Abstract
Aims
Developmental dysplasia of the hip (DDH) is a complex musculoskeletal disease that occurs mostly in children. This study aimed to investigate the molecular changes in the hip joint capsule of patients with DDH.
Methods
High-throughput sequencing was used to identify genes that were differentially expressed in hip joint capsules between healthy controls and DDH patients. Biological assays including cell cycle, viability, apoptosis, immunofluorescence, reverse transcription polymerase chain reaction (RT-PCR), and western blotting were performed to determine the roles of the differentially expressed genes in DDH pathology.
Results
More than 1,000 genes were differentially expressed in hip joint capsules between healthy controls and DDH. Both gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses revealed that extracellular matrix (ECM) modifications, muscle system processes, and cell proliferation were markedly influenced by the differentially expressed genes. Expression of Collagen Type I Alpha 1 Chain (COL1A1), COL3A1, matrix metalloproteinase-1 (MMP1), MMP3, MMP9, and MMP13 was downregulated in DDH, with the loss of collagen fibres in the joint capsule. Expression of transforming growth factor beta 1 (TGF-β1) was downregulated, while that of TGF-β2, Mothers against decapentaplegic homolog 3 (SMAD3), and WNT11 were upregulated in DDH, and alpha smooth muscle actin (αSMA), a key myofibroblast marker, showed marginal increase. In vitro studies showed that fibroblast proliferation was suppressed in DDH, which was associated with cell cycle arrest in G0/G1 and G2/M phases. Cell cycle regulators including Cyclin B1 (CCNB1), Cyclin E2 (CCNE2), Cyclin A2 (CCNA2), Cyclin-dependent kinase 1 (CDK1), E2F1, cell division cycle 6 (CDC6), and CDC7 were downregulated in DDH.
Conclusion
DDH is associated with the loss of collagen fibres and fibroblasts, which may cause loose joint capsule formation. However, the degree of differentiation of fibroblasts to myofibroblasts needs further study.
Cite this article: Bone Joint Res 2021;10(9):558–570.
Keywords: Developmental dysplasia of the hip, High-throughput sequencing, Extracellular matrix, joint capsules, Developmental dysplasia of the hip (DDH), fibroblasts, collagens, cell cycle, Alpha smooth muscle actin, Myofibroblasts, Western Blotting, MMP13, Collagen fibres
Article focus
Exploring gene difference in the hip joint capsule between health controls and patients with developmental dysplasia of the hip (DDH).
Exploring pathological changes in extracellular matrix in the hip joint capsule patients with DDH.
Study of the proliferation of fibroblast isolated from the hip joint capsule between health controls and patients with DDH.
Key messages
More than 1,000 genes were differentially expressed in hip joint capsules between healthy controls and DDH. Both gene ontology and Kyoto Encyclopedia of Genes and Genomes analyses revealed that extracellular matrix modifications, muscle system processes, and cell proliferation were markedly influenced by the differentially expressed genes.
Expression of Collagen Type I Alpha 1 Chain (COL1A1), COL3A1, matrix metalloproteinase-1 (MMP1), MMP3, MMP9, and MMP13 was downregulated in DDH, with the loss of collagen fibres in the joint capsule. Expression of transforming growth factor beta 1 (TGF-β1) was downregulated, while that of TGF-β2, Mothers against decapentaplegic homolog 3 (SMAD3), and WNT11 were upregulated in DDH.
In vitro studies showed that fibroblast proliferation was suppressed in DDH, which was associated with cell cycle arrest in G0/G1 and G2/M phases. Cell cycle regulators including Cyclin B1 (CCNB1), Cyclin E2 (CCNE2), Cyclin A2 (CCNA2), Cyclin-dependent kinase 1 (CDK1), E2F1, cell division cycle 6 (CDC6), and CDC7 were downregulated in DDH.
Strengths and limitations
This is the first study to explore gene differences in the hip joint capsule between health controls and patients with DDH.
This was also the first study to research the proliferation of fibroblasts isolated from the hip joint capsule between health controls and patients with DDH.
This study did not perform an animal study to identify the data from clinical and cell studies.
Introduction
Developmental dysplasia of the hip (DDH) is a complex musculoskeletal disease with an incidence rate of 1.4% to 3.5%.1,2 Important risk factors for DDH include previous family history of DDH, breech position, female sex (the incidence rate in females is approximately five times that in males), and incorrect lower limb swaddling. DDH is associated with incompatibility of the femoral head and abnormal joint socket.3 Normal development of the femoral head and acetabulum are codependent; the head must be stable in the hip socket for both to form spherically and concentrically. If either component is deficient, the head becomes loose in the acetabulum, which results in joint instability, subluxation, and irreducible hip dislocation.4 Untreated DDH can cause secondary damage to the femur, destruction of the joint cartilage, and even severe movement impairment and osteoarthritis (OA) at any age.
Currently, most studies on the pathogenesis of DDH focus on the role of defects in the femoral head and acetabulum, and very few studies have examined the pathological changes in the soft tissue around the hip joint. The soft-tissue structures consist of the labrum, capsule, and muscles. Besides the bones, the hip capsule is one of the most important static stabilizers of the hip joint, whereas the muscles serve as dynamic hip stabilizers. Articular capsule belongs to connective tissue, which mainly consists of fibroblasts and collagen fibres. Some researchers deem that a lax joint capsule is also an important contributor to hip instability in DDH.5,6 This pathological change is probably associated with various hereditary factors, secondary adaptation to the dislocation, and abnormal movement of the femoral head. In this study, we conducted high-throughput sequencing to gain insight into the molecular changes in the hip joint capsule of patients with DDH. Furthermore, a series of other assays were performed to determine which abnormally expressed genes and biological processes are associated with DDH pathology.
Methods
Clinical assessment and sample collection
A total of 15 patients with DDH and 12 control subjects were enrolled in this study. There was no statistical difference between patients with DDH and control subjects in age and sex ratio (female/male). More detailed patient data are shown in Table I. The diagnosis of the patients with DDH was confirmed by expert medical examination with nuclear MRI and radiological evidence, and they all had unilateral or bilateral DDH. The inclusion criterion was a diagnosis of a subluxated or dislocated hip. Exclusion criteria included diagnosis of neuromuscular disease or teratological dislocations, history of open reduction as initial management, incomplete radiological data, history of multiple closed reductions, and onset of avascular necrosis after subsequent surgery. Control subjects were enrolled from patients with femoral neck fracture due to an accidental fall. Despite the femoral neck fracture, the controls had no symptoms or history of congenital dislocation of the hip or other diseases. Control subjects were identified by taking a detailed history and physical examination. Tissues from the hip joint capsule of patients with DDH and control subjects were obtained from Colonna arthroplasty and open reduction of fracture surgeries, respectively. All subjects signed an informed consent form before enrolment. The joint capsule tissues were washed by phosphate-buffered saline (Thermofisher, China), and then synovium tissues were removed from the joint capsules. The isolated joint capsule tissues were used in the study.
Table I.
Participants' clinical information.
| Group | Mean age, yrs (range) | Female/male, n | Mean weight, kg (SD) | Mean height, cm (SD) |
|---|---|---|---|---|
| Control | 20.5 (12 to 36) | 8/2 | 42.65 (18.54) | 145.26 (24.08) |
| DDH | 14.8 (3 to 30) | 12/3 | 35.42 (22.17) | 112.35 (67.25) |
DDH, developmental dysplasia of the hip; SD, standard deviation.
