon 96 well microplate and Corning 384 low volume-well microplate to measure the absorption spectra of GNPs and their corresponding color. The wavelength range of the UV–Visible spectrometry was 400 nm–700 nm with a 1 nm resolution. A Scanning Electron Microscopy (SEM) FEI/Philips XL30 was used to take photos of GNPs at dispersed and aggregated states. We collected the secondary electrons with the Everhart-Thornley detector to get the images with best a resolution of 3.5 nm at 30 kV. The Direct Detect Spectrometer is also used for the concentration measurement of batch samples.
2.6. Numerical modeling
The LSPR of GNPs in dispersed and aggregated states was modeled by COMSOL Multiphysics (V.5.6). Maxwell's equations for electromagnetic waves were solved in the scatter field and the surrounding environment was set as water. The outer layer was defined as perfectly matched layer (PML) with the scattering boundary condition. The background electromagnetic wave was defined with a constant amplitude sinusoidal function, polarized in the z-axis direction and the optical absorption was computed in the whole spectrum. A variety of different configurations of GNPs were simulated in different orientations relative to the linearly polarized incident light at the center and modeled for each case through the frequency domain by solving Maxwell's equations (see Fig. S17). In order to capture the near-field enhancement of the EM waves, the minimum mesh size is used. The mesh size around the aggregates is much smaller than the wavelength of the incoming EM wave and the mesh size gradually increases (with the maximum growth rate of 1.5 as limited by the maximum mesh size of GNPs and the antibody layer) from the aggregate position towards the PML. We have used the McPeak et al. model (McPeak et al., 2015) for the gold refractive index. However, for the sake of comparison, we have also used Brendel-Bormann (Rakic et al., 1998) and Lorentz-Drude (Rakic et al., 1998) models (see Figs. S1 and S12).
2.7. Estimation of number of antibodies per GNP
The conjugation of GNPs with antibodies results in adding a layer around each GNP. To find the thickness of the antibody layer, we have used both theoretical and numerical models. For the numerical method, we have added a layer of protein around a single GNP with the refractive index of and the wavelength shift for each case has been calculated (see Fig. S4). From the theoretical model, the thickness has been estimated by incorporating the observed wavelength shift between uncoated and coated GNPs, , into the Rayleigh scattering approximation (Kim et al., 2017) in Eq. (1).
| (1) |
where is the free electron oscillation wavelength (131 nm for gold (Iarossi et al., 2018)); is the maximum wavelength of the optical absorption spectrum of uncoated GNPs (present work ∼ 524 nm); and are relative permittivity of the antibody layer and water, respectively ( ∼ 2.02, ∼ 1.77 (Bell et al., 2013; Iarossi et al., 2018)); is the GNPs diameter (40 nm); and is the thickness of the antibody layer. Finally, we have estimated the number of antibodies per GNP from calculated and the De Feijter formula (De Feijter et al., 1978) in Eq. (2).
| (2) |
where is the concentration of antibodies on GNPs; is the refractive index of water (∼1.33 (Iarossi et al., 2018)) and is the rate of refractive index increment with respect to concentration (for antibody ∼ 0.19 ml/g (Zhao et al., 2011)). To convert into the number of antibodies, the mass of the N protein antibody is assumed to be 150 kDa (Zeng et al., 2020). By using these values, the number of antibodies per GNP is found to be 233 and 168 from the numerical modeling and Eq. (2), respectively (see Fig. S4). Therefore, each GNP can attach about 200 GNPs. Polyclonal antibodies have multiple epitopes that can attach to different parts of the N proteins, which increases the amplification factor of the biosensor. Furthermore, the zeta potential of coated GNPs is about −32 mV (provided by Nanocomposix company), which is strong to electrostatically repel GNPs for non-specific aggregations.
2.8. Color extracting algorithm
The color of GNPs at different aggregated configurations was characterized by a code developed in Matlab (V.R2019b) to convert input optical intensity spectrum into RGB values based on the CIE 1964 converting table (Schanda, 2015). The spectrum was first converted into averaged XYZ values, and then the equivalent RGB values. This algorithm was also used to reconstruct the color of the experimental samples. For the sample solutions, we measured the optical absorption spectrum of the solution and characterized the reflection spectrum by assuming there is negligible transmission.
2.9. Model optimization
Reconstruction of experimental data with the numerical model was conducted by implementing different individual elements into the Genetic algorithm in Matlab. We found out the best combination of different elements to generate experimentally derived optical absorption intensity spectrum. In each case, we set the number of population equal to 100 times the number of variables. The square of deviation of the numerically predicted spectrum is minimized from the experimental value as the objective function.
