Abstract
The correct organization of mitochondrial DNA (mtDNA) in nucleoids and the contacts of mitochondria with the ER play an important role in maintaining the mitochondrial genome distribution within the cell. Mitochondria-associated ER membranes (MAMs) consist of interacting proteins and lipids located in the outer mitochondrial membrane and ER membrane, forming a platform for the mitochondrial inner membrane-associated genome replication factory as well as connecting the nucleoids with the mitochondrial division machinery. We show here that knockdown of a core component of mitochondrial nucleoids, TFAM, causes changes in the mitochondrial nucleoid populations, which subsequently impact ER-mitochondria membrane contacts. Knockdown of TFAM causes a significant decrease in the copy number of mtDNA as well as aggregation of mtDNA nucleoids. At the same time, it causes significant upregulation of the replicative TWNK helicase in the membrane-associated nucleoid fraction. This is accompanied by a transient elevation of MAM proteins, indicating a rearrangement of the linkage between ER and mitochondria triggered by changes in mitochondrial nucleoids. Reciprocal knockdown of the mitochondrial replicative helicase TWNK causes a decrease in mtDNA copy number and modifies mtDNA membrane association, however, it does not cause nucleoid aggregation and considerable alterations of MAM proteins in the membrane-associated fraction. Our explanation is that the aggregation of mitochondrial nucleoids resulting from TFAM knockdown triggers a compensatory mechanism involving the reorganization of both mitochondrial nucleoids and MAM. These results could provide an important insight into pathological conditions associated with impaired nucleoid organization or defects of mtDNA distribution.
Keywords: Mitochondrial DNA, Nucleoids, Mitochondrial transcription factor A (TFAM), Organellar membranes, TWNK helicase
Highlights
-
•
Mitochondrial DNA (mtDNA) is packaged with non-histone proteins into nucleoids.
-
•
TFAM covers and packs mtDNA; knockdown of TFAM results in nucleoid aggregation.
-
•
Mitochondrial dynamics and mtDNA segregation are tightly linked.
-
•
Replicating mitochondrial nucleoids are associated with ER-mitochondria tethers (MAM).
-
•
Loss of nucleoid integrity triggers MAM reorganization.
1. Introduction
Mitochondria are a central hub of the cellular metabolism [1]. Besides being responsible for the majority of ATP production, they are involved in the production of fatty acids, amino acids, nucleotides and other anabolites, while at the same time regulating energy production, intracellular calcium levels, immune responses as well as cell cycle [2] and apoptosis [3,4]. Mitochondria are no static structures, but form a dynamic network that is intertwined with other cellular organelles [5]. The dynamic nature of mitochondria is of great importance for mitochondrial quality control and is altered in various pathologies, especially in neurodegenerative diseases [5,6].
The mitochondrial genome (mtDNA) together with several protein components is organized into units called nucleoids [7,8]. As mtDNA encodes for essential subunits of the electron transfer chain (ETC), its maintenance is a prerequisite to ensure the proper function of mitochondria. A plethora of proteins is known to be involved in mtDNA maintenance. Among them is the so-called minimal replisome, consisting of the DNA polymerase γ (POLG), the helicase TWNK (TWNK) and the single-strand binding protein mtSSB [9]. Of great importance is TFAM, a protein initially described as transcription factor in mitochondria, but found to have an important function in binding, bending and packaging mtDNA, in analogy to the function of histones in the nucleus [10]. In vitro, TFAM appears to regulate replication and transcription by its binding form [11]. This is concordant with numerous works showing that changes in TFAM protein levels cause a decrease in mtDNA copy number and impair nucleoid organization and distribution [[12], [13], [14]].
During the past years, we and others were able to characterize a number of mtDNA nucleoid proteins (for a review see e.g. Ref. [15]). The steadiest components of nucleoids are the mtDNA itself and TFAM, while other nucleoid-associated proteins (mtNAPs) co-occur most likely depending on metabolic requirements. We were able to identify two different pools of nucleoids, one of which is defined through the presence of replicative helicase TWNK and mtSSB [16]. While all observed nucleoids in different cell lines show the presence of TFAM and mtDNA, only roughly half of them are positive for TWNK [16]. These observations were further confirmed through sub-mitochondrial fractionation, leading to the identification of a specialized mitochondrial microdomain specifically harboring replicating nucleoids [16,17].
Physical connections between mitochondria and endoplasmic reticulum (ER) were visualized by EM already in the 1950s in rat liver [18,19] and later termed as mitochondria-associated ER membranes (MAM) [20]. The constantly changing interaction of the two organelles serves several functions, ranging from synthesis and transport of metabolites (phospholipids, cholesterol, Ca2+ etc.) to the maintenance and segregation of mtDNA. Early observations made in Saccharomyces cerevisiae [21,22] showed that the mtDNA polymerase Mip1 is found in close vicinity of the MAM-analogue in yeasts, ERMES (ER-mitochondria encounter structure). The finding was further corroborated by studies showing that the constriction of mitochondria by the ER, facilitating the fission of the mitochondrial network [23] involves replicating nucleoids attaching to either side of the constriction [24]. However, the precise mechanism remains to be clarified. Gerhold et al. described markers for MAM in close vicinity to the replication microdomain [17]. These proteins could serve the purpose of positioning faithfully replicated mtDNA to ensure appropriate segregation during fission of the mitochondrial network. Here we show that the alteration of mitochondrial nucleoid integrity through knockdown of one of its core components, TFAM, not only induces rearrangements in nucleoid populations, but also leads to an evident elevation of MAM proteins. This effect seems to be reversible upon recovery of TFAM levels. While also the gene silencing of TWNK helicase causes a reduction in mtDNA copy number and changes in mtDNA membrane association, the impact on MAM proteins in the membrane-associated fraction is weaker compared to TFAM knockdown.
2. Materials and methods
2.1. Cell culture
Human embryonic kidney cells (HEK293) and human osteosarcoma cells (U2OS) were cultured in Dulbecco's modified Eagle's medium (Lonza DMEM (12–604F) with 4.5 g/L Glucose, l-Glutamine and Sodium Pyruvate) supplemented with 10% FBS (Capricorn) at 37 °C and 5% CO2.