High-throughput sequencing
For RNA extraction, tissues from the hip joint capsule obtained from healthy controls (n = 6) and patients with DDH (n = 6) were homogenized in TRIzol reagent (Thermo Fisher Scientific, USA) using a 1600 MiniG-Automated Tissue Homogenizer (Metuchen, USA). Agilent 2100 Bioanalyzer was used to test the quality of the RNA (Supplementary Material). Only RNAs with OD260/280 values between 1.8 and 2.0 were used in subsequent studies. One microgram of total RNA was used as input for messenger RNA (mRNA) synthesis. mRNAs were purified using oligo-dTs covalently coupled to magnetic beads, and fragmented into smaller pieces of 180 nt to 250 nt in length by heating. First-strand complementary DNA (cDNA) was synthesized using RNAs as templates. A DNA library was constructed through end-repair, adaptor-ligation, and polymerase chain reaction (PCR) amplification using the Illumina NovaSeq 600 platform (Illumina, USA).
Enrichment analysis
RNA-seq reads were first mapped to the rRNA sequences using STAR (v2.7.2b) with the default parameter to remove potential rRNA reads. Functional enrichment analysis was performed on the differentially expressed genes (adjusted p < 0.05) using the ClusterProfiler package (v 3.14.3, Bioconductor, USA). The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation were downloaded from gene ontology resource (last updated 22 February 2020) and KEGG database (last updated 14 January 2020), respectively. They were used to classify the significantly differentially expressed genes in specific GO terms and KEGG pathways. Only significantly enriched KEGG functional categories (p < 0.05) are depicted according to their p-values (-log 10 (p-value)).
Histological examination
Hip joint capsule specimens were fixed with 10% neutral-buffered formalin, followed by gradient ethanol dehydration and embedding in paraffin. Sections of 5 µm thickness were prepared for haematoxylin and eosin (HE) staining.
For Masson’s staining, the sections were first stained with Harris haematoxylin for five minutes, and then flushed with a copious amount of 1% (v/v) acetic acid solution. Then the sections were further stained with a Ponceau 2 R/Acid Fuchsin mixture, Light Green SF Yellowish, and phosphomolybdic acid intermediate solution for two minutes, and washed with acetic acid solution to remove the unbound dyes. After washing, dehydration, and mounting, collagen staining intensity in the sections was visualized under a light microscope.
For immunocytochemistry, paraffin-embedded sections of the tissue specimens were deparaffinized and heated at 97°C in 10 mM citrate buffer (pH 6.0) for 20 minutes for antigen retrieval. Tissue sections were incubated with anti-type I collagen (1:1,000; Abcam, USA, ab34710), anti-type III collagen (1:1,000; Abcam, ab7778), anti-MMP1 (1:1,000; Abcam, ab52631), anti-MMP3 (1:1,000; Abcam, ab52915), anti-MMP9 (1:500; Abcam, ab76003), and anti-MMP13 (1:500; Abcam, ab219620) antibodies overnight at 4°C, followed by incubation with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (1:200; Abcam) at 37°C for 30 minutes. The sections were then incubated with 500 µl of diaminobenzidine (working solution) at room temperature for three to ten minutes.
Isolation and culture of human fibroblasts
Briefly, tissues from the hip joint capsule were minced into small pieces (1 mm3) and digested with 0.2% collagenase for four hours at 37°C. The isolated cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Thermo Fisher Scientific, USA) containing 10% fetal bovine serum (MilliporeSigma, USA) and 1% antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin sulphate). The second passage of primary fibroblasts was used in the study.
Immunofluorescence
Primary fibroblasts growing as a monolayer on glass cover slides were fixed with 4% paraformaldehyde, blocked with 5% normal goat serum, and then subjected to overnight incubation with anti-human type I collagen antibody (1:100) at 4°C. The cells were washed three times with phosphate-buffered saline (PBS) containing the detergent tween (PBST) and incubated with Alexa Fluor 488-conjugated secondary antibody for one hour at room temperature.
Cell viability assay
Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was used to assess cell viability. As per the manufacturer’s instructions, cells were washed with PBS and then cultured in a culture medium containing 10% CCK-8 solution at 37°C for three to six hours. Optical density was measured at 450 nm using a microplate reader (BioTek, USA).
Apoptosis and cell cycle assays
Apoptosis was evaluated using the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining Kit (Dojindo Molecular Technologies, China). After the indicated treatments, cells were harvested and resuspended in PBS at a concentration of 1 × 106 cells/ml. A 100 µl sample of the cell suspension was mixed with 5 µl Annexin V-FITC and 5 μl PI. The mixture was incubated for 15 minutes at room temperature in the dark and analyzed using a FACSCalibur Flow Cytometer (Beckman Coulter, CytoFLEX S, USA).
Cell cycle progression was determined using PI (BD Biosciences) staining, using a flow cytometer. Briefly, cells were fixed with 70% cold ethanol at 4°C overnight. The cells were rehydrated, washed twice with ice-cold PBS, and incubated with 10 mg/ml RNase (Fermentas, China) at 37°C. Subsequently, cell cycle was assessed by analyzing the PI-stained nuclei using FACSCalibur flow cytometer (Becton Dickinson, USA).
5-ethylnyl-2ʹ-deoxyuridine assay
EdU assay was performed using a Cell-Light EdU DNA Cell Proliferation kit (RiboBio, China). Cells (1 × 104) were plated in each well of a 96-well plate. After incubation with 50 mM EdU for two hours, the cells were fixed in 4% paraformaldehyde and stained with Apollo Dye Solution. DAPI was used to stain the nucleic acids within the cells. Images were obtained with a fluorescence microscope, and the number of EdU-positive cells was counted.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from the joint capsular tissues or primary fibroblasts using TRIzol reagent (Invitrogen), and reverse-transcribed using PrimeScript RT reagent kit (Takara, China). The sequences of the primers used are shown in Table II. Quantitative PCR was then performed using SYBR Green Real-Time PCR Master Mix (Thermo Fisher Scientific) in an ABI StepOnePlus Real-Time PCR System (Applied Biosystems, USA).
Table II.
The primer information.
