Abstract
Background & Aims
The transcription factor GATA4 is broadly expressed in nascent foregut endoderm. As development progresses, GATA4 is lost in the domain giving rise to the stratified squamous epithelium of the esophagus and forestomach (FS), while it is maintained in the domain giving rise to the simple columnar epithelium of the hindstomach (HS). Differential GATA4 expression within these domains coincides with the onset of distinct tissue morphogenetic events, suggesting a role for GATA4 in diversifying foregut endoderm into discrete esophageal/FS and HS epithelial tissues. The goal of this study was to determine how GATA4 regulates differential morphogenesis of the mouse gastric epithelium.
Methods
We used a Gata4 conditional knockout mouse line to eliminate GATA4 in the developing HS and a Gata4 conditional knock-in mouse line to express GATA4 in the developing FS.
Results
We found that GATA4-deficient HS epithelium adopted a FS-like fate, and conversely, that GATA4-expressing FS epithelium adopted a HS-like fate. Underlying structural changes in these epithelia were broad changes in gene expression networks attributable to GATA4 directly activating or repressing expression of HS or FS defining transcripts. Our study implicates GATA4 as having a primary role in suppressing an esophageal/FS transcription factor network during HS development to promote columnar epithelium. Moreover, GATA4-dependent phenotypes in developmental mutants reflected changes in gene expression associated with Barrett’s esophagus.
Conclusions
This study demonstrates that GATA4 is necessary and sufficient to activate the development of simple columnar epithelium, rather than stratified squamous epithelium, in the embryonic stomach. Moreover, similarities between mutants and Barrett’s esophagus suggest that developmental biology can provide insight into human disease mechanisms.
Keywords: Transcriptional Regulation, Gastric Development, Barrett’s Esophagus, Epithelium
Abbreviations used in this paper: BE, Barrett’s esophagus; ChIP-seq, chromatin immunoprecipitation sequencing; cKI, conditional knock-in; cKO, conditional knockout; DEG, differentially expressed gene; FOG, Friend of GATA; FS, forestomach; FSE, forestomach enriched; GI, gastrointestinal; H&E, hematoxylin and eosin; HS, hindstomach; HSE, hindstomach enriched; IF, immunofluorescence; IHC, immunohistochemistry; mRNA, messenger RNA; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; RNA-seq, RNA-sequencing; TSS, transcription start site
Graphical abstract
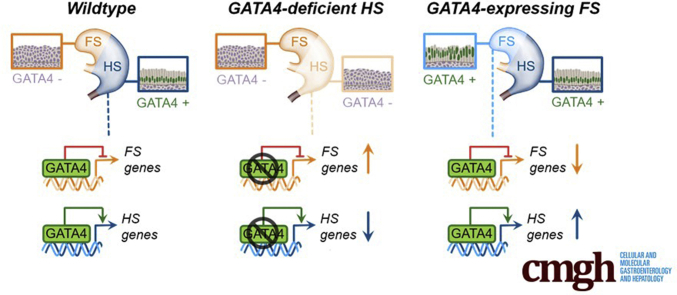
Summary.
By eliminating GATA4 in mouse hindstomach or maintaining GATA4 in the mouse forestomach during development, we identified GATA4 as an essential, principal regulator of simple columnar epithelial morphogenesis within the stomach.
The mouse stomach consists of two functional domains: a forestomach (FS) comprising a keratinized stratified squamous epithelium analogous to the esophagus and a hindstomach (HS) containing a simple columnar epithelium. The HS consists of a glandular body (corpus) and an antrum. The epithelium of the body contains abundant parietal cells and zymogenic cells, whereas the epithelium of the antrum primarily consists of mucous cells and gastrin-secreting G cells.1,2 Although development of distinct HS and FS epithelial architectures, each containing specialized cell types, is critical for digestion, a gap remains regarding the molecular pathways regulating regionalization. The establishment and maintenance of these domains is crucial for gastrointestinal (GI) tract homeostasis. For example, in Barrett’s esophagus (BE), the esophageal stratified squamous epithelium converts to a simple columnar epithelium with intestinal and gastric properties.3 It is clear that defining the molecular mechanisms required for regionalization and maintenance of discrete domains within the GI tract is an essential step toward understanding GI diseases, including BE.
During embryonic development, the GI epithelium emerges from endoderm. Morphogenetic events driven by evolutionarily conserved signaling pathways direct anteroposterior regionalization of the gut tube into SOX2+ foregut and CDX2+ midgut and hindgut.4 As development progresses, the foregut is patterned into primitive organ domains, and diversification of the epithelial linings of the esophagus, glandular stomach, and proximal intestine becomes morphologically apparent. Although not fully understood, specification and differentiation of the epithelia of the esophagus, glandular stomach, and intestines are driven by gene regulatory networks involving lineage-restricted transcription factors commonly found to be silenced or amplified in GI diseases, including CDX2, SOX2, TRP63, and GATA factors 4 and 6.4, 5, 6, 7, 8, 9, 10
The zinc finger containing transcription factor GATA4 is critical for development of many GI organs, including the glandular stomach and small intestine, and is aberrantly expressed in BE and esophageal, gastric, and colorectal cancers.11, 12, 13, 14, 15, 16, 17, 18, 19, 20 Binding of GATA4’s zinc finger domains to consensus (A/T)GATA(A/G) sequences in promoters and enhancers regulates gene expression.21 Protein-protein interactions between GATA4 and other transcriptional regulators further influence tissue-specific and cell-specific gene expression programs. For example, interactions between the GATA4 N-terminal zinc finger and the non–DNA binding co-factor Friend of GATA (FOG) proteins contribute to gene expression programs in a multitude of tissues including the heart, GI tract, reproductive tract, and lung.22 Disruption of GATA4-FOG interactions can disturb organ development and function.22
Between E8.5 and E10.5 of mouse development, GATA4 is expressed throughout the caudal foregut endoderm.18,23,24 By E11.5–E12.5, however, GATA4 is lost in the foregut endoderm giving rise to the stratified squamous epithelium of the esophagus and FS, while it is maintained in the foregut endoderm giving rise to the simple columnar epithelium of the HS.18 Differential GATA4 expression within these domains coincides with the onset of divergent tissue morphogenesis, suggesting a role for GATA4 in this developmental process. Further supporting a role for GATA4 as a patterning factor is our previous work demonstrating that GATA4 is essential to define the intestinal jejunal/ileal junction.17,25 Moreover, histological characterization of chimeric mouse stomachs generated with wild-type and Gata4–/– embryonic stem cells shows that GATA4-null regions display features of stratified epithelium and lose expression of parietal cell, zymogenic chief cell, and neck cell marker genes.18 Additional studies using a mouse expressing a FOG-binding deficient GATA4 protein show that the GATA4-FOG interaction correlates with repression of distal ileal gene expression in the proximal intestine.26 Another study with the same GATA4 mutant protein demonstrates an important role for FOG1 in gastric development.27 Conditional GATA4 elimination primarily in the distal region of the mouse stomach during late stages of development using Pdx1-Cre results in an antrum expressing FS and pancreatic genes.19 These mouse models provide insight into the requirement of GATA4 during late stages of glandular stomach development. The role of GATA4 during the earliest stages of foregut development, when domains with different epithelial structures are first delineated has yet to be comprehensively determined.
This study explored the idea that GATA4, acting as a regionalizing factor, is necessary and sufficient to direct morphogenesis of simple columnar epithelium, rather than stratified squamous epithelium during early gastric development. We used a conditional knockout (cKO) approach to eliminate GATA4 early in the process of HS development and a conditional knock-in (cKI) approach to maintain GATA4 expression in the developing FS. We found that GATA4 drives a molecular program in the developing HS by activating expression of a gene network essential for simple columnar glandular epithelial cell fate while repressing a network of genes essential for stratified epithelial cell identity, including many esophageal-enriched transcription factors. Parallel studies of GATA6 mutants during this developmental window revealed that the phenotype observed in GATA4 mutants was specifically attributable to GATA4. Finally, we observed changes in the global transcriptome of GATA4 mutants that overlapped with the metaplastic gene signature of BE suggesting that identifying direct targets of GATA4 in developing epithelium can provide insight into BE.
Results
GATA4 Is Essential for Simple Columnar Epithelial Tissue Development in the Mouse HS
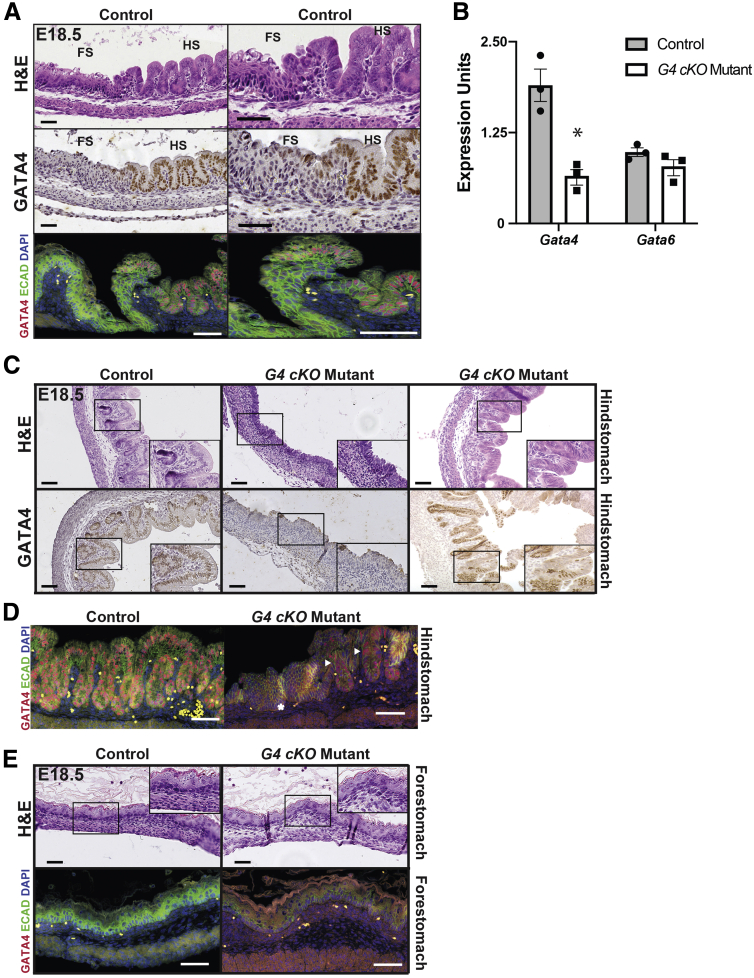
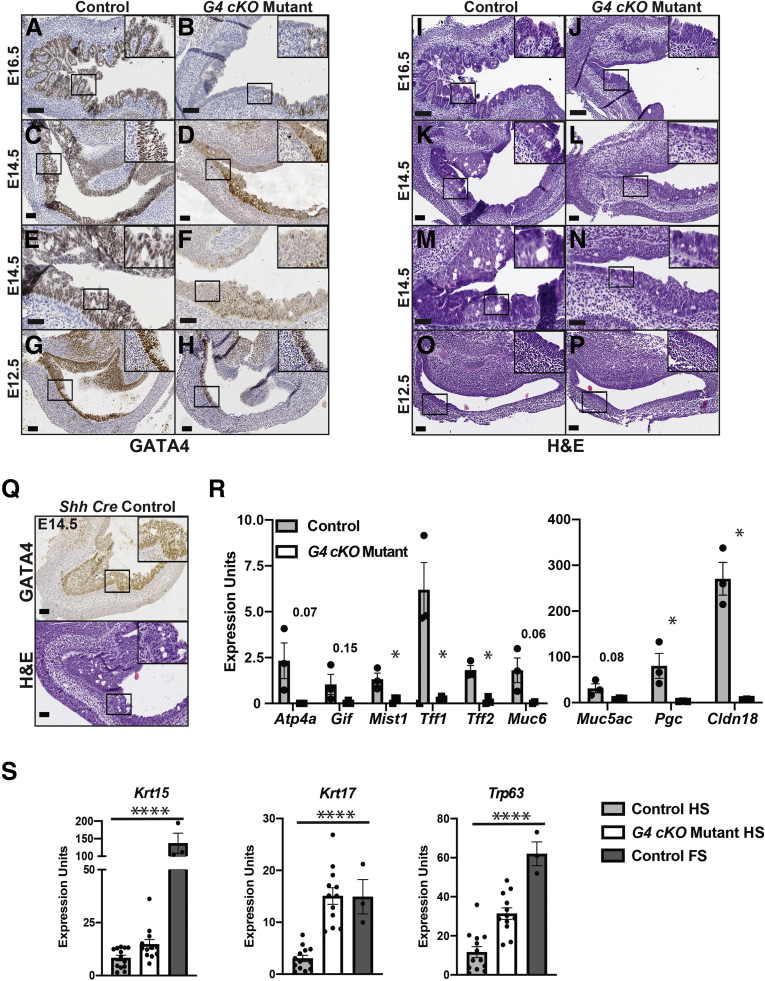
GATA4 protein distinguishes the stratified squamous FS and simple columnar HS boundary with HS epithelium expressing GATA4 (Figure 1A). We established a Gata4 cKO (G4 cKO) model to determine the extent to which GATA4 is necessary to drive simple columnar epithelial tissue morphogenesis. We compared stomachs across a spectrum of developmental stages among Gata4loxP/loxP, Gata4loxP/+, or Gata4loxP/- control and G4 cKO Gata4loxP/-::ShhCre mutant embryos. At E18.5, Gata4 messenger RNA (mRNA) was reduced in G4 cKO embryonic HS epithelium compared with control epithelium (Figure 1B). Expression of the closely related Gata6 gene was unchanged (Figure 1B). Hematoxylin and eosin (H&E) staining revealed that control, GATA4-expressing HS epithelium was appropriately organized into primordial glandular buds lined by simple columnar epithelium (Figure 1C, left). In contrast, GATA4-deficient HS epithelial tissue architecture was severely disrupted (Figure 1C, center). The GATA4-deficient HS epithelium lacked columnar morphology, and primordial glandular buds were not present. Mutant epithelium was instead organized similar to a stratified squamous epithelium (Figure 1C, center). Reflective of the reduced but not completely extinguished Gata4 expression in mutant HS (Figure 1B), we observed that most mutant stomachs contained regions that maintained GATA4 expression, likely owing to Cre inefficiency (Figure 1C, right). Areas with GATA4+ cell patches appeared to have a more columnar, glandular-like morphology (Figure 1C, right). Costaining for the epithelial cell membrane marker E-cadherin and GATA4 confirmed that mutant HS epithelium regions expressing GATA4 maintained a columnar, glandular morphology (Figure 1D, arrowheads) compared with regions lacking GATA4, which appeared stratified squamous (Figure 1D, starred). These data suggest that GATA4 acts in a cell autonomous manner to regulate HS morphogenesis during development. As expected, control and G4 cKO embryonic FS were normal containing keratinized stratified squamous epithelium (Figure 1E).