3. Results and discussions
3.1. Modeling of GNPs in different configurations
COMSOL Multiphysics (V.5.6) was utilized with three different refractive index models (McPeak et al. (2015), Brendel-Bormann (Rakic et al., 1998) and Lorentz-Drude (Rakic et al., 1998)) with the Mie scattering theory (Fan et al., 2014) (see Fig. S1a). Results show that the McPeak et al. (McPeak et al., 2015) model provides the best prediction for the peak wavelength of a single GNP (∼524 nm). The complex permittivity values of these models with respect to the optical spectrum are shown in Figs. S1b and c. Upon the interaction with antigens, GNPs may form different configurations, such as linear, pyramidal, planar, and cubic shapes. From simulation results, the elongation in the direction of incident light shifts the absorption spectrum towards the red zone due to electric field amplifications at the corners (see Fig. S2). The elongation can be a representative of aggregation in one direction, which accompanies the increase in J1 and decrease in J2, as depicted in Fig. S2. The effect of interspace distance between two GNPs has been also studied with respect to their spectra, as shown in Fig. S3. It reveals that the reduction in the interspace distance leads to the red shift and the generation of a second peak. GNPs are coated with molecular linkers and antibodies such that a thin covering shell (antibody layer) is adopted in our model. The thickness of this antibody layer is estimated by both the Rayleigh scattering approximation in Eq. (1), and numerical modeling as around 5 nm (see Fig. S4). This number is calculated based on the 3 nm wavelength shift from the experiments between uncoated and coated GNPs (see Fig. S19) and it is consistent with simulation results. It is worth noting that using Fig. S4 and Eq. (2), one can predict the number of antibodies per GNP as 200 or each GNP has about 200 sites to interact with N proteins. The study on antibody coated GNPs has also shown the red shift characteristics with smaller amounts and no second peak since the protein layer limits the GNPs spacing to 5 nm (see Fig. S5). J1 and J2 simulation results show that the linear-shape aggregation would result in higher absorptions at the bottom (400 nm) and top (700 nm) visible-light band wavelengths, respectively (see Fig. S6). By increasing the number of GNPs in an array with a fix spacing would amplify the incident beam in the array axis and cause the red shift. However, alignment in the orthogonal direction to the light ends up with a subtle increase in the absorption in the bottom band (400 nm) without any significant red shift in comparison to those of a single GNP (see Figs. S6 and S7).
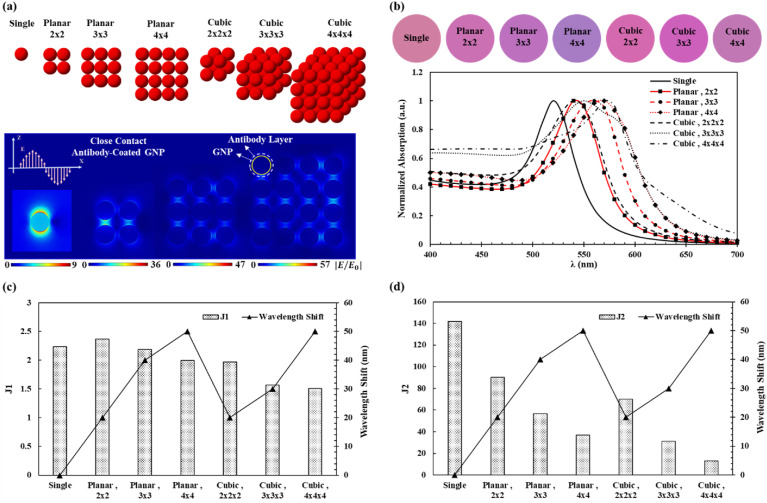
2D and 3D configurations of GNPs as shown in Fig. 2 a have been analyzed. It is found that both planar and cubic arrangements of antibody coated GNPs would lead to the red shift in the absorption spectrum as the size of the aggregates increases. At the same time, J1 decreases slightly and J2 decreases more significantly (see Fig. 2b, c and d). In the 2D and 3D cases, an extra amplification is found in the light propagation direction (x-axis), as depicted in Fig. 2a and Fig. S8. Studies have also been performed on uncoated GNPs with different spacings with similar results, except for the 2 nm case (see Fig. S9). Fig. 2b shows that larger aggregates can absorb more light at the wavelength of 400 and 700 nm range, and the peak wavelength shifts towards the red zone - consistent with earlier results for the linear arrangements. It is worth noting that the cubic configuration has higher absorptions at regions close to the 400 nm wavelength range than those of their equivalent planar configurations. Furthermore, it is found that the J2 parameter is a better indicator in revealing the increase of the aggregate sizes than the shifts of the peak wavelength. Specifically, the transition from the linear arrangement to 3D pyramidal shape structure clearly illustrates the effectiveness of J2 parameter as the 3D structure reverts the spectrum back from the linear array to its 60° counterpart (see Fig. S10). To capture the non-equal spacing case and its corresponding effect on the light absorptions, two of already investigated cases, planar 4x4 and three GNPs in a linear shape, have been studied as depicted in Fig. S11. Outcomes of these simulations show that the heterogenous spacing can result in the spectral shifts. In the 2D configuration, spectra of the heterogenous cases are close to their linear counterpart. It is also observed that the J2 values of heterogeneous cases are between those of the homogeneous cases, which suggests that minimum and maximum spacings exist inside the aggregates.
Fig. 2.