| Gene names | Primer direction | Sequence (5' - 3') | Tm(°C) | Location | Amplification size |
|---|---|---|---|---|---|
| COL1A1 | Forward | GAGGGCCAAGACGAAGACATC | 62.5 | 85 to 105 | 140 |
| Reverse | CAGATCACGTCATCGCACAAC | 61.6 | 224 to 204 | ||
| COL3A1 | Forward | GGAGCTGGCTACTTCTCGC | 62.2 | 23 to 41 | 78 |
| Reverse | GGGAACATCCTCCTTCAACAG | 60 | 100 to 80 | ||
| MMP1 | Forward | AAAATTACACGCCAGATTTGCC | 60 | 356 to 377 | 82 |
| Reverse | GGTGTGACATTACTCCAGAGTTG | 60.5 | 437 to 415 | ||
| MMP3 | Forward | AGTCTTCCAATCCTACTGTTGCT | 61 | 7 to 29 | 226 |
| Reverse | TCCCCGTCACCTCCAATCC | 63 | 232 to 214 | ||
| MMP9 | Forward | TGTACCGCTATGGTTACACTCG | 61.5 | 146 to 167 | 97 |
| Reverse | GGCAGGGACAGTTGCTTCT | 61.9 | 242 to 224 | ||
| MMP13 | Forward | ACTGAGAGGCTCCGAGAAATG | 61.3 | 193 to 213 | 103 |
| Reverse | GAACCCCGCATCTTGGCTT | 62.7 | 295 to 277 | ||
| TGF-β1 | Forward | GGCCAGATCCTGTCCAAGC | 62.4 | 151 to 169 | 201 |
| Reverse | GTGGGTTTCCACCATTAGCAC | 61.2 | 351 to 331 | ||
| TGF-β2 | Forward | CAGCACACTCGATATGGACCA | 61.6 | 72 to 92 | 113 |
| Reverse | CCTCGGGCTCAGGATAGTCT | 62.1 | 184 to 165 | ||
| SMAD3 | Forward | TGGACGCAGGTTCTCCAAAC | 62.4 | 284 to 303 | 90 |
| Reverse | CCGGCTCGCAGTAGGTAAC | 62.1 | 373 to 355 | ||
| WNT11 | Forward | GGAGTCGGCCTTCGTGTATG | 62.3 | 327 to 346 | 167 |
| Reverse | GCCCGTAGCTGAGGTTGTC | 62.4 | 493 to 475 | ||
| CCNB1 | Forward | TTGGGGACATTGGTAACAAAGTC | 60.7 | 134 to 156 | 226 |
| Reverse | ATAGGCTCAGGCGAAAGTTTTT | 60.2 | 359 to 338 | ||
| CCNE2 | Forward | TCAAGACGAAGTAGCCGTTTAC | 60 | 4 to 25 | 115 |
| Reverse | TGACATCCTGGGTAGTTTTCCTC | 61.3 | 118 to 96 | ||
| CCNA2 | Forward | CGCTGGCGGTACTGAAGTC | 62.7 | 146 to 164 | 120 |
| Reverse | GAGGAACGGTGACATGCTCAT | 62.1 | 265 to 245 | ||
| CDK1 | Forward | AAACTACAGGTCAAGTGGTAGCC | 61.8 | 71 to 93 | 148 |
| Reverse | TCCTGCATAAGCACATCCTGA | 60.9 | 218 to 198 | ||
| E2F1 | Forward | ACGCTATGAGACCTCACTGAA | 60.3 | 378 to 398 | 249 |
| Reverse | TCCTGGGTCAACCCCTCAAG | 63 | 626 to 607 | ||
| CDC6 | Forward | CCAGGCACAGGCTACAATCAG | 62.8 | 18 to 38 | 116 |
| Reverse | AACAGGTTACGGTTTGGACATT | 60.1 | 133 to 112 | ||
| CDC7 | Forward | GAGGCGTCTTTGGGGATTCAG | 62.7 | 4 to 24 | 245 |
| Reverse | GGTCCTACTTGTAACTGTGCTG | 60.3 | 248 to 227 | ||
| ACTB | Forward | GGCTGTATTCCCCTCCATCG | 61.8 | 84 to 103 | 154 |
| Reverse | CCAGTTGGTAACAATGCCATGT | 61.1 | 237 to 216 |
ACTB, actin beta; CCNB1, Cyclin B1; CDC6, cell division cycle 6; CDK1, cyclin dependent kinase 1; COL1A1, collagen, type 1, alpha 1; COL3A1, collagen, type 3, alpha 1; MMP, matrix metalloproteinase; SMAD3, Mothers against decapentaplegic homolog 3; TGF-β1, tumour growth factor beta 1.
Western blotting
Primary fibroblasts were lysed with RIPA buffer supplemented with a protease inhibitor cocktail (Beyotime Institute of Biotechnology, China). Cell lysates were subjected to ultrasonication at 4°C. Based on the molecular weight of the target proteins, equal amounts of total proteins from each sample were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a gel of the appropriate concentration and electroblotted onto a polyvinylidene difluoride (PVDF) membrane (Invitrogen, USA). The membrane was incubated with the primary antibodies overnight at 4°C. Detailed information about the antibodies is shown in Table III. After the membrane was incubated with the second antibody (Abcam) and Pierc ECL Western Blotting Substrate, the protein bands were imaged using a Gel Imaging System (Life Science, USA), and the band intensity was determined using ImageJ software (National Institutes of Health, USA).
Table III.
The manufacturer and catalogue numbers of antibodies used in western blot assay.
| Protein name | Manufacturer | Catalogue numbers | Dilution |
|---|---|---|---|
| Type I collagen | Abcam | ab34710 | 1:1,000 |
| Type III collagen | Abcam | ab7778 | 1:500 |
| MMP1 | Abcam | ab134184 | 1:500 |
| MMP3 | Abcam | ab52915 | 1:500 |
| MMP9 | Abcam | ab76003 | 1:500 |
| MMP13 | Abcam | ab51072 | 1:250 |
| TGF-β1 | Abcam | ab215715 | 1:250 |
| TGF-β2 | Abcam | ab113670 | 1:500 |
| SMAD3 | Abcam | ab40854 | 1:500 |
| WNT11 | Abcam | ab96730 | 1:500 |
| αSMA | Abcam | ab5694 | 1:500 |
| β-actin | Abcam | ab8226 | 1:1,000 |
MMP, matrix metalloproteinase; αSMA, smooth muscle alpha-actin; SMAD3, Mothers against decapentaplegic homolog 3; TGF-β1, tumour growth factor beta 1.
Statistical analysis
Data are expressed as the mean and standard deviation of three independent experiments. Experimental data were examined using an unpaired two-tailed t-test with GraphPad Prism software (GraphPad Software, USA). Statistical significance was set at p < 0.05.
Results
Differences in gene expression profile in the hip joint capsule between healthy controls and patients with DDH
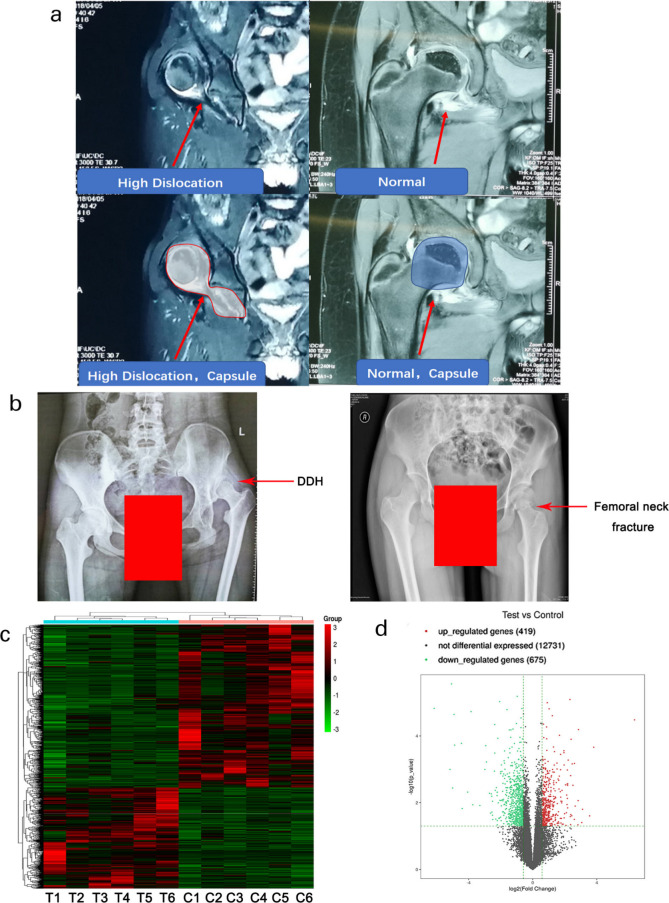
Nuclear magnetic resonance and radiograph detection were used for the diagnosis of DDH disease. Nuclear magnetic resonance results showed the hip capsule tissue was stretched in patients with DDH compared to healthy controls (Figure 1a). Radiographs showed that the femoral head slipped off from acetabulum in patients with DDH compared to healthy control (Figure 1b). Hip joint capsule tissues from patients with DDH and control subjects were obtained from Colonna arthroplasty and open reduction of fracture surgeries, respectively. RNA-seq analysis showed 419 upregulated and 675 downregulated genes (p < 0.05 and log2 (fold changes) > 1 or log2 (fold changes) < -1) in hip joint capsule from patients with DDH compared to healthy controls. The expression of these genes is presented as a clustering heatmap (Figure 1c) and scatter plot (Figure 1d). Moreover, the top 30 up- and downregulated genes are listed in Table IV and Table V, respectively. MMP1, MMP3, MMP9, and MMP13 were among the top 30 downregulated genes belonging to matrix metallopeptidase family of proteins. All the RNA-seq data have been deposited on the GEO database.