Figure 1.
GATA4 is required for morphogenesis of the HS epithelium. (A) H&E and GATA4 IHC staining of E18.5 control mouse stomach showed differential GATA4 expression between FS and HS epithelial cells. E-cadherin protein (IF, green) marks the differing epithelial tissue and cell architectures between the FS and HS. GATA4 protein (IF, red) was restricted to the simple columnar HS epithelium. (B) Gata4 mRNA was depleted in G4 cKO HS compared with control HS; Gata6 mRNA was unchanged (n = 3 control, 3 mutant). (C) H&E and GATA4 staining of E18.5 control and G4 cKO HS (n = 7 control, 7 mutant) identified GATA4 expressing simple columnar epithelium with emerging gastric units and glands in control embryos (left). G4 cKO HS generally lacked GATA4 protein and resembled stratified squamous epithelium (center). Regions in mutants maintaining GATA4 protein were morphologically similar to control embryos (right) suggesting a cell autonomous phenotype. (D) IF co-staining of control and mutant HS (n = 3 control, 3 mutant) was used to identify expression of GATA4 protein (red) in epithelial cells (E-cadherin, green). Control tissue consisted of a glandular epithelium with E-cadherin positive epithelial cells expressing nuclear GATA4 protein. IF staining of mutant HS showed that E-cadherin–positive, GATA4-negative epithelial cells were arranged similar to a stratified epithelium (star, compare with FS in A and E), whereas regions of E-cadherin positive cells maintaining nuclear GATA4 protein more closely resemble a normal, glandular epithelium (arrowheads). These data further support that the phenotype observed in GATA4 mutant tissue is cell autonomous. (E) H&E and GATA4 (red)/E-cadherin (green) IF costaining showed identical morphology of GATA4-negative FS from E18.5 control and G4 cKO embryos (n = 7 control, 7 mutant H&E; 3 control, 3 mutant IF). All scale bars = 50 μm. Higher magnification of boxed regions are found in insets. DAPI (blue) was used in all IF staining to mark nuclei. Autofluorescent blood cells can be seen in the lamina propria of both control and mutant tissues.
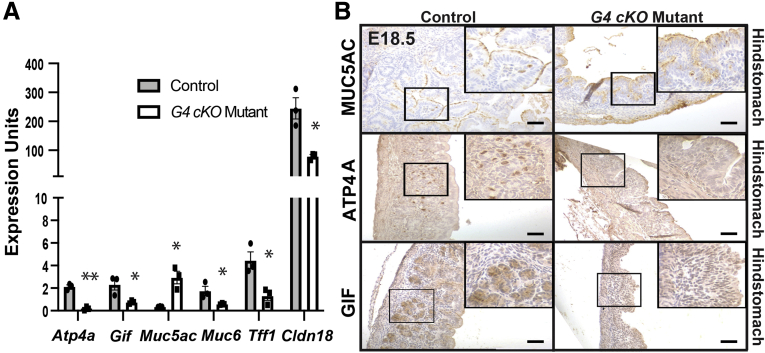
We next addressed whether the morphological changes in mutant HS reflected changes in the molecular identity of GATA4-deficient cells. To determine the extent to which HS cytodifferentiation was disrupted in E18.5 G4 cKO embryos, we measured gastric cell marker expression in control and mutant HS epithelia. Transcripts for genes encoding markers of gastric epithelial cells (Cldn18), neck cells (Muc6, Tff1), parietal cells (Atp4a), and zymogenic chief cells (Gif) were significantly reduced in epithelial cells isolated from mutants compared with control embryos; Muc5ac (surface mucus cell) transcript was higher in mutants compared with control embryos (Figure 2A). MUC5AC, ATP4A, and GIF proteins were present in control HS, while all but MUC5AC were lacking or diminished in mutant HS (Figure 2B). These data provide evidence that GATA4 is necessary for HS epithelial morphogenesis and development of the majority of differentiated HS epithelial cell types.
Figure 2.
GATA4-deficient HS epithelium lacks expression of differentiated gastric cell markers. (A) Differentiated HS epithelial cell transcripts detected by qRT-PCR were changed in G4 cKO HS compared with control HS (n = 3 mutant, 3 control). All except Muc5ac were depleted. Muc5ac increased. (B) MUC5AC protein was similar between E18.5 G4 cKO mutant and control HS (n = 4 control, 4 mutant). ATP4A (n = 3 control, 3 mutant) and GIF (n = 4 control, 4 mutant) proteins were depleted in G4 cKO mutant HS compared with control HS. Boxed regions are shown at higher magnification in upper-right corners. All scale bars = 50 μm. All qRT-PCR used HS epithelial cells. Error bars show SEM. P values determined by a Student’s t test, ∗P ≤ .05, ∗∗P ≤ .001.
GATA4 Depleted HS Acquires Expression of Genes Marking Stratified Squamous Epithelium
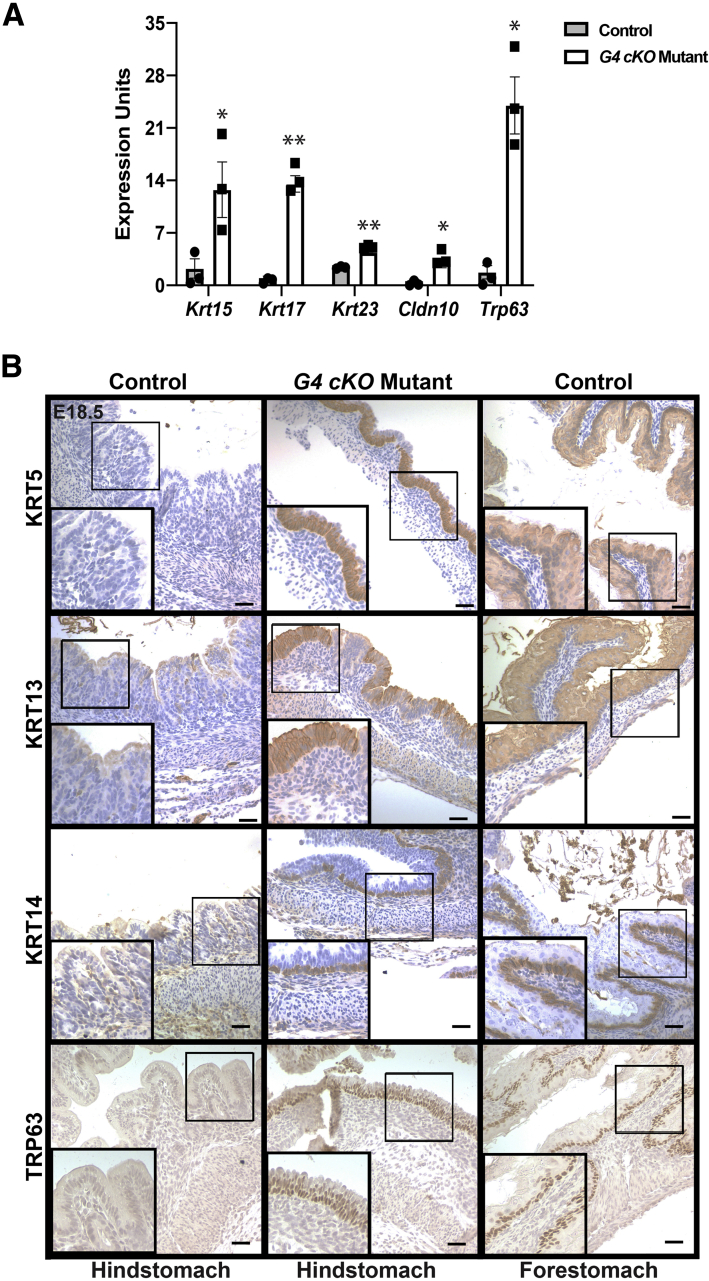
The loss of appropriate HS differentiated cell markers paired with the morphological similarity to FS suggested that GATA4-deficient HS epithelium had taken on FS epithelial cell identity. To ascertain if G4 cKO HS epithelium expressed markers of stratified squamous epithelium, we measured transcript and protein levels of key markers of stratified squamous epithelium. We found that transcripts for all FS markers examined—Krt15, Krt17, Krt23, Cldn10, and Trp63—were ectopically induced in GATA4-deficient HS epithelium compared with control epithelium (Figure 3A). Induction of FS marker gene expression correlated with ectopic protein expression; KRT5, KRT13, KRT14, and TRP63 were detected in GATA4-deficient, but not control, HS (Figure 3B). These data support the idea that GATA4 is necessary for HS columnar cell development because in its absence, abnormal squamous cell development occurs. Notably, Trp63, a master regulator of stratified squamous epithelial cell differentiation, was induced in GATA4-deficient HS epithelium.
Figure 3.
GATA4 depleted HS acquires stratified squamous epithelial cell gene expression. (A) qRT-PCR with epithelial cells from E18.5 control and G4 cKO mutant HS showed induced expression of genes associated with stratified squamous epithelium in mutants (n = 3 control, 3 mutant). Error bars show SEM. P values determined by a Student’s t test, ∗P ≤ .05, ∗∗P ≤ .001. (B) FS associated stratified squamous epithelial cell proteins, absent in E18.5 control HS, were induced in G4 cKO mutant HS. FS staining is shown as a reference for normal marker expression. (KRT5: n = 3 control, 5 mutant; KRT13: n = 6 control, 5 mutant; KRT14: n = 4 control, n = 5 mutant; TRP63: n = 5 control, 5 mutant) Scale bars = 50 μm. Boxed regions are shown at higher magnification in lower left corners.
Developmental Morphological Milestones Are Absent in GATA4 cKO HS by E14.5
Because ShhCre drives gene deletion as early as E8.5–E9.5, we traced back GATA4 depletion in G4 cKO embryonic HS to identify when both GATA4 was depleted and phenotypic changes were observable (Figure 4A–P). At E16.5, GATA4 loss was apparent in mutants, and GATA4-deficient epithelium was disorganized lacking primordial gastric glands and appearing mainly stratified (Figure 4A, B, I, and J). Similar to E18.5, GATA4+ cell patches were present in E16.5 G4 cKO HS tissue, and the morphology of these regions more closely resembled the normal emerging columnar, glandular epithelium (Figure 4B). At E14.5, disrupted GATA4 expression was observed in mutant HS (Figure 4C–F). Control HS epithelium displayed increased apicobasal thickness (compared with E12.5 stomachs), and intraepithelial spaces were present, which have been identified as precursors to gland and gastric unit development (Figure 4K and M).28,29 In contrast, mutant HS epithelial thickness was reduced compared with control embryos, and there were fewer, smaller intraepithelial spaces (Figure 4L and N). Again, in GATA4+ regions, HS architecture more closely resembled that of control embryos (Figure 4D and F). Finally, although GATA4 protein was reduced in E12.5 HS (Figure 4G and H), the morphology of control and mutant epithelia was comparable (Figure 4O and P).
Figure 4.
GATA4-deficient HS architecture and gene expression are disrupted at E14.5. (A–H) GATA4 IHC of control and G4 cKO stomachs from E16.5 (n = 6 control, 6 mutant), E14.5 (n = 7 control, 11M) and E12.5 (n = 5 control, 6 mutant) embryos showed depleted GATA4 protein in mutants. Small regions of GATA4+ cells were evident in mutant HS. GATA4 protein loss became more evident in mutants between E12.5 to E14.5. (I–P) H&E staining of E16.5 (n = 6 control, 6 mutant), E14.5 (n = 5 control, 8 mutant), and E12.5 (n = 4 control, 4 mutant) control and G4 cKO HS showed disrupted epithelial structure at E16.5 and E14.5. E16.5 control HS epithelium was columnar and lined with primordial buds, whereas G4 cKO HS epithelium appeared stratified, containing few abnormal primordial buds. E14.5 control HS epithelium had a typical morphology containing intraepithelial spaces, which are associated with glandular morphogenesis, whereas mutant HS epithelium was thinner with sparse, smaller intraepithelial spaces. (Q) GATA4 protein expression and epithelial structure were normal in HS from Gata4+/+Shh Cre E14.5 embryos (n = 2). Boxed regions are shown at higher magnification in insets (A–Q). (R) Differentiated HS epithelial cell transcripts detected by qRT-PCR were decreased in E14.5 G4 cKO HS compared with control HS (n = 3 mutant, 3 control). (S) Expression of stratified squamous epithelial cell transcripts detected by qRT-PCR was increased in E14.5 G4 cKO HS compared with control HS. All were expressed in control FS (n = 13 control HS, 12 mutant HS, 3 control FS). All scale bars = 50 μm. All qRT-PCR used HS or FS epithelial cells. Error bars show SEM. P values determined by a Student’s t test or 1-way analysis of variance as appropriate. ∗P ≤ .05, ∗∗P ≤ .001, ∗∗∗∗P ≤ .0001.