Numerical modeling of aggregated GNPs. (a) Different configurations of GNPs with electric field enhancement contours of different cases (antibody-coated GNP in close contact) under the z-axis polarized incident light. (b) The optical absorption spectra of different GNP aggregates with respect to the wavelength showing the peak wavelength shifts. Extracted colors of the simulations showing an obvious color change in various configurations. (c–d) Wavelength shifts and J1 and J2 values for different 2D and 3D configurations, respectively. Increasing the size of the aggregates causes the reduction in the values of J1 and J2, while the changes in J2 are more significant. In general, the peak wavelength shifts increase as the GNP aggregates size increases, while J1 and J2 values decrease. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Experimental data for numerical modeling
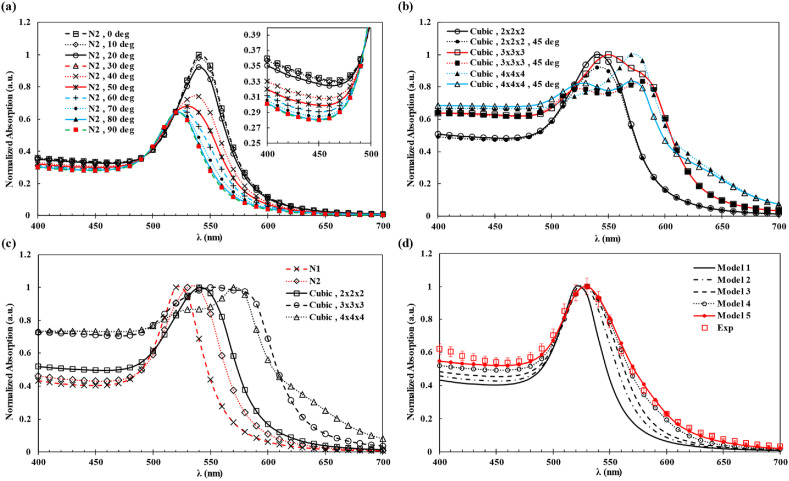
The light absorption spectra of GNP aggregates have been simulated based on linear, cubic 2 × 2 × 2, cubic 3 × 3 × 3 and cubic 4 × 4 × 4 cases. It should be noted that the cubic structures could be used as a simplified configuration for GNPs aggregates at low N protein concentrations as observed in SEM images of the samples (Fig. S13). Figs. S7 and S10 show simulation results on the orientation of GNPs aggregates (relative to the polarization direction of the incident light) and light absorption spectra. Simulations at different angles from 0 to 90° have also been conducted and the average spectra for each case are shown in Fig. 3 , where the absorption spectra are normalized to the value at the peak wavelength.
Fig. 3.
Numerical simulation results based on the McPeak et al. model (McPeak et al., 2015). (a) Optical absorption spectra of two antibody-coated GNPs with a 10 nm gap under various light irradiation orientations relative to the polarization direction. Increasing the polarization direction angle causes the red shift of the peak wavelength and the amplification in optical absorptions. All spectra were normalized with respect to the absorption value at the peak wavelength. (b) Optical spectra of the antibody-coated GNPs in the cubic arrangement with a 10 nm gap of different sizes and orientations. For each case, two extreme orientations of 0- and 45-degree are simulated. (c) Five different types of GNP aggregates. The increase in the aggregates size leads to red shift of the peak wavelength and higher absorption in the regions close to the 400 nm and 700 nm band regions. (d) Experimental data for the case of 150 ng/ml together with the simulation results. The definition of each model is given in Table 1. It is noted that model 5 results have the best match with experimental data implying larger aggregates are formed in the experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The linear array of two GNPs (dimer GNPs) shows obvious changes of spectra with respect to the incident light orientations, as shown in Fig. 3a and the cubic configurations illustrate smaller changes around the peak absorption wavelength in Fig. 3b. Since GNPs aggregates are expected to have random orientations, the results are calculated and averaged over different orientations. By including aggregates in different compositions as shown Table 1 , it is observed that the numerical model can match relatively well to the experimental data in Fig. 3d. The best result is from model 5, which includes high order aggregates with simulation results as shown in Fig. 3c. It is observed that dimer case is only present in model 2. In the rest of models, cubic arrangements can give rise to the red shift of the peak wavelength and change in the optical response of the solution. We also used the Brendel-Bormann model (Rakic et al., 1998) as shown in Fig. S12 and Table S2, which resulted in significant deviations from the results of the McPeak et al. model. As the Brendel-Bormann model results in large red shift at the peak wavelength with small aggregates, only single GNP was used by Genetic algorithm without high order aggregates. It is well-known that GNP has the strong chemical reaction characteristics. After the GNP modifications, its stability can be improved, while the molecular size of the interface modifier will affect its chargeability and stability, including non-specific polymerization such as electrostatic adsorption, and molecular precipitation. The goal of this work is to study the effects of aggregation with respect to spectral changes and the optical responses for potential naked eye detections of the corona viruses and further studies are required to include these aforementioned factors.
Table 1.
