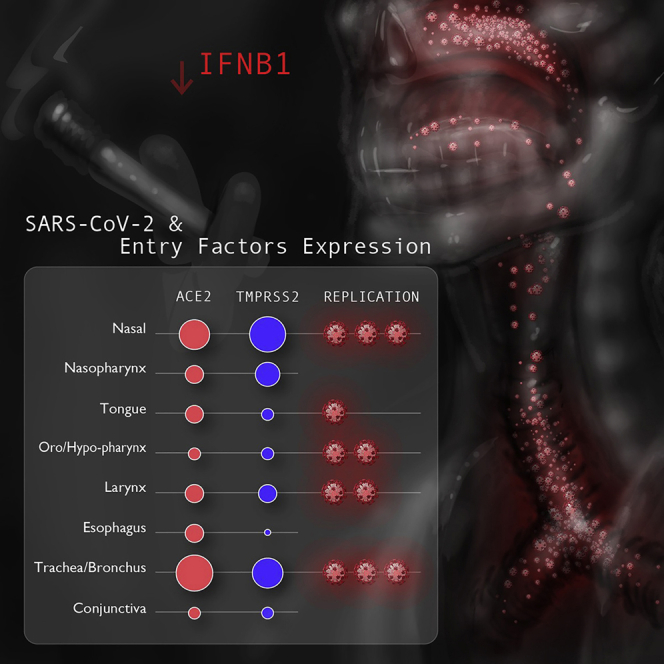
Summary
Understanding viral tropism is an essential step toward reducing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission, decreasing mortality from coronavirus disease 2019 (COVID-19) and limiting opportunities for mutant strains to arise. Currently, little is known about the extent to which distinct tissue sites in the human head and neck region and proximal respiratory tract selectively permit SARS-CoV-2 infection and replication. In this translational study, we discover key variabilities in expression of angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2), essential SARS-CoV-2 entry factors, among the mucosal tissues of the human proximal airways. We show that SARS-CoV-2 infection is present in all examined head and neck tissues, with a notable tropism for the nasal cavity and tracheal mucosa. Finally, we uncover an association between smoking and higher SARS-CoV-2 viral infection in the human proximal airway, which may explain the increased susceptibility of smokers to developing severe COVID-19. This is at least partially explained by differences in interferon (IFN)-β1 levels between smokers and non-smokers.
Keywords: SARS-CoV-2, COVID-19, ACE2, TMPRSS2, ciliated epithelial cell, IFN-β1, nasal cavity, trachea, smoking, upper airway
Graphical abstract
Highlights
-
•
ACE2 and TMPRSS2 are variably expressed among mucosal surfaces of the head and neck
-
•
Enriched TMPRSS2 expression may drive SARS-CoV-2 tropism for the nasal cavity and trachea
-
•
SARS-CoV-2 preferentially replicates in the nasal cavity and trachea of individuals with COVID-19
-
•
Smoking is associated with increased proximal airway SARS-CoV-2, in part because of IFN-β1
Nakayama et al. discover key variabilities in expression of SARS-CoV-2 entry factors among human head and neck mucosal tissues. They demonstrate that SARS-CoV-2 preferentially infects the nasal cavity and trachea. They uncover an association between smoking and higher SARS-CoV-2 in the human proximal airway, in part because of differences in IFN-β1 expression.
Introduction
The causative agent of coronavirus disease 2019 (COVID-19), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), infects host cells via initial attachment to angiotensin-converting enzyme 2 (ACE2) and subsequent priming by host entry factors, including transmembrane serine protease 2 (TMPRSS2).2 Mucosal surfaces of the head and neck, comprising the nasal and oral cavities and ocular conjunctiva, are the primary portals of entry for most respiratory viruses, including SARS-CoV-2. We and others have shown previously that nasal mucosal surfaces are a primary site for SARS-CoV-2 infection.3,1 On the other hand, the infectivity of SARS-CoV-2 in the oral mucosa is unknown. Although saliva specimens are used widely in diagnostic testing, it is uncertain whether detected viral RNA in the saliva originates from retronasal transit of secretions or from active viral replication within the oral epithelium. By contrast, the ocular mucosa does not appear to be a major transmission route for SARS-CoV-2, given the limited viral loads and low rate of positive SARS-CoV-2 detection from conjunctival swabs,4,5 although inoculation through the nasolacrimal duct (ocular-to-nasal route) has been shown to result in SARS-CoV-2 infection in a limited number of non-human primates.6
Understanding the tissue-specific tropism of SARS-CoV-2 for the distinct mucosal surfaces of the head and neck region is fundamental for prevention and treatment of viral infection. First, knowledge of SARS-CoV-2 transmission dynamics has guided use of non-pharmaceutical interventions such as face masks and physical distancing to a great effect.7 Second, aspiration of SARS-CoV-2 from nasal and/or oral secretions into the lungs is the likely initiator of viral pathogenesis and subsequent acute lung injury, the primary cause of mortality in COVID-19.3,8 Reduction of viral transmission from the upper into the lower airway will likely improve clinical outcomes. Third, variants of SARS-CoV-2 with more efficient transmission have arisen during the process of viral replication. Available evidence suggests that recent mutant strains, including the Delta variant, are associated with higher viral loads from nasal/oral swab specimens and increased efficiency of transmission.9, 10, 11 Strategies to reduce viral replication at targeted anatomic sites will likely be effective in decreasing SARS-CoV-2 mortality and transmission because viral load correlates with disease severity and infectivity.12, 13, 14 Additionally, several antiviral intranasal15,16, inhaled17,18, and oral rinse19,20 formulations targeting the upper respiratory mucosa are currently under investigation and may become important measures to reduce COVID-19 disease burden/transmission.
In this study, we comparatively analyzed the expression of essential SARS-CoV-2 entry factors, ACE2 and TMPRSS2, across various normal human proximal mucosal tissues of the head and neck (nasal, oral, pharyngeal, laryngeal, tracheal, and ocular) to gain insight into the tissue tropism and host-virus interactions of SARS-CoV-2. We next quantified the intracellular levels of SARS-CoV-2 viral infection in head and neck mucosal tissues from individuals with COVID-19 and discovered that an array of distinct proximal airway tissues serves as infection sites for SARS-CoV-2. Notably, the nasal cavity and trachea were the most prominent sites of viral infection, harboring significant quantities of viral nucleic acids even after individuals succumbed to COVID-19. Finally, comparative analysis of SARS-CoV-2 infection among individuals with COVID-19 revealed clinical characteristics, such as smoking status, that were associated with heightened intracellular SARS-CoV-2 propagation.
Results
ACE2 and TMPRSS2 are expressed in human nasal, oral, and ocular mucosal surfaces
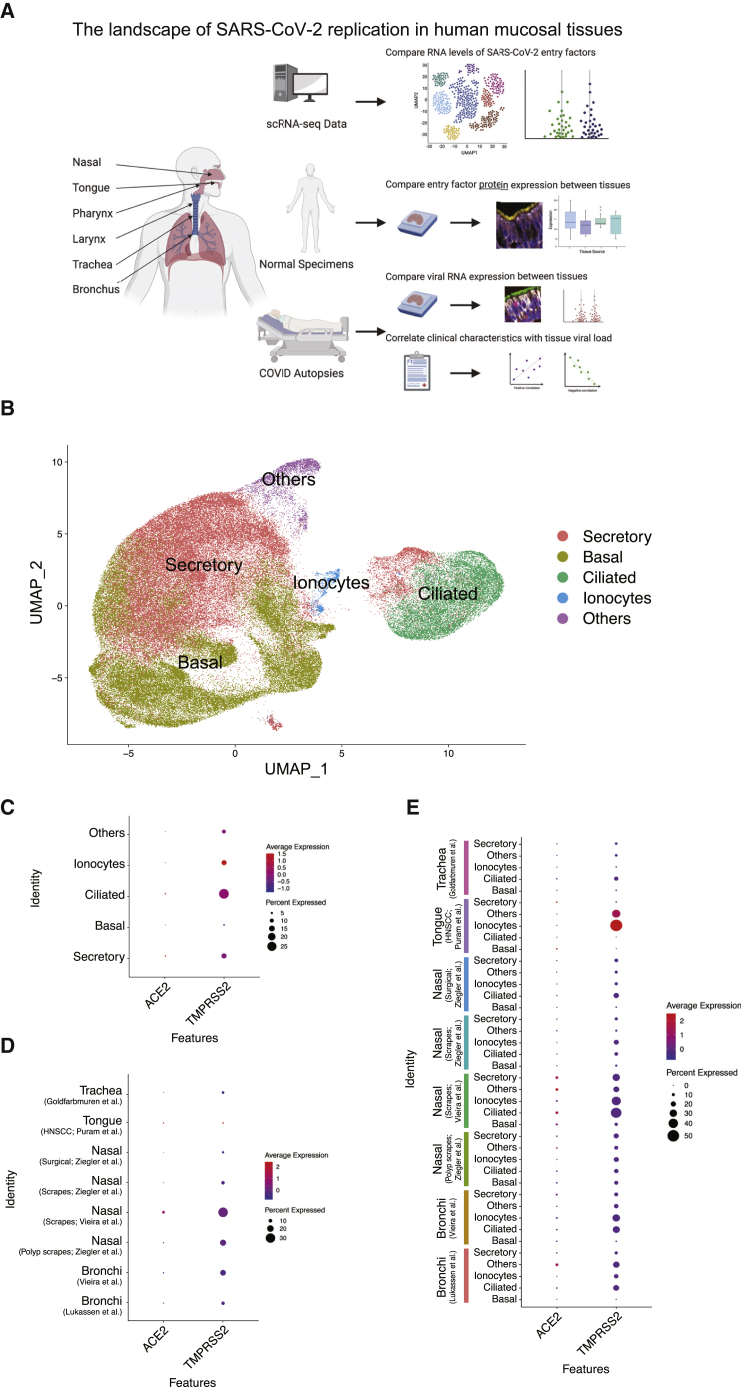
The SARS-CoV-2 entry factors ACE2 and TMPRSS2 have been reported previously to be expressed in human head and neck mucosal surfaces, including the nasal cavity,1,3,21 oral cavity,22,23 and eyes.24 A comprehensive and comparative analysis of ACE2 and TMPRSS2 protein expression across these primary barrier tissues is lacking and of paramount importance for understanding the various mucosal routes of entry for SARS-CoV-2. We therefore defined the landscape of ACE2 and TMPRSS2 expression in head and neck mucosal tissues from normal control and COVID-19 individuals (Figure 1A).
Figure 1.
Integrative analysis of ACE2 and TMPRSS2 expression in human head and neck tissues
(A) Cartoon depiction of the study design to determine the RNA and protein expression levels of ACE2 and TMPRSS2 as well as tissues harboring SARS-CoV-2 RNA in human head and neck mucosal tissues. The figure was generated using BioRender.
(B) A UMAP dimensional reduction representation of the single-cell gene expression data from 8 tissue sources and their annotated cell types. See additional details in STAR Methods.
(C) RNA expression levels of ACE2 and TMPRSS2 within each epithelial cell type.
(D) RNA expression levels of ACE2 and TMPRSS2 across the 8 tissue sources.
(E) RNA expression levels of ACE2 and TMPRSS2 in each cell type across the 8 tissue sources.
Datasets were derived from a variety of tissues from healthy individuals25, 26, 27,28 and individuals with chronic rhinosinusitis,26,27 head and neck squamous cell carcinoma (HNSCC),29 and unclear disease history.30 See also Figure S1.