2.2. RNA-interference
Knockdown of TFAM was achieved by small interfering RNA-mediated gene silencing using 25 nM of target specific siRNA (Dharmacon™ siGENOME Human TFAM (M-019734-00-0020) and C10orf2 siRNA SMARTpool (M-017815-01)) and Dharmafect1 transfection reagent (Dharmacon™) in serum-free medium according to the manufacturer's instructions. As a control, a non-targeting siRNA control pool (Dharmacon™; D-001206-14-05) was used. Cells were transfected at a confluence of ∼20–30%. 3, 4, 5 and 6 days after transfection cells were harvested for mitochondrial and mtDNA isolation.
2.3. Confocal microscopy
U2OS cells were plated on coverslips and transfected with siRNAs. 3 days after transfection cells were fixed with 3,3% paraformaldehyde for 15 min, carefully washed thrice with PBS and permeabilized with 10% FBS in PBS +0,5% Triton X-100 for 20 min. Cells were then incubated with primary (anti-TFAM, 1:500; anti-DNA, 1:500; anti-FACL4, 1:250; anti-SSB, 1:500 or anti-TOM20, 1:500) and secondary (Alexa Fluor 488 anti-mouse IgG and anti-rabbit IgG, 1:1000; Alexa Fluor 568 anti-mouse and anti-rabbit IgG, 1:1000) antibodies for at least 1 h. DNA was stained using ProLong™ Gold Antifade Mountant with DAPI (Thermo Scientific). Coverslips were imaged using a Zeiss LSM710 confocal microscope with a 63× oil immersion objective.
2.4. Mitochondrial isolation and digitonin fractionation
Confluent cells were harvested and washed with PBS. Next, cells were swollen in hypotonic buffer (4 mM Tris-HCl, pH 7.4, 2.5 mM NaCl, 0.5 mM MgCl2 including protease inhibitors (Applichem)), followed by a disruption of cells with a Glass/Teflon Potter Elvehjem homogenizer until ∼80% of cells were broken. The cell suspension was adjusted to be isotonic and centrifuged thrice to pellet cell debris and broken nuclei for 5 min at 1200 g and 4 °C. Crude mitochondria were pelleted at 14000 g and 4 °C for 10 min. Crude intact mitochondria were re-suspended in PBS and lysed with digitonin, using a protein (μg) to digitonin (μg) ratio of 1:2 in PBS supplemented with protease inhibitors (Applichem). The mitochondrial suspension was incubated on ice for 10 min, followed by a centrifugation at 14000g and 4 °C for 10 min to separate soluble and insoluble mitochondrial protein fractions.
2.5. MAM and ER isolation
Was carried out from crude mitochondrial preparations as described [25]. Briefly, mitochondria were re-suspended in MRB buffer containing 225 mM mannitol, 75 mM sucrose and 30 mM Tris-HCl pH 7.4, layered onto a Percoll gradient (225 mM mannitol, 25 mM HEPES pH 7.4, 1 mM EDTA and 30% Percoll) and centrifuged at 95 000 g for 30 min at 4 °C. The distinct layers of mitochondria (lower band) and MAM (upper band) were collected from the ultracentrifuge tube and diluted with re-suspension buffer. The supernatant of crude MAM was again centrifuged at 100 000 g for 60 min at 4 °C and the purified MAM pellet at the bottom of the ultracentrifuge tube was re-suspended in MRB.
A pure ER preparation was sub-fractioned from the supernatant obtained after crude mitochondrial isolation by a centrifugation at 20 000 g for 30 min at 4 °C to first remove lysosomes and plasma membrane. The supernatant was centrifuged at 100 000 g for 60 min at 4 °C and the pellet containing pure ER was collected.
2.6. Iodixanol gradients of fractioned mitochondria
Were created as described in Ref. [16]. Shortly, 1 mg of crude mitochondrial protein was subjected to digitonin lysis for the separation of ‘pellet’ and ‘supernatant’ (see digitonin fractionation). To both fractions ice-cold Optiprep™ (Axis-Shield PoC AS) was added to a final concentration of 42,5%, pipetted on the bottom of polycarbonate tubes and overlayered with solutions with different iodixanol (v/v) concentrations (40 %–0%) in 1x TN buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM DTT, protease inhibitor cocktail, 10% sucrose, 1% Triton X-100). The gradients were subjected to centrifugation at 100 000 g for 14 h at 4 °C, after which fractions of 400 μl (top-to-bottom) were collected.
2.7. Protein and DNA analyses
of digitonin fractionations, MAM and ER preparations, total cell extracts and floatation gradients were carried out using Western and Southern analyses. For Western blots, samples were separated by SDS-PAGE (Bio-Rad Mini-PROTEAN® Tetra Cell) and transferred onto positively charged nitrocellulose membrane (GE Healthcare Amersham™ Protran™ 0,45 μm). Primary antibodies used were mouse-anti-TWNK (a gift from Prof. Anu Suomalainen-Wartiovaara), rabbit-anti-TFAM (a gift from Prof. Rudolf Wiesner), rabbit-anti-COX2 from Proteintech, mouse-anti-VDAC1 from Abcam, mouse-anti-HSP60, mouse-anti-FACL4, mouse-anti-MFN1 and -2 from Santa Cruz Biotechnology, mouse-anti-α-tubulin and mouse-anti-β-actin from Abcam, rabbit-anti-SSBP from Sigma Aldrich, rabbit-anti-TOM40 from Santa Cruz Biotechnology, rabbit-anti-ATAD3 [26], rat-anti-ATAD3A (a gift from Dr. Alexander Wolf) and rabbit-anti-mitofilin from Abcam. Quantifications of resulting ECL signals were performed with Image Studio™ (LI-COR) software.