Epithelial morphogenesis begins at E13.5–E14.5 with increased apical-basal epithelial thickness, the emergence of intraepithelial spaces, and the onset of cytodifferentiation.1,28, 29, 30, 31 We observed the most consistent loss of GATA4 protein in E14.5 stomachs compared with earlier time points. We further observed a morphological defect at E14.5. Therefore, we chose to perform subsequent molecular analyses at E14.5. To confirm that phenotypes were independent of the disruption of one Shh allele by Cre insertion, we examined GATA4 protein levels and morphology of HS epithelium from Gata4+/+ShhCre E14.5 embryos and found these to be indistinguishable from control HS (Figure 4Q). To examine the extent of cytodifferentiation in E14.5 G4 cKO HS, we measured expression of genes marking specific glandular epithelial cell types. We detected decreased levels of parietal cell (Atp4a, Pgc), zymogenic chief cell (Gif, Mist1), neck cell (Tff1, Tff2, Muc6), and surface mucous cell (Muc5Ac) transcripts in mutants compared with control embryos (Figure 4R). Furthermore, to determine if FS-specific gene expression was induced in HS epithelium of E14.5 G4 cKOs, we performed quantitative reverse-transcription polymerase chain reaction (qRT-PCR) for Krt15, Krt17, and Trp63 and found these to be ectopically expressed in GATA4-deficient HS epithelium compared with control epithelium (Figure 4S). These data further support the idea that GATA4 is necessary for HS epithelial cell differentiation and to suppress FS cell differentiation in the HS.
Loss of GATA6 in the HS Does Not Disrupt Epithelial Morphogenesis
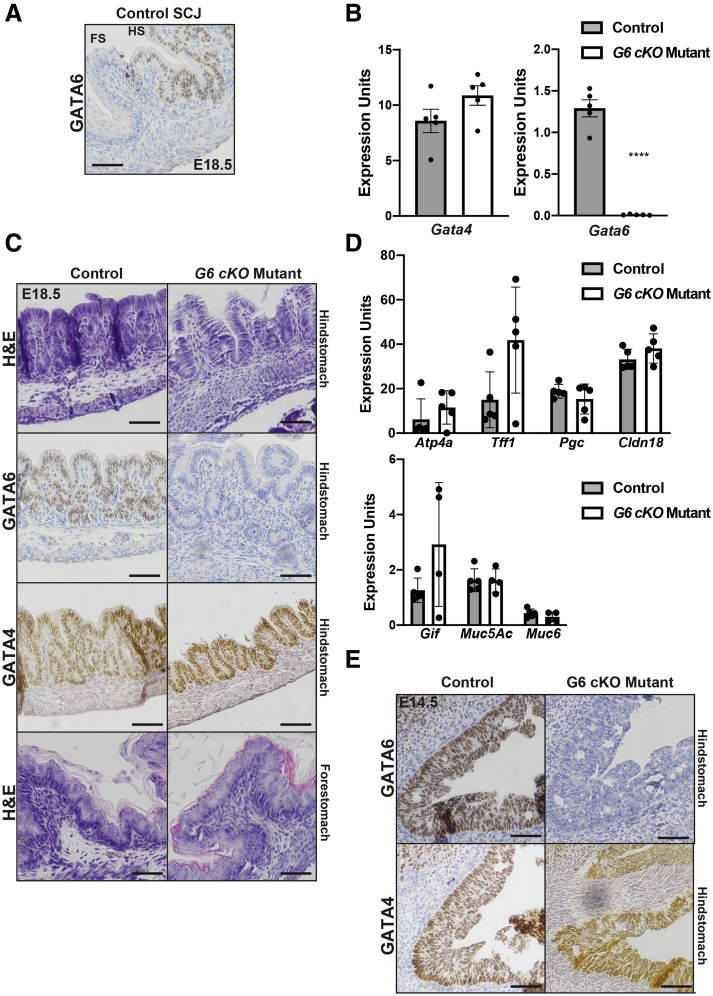
Because both GATA4 and GATA6 are expressed in the developing HS epithelium (Figures 1A and 5A–C), we considered the possibility that GATA4 mutant phenotypes reflected an imbalance of total GATA protein, rather than a loss of GATA4 itself. Therefore, we examined control Gata6loxP/+ and mutant Gata6loxP/-::ShhCre (G6 cKO) HS at E18.5 and E14.5. At E18.5, GATA4 expression was similar between control and G6 cKO HS epithelium while GATA6 was depleted in G6 cKO HS epithelium (Figure 5B and C). H&E staining showed comparable epithelial architecture among all genotypes (Figure 5C). Levels of HS cell marker transcripts were assessed, and no expression changes were detected between control and G6 cKO HS epithelium (Figure 5D). Finally, we validated that GATA6 protein was efficiently deleted at E14.5 (Figure 5E). GATA4 protein was present throughout control and G6 cKO HS, whereas GATA6 protein was depleted in mutants. Therefore, we concluded that phenotypes evident in G4 cKO HS were specific to GATA4 deletion, rather than to an overall GATA protein reduction.
Figure 5.
GATA6-depleted HS epithelium develops normally. (A) GATA6 staining of E18.5 the mouse squamocolumnar junction (SCJ) showed differential GATA6 expression between HS and FS with GATA6 expression being restricted to HS epithelial cells. (B) Gata6 mRNA was depleted in G6 cKO mutant HS compared with control HS; Gata4 mRNA was unchanged (n = 5 control, 5 mutant). (C) H&E, GATA6, and GATA4 staining of E18.5 control and G6 cKO mutant HS showed GATA6 protein depletion in mutants, yet epithelial structure remained normal. GATA4 protein was unchanged. FS morphology was comparable between control embryos and mutants (n = 4 control, 4 mutant). (D) Differentiated HS epithelial cell transcripts were unchanged in G6 cKO mutant HS compared with control HS (n = 5 mutant, 5 control). (E) GATA4 and GATA6 staining of control and G6 cKO mutant HS at E14.5 (n = 6 control, 6 mutant) showed comparable morphology between tissues with GATA6 protein depleted and GATA4 protein unchanged. All scale bars = 50 μm. All qRT-PCR used HS epithelial cells. Error bars show SEM. P values determined by a Student’s t test, ∗∗∗∗P ≤ .0001.
Induction of GATA4 Expression in the Developing FS Epithelium Alters Epithelial Architecture Toward Columnar
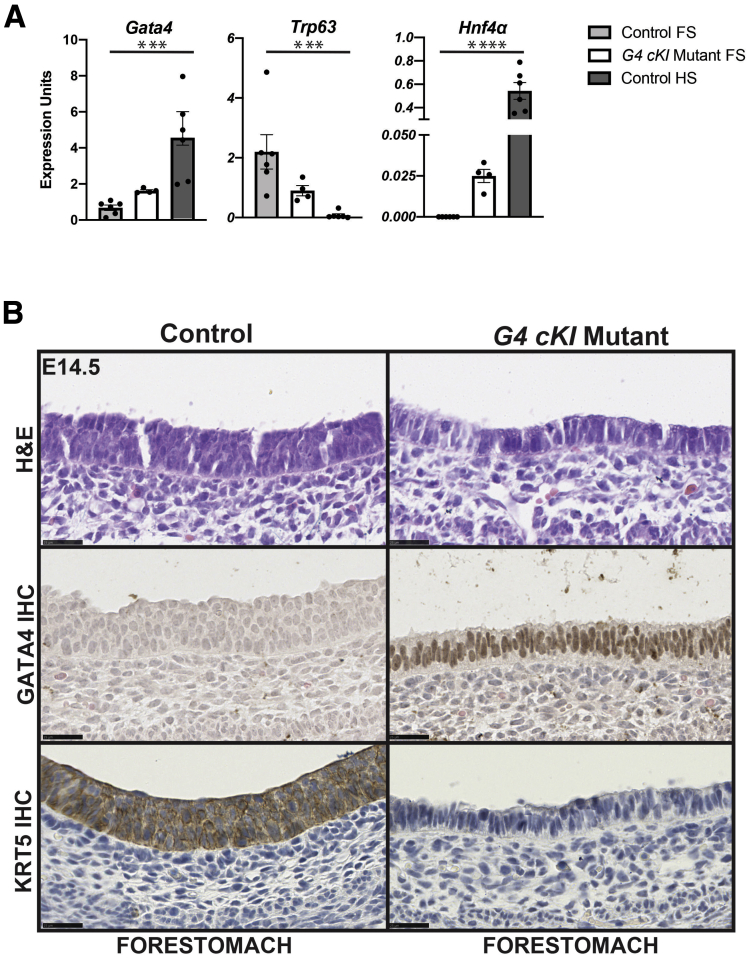
Observing that GATA4 expression was necessary to drive columnar cell fate in the developing HS epithelium, we sought to determine whether GATA4 was sufficient to induce columnar fate in the developing FS epithelium. Using ShhCre to drive expression of a Gata4 cKI allele, Rosa26 lnlG4 (G4 cKI),17 in the developing FS, we analyzed the effects of ectopic GATA4 expression on FS epithelial morphogenesis at E14.5, parallel to studies of G4 cKO mutants. We detected Gata4 mRNA in E14.5 G4 cKI FS epithelium (Figure 6A). Although the amount of Gata4 mRNA detected in G4 cKI FS epithelium was ~3-fold lower than that of control HS, GATA4 protein was uniformly expressed in mutant FS epithelium (Figure 6B). As expected, GATA4 protein was undetectable in E14.5 control FS epithelium. Examination of the epithelial architecture of control and G4 cKI FS indicated that control epithelium developed as a stratified epithelium while the GATA4-expressing FS epithelium appeared columnar-like (Figure 6B). Moreover, ectopic GATA4 expression in the FS was sufficient to inhibit Trp63 expression (Figure 6A). This was the inverse of what occurred in G4 cKO HS mutants in which Trp63 expression was induced upon GATA4 depletion (Figure 3A). The finding that Trp63, a stratified squamous epithelium master transcription factor, was reduced in G4 cKI mutant FS suggested a change in cell identity from stratified to columnar. Therefore, as a preliminary examination for the emergence of HS gene expression in mutant FS epithelium, we measured Hnf4α mRNA, a factor specifically present in developing HS but absent in developing FS. We detected Hnf4α mRNA in G4 cKI FS but not in control FS (Figure 6A). To further probe the character of the developing GATA4-expressing FS epithelium, we used immunohistochemistry (IHC) to compare expression of the stratified squamous cell cytokeratin KRT5 between control and mutant embryos. Although present in control FS epithelium, KRT5 protein was uniformly depleted in GATA4-expressing FS epithelium (Figure 6B). These data suggest that GATA4 protein is sufficient to drive a columnar epithelial cell fate and morphogenesis in the developing murine stomach.
Figure 6.
Maintained GATA4 expression in the developing FS epithelium alters epithelial architecture toward columnar. (A) Gata4, Trp63, and Hnf4a mRNA levels were measured in epithelial cells from control FS, HS, and G4 cKI mutant FS at E14.5 (n = 6 control FS and HS, 4 mutant FS). Gata4 was upregulated in G4 cKI mutant FS. Trp63 was highly expressed in control FS and diminished in G4 cKI mutant FS. Hnf4a mRNA expression was induced in G4 cKI mutant FS. Error bars show SEM. P-values were calculated using a 1-way analysis of variance, ∗∗∗P ≤ .005, ∗∗∗∗P ≤ .0001. (B) H&E, GATA4, and KRT5 staining of E14.5 control and G4 cKI FS (n = 3 control, 3 mutant) showed that control FS contained a multilayer epithelium, whereas G4 cKI FS appeared columnar. GATA4 protein was ectopically expressed in E14.5 G4 cKI FS epithelial cells. Control FS epithelial cells expressed KRT5 protein that was completely absent in FS of G4 cKIs. Scale bars = 25 μm.