Description of five different models to reconstruct the experimental optical intensity spectrum. The normalized optical absorption spectrum of each component is weighted by the given factor in each model.
| Model | Single GNP |
Dimer GNPs |
Cubic 2 × 2 × 2 |
Cubic 3 × 3 × 3 |
Cubic 4 × 4 × 4 |
|---|---|---|---|---|---|
| (Weight = 1) | (Weight = 1/2) | (Weight = 1/8) | (Weight = 1/27) | (Weight = 1/64) | |
| 100% | 0% | 0% | 0% | 0% | |
| 2 | 2.1% | 97.9% | 0% | 0% | 0% |
| 3 | 24.2% | 0% | 75.8% | 0% | 0% |
| 4 | 31.2% | 4.9% | 24.4% | 39.5% | 0% |
| 5 | 30.5% | 0.5% | 30.1% | 21.3% | 17.5% |
3.3. Experimental results and concentration detections
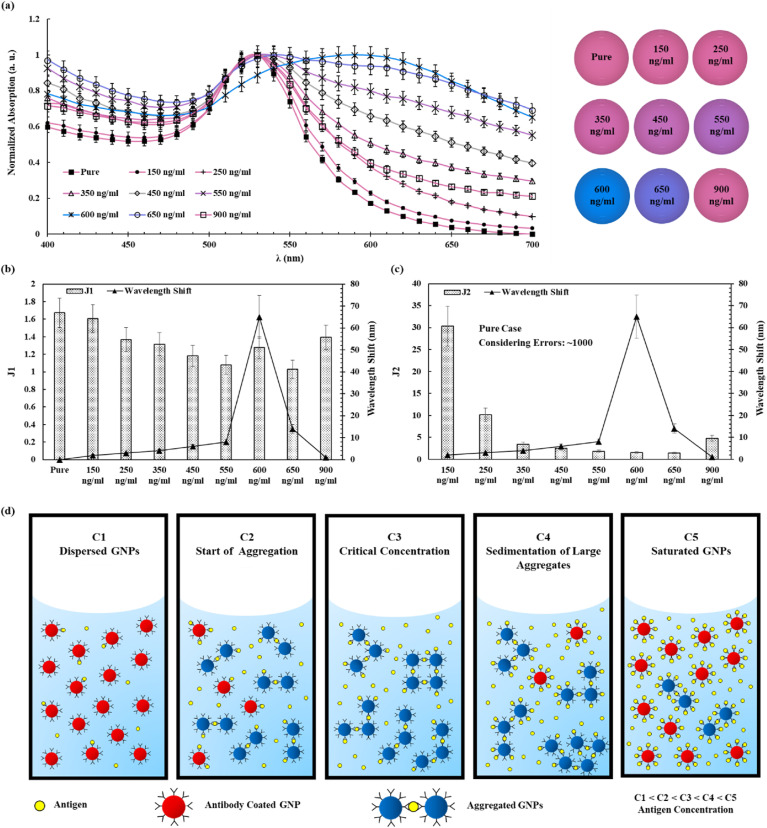
Higher concentrations of N proteins which can be potentially obtained in patients with clear symptoms (Leung et al., 2020; Wölfel et al., 2020; Xu et al., 2020) have been used in tests as shown in Fig. 4 a.
Fig. 4.
Experimental results of optical responses of antibody-coated GNPs solutions with respect to the N protein concentration. (a) The optical absorption spectrum of GNPs solutions at different N protein concentrations. The increase in the N protein concentration leads to the red shift of the peak wavelength around the critical concentration. Aggregated GNPs can also result in the high optical absorption at regions around the 400 nm and 700 nm wavelength bands. The red-shift color changes with respect to the increase of N protein concentration increase the most at a critical concentration value and revert back to the antigen-free case afterwards. The CIE chromaticity plot in Fig. S18 shows the clear views of the color changes. All spectra results are normalized with respect to their maximum absorption values. (b–c) The peak wavelength-shift together with J1 and J2 with respect to the N protein concentration, respectively. The peak wavelength-shift increases before the critical concentration value and reduces afterwards to the value close to the antigen-free case. Both J1 and J2 values decrease as the protein concentration increases before the critical concentration value and increase afterwards. The variations in J2 are more significant than those of J1. (d) The aggregating process of antibody coated GNPs at different antigen concentrations. High antigen concentration increases the probability of antibody-antigen interactions to result in large GNPs aggregates. This trend continues up to a critical antigen concentration which provides the optimum condition for the aggregate-interaction. At concentrations higher than the critical concentration value, the antibodies on GNPs can saturate the binding sites to reduce the aggregate-interactions and result in the combination of small aggregates and dispersed GNPs in the solution. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The color change of samples containing N protein is visible to naked as shown in Fig. 4a. However, for concentration values higher than the critical value of 600 ng/ml in this work, the color change starts to revert to the pure case (antigen-free). In general, the presence of aggregated GNPs causes the increases in the light absorptions near the 400 nm and 700 nm wavelength bands and the red shift of the peak wavelength spectrum. It is found that the aggregation size increases as the N protein concentration increases up to the critical concentration (see Fig. S13). Due to the settlement of very large aggregates, the color of the solution around the critical concentration value would be pale (see Fig. S14). To characterize the effect of the N protein concentration on the optical spectra of GNPs solutions, the peak wavelength-shift together with J1 and J2 at different concentrations are plotted in Fig. 4b and c.
In general, the peak wavelength-shift increases monotonically with respect to the N protein concentration up to the critical concentration and reduces afterwards for cases with higher N protein concentrations. This behavior is expected based on a prior study of a different biosensor using GNPs (Iarossi et al., 2018). Fig. 4b shows that J1 reduces as the N protein concentration increases before the critical concentration and increases afterwards for cases of higher N protein concentrations. Since both the peak wavelength-shift and J1 revert to their antigen-free values, it is problematic to use these two parameters for positive virus detections. On the other hand, J2 decreases significantly as the N protein concentration increases before the critical concentration value and it increases slightly afterwards for cases at high N protein concentrations and won't revert to the value of the antigen-free case. As such, J2 is a better parameter to detect the existence of the virus.
