The contribution of obesity and insulin resistance to nonalcoholic fatty liver disease (NAFLD) is undeniable [1], and NAFLD has even been called the liver manifestation of metabolic syndrome [2]. However, intrahepatic triglyceride accumulation is an early sign of metabolic disease, and it oftentimes predates the development of metabolic syndrome [3]. In recognition of this close link between NAFLD and metabolic disease, a panel of experts proposed a new term and definition for this condition [4]. Metabolic (dysfunction)-associated fatty liver disease (MAFLD) is defined as the evidence of hepatic steatosis in the presence of (a) overweight/obesity, (b) type 2 diabetes (T2D) or (c) evidence of metabolic dysregulation [4].
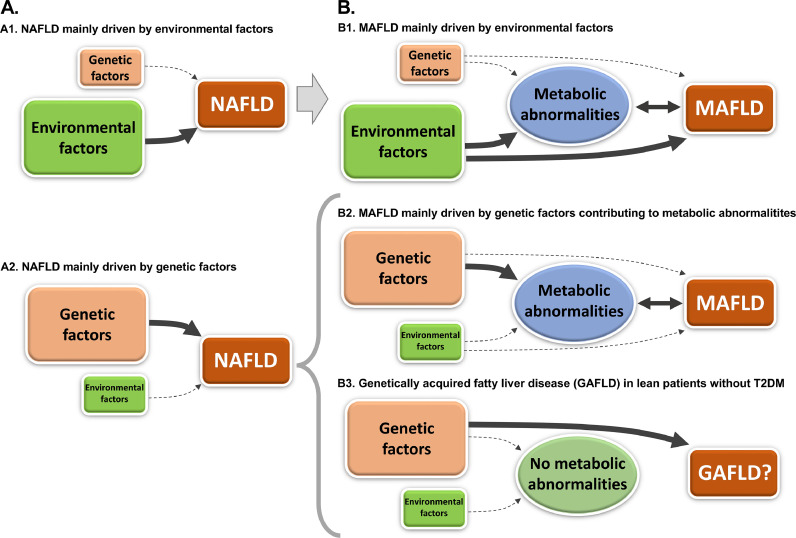
In light of this “metabolic” definition, the role of genetic determinants has been relegated to that of a “disease-modifier”, leaving environmental factors at center stage. Figure 1 summarizes different scenarios on how environmental and genetic factors could interact to promote liver disease. The prior definition of NAFLD could encompass any degree of contribution from environmental and/or genetic factors (panel A). However, the new definition of MAFLD inevitably requires the presence of metabolic abnormalities. While there is still a possibility for different contributions from environmental and genetic factors to MAFLD (panels B1-2), there is now an orphan scenario that cannot be included within the spectrum of MAFLD (panel B3). This corresponds to genetically-driven hepatic steatosis without a metabolic correlate, i.e., genetically acquired fatty liver disease (GAFLD).
Figure 1.
Environmental and genetic contributions to NAFLD (panel A) and MAFLD (panel B). The transition from one definition to the other, leaves a subset of patients undiagnosed: genetically acquired fatty liver disease (GAFLD).
As we transition towards ‘precision medicine’ in metabolic disorders [5], it is evident that NAFLD was too broad of a term. Luukkonen et al [6] identified distinctive subtypes of NAFLD based on different insulin resistance measurements and genetic variants. Patients with PNPLA3 I148M gene variant have higher fat accumulation, higher prevalence of NASH and more progression to advanced fibrosis, in the absence of worse insulin resistance [7]. Moreover, PNPLA3 I148M gene variant plays a key role in NAFLD among lean patients without T2D (i.e., those unlikely to qualify as MAFLD) [8]. This supports the concept that genetic variations may play a key role in lean patients with NAFLD, who may have GAFLD without fulfilling criteria for MAFLD.
In this article of EBioMedicine, Metwally et al provided yet another proof that genetic variants may play an important role in some patients with hepatic steatosis [9]. The authors examined exportin 4 (XPO4) copy number variations (CNVs) in the context of liver disease. Only few prior studies had looked at the role of CNVs in the development or progression of NAFLD, and XPO4 CNVs had only been previously examined in an Asian population [10].
A large cohort of 646 MAFLD patients and 170 controls were included in the study. There was a significant difference in the distribution of XPO4 CNVs between MAFLD and control patients. While 95.3% of patients were copy number neutral in the control group, only 41.3% of MAFLD patients expressed this pattern, with the majority showing duplications. While this may imply that XPO4 CNVs plays a critical role in the development of MAFLD, prior studies did not show these marked differences in the CNVs distribution [10]. Unfortunately, the authors did not provide much information about control group characteristics, so it is hard to extract any significant conclusions from this observation. It is also difficult to assess the representativeness of their MAFLD cohort, as little information is provided regarding recruitment strategy. Yet, the sample size is compelling, and baseline characteristics are not far from what we would expect of a MAFLD cohort randomly selected. Information on comorbidities and medication use would have also contributed to a better interpretation of the results.