The Transcriptome of GATA4 Gastric Mutants Reflects Global Shifts in Epithelial Cell Identities
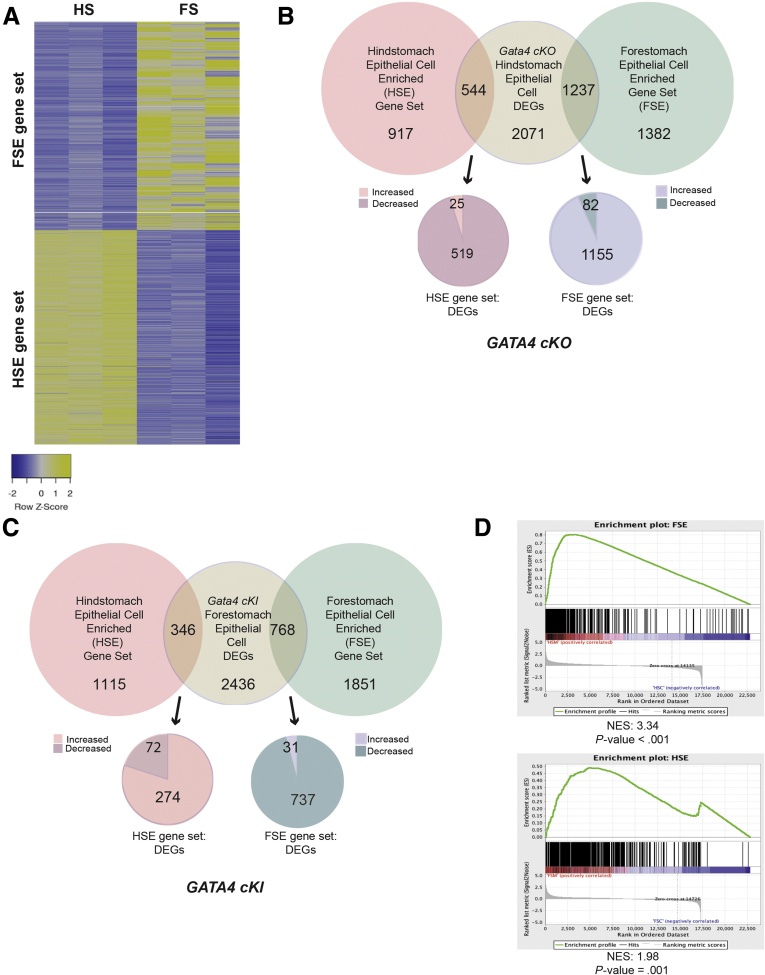
Our morphological analyses of GATA4-depleted HS epithelium and GATA4-expressing FS epithelium support the idea that GATA4 is necessary and sufficient to promote columnar epithelium development and inhibit stratified squamous epithelium development in the stomach. Given that GATA4 has been shown to be a critical transcription factor in regulating jejunal-ileal boundaries and cell fates in the small intestine,17,25 we reasoned that global shifts in the transcriptomes of these developing gastric tissues must be occurring when GATA4 was altered. Therefore, we performed RNA-sequencing (RNA-seq) using HS and FS epithelial cells to compare transcriptomes among control and mutant E14.5 embryos. We used gene expression profiles of control FS and HS to construct HS-enriched (HSE) and FS-enriched (HSE) transcript sets by selecting transcripts with ≥ 2-fold differential expression (P ≤ .05) between HS and FS (Figure 7A). We found 1461 genes encoding HSE transcripts and 2619 genes encoding FSE transcripts (Figure 7A). We compared genes differentially expressed between control HS and GATA4-depleted HS or control FS and GATA4-expressing FS with HSE and FSE gene sets. Of the differentially expressed genes (DEGs) identified by comparing G4 cKO HS epithelium vs control epithelium, 46% overlapped with the HSE and FSE gene sets (Figure 7B). Of the DEGs identified by comparing control and G4 cKI FS epithelia, 29% overlapped with the HSE and FSE gene sets (Figure 7C). Reflecting the HS to FS transition in GATA4-deficient HS mutants, 95% (519 of 544) of the DEGs overlapping with the HSE set were downregulated, and 93% (1155 of 1237) of the DEGs overlapping with the FSE set were upregulated (Figure 7B). Similarly, in GATA4-expressing FS mutants, in which FS transitioned to HS, 96% (737 of 768) of the DEGs overlapping with the FSE set were downregulated, and 79% (274 of 346) of the DEGs overlapping with the HSE set were upregulated (Figure 7C). We further used gene set enrichment analysis to compare control and mutant gene expression profiles.32 We performed gene set enrichment analysis using HSE (1461) and FSE (2619) gene sets (Figure 7A) and RNA-seq data profiling gene expression of HS epithelium from G4 cKO and control embryos and of FS epithelium from G4 cKI and control embryos. The global gene expression profile of G4 cKO HS epithelium, unlike the control profile, aligned with the FSE gene set (Figure 7D, top). Similarly, the global gene expression profile of G4 cKI FS epithelium, unlike the control profile, aligned with the HSE gene set (Figure 7D, bottom). These data further support the idea that GATA4 is necessary and sufficient to promote development of columnar epithelium in the embryonic stomach. Loss of GATA4 in the HS epithelium causes a columnar-to-stratified switch and gain of GATA4 in the FS epithelium causes a stratified-to-columnar switch. Moreover, changes are not simply morphological, but rather represent global shifts in foundational gene expression patterns that underlie columnar vs stratified epithelial tissue morphogenesis in the developing stomach.
Figure 7.
GATA4 mutant HS and FS epithelial cells undergo broad shifts in gene expression correlating with tissue morphology. (A) HSE and FSE epithelial cell gene expression patterns were determined by comparing gene expression between E14.5 control HS and FS (RNA-seq, n = 3 control HS, 3 control FS; ≥2-fold change, P ≤ .05). Heatmap displays FPKM for HSE and FSE gene set expression in control HS and FS epithelial cells. (B) DEGs between E14.5 G4 cKO mutant and control HS epithelial cells were identified (≥2-fold, P ≤ .05) (n = 3 control, 5 mutant) and overlaid with HSE and FSE sets defined in A. Genes within the intersection of these groups were discriminated by direction of gene expression change. Most DEGs from the HSE gene set were downregulated in mutant tissue, and most DEGs from the FSE gene set were upregulated in mutant tissue corresponding with morphological changes. (C) DEGs between in E14.5 G4 cKI mutant FS and control FS epithelial cells mapping to HSE or FSE gene sets were determined as in (B) (n = 3 control, 3 mutant). Genes within the intersection of these groups were discriminated by direction of gene expression change. Most DEGs from the FSE gene set were downregulated in mutant tissue, and most DEGs from the HSE gene set were upregulated in mutant tissue corresponding with morphological changes. (D) Gene set enrichment analysis performed using HSE and FSE gene sets and control and mutant RNA-seq profiles indicated that the G4 cKO mutant HS epithelial cell gene expression aligned with FS while G4 cKI mutant FS epithelial cell gene expression aligned with HS. Gene lists provided in Supplementary Table 1.
Further focusing on disrupted patterning in G4 cKO HS and G4 cKI FS, we examined mRNA and/or protein expression of FS, HS, and intestinal lineage defining transcription factors Sox2, Pdx1, Trp63, and Cdx2 in GATA4-expressing FS and GATA4-deficient HS. In development, SOX2/TRP63 define FS, SOX2/GATA4 define gastric corpus, SOX2/GATA4/PDX1 define gastric antrum, and CDX2/PDX1/GATA4 define duodenum.1,4 Although Sox2 was increased in GATA4-expressing FS compared with control FS, the stratified squamous-specific marker Trp63 was downregulated (Figure 6A; Supplementary Table 1), supporting the idea that FS cell fate was lost in mutants. To further define the columnar character of mutant FS, we examined Pdx1 and Cdx2 expression. Both Pdx1 and Cdx2 were absent in control and mutant FS epithelium (FPKM: Pdx1 control FS and mutant FS = 0; Cdx2 control FS = 0, mutant FS = 0.009; FPKM > 0.1 considered expressed). We further detected no CDX2 protein in either control or G4 cKI mutant FS (data not shown). The presence of Sox2 and Gata4, Trp63 downregulation, and the absence of Pdx1 and Cdx2 in GATA4-expressing FS epithelium supported our conclusion that FS was patterned toward the developing columnar gastric corpus (GATA4+SOX2+), rather than toward antrum or intestine. Moreover, corpus markers Irx2, Irx3, and Irx533 were all upregulated in G4 cKI mutant FS compared with control FS (Supplementary Table 1). In GATA4-deficient HS epithelium, the FS-specific marker Trp63 was upregulated, and Sox2 remained expressed (Figures 3 and 4S; Supplementary Table 1). Pdx1 expression was greatly reduced in G4 cKO HS mutants (FPKM: Pdx1 control HS = 1.6, mutant HS = 0.2). As expected, Cdx2 mRNA was absent in both control and mutant HS epithelium (FPKM: Cdx2 control HS = 0.03, mutant HS = 0.08, FPKM > 0.1 considered expressed). The presence Sox2, Trp63 upregulation and the absence of Pdx1 and Cdx2 in GATA4-deficient HS epithelium supported our conclusion that HS was patterned toward the developing stratified squamous FS (SOX2+TRP63+), rather than toward corpus, antrum, or intestine.
GATA4 Likely Directly Activates and Represses Gene Expression to Control Epithelial Morphogenesis in the Developing HS
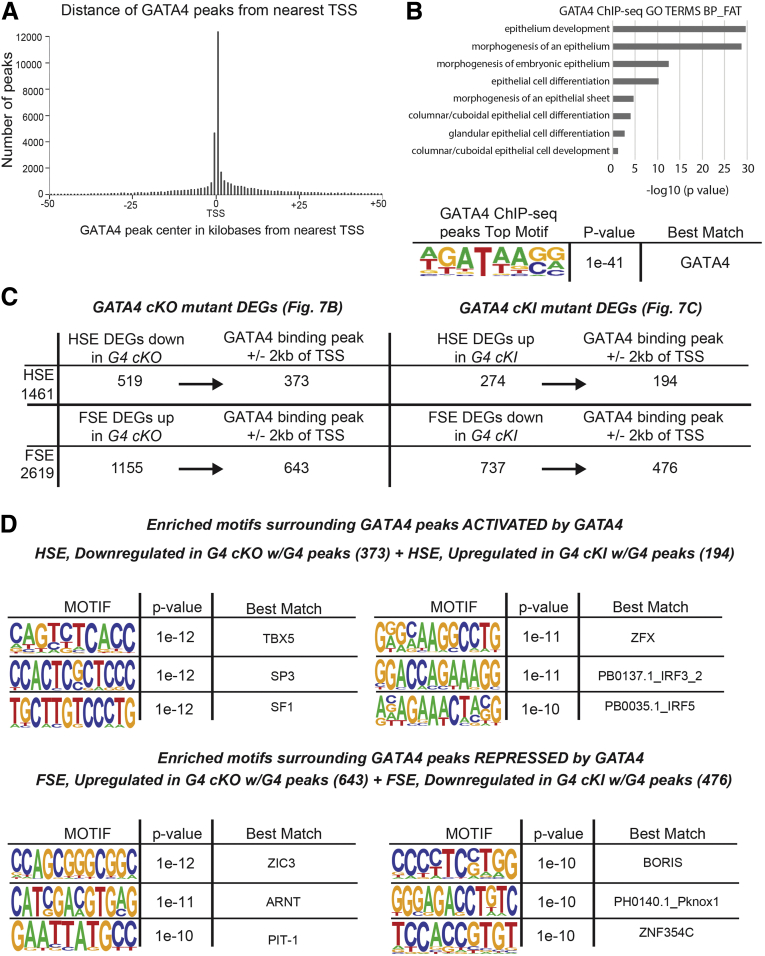
Our next goal was to identify genes, among those altered in GATA4-deficient HS and GATA4-expressing FS, most likely to be directly regulated by GATA4 to establish simple columnar epithelial cell fate and repress stratified squamous epithelial cell fate. We took advantage of GATA4-Bio-ChIP-seq data that we generated using adult mouse HS epithelial cells to identify those DEGs that contained experimentally validated GATA4 binding sites. We identified 50,869 total GATA4 binding peaks in HS epithelium with 41,332 peaks within ±50 kb of the nearest transcription start site (TSS) and 18,658 peaks within ±2 kb of the nearest TSS (Figure 8A). We used DAVID functional annotation to identify biological themes among genes with identified GATA4 binding sites finding epithelium development and morphogenesis, columnar epithelial cell differentiation, and glandular epithelial cell development enriched (Figure 8B), agreeing with phenotypic data showing that GATA4 promotes columnar cell development. Analysis of the DNA sequence of GATA4 chromatin immunoprecipitation sequencing (ChIP-seq) peaks within ±2 kb of the nearest TSS using HOMER showed that the top motif identified matched the GATA4 consensus motif (Figure 8B).
Figure 8.
Putative GATA4 direct targets identified by cross-referencing RNA-seq and ChIP-seq datasets. (A) GATA4 ChIP-seq was performed with HS epithelial cells from adult mice. Peaks were mapped to the nearest TSS. The spatial distribution of GATA4 peaks within ±50 kb of the nearest TSS is shown. (B) DAVID analysis identified enriched gene ontology terms associated with peaks shown in A. The GATA4 consensus sequence was the top motif present in peaks shown in A, determined by de novo HOMER motif analysis. (C) GATA4 binding peaks from A were filtered to identify those within proximal regulatory regions of genes (±2 kb TSS). The filtered peak set was cross-referenced with G4 cKO mutant HS and G4 cKI mutant FS DEGs corresponding to the HSE or FSE gene sets (Figure 7B and C) to identify DEGs with HSE and FSE expression likely directly regulated by GATA4. (D) HOMER de novo motif analysis tool was used to scan ±100 bp of each GATA4 binding peak center among genes identified in C that were predicted to be activated (HSE expression, downregulated in HS mutants, upregulated in FS mutants) or repressed by GATA4 (FSE expression, upregulated in HS mutants, downregulated in FS mutants). The top six enriched motifs are shown. Gene lists provided in Supplementary Table 2.