From experimental and numerical studies, the underlying physical phenomenon of aggregations of GNPs in the presence of different antigen concentrations is illustrated in Fig. 4d. In general, GNPs can aggregate with the help of antigens to increase the size. If the antigen concentration is higher than a critical value, antibodies on GNPs can saturate the binding sites and prevent further aggregations, which results in smaller GNPs aggregates. A similar phenomenon was reported and modeled by Iarossi et al. (Iarossi et al., 2018). In general, the solution has functionalized GNPs combined with N protein already at a concentration of [GNPSaturated] and free GNPs without the N protein at a concentration of [GNPFree]. The concentrations of GNPs should follow the law of conservation as: [GNPFree] + [GNPSaturated] = [GNP]0 (total or initial GNP concentration). At equilibrium, one can derive the following equations [25]:
| (3) |
| (4) |
where [K] is the equilibrium constant of the reaction. The N protein concentration can shift the balance between [GNPFree] and [GNPSaturated] in a complex way. For example, as the N protein concentration increases initially, the probability of making large aggregates increases and the saturated GNP concentration increases to result in the peak wavelength-shift. However, as the N protein concentration is higher than a critical value, aggregation of GNPs can saturate to result in smaller GNPs aggregates and the reversion of the peak wavelength-shift. A power-law model is derived to describe this behavior based on the difference between saturated and free GNPs concentrations with experimental fittings in Fig. 5 a:
| (5) |
where C is a constant to include all factors. The best fitting parameters from the experimental result in Fig. 5a are: ng/ml, ng/ml, and .
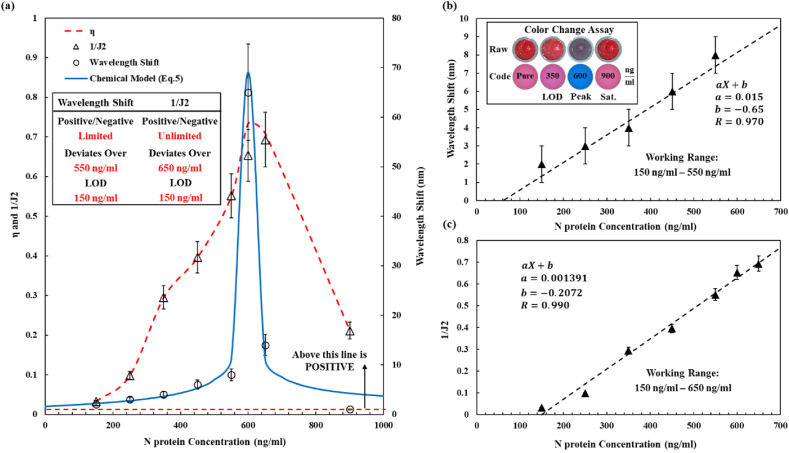
Fig. 5.
Concentration measurements of SARS-CoV-2 N proteins. (a) Comparison of the , 1/J2 and peak wavelength-shift in the full range of tested N protein concentrations. The chemical model (Eq. (5)) matches well with experimental results on the peak wavelength-shift. The peak wavelength-shift parameter has a maximum around 600 ng/ml and reverts to its antigen-free case at high concentrations of 900 ng/ml. The 1/J2 parameter has a maximum value at 650 ng/ml and reduces at high N protein concentrations with values much higher than that of the antigen-free case. (b–c) Linear approximation of the wavelength-shift and 1/J2 with respect to the N protein concentration, respectively. Experimental results on the color changes of various concentrations are also shown for both optical photos (top) and spectrum extracted color (bottom) images. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
According to Fig. 5a, at high antigen concentrations, the peak wavelength-shift reverts toward the antigen-free case. On the other hand, the small aggregates can still induce optical absorptions in regions close to the 400 nm and 700 nm wavelength bands and the optical adsorption in the 700 nm wavelength region increases more as compared to those in the 400 nm wavelength region. As such, J2 is a better parameter to detect the existence of the virus. First, Fig. 5b shows the peak wavelength-shift monotonically increases from 150 ng/ml to 550 ng/ml before the critical concentration around 600 ng/ml with a good linear relationship with a tested LOD at 150 ng/ml. To further improve the LOD, one can optimize the shape, size, and surface chemistry of GNPs, and some of these approaches have been proposed by Guo et al. (Guo et al., 2015). Furthermore, there are various GNP-based biosensing methods with very low LOD in the range of fg/ml to μg/ml levels by using complex testing setups (Aldewachi et al., 2018; Draz and Shafiee, 2018; Jazayeri et al., 2018; Verma et al., 2015). This work demonstrates a less sensitive LOD based on a very simple experimental setup for direct naked-eye detections.