For dot blot analysis of mtDNA, a radioactive hybridization was applied. Briefly, 20 μl of sample was first diluted 20-fold with 2x SSC, denatured at 95 °C for 15 min and spotted onto positively charged nylon membrane (GE Healthcare Amersham Hybond™-N+) using a Bio-Rad Bio-Dot® blotter. Membranes were hybridized in Church buffer using a radioactively labelled CYTB (nt 14837–15367 of human mtDNA) probe at 65 °C, and signals were detected using a phosphor screen, Amersham Typhoon biomolecular imager and quantified using ImageQuant (GE Healthcare) software.
2.8. Quantitative PCR
for the determination of relative copy numbers was carried out in 384-well qPCR plates. Total DNA was extracted from control and knockdown cells using standard proteinase K digest and phenol:chloroform (pH 6,8) extraction.
For the subsequent copy number estimation, two specific primers pairs were used: one for the mtDNA encoded cytochrome b gene (CYTB forward: 5′-GCCTGCCTGATCCTCCAAAT-3’; CYTB reverse: 5′-AAGGTAGCGGATGATTCAGCC-3′) and the second for the amyloid precursor protein (APP) gene (APP forward: 5′-TTTTTGTGTGCTCTCCCAGGTCT-3’; APP reverse: 5′-TGGTCACTGGTTGGTTGGC-3′) encoded by nuclear DNA. For amplification a HOT FIREPol® EvaGreen® qPCR Mix Plus (ROX) (Solis Biodyne) was used according to the manufacturers' guidelines. Each reaction contained 25 ng of total DNA. The amplification program (Applied Biosystems 7900 HT) for both genes was as follows: initial denaturation at 95 °C for 12 min, 46 cycles of denaturation at 95 °C for 15 s and primers annealing at 60 °C for 60 s. Changes in relative copy number of mtDNA were analyzed using the -ΔΔCt algorithm.
3. Results
3.1. Knockdown of TFAM causes mtDNA-nucleoid aggregation, an increased association of nucleoids with the inner mitochondrial membrane and an increase of MAM markers
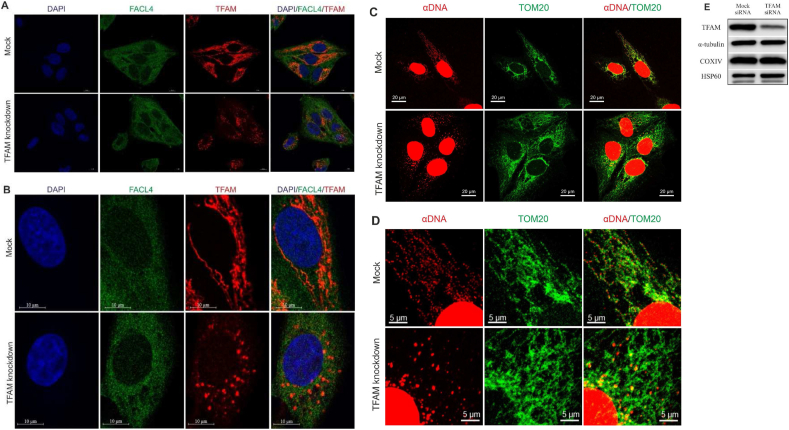
Confocal images showed that in control cells TFAM is highly abundant throughout the mitochondrial network (Fig. 1fig1A–B). Mitochondrial nucleoids stained with an anti-DNA antibody showed a relatively even distribution, with similar fluorescence intensities within the mitochondrial network (Fig. 1C–D). Upon a 3-day TFAM knockdown (Fig. 1E), the TFAM signal concentrates at defined large foci containing both mtSSB and DNA (Fig. 1A; Fig. S1), indicating nucleoid aggregation. The overall signal of mtDNA, however, was reduced, implying a declined copy number. This was confirmed by quantitative PCR, showing a ∼50% decrease in mtDNA (Fig. 2D).
Fig. 1.
TFAM knockdown causes aggregation of nucleoids. Confocal microscopy of control and TFAM siRNA-treated U2OS cells 3 days after transfection. The cells were immunolabelled with α-TFAM (mtDNA-nucleoid marker), α-FACL4 (ER network marker) (a-b), α-DNA and α-TOM20 (mitochondrial network marker) (c-d) antibodies. The nuclei were stained with DAPI (a-b). Images were obtained using a Zeiss LSM710 confocal microscope with a 63× oil immersion objective and adjusted using ZEN Imaging Software. Western blots of total cell extracts from 3-day TFAM knockdown cells showing ∼70% decrease in TFAM protein level (e).
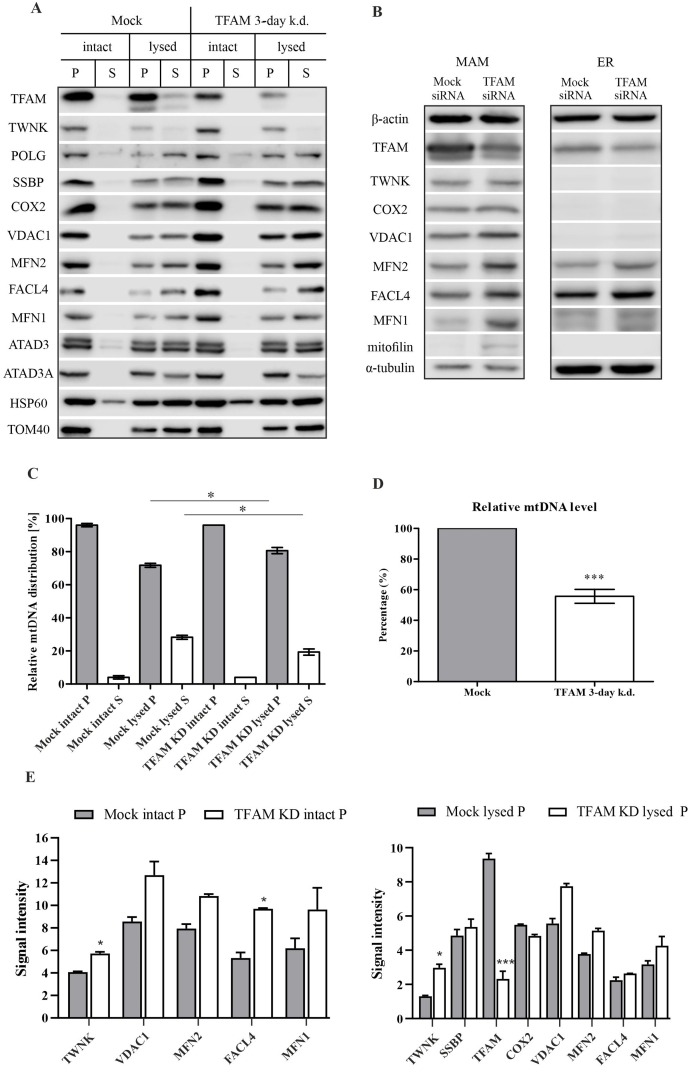
Fig. 2.