We first integrated RNA expression from a number of published single-cell RNA sequencing (scRNA-seq) datasets (Figure 1B).25, 26, 27, 30, 28, 29, 31 We focused our analysis on epithelial cells, given that this population is the most accessible and plausible initial target of viral entry and also the most abundant cell population in head and neck mucosal tissues.21,32,33 We annotated five major epithelial cell types using a number of well-established markers, including those described here (Figures 1B, S1A, and S1B).34
ACE2 expression was generally sparse in the reported transcriptome datasets (Figures 1C–1E) but highly expressed when present in the ciliated and secretory epithelial cells of head and neck tissues (Figure 1C). We observed variable levels of ACE2 expression across the nasal, tracheal, and bronchial tissues with a subset of epithelial cells from each tissue type demonstrating elevated ACE2 RNA expression (Figures 1D and 1E), although anatomy-specific comparisons were inconclusive because of sparse expression of ACE2 RNA and the low percentage of ACE2-positive cells (Figures 1C–1E). Our analysis found TMPRSS2 expression to be more robust and detectable in epithelial cells, especially in ciliated cells, of these head and neck tissues (Figures 1C–1E). Comparison of TMPRSS2 expression levels across these tissue types suggested higher expression in the nasal cavity, trachea, and bronchus but lower expression in the tongue (Figure 1D). We also analyzed the expression level of other putative SARS-CoV-2 entry factors, TMPRSS4,35 transferrin receptor (TFRC),36 and neuropilin 1 (NRP1).37,38 TMPRSS4 was expressed in all epithelial cells, particularly secretory and basal cells of the nasal cavity. TFRC expression was highest in the tongue, and NRP1 was very sparsely expressed overall (Figures S1C–S1E). These analyses were notably limited, given that these RNA datasets largely derived from diseased tissues rather than healthy donor controls and the limitations of detecting low-abundance transcripts, such as ACE2, via single-cell transcriptomics studies.
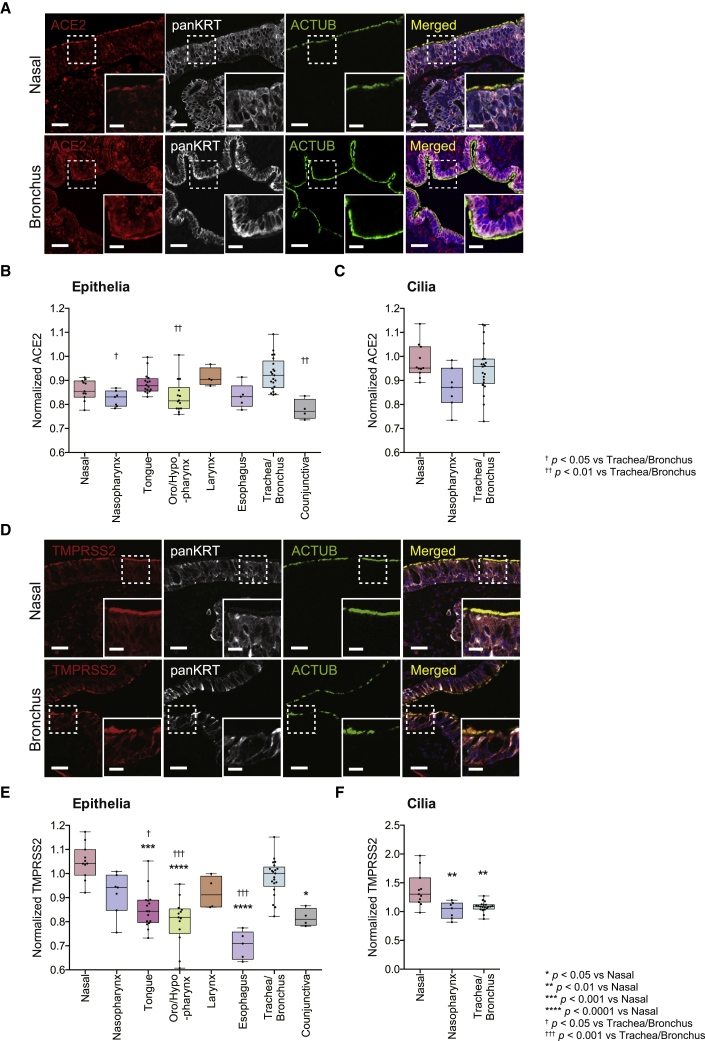
Because proteins are the functional products of RNA transcripts, we next investigated the protein expression of the two most established viral entry factors, ACE2 and TMPRSS2, across an array of normal human mucosal epithelia to further define the potential relevance of each tissue to SARS-CoV-2 transmission dynamics. We performed immunofluorescence staining of these tissues simultaneously using previously validated antibodies against ACE21 (see Figure 2A for further antibody validation), the epithelial marker pan-cytokeratin (panKRT), and the cilium marker acetylated α-tubulin (ACTUB). We observed robust ACE2 expression within the epithelia of all stained tissues (Figures 3A and S2). Consistent with our prior findings,1 this protein expression was prominent in the cilium organelle of the ciliated epithelial tissues in the nasal cavity, nasopharynx, and trachea/bronchus (Figures 3A and S2). Quantitative analysis of these data showed ACE2 expression to be higher in the trachea/bronchus compared with the nasopharynx, oro/hypopharynx, and conjunctiva (Figure 3B; p < 0.05). There were no statistical differences in ACE2 expression between the trachea/bronchus, nasal cavity, tongue, larynx, and esophagus (Figure 3B). We also did not observe any significant differences in ACE2 expression within the motile cilia of the analyzed ciliated tissues (Figure 3C). ACE2 is expressed among numerous mucosal surfaces of the head and neck with modest inter-regional variance.
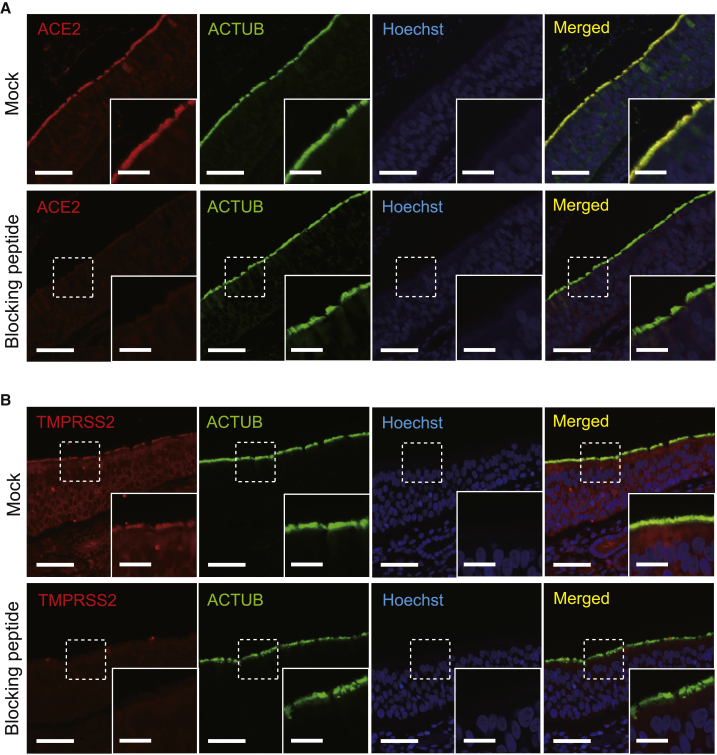
Figure 2.
Pre-incubation with immunizing peptides blocks ACE2 and TMPRSS2 antibody staining in the cilia and epithelia
(A) ACE2 protein expression in the cilia and epithelia (mock treated) is abolished when an anti-ACE2 antibody is pre-incubated with the immunizing peptide prior to immunofluorescence staining. Acetylated α-tubulin (ACTUB) is a marker of motile cilia. The nuclei were stained using Hoechst (blue) as a counterstain.
(B) TMPRSS2 protein expression in the cilia and epithelia (mock treated) is abolished when an anti-TMPRSS2 antibody is pre-incubated with the immunizing peptide prior to immunofluorescence staining. ACTUB is a marker of motile cilia. The nuclei were stained using Hoechst (blue) as a counterstain.
Scale bars, 50 μm and 20 μm (insets).
Figure 3.
Comparative analysis of ACE2 and TMPRSS2 protein expression in normal human head and neck tissues
(A) Representative immunofluorescence staining of ACE2, the epithelial marker pan-cytokeratin (panKRT), and the cilium marker ACTUB on normal human nasal cavity tissue (top row) and bronchus tissue (bottom row). Note co-localization of ACE2 and panKRT/ACTUB staining in the epithelia/cilia, respectively. Nuclei were stained using Hoechst (blue) as a counterstain.
(B) Quantification of ACE2 in the epithelia of normal human mucosal tissues of the head and neck. ACE2 protein expression was statistically significantly higher in the trachea/bronchus (0.93 ± 0.07, n = 21) compared with the nasopharynx (0.82 ± 0.03, n = 7), oro/hypopharynx (0.83 ± 0.07, n = 14), and conjunctiva (0.78 ± 0.04, n = 4). (Kruskal-Wallis test p < 0.0001; Dunn’s multiple comparison post hoc test, †adjusted p = 0.0130, ††adjusted p = 0.0015, and ††adjusted p = 0.0025, respectively).
(C) Quantification of ACE2 within the cilia of normal human mucosal tissues of the head and neck. No significant differences in ACE2 expression were detected between cilia of the nose (n = 11), nasopharynx (n = 7), and trachea/bronchus (n = 21). (Kruskal-Wallis test, p = 0.0624). The larynx tissue used in Figure 3B did not contain cilia.
(D) Representative immunofluorescence staining of TMPRSS2, panKRT, and ACTUB in normal human nasal (top row) and bronchial tissues (bottom row). Nuclei were stained using Hoechst (blue) as a counterstain. Similar to ACE2, note co-localization of TMPRSS2 and panKRT/ACTUB staining in the epithelia/cilia, respectively.
(E) Quantification of TMPRSS2 in the epithelia of normal human mucosal tissues. TMPRSS2 protein expression was statistically significantly higher in the nose (1.05 ± 0.07, n = 11) compared with the tongue (0.85 ± 0.08, n = 16), oro/hypopharynx (0.80 ± 0.10, n = 13), esophagus (0.70 ± 0.06, n = 5), and conjunctiva (0.82 ± 0.04, n = 4) (Dunn’s multiple comparison post hoc test, ∗∗∗adjusted p = 0.0007, ∗∗∗∗adjusted p < 0.0001, ∗∗∗∗adjusted p < 0.0001, and ∗adjusted p = 0.0167, respectively). TMPRSS2 in the trachea/bronchus (0.98 ± 0.08, n = 21) was statistically significantly higher compared with the tongue, oro/hypopharynx, and esophagus (Kruskal-Wallis test p < 0.0001; Dunn’s multiple comparison post hoc test, †adjusted p = 0.0149, †††adjusted p = 0.0006, and †††adjusted p = 0.0004, respectively).
(F) Quantification of TMPRSS2 in the cilia of normal human mucosal tissues. TMPRSS2 protein expression was statistically significantly higher in nasal tissues (1.39 ± 0.30, n = 11) compared with the nasopharynx (1.02 ± 0.15, n = 7) and bronchus/trachea (1.09 ± 0.08, n = 20) (Kruskal-Wallis test p = 0.0008; Dunn’s multiple comparison post hoc test, ∗∗adjusted p = 0.0048 and ∗∗adjusted p = 0.0020, respectively).
Values in parentheses are presented as mean ± SD. The bands within the boxplot show the median value. The bottom and top of the boxplots represent the 25th and 75th percentiles, respectively. The whiskers extending from both ends of the boxes are minimum and maximum values. Each dot represents one individual. Scale bars, 50 μm and 20 μm (insets). See also Figures S2 and S3.