Fig. 1.
High-throughput sequencing assay reveals genes differentially expressed in hip joint capsules between healthy control and developmental dysplasia of the hip (DDH). Tissues from the hip joint capsule of patients with DDH (n = 6) and healthy controls (n = 6) were used for the high-throughput sequencing assay. Control subjects are enrolled from patients with femoral neck fracture. Despite the femoral neck fracture, the controls had no symptoms or history of congenital dislocation of the hip and other diseases. a) Nuclear magnetic resonance and b) radiograph detection were used for the diagnosis of DDH. c) Gene expression levels are shown in a heat map, in which each row represents a gene and each column represents a sample of hip joint capsule tissue. The variation in colour from red to green represents the signal level, with red indicating high signal or upregulation, green indicating low signal or downregulation, and black representing unchanged expression. T: Test group in which the samples were obtained from patients with DDH; C: Control group in which the samples were obtained from healthy controls. d) Scatter plot showing the distribution of differentially expressed genes between healthy controls and patients with DDH. The x and y axes represent log2 (fold change) and -log10 (p-value), respectively. Red dots indicate upregulated genes and green dots indicate downregulated genes. Gray dots represent genes with no difference in expression. The dashed line defines the up- and downregulation boundaries (p < 0.05 and log2 (fold changes) > 1 or log2 (fold changes) < -1).
Table IV.
Top 30 upregulated genes (Test group vs control group).
| Gene_Name | Locus | Fold_Change | p_value |
|---|---|---|---|
| MYOC | chr1:171604557 to 171621823 | 82.09671149 | 3.30628E-05 |
| FAM180B | chr11:47608245 to 47610746 | 14.03528461 | 0.000218204 |
| CCL21 | chr9:34709002 to 34710121 | 11.75325576 | 0.024751568 |
| CLIC5 | chr6:45866188 to 46048132 | 8.637746402 | 0.015670664 |
| SLN | chr11:107578104 to 107590419 | 8.366523839 | 0.039283577 |
| ADH1C | chr4:100257649 to 100274184 | 7.253611803 | 6.46588E-05 |
| FGL2 | chr7:76822688 to 76829165 | 7.140147496 | 0.008662333 |
| GPR1 | chr2:207040040 to 207082771 | 7.088187725 | 0.046015133 |
| PLN | chr6:118869461 to 118881893 | 6.287545514 | 0.012610094 |
| FRZB | chr2:183698002 to 183731890 | 6.102363994 | 0.000664237 |
| HSPB7 | chr1:16340523 to 16346089 | 6.05151459 | 0.031512454 |
| FHL1 | chrX:135228861 to 135293518 | 5.704869898 | 0.00300196 |
| NOV | chr8:120428546 to 120436593 | 5.528264771 | 0.000687254 |
| CHAD | chr17:48541857 to 48546327 | 5.429806708 | 0.024503997 |
| GSN | chr9:123970072 to 124095121 | 5.27406107 | 0.000125852 |
| ATP1A2 | chr1:160085549 to 160113381 | 5.198570136 | 0.022687057 |
| IGFBP6 | chr12:53491220 to 53496129 | 5.174229594 | 0.00154529 |
| PPP1R1A | chr12:54969171 to 54982443 | 5.17016258 | 0.045299859 |
| CHRDL1 | chrX:109917084 to 110039047 | 5.078743134 | 0.02615956 |
| TSPAN7 | chrX:38420623 to 38548174 | 5.011818796 | 8.14523E-06 |
| PNMT | chr17:37824234 to 37826728 | 4.649526067 | 0.000528959 |
| FXYD1 | chr19:35629712 to 35634013 | 4.550435969 | 0.002708206 |
| MFAP5 | chr12:8789942 to 8815484 | 4.538935719 | 0.014269679 |
| ANGPTL1 | chr1:178818840 to 178840187 | 4.537908408 | 0.003011885 |
| C1QTNF4 | chr11:47611216 to 47616211 | 4.487543744 | 0.007462901 |
| VIT | chr2:36923833 to 37041935 | 4.481259104 | 0.001045967 |
| PAMR1 | chr11:35453370 to 35551848 | 4.445844634 | 0.002758877 |
| HSPB6 | chr19:36245470 to 36248980 | 4.413947554 | 0.011363067 |
| HRCT1 | chr9:35906189 to 35907138 | 4.357810059 | 0.00587893 |
| ANGPTL5 | chr11:101761405 to 101787253 | 4.293532067 | 0.019980743 |
ADH1C, alcohol dehydrogenase 1C (Class I); ANGPTL1, angiopoietin like 1; ATP1A2, ATPase Na+/K+ transporting subunit alpha 2; CCL21, C-C Motif Chemokine Ligand 21; CHAD, chondroadherin; CHRDL1, chordin like 1; CLIC5, chloride intracellular channel 5; C1QTNF4, C1q and tumor necrosis factor related protein 4; FAM180B, Family With Sequence Similarity 180 Member B; FGL2, fibrinogen Like 2; FHL1, four and a half LIM domains 1; FRZB, frizzled related protein; FXYD1, FXYD domain containing ion transport regulator 1; GPR1, G protein-coupled receptor 1; GSN, gelsolin; HRCT1, histidine rich carboxyl terminus 1; HSPB7, heat shock protein family B (small) member 7; IGFBP6, insulin like growth factor binding protein 6; MFAP5, microfibril associated protein 5; MYOC, myocilin; NOV, nephroblastoma overexpressed; PAMR1, peptidase domain containing associated with muscle regeneration 1; PLN, phospholamban; PMNT, phenylethanolamine N-Methyltransferase; PPP1R1A, protein phosphatase 1 regulatory inhibitor subunit 1A; SLN, sarcolipin; TSPAN7, tetraspanin 7; VIT, vitrin.
Table V.
Top 30 downregulated genes (Test group vs control group).