TFAM knockdown alters the levels of both nucleoid and MAM marker proteins. (a) Western blots of digitonin-based isolation of matrix (lysed S; supernatant) and membrane-associated (lysed P; pellet) mitochondrial protein fractions. Intact mitochondrial protein fractions represent whole mitochondria and their wash solution following centrifugation (intact P and S). (b) Sub-fractioned MAM and ER from HEK293 cells analyzed by Western blot. (c) Quantification of relative mtDNA levels in the pellet (gray bars) and supernatant fractions (white bars) of whole and lysed mitochondria using dot-blot analysis. TFAM knockdown induces a shift of mtDNA towards the membrane-associated fraction. Error bars indicate±SEM over six biological repeats (n = 6) (d) Quantification by real-time PCR shows a ∼40% decrease of mtDNA 3 days after TFAM knockdown. Error bars indicate±SEM (n = 6). (e) The quantification of the pellet fractions of three biological repeats on Western blots, for which the band intensities were determined and corrected for loading based on the HSP60 signal. All statistical significances were calculated using a two-tailed paired Student's t-test comparing knockdowns with the respective controls. Error bars indicate±SEM. * indicates a p-value ≤ 0.05, *** indicates a p-value ≤ 0.001. The representative Western blot panels are cropped versions of the original exposure pictures.
TWNK helicase is a mtDNA replication factor and a constituent of replicating nucleoids [16,17]. Previous experiments have reported up to 60% decrease in mtDNA copy number and an impaired mtDNA metabolism upon TWNK knockdown [27]. Hence, we here tested whether a reduction in mtDNA levels caused by TWNK knockdown induces similar changes in nucleoid organization as TFAM knockdown. We observed that a 3-day TWNK knockdown does not cause nucleoid aggregation (Fig. S1).
To test whether TFAM knockdown alters the mtDNA nucleoid populations, we isolated a crude mitochondrial fraction from HEK293 cells. Mitochondrial nucleoid pools were further separated by digitonin, solubilizing the mitochondrial outer membrane and fragmenting the mitochondrial inner membrane, leading to the separation of two pools of mtDNA nucleoids [16,17]. The more firmly mitochondrial inner membrane-associated nucleoid type, remaining largely in the insoluble (‘pellet’, P) fraction, is enriched with mitochondrial replication proteins such as TWNK, mtSSB and POLG, and associates with a cholesterol-rich membrane platform, while the other (‘supernatant’, S) is less membrane-associated and contains matrix-soluble components (e.g. mitochondrial ribosomal proteins and mitochondrial RNA granules) [17,27].
Upon TFAM knockdown, TWNK levels in the membrane-associated pool significantly increase, and also total mitochondrial TWNK levels are significantly higher, indicating an overall increase in TWNK upon TFAM depletion (Fig. 2A–E). For mtSSB and COX2 no significant change in total signals were observed in both soluble (‘supernatant’) and membrane-associated fractions (Fig. 2A–E).
Interestingly, the mitochondrial structural and nucleoid-organizing protein ATAD3, which interacts with the mitochondrial D-loop and forms direct contacts with mtDNA [28], shows no clear alteration upon TFAM knockdown (Fig. 2A). The downregulation of ATAD3 itself disrupts the membrane-associated cholesterol-rich microdomain and leads to an impaired mtDNA maintenance [17,26,28]. The lack of change here might indicate that ATAD3 influences nucleoid organization only indirectly through mitochondrial cristae organization [29], and a 3-day knockdown of TFAM fails to cause such significant changes in mitochondrial ultrastructure.
TFAM knockdown leads to an overall decrease in mtDNA levels by ∼50% (Fig. 2D). Analysis of mtDNA content by dot blot showed that in control mitochondria ∼30% of the mtDNA located to the soluble pool, while in TFAM knockdown mitochondria only ∼20% of the remaining mtDNA was consistently found in the soluble fraction (Fig. 2C), indicating a shift in mtDNA distribution from the soluble to the membrane-associated nucleoid fraction. Our data showing an increase of both TWNK protein levels (Fig. 2A–E) and nucleoid membrane association corroborates previous findings that mtDNA membrane association increases at higher TWNK levels [16].
TFAM knockdown caused not only changes in TWNK, but also in the MAM proteins MFN1/2, VDAC1 and FACL4. The levels of all three proteins increased in crude mitochondrial preparations upon TFAM depletion (Fig. 2A–E). As described above, mitochondrial nucleoids localize at foci, where also MAM are found, and specifically replicating mtDNA-nucleoids are spatially linked with ER-mitochondria membrane structures and influence the formation of sites for ER-directed mitochondria constriction by ER tubules [24].
3.2. TFAM knockdown causes changes in fractioned MAM
Based on the findings presented here and previously by others, we set out to investigate the levels of different MAM proteins upon the knockdown of TFAM by sub-fractioning HEK293 cells into mitochondria, ER and MAM. The results show that a 3-day knockdown of TFAM causes a significant increase in the MAM-forming proteins MFN1/2, VDAC1 and FACL4 (Fig. 2B; Fig. S2) in the fractioned MAM of TFAM knockdown cells. The greatest increase in fractioned MAM of knockdown cells compared to control was found for mitofusin 2 (3-fold), followed by VDAC1 (1.8-fold) and FACL4 (1.5-fold) (Fig. S2). This increase of MAM protein signals likely indicates increased interactions between mitochondria and ER.