Next we evaluated TMPRSS2 expression within the epithelial cells and cilia of these mucosal tissues using validated antibodies against TMPRSS239 (see Figure 2B for antibody validation), panKRT, and ACTUB. Similar to ACE2 expression, TMPRSS2 protein expression was also observed in the epithelia and cilium organelles of the ciliated epithelium (when anatomically present) of all stained tissues (Figures 3D and S3). However, we found TMPRSS2 expression to be significantly higher in the mucosa of the nasal cavity and trachea/bronchus compared with the tongue, oro/hypopharynx, esophagus, and conjunctiva (Figure 3E; p < 0.05). Within the motile cilia, TMPRSS2 expression was notably elevated in the nasal cavity compared with the nasopharynx and trachea/bronchus (Figure 3F; p < 0.01). These patterns of TMPRSS2 expression within the epithelia suggest that, although proximal airway and ocular tissues are susceptible to SARS-CoV-2, greater TMPRSS2 expression in the nasal cavity and trachea/bronchus may render these airway sites particularly conducive to SARS-CoV-2 infection and proliferation.
SARS-CoV-2 replicates in human respiratory and oral mucosae
Diagnostic swab sampling of the nasal and oral cavities detects shedded SARS-CoV-2 virus in the respective mucus/saliva secretions and is the basis of most RNA-based COVID-19 testing methods.40 However, nasal secretions physiologically drain into the oropharyngeal space, rendering infeasible the use of swab samples to localize the primary sites of major viral infection and intracellular replication. To test the hypothesis that SARS-CoV-2 infects and replicates intracellularly in multiple head and neck mucosal epithelial surfaces, we evaluated SARS-CoV-2-infected human proximal airway (nasal cavity, tongue, pharynx, larynx, and trachea) tissues collected postmortem at autopsy from 27 individuals with COVID-19 between March and July 2020. All 27 individuals (n = 18 male, n = 9 female) were diagnosed with COVID-19 by a positive reverse-transcriptase polymerase chain reaction (RT-PCR) test for SARS-CoV-2. The median age at death was 74.7 years (interquartile range, 66–82 years; range, 53–96 years), and the time from onset of known COVID-19 symptoms to death ranged from 4–44 days, with a median of 19 days (Table S1). Cases were not preselected with regard to clinical characteristics because of the extreme challenges of tissue procurement in the pandemic setting. Because of the potential for body disfigurement and unavailability of an ophthalmic pathologist during the time of autopsy, we were unable to obtain a meaningful number of ocular tissues from individuals with COVID-19. Ocular tissues were obtained from 3 individuals in our cohort, but only one specimen had detectable low levels of SARS-CoV-2 RNA.41
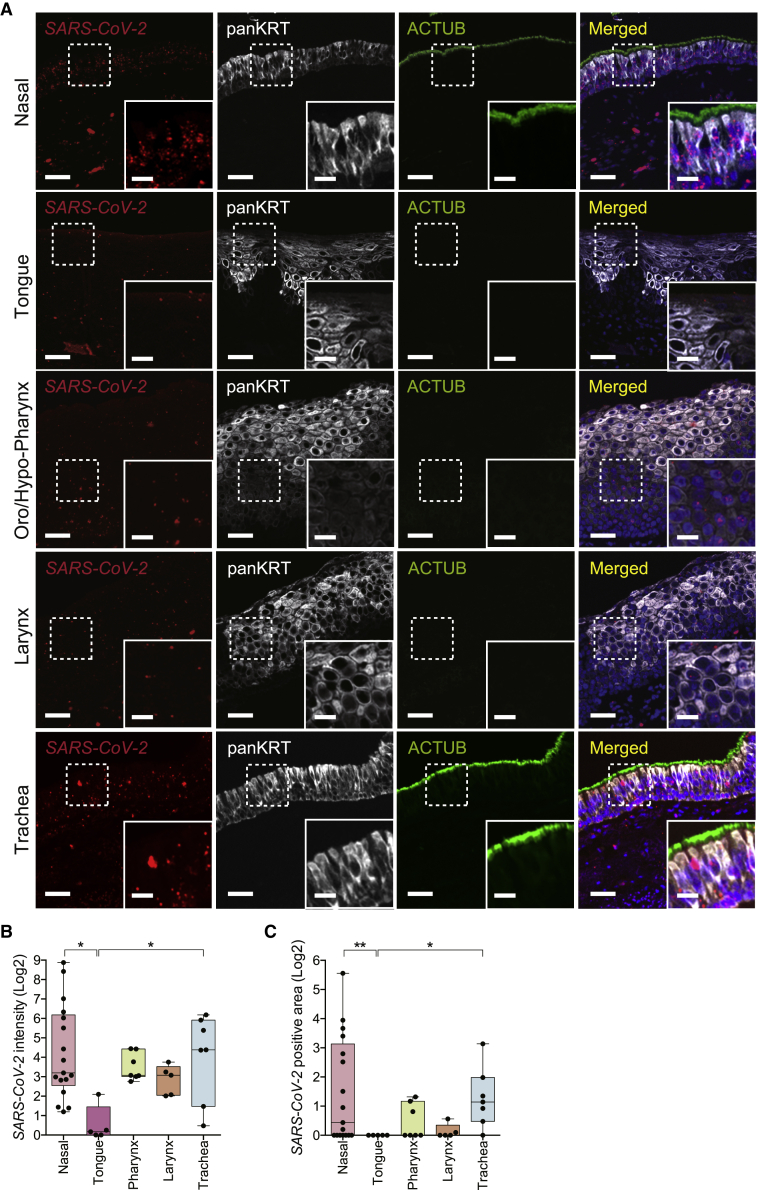
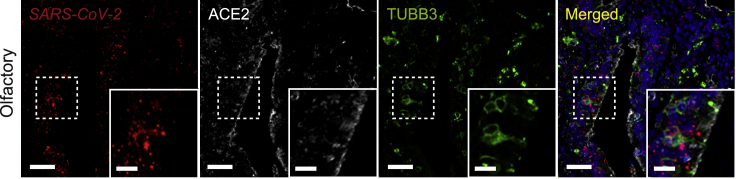
We performed in situ hybridization (ISH) using a robustly validated SARS-CoV-2 Spike mRNA probe in combination with antibodies against panKRT and ACTUB on these proximal respiratory tract and oral tissues (see Figures S4A–S4C for independent SARS-CoV-2 probe validation).42 The rationale for quantifying SARS-CoV-2 Spike RNA rather than Spike protein was to minimize confounding or artifactual signal because of partially degraded Spike protein fragments detected in the extracellular space or taken up by cells, leading to misrepresentation of SARS-CoV-2 localization.43 Viral RNA also has the additional benefit of being present as part of the virion as well as being an accurate indicator of viral replication and transcription in an infected cell.44 As above, we focused on SARS-CoV-2 infection in the head and neck mucosal epithelium, which faces the airway lumen and is relevant to the dynamics of respiratory viral transmission. Interestingly, we observed SARS-CoV-2 viral RNA within the epithelia of all 5 distinct proximal airway tissue sites (Figure 4A). The olfactory cleft, part of the superior nasal cavity along the skull base, also stained positive for SARS-CoV-2 RNA (Figure 5), as demonstrated similarly by Meinhardt et al.45 Comparatively, SARS-CoV-2 RNA was significantly more abundant in the nasal and tracheal epithelia compared with the lingual (tongue) epithelium with respect to infected surface area and copies of RNA within the infected area (Figures 4B and 4C; p < 0.05). Intermediate levels of SARS-CoV-2 RNA transcripts were detected in the epithelia of the pharynx and larynx compared with the nasal cavity/trachea and the tongue (Figures 4B and 4C). These results demonstrate that, in individuals with COVID-19, SARS-CoV-2 infects and propagates within multiple regions of the proximal respiratory tract from the nasal cavity to the tongue. Our findings further implicate the nose and trachea as primary sites of SARS-CoV-2 infection among the mucosal surfaces of the head and neck region.
Figure 4.
SARS-CoV-2 Spike mRNA expression in mucosal tissues of the head and neck from individuals with COVID-19
(A) Representative multiplexed images of ISH against the SARS-CoV-2 Spike mRNA in combination with immunofluorescence staining of the epithelial marker panKRT and the cilium marker ACTUB in mucosal tissues from individuals with COVID-19.
(B) Quantification of SARS-CoV-2 intensity in mucosal tissues from individuals with COVID-19. SARS-CoV-2 intensity in nasal (n = 17) and tracheal (n = 7) tissues was significantly higher than in the tongue (n = 5) (Kruskal-Wallis test p = 0.0171; Dunn’s multiple comparison post hoc test, ∗adjusted p = 0.0117 and ∗adjusted p = 0.0310, respectively).
(C) Quantification of SARS-CoV-2-positive area in mucosal tissues from individuals with COVID-19. SARS-CoV-2-positive areas in nasal (n = 17) and trachea (n = 7) tissues were significantly higher than in the tongue (n = 5) (Kruskal-Wallis test p = 0.0091; Dunn’s multiple comparison post hoc test, ∗∗adjusted p = 0.0094 and ∗adjusted p < 0.0115, respectively).
The bands within the boxplot show the median value. The bottom and top of the boxplots represent the 25th and 75th percentiles, respectively. The whiskers extending from both ends of the boxes are minimum and maximum values. Each dot represents one individual. See also Table S1.
Figure 5.
SARS-CoV-2 Spike mRNA expression in human olfactory tissues from an individual with COVID-19
Representative multiplexed images of ISH against the SARS-CoV-2 Spike mRNA in combination with immunofluorescence staining of ACE2 and a neuronal marker, tubulin β3 (TUBB3), in the olfactory cleft from the nasal cavity of an individual with COVID-19. Scale bars, 50 μm and 20 μm (insets).
SARS-CoV-2 viral infectivity is elevated in smokers
We next identified clinical characteristics that could potentially account for the variability in SARS-CoV-2 transcription noted among nasal and tracheal tissues from individuals with COVID-19. Minimal inter-subject viral load variance was seen in the tongue, pharyngeal, and laryngeal tissues; thus, only nasal and tracheal samples (n = 24 individuals) were compared (Figures 4B and 4C). We first determined the extent to which epithelial SARS-CoV-2 RNA infection differs by age, gender, body mass index (BMI), smoking status, hypertension, and diabetes—covariates reported to be associated with COVID-19 disease severity. Demographics and clinical comorbidities are listed in Table S1. Known coexisting medical conditions included obesity (n = 9), diabetes mellitus (n = 10), and hypertension (n = 15).
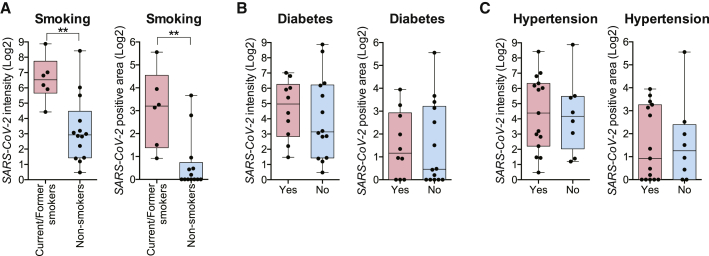
We found no significant differences in SARS-CoV-2 RNA abundance based on age, gender, or BMI (Figures S5A–S5C). There was also no correlation between SARS-CoV-2 RNA and duration of disease (Figure S5D; e.g., days between initial symptom onset and death), similar to the findings of Silva et al.46 Interestingly, SARS-CoV-2 RNA levels were significantly higher among smokers (current and former) compared with non-smokers (Figure 6A; p < 0.01). We also identified a trend toward higher SARS-CoV-2 RNA levels in individuals with diabetes mellitus (Figure 6B), but these results were not statistically significant because of the low sample size. No significant differences were noted between individuals with or without hypertension (Figure 6C). We then compared SARS-CoV-2 RNA levels between individuals who received systemic steroids, antivirals, or interleukin-6 (IL-6) inhibitors during the course of their hospitalization for COVID-19. Not surprisingly, neither steroid nor tocilizumab interventions were associated with changes in SARS-CoV-2 RNA expression in the proximal airways (Figures S5E and S5F), but, paradoxically, individuals who received antiviral medications displayed statistically significantly higher tissue levels of SARS-CoV-2 RNA (Figure S5G; Discussion). Overall, these results indicate that smoking status (current or former) is associated with higher SARS-CoV-2 propagation in the proximal respiratory tract and may explain why smokers are at higher risk for heightened severity of COVID-19 disease.47,48 Further studies with larger sample sizes will be necessary for validation.