| Gene_Name | Locus | Fold_Change | p_value |
|---|---|---|---|
| MMP3 | chr11:102706532 to 102714534 | 0.014042996 | 1.50127E-05 |
| MMP1 | chr11:102660651 to 102668891 | 0.027904979 | 0.001006739 |
| CHI3L2 | chr1:111743393 to 111786062 | 0.029793628 | 2.76304E-06 |
| AMTN | chr4:71384257 to 71398459 | 0.031193966 | 0.003623208 |
| CHI3L1 | chr1:203148059 to 203155877 | 0.033144309 | 2.27443E-05 |
| MMP13 | chr11:102813724 to 102826463 | 0.034096806 | 0.000188804 |
| SPP1 | chr4:88896802 to 88904578 | 0.045835679 | 0.000167266 |
| CSN1S1 | chr4:70796799 to 70812292 | 0.047230259 | 0.014938042 |
| RPS4Y1 | chrY:2709527 to 2800041 | 0.059853056 | 0.004809868 |
| MT1G | chr16:56700643 to 56701977 | 0.069314334 | 1.87297E-05 |
| MMP9 | chr20:44637547 to 44645200 | 0.074788099 | 0.011522448 |
| CCL18 | chr17:34391640 to 34399392 | 0.101491719 | 0.012068675 |
| RGS1 | chr1:192544857 to 192549161 | 0.126175485 | 0.001007045 |
| POSTN | chr13:38136720 to 38172981 | 0.131198599 | 0.000195461 |
| TREM1 | chr6:41235664 to 41254457 | 0.135158542 | 0.000603453 |
| LRRC15 | chr3:194075976 to 194090472 | 0.154291164 | 0.005223527 |
| LBP | chr20:36974759 to 37005665 | 0.160979678 | 0.020468865 |
| DDX3Y | chrY:15016019 to 15032390 | 0.166462769 | 0.005450236 |
| CPXM1 | chr20:2774715 to 2781283 | 0.170064956 | 0.006423577 |
| AL645922.1 | chr6:31895475 to 31919825 | 0.171054864 | 0.019080061 |
| S100A9 | chr1:153330330 to 153333503 | 0.17538335 | 0.023651438 |
| CA9 | chr9:35673853 to 35681156 | 0.175724451 | 0.009030186 |
| HBA1 | chr16:226679 to 227521 | 0.177789082 | 0.031770506 |
| ARL4C | chr2:235401685 to 235405697 | 0.19074171 | 0.001131891 |
| MT1F | chr16:56691606 to 56694610 | 0.193573717 | 4.55998E-05 |
| ANGPTL4 | chr19:8428173 to 8439257 | 0.193679538 | 0.000326219 |
| TMIGD3 | chr1:112025970 to 112106584 | 0.197980927 | 0.004255273 |
| HBB | chr11:5246694 to 5250625 | 0.202378973 | 0.030887531 |
| CXCL8 | chr4:74606223 to 74609433 | 0.207242113 | 0.003540663 |
| ALPL | chr1:21835865 to 21904905 | 0.213662582 | 0.00609762 |
ALPL, alkaline phosphatase, biomineralization associated; AMTN, amelotin; ANGPTL4, angiopoietin like 4; ARL4C, ADP ribosylation factor like GTPase 4C; CA9, carbonic anhydrase 9; CCL18, C-C motif chemokine ligand 18; CHI3L1, chitinase 3 like 1; CPXM1, carboxypeptidase X, M14 family member 1; CSN1S1, casein alpha S1; CXCL8, C-X-C motif chemokine ligand 8; DDX3Y, DEAD-Box Helicase 3 Y-Linked; HBA1, haemoglobin subunit alpha 1; HBB, haemoglobin subunit beta; LBP, lipopolysaccharide binding protein; LRRC15, leucine rich repeat containing 15; MMP, matrix metalloproteinase; MT1F, metallothionein 1F; MT1G, metallothionein 1G; POSTN, periostin; RGS1, regulator of G protein signaling 1; RPS4Y1, ribosomal protein S4 Y-Linked 1; S100A9, S100 calcium binding protein A9; SPP1, secreted phosphoprotein 1; TMIGD3, transmembrane and immunoglobulin domain containing 3; TREM1, triggering receptor expressed on myeloid cells 1.
Differences in biological functions in hip joint capsule between healthy controls and patients with DDH
To gain insights into the biological functions influenced by the differentially expressed genes, we conducted GO enrichment analysis based on biological processes (BPs) and molecular functions (MFs). The upregulated genes primarily influenced BPs, including anatomical structure morphogenesis/development, system development, development process, and muscle system process (Supplementary Figure aa). The major MFs influenced by the upregulated genes were implicated in growth factor binding, cytoskeletal protein binding, amide binding, and actin binding (Supplementary Figure ab). These MFs were important in the above-mentioned BPs. The downregulated genes influenced multiple BPs implicated in cell proliferation, including mitotic cell cycle, cell division, chromosome segregation, cell cycle and nuclear division, and in extracellular matrix (ECM) modifications, including ECM organization, extracellular structure organization, and collagen metabolic process (Supplementary Figure ac). Some MFs influenced by the downregulated genes are also involved in ECM modification, such as metalloendopeptidase activity, metallopeptidase activity, and collagen binding (Supplementary Figure ad).
We further performed KEGG pathway analysis to identify the signalling pathways influenced by the differentially expressed genes. The upregulated genes primarily affected the TGF-β and Wnt signalling pathways that play important roles in tissue development and differentiation (Supplementary Figure ba). The upregulated genes implicated in these two pathways are marked in red (Supplementary Figure bc). In agreement with the results of the GO analysis, the downregulated genes primarily affected cell cycle and ECM-receptor interaction pathways (Supplementary Figure ca). The downregulated genes implicated in these two pathways are marked in green and shown in Supplementary Figure cc.