Overall TWNK levels as well as TWNK levels in the fractioned MAM increase upon TFAM knockdown (Fig. 2B, Fig. S3), further indicating an enhanced association of replicating nucleoids with MAM.
As expected, purified ER does not contain the mitochondrial markers mitofilin, COX2 and VDAC1, but instead FACL4 and MFN2, that show stronger signals in the TFAM knockdown fraction (Fig. 2B).
The movement of ER tubules is dependent on the microtubular network [30,31], and fittingly, the signals of tubulin in purified ER fractions are clearly stronger than in MAM (Fig. 2B). Actin filaments are responsible for motility, network homeostasis and for mitochondrial nucleoid segregation. As β-actin also localizes to mitochondria [32], it was used here as a loading control over all fractions.
3.3. TFAM knockdown induced changes in MAM are transient
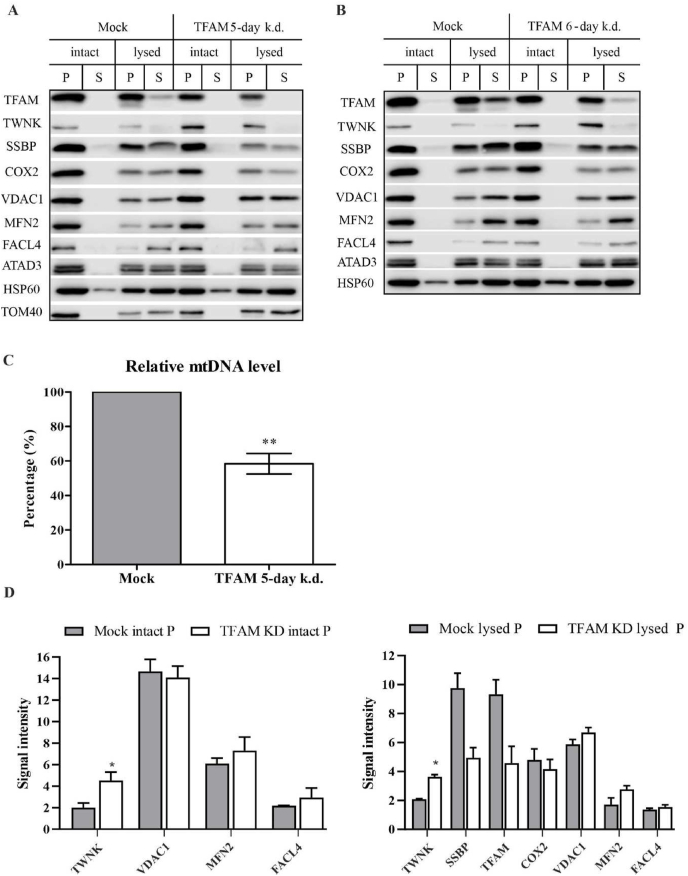
Earlier studies have demonstrated that the close linkage between ER and mitochondria is reversible and ER-mitochondria contacts are constantly changing [33]. To test whether there is a possible turnover in the alteration of MAM and nucleoid proteins, the TFAM knockdown cells were grown for up to 6 days from the time of transfection and sub-fractioned. The results indicate that 5 days after transfection cells start to recover from the decrease in TFAM (Fig. 3A, B and 3D).
Fig. 3.
5 and 6 days after TFAM knockdown the MAM protein levels start to recover. (a-b) Western blots of ER and mitochondrial proteins 5 and 6 days after TFAM knockdown. (a-b) TWNK signals remain elevated in the TFAM knockdown mitochondria compared to control, but COX2 and SSB levels have decreased in TFAM knockdown membrane-associated pellet fraction P. (a-b) The levels of the MAM marker proteins VDAC1, MFN2 and FACL4 are comparable in both knockdown and control mitochondria. (c) Both relative mtDNA copy number and TFAM protein levels are decreased after 5 days. Error bars indicate±SEM (n = 3). (d) The quantification of the pellet fractions of three biological repeats on Western blots, for which the band intensities were determined and corrected for loading based on the HSP60 signal. All statistical significances were calculated using a two-tailed paired Student's t-test comparing knockdowns with the respective controls. Error bars indicate±SEM. * indicates a p-value ≤ 0.05, ** indicates a p-value ≤ 0.01. The representative Western blot panels are cropped versions of the original exposure pictures.
The relative mtDNA level 5 days after TFAM knockdown remains significantly lower than in control cells (Fig. 3C), and TWNK signals remain significantly elevated both in whole mitochondria and the membrane-associated protein fraction even 6 days after TFAM knockdown (Fig. 3A, B and 3D). This could imply that the tight inner membrane association of TWNK persists as long as mtDNA levels are decreased. It has been previously shown that this tight membrane association of nucleoids remains even in the absence of mtDNA [16,17]. The levels of mtSSB and COX2 in the membrane-associated fraction decrease compared to control cells (Fig. 3A, B and 3D), while they are unaltered 3 days after TFAM knockdown. As TFAM is required for mitochondrial transcription [34], the decrease in COX2 is likely caused by the lack of mtDNA transcription due to the knockdown. Similarly, reduced mtSSB levels could indicate that mtSSB is required in mtDNA replication foci to a much lesser extent, possibly due to a decrease in mtDNA replication initiation that is dependent on primers provided by the transcription machinery [35,36]. At the same time the steady-state proportion of TWNK is high in the membrane-associated replication foci (‘pellet fractions’).
5 and 6 days after transfection, the overall levels of the MAM proteins VDAC1, FACL4 and MFN2 in TFAM knockdown mitochondria have decreased to the level of control cells (Fig. 3A, B and 3D), indicating that the observed alterations in MAM protein levels are transient.