Figure 6.
Comparison of SARS-CoV-2 Spike mRNA epithelial expression by clinical comorbidities
(A) SARS-CoV-2 intensity (left) and positive areas (right) are statistically significantly higher among smokers (current/former) (n = 6) compared with non-smokers (n = 14) (two-tailed Mann-Whitney test, ∗∗p = 0.0033 and ∗∗p = 0.0032, respectively).
(B) No statistically significant differences were seen in SARS-CoV-2 intensity (left) or positive areas (right) among individuals with or without pre-existing type II diabetes mellitus (n = 10 and n = 14, respectively) (two-tailed Mann-Whitney test, p = 0.4716 and p = 0.7521, respectively).
(C) No statistically significant differences were seen in SARS-CoV-2 intensity (left) or positive areas (right) among individuals with or without pre-existing hypertension (n = 15 and n = 8, respectively) (two-tailed Mann-Whitney test, p = 0.7284 and p = 0.9246, respectively).
See also Figure S5.
Cigarette Smoke (CS) exposure reduces IFNB1 expression and increases SARS-CoV-2 infectivity in the airway epithelium
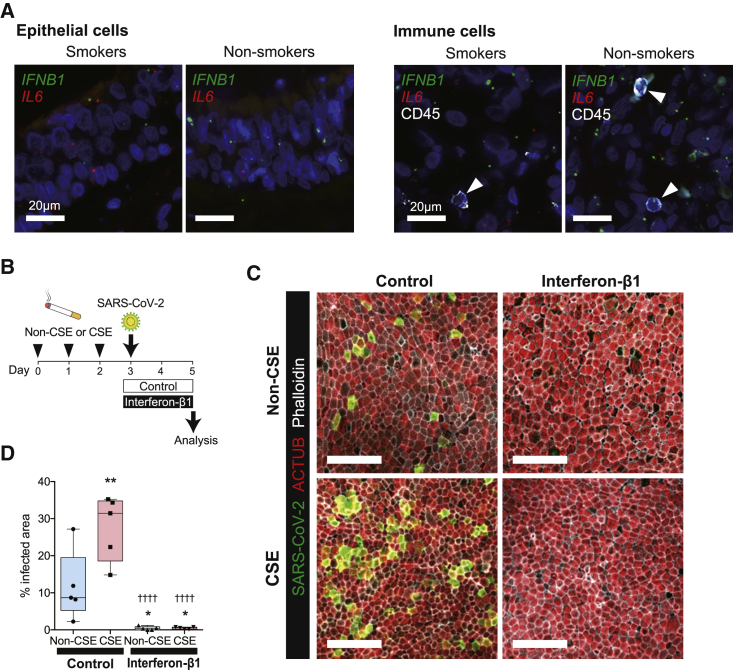
To further elucidate potential mechanisms of elevated SARS-CoV-2 infection in smokers, we first assessed ACE2 and TMPRSS2 expression in these tissues because higher levels of entry factors may facilitate SARS-CoV-2 infection. Epithelial and ciliary ACE2 and TMPRSS2 protein expression were not significantly different between upper airway tissues of smokers and non-smokers (Figures S6A and S6B). We also cultured human nasal epithelial cells (HNEpCs) in air-liquid interface (ALI) and found no significant differences in ACE2 and TMPRSS2 expression between CS extract (CSE)-exposed cells and non-exposed controls (Figure S6C). We next compared interferon β1 (IFNB1) expression between upper airway tissues of smokers and non-smokers because CS exposure has been shown to decrease type 1 IFN signaling,49 and IFN-β1 has been demonstrated to play an important role in abrogating SARS-CoV-2 infection in CS-exposed ALI cultures.50 We performed ISH using probes against IFNB1 and another inflammatory cytokine, IL6, in combination with antibodies against CD45. IFNB1 expression within epithelial cells and CD45+ immune cells were significantly lower in upper airway tissues from smokers, but IL6 expression did not differ (Figures 7A and S6E). Similarly, ex vivo, IFNB1 expression was significantly lower in CSE-exposed HNEpCs in ALI culture compared with controls (Figure S6D).
Figure 7.
CS exposure reduces the interferon-β1 (IFN-β1) response and results in higher SARS-CoV-2 infectivity
(A) Representative immunofluorescence images of ISH against IFNB1 (green) and IL6 (red) in combination with immunofluorescence staining of the immune cell marker CD45 (white) in airway autopsy tissues. Arrowheads indicate CD45+ cells.
(B) Experimental design of the ex vivo experiment using air-liquid interface (ALI)-cultured human nasal epithelial cells (HNEpCs).
(C) Representative immunofluorescence staining of SARS-CoV-2 (green), ACTUB (red), and phalloidin (white), an actin filament marker, in SARS-CoV-2-infected cells across four different conditions. Scale bars, 50 μm.
(D) Quantification of infected area of ALI-cultured HNEpCs under four different exposure conditions as shown in (C). The SARS-CoV-2-infected area was statistically significantly higher in the CS extract (CSE) group (27.6 ± 8.80) compared with the non-CSE control group (11.6 ± 9.35) (one-way ANOVA, p < 0.0001; Holm-Sidak’s multiple comparisons post hoc test, ∗∗adjusted p = 0.0047). Addition of IFN-β1 suppresses SARS-CoV-2 infection in the non-CSE (0.42 ± 0.49) and CSE groups (0.43 ± 0.31) compared with the non-CSE group without IFN-β1 (Holm-Sidak’s multiple comparisons post hoc test, ∗adjusted p = 0.0412 and ∗adjusted p = 0.0412, respectively) and CSE group without IFN-β1 (Holm-Sidak’s multiple comparisons post hoc test, ††††adjusted p < 0.0001, ††††adjusted p < 0.0001, respectively).
Values in parentheses are presented as mean ± SD. The bands within the boxplot show the median value. The bottom and top of the boxplots represent the 25th and 75th percentiles, respectively. The whiskers extending from both ends of the boxes are minimum and maximum values. Each dot represents one individual sample (n = 5). See also Figure S6.
To show the causative effects of smoking on IFNB1 expression and subsequent increased susceptibility to SARS-CoV-2 infection, we exposed HNEpCs to CSE in ALI culture prior to infection with SARS-CoV-2 ex vivo (Figure 7B). We observed that CSE exposure significantly increased the number of cells productively infected by SARS-CoV-2 compared with non-CSE exposed controls (Figures 7C and 7D). Furthermore, addition of recombinant IFN-β1 to the ALI culture abrogated SARS-CoV-2 replication in CSE-exposed HNEpCs and controls (Figures 7C and 7D). These results are consistent with prior findings of reduced IFN response after CS exposure and abrogation of SARS-CoV-2 infection by exogenous IFN-β1 in human bronchial/lung tissues.50 Our results suggest that downregulation of IFN-β1 is one mechanism that at least partially explains the higher SARS-CoV-2 infection in smokers.
Discussion
We report several notable discoveries in this human translational research study. First, we found that ACE2 and TMPRSS2, the primary entry factors of SARS-CoV-2, are expressed in a wide range of human mucosal surfaces of the head and neck, including the tongue, pharynx, larynx, conjunctiva, and, most prominently, in the nasal cavity and trachea. Importantly, antibody-based protein staining consistently yielded robust information compared with single-cell gene expression analyses,1 which are likely confounded by the low expression levels of ACE2. Second, in line with these results, we observed that SARS-CoV-2 can infect and propagate in disparate head and neck mucosal tissues but that viral infection is particularly enriched in the nasal cavity and trachea. This tissue tropism may be explained by our findings of enriched TMPRSS2 gene and protein expression in the epithelia of the nasal cavity and the trachea. Another non-mutually exclusive explanation is that the nasal cavity and trachea contain high proportions of ciliated epithelium, in contrast to the squamous and/or pseudostratified epithelium of the other mucosal tissues examined. We and others have shown previously that ciliated epithelial cells of the nose and trachea are the initial/early sites of entry for SARS-CoV-2,3,1 in large part because ACE2 and TMPRSS2 show robust expression in the motile cilia of the airway lumen compared with the epithelial cell body (Figure 3),1 which is supported by the scRNA-seq results in this study. Notably, because the head and neck tissue subsites shown here were obtained at autopsy from individuals who succumbed to COVID-19, the SARS-CoV-2 Spike RNA detected via ISH represents RNA replication throughout the course of the disease rather than early viral transmission. Given our results, it is possible that the degree of viral propagation is a consequence of early viral entry and sustained epithelial cell infection because the nose and trachea are also early sites of infection. Finally, other entry factors, such as TMPRSS4, TFRC, and NRP1, may also play a role in determining SARS-CoV-2 tissue tropism. Further studies are underway to determine the protein expression of these entry factors within head and neck tissues.
Our findings of robust SARS-CoV-2 transcription in the nasal cavity supports the idea that the nasal epithelium is not only an early site of entry3,1 but also a prominent site of ongoing SARS-CoV-2 replication. In line with our findings, loss of the sense of smell, termed anosmia, is a common and often lasting symptom of COVID-1951 and has been attributed to SARS-CoV-2 infection of the olfactory cleft in the nasal cavity.45,52 Ageusia, the loss of taste, is another symptom frequently associated with COVID-19,51 with possible mechanisms being viral damage of gustatory receptors in the tongue or as a secondary consequence of anosmia because olfaction strongly contributes to taste perception. Our findings of relatively lower levels of SARS-CoV-2 infection in the tongue, in contrast to the nose (Figures 4B and 4C), suggest the latter hypothesis as the more likely mechanism for dysgeusia and would be an interesting future area of research.
The observation in this study that SARS-CoV-2 can infect and replicate in the oropharynx and the tongue suggests that the oral mucosa can serve as an additional reservoir or an indicator of host SARS-CoV-2 infection status. Recent studies have reported that saliva and nasopharyngeal viral load correlate with COVID-19 severity, with saliva viral load as a potentially better indicator of cumulative viral load because of admixed secretions from the nose and the lower respiratory tract.12, 13, 14,46 Whether SARS-CoV-2 directly infects the oral cavity upon aerosol/droplet exposure or indirectly via the nasal-to-oral route is uncertain. Nasal secretions drain into the pharynx, larynx, oral cavity, and even trachea, allowing viral particles from the nasal cavity to potentially seed the remaining downstream mucosal tissues and prime the oral route of transmission to new hosts. Our finding of relatively lower SARS-CoV-2 transcription in the tongue compared with the nose is in support of an indirect route of oral tissue infection via the nose. However, in a recent study of the influenza virus using a ferret model, throat swabs of flu-infected ferrets sometimes tested positive for influenza before nose swabs, suggesting that viral deposition and infection can also be initiated in the oropharyngeal cavity for other respiratory viruses.53 Studies of SARS-CoV-2 in animal models are needed to deduce the mechanisms of viral replication and deposition in the oral mucosa, which will have important implications for understanding and preventing transmission of COVID-19.