Slower remodelling of the ECM in hip joint capsule of patients with DDH
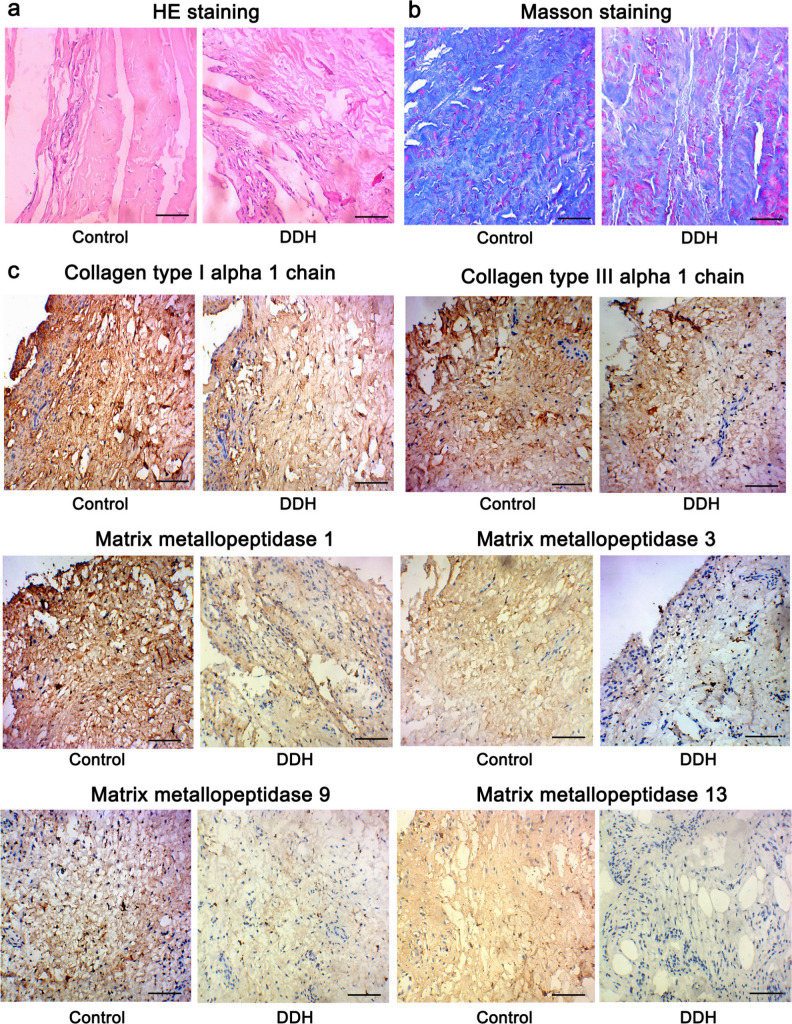
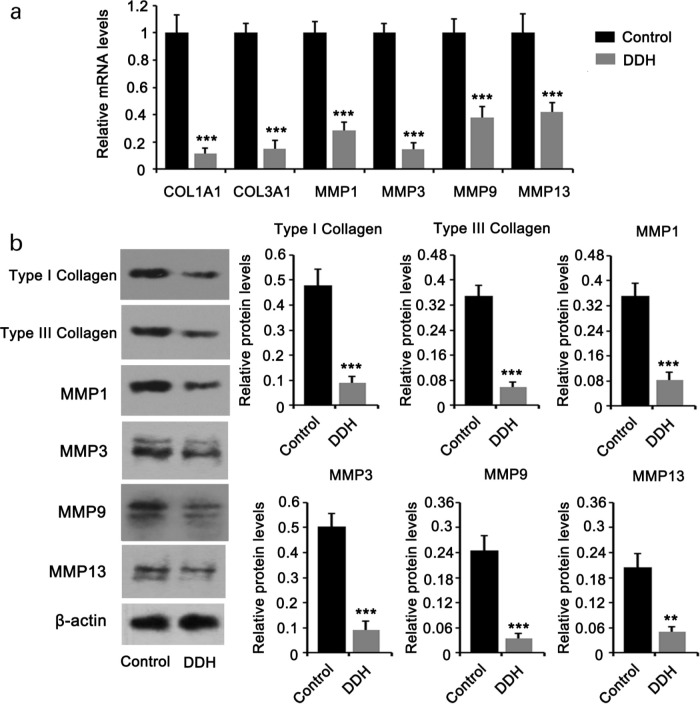
Enrichment analysis suggested that the ECM was notably altered in the hip joint capsule in patients with DDH. To further validate the changes in ECM, we performed HE, Masson’s staining, and immunocytochemistry. HE staining showed that the joint capsule tissue was disordered in patients with DDH compared to healthy controls (Figure 2a). In addition, Masson’s staining revealed that the degree of fibrosis was lower in the hip joint capsule tissues of patients with DDH (Figure 2b). As indicated by immunocytochemistry, the staining of type I and III collagens was weaker in the joint capsule tissues of patients with DDH compared to that in healthy controls (Figure 2c). MMP1, MMP3, MMP9, and MMP13 proteins are responsible for the degradation of diverse collagen fibres. The staining intensity of these proteins was decreased in the hip joint capsule tissues of patients with DDH. Moreover, we calculated the positive rate of cells expressing MMP1, MMP3, MMP9, and MMP13 in capsule tissues. The positive rates of cells expressing MMP1, MMP3, MMP9, and MMP13 were > 90%, 50% to 60%, 60% to 70%, and 10% to 20%, respectively, in controls. However, their positive rates in patients with DDH were lower, which were 50% to 60%, 30% to 40%, 10% to 20% and < 10%, respectively. To more accurately evaluate their enrichment in capsule tissues of patients with DDH and healthy controls, we also performed PCR and western blotting assays. As indicated by PCR assay, the reduction of COL1A1, COL3A1, MMP1, MMP3, MMP9, and MMP13 expression was shown in hip joint capsule tissues of patients with DDH (Figure 3a). Consistent with the PCR results, the protein levels of type I and III collagens, MMP1, MMP3, MMP9, and MMP13 were also decreased in patients with DDH (Figure 3b). The reduced production of type I and III collagens and the inhibition of degradation of collagen fibres suggest slow remodelling and restoration in the ECM.
Fig. 2.
Histological examinations using haematoxylin and eosin (HE), Masson’s, and immunohistochemistry staining (200×). Tissues from the hip joint capsule from patients with developmental dysplasia of the hip (DDH) and healthy controls were subjected to a) HE, b) Masson’s, and c) immunohistochemistry staining. Bar: 10 μm.
Fig. 3.
Expression of collagen and matrix metallopeptidase family of proteins was reduced in developmental dysplasia of the hip (DDH). Tissues from the hip joint capsule from patients with DDH and healthy controls were used for a) polymerase chain reaction (PCR), and b) western blotting. Expression of collagen, type 1, alpha 1 (COL1A1), COL3A1 (type III collagen), matrix metalloproteinase (MMP)1, MMP3, MMP9, and MMP13 was downregulated in patients with DDH. Experimental data (n = 5) were examined using an unpaired two-tailed t-test. **p < 0.01 and ***p < 0.001 versus control. mRNA, messenger RNA.
Differentiation of fibroblasts to myofibroblasts in the hip joint capsule of patients with DDH
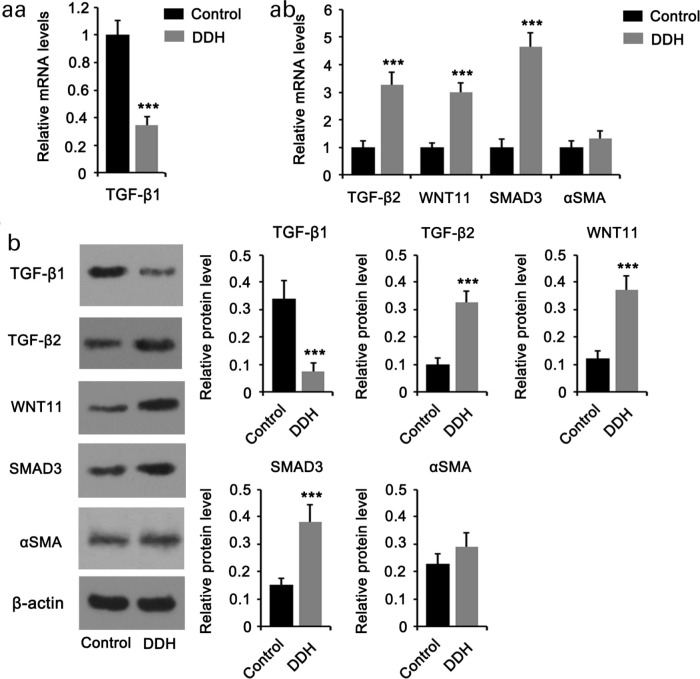
GO enrichment analysis revealed that the muscle system process is one of the primary BPs influenced by the upregulated genes in patients with DDH. Furthermore, according to KEGG pathway analysis, the upregulated genes markedly influenced TGF-β and Wnt signalling pathways, the key signalling pathways that induce the differentiation of fibroblasts to myofibroblasts. Thus, we hypothesized that the fibroblasts in the hip joint capsule of patients with DDH are inclined to differentiate into myofibroblasts. As indicated by PCR (Figures 4aa and 4ab ) and western blotting (Figure 4b), the expression levels of TGF-β2, SMAD3, and WNT11 were increased, whereas TGF-β1 expression was decreased in the hip joint capsule of patients with DDH compared to healthy controls. Alpha smooth muscle actin (αSMA), also known as ACTA2 and ACTVS, is a key marker for the differentiation of fibroblasts to myofibroblasts. However, the increase in αSMA expression in the hip joint capsule of patients with DDH was marginal.
Fig. 4.
Expression of signalling molecules in tumour growth factor beta (TGF-β) and Wnt pathways. Tissues from the hip joint capsule of patients with developmental dysplasia of the hip (DDH) and healthy controls were used for a) polymerase chain reaction (PCR), and b) western blotting. Expression of TGF-β1 was downregulated, whereas that of TGF-β2, Mothers against decapentaplegic homolog 3 (SMAD3), and WNT11 was upregulated in patients with DDH. However, the increase in alpha smooth muscle actin (αSMA), a key myofibroblast marker, was marginal. Experimental data (n = 5) were examined using an unpaired two-tailed t-test. p < 0.001 versus control.