By using the peak wavelength-shift detection scheme within the linear range (150 ng/ml to 550 ng/ml), a good linear response has been recorded. However, at high concentrations (over 650 ng/ml), the responses start reverting backwards. At very high concentrations (900 ng/ml), the response is the same as the antigen-free case. On the other hand, by using the 1/J2 parameter, it has a good linear approximation up to 650 ng/ml and it can detect the existence of N-proteins over 900 ng/ml (Fig. 5a and c) as this parameter is always much larger than that of the antigen-free case. It is further validated that one can detect SARS-CoV-2 N proteins with naked eye by color changes within a range approximately 350–650 ng/ml. The measured optical intensity spectra at different N protein concentrations as depicted in Fig. 4a reveal that the peak wavelength of the optical absorption for most cases is close to 530 nm. Instead of using 1/J2, a new parameter is defined as , which is the ratio of the light absorption intensity at the 700 nm wavelength (top band) to the absorption intensity at the 530 nm wavelength. It is found that can follow the trajectory of 1/J2 very closely in nearly all concentrations, except at the critical concentration for about 12.55% deviations (see Fig. 5a). The comparison of 1/J2 and at different N protein concentrations is shown in Table S3 with standard deviations. It is observed that can be utilized to determine positive/negative detection results as well as the virus concentration. Specifically in real practice, one only needs to measure the optical absorptions at two wavelengths (530 nm and 700 nm) to calculate η to simplify the detection procedure. Furthermore, based on the information from the Sino biological company and studies conducted by Abbot company on the genetic similarities of SARS-CoV-2 and other common respiratory viruses (Almaghaslah et al., 2020; Diao et al., 2021; Hu et al., 2020), the antibodies of the SARS-CoV-2 N protein used in this work have no cross-reactivity with other respiratory viruses, except for SARS-CoV and MERS-CoV. Since SARS-CoV-2 is the only widespread virus now, positive results only from SARS-CoV-2 infections are expected. Non-specific binding can also happen due to the change in the composition of the sample solution, such as changes in pH values (Jazayeri et al., 2016). Here, covalent conjugation which is less sensitive to pH values has been used (controlled pH value between 7 and 8 in tests) to reduce the possible influences due to non-specific binding. Finally, to determine the response time of the biosensor, the optical absorptions of GNPs solution at two different concentrations (see Fig. S15) have been recorded. It is observed that normalized adsorption values change initially. After 2 min, some of the aggregations may settle down and after 10 min, all systems are at steady state. The observable results such as positive/negative or color change, can be obtained in 5 min in general by the naked eye, while the concentration value can be obtained by the optical spectrometer after 10 min.
4. Conclusions
In this article, a new diagnostic method based on the aggregation of GNPs is presented to detect low concentration of SARS-CoV-2 N proteins with LOD of 150 ng/ml in less than 5 min by the naked eye. With the help of an optical spectrometer, the viral sample concentration can also be estimated. To understand the underlying physics of the aggregated GNPs in the LSPR process, numerical simulations on different GNPs aggregation configurations have been conducted. Two parameters, J1 and J2, have been defined as the ratios of the peak optical absorption intensity of the sample solution to the optical absorption of the sample solution at 400 nm and 700 nm, respectively. Experimentally measured optical absorption spectra of aggregated GNPs with different N protein concentrations reveal the red-shift of the peak wavelength optical adsorption intensity as predicted by the simulation results. For N protein concentrations higher than the critical value, this peak wavelength shift is reverted toward the N protein-free case. To solve this problem of the biosensor, it is found that J2 is the more suitable parameter and 1/J2 is utilized to detect the existence of the N protein. Furthermore, it is found that the N-protein concentration has a good linear relationship with respect to 1/J2 in the range between 150 and 650 ng/ml. A parameter, , which is the ratio of the optical absorption intensity at the 700 nm wavelength to that of the 530 nm wavelength, has been utilized. Without the need to a full optical spectra measurement, one can detect and determine SARS-CoV-2 concentration by measuring the optical absorption intensity of the sample only at these two wavelengths. In the prior reports, GNP-based sensing methods are limited to small working ranges and encounter issues for the cases of high concentration detections and the new parameter used in this work help alleviating the problem. To enhance the sensitivity and specificity of current method, one can optimize shape, size and composition of individual GNPs, and antibody, respectively, for broad applications. As such, this new POC method could be utilized to detect SARS-CoV-2 N proteins for the current pandemic and possible viral infections for future pandemics.
Source of funding
This work was supported by 2020 Seed Fund Award 2020-0000000157 from CITRIS and the Banatao Institute at the University of California.