Although it is possible that TFAM knockdown induces morphological changes in ER-mitochondria contacts, we were unable to judge any alterations in FACL4 localization by confocal microscopy due to the abundance of the protein in the ER (Fig. 1A–B). Thus, we tested whether TFAM knockdown triggers an early ER stress by quantification of the ER chaperone calnexin as well as IRE1α, PERK and ATF6, since an activation of these proteins is orchestrated by the unfolded protein response during ER stress [37]. Our results show that TFAM knockdown does not induce clear changes in ER stress-related proteins (Fig. S4), and the described effect on MAM proteins is therefore likely related to changes in nucleoid organization.
3.4. TWNK knockdown has a minor effect on MAM
To compare whether the changes in nucleoid membrane association and MAM proteins upon TFAM knockdown are induced directly by the reduction in TFAM protein or indirectly by the consecutive mtDNA depletion, we first tested whether a decrease in mtDNA levels by knockdown of TWNK has a similar effect on MAM proteins. Previously, it was established that only ∼50% of mtDNA is associated with endogenous TWNK in steady-state, suggesting that TWNK is not a constitutive nucleoid component, but rather dynamically associates with mtDNA nucleoids at specialized membrane domains to participate in mtDNA replication [16,17].
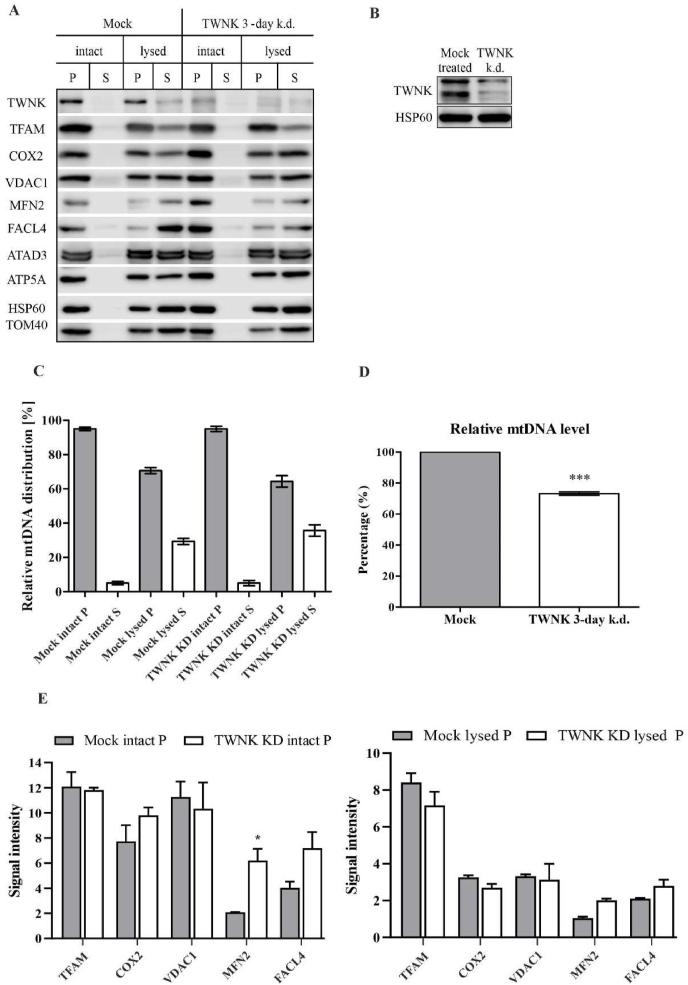
Our TWNK knockdown caused a decrease in relative mtDNA levels by ∼30% (Fig. 4D), as previously described [27,38]. At the same time, also TFAM levels decreased slightly in the digitonin-fractioned mitochondria, most likely caused by the decline in mtDNA level (Fig. 4A and E).
Fig. 4.
TWNK knockdown induces milder changes to MAM than TFAM. (a) Western blots of digitonin based isolation of matrix (lysed S) and membrane-associated (lysed P) mitochondrial fractions from TWNK knockdown cells. Intact mitochondrial fractions represent intact mitochondria and their wash solution following centrifugation (intact P and S). Unlike TFAM knockdown, the MAM marker proteins VDAC1, MFN2 and FACL4 showed little to no change in the membrane-associated fractions after TWNK knockdown indicating no re-organization of MAM. (b) A Western blot of HEK293 control-transfected and 3-day TWNK knockdown total cell extracts. HSP60 is used as a loading control. (c) Dot-blot analysis of relative mtDNA distribution in digitonin-lysed and non-lysed mitochondria shows an increase towards soluble fraction in TWNK knockdown mitochondria (10% difference of ‘insoluble: soluble’ ratio compared to the control mitochondria), causing the remaining mtDNA to become less membrane-associated. Error bars indicate±SEM over three biological repeats (n = 3). (d) Knockdown of TWNK causes a decrease in mtDNA levels by ∼30%. Error bars indicate±SEM (n = 4). (e) The quantification of the pellet fractions of three biological repeats on Western blots, for which the band intensities were determined and corrected for loading based on the HSP60 signal. All statistical significances were calculated using a two-tailed paired Student's t-test comparing knockdowns with the respective controls. Error bars indicate±SEM. * indicates a p-value ≤ 0.05, *** indicates a p-value ≤ 0.01. The representative Western blot panels are cropped versions of the original exposure pictures.
The relative mtDNA distribution between supernatant and membrane-associated fractions showed a modest shift towards more soluble nucleoids upon TWNK knockdown (Fig. 4C), indicating less nucleoids to be attached to the mitochondrial inner membrane.
As in the TFAM 3-day knockdown, also upon TWNK knockdown COX2 levels were by and large unaltered (Fig. 4A and E).
Compared to the TFAM knockdown, a slightly weaker impact on MAM proteins upon the loss of TWNK was observed in the digitonin membrane-associated fractions, which could be attributed to the weaker mtDNA depletion. VDAC1 signals remained unaltered in both intact and sub-fractioned mitochondria compared to controls (Fig. 4A and E). Both MFN2 and FACL4, however, show stronger signals in intact mitochondria (Fig. 4A and E), although the effect is less clear in soluble and membrane-associated fractions of mitochondria (Fig. 4A and E).