Suppression of proliferation of fibroblasts in the hip joint capsule of patients with DDH
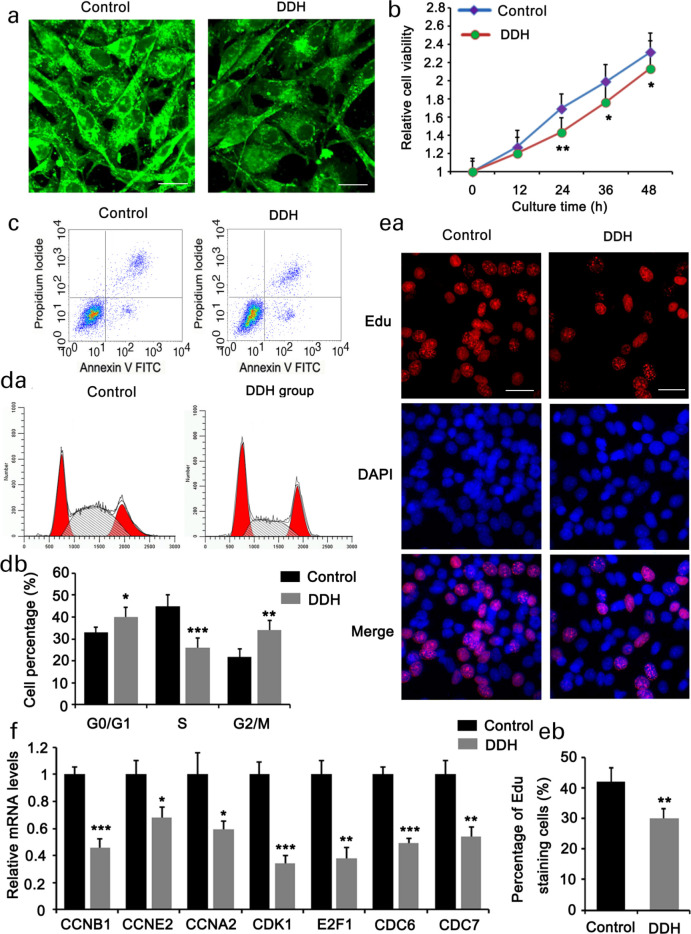
Based on the results from GO enrichment analysis, the downregulated genes influenced multiple BPs implicated in cell proliferation, including mitotic cell cycle, cell division, chromosome segregation, cell cycle, and nuclear division. In addition, KEGG analysis also showed the influence of the downregulated genes on cell cycle signalling. To validate the results of the bioinformatics analysis, we isolated the primary fibroblasts from the hip joint capsule of both patients with DDH and healthy controls. IF staining of type I collagen was used to identify primary fibroblasts. Type I collagen staining in the primary fibroblasts was weaker in patients with DDH than in healthy controls (Figure 5a). These results are consistent with those of the immunocytochemistry assays. Primary fibroblasts from patients with DDH showed reduced cell viability compared to cells from healthy controls (Figure 5b), but no significant difference was observed in the apoptotic rate (Figure 5c). Compared to healthy controls, the primary fibroblasts from patients with DDH showed an increased percentage in the G2/M phase, but decreased percentage in the S phase (Figures 5da and 5db). The percentage of cells with EdU-positive staining was also reduced in the DDH group (Figures 5ea and 5eb). Using PCR, we measured the expression of cell cycle-related genes that were downregulated in the DDH group according to high-throughput sequencing. The expression levels of CCNB1, CCNE2, CCNA2, cyclin-dependent kinase 1 (CDK1), E2F1, cell division cycle 6 (CDC6), and CDC7 were decreased in primary fibroblasts from patients with DDH (Figure 5f).
Fig. 5.
Fibroblast proliferation was suppressed in the hip joint capsule of patients with developmental dysplasia of the hip (DDH). Primary fibroblasts were isolated from the hip joint capsules of patients with DDH and healthy controls. a) Immunofluorescence staining (×200) of type I collagen was used to identify the primary fibroblasts. The primary fibroblasts were further analyzed by b) cell viability, c) apoptosis, d) cell cycle, and e) 5-ethylnyl-2ʹ-deoxyuridine (EdU) staining assays (×200). f) Polymerase chain reaction (PCR) was used to detect the expression of cell cycle regulators, including cyclin B1 (CCNB1), CCNE2, CCNA2, cyclin-dependent kinase 1 (CDK1), E2F1, cell division cycle 6 (CDC6), and CDC7. Experimental data (n = 3) were examined using an unpaired two-tailed t-test. p < 0.05, p < 0.01, and p < 0.001 versus control. DAPI, 4′,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate; mRNA, messenger RNA.
Discussion
This study is the first to comprehensively analyze the differences in gene expression profiles in hip joint capsules between healthy controls and patients with DDH. More than 1,000 genes were differentially expressed in hip joint capsules between healthy controls and patients with DDH. Although it is unclear whether the genetic difference is caused by a congenital element or an acquired factor, the differentially expressed genes very likely cause changes in many biological functions in the hip joint capsule. Indeed, GO enrichment analysis revealed that the ECM structure, tissue development, and fibroblast proliferation were markedly affected by the differentially expressed genes in DDH.
The hip joint capsule is composed of fibroblasts and various kinds of ECM. Type I and III collagens are the major components of the collagen in the joint capsule. Collagen also forms fibres and networks that constitute the structural element for the ECM to anchor, support cells, and provide an appropriate microenvironment for cell growth. This study showed that COL1A1 and COL3A1 expression was decreased in the hip joint capsule of patients with DDH. A previous study found that alleles at the polymorphisms (T-139C, C-106T, and C-35T) in COL1A1 gene promoter are more common in Chinese female patients with DDH than in controls, which probably affects cartilage formation and chondrocyte development.7 Interestingly, the expression of MMP1, MMP3, MMP9, and MMP13 were also decreased in the hip joint capsule of patients with DDH, which suggests that the degradation of collagen and other components of the ECM is suppressed. This result is different from that in the cartilage, where the expression of MMP13 is increased in patients with DDH.8-10 Increased expression of MMP13 accelerates cartilage degradation. Studies also found higher levels of MMP3 in the serum and synovial fluid of patients with hip OA secondary to DDH.11,12 Therefore, the expression of the MMP family of proteins may differ among different tissues in patients with DDH and during different pathological stages.
The consequence of the reduction of type I and III collagens, and these MMP proteins is the loss of collagen fibres in the hip joint capsule of patients with DDH, as revealed by Masson’s staining. It is likely that the reduction of type I and III collagens contributed to the loss of collagen fibres. Given that fibrillar collagens enable tissues to tolerate and distribute compressive and tensile forces, this pathological change in collagen fibres probably results in a lax joint capsule that warrants further validation. Collagen fibres are continuously remodelled by resident cells (such as fibroblasts) to perform these functions throughout life. Collagen remodelling comprises of distinct and highly coordinated processes such as synthesis, degradation, fibrillar reorientation, and cross-linking to maintain connective tissue homeostasis.13,14 Given that both the production and degradation of collagen were suppressed, remodelling of the ECM is likely to slow down in the joint capsule in patients with DDH.
Both GO and KEGG analyses implied that fibroblasts in the hip joint capsule of patients with DDH were inclined to differentiate into myofibroblasts. Myofibroblasts are a type of cell with characteristics of both smooth muscle cells and fibroblasts. Myofibroblasts exhibit strong contraction by developing muscle-like features, including the formation of contractile actin-myosin bundles.15 Owing to this property, myofibroblasts play an important role in wound healing, to restore tissue integrity. Myofibroblasts also maintain the characteristic of fibroblasts by secreting collagen; therefore, myofibroblasts participate in the fibrosis of diverse organs, such as the heart, lung, kidney, and liver.16-19 The differentiation of fibroblasts to myofibroblasts is driven by activation of TGF-β and Wnt signals as well as stimulation of chronic mechanical stretch.20,21 Indeed, hip dislocation in DDH probably more easily or frequently induces mechanical stretch in the hip joint capsule. The differentiation of fibroblasts into myofibroblasts enhances the contractile property of the joint capsule and the stability of the hip joint. Except for the downregulation of TGF-β1, TGF-β2, SMAD3, and WNT11 were upregulated in the hip joint capsule of patients with DDH. However, the increase in αSMA, a key myofibroblast marker, was marginal. The degree of differentiation of fibroblasts to myofibroblasts needs further studies.