CRediT authorship contribution statement
Kamyar Behrouzi: Writing - original draft, Investigation, Formal analysis, Visualization, Validation. Liwei Lin: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge Dr. Paul Lum, manager of QB3 Berkeley Nanotechnology Center (BNC) and Dr. Mary G. West, manager of QB3 Cell and Tissue Analysis Facility (CTAF) for providing instruments and materials. COMSOL Multiphysics (V.5.6) license was provided by Molecular Graphics and Computation Facility (MGCF) through NIH S10OD023532.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113669.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahmadivand A., Gerislioglu B., Ahuja R., Kumar Mishra Y. Mater. Today. 2020;32:108–130. doi: 10.1016/j.mattod.2019.08.002. [DOI] [Google Scholar]
- Ahmadivand A., Gerislioglu B., Ramezani Z., Kaushik A., Manickam P., Ghoreishi S.A. Biosens. Bioelectron. 2021;177:112971. doi: 10.1016/j.bios.2021.112971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alafeef M., Dighe K., Moitra P., Pan D. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldewachi H., Chalati T., Woodroofe M.N., Bricklebank N., Sharrack B., Gardiner P. Nanoscale. 2018;10:18–33. doi: 10.1039/c7nr06367a. [DOI] [PubMed] [Google Scholar]
- Almaghaslah D., Kandasamy G., Almanasef M., Vasudevan R., Chandramohan S. Int. J. Clin. Pract. 2020;74:1–9. doi: 10.1111/ijcp.13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrouzi Kamyar, Lin Liwei. 2021 21st International Conference on Solid-State Sensors, Actuators and Microsystems (Transducers) 2021. pp. 381–384. [DOI] [Google Scholar]
- Bell N.C., Minelli C., Shard A.G. Anal. Methods. 2013;5:4591–4601. doi: 10.1039/c3ay40771c. [DOI] [Google Scholar]
- Bielecki M., Crameri G.A.G., Schlagenhauf P., Buehrer T.W., Deuel J.W. Travel Med. Infect. Dis. 2020;37:101832. doi: 10.1016/j.tmaid.2020.101832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong J., Yu H., Lee C.Y., Lee J. uk, Choi H.J., Lee J.H., Lee H., Cheon J. Nat. Biomed. Eng. 2020;4:1159–1167. doi: 10.1038/s41551-020-00654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Feijter J.A., Benjamins J., Veer F.A. Biopolymers. 1978;17:1759–1772. doi: 10.1002/bip.1978.360170711. [DOI] [Google Scholar]
- Diao B., Wen K., Zhang J., Chen J., Han C., Chen Y., Wang S., Deng G., Zhou H., Wu Y. Clin. Microbiol. Infect. 2021;27 doi: 10.1016/j.cmi.2020.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draz M.S., Shafiee H. 2018. Theranostics 8, 1985–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Zheng W., Singh D.J. Light Sci. Appl. 2014;3:1–14. doi: 10.1038/lsa.2014.60. [DOI] [Google Scholar]
- Ghosh S.K., Pal T. Chem. Rev. 2007;107:4797–4862. doi: 10.1021/cr0680282. [DOI] [PubMed] [Google Scholar]
- Gomes A.S.L., Maldonado M., de S., Menezes L., Acioli L.H., de Araújo C.B., Dysart J., Doyle D., Johns P., Naciri J., Charipar N., Fontana J. Nanophotonics. 2020;9:725–740. doi: 10.1515/nanoph-2019-0521. [DOI] [Google Scholar]
- Guo L., Jackman J.A., Yang H.H., Chen P., Cho N.J., Kim D.H. Nano Today. 2015;10:213–239. doi: 10.1016/j.nantod.2015.02.007. [DOI] [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.L. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00459-7. [DOI] [Google Scholar]
- Iarossi M., Schiattarella C., Rea I., De Stefano L., Fittipaldi R., Vecchione A., Velotta R., Ventura B. Della. ACS Omega. 2018;3:3805–3812. doi: 10.1021/acsomega.8b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iravani S. Mater. Adv. 2020;1:3092–3103. doi: 10.1039/d0ma00702a. [DOI] [Google Scholar]
- Jazayeri M.H., Aghaie T., Avan A., Vatankhah A., Ghaffari M.R.S. Sens. Bio-Sensing Res. 2018;20:1–8. doi: 10.1016/j.sbsr.2018.05.002. [DOI] [Google Scholar]
- Jazayeri M.H., Amani H., Pourfatollah A.A., Pazoki-Toroudi H., Sedighimoghaddam B. Sens. Bio-Sensing Res. 2016;9:17–22. doi: 10.1016/j.sbsr.2016.04.002. [DOI] [Google Scholar]
- Kim J.-H., Park J.-E., Lin M., Kim S., Kim G.-H., Park S., Ko G., Nam J.-M. Adv. Mater. 2017;29:1702945. doi: 10.1002/adma.201702945. [DOI] [PubMed] [Google Scholar]
- Kim S.K., Cho H., Jeong J., Kwon J.N., Jung Y., Chung B.H. Chem. Commun. 2010;46:3315–3317. doi: 10.1039/b926940a. [DOI] [PubMed] [Google Scholar]
- Leung N.H.L., Chu D.K.W., Shiu E.Y.C., Chan K.H., McDevitt J.J., Hau B.J.P., Yen H.L., Li Y., Ip D.K.M., Peiris J.S.M., Seto W.H., Leung G.M., Milton D.K., Cowling B.J. Nat. Med. 2020;26:676–680. doi: 10.1038/s41591-020-0843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link S., Mohamed M.B., El-Sayed M.A. J. Phys. Chem. B. 1999;103:3073–3077. doi: 10.1021/jp990183f. [DOI] [Google Scholar]