4. Discussion
In the presented study we have shed light onto the interplay between mitochondrial nucleoid and MAM, using the knockdown of the mtDNA nucleoid proteins TFAM and TWNK. We first confirmed by confocal microscopy that TFAM knockdown causes mtDNA nucleoid aggregation, as previously observed by others [14,39]. The aggregation phenotype upon TFAM knockdown has been further associated with an impaired mtDNA distribution via segregation [14].
Our results from digitonin-based mitochondrial fractionation show that upon TFAM knockdown more mtDNA is localized in the membrane-associated nucleoid fraction (Fig. 2C), while also TWNK increasingly associates with this fraction (Fig. 2A–E). A plausible explanation for this observation is that the cells try to compensate the decreased mtDNA copy number by recruiting more nucleoids to the TWNK-associated replication centers on the inner membrane. Further nucleoid fractionations by iodixanol gradients also confirm that the replication platform persists in the pellet fraction upon depletion of TFAM (Fig. S5).
As TFAM affects the proportion of DNA molecules available for transcription and replication via mtDNA compaction, an upregulation of TWNK helicase might indicate that it is recruited more to the replisomes without nucleoid remodeling, as the knockdown of TFAM decreases the compaction of mitochondrial DNA. This itself might indicate an enhanced mtDNA replication as a rescue attempt for the loss of mtDNA upon TFAM knockdown. However, further investigation of mitochondrial replication is required to address these issues.
Alternatively, our results here could also reflect that increased TWNK levels (together with unaltered mtSSB) are associated with mtRNA metabolism in the vicinity of nucleoids.
Interestingly, while TWNK levels remained clearly elevated for 5–6 days after TFAM knockdown, there was a delayed reduction in COX2 and mtSSB, likely caused by defective mtDNA transcription and mitochondrial translation (Fig. 3A, B and 3D). One the other hand, reduced mtSSB levels could also indicate that mtSSB is required in mtDNA replication foci to a much lesser extent, possibly due to a decrease in mtDNA replication initiation that is dependent on primers provided by the transcription machinery [35,36].
Additionally, we saw that TFAM knockdown leads to an increase in the MAM proteins VDAC1, MFN1/2 and FACL4, indicating an upregulation of MAM upon loss of nucleoid integrity (Fig. 2A, B and 2E). Thus, our findings here show that upon the loss of TFAM and subsequent reorganization of nucleoids also the MAM and its protein components are upregulated.
To identify whether the observed alterations are caused by reduced TFAM protein levels or rather by the following mtDNA depletion, we analyzed mitochondrial subfractions obtained from TWNK knockdown cells, as also this induces a decrease in mtDNA level [27]. The knockdown of TWNK caused a ∼30% decrease in relative mtDNA level without altering the levels COX2 protein, inducing a milder effect than TFAM knockdown. This also shows that the COX2 levels are not dependent on mtDNA copy number, but are regulated by transcription via TFAM. Also, the total mitochondrial levels of the MAM markers VDAC1, MFN2 and FACL4 were upregulated, although no clear change in the membrane-associated protein fraction was observed (Fig. 4A and E). This allows us to propose that MAM upregulation is predominantly triggered by a decrease in mtDNA levels. The reduction of TWNK prevents the membrane association of nucleoids despite an upregulation of MAM proteins, indicating that TWNK itself might be required for the membrane association as previously reported [16].
Hence, we propose that early changes in the integrity of mitochondrial nucleoids caused by depletion of the mtDNA packaging and regulatory protein TFAM causes aggregation of mtDNA nucleoids and a consequential decrease in mtDNA copy number. The reduction of mtDNA levels via TFAM knockdown triggers a more pronounced compensatory enhancement of the MAM sub-compartment required for mtDNA segregation than that of the TWNK knockdown. This MAM stimulation is transient and fades off before mtDNA and TFAM protein levels have completely recovered.
To conclude, our data suggests an intricate liaison between nucleoid organization and MAM regulation. While we found a clear indication of TFAM regulating nucleoid structure, with reduced levels of TFAM causing both mtDNA depletion and severe nucleoid aggregation, we did not find evidence for a direct connection of TFAM protein to MAM organization, as similar, although milder effects on MAM were produced also by knockdown of TWNK.