Notably, in the present study, we found that current and former smokers have higher SARS-CoV-2 infection in the proximal respiratory tract. This result is supported by the finding that CS exposure increases the number of SARS-CoV-2-infected airway epithelial cells ex vivo (Figure 7D).50 Although meta-analyses and our own data here suggest that smokers may not be at a higher risk for SARS-CoV-2 initial infection, given the absence of ACE2 and TMPRSS2 upregulation in smokers (Figures S6A–S6C),1,47,48 when infected with the virus, smokers are at an increased risk of severe COVID-19.47,48 Our findings of reduced IFN-β1 expression in upper airway tissues from smokers and CSE-exposed HNEpCs as well as abrogation of SARS-CoV-2 infection by recombinant IFN-β1 suggest that a defective IFN response from cigarette smoking49 may compromise cellular antiviral defense, leading to higher viral replication. Alternatively, impairment of mucociliary clearance in the proximal airway, seen in current and former smokers,54,55 may perpetuate local infection of neighboring cells from infected cells, leading to overall greater or sustained SARS-CoV-2 replication, which, in turn, leads to higher susceptibility to severe COVID-19. Indeed, of all clinical characteristics available for our analyses, smoking is the only variable that directly affects the airway tissue. In this study, we also observed a tendency toward higher SARS-CoV-2 infection in the airway of individuals with diabetes (Figure 6B). Although statistical significance was not achieved, this finding is consistent with a recent study demonstrating a trend of prolonged viral shedding in the lungs of diabetics.56 Additionally, we unexpectedly found that individuals who had received antiviral therapy had higher SARS-CoV-2 infection in the proximal airway (Figure S5G). Aside from the small sample size of this cohort, one major caveat is that 4 of the 5 individuals in this latter group had only received a 2-day course of remdesivir or lopinavir-ritonavir in the 2 days preceding death, and one of the individuals received remdesivir very late in the disease course (20 days following symptom onset). Although early trials of lopinavir-ritonavir and remdesivir did not demonstrate a reduction in viral load from upper airway swab specimens,57,58 more recent larger trials have not provided viral load data.59,60 Whether a complete and earlier course of antiviral treatment may decrease SARS-CoV-2 replication in the proximal airways is not established. It also remains to be seen whether antiviral agents can decrease viremia (regardless of their clinical efficacy). We caution against overinterpretation of our findings because of these possible confounders as well as the study limitations discussed below. Lastly, our finding that the use of steroids or tocilizumab was not associated with higher SARS-CoV-2 infection in the proximal airway is consistent with recent studies measuring viral load from nasopharyngeal swabs.61,62
Limitations of the study
This study has expected limitations in the setting of the ongoing pandemic. The sample size of our COVID-19 cohort has been constrained by the logistical challenges and precautions required when harvesting SARS-CoV-2 infected head and neck tissues. Similarly, the sample size of our HNEpCs was also limited by tissue availability and challenges of conducting experiments under biosafety level 3 (BSL3). Our study was likely underpowered to detect statistically significant differences in SARS-CoV-2 infection in individuals with diabetes, hypertension, and obesity—comorbidities that may predispose to higher SARS-CoV-2 viral replication, given their association with greater disease severity. However, the cohort presented here contains the largest published collection of head and neck and proximal airway tissues from individuals with COVID-19 to date. An additional limitation is that, as is the case with any observational study, we cannot infer a causal relationship between SARS-CoV-2 infection and clinical characteristics, including smoking and antiviral agent use. There may be potential confounders that can independently influence levels of viral replication in these individuals. Treatment selection bias, such as use of antiviral agents in individuals deemed to have more severe disease, is possible. Additionally, because we analyzed only tissues procured at autopsy from individuals who died of COVID-19, we are unable to associate viral replication with clinical outcome or derive a more mechanistic understanding of SARS-CoV-2 pathogenesis. Regardless of the differing amounts of SARS-CoV-2 replication, all of these individuals had succumbed to COVID-19. However, it is conceivable that a therapeutic intervention that was unavailable earlier in the pandemic would have prevented their death. The REGN-COV2 antibody cocktail, for example, has been shown to be more effective in outpatients with a higher baseline viral load.63 Finally, although SARS-CoV-2 has been infrequently isolated from conjunctival swabs or tears,4,5 we were unable to comparatively assess SARS-CoV-2 propagation in ocular tissues. Likewise, we confined our analysis to head and neck and proximal airway tissues and did not examine lung tissues.
Here we describe how the diverse human head and neck mucosal tissues, particularly the nasal cavity and trachea, serve as active sites of SARS-CoV-2 infection. Measures directed at reducing SARS-CoV-2 propagation in these anatomic regions will likely not only reduce mortality and transmission but also limit opportunities for SARS-CoV-2 mutant strains to arise. There is emerging evidence showing that novel SARS-CoV-2 variants can arise from the upper respiratory tract of immunocompromised hosts.64,65 Therapeutic agents administered through the upper airway have the potential to hamper SARS-CoV-2 evolution, especially in persistently infected individuals. Nasally administered povidone-iodine,16 inhaled ciclesonide,17 and hydrogen peroxide mouth rinses20 are currently under investigation as antiviral/virucidal agents that may serve this purpose. Future studies should also explore use of neutralizing antibodies63,66 or recombinant ACE267 via nasal administration or inhalation in individuals with COVID-19 or in the setting of prophylaxis against viral transmission.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-ACE2 (Lot GR3333640) | Abcam | Cat# ab15348; RRID:AB_301861 |
| Mouse monoclonal anti-TMPRSS2 | Millipore Sigma | Cat# MABF2158 |
| Mouse monoclonal anti-Acetylated α Tubulin | Santa Cruz | Cat# sc-23950; RRID:AB_628409 |
| Rabbit monoclonal anti-Acetyl-α-Tubulin | Cell Signaling Technology | Cat# 5335; RRID:AB_10544694 |
| Guinea pig polyclonal anti-Pan Cytokeratin | LSBio | Cat# LS-B16812 |
| Mouse monoclonal anti-Tubulin β 3 | Biolegend | Cat# MMS-435P; RRID:AB_2313773 |
| Mouse monoclonal anti-CD45 | Abcam | Cat# ab781; RRID:AB_306098 |
| Mouse monoclonal anti-SARS-CoV-2 spike | GeneTex | Cat# GTX632604; RRID:AB_2864418 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Absorbed Secondary Antibody, Alexa Flour Plus 647 | Thermo Fisher Scientific | Cat# A32795; RRID:AB_2762835 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Absorbed Secondary Antibody, Alexa Flour Plus 555 | Thermo Fisher Scientific | Cat# A32773; RRID:AB_2762848 |
| Goat anti-Guinea Pig IgG (H+L) Highly Cross-Absorbed Secondary Antibody, Alexa Flour 488 | Thermo Fisher Scientific | Cat# A11073; RRID:AB_2534117 |
| Phalloidin-Atto 647N | Millipore Sigma | Cat# 65906 |
| Bacterial and virus strains | ||
| SARS-CoV-2 | Joe DeRisi lab, UCSF | SARS-CoV-2/human/USA/CA-UCSF-0001C/2020 |
| Biological samples | ||
| Human nasal, nasopharynx, oro/hypo-pharynx, larynx, bronchus, uvula, tonsil, and conjunctiva tissues | Stanford University School of Medicine | N/A |
| Human Tissue Microarray | US Biolab | Cat# XHN801 |
| Human Tissue Microarray | US Biolab | Cat# XHN803 |
| Human Paraffin Embedded Tissue Array | US Biomax | Cat# OR208a |
| Human Paraffin Embedded Tissue Array | US Biomax | Cat# BN1002b |
| Human Paraffin Embedded Tissue Array | US Biomax | Cat# LP208 |
| Human nasal epithelial cells | Stanford University School of Medicine | N/A |
| MucilAir | Epithelix | Cat# EP01MD |
| Chemicals, peptides, and recombinant proteins | ||
| The ACE2 peptide for blocking assay: KGENNPGFQNTDDVQTSF | GenScript | N/A |
| The TMPRSS2 peptide for blocking assay: (sequence provided in confidence by the manufacturer) | GenScript | N/A |
| RNAscope Hydrogen Peroxide Reagent | Bio-Techne | Cat# 322335 |
| Dako Target Retrieval Solution, pH9 | Dako Agilent | Cat# S236784-2 |
| Recombinant Human Interferon-β1 | Peprotech | Cat# 300-02BC |
| RNeasy Lipid Tissue Kit | QIAGEN | Cat# 74804 |
| M-MLV Reverse Transcriptase | Invitrogen | Cat# 28025-013 |
| TaqMan Gene Expression Master Mix | Applied Biosystems | Cat# 4369016 |
| Critical commercial assays | ||
| RNAscope Multiplex Fluorescent Reagent Kit v2 | Bio-Techne | Cat# 323100 |
| TSA Cyanine 3 | Akoya Biosciences | Cat# NEL744001KT |
| TSA Cyanine 5 | Akoya Biosciences | Cat# NEL745001KT |
| Deposited data | ||
| Single cell RNA-seq data30 | NCBI Gene Expression Omnibus | GEO: GSE134174 |
| Single cell RNA-seq data28 | European Genome-Phenome Archive | EGAD00001006185 |
| Single cell RNA-seq data29 | NCBI Gene Expression Omnibus | GEO: GSE103322 |
| Single cell RNA-seq data25 | European Genome-Phenome Archive | EGAS00001001755 |
| Single cell RNA-seq data26 | Single Cell Portal | https://singlecell.broadinstitute.org/single_cell/study/SCP253/allergic-inflammatory-memory-in-human-respiratory-epithelial-progenitor-cells#study-download |
| Oligonucleotides | ||
| SARS-CoV-2 Spike mRNA | Bio-Techne | Cat# 848561 |
| IFNB1 mRNA | Bio-Techne | Cat# 417071 |
| IL6 mRNA | Bio-Techne | Cat# 310371 |
| ACE2 (Hs00222343_m1) | Life Technologies | Cat# 4331182 |
| TMPRSS2 (Hs00237175_m1) | Life Technologies | Cat# 4331182 |
| IFNB1 (Hs01077958_s1) | Life Technologies | Cat# 4331182 |
| Software and algorithms | ||
| GraphPad Prism 6 | GraphPad | https://www.graphpad.com/ |
| IBM SPSS Statistics version 23 | IBM | https://www.ibm.com/support/pages/downloading-ibm-spss-statistics-23 |
| Seurat v4.0 | Seurat31 | https://satijalab.org/seurat/ |
| Single R | Aran et al.68 | https://github.com/dviraran/SingleR |
| ImageJ v1.52t | NIH | https://imagej.net/downloads |
| deepcell-tf 0.6.0 | Greenwald et al.69; Van Valen et al.70 | https://github.com/vanvalenlab/deepcell-tf |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jayakar V. Nayak (jnayak@stanford.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Normal human tissue specimen collection
Normal tissues from the nasal cavity and the paranasal sinuses (collectively termed “nasal” tissues) were collected from healthy control donors (range 12-82 years; n = 4 male and n = 7 female) from the Stanford Sinus Center within the Stanford University School of Medicine. These controls were patients without history, endoscopic, or radiographic evidence of sinus disease, but underwent sinus procedures for surgical treatment of adjacent anatomic sites, such as for repair of cerebrospinal fluid leaks at the skull base. All tissue specimens were collected under approved Institutional Review Board (IRB) protocols in accordance with the regulations of the Research Compliance Office (protocol ID #18981 at Stanford University). All subjects provided informed consent. Following surgical excision, sinonasal specimens were placed in physiologic saline, immediately transported to the lab, and placed in 10% neutral buffered formalin for 24–48 h before paraffin embedding. Sinonasal tissues that contain bone were placed into EDTA for bone decalcification prior to embedding into tissue blocks. Normal formalin-fixed, paraffin-embedded (FFPE) human tissues of the nasopharynx (range 21-67 years; n = 3 male and n = 4 female), oro/hypo-pharynx (range 38-71, n = 6 male and n = 4 female), trachea/bronchus (range 19-68 years; n = 12 male and n = 9 female), and conjunctiva (range 52-85 years; n = 4 male and n = 0 female) were selected from Stanford University Pathology archives based on specific notation of disease absence and normal histology. Additional utilized tissue sections include normal human tissue microarrays obtained from US Biolab XHN801 (tongue) and XHN803 (tongue) as well as US Biomax OR208a (tongue) (range 22-73 years; n = 12 male and n = 4 female), BN1002b (esophagus) (range 22-42 years; n = 4 male and n = 1 female), and LP208 (Larynx (range 16-48 years; n = 3 male and n = 1 female), oro/hypo-pharynx (range 15-43 years; n = 2 male and n = 2 female)). Normal histology was independently confirmed again by board-certified pathologists, M.S.M., C.M.S., and A.T., using H&E stained images for evaluation.