Of the DEGs with HSE expression downregulated in E14.5 G4 cKO HS, 72% had GATA4 binding peaks within their proximal regulatory regions (Figure 8C). Similarly, of the DEGs with HSE expression upregulated in E14.5 G4 cKI FS, 71% had GATA4 binding peaks within their proximal regulatory regions (Figure 8C). These represent likely direct targets that are positively regulated by GATA4 such that expression is lost when GATA4 is deleted and induced when GATA4 is ectopically expressed. Looking at those genes likely to be direct targets negatively regulated by GATA4, such that expression is induced when GATA4 is deleted and lost when GATA4 is ectopically expressed, we found that 56% of the DEGs with FSE expression upregulated in E14.5 G4 cKO HS had GATA4 binding peaks within their proximal regulatory regions and that 65% of the DEGs with FSE expression downregulated in E14.5 G4 cKI FS contained GATA4 binding peaks within their proximal regulatory regions (Figure 8C). These analyses allowed us to identify gene sets that we predict are directly induced or repressed by GATA4 in the developing stomach to regulate phenotypic changes.
An outstanding question in GATA biology is the mechanism by which GATA factors function to activate or repress genes within a given tissue. To gain insight into this question in the context of gastric development, we examined the DNA sequence neighborhoods surrounding GATA4 binding peaks within the proximal regulatory regions of genes positively or negatively regulated in G4 cKO HS and G4 cKI FS using HOMER de novo motif analysis. We identified different sets of transcription factor binding motifs statistically enriched around GATA4 binding sites (±100 bp of the peak) in the HSE and FSE gene sets differentially expressed in GATA4 mutants (Figure 8D). Several motifs predicted in the GATA4 binding neighborhoods of activated and repressed gene sets identified transcription factors previously shown to cooperate with GATA4 to direct gene regulatory networks in other organ systems including TBX5, SF1, and SP3.34, 35, 36, 37, 38, 39 Others, although not yet definitively linked with GATA4, have transcriptional activities aligning with the GATA4 neighborhoods in which they were identified (i.e., positive regulators in activated genes or negative regulators in repressed genes).40, 41, 42, 43, 44, 45, 46, 47
GATA4 May Regulate a Transcription Factor Network to Repress Stratified Squamous Epithelial Morphogenesis
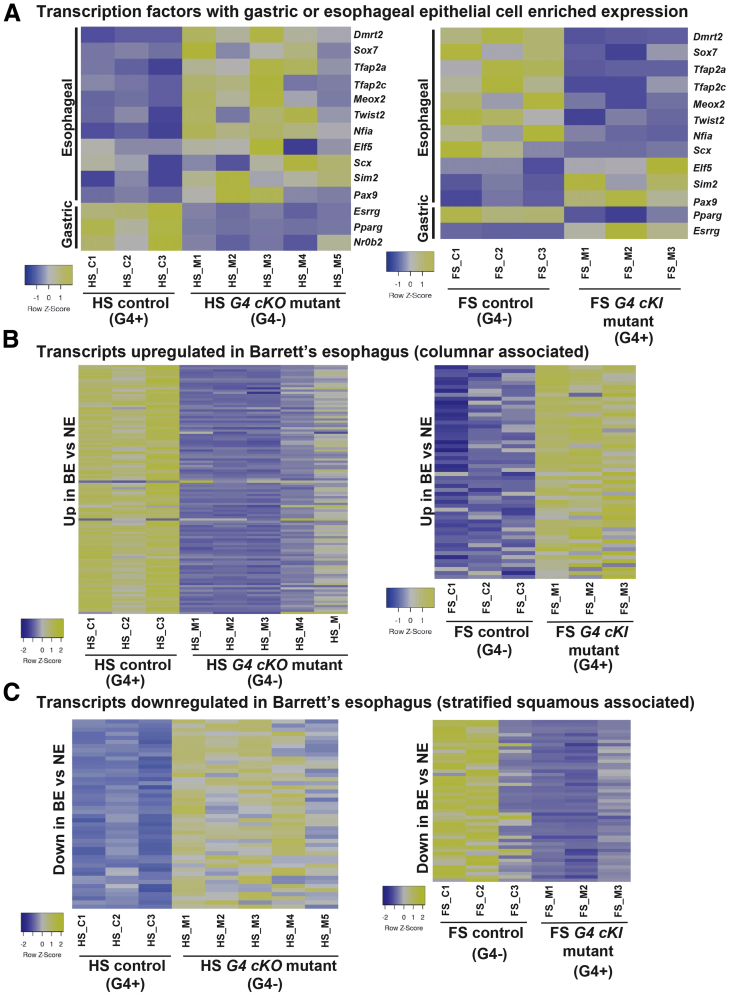
To better understand the mechanism through which GATA4 promotes columnar epithelial cell development over stratified squamous epithelial cell development, we examined the status of known tissue-enriched transcription factors in GATA4 mutants. Previously, Sulahian et al48 determined the expression of 1880 known and putative DNA-binding proteins in esophageal, gastric, and intestinal epithelial cells from 1-month-old mice to identify those with tissue-enriched expression. This analysis yielded 21 factors with enriched expression in esophagus vs stomach and intestine and 9 factors with enriched expression in the stomach vs the intestine and esophagus. Three of the 9 with gastric enriched expression and 15 of the 21 with esophageal enriched expression had similarly enriched expression patterns in E14.5 HS or FS, respectively. All 3 gastric factors and 11 of the 15 esophageal factors contained experimentally validated GATA4 binding peaks within ±2 kb of their TSS. Of the 3 gastric factors, only Esrrg was coordinately regulated in GATA4 mutants with its expression decreasing in G4 cKO HS epithelium and increasing in G4 cKI FS epithelium (Figure 9A). With respect to the esophageal factors, 8 of the 11 with GATA4 binding peaks were coordinately regulated in GATA4 mutants with their expression decreasing in G4 cKI FS epithelium and increasing in G4 cKO HS epithelium (Figure 9A). These results suggest a mechanism through which GATA4 promotes columnar fate in the HS epithelium by repressing expression of a network of stratified squamous enriched transcription factors.
Figure 9.
Transcriptional programs associated with esophageal/FS identity and human disease are altered in GATA4 mutant HS and FS epithelial cells. (A) Expression of a network of transcription factors with esophageal-enriched expression and containing functional GATA4 binding sites ±2 kb of the TSS were inversely regulated in E14.5 G4 cKO HS and G4 cKI FS compared with control embryos. Heatmaps shown compare FPMK values for these factors between control (HS_C1 through HS_C3) and G4 cKO HS (HS_M1 through HS_M5) or control (FS_C1 through FS_C3) and G4 cKI FS (FS_M1 through FS_M3). Esophageal transcription factor expression was induced in G4 cKO HS and suppressed in G4 cKI FS. Of a set of three gastric-specific factors with functional GATA4 binding sites ±2 kb of the TSS, expression of all was suppressed in G4 cKO HS, but only Esrrg was induced in G4 cKI FS. (B, C) Genes with differential expression in BE epithelium vs normal esophageal epithelium, identified from published datasets,49, 50, 51 were cross-referenced with GATA4 ChIP-seq data to generate sets of DEGs in BE with functional GATA4 binding sites within ±2 kb of the TSS: 527 DEGs upregulated in BE compared with control esophagus and 380 DEGs downregulated in BE compared with control esophagus. These sets of putative GATA4 targets up or downregulated in BE were cross-referenced with DEGs in E14.5 G4 cKO HS or G4 cKI FS (Figures 7B and C). Heatmaps show comparisons of FPMK values for BE associated GATA4 target genes between control HS (HS_C1 through HS_C3) and G4 cKO mutant HS (HS_M1 through HS_M5) or control FS (FS_C1 through FS_C3) and G4 cKI mutant FS (FS_M1 through FS_M3). Expression profiles for genes with GATA4 binding sites upregulated in BE epithelium compared with normal esophageal epithelium (columnar-associated transcripts) among control embryos and GATA4 mutants are shown in B. Expression profiles for genes with GATA4 binding sites downregulated in BE epithelium compared with normal esophageal epithelium (stratified squamous associated) among control and GATA4 mutants are shown in C. Overall, ~30% of the DEGs in BE we identified as containing functional GATA4 binding sites overlapped with GATA4 mutants. Supplementary Table 3 provides the gene identifiers and binding site locations for transcripts shown in heatmaps in panels B and C.
Genes Regulated by GATA4 During Gastric Epithelial Morphogenesis Overlap With Those Dysregulated in BE
The columnar-like, rather than stratified squamous, epithelium observed in GATA4-expressing FS was reminiscent of BE, a premalignant stratified-to-columnar esophageal metaplasia. We and others have shown that GATA4 is abnormally expressed in BE.12,13,20 To determine whether altered gene expression patterns observed in GATA4-expressing embryonic FS epithelium and GATA4-deficient embryonic HS epithelium correlated with patterns observed in BE, we compiled sets of genes with known up- or downregulated expression in BE vs normal esophageal tissue from published datasets.49, 50, 51 Among these BE-associated genes, we identified those with GATA4 binding peaks in their proximal regulatory regions: 527 presumptive GATA4 targets with upregulated expression in BE and 380 presumptive GATA4 targets with downregulated expression in BE. We cross-referenced BE-associated DEGs containing functional GATA4 binding sites with HS and FS DEGs identified by comparing control HS with G4 cKO HS and control FS with G4 cKI FS (Figure 7B and C). Heatmaps shown in Figures 9B and 9C show correlations among DEGs in GATA4 mutant embryonic tissues and DEGs with GATA4 binding sites in BE. Among the upregulated BE-associated DEGs with functional GATA4 binding sites (columnar associated genes), 112 overlapped with DEGs downregulated in Gata4 cKO HS and 54 overlapped with DEGs upregulated in Gata4 cKI FS (Figure 9B). Together, 31% of the upregulated, putative GATA4 bound BE gene set overlapped with DEGs up or downregulated in GATA4 mutant FS or HS epithelium (Figure 9B). Among the downregulated BE-associated DEGs with functional GATA4 binding sites (squamous associated genes), 46 overlapped with DEGs upregulated in Gata4 cKO HS and 49 overlapped with DEGs downregulated in Gata4 cKI FS (Figure 9C). Together, 25% of the downregulated, putative GATA4 bound BE gene set overlapped with up or downregulated DEGSs in GATA4 mutant HS or FS epithelium. Overall, almost 30% of presumptive GATA4 targets misregulated in BE overlapped with genes found to be differentially expressed in our developmental models. Among these putative GATA4-regulated BE-associated genes, it was notable that the GI stem cell marker Lgr5 was strongly induced in GATA4-expressing FS epithelium (+50-fold G4 cKI) (Supplementary Table 1). In the adult stomach, Lgr5 marks antral stem cells as well as reserve stem cells in the corpus.52,53 In the embryonic stomach, however, Lgr5 expression is highly correlated with stem/progenitor cells of the developing corpus.31 Highly upregulated Lgr5 expression in GATA4-expressing FS epithelium suggests a pivotal change in stem cell fate in mutant FS toward that of the corpus. This agrees with our finding that the corpus-enriched transcription factors Irx2, Irx3, and Irx5 were upregulated in GATA4-expressing FS epithelium while the antral marker Pdx1 was absent. Taken together, these data correlate findings of a developmental model of GATA4 function in epithelial cell biology with a human disease in which GATA4 protein is misregulated.
Discussion
During embryonic development, gene regulatory networks consisting of regionally restricted and lineage-specific transcription factors regulate specification, morphogenesis, and differentiation of the GI epithelium. Our goal was to understand how GATA4, a key developmentally expressed transcription factor, participates in development and differential morphogenesis of the mouse stomach epithelium. Our studies using Gata4 conditional knockout and knock-in mouse lines support the conclusion that GATA4 is necessary and sufficient for columnar epithelial morphogenesis in the stomach. Gastric epithelial morphogenesis was normal in Gata6 conditional knockout embryos indicating that GATA4 uniquely controls this process. GATA4 must be expressed in the developing HS to permit columnar gastric epithelial cell fate and repress stratified squamous epithelial cell fate. Conversely, GATA4 must be silenced in the developing FS to permit stratified squamous epithelial cell fate and block columnar gastric epithelial cell fate. In this way, the establishment of domains with differential GATA4 expression in the developing mouse stomach determines gastric epithelial morphogenesis.
Our study agrees with and extends on the previously published findings of Jacobsen et al18 and Rodriguez-Seguel et al.19 Both studies provide a primarily histologic characterization of GATA4-dependent phenotypes in the developing stomach and show that glandular columnar epithelial architecture and mature HS cell types are abnormal in GATA4 mutant stomachs. Moving these studies forward, our study identified gastric defects earlier during development (E14.5 vs E17.5–E18.5), used both cKO and cKI models to demonstrate necessity and sufficiency of GATA4 in columnar HS development, and added in-depth global transcriptomic and DNA binding analyses to augment the understanding of GATA4’s role in gastric biology. One caveat to our study, however, is that although we based the timing of our analysis on efficient GATA4 protein loss and the appearance of a morphological phenotype, we cannot dismiss the possibility that molecular changes prior to E14.5 influence the phenotype.