GO and KEGG analyses indicated that cell proliferation was suppressed in the hip joint capsule of patients with DDH. Through a series of in vitro studies, we found that the decreased fibroblast proliferation was associated with cell cycle arrest in the G2/M phase, and not with apoptosis. Consistent with this finding, several cell cycle regulators were downregulated. These included the cyclin family of proteins (including CCNB1, CCNE2, and CCNA2), CDK1, E2F1, CDC6, and CDC7. The interaction between cyclin family of proteins and CDKs is necessary to drive the progression of mitosis. For example, the interaction between cyclins and CDKs disrupts the inhibitory effect of Rb on the transcription factor E2F1, following which E2F1 initiates the expression of genes related to mitosis.22 Deficiency in either cyclin or CDK proteins results in cell cycle arrest.23,24 CDC6 and CDC7 are essential regulators of DNA replication, during cell division.25 Therefore, any reduction in these cell cycle regulators would inevitably suppress fibroblast proliferation. The loss of fibroblasts and collagen may affect the mechanical and biological properties of the joint capsule.
In conclusion, this study identified several genes that were differentially expressed in hip joint capsules between healthy controls and patients with DDH. Some of these genes have been confirmed to be related to the loss of collagen fibres and fibroblasts, which may lead to the loss of the joint capsule. However, the degree of differentiation of the fibroblasts to myofibroblasts requires further studies.
Author contributions
C. Li: Investigation, Writing – original draft, Methodology, Funding acquisition.
Z. Peng: Investigation, Writing – original draft, Methodology.
Y. Zhou: Investigation, Methodology, Resources.
Y. Su: Investigation, Methodology, Resources.
P. Bu: Methodology.
X. Meng: Investigation.
B. Li: Investigation.
Y. Xu: Writing – review and editing.
Funding statement
This study (including open access funding) was funded by The Major Science and Technology Special Project of Yunnan Province Science and Technology Plan Project (2018ZF008) and The Key project of Construction of Clinical Center of Yunnan Provincial Health Commission (ZX20191001).
Ethical review statement
All subjects signed an informed consent form before enrolment. This study was approved by the ethics committee of Children's Hospital of Kunming Medical University.
Supplementary material
Gene ontology and Kyoto Encyclopedia of Genes and Genomes analyses.
References
- 1.Schwend RM, Shaw BA, Segal LS. Evaluation and treatment of developmental hip dysplasia in the newborn and infant. Pediatr Clin North Am. 2014;61(6):1095–1107. [DOI] [PubMed] [Google Scholar]
- 2.Walton S, Schaeffer E, Mulpuri K, Cundy P, Williams N. Evaluating the role of prereduction hip traction in the management of infants and children with developmental dysplasia of the hip (DDH): protocol for a systematic review and planned meta-analysis. BMJ Open. 2018;8(1):e019599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhodes AM, Clarke NM. A review of environmental factors implicated in human developmental dysplasia of the hip. J Child Orthop. 2014;8(5):375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaquero-Picado A, González-Morán G, Garay EG, Moraleda L. Developmental dysplasia of the hip: update of management. EFORT Open Rev. 2019;4(9):548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolia I, Chahla J, Locks R, Briggs K, Philippon MJ. Microinstability of the hip: a previously unrecognized pathology. Muscles Ligaments Tendons J. 2016;6(3):354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai J, Shi D, Zhu P, et al. Association of a single nucleotide polymorphism in growth differentiate factor 5 with congenital dysplasia of the hip: a case-control study. Arthritis Res Ther. 2008;10(5):R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao L, Tian W, Pan H, et al. Variations of the COL1A1 gene promoter and the relation to developmental dysplasia of the hip. Genet Test Mol Biomarkers. 2013;17(11):840–843. [DOI] [PubMed] [Google Scholar]
- 8.Ning B, Jin R, Wan L, Wang D. Cellular and molecular changes to chondrocytes in an in vitro model of developmental dysplasia of the hip‑an experimental model of DDH with swaddling position. Mol Med Rep. 2018;18(4):3873–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng WJ, Wang H, Shen C, Zhu JF, Chen XD. Severe cartilage degeneration in patients with developmental dysplasia of the hip. IUBMB Life. 2017;69(3):179–187. [DOI] [PubMed] [Google Scholar]
- 10.Ning B, Sun J, Yuan Y, Yao J, Wang P, Ma R. Early articular cartilage degeneration in a developmental dislocation of the hip model results from activation of β-catenin. Int J Clin Exp Pathol. 2014;7(4):1369–1378. [PMC free article] [PubMed] [Google Scholar]
- 11.Yasuda T, Matsunaga K, Hashimura T, et al. Characterization of rapidly progressive osteoarthritis of the hip in its early stage. Eur J Rheumatol. 2020;7(3):130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi R, Yamamoto T, Motomura G, et al. Bone and cartilage metabolism markers in synovial fluid of the hip joint with secondary osteoarthritis. Rheumatology. 2014;53(12):2191–2195. [DOI] [PubMed] [Google Scholar]
- 13.Coelho NM, McCulloch CA. Contribution of collagen adhesion receptors to tissue fibrosis. Cell Tissue Res. 2016;365(3):521–538. [DOI] [PubMed] [Google Scholar]
- 14.Karamanos NK, Theocharis AD, Neill T, Iozzo RV. Matrix modeling and remodeling: a biological interplay regulating tissue homeostasis and diseases. Matrix Biol. 2019;75-76:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinz B, McCulloch CA, Coelho NM. Mechanical regulation of myofibroblast phenoconversion and collagen contraction. Exp Cell Res. 2019;379(1):119–128. [DOI] [PubMed] [Google Scholar]
- 16.Yong KW, Li Y, Huang G, et al. Mechanoregulation of cardiac myofibroblast differentiation: implications for cardiac fibrosis and therapy. Am J Physiol Heart Circ Physiol. 2015;309(4):H532–H542. [DOI] [PubMed] [Google Scholar]
- 17.Hinz B. Mechanical aspects of lung fibrosis: a spotlight on the myofibroblast. Proc Am Thorac Soc. 2012;9(3):137–147. [DOI] [PubMed] [Google Scholar]
- 18.Yuan Q, Tan RJ, Liu Y. Myofibroblast in kidney fibrosis: origin, activation, and regulation. Adv Exp Med Biol. 2019;1165:253–283. [DOI] [PubMed] [Google Scholar]
- 19.Seki E, Brenner DA. Recent advancement of molecular mechanisms of liver fibrosis. J Hepatobiliary Pancreat Sci. 2015;22(7):512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roche PL, Nagalingam RS, Bagchi RA, et al. Role of scleraxis in mechanical stretch-mediated regulation of cardiac myofibroblast phenotype. Am J Physiol Cell Physiol. 2016;311(2):C297–C307. [DOI] [PubMed] [Google Scholar]
- 21.Hinz B. Myofibroblasts. Exp Eye Res. 2016;142:56–70. [DOI] [PubMed] [Google Scholar]
- 22.Madan E, Gogna R, Kuppusamy P, Bhatt M, Pati U, Mahdi AA. Tigar induces p53-mediated cell-cycle arrest by regulation of RB-E2F1 complex. Br J Cancer. 2012;107(3):516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paruthiyil S, Cvoro A, Tagliaferri M, Cohen I, Shtivelman E, Leitman DC. Estrogen receptor β causes a G2 cell cycle arrest by inhibiting Cdk1 activity through the regulation of cyclin B1, Gadd45a, and BTG2. Breast Cancer Res Treat. 2011;129(3):777–784. [DOI] [PubMed] [Google Scholar]
- 24.Deota S, Rathnachalam S, Namrata K, et al. Allosteric regulation of Cyclin-B binding by the charge state of catalytic lysine in Cdk1 is essential for cell-cycle progression. J Mol Biol. 2019;431(11):2127–2142. [DOI] [PubMed] [Google Scholar]
- 25.Nishitani H, Lygerou Z. Control of DNA replication licensing in a cell cycle. Genes Cells. 2002;7(6):523–534. [DOI] [PubMed] [Google Scholar]