COVID-19 human tissue specimen collection
SARS-CoV-2 infected proximal respiratory tract tissues were obtained during autopsy at the University Hospital Basel Switzerland, University of California San Diego, California, USA, and Columbia University School of Medicine, New York City, USA. Tissues were processed in a similar manner as previously described,71 and collection was approved by the ethics commission of Northern Switzerland (EKNZ; study ID #2020-00969) and the Institutional Review Board (IRB) in accordance with the regulations of the Research Compliance Office (protocol ID #200485X at University of California San Diego, #AAAT0689 at Columbia University School of Medicine). All patients with COVID-19 or their relatives consented to the use of tissue for research purposes. Smoking status (current/former/never) was defined based on self-reports in the electronic medical record. Patients were considered to have hypertension and/or diabetes mellitus based on documented pre-existing medical history and the use of medications for hypertension and/or diabetes mellitus on admission. Patients whose co-morbidities or use of medications could not be confirmed were excluded from data analyses in Figures 6 and S5 (see Table S1).
Method details
Single cell gene expression analysis
Publicly available single cell RNA-seq datasets pertaining to nasal tissues,25, 26, 27 bronchial tissues,25,28 trachea,30 and tongue29 were downloaded and processed with Seurat,31 where the data was processed in parallel by normalization with the NormalizeData() function, followed by variable feature identification with FindVariableFeatures(), and data scaling subsequently with ScaleData().
Automated cell type annotation was performed with SingleR68 using the Human Primary Cell Atlas data in celldex,68,72 with the primary focus of isolating epithelial cells. Since the datasets were of different library preparation methods and tissue sources, we performed data integration in Seurat using the FindIntegrationAnchors() and IntegrateData() functions, as previously described.31 Five major epithelial cell types identified here, namely secretory cells (SCGB1A1, KRT15, LYPD2, KRT4, KRT13), basal cells (KRT5, DAPL1, TP64), ciliated cells (FOXJ1, CCDC153, CCDC113, MLF1, LZTFL1), ionocytes (FOXI1, CFTR, ASCL3), and others. We refer readers to a more detailed annotation list described here.34 Plots were generated in Seurat or ggplot directly.73
Immunofluorescence immunohistochemistry (IF IHC) and imaging
Sections were cut to 4 μm thickness and mounted on frosted glass slides. Deparaffinization, rehydration, and heat-induced epitope retrieval (HIER) were performed on a ST4020 small linear stainer (Leica). For deparaffinization, slides were baked at 70°C for 1–1.5 h, followed by rehydration in descending concentrations of ethanol (100% twice, 95% twice, 80%, 70%, ddH2O twice; each step for 30 s). Washes were performed using a Leica ST4020 Linear Stainer (Leica Biosystems, Wetzlar, Germany) programmed to three dips per wash for 30 s each. HIER was performed in a Lab VisionTM PT module (Thermo Fisher) using Dako Target Retrieval Solution, pH 9 (S236784-2, DAKO Agilent) at 97°C for 10 min and cooled down to 65°C. After further cooling to room temperature for 30 min, slides were washed for 10 min three times in Tris-Buffered Saline (TBS), containing 0.1% Tween 20 (Cell Marque; TBS-T). Sections were then blocked in 5% normal donkey serum in TBS-T at room temperature for 1 h, followed by incubation with primary antibodies in the blocking solution. After one overnight incubation of primary antibodies in 4°C, sections were washed three times with TBS-T and stained with the appropriate secondary antibodies in PBS with 3% bovine serum albumin, 0.4% saponin, and 0.02% sodium azide at room temperature for 1 h. Following this, sections were washed three times with TBS-T and mounted with ProLong Gold Antifade mounting medium (Invitrogen). Hoechst 33342 (Thermo) was also used in the second to last TBS-T wash for nuclear staining. The primary antibodies and final titrations used were rabbit anti-ACE2 (1:100; Abcam ab15348), mouse anti-TMPRSS2 (1:50; Millipore MABF2158), mouse anti-acetylated α Tubulin (1:300; Santa Cruz sc-23950; for immunofluorescence staining with anti-ACE2), rabbit anti-acetylated α Tubulin (1:800, CST 5335S; for immunofluorescence staining with anti-TMPRSS2), guinea pig anti-pan-cytokeratin (1:250; LifeSpan Biosciences LS-B16812-50), mouse anti-Tubulin β3 (TUBB3) Antibody (1:500; Biolegend 801202), mouse anti-CD45 (1:100; Abcam ab781; PD7/26 + 2B11), and mouse anti-SARS-CoV-2 spike (1:1000, Genetex 1A9). Secondary antibodies include highly cross-adsorbed donkey anti-rabbit Alexa Fluor Plus 647 (1:500; Thermo A32795), highly cross-adsorbed donkey anti-mouse Alexa Fluor Plus 555 (1:500; Thermo A32773), highly cross-adsorbed goat anti-guinea pig Alexa Fluor 488 (1:500; Thermo A11073). Phalloidin (Sigma-Aldrich 65906) was used to stain actin filaments. Fluorescence-immunolabeled images were acquired using an Andor Dragonfly 302 Spinning Disk Confocal (Oxford Instruments) attached to a Nikon Eclipse Ti microscope and a Keyence BZ-X710 microscope. Post-imaging processing was performed using FIJI package of ImageJ 1.52t. Figures were organized using Adobe Illustrator.
Blocking with immunizing peptide (peptide competition)
Using peptide sequences generously provided by the manufacturers of anti-ACE2 (Abcam ab15348) and anti-TMPRSS2 (Millipore MABF2158) antibodies, we custom synthesized the immunizing ACE2 and TMPRSS2 peptides, respectively, through GenScript. The sequence of the synthesized immunizing ACE2 peptide is: KGENNPGFQNTDDVQTSF. The sequence of the synthesized immunizing TMPRSS2 peptide was provided in confidence by the manufacturer. Prior to primary antibody staining, the primary antibody was mixed and incubated with a 20-fold molar excess of immunizing peptide or an unrelated mock peptide for 3 h at room temperature. Other staining procedures were performed as described above.
In situ hybridization
Rehydration and HIER of tissue sections were performed as described above and in Lee et al.1 After cooling to room temperature, slides were washed for 2 × 2 min with ddH2O before a 15 min H2O2 block at 40°C (322335, Bio-Techne). Slides were then washed for 2 × 2 min ddH2O before an overnight hybridization at 40°C with probes against the SARS-CoV-2 Spike mRNA (848561, Bio-Techne), IFNB1 (417071, Bio-Techne), and IL6 (310371, Bio-Techne). Amplification of the ISH probes was performed the next day according to manufacturer’s protocol (323100, Bio-Techne), with the final deposition of Cyanine 3 (NEL744001KT, Akoya Biosciences) or Cyanine 5 (NEL745001KT, Akoya Biosciences). Slides were then processed as described above for IF IHC staining.
Validation of assay specificity was performed using control slides in parallel to the experimental slides. The control slides contained either 1) Vero E6 cells infected with SARS-CoV-2 or mock infected (generated in-house and also via a generous gift from Dr. Xiankun Zeng), or 2) COVID-19 lung or non-COVID-19 lung autopsy samples, or 3) COVID-19 nasal or non-COVID-19 nasal autopsy samples. These autopsy samples were also subject to qRT-PCR validation to confirm viral presence.
Air liquid interface (ALI) cultures
3.3 × 104 human nasal epithelial cells (HNEpCs) were seeded in 24-well 6.5mm Transwell inserts (StemCell technologies) with PneumaCult-EX Plus Medium. After confluence was reached, PneumaCult-ALI Maintenance Medium was added to the basal chamber only and maintained 21-28 days to achieve differentiation into pseudostratified mucociliary epithelium. Media was changed every other day and cultures were maintained at 37°C and 5% CO2.
Ex vivo SARS-CoV-2 infection
The clinical SARS-CoV-2 isolate of the pandemic D614G variant (SARS-CoV-2/human/USA/CA-UCSF-0001C/2020) was kindly provided by Sara Sunshine and Dr. Joe DeRisi. 100 μL of SARS-CoV-2 suspension (5-10x10ˆ6 PFU per ml) was added to the top chamber of the ALI-cultured HNEpCs for ex vivo infection. Media containing the virus were removed after 2 h of inoculation, and the ALI cultures were incubated at 37°C for the indicated time.
Cigarette smoke extract (CSE) treatment
CSE was made by bubbling the cigarette smoke using 3R4F University of Kentucky standard reference cigarettes with 9.4 mg tar/0.726 mg nicotine (University of Kentucky, Lexington, KY) through 40 mL of normal saline. A 35 mL cigarette puff drawn at 1L/min over 2 s was delivered to normal saline, followed by 58 s of filtered air at 2L/min rate, every minute, as per FTC/ISO protocol. A single cigarette was consumed in 9 puffs and thus combusted for a total of 9 min. The saline was exposed to 4 cigarettes in the inExpose smoking system (SCIREQ, Montreal, QB, Canada), and then filtered through a 0.22 μm filter. CSE was diluted with the medium to 5% final concentration. The cells were treated with CSE 1 h per day for 3 days before the SARS-CoV-2 inoculation.
Interferon-β1 treatment
Interferon-β1 (200 ng/ml) (Peprotech) was added to the ALI cultures 1 h before the SARS-CoV-2 infection. Vehicle was added to controls at the same time point in parallel.
Quantitative polymerase chain reaction (qPCR)
At 48 h post infection, the cells were fixed or lysed for the immunofluorescence staining and gene expression level analysis. Cells were lysed in TRIzol (Invitrogen) and total RNA was isolated using RNeasy Lipid Tissue Kit (QIAGEN) and cDNA was synthesized using M-MLV Reverse Transcriptase (Invitrogen, 28025-013). qPCR was performed using TaqMan Probes (Invitrogen) and the TaqMan Gene Expression Master Mix (Applied Biosystems, 4369016) in 96-well Micro Amp Optical reaction plates (Applied Biosystems, N8010560). Life Technologies TaqMan probe: ACE2: Hs00222343_m1, TMPRSS2: Hs00237175_m1, IFNB1: Hs01077958_s1.