- Liu Huiqiao, Rong P., Jia H., Yang J., Dong B., Dong Q., Yang C., Hu P., Wang W., Liu Haitao, Liu D. Theranostics. 2016;6:54–64. doi: 10.7150/thno.13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang L., Wei W., Zhao H., Zhou Z., Zhang Y., Liu S. Analyst. 2015;140:3989–3995. doi: 10.1039/c5an00407a. [DOI] [PubMed] [Google Scholar]
- Mayer K.M., Hafner J.H. Chem. Rev. 2011;111:3828–3857. doi: 10.1021/cr100313v. [DOI] [PubMed] [Google Scholar]
- McPeak K.M., Jayanti S.V., Kress S.J.P., Meyer S., Iotti S., Rossinelli A., Norris D.J. ACS Photonics. 2015;2:326–333. doi: 10.1021/ph5004237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miripour Z.S., Sarrami-Forooshani R., Sanati H., Makarem J., Taheri M.S., Shojaeian F., Eskafi A.H., Abbasvandi F., Namdar N., Ghafari H., Aghaee P., Zandi A., Faramarzpour M., Hoseinyazdi M., Tayebi M., Abdolahad M. Biosens. Bioelectron. 2020;165:112435. doi: 10.1016/j.bios.2020.112435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra P., Alafeef M., Alafeef M., Alafeef M., Dighe K., Frieman M.B., Pan D., Pan D., Pan D. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myroshnychenko V., Rodríguez-Fernández J., Pastoriza-Santos I., Funston A.M., Novo C., Mulvaney P., Liz-Marzán L.M., García de Abajo F.J. Chem. Soc. Rev. 2008;37:1792–1805. doi: 10.1039/B711486A. [DOI] [PubMed] [Google Scholar]
- Nie X.M., Huang R., Dong C.X., Tang L.J., Gui R., Jiang J.H. Biosens. Bioelectron. 2014;58:314–319. doi: 10.1016/j.bios.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Petryayeva E., Krull U.J. Anal. Chim. Acta. 2011;706:8–24. doi: 10.1016/j.aca.2011.08.020. [DOI] [PubMed] [Google Scholar]
- Pfefferle S., Reucher S., Nörz D., Lütgehetmann M. Euro Surveill. 2020;25:1–5. doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Rakic A.D., Djuris A.B., Elazar J.M., Majewski M.L. Appl. Opt. 1998;37:5271–5283. doi: 10.1364/AO.37.005271. [DOI] [PubMed] [Google Scholar]
- Schanda J. Encyclopedia of Color Science and Technology. Springer Science & Business Media; New York: 2015. pp. 3–7. [DOI] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S. Il. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shabaninezhad M., Ramakrishna G. J. Chem. Phys. 2019;150 doi: 10.1063/1.5090885. [DOI] [PubMed] [Google Scholar]
- Shukla R., Bansal V., Chaudhary M., Basu A., Bhonde R.R., Sastry M. Langmuir. 2005;21:10644–10654. doi: 10.1021/la0513712. [DOI] [PubMed] [Google Scholar]
- Smyrlaki I., Ekman M., Lentini A., Rufino de Sousa N., Papanicolaou N., Vondracek M., Aarum J., Safari H., Muradrasoli S., Rothfuchs A.G., Albert J., Högberg B., Reinius B. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J., Chan W.C.W. 2020. ACS Nano. [DOI] [PubMed] [Google Scholar]
- Valentini P., Fiammengo R., Sabella S., Gariboldi M., Maiorano G., Cingolani R., Pompa P.P. ACS Nano. 2013;7:5530–5538. doi: 10.1021/nn401757w. [DOI] [PubMed] [Google Scholar]
- Verma M.S., Rogowski J.L., Jones L., Gu F.X. Biotechnol. Adv. 2015;33:666–680. doi: 10.1016/j.biotechadv.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Wang D., He S., Wang X., Yan Y., Liu J., Wu S., Liu S., Lei Y., Chen M., Li L., Zhang J., Zhang L., Hu X., Zheng X., Bai J., Zhang Yulong, Zhang Yitong, Song M., Tang Y. Nat. Biomed. Eng. 2020;4:1150–1158. doi: 10.1038/s41551-020-00655-z. [DOI] [PubMed] [Google Scholar]
- Wang Y., Gao Z., Han Z., Liu Y., Yang H., Akkin T., Hogan C.J., Bischof J.C. Sci. Rep. 2021;11:1–12. doi: 10.1038/s41598-020-79393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Xu R., Cui B., Duan X., Zhang P., Zhou X., Yuan Q. Int. J. Oral Sci. 2020;12 doi: 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Hallinan D.T. Sci. Rep. 2016;6:1–17. doi: 10.1038/srep35339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Liu G., Ma H., Zhao D., Yang Yunru, Liu M., Mohammed A., Zhao C., Yang Yun, Xie J., Ding C., Ma X., Weng J., Gao Y., He H., Jin T. Biochem. Biophys. Res. Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang L., Xu W. J. Phys. D Appl. Phys. 2012;45:113001. doi: 10.1088/0022-3727/45/11/113001. [DOI] [Google Scholar]
- Zhao H., Brown P.H., Schuck P. Biophys. J. 2011;100:2309–2317. doi: 10.1016/j.bpj.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong C., Xu M., Xu L.-J., Wei T., Ma X., Zheng X.-S., Hu R., Ren B. Chem. Rev. 2018;118:4946–4980. doi: 10.1021/acs.chemrev.7b00668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.