In order to reach a complete understanding of the regulatory mechanism of the interaction between ER-mitochondria coupling via MAM and mitochondrial nucleoids, especially in the context of human pathologies, further work is required.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors wish to thank E. Dufour for the valuable technical support. This work was funded by the Estonian Research Council (PUT610), the Estonian Ministry of Science and Education (PUTPRG433) and European Regional Development Fund (Centre of Excellence in Molecular Cell Engineering CEMCE).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101142.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Mishra P., Chan D.C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 2016;212:379–387. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakaki N., Nishihama T., Owaki H., Kuramoto Y., Suenaga M., Miyoshi E., Emoto Y., Shibata H., Shono M., Higuti T. Dynamics of mitochondria during the cell cycle. Biol. Pharm. Bull. 2006;29:1962–1965. doi: 10.1248/bpb.29.1962. [DOI] [PubMed] [Google Scholar]
- 3.Suen D.-F., Norris K.L., Youle R.J. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tait S.W.G., Green D.R. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 5.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu B. Mitochondrial dynamics and neurodegeneration. Curr. Neurol. Neurosci. Rep. 2009;9:212–219. doi: 10.1007/s11910-009-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X.J., Butow R.A. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 8.Spelbrink J.N. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life. 2009:n. doi: 10.1002/iub.282. /a-n/a. [DOI] [PubMed] [Google Scholar]
- 9.Korhonen J.A., Pham X.H., Pellegrini M., Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanki T., Nakayama H., Sasaki N., Takio K., Alam T.I., Hamasaki N., Kang D. Mitochondrial nucleoid and transcription factor A. Ann. N. Y. Acad. Sci. 2004;1011:61–68. doi: 10.1196/annals.1293.007. [DOI] [PubMed] [Google Scholar]
- 11.Farge G., Laurens N., Broekmans O.D., van den Wildenberg S.M.J.L., Dekker L.C.M., Gaspari M., Gustafsson C.M., Peterman E.J.G., Falkenberg M., Wuite G.J.L. Protein sliding and DNA denaturation are essential for DNA organization by human mitochondrial transcription factor A. Nat. Commun. 2012;3:1013. doi: 10.1038/ncomms2001. [DOI] [PubMed] [Google Scholar]
- 12.Ekstrand M.I. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 13.Kanki T., Ohgaki K., Gaspari M., Gustafsson C.M., Fukuoh A., Sasaki N., Hamasaki N., Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol. Cell Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasashima K., Sumitani M., Endo H. Human mitochondrial transcription factor A is required for the segregation of mitochondrial DNA in cultured cells. Exp. Cell Res. 2011;317:210–220. doi: 10.1016/j.yexcr.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Hensen F., Cansiz S., Gerhold J.M., Spelbrink J.N. To be or not to be a nucleoid protein: a comparison of mass-spectrometry based approaches in the identification of potential mtDNA-nucleoid associated proteins. Biochimie. 2014;100:219–226. doi: 10.1016/j.biochi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 16.Rajala N., Gerhold J.M., Martinsson P., Klymov A., Spelbrink J.N. Replication factors transiently associate with mtDNA at the mitochondrial inner membrane to facilitate replication. Nucleic Acids Res. 2014;42:952–967. doi: 10.1093/nar/gkt988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerhold J.M., Cansiz-Arda Ş., Lõhmus M., Engberg O., Reyes A., van Rennes H., Sanz A., Holt I.J., Cooper H.M., Spelbrink J.N. Human mitochondrial DNA-protein complexes attach to a cholesterol-rich membrane structure. Sci. Rep. 2015;5:15292. doi: 10.1038/srep15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernhard W., Rouiller C. Close topographical relationship between mitochondria and ergastoplasm of liver cells in a definite phase of cellular activity. J. Biophys. Biochem. Cytol. 1956;2:73–78. doi: 10.1083/jcb.2.4.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernhard W., Haguenau F., Gautier A., Oberling C. [Submicroscopical structure of cytoplasmic basophils in the liver, pancreas and salivary gland; study of ultrafine slices by electron microscope] Z. Zellforsch. Mikrosk. Anat. Vienna Austria. 1948;37(1952):281–300. [PubMed] [Google Scholar]
- 20.Vance J.E. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- 21.Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meeusen S., Nunnari J. Evidence for a two membrane–spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 2003;163:503–510. doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis S.C., Uchiyama L.F., Nunnari J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 2016;353:aaf5549. doi: 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wieckowski M.R., Giorgi C., Lebiedzinska M., Duszynski J., Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 26.He J., Cooper H.M., Reyes A., Di Re M., Sembongi H., Litwin T.R., Gao J., Neuman K.C., Fearnley I.M., Spinazzola A., Walker J.E., Holt I.J. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 2012;40:6109–6121. doi: 10.1093/nar/gks266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hensen F., Potter A., van Esveld S.L., Tarrés-Solé A., Chakraborty A., Solà M., Spelbrink J.N. Mitochondrial RNA granules are critically dependent on mtDNA replication factors Twinkle and mtSSB. Nucleic Acids Res. 2019;47:3680–3698. doi: 10.1093/nar/gkz047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J., Mao C.-C., Reyes A., Sembongi H., Di Re M., Granycome C., Clippingdale A.B., Fearnley I.M., Harbour M., Robinson A.J., Reichelt S., Spelbrink J.N., Walker J.E., Holt I.J. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peralta S., Goffart S., Williams S.L., Diaz F., Garcia S., Nissanka N., Area-Gomez E., Pohjoismäki J., Moraes C.T. ATAD3 controls mitochondrial cristae structure in mouse muscle, influencing mtDNA replication and cholesterol levels. J. Cell Sci. 2018;131:jcs217075. doi: 10.1242/jcs.217075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gurel P.S., Hatch A.L., Higgs H.N. Connecting the cytoskeleton to the endoplasmic reticulum and golgi. Curr. Biol. 2014;24:R660. doi: 10.1016/j.cub.2014.05.033. –R672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterman-Storer C.M., Salmon E.D. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol. 1998;8:798–807. doi: 10.1016/S0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- 32.Xie X., Venit T., Drou N., Percipalle P. In mitochondria β-actin regulates mtDNA transcription and is required for mitochondrial quality control. IScience. 2018;3:226–237. doi: 10.1016/j.isci.2018.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowland A.A., Voeltz G.K. Endoplasmic reticulum–mitochondria contacts: function of the junction. Nat. Rev. Mol. Cell Biol. 2012;13:607–615. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ngo H.B., Lovely G.A., Phillips R., Chan D.C. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun. 2014;5:3077. doi: 10.1038/ncomms4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agaronyan K., Morozov Y.I., Anikin M., Temiakov D. Mitochondrial biology. Replication-transcription switch in human mitochondria. Science. 2015;347:548–551. doi: 10.1126/science.aaa0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kühl I., Miranda M., Posse V., Milenkovic D., Mourier A., Siira S.J., Bonekamp N.A., Neumann U., Filipovska A., Polosa P.L., Gustafsson C.M., Larsson N.-G. POLRMT regulates the switch between replication primer formation and gene expression of mammalian mtDNA. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1600963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oslowski C.M., Urano F. Methods Enzymol. Elsevier; 2011. Measuring ER stress and the unfolded protein response using mammalian tissue culture system; pp. 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyynismaa H., Sembongi H., Bokori-Brown M., Granycome C., Ashley N., Poulton J., Jalanko A., Spelbrink J.N., Holt I.J., Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 39.West A.P., Khoury-Hanold W., Staron M., Tal M.C., Pineda C.M., Lang S.M., Bestwick M., Duguay B.A., Raimundo N., MacDuff D.A., Kaech S.M., Smiley J.R., Means R.E., Iwasaki A., Shadel G.S. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.