Quantification and statistical analysis
Quantification of fluorescence intensity
Samples within each cohort for comparative analysis were stained simultaneously using the same master mixes, washes, and identical incubation times under staining conditions described above. Exposure times under confocal microscopy were identical for samples within the same cohort. Quantification was performed using a custom script developed in the FIJI package of ImageJ as previously described.1 Briefly, a binary mask was created by thresholding the pan-cytokeratin (Pan-KRT) and acetylated α-Tubulin (ACTUB) channels using selected cutoff values to generate a comprehensive outline of each channel. Pan-KRT- and ACTUB-positive regions were segmented using continuity of high signal regions on a binary mask. The signal within the segmented areas was computed for fluorescent channels that represent ACE2 or TMPRSS2 protein expression. For the sake of cross-sample normalization for ACE2 and TMPRSS2, the ratio of the ACE2 or TMPRSS2 signal divided by the epithelial Pan-KRT (Figures 3B and 3E) or cilia ACTUB signal (Figures 3C or 3F; “normalized ACE2” or “normalized TMPRSS2”) for each tissue sample was used for further comparative analysis. For SARS-CoV-2 mRNA, the area and intensity within the areas segmented by thresholding SARS-CoV-2 signal (within the Pan-KRT positive area) was used in quantitative analyses.
Images of in situ hybridization of IFNB1 and IL6 in SARS-CoV-2 infected autopsy samples were collected using a Keyence BZ-X710 with Nikon PlanFluor 40x NA 1.3 oil immersion lens. Since the expression levels of IFNB1 and IL6 were lower compared to the expression of SARS-CoV-2 Spike mRNA, we applied the 40X objective lens to count the dots where a single dot represents a single copy of RNA. The number of dots was counted by thresholding the IFNB1 and IL6 signals into binary masks using the FIJI Analyze Particles function within regions of cell segmentation. To segment out the CD45+ immune cells, Mesmer, a deep learning-based algorithm utilizing the DeepCell library (deepcell-tf 0.6.0) was locally implemented to first segment out all the cells.70,69 The prediction was performed with the multiplex_segmentation.py within deepcell-tf, and the neural network weights were imported from https://deepcell-data.s3-us-west-1.amazonaws.com/model-weights/Multiplex_Segmentation_20200908_2_head.h5. Signals from the nuclear hoechst channel were first capped at the 99.7th percentile, then input as the nuclear channel for all the FOVs, while a dummy all-zero image was generated and used as the membrane channel. model_mpp = 0.6 was used in the multiplex_segmentation.py script for all FOVs to generate the nuclear segmentation.
To remove confounding RBC background, segmented cells with an area smaller than 200 pixels were first excluded. Next, the signal ratio between hoechst and CD45 were calculated for each cell, and the cells with a number smaller than 0.15 were excluded, as RBCs do not contain nuclei. To further remove confounding non-specific CD45 signals, Otsu’s thresholding was applied to the distances between the cell centroids and their CD45 signal weighted centroids.
Statistics and reproducibility
Analyses were performed with GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA) and IBM SPSS Statistics version 23 (IBM, Armonk, NY) software. Detailed information on the statistical analysis used can be found in figure legends. A p value of < 0.05 was considered statistically significant.
Acknowledgments
The authors sincerely appreciate the generosity of tissue donation from the deceased individuals and their families which enabled this study. We thank members of the Nayak and Nolan labs for critical and thoughtful discussions. This work was supported by the California Institute for Regenerative Medicine (DISC2-09637 to J.V.N.), the Defense Advanced Research Projects Agency (HR001118S0037-PREPARE-FP-001 to J.V.N.), the Stanford Initiative to Cure Hearing Loss (SICHL) (to J.V.N.), the PDev Foundation (to J.V.N.), the Parker Institute for Cancer Immunotherapy (to G.P.N.), the Food and Drug Administration (HHSF223201610018C and DSTL/AGR00980 to G.P.N.), Fast Grant Funding for COVID-19 Science (to G.P.N.), Botnar Research Centre for Child Health Emergency Response to COVID-19 grant (to S.J., C.M.S., D.R.M., T.M., G.P.N., M.S.M., and A.T.), Bill and Melinda Gates Foundation COVID-19 Pilot Award (to S.J., D.R.M., and G.P.N.), the National Institutes of Health (R01HL151677 to J.V.N., 1R01AI149672-01 to G.P.N., and U54-CA209971 to G.P.N.), the Rachford and Carlotta A. Harris Endowed Chair (to G.P.N.), Stanford Translational Research and Applied Medicine (TRAM) Pilot Grant (to I.T.L.), Thrasher Research Fund Early Career Award (to I.T.L.), Stanford Maternal and Child Health Research Institute (MCHRI) Clinical (MD) Trainee Support Award (to I.T.L., an Ernest and Amelia Gallo Endowed Postdoctoral Fellow), the Leukemia and Lymphoma Society Career Development Program (to S.J.), Stanford COVID-19 Crisis Response grant (to I.T.L., S.J., C.-T.W., and T.N.), and the Swiss National Science Foundation (SNSF; 320030_189275 to M.S.M.).
Author contributions
I.T.L. conceived and coordinated the study. T.N., I.T.L., and S.J. designed and performed the experiments. S.J. performed the single-cell RNA-seq analysis. T.N. performed the microscopy imaging. M.S.M., C.H.Y., J.B.O., P.C., L.X., A.K.S., J.M.D., K.D.M., J.D.H., T.M., P.D.C., A.S.D., and A.T. acquired informed consent, acquired and processed COVID-19 tissue specimen, and/or collected clinical data. T.N., I.T.L., L.-C.S., C.-K.L., D.Z., A.Y., P.C., N.A.B., P.A.G., S.S.D., W.L., C.-J.T., T.-H.Y., Z.M.P., P.H.H., and J.V.N. acquired informed consent and acquired and/or processed normal human tissue specimens. M.S.M., C.M.S., and A.T. evaluated the histology of tissue specimens. C.-T.W., P.L., Y.X., P.K.J., and R.A. performed/supervised ex vivo infection experiments and contributed validation reagents. Y.G. developed and performed computational image processing. T.N., I.T.L., S.J., and Y.B. analyzed the data. T.N. conducted statistical analyses. B.Z., D.Z., M.E., H.C., and E.E.-D. provided experimental assistance. T.N., I.T.L., and S.J. designed and prepared the final figures. I.T.L. and S.J. wrote the manuscript with significant contributions from T.N., M.S.M., C.H.Y., J.B.O., A.T., G.P.N., and J.V.N. Funding and supervision were provided by T.N., I.T.L., S.J., M.S.M., C.-T.W., D.R.M., P.K.J., A.T., G.P.N., and J.V.N. All authors reviewed and agreed with the content of this manuscript.
Declaration of interests
I.T.L. is currently an employee and shareholder of Moderna, although this work was conducted prior to/independent of his employment. I.T.L. had also received research support unrelated to this study from Genentech (Roche). Moderna did not fund or participate in this study in any form.
Published: September 28, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100421.
Contributor Information
Ivan T. Lee, Email: ivantlee@gmail.com.
Alexandar Tzankov, Email: alexandar.tzankov@usb.ch.
Garry P. Nolan, Email: gnolan@stanford.edu.
Jayakar V. Nayak, Email: jnayak@stanford.edu.
Supplemental information
Data and code availability
This paper analyzes existing, publicly available RNA datasets. The accession numbers for these datasets are listed in the Key resources table. The analysis codes used to support additional findings of this study were minimally modified from previously published codes1 and are available at: GitHub:
https://github.com/bmyury/membrane_ACE2_quantitation. Additional data reported in this paper will be shared by the Lead Contact upon reasonable request.
References
- 1.Lee I.T., Nakayama T., Wu C.-T., Goltsev Y., Jiang S., Gall P.A., Liao C.K., Shih L.C., Schürch C.M., McIlwain D.R. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat. Commun. 2020;11:5453. doi: 10.1038/s41467-020-19145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xia J., Tong J., Liu M., Shen Y., Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020;92:589–594. doi: 10.1002/jmv.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo D., Xia J., Shen Y., Tong J. SARS-CoV-2 may be related to conjunctivitis but not necessarily spread through the conjunctiva SARS-CoV-2 and conjunctiva. J. Med. Virol. 2020;92:1757–1758. doi: 10.1002/jmv.25856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng W., Bao L., Gao H., Xiang Z., Qu Y., Song Z., Gong S., Liu J., Liu J., Yu P. Ocular conjunctival inoculation of SARS-CoV-2 can cause mild COVID-19 in rhesus macaques. Nat. Commun. 2020;11:4400. doi: 10.1038/s41467-020-18149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu D.K., Akl E.A., Duda S., Solo K., Yaacoub S., Schünemann H.J., COVID-19 Systematic Urgent Review Group Effort (SURGE) study authors Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973–1987. doi: 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallo O., Locatello L.G., Mazzoni A., Novelli L., Annunziato F. The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol. 2021;14:305–316. doi: 10.1038/s41385-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 10.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á. Transmission of SARS-CoV-2 Lineage B. 1.1. 7 in England: Insights from linking epidemiological and genetic data. medRxiv. 2021 doi: 10.1101/2020.12.30.20249034. [DOI] [Google Scholar]
- 11.Li B., Deng A., Li K., Hu Y., Li Z., Xiong Q., Liu Z., Guo Q., Zou L., Zhang H. Viral infection and transmission in a large well-traced outbreak caused by the Delta SARS-CoV-2 variant. medRxiv. 2021 doi: 10.1101/2021.07.07.21260122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magleby R., Westblade L.F., Trzebucki A., Simon M.S., Rajan M., Park J., Goyal P., Safford M.M., Satlin M.J. Impact of Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load on Risk of Intubation and Mortality Among Hospitalized Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa851. Published online June 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westblade L.F., Brar G., Pinheiro L.C., Paidoussis D., Rajan M., Martin P., Goyal P., Sepulveda J.L., Zhang L., George G. SARS-CoV-2 Viral Load Predicts Mortality in Patients with and without Cancer Who Are Hospitalized with COVID-19. Cancer Cell. 2020;38:661–671.e2. doi: 10.1016/j.ccell.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y., Yan L.-M., Wan L., Xiang T.-X., Le A., Liu J.-M., Peiris M., Poon L.L.M., Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura K.S., Freeman M.H., Wessinger B.C., Gupta V., Sheng Q., Huang L.C., Von Wahlde K., Das S.R., Chowdhury N.I., Turner J.H. Interim analysis of an open-label randomized controlled trial evaluating nasal irrigations in non-hospitalized patients with coronavirus disease 2019. Int. Forum Allergy Rhinol. 2020;10:1325–1328. doi: 10.1002/alr.22703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank S., Brown S.M., Capriotti J.A., Westover J.B., Pelletier J.S., Tessema B. In Vitro Efficacy of a Povidone-Iodine Nasal Antiseptic for Rapid Inactivation of SARS-CoV-2. JAMA Otolaryngol. Head Neck Surg. 2020;146:1054–1058. doi: 10.1001/jamaoto.2020.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuyama S., Kawase M., Nao N., Shirato K., Ujike M., Kamitani W., Shimojima M., Fukushi S. The Inhaled Steroid Ciclesonide Blocks SARS-CoV-2 RNA Replication by Targeting the Viral Replication-Transcription Complex in Cultured Cells. J. Virol. 