Simply identifying genes with upregulated or downregulated expression in GATA4 mutants, however, does not in itself indicate direct regulation of a gene’s transcription by GATA4. Identification of functional GATA4 binding sites within regulatory regions of DEGs can establish likely direct regulatory relationships between GATA4 and its putative targets. Overlapping expression data with GATA4 DNA binding data enabled us to establish connections between experimentally validated GATA4 binding sites and DEGs in our models to identify candidate genes most likely to be directly regulated by GATA4 in the developing murine stomach. One caveat to this analysis is that we used binding data from adult mouse HS epithelial cells rather than from E14.5 embryos. Although ideal to use embryonic cells, we encountered technical challenges. It is possible that our analyses identified targets bound by GATA4 in the adult HS that aren’t similarly bound by GATA4 in the developing embryonic tissue (false positives) and that GATA4 binding peaks were missed because some targets are only bound by GATA4 during development (false negatives). Nevertheless, data from adult tissue can serve as a surrogate to demonstrate that GATA4 has the ability to bind to specific chromatin domains in simple columnar HS epithelium. Supporting the efficacy of using ChIP-seq data from adult tissue to probe GATA4 function in embryonic tissue was the finding that GO terms for genes with GATA4 binding sites aligned with developmentally relevant biological functions. Association of GATA4 binding sites with these themes further supports the idea that GATA4 is a critical regulator of columnar epithelium development in the glandular HS. The correlation between HSE and FSE genes differentially expressed in mutants and genes with experimentally validated GATA4 binding sites places GATA4 directly upstream of large gene networks essential in mediating columnar HS development.
GATA factors generally, and GATA4 specifically, have been implicated as master regulators in other developmental systems. GATA4 and GATA6 are key regulators of intestinal development.14 GATA4 functions as an intestinal regionalization factor by activating expression of jejunal cell associated genes and repressing expression of ileal cell associated genes.17 GATA4 activates expression of key proteins in a transcription factor network that guides cardiac development.54 In terms of repressive functions, GATA2/3 regulate a transcription factor network that promotes trophoblast differentiation by repressing pluripotency transcription factors in stem cells.55 GATA1 promotes erythroid differentiation by repressing opposing upstream transcriptional activators of myelolymphoid differentiation while also suppressing expression of opposing factors’ downstream targets.56 These dual mechanisms of GATA1 function have a synergistic effect on lineage specification resulting in erythroid rather than myelolymphoid differentiation. The facts that functional GATA4 binding sites were identified in the promoters of more than half of an essential esophageal/FSE transcription factor set and that the expression of these transcription factors correlated with GATA4 presence or absence suggest that GATA4 directly represses expression of a transcription factor network in the developing HS to suppress stratified squamous epithelium development. Furthermore, downstream targets of these esophageal/FS transcription factors also contained GATA4 binding sites and changed expression state depending on the presence or absence of GATA4. Together, these data support the idea that GATA4 works through synergistic mechanisms to repress stratified squamous epithelium development in the HS.
Using information gained from cross-referencing expression, chromatin binding, and motif analyses, we identified GATA4 cofactors that may contribute to GATA4’s ability to specifically activate or repress transcription. Among these were several transcription factors that act with GATA factors to direct gene expression programs in other developmental systems.34, 35, 36, 37, 38, 39 For example, TBX5 and GATA4 co-bind genes in cardiac cells to activate gene expression.34, 35, 36 SF1 and GATA4 function cooperatively to activate Sertoli cell gene expression.37,38 Although SP3, like GATA4, can function to activate or repress gene expression,57,58 GATA4 and SP3 can interact to activate cardiac Carp1 expression.39 Because SP1 and SP3 bind the same consensus sequence,58 it is conceivable that SP1, typically a transactivating factor, serves as a GATA4 co-factor in the developing stomach. SP1 and GATA4 positively regulate cardiac and intestinal gene expression.59, 60, 61 Although not yet linked with GATA4, ZNF354C (ZFP354C) contains a KRAB transcriptional repression domain.42,43 PKNOX1, which preferentially binds to promoter sequences, represses adipogenic differentiation and is implicated in negative regulation of the GLUT4 gene.44, 45, 46 BORIS (brother of the regulator of imprinted sites) negatively regulates an androgen receptor regulatory network in ovarian cancer.47 Finally, PIT-1 (pituitary-specific positive transcription factor 1) functions with nuclear receptor corepressor N-CoR to repress gene expression.41,62 It is also possible that GATA4 binding to promoters disrupts binding of other essential activating transcription factors to interfere with gene expression. We proposed this mechanism as a way in which GATA4 represses the Fgf15 gene in the mouse jejunum.17
The morphological and molecular shifts from stratified-to-columnar and columnar-to-stratified epithelia observed in GATA4 mutant embryonic stomachs are reminiscent of tissue metaplasia.63 Stratified-to-columnar metaplasia is particularly relevant to the GI tract because Barrett’s metaplasia in the esophagus is a premalignant risk factor for esophageal adenocarcinoma. Our previous work and that of others show GATA4 to be aberrantly expressed in BE and esophageal adenocarcinoma.12,13,20 Our recent in vitro studies with human esophageal cell lines demonstrate that GATA4 expression responds to acid and bile and that, when over-expressed, GATA4 represses stratified cell associated gene expression including Trp63, Krt5, and Krt15.20 The correlation between dysregulated gene expression patterns in BE and GATA4 mutants, as well as our finding that many of these genes have functional GATA4 binding sites, support the idea that GATA4 does not merely serve as a disease marker but that GATA4 plays a functional role in disease perhaps by supporting maintenance of columnar epithelium in BE. We propose that GATA4 regulates multiple BE-associated pathways by activating or repressing the expression of essential columnar and stratified squamous cell–associated transcription factors and their targets. For example, FOXA2, a columnar cell–associated transcription factor upregulated in BE, and TFAP2C, a stratified squamous cell–associated transcription factor downregulated in BE, both contain functional GATA4 binding sites and were misregulated in GATA4-expressing FS, with FOXA2 being induced and TFAP2C being repressed.64,65 These particular transcription factors were recently shown to be consistently altered across multiple BE expression studies with FOXA2 being activated and TFAP2C being suppressed in metaplastic cells.64, 65 Given that GATA4 mutant HS and FS epithelial cells more closely aligned with gastric corpus identity, rather than with intestinal identity and that we used GATA4 chromatin binding data gathered from HS epithelium, the correlations drawn related to BE likely align more closely with gastric-type rather than intestinal-type metaplasia. Although we propose that developmental studies like ours inform our understanding of disease, we cannot with certainty conclude that GATA4 contributes to BE pathogenesis. Further studies using mice with GATA4 induction in the adult mouse esophagus or with alternative models such as human cell or organoid cultures from normal and BE tissues in which GATA4 is modulated will be crucial to understanding the role of GATA4 in BE.
While our paper was being revised, the Kawaguchi group published a study complementary to ours examining the relationship between SOX2 and GATA4 during mouse gastric development.66 Their study supports the findings we report here, demonstrating an essential role for GATA4 in specifying columnar epithelium in the developing stomach. Our study adds molecular mechanistic depth to the understanding of how GATA4 acts as an essential patterning factor during gastric epithelium development. By combining morphological tissue analyses with unbiased, global gene expression and chromatin binding approaches, we place GATA4 at the top of a transcriptional cascade that defines columnar epithelial cell identity. GATA4 activates expression of a large cohort of genes essential to direct columnar epithelial cell morphogenesis and differentiation to mature epithelial cell types. Equally important, but not previously well recognized, is GATA4’s function to enforce columnar epithelial cell fate by directly repressing expression of essential squamous cell patterning transcription factors and their targets. Overall, our data support a role for GATA4 as a vital, cell autonomous transcriptional regulator that patterns the HS epithelium during development.
Material and Methods
Mice
Mouse lines used to generate control and experimental samples were ShhCre, Gata4loxP, Gata4–, Rosa26lnlG4, Gata6loxP, Gata6−, Gata4flbio/flbio, and Rosa26BirA.17,67, 68, 69, 70, 71, 72 Genotypes of embryos compared in Gata4 cKO studies were Gata4loxP/loxP, Gata4loxP/+, Gata4loxP/–, or Gata4+/+::ShhCre control embryos and G4 cKO Gata4loxP–-::ShhCre mutants. Genotypes of embryos compared in Gata4 cKI studies were Rosa26 lnlG4/+ and Rosa26lnlG4/lnlG4 control embryos and G4 cKI Gata4lnlG4/lnlG4::ShhCre mutants. Genotypes of embryos compared in Gata6 cKO studies were Gata6loxP/+ control and Gata6loxP/-::ShhCre mutants. For ChIP studies, Gata4flbio/flbio::Rosa26BirA/BirA and control Rosa26BirA/BirA mice were compared. Embryos were derived via timed breeding with a vaginal plug at noon considered E0.5. Embryos at E12.5, E14.5, E16.5, or E18.5 were analyzed in G4 cKO, G4 cKI, and G6 cKO studies. Tail tip, ear punch, or yolk sac DNA were used for PCR genotyping (primers listed in Table 1). Sample sizes for each comparison are reported in figure legends. The Medical College of Wisconsin’s Institutional Animal Care and Use Committee approved all animal procedures.
Table 1.
Genotyping Primers
| Gene | Forward | Reverse | Size (bp) |
|---|---|---|---|
| ShhCre | gggacagctcacaagtcctc | ggtgcgctcctggacgta | 350 |
| Gata4 cKO alleles | |||
| Gata4loxP | cccagtaaagaagtcagcacaaggaaac | agactattgatcccggagtgaacatt | 355, wt 455, loxP |
| Gata4null | ggggcaggacagcaagggggaggatt | gtgagacctgcagaatgggagtggagaatg | 207 |
| Gata4 cKI alleles | |||
| Rosa26G4 | aaagtcgctctgagttgttat | gcgaagagtttgtcctcaacc | 311 |
| Rosa26WT | aaagtcgctctgagttgttat | ggagcgggagaaatggatatg | 603 |
| Gata6 cKO alleles | |||
| Gata6loxP | gtggttgtaaggcggtttgt | acgcgagctccagaaaaagt | 159, wt 250, loxP |
| Gata6null | gctccaccctactatgaccaattcc | cccggtttaaaaatctgcttgagtc | 400 |
| Gata4flbio and BirA alleles | |||
| Gata4Flbio | cagtgctgtctgctctgaagctgt | ccaaggtgggcttctctgtaagaac | 378, G4wt 550, G4Flbio |
| BirAJax14/15 | ttcagacactgcgtgact | ggctccaatgactatttgc | 500 (BirA+) |
| BirAJax16/17 | gtgtaactgtggacagaggag | gaacttgatgtgtagaccagg | 400 (BirA-) |
Histochemistry, Immunohistochemistry, and Immunofluorescence Tissue Staining
Embryonic stomachs (E12.5–E18.5) were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 5 μm. H&E staining was performed per standard procedures. Citric acid antigen retrieval was used for immunohistochemistry (IHC) and immunofluorescence (IF) with antibodies listed in Table 2. IHC staining was visualized using R.T.U Vectastain Elite ABC reagent (Vector Labs, Burlingame, CA) and Metal Enhanced DAB substrate kit (Thermo Fisher Scientific, Waltham, MA). H&E and IHC images were obtained using a NanoZoomer slide scanner and NDP.view 2 software (Hamamatsu, Hamamatsu, Japan), a Nikon Eclipse 80i microscope (Nikon Instruments Inc., Melville, NY) with a Nikon digital smart DS-QiMc camera and SPOT Advanced Imaging software version 5.1, or a Nikon Ti2 U microscope with a Nikon Digital Sight color camera and NIS Elements software. IF images were obtained using a Zeiss LSM980 laser scanning confocal microscope (Zeiss, Oberkochen, Germany). Files were converted to tif files using the Fiji interface for ImageJ version 2.1.0/1.53c; Java 1.8.0_202 software (National Institutes of Health, Bethesda, MD). All H&E, IHC, and IF images were processed for presentation in figures using Photoshop (Adobe, San Jose, CA). Control and mutant images for each stain were processed identically.
Table 2.