Authors later focused on assessing the role of XPO4 CNVs in the development of fibrosis in patients with MAFLD. They extensively complemented their human observations, with in vitro and animal studies. Mechanistically, they showed strong evidence regarding their hypothesized mechanism by which XPO4 CNVs would influence fibrosis progression in MAFLD. However, their human data showed significant overlap in the degrees and stages of liver disease when patients with MAFLD were divided based on CNV gain (duplication) or non-gain (CNV neutral or deletion), reinforcing the concept that this is likely a multifactorial disease.
The strength of their work resides in the combination of human observations in a large cohort of MAFLD patients together with comprehensive mechanistic studies to assess the pathophysiology behind those observations. While authors suggested that XPO4 CNVs may be used as a risk biomarker or therapeutic target, future studies are needed to establish the significance of this association. Based on their results, there may only be a modest role of this genetic variation in humans. Nevertheless, as evidence accumulates regarding the role of genetic variants, especially in lean NAFLD [8], we need to celebrate every effort and novel strategy to detect new genetic determinants. The answer may not dwell in just one, but in the combination of different genetic variants.
Contributors
Fernando Bril: writing, editing and final revision of manuscript.
Declaration of Competing Interest
No conflict of interests to disclose.
Footnotes
Conflict of interests
Nothing to disclose.
References
- 1.Godinez-Leiva E, Bril F. Nonalcoholic Fatty Liver Disease (NAFLD) for Primary Care Providers: Beyond the Liver. Curr Hypertens Rev. 2020 doi: 10.2174/1573402116999201209203534. [DOI] [PubMed] [Google Scholar]
- 2.Kim CH, Younossi ZM. Nonalcoholic fatty liver disease: a manifestation of the metabolic syndrome. Cleve Clin J Med. 2008;75:721–728. doi: 10.3949/ccjm.75.10.721. [DOI] [PubMed] [Google Scholar]
- 3.Bril F, Barb D, Portillo-Sanchez P, Biernacki D, Lomonaco R, Suman A, Weber MH. Metabolic and histological implications of intrahepatic triglyceride content in nonalcoholic fatty liver disease. Hepatology. 2017;65:1132–1144. doi: 10.1002/hep.28985. [DOI] [PubMed] [Google Scholar]
- 4.Eslam M, Newsome PN, Sarin SK, Anstee QM, Targher G, Romero-Gomez M, Zelber-Sagi S. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J Hepatol. 2020;73:202–209. doi: 10.1016/j.jhep.2020.03.039. [DOI] [PubMed] [Google Scholar]
- 5.Lonardo A, Byrne CD, Targher G. Precision medicine approaches in metabolic disorders and target organ damage: where are we now, and where are we going? Metab Target Organ Damage. 2021;1:3. [Google Scholar]
- 6.Luukkonen PK, Zhou Y, Sadevirta S, Leivonen M, Arola J, Oresic M, Hyotylainen T. Hepatic ceramides dissociate steatosis and insulin resistance in patients with non-alcoholic fatty liver disease. J Hepatol. 2016;64:1167–1175. doi: 10.1016/j.jhep.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Krawczyk M, Liebe R, Lammert F. Toward Genetic Prediction of Nonalcoholic Fatty Liver Disease Trajectories: PNPLA3 and Beyond. Gastroenterology. 2020;158:1865–1880. doi: 10.1053/j.gastro.2020.01.053. e1861. [DOI] [PubMed] [Google Scholar]
- 8.Krawczyk M, Bantel H, Rau M, Schattenberg JM, Grunhage F, Pathil A, Demir M. Could inherited predisposition drive non-obese fatty liver disease? Results from German tertiary referral centers. J Hum Genet. 2018;63:621–626. doi: 10.1038/s10038-018-0420-4. [DOI] [PubMed] [Google Scholar]
- 9.Metwally M, Bayoumi A, Khan A, Adams LA, Aller R, Garcia-Monzon C, Arias-Loste MT. Copy number variation and expression of exportin-4 associates with severity of fibrosis in metabolic associated fatty liver disease. EBioMedicine. 2021;70 doi: 10.1016/j.ebiom.2021.103521. 10.1016/j.ebiom.2021.103521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zain SM, Mohamed Z, Pirmohamed M, Tan HL, Alshawsh MA, Mahadeva S, Chan WK. Copy number variation in exportin-4 (XPO4) gene and its association with histological severity of non-alcoholic fatty liver disease. Sci Rep. 2015;5:13306. doi: 10.1038/srep13306. [DOI] [PMC free article] [PubMed] [Google Scholar]