2020;95:e01648-20. doi: 10.1128/JVI.01648-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monk P.D., Marsden R.J., Tear V.J., Brookes J., Batten T.N., Mankowski M., Gabbay F.J., Davies D.E., Holgate S.T., Ho L.P., Inhaled Interferon Beta COVID-19 Study Group Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir. Med. 2021;9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrouel F., Gonçalves L.S., Conte M.P., Campus G., Fisher J., Fraticelli L., Gadea-Deschamps E., Ottolenghi L., Bourgeois D. Antiviral Activity of Reagents in Mouth Rinses against SARS-CoV-2. J. Dent. Res. 2021;100:124–132. doi: 10.1177/0022034520967933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan F.R., Kazmi S.M.R., Iqbal N.T., Iqbal J., Ali S.T., Abbas S.A. A quadruple blind, randomised controlled trial of gargling agents in reducing intraoral viral load among hospitalised COVID-19 patients: A structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21:785. doi: 10.1186/s13063-020-04634-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakaguchi W., Kubota N., Shimizu T., Saruta J., Fuchida S., Kawata A., Yamamoto Y., Sugimoto M., Yakeishi M., Tsukinoki K. Existence of SARS-CoV-2 Entry Molecules in the Oral Cavity. Int. J. Mol. Sci. 2020;21:E6000. doi: 10.3390/ijms21176000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou L., Xu Z., Castiglione G.M., Soiberman U.S., Eberhart C.G., Duh E.J. ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. Ocul. Surf. 2020;18:537–544. doi: 10.1016/j.jtos.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vieira Braga F.A., Kar G., Berg M., Carpaij O.A., Polanski K., Simon L.M., Brouwer S., Gomes T., Hesse L., Jiang J. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 2019;25:1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 26.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., HCA Lung Biological Network. Electronic address: lung-network@humancellatlas.org. HCA Lung Biological Network SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ordovas-Montanes J., Dwyer D.F., Nyquist S.K., Buchheit K.M., Vukovic M., Deb C., Wadsworth M.H., 2nd, Hughes T.K., Kazer S.W., Yoshimoto E. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature. 2018;560:649–654. doi: 10.1038/s41586-018-0449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171:1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldfarbmuren K.C., Jackson N.D., Sajuthi S.P., Dyjack N., Li K.S., Rios C.L., Plender E.G., Montgomery M.T., Everman J.L., Bratcher P.E. Dissecting the cellular specificity of smoking effects and reconstructing lineages in the human airway epithelium. Nat. Commun. 2020;11:2485. doi: 10.1038/s41467-020-16239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamage A.M., Tan K.S., Chan W.O.Y., Liu J., Tan C.W., Ong Y.K., Thong M., Andiappan A.K., Anderson D.E., Wang Y., Wang L.F. Infection of human Nasal Epithelial Cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog. 2020;16:e1009130. doi: 10.1371/journal.ppat.1009130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aguiar J.A., Tremblay B.J.-M., Mansfield M.J., Woody O., Lobb B., Banerjee A., Chandiramohan A., Tiessen N., Cao Q., Dvorkin-Gheva A. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur. Respir. J. 2020;56:2001123. doi: 10.1183/13993003.01123-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravindra N.G., Alfajaro M.M., Gasque V., Huston N.C., Wan H., Szigeti-Buck K., Yasumoto Y., Greaney A.M., Habet V., Chow R.D. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol. 2021;19:e3001143. doi: 10.1371/journal.pbio.3001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang X., Yang M., Duan Z., Liao Z., Liu L., Cheng R., Fang M., Wang G., Liu H., Xu J. Transferrin receptor is another receptor for SARS-CoV-2 entry. bioRxiv. 2020 doi: 10.1101/2020.10.23.350348. [DOI] [Google Scholar]
- 37.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daly J.L., Simonetti B., Klein K., Chen K.-E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucas J.M., True L., Hawley S., Matsumura M., Morrissey C., Vessella R., Nelson P.S. The androgen-regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J. Pathol. 2008;215:118–125. doi: 10.1002/path.2330. [DOI] [PubMed] [Google Scholar]
- 40.McAdam A.J. Special Issue on Diagnostic Testing for Severe Acute Respiratory Syndrome Coronavirus 2 and Lessons from This Pandemic. J. Clin. Microbiol. 2020;58:e01324-20. doi: 10.1128/JCM.01324-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhold A., Tzankov A., Matter M., Mihic-Probst D., Scholl H.P.N., Meyer P. Ocular pathology and occasionally detectable intraocular SARS-CoV-2 RNA in five fatal COVID-19 cases. Ophthalmic Res. 2021 doi: 10.1159/000514573. Published online January 20, 2021. [DOI] [PubMed] [Google Scholar]
- 42.Liu J., Babka A.M., Kearney B.J., Radoshitzky S.R., Kuhn J.H., Zeng X. Molecular detection of SARS-CoV-2 in formalin-fixed, paraffin-embedded specimens. JCI Insight. 2020;5:139042. doi: 10.1172/jci.insight.139042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ko C.J., Harigopal M., Gehlhausen J.R., Bosenberg M., McNiff J.M., Damsky W. Discordant anti-SARS-CoV-2 spike protein and RNA staining in cutaneous perniotic lesions suggests endothelial deposition of cleaved spike protein. J. Cutan. Pathol. 2021;48:47–52. doi: 10.1111/cup.13866. [DOI] [PubMed] [Google Scholar]
- 44.Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meinhardt J., Radke J., Dittmayer C., Franz J., Thomas C., Mothes R., Laue M., Schneider J., Brünink S., Greuel S. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2021;24:168–175. doi: 10.1038/s41593-020-00758-5. [DOI] [PubMed] [Google Scholar]
- 46.Silva J., Lucas C., Sundaram M., Israelow B., Wong P., Klein J., Tokuyama M., Lu P., Venkataraman A., Liu F. Saliva viral load is a dynamic unifying correlate of COVID-19 severity and mortality. medRxiv. 2021 doi: 10.1101/2021.01.04.21249236. [DOI] [Google Scholar]
- 47.Reddy R.K., Charles W.N., Sklavounos A., Dutt A., Seed P.T., Khajuria A. The effect of smoking on COVID-19 severity: A systematic review and meta-analysis. J. Med. Virol. 2020;93:1045–1046. doi: 10.1002/jmv.26389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farsalinos K., Barbouni A., Poulas K., Polosa R., Caponnetto P., Niaura R. Current smoking, former smoking, and adverse outcome among hospitalized COVID-19 patients: a systematic review and meta-analysis. Ther. Adv. Chronic Dis. 2020;11 doi: 10.1177/2040622320935765. 2040622320935765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.HuangFu W.C., Liu J., Harty R.N., Fuchs S.Y. Cigarette smoking products suppress anti-viral effects of Type I interferon via phosphorylation-dependent downregulation of its receptor. FEBS Lett. 2008;582:3206–3210. doi: 10.1016/j.febslet.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purkayastha A., Sen C., Garcia G., Jr., Langerman J., Shia D.W., Meneses L.K., Vijayaraj P., Durra A., Koloff C.R., Freund D.R. Direct Exposure to SARS-CoV-2 and Cigarette Smoke Increases Infection Severity and Alters the Stem Cell-Derived Airway Repair Response. Cell Stem Cell. 2020;27:869–875.e4. doi: 10.1016/j.stem.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agyeman A.A., Chin K.L., Landersdorfer C.B., Liew D., Ofori-Asenso R. Smell and Taste Dysfunction in Patients With COVID-19: A Systematic Review and Meta-analysis. Mayo Clin. Proc. 2020;95:1621–1631. doi: 10.1016/j.mayocp.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brann D.H., Tsukahara T., Weinreb C., Lipovsek M., Van den Berge K., Gong B., Chance R., Macaulay I.C., Chou H.J., Fletcher R.B. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020;6:eabc5801. doi: 10.1126/sciadv.abc5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richard M., van den Brand J.M.A., Bestebroer T.M., Lexmond P., de Meulder D., Fouchier R.A.M., Lowen A.C., Herfst S. Influenza A viruses are transmitted via the air from the nasal respiratory epithelium of ferrets. Nat. Commun. 2020;11:766. doi: 10.1038/s41467-020-14626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koblizek V., Tomsova M., Cermakova E., Papousek P., Pracharova S., Mandalia R.A.A., Ceral J., Novosad J., Fila L., Sedlak V. Impairment of nasal mucociliary clearance in former smokers with stable chronic obstructive pulmonary disease relates to the presence of a chronic bronchitis phenotype. Rhinology. 2011;49:397–406. doi: 10.4193/Rhino11.051. [DOI] [PubMed] [Google Scholar]
- 55.Xavier R.F., Ramos D., Ito J.T., Rodrigues F.M.M., Bertolini G.N., Macchione M., de Toledo A.C., Ramos E.M. Effects of cigarette smoking intensity on the mucociliary clearance of active smokers. Respiration. 2013;86:479–485. doi: 10.1159/000348398. [DOI] [PubMed] [Google Scholar]
- 56.Buetti N., Trimboli P., Mazzuchelli T., Lo Priore E., Balmelli C., Trkola A., Conti M., Martinetti G., Elzi L., Ceschi A. Diabetes mellitus is a risk factor for prolonged SARS-CoV-2 viral shedding in lower respiratory tract samples of critically ill patients. Endocrine. 2020;70:454–460. doi: 10.1007/s12020-020-02465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., ACTT-1 Study Group Members Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horby P.W., Mafham M., Bell J.L., Linsell L., Staplin N., Emberson J., RECOVERY Collaborative Group Lopinavir-ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2020;396:1345–1352. doi: 10.1016/S0140-6736(20)32013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masiá M., Fernández-González M., Padilla S., Ortega P., García J.A., Agulló V., García-Abellán J., Telenti G., Guillén L., Gutiérrez F. Impact of interleukin-6 blockade with tocilizumab on SARS-CoV-2 viral kinetics and antibody responses in patients with COVID-19: A prospective cohort study. EBioMedicine. 2020;60:102999. doi: 10.1016/j.ebiom.2020.102999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spagnuolo V., Guffanti M., Galli L., Poli A., Querini P.R., Ripa M., Clementi M., Scarpellini P., Lazzarin A., Tresoldi M., COVID-BioB study group Viral clearance after early corticosteroid treatment in patients with moderate or severe covid-19. Sci. Rep. 2020;10:21291. doi: 10.1038/s41598-020-78039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., Trial Investigators REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell. 2020;183:1901–1912.e9. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.H., Boucau J., Bowman K. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., BLAZE-1 Investigators SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zoufaly A., Poglitsch M., Aberle J.H., Hoepler W., Seitz T., Traugott M., Grieb A., Pawelka E., Laferl H., Wenisch C. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir. Med. 2020;8:1154–1158. doi: 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greenwald N.F., Miller G., Moen E., Kong A., Kagel A., Camacho Fullaway C., McIntosh B.J., Leow K., Schwartz M.S. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. bioRxiv. 2021 doi: 10.1101/2021.03.01.431313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Valen D.A., Kudo T., Lane K.M., Macklin D.N., Quach N.T., DeFelice M.M., Maayan I., Tanouchi Y., Ashley E.A., Covert M.W. Deep Learning Automates the Quantitative Analysis of Individual Cells in Live-Cell Imaging Experiments. PLoS Comput. Biol. 2016;12:e1005177. doi: 10.1371/journal.pcbi.1005177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Menter T., Haslbauer J.D., Nienhold R., Savic S., Hopfer H., Deigendesch N., Frank S., Turek D., Willi N., Pargger H. Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology. 2020;77:198–209. doi: 10.1111/his.14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mabbott N.A., Baillie J.K., Brown H., Freeman T.C., Hume D.A. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics. 2013;14:632. doi: 10.1186/1471-2164-14-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wickham H. Springer; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available RNA datasets. The accession numbers for these datasets are listed in the Key resources table. The analysis codes used to support additional findings of this study were minimally modified from previously published codes1 and are available at: GitHub:
https://github.com/bmyury/membrane_ACE2_quantitation. Additional data reported in this paper will be shared by the Lead Contact upon reasonable request.