Antibodies
| Antibody | Dilution | Manufacturer | Catalog Number |
|---|---|---|---|
| ATP4A (mouse polyclonal) | 1:100 | Gift from Dr Eunyoung Choi | n/a |
| E-cadherin (mouse monoclonal) | 1:4000 | BD Biosciences (Franklin Lakes, NJ) | 610181 |
| GATA4 (rabbit monoclonal) | 1:500 | Cell Signaling Technology (Danvers, MA) | 36966 |
| GATA4 C-20 (goat polyclonal) | 1:250 | Santa Cruz Biotechnology (Santa Cruz, CA) | sc-1237 |
| GATA6 (rabbit monoclonal) | 1:500 | Cell Signaling Technology (Danvers, MA) | 5851 |
| GIF (goat polyclonal) | 1:2000 | Gift from Dr Jason Mills | n/a |
| KRT5 (rabbit polyclonal) | 1:4000 | Biolegend (formerly Covance) (Dedham, MA) | 905501 |
| KRT13 (rabbit monoclonal) | 1:250 | Abcam (Cambridge, United Kingdom) | AB92551 |
| KRT14 (LL002) (mouse monoclonal) | 1:1600 | Thermo Fisher Scientific (Rockford, IL) | MA5-11599 |
| MUC5AC 45M1 (mouse monoclonal) | 1:100 | Thermo Fisher Scientific(Rockford, IL) | MA1-38223 |
| TRP63 (rabbit polyclonal) | 1:250 | Abcam (Cambridge, United Kingdom) | AB53039 |
| TRP63 (rabbit monoclonal) | 1:500 | Cell Signaling Technology (Danvers, MA) | 13109 |
| Biotinylated horse-anti mouse IgG (H+L) | 1:66 | Vector Laboratories (Burlingame, CA) | BA-2000 |
| Biotinylated goat anti-rabbit IgG (H+L) | 1:66 | Vector Laboratories (Burlingame, CA) | BA-1000 |
| Biotinylated rabbit anti-goat IgG (H+L) | 1:66 | Vector Laboratories (Burlingame, CA) | BA-5000 |
| Alexa-Fluor 488 Donkey anti-mouse | 1:133 | Thermo Fisher Scientific (Rockford, IL) | A-21202 |
| Alexa-Fluor 594 Donkey anti-rabbit | 1:133 | Thermo Fisher Scientific (Rockford, IL) | A-21207 |
Quantitative RT-PCR
HS or FS epithelial cells were isolated from E14.5 or E18.5 HS or FS,73 and total RNA from cells was DNase-treated as in previous studies or with EZ-DNase (Invitrogen, Waltham, MA).73,74 MMLV reverse transcriptase (Invitrogen) or VILO master mix (Invitrogen) were used to synthesize complementary DNA. TaqMan assays (Table 3) were used for qRT-PCR with TaqMan Gene Expression Master Mix (Applied Biosystems, Waltham, MA). For each gene assayed, at least 3 biological replicates per genotype were assayed in 3 independent experiments. Gapdh was used to normalize. For each target, expression units were calculated using the formula [2(–dCq)]×1000.15 Error bars show SEM. Statistical tests used were either a Student’s t test or 1-way analysis of variance, as appropriate. Figure legends specify which statistical tests were used for each analysis.
Table 3.
TaqMan Assay Identifiers
| Gene | TaqMan ID |
|---|---|
| Atp4a | Mm00444417_m1 |
| Cldn10 | Mm01226326_g1 |
| Cldn18 | Mm00517321_m1 |
| Gata4 | Mm00484689_m1 |
| Gata6 | Mm00802632_m1 |
| Gif | Mm00433596_m1 |
| Hnf4a | Mm00433964_m1 |
| Krt15 | Mm00492972_m1 |
| Krt17 | Mm00495207_m1 |
| Krt23 | Mm00840789_m1 |
| Mist1 | Mm00627532_s1 |
| Muc5ac | Mm01276718_m1 |
| Muc6 | Mm00725165_m1 |
| Pgc | Mm00482488_m1 |
| Tff1 | Mm00436945_m1 |
| Tff2 | Mm00447491_m1 |
| Trp63 | Mm00495793_m1 |
| Gapdh | 4351309 |
RNA-Sequencing
Epithelial cells were isolated from E14.5 control and mutant embryos as previously described.73 Total epithelial cell RNA (20 ng) per biological replicate spiked with ERCC RNA Spike-In Controls (Thermo Fisher Scientific) was subjected to Poly-A mRNA selection (New England Biolabs, Ipswich, MA) (n = 3 control HS, 3 control FS, 5 G4 cKO HS, 3 G4 cKI FS). The NEBnext Ultra II Directional RNA library prep kit for Illumina (New England Biolabs) was used to generate libraries. All libraries were sequenced as paired-end (38 × 2, total of 76 cycles) using an Illumina NextSeq 500 (Illumina, San Diego, CA). The raw RNA sequence reads were mapped to mouse reference genome (build mm9) using STAR in Base Pair Tech (basepairtech.com) (default parameters) and normalized with ERCC spike-ins. Differential expression analysis was completed using the DESeq package. We performed DESeq between FS and HS control data sets to generate HSE and FSE gene sets (P ≤ .05, fold change ≥2). Genes up- or downregulated in G4 cKO HS mutants and G4 cKI FS mutants were defined similarly (P ≤ .05, fold change ≥2 vs control embryos). Heatmaps were generated using RNA-seq FPKM values imported into Heatmapper.75
ChIP-Sequencing
GATA4-Bio-ChIP using HS epithelial cells from adult Gata4flbio/flbio::Rosa26BirA/BirA experimental and Gata4wt/wt::Rosa26BirA/BirA negative control animals was performed as previously described.17,70,71 The NEBNext Ultra II DNA Library Prep Kit (New England Biolabs) was used to make libraries. All libraries were sequenced using an Illumina NextSeq 500. The sequencing reads were aligned to the mouse genome (mm9 build) using Bowtie2 (default parameters) via Base Pair Tech. Peak calling was done using in-house script employing MACS2 algorithm with a q-value cutoff of 0.05 and model fold of 5,50. A total of 50,869 GATA4 peaks were called and annotated with nearest refseq genes using annotatePeaks.pl script in HOMER v4.11.1 (default settings). The distribution of number of peaks across center of TSS was plotted. The genes were imported into DAVID pathway analysis. GO TERM: BP_FAT was reported.
ChIP-Seq and RNA-Seq Combined Analyses
A list of genes with GATA4 binding peaks in proximal regulatory regions (±2 kb of TSS) was constructed from the genes identified as having HSE or FSE expression and differential expression in G4 cKO or G4 cKI compared with control embryos. Transcription factor motifs were identified within ±100 bp of GATA4 peak center using findMotifsGenome.pl script in HOMER v4.11.1 with default settings.
Acknowledgments
A preprint of this work was published on bioRxiv at doi: https://doi.org/10.1101/2020.08.18.251587.
All data in this publication have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus and are accessible through GEO Series accession number GSE156324 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156324).
The authors thank Oscar Rosas Mejia and Rebekah Mokry for technical assistance and Dr. Teresa Patitucci for assistance with illustration.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (DK111822 [to MAB]); the Medical College of Wisconsin Cancer Center (to MAB) and Digestive Diseases Center (to MAB); the National Institutes of Health, National Cancer Institute (CA204231 [to SR]); and the American Cancer Society (PF-19-023-01-DDC [to AD]).
Supplementary Material
References
- 1.Willet S.G., Mills J.C. Stomach organ and cell lineage differentiation: from embryogenesis to adult homeostasis. Cell Mol Gastroenterol Hepatol. 2016;2:546–559. doi: 10.1016/j.jcmgh.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi E., Roland J.T., Barlow B.J., O’Neal R., Rich A.E., Nam K.T., Shi C., Goldenring J.R. Cell lineage distribution atlas of the human stomach reveals heterogeneous gland populations in the gastric antrum. Gut. 2014;63:1711–1720. doi: 10.1136/gutjnl-2013-305964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Que J., Garman K.S., Souza R.F., Spechler S.J. Pathogenesis and cells of origin of Barrett’s esophagus. Gastroenterology. 2019;157:349–364.e1. doi: 10.1053/j.gastro.2019.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson C.A., DeLaForest A., Battle M.A. Patterning the gastrointestinal epithelium to confer regional-specific functions. Dev Biol. 2018;435:97–108. doi: 10.1016/j.ydbio.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim T.H., Shivdasani R.A. Stomach development, stem cells and disease. Development. 2016;143:554–565. doi: 10.1242/dev.124891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis R., Guo H., Streutker C., Ahmed M., Yung T., Dirks P.B., He H.H., Kim T.H. Gastrointestinal transcription factors drive lineage-specific developmental programs in organ specification and cancer. Sci Adv. 2019;5 doi: 10.1126/sciadv.aax8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nekulova M., Holcakova J., Coates P., Vojtesek B. The role of P63 in cancer, stem cells and cancer stem cells. Cell Mol Biol Lett. 2011;16:296–327. doi: 10.2478/s11658-011-0009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng R., Blobel G.A. GATA transcription factors and cancer. Genes Cancer. 2010;1:1178–1188. doi: 10.1177/1947601911404223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Z., Yan B. Multiple roles and regulatory mechanisms of the transcription factor GATA6 in human cancers. Clin Genet. 2020;97:64–72. doi: 10.1111/cge.13630. [DOI] [PubMed] [Google Scholar]
- 10.Ayanbule F., Belaguli N.S., Berger D.H. GATA factors in gastrointestinal malignancy. World J Surg. 2011;35:1757–1765. doi: 10.1007/s00268-010-0950-1. [DOI] [PubMed] [Google Scholar]
- 11.Dulak A.M., Schumacher S.E., van Lieshout J., Imamura Y., Fox C., Shim B., Ramos A.H., Saksena G., Baca S.C., Baselga J., Tabernero J., Barretina J., Enzinger P.C., Corso G., Roviello F., Lin L., Bandla S., Luketich J.D., Pennathur A., Meyerson M., Ogino S., Shivdasani R.A., Beer D.G., Godfrey T.E., Beroukhim R., Bass A.J. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–4393. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller C.T., Moy J.R., Lin L., Schipper M., Normolle D., Brenner D.E., Iannettoni M.D., Orringer M.B., Beer D.G. Gene amplification in esophageal adenocarcinomas and Barrett’s with high-grade dysplasia. Clinical Cancer Research. 2003;9:4819–4825. [PubMed] [Google Scholar]
- 13.Haveri H., Westerholm-Ormio M., Lindfors K., Mäki M., Savilahti E., Andersson L.C., Heikinheimo M. Transcription factors GATA-4 and GATA-6 in normal and neoplastic human gastrointestinal mucosa. BMC Gastroenterol. 2008;8:9. doi: 10.1186/1471-230X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker E.M., Thompson C.A., Battle M.A. GATA4 and GATA6 regulate intestinal epithelial cytodifferentiation during development. Dev Biol. 2014;392:283–294. doi: 10.1016/j.ydbio.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker E.M., Thompson C.A., Kohlnhofer B.M., Faber M.L., Battle M.A. Characterization of the developing small intestine in the absence of either GATA4 or GATA6. BMC Res Notes. 2014;7:902. doi: 10.1186/1756-0500-7-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohlnhofer B.M., Thompson C.A., Walker E.M., Battle M.A. GATA4 regulates epithelial cell proliferation to control intestinal growth and development in mice. Cell Mol Gastroenterol Hepatol. 2016;2:189–209. doi: 10.1016/j.jcmgh.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson C.A., Wojta K., Pulakanti K., Rao S., Dawson P., Battle M.A. GATA4 is sufficient to establish jejunal versus ileal identity in the small intestine. Cell Mol Gastroenterol Hepatol. 2017;3:422–446. doi: 10.1016/j.jcmgh.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobsen C.M., Narita N., Bielinska M., Syder A.J., Gordon J.I., Wilson D.B. Genetic mosaic analysis reveals that GATA-4 is required for proper differentiation of mouse gastric epithelium. Dev Biol. 2002;241:34–46. doi: 10.1006/dbio.2001.0424. [DOI] [PubMed] [Google Scholar]
- 19.Rodríguez-Seguel E., Villamayor L., Arroyo N., de Andrés M.P., Real F.X., Martín F., Cano D.A., Rojas A. Loss of GATA4 causes ectopic pancreas in the stomach. J Pathol. 2020;250:362–373. doi: 10.1002/path.5378. [DOI] [PubMed] [Google Scholar]
- 20.Stavniichuk R., DeLaForest A., Thompson C.A., Miller J., Souza R.F., Battle M.A. GATA4 blocks squamous epithelial cell gene expression in human esophageal squamous cells. Sci Rep. 2021;11:3206. doi: 10.1038/s41598-021-82557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molkentin J.D. The zinc finger-containing transcription factors GATA-4, -5, and -6: Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 22.Chlon T.M., Crispino J.D. Combinatorial regulation of tissue specification by GATA and FOG factors. Development. 2012;139:3905–3916. doi: 10.1242/dev.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rojas A., Schachterle W., Xu S.M., Martín F., Black B.L. Direct transcriptional regulation of Gata4 during early endoderm specification is controlled by FoxA2 binding to an intronic enhancer. Dev Biol. 2010;346:346–355. doi: 10.1016/j.ydbio.2010.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watt A.J., Zhao R., Li J., Duncan S.A. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev Biol. 2007;7:37. doi: 10.1186/1471-213X-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Battle M.A., Bondow B.J., Iverson M.A., Adams S.J., Jandacek R.J., Tso P., Duncan S.A. GATA4 is essential for jejunal function in mice. Gastroenterology. 2008;135:1676–1686. doi: 10.1053/j.gastro.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beuling E., Bosse T., aan de Kerk D.J., Piaseckyj C.M., Fujiwara Y., Katz S.G., Orkin S.H., Grand R.J., Krasinski S.D. GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors. Dev Biol. 2008;322:179–189. doi: 10.1016/j.ydbio.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobsen C.M., Mannisto S., Porter-Tinge S., Genova E., Parviainen H., Heikinheimo M., Adameyko I.I., Tevosian S.G., Wilson D.B. GATA-4:FOG interactions regulate gastric epithelial development in the mouse. Dev Dyn. 2005;234:355–362. doi: 10.1002/dvdy.20552. [DOI] [PubMed] [Google Scholar]
- 28.Kataoka K., Sakano Y., Miura J. Histogenesis of the mouse gastric mucosa, with special reference to type and distribution of proliferative cells. Arch Histol Jpn. 1984;47:459–474. doi: 10.1679/aohc.47.459. [DOI] [PubMed] [Google Scholar]
- 29.Karam S.M., Leblond C.P. Identifying and counting epithelial cell types in the “corpus” of the mouse stomach. Anat Rec. 1992;232:231–246. doi: 10.1002/ar.1092320208. [DOI] [PubMed] [Google Scholar]
- 30.Kim T.-H., Shivdasani R. In: Translational Research and Discovery in Gastroenterology: Organogenesis to Disease. Gumucio D., Samuelson L., Spence J., editors. Wiley-Blackwell; London, United Kingdom: 2014. Basic science of stomach development; pp. 43–56. [Google Scholar]
- 31.Sayols S., Klassek J., Werner C., Möckel S., Ritz S., Mendez-Lago M., Soshnikova N. Signalling codes for the maintenance and lineage commitment of embryonic gastric epithelial progenitors. Development. 2020;147 doi: 10.1242/dev.188839. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCracken K.W., Aihara E., Martin B., Crawford C.M., Broda T., Treguier J., Zhang X., Shannon J.M., Montrose M.H., Wells J.M. Wnt/β-catenin promotes gastric fundus specification in mice and humans. Nature. 2017;541:182–187. doi: 10.1038/nature21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maitra M., Schluterman M.K., Nichols H.A., Richardson J.A., Lo C.W., Srivastava D., Garg V. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol. 2009;326:368–377. doi: 10.1016/j.ydbio.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nadeau M., Georges R.O., Laforest B., Yamak A., Lefebvre C., Beauregard J., Paradis P., Bruneaud B.G., Andelfinger G., Nemera M. An endocardial pathway involving tbx5, gata4, and nos3 required for atrial septum formation. Proc Natl Acad Sci U S A. 2010;107:19356–19361. doi: 10.1073/pnas.0914888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ang Y.S., Rivas R.N., Ribeiro A.J.S., Srivas R., Rivera J., Stone N.R., Pratt K., Mohamed T.M.A., Fu J.D., Spencer C.I., Tippens N.D., Li M., Narasimha A., Radzinsky E., Moon-Grady A.J., Yu H., Pruitt B.L., Snyder M.P., Srivastava D. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. 2016;167:1734–1749.e22. doi: 10.1016/j.cell.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viger R.S., Guittot S.M., Anttonen M., Wilson D.B., Heikinheimo M. Role of the GATA family of transcription factors in endocrine development, function, and disease. Mol Endocrinol. 2008;22:781–798. doi: 10.1210/me.2007-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schteingart H.F., Picard J.Y., Valeri C., Marshall I., Treton D., di Clemente N., Rey R.A., Josso N. A mutation inactivating the distal SF1 binding site on the human anti-Müllerian hormone promoter causes persistent Müllerian duct syndrome. Hum Mol Genet. 2019;28:3211–3218. doi: 10.1093/hmg/ddz147. [DOI] [PubMed] [Google Scholar]
- 39.van Loo P.F., Mahtab E.A.F., Wisse L.J., Hou J., Grosveld F., Suske G., Philipsen S., Gittenberger-de Groot A.C. Transcription factor Sp3 knockout mice display serious cardiac malformations. Mol Cell Biol. 2007;27:8571–8582. doi: 10.1128/MCB.01350-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni W., Perez A.A., Schreiner S., Nicolet C.M., Farnham P.J. Characterization of the ZFX family of transcription factors that bind downstream of the start site of CpG island promoters. Nucl Acids Res. 2020;48:5986–6000. doi: 10.1093/nar/gkaa384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sporici R.A., Hodskins J.S., Locasto D.M., Meszaros L.B., Ferry A.L., Weidner A.M., Rinehart C.A., Bailey J.C., Mains I.M., Diamond S.E. Repression of the prolactin promoter: a functional consequence of the heterodimerization between Pit-1 and Pit-1 β. J Mol Endocrinol. 2005;35:317–331. doi: 10.1677/jme.1.01678. [DOI] [PubMed] [Google Scholar]
- 42.Kasaai B., Gaumond M.H., Moffatt P. Regulation of the bone-restricted IFITM-like (Bril) gene transcription by Sp and Gli family members and CpG methylation. J Biol Chem. 2013;288:13278–13294. doi: 10.1074/jbc.M113.457010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson R.P., Tekki-Kessaris N., Boulter C.A. Characterisation, chromosomal localisation and expression of the mouse Kid3 gene. Biochim Biophys Acta. 2000;1490:153–158. doi: 10.1016/s0167-4781(99)00239-0. [DOI] [PubMed] [Google Scholar]
- 44.Longobardi E., Penkov D., Mateos D., de Florian G., Torres M., Blasi F. Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Dev Dyn. 2014;243:59–75. doi: 10.1002/dvdy.24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maroni G., Tkachuk V.A., Egorov A., Morelli M.J., Luongo R., Levantini E., Blasi F., Magli M.C., Penkov D. Prep1 prevents premature adipogenesis of mesenchymal progenitors. Sci Rep. 2017;7:15573. doi: 10.1038/s41598-017-15828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciccarelli M., Vastolo V., Albano L., Lecce M., Cabaro S., Liotti A., Longo M., Oriente F., Russo G.L., Macchia P.E., Formisano P., Beguinot F., Ungaro P. Glucose-induced expression of the homeotic transcription factor Prep1 is associated with histone post-translational modifications in skeletal muscle. Diabetologia. 2016;59:176–186. doi: 10.1007/s00125-015-3774-6. [DOI] [PubMed] [Google Scholar]
- 47.Salgado-Albarrán M., González-Barrios R., Guerra-Calderas L., Alcaraz N., Estefanía Sánchez-Correa T., Castro-Hernández C., Sánchez-Pérez Y., Aréchaga-Ocampo E., García-Carrancá A., Cantú de León D., Herrera L.A., Baumbach J., Soto-Reyes E. The epigenetic factor BORIS (CTCFL) controls the androgen receptor regulatory network in ovarian cancer. Oncogenesis. 2019;8:41. doi: 10.1038/s41389-019-0150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sulahian R., Chen J., Arany Z., Jadhav U., Peng S., Rustgi A.K., Bass A.J., Srivastava A., Hornick J.L., Shivdasani R.A. SOX15 governs transcription in human stratified epithelia and a subset of esophageal adenocarcinomas. Cell Mol Gastroenterol Hepatol. 2015;1:598–609.e6. doi: 10.1016/j.jcmgh.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hyland P.L., Hu N., Rotunno M., Su H., Wang C., Wang L., Pfeiffer R.M., Gherman B., Giffen C., Dykes C., Dawsey S.M., Abnet C.C., Johnson K.M., Acosta R.D., Young P.E., Cash B.D., Taylor P.R. Global changes in gene expression of Barrett’s esophagus compared to normal squamous esophagus and gastric cardia tissues. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q., Agoston A.T., Pham T.H., Zhang W., Zhang X., Huo X., Peng S., Bajpai M., Das K., Odze R.D., Spechler S.J., Souza R.F. Acidic bile salts induce epithelial to mesenchymal transition via VEGF signaling in non-neoplastic Barrett’s cells. Gastroenterology. 2019;156:130–144.e10. doi: 10.1053/j.gastro.2018.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quante M., Bhagat G., Abrams J.A., Marache F., Good P., Lee M.D., Lee Y., Friedman R., Asfaha S., Dubeykovskaya Z., Mahmood U., Figueiredo J.L., Kitajewski J., Shawber C., Lightdale C.J., Rustgi A.K., Wang T.C. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell. 2012;21:36–51. doi: 10.1016/j.ccr.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leushacke M., Tan S.H., Wong A., Swathi Y., Hajamohideen A., Tan L.T., Goh J., Wong E., Denil S.L.I.J., Murakami K., Barker N. Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat Cell Biol. 2017;19:774–786. doi: 10.1038/ncb3541. [DOI] [PubMed] [Google Scholar]
- 53.Han S., Fink J., Jörg D.J., Lee E., Yum M.K., Chatzeli L., Merker S.R., Josserand M., Trendafilova T., Andersson-Rolf A., Dabrowska C., Kim H., Naumann R., Lee J.H., Sasaki N., Mort R.L., Basak O., Clevers H., Stange D.E., Philpott A., Kim J.K., Simons B.D., Koo B.K. Defining the identity and dynamics of adult gastric isthmus stem cells. Cell Stem Cell. 2019;25:342–356.e7. doi: 10.1016/j.stem.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stefanovic S., Christoffels V.M. GATA-dependent transcriptional and epigenetic control of cardiac lineage specification and differentiation. Cell Mol Life Sci. 2015;72:3871–3881. doi: 10.1007/s00018-015-1974-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krendl C., Shaposhnikov D., Rishko V., Ori C., Ziegenhain C., Sass S., Simon L., Müller N.S., Straub T., Brooks K.E., Chavez S.L., Enard W., Theis F.J., Drukker M. GATA2/3-TFAP2A/C transcription factor network couples human pluripotent stem cell differentiation to trophectoderm with repression of pluripotency. Proc Natl Acad Sci U S A. 2017;114:E9579–E9588. doi: 10.1073/pnas.1708341114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wontakal S.N., Guo X., Smith C., MacCarthy T., Bresnick E.H., Bergman A., Snyder M.P., Weissman S.M., Zheng D., Skoultchi A.I. A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proc Natl Acad Sci U S A. 2012;109:3832–3837. doi: 10.1073/pnas.1121019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Majello B., de Luca P., Lania L. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J Biol Chem. 1997;272:4021–4026. doi: 10.1074/jbc.272.7.4021. [DOI] [PubMed] [Google Scholar]
- 58.Lania L., Majello B., de Luca P. Transcriptional regulation by the Sp family proteins. Int J Biochem Cell Biol. 1997;29:1313–1323. doi: 10.1016/s1357-2725(97)00094-0. [DOI] [PubMed] [Google Scholar]
- 59.Belaguli N.S., Zhang M., Rigi M., Aftab M., Berger D.H. Cooperation between GATA4 and TGF-β signaling regulates intestinal epithelial gene expression. Am J Physiol Gastrointest Liver Physiol. 2007;292:1520–1533. doi: 10.1152/ajpgi.00236.2006. [DOI] [PubMed] [Google Scholar]
- 60.Hu X., Li T., Zhang C., Liu Y., Xu M., Wang W., Jia Z., Ma K., Zhang Y., Zhou C. GATA4 regulates ANF expression synergistically with Sp1 in a cardiac hypertrophy model. J Cell Mol Med. 2011;15:1865–1877. doi: 10.1111/j.1582-4934.2010.01182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.von Salisch S., Klar M., Thurisch B., Bungert J., Dame C. Gata4 and Sp1 regulate expression of the erythropoietin receptor in cardiomyocytes. J Cell Mol Med. 2011;15:1963–1972. doi: 10.1111/j.1582-4934.2010.01193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scully K.H., Jacobson E.M., Jepsen K., Lunyak V., Viadiu H., Carriere C., Rose D.W., Hooshmand F., Aggarwal A.K., Rosenfeld M.G. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 63.Giroux V., Rustgi A.K. Metaplasia: Tissue injury adaptation and a precursor to the dysplasia-cancer sequence. Nat Rev Cancer. 2017;17:594–604. doi: 10.1038/nrc.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lv J., Guo L., Wang J.H., Yan Y.Z., Zhang J., Wang Y.Y., Yu Y., Huang Y.F., Zhao H.P. Biomarker identification and trans-regulatory network analyses in esophageal adenocarcinoma and Barrett’s esophagus. World J Gastroenterol. 2019;25:233–244. doi: 10.3748/wjg.v25.i2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang D.H., Spechler S.J., Souza R.F., Wang D.H., Tiwari A., Kim M.E., Clemons N.J., Regmi N.L., Hodges W.A., Berman D.M., Montgomery E.A., Watkins D.N., Zhang X., Zhang Q., Jie C. Hedgehog signaling regulates FOXA2 in esophageal embryogenesis and Barrett’s metaplasia. J Clin Invest. 2014;124:3767–3780. doi: 10.1172/JCI66603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sankoda N., Tanabe W., Tanaka A., Shibata H., Woltjen K., Chiba T., Haga H., Sakai Y., Mandai M., Yamamoto T., Yamada Y., Uemoto S., Kawaguchi Y. Epithelial expression of Gata4 and Sox2 regulates specification of the squamous–columnar junction via MAPK/ERK signaling in mice. Nat Commun. 2021;12:560. doi: 10.1038/s41467-021-20906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watt A.J., Battle M.A., Li J., Duncan S.A. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sodhi C.P., Li J., Duncan S.A. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC Dev Biol. 2006;6:19. doi: 10.1186/1471-213X-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Molkentin J.D., Lin Q., Duncan S.A., Olson E.N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 70.Driegen S., Ferreira R., van Zon A., Strouboulis J., Jaegle M., Grosveld F., Philipsen S., Meijer D. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgen Res. 2005;14:477–482. doi: 10.1007/s11248-005-7220-2. [DOI] [PubMed] [Google Scholar]
- 71.He A., Pu W.T. Genome-wide location analysis by pull down of in vivo biotinylated transcription factors. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb2120s92. Chapter21:Unit 21.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harfe B.D., Scherz P.J., Nissim S., Tian H., McMahon A.P., Tabin C.J. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 73.Bondow B.J., Faber M.L., Wojta K.J., Walker E.M., Battle M.A. E-cadherin is required for intestinal morphogenesis in the mouse. Dev Biol. 2012;371:1–12. doi: 10.1016/j.ydbio.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duncan S.A., Nagy A., Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4(–/–) embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- 75.Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., Wishart D.S. Heatmapper: web-enabled heat mapping for all. Nucl Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.