Abstract
Motivation
Organization of the organoid models, imaged in 3D with a confocal microscope, is an essential morphometric index to assess responses to stress or therapeutic targets. In fact, differentiating malignant and normal cells is often difficult in monolayer cultures. But in 3D culture, colony organization can provide a clear set of indices for differentiating malignant and normal cells. The limiting factors are delineating each cell in a 3D colony in the presence of perceptual boundaries between adjacent cells and heterogeneity associated with cells being at different cell cycles.
Results
In a previous paper, we defined a potential field for delineating adjacent nuclei, with perceptual boundaries, in 2D histology images by coupling three deep networks. This concept is now extended to 3D and simplified by an enhanced cost function that replaces three deep networks with one. Validation includes four cell lines with diverse mutations, and a comparative analysis with the UNet models of microscopy indicates an improved performance with the F1-score of 0.83.
Availability and implementation
All software and annotated images are available through GitHub and Bioinformatics online. The software includes the proposed method, UNet for microscopy that was extended to 3D and report generation for profiling colony organization.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Organoid models of normal and malignant cells play an essential role in studying biological processes, response to stress and drug screening (Rossi et al., 2018). The organoid models are functional assays, where their patterns and response to therapy are different than the 2D models (Simian and Bissell, 2017). More importantly, the 3D colony organization is an important phenotypic endpoint. Although it is almost impossible to differentiate malignant and normal cells based on their 2D images, the 3D organization of malignant cells can be differentiated using confocal microscopy (Bilgin et al., 2016). More importantly, specific mutations, such as ERBB2+ versus triple-negative in breast cancer cell lines, can be deduced based on the structure of the 3D colony organization. Conversely, aberrant colony organization of normal cells (e.g. as a result of external stress) can be interpreted as dysplasia and a potential precursor to malignancy (Cheng and Parvin, 2020). Hence, efficient characterization of colony organization, when imaged with confocal microscopy, is a vital endpoint for investigating aberrant biological processes or evaluating the efficacy of therapeutic targets. In this paper, we extend a previously developed (Khoshdeli et al., 2019) deep learning method for delineating nuclei in histology sections to 3D for characterizing the 3D colony organization. In addition, we simplify the system by replacing three deep learning models with one but with an enhanced loss function. The method is validated with four breast cancer cell lines with a diverse set mutation and compared with the variations of UNet (Ronneberger et al., 2015) that has been developed for delineating cells that are imaged with phase-contrast microscopy. This particular flavor of UNet has also been extended to 3D and released online.
2 Materials and methods
In this section, we summarize the (i) proposed loss function, (ii) training and testing data and (iii) model architecture, training parameters and experiments conducted.
The proposed loss function includes an additional term that specifically accentuates perceptual boundaries between adjacent nuclei, as shown in Equation 1. Where M, N and O are image dimensions; is the potential field at location accentuating the perception boundaries (Supplementary SA), and λ is the regularization term between potential field and data fitness term. The first cross-entropy weighs the similarities between the ground truth and prediction. The second cross-entropy term weights each pixel by its potential field defined earlier (Khoshdeli et al., 2019), but is computed and visualized in the 3D space as shown in Supplementary Figure S1. Delete
| (1) |
Training data consisted of annotated organoid models of four cell lines with diverse mutations that were imaged with a confocal microscope: MCF10A (non-transformed), MCF7 (ER+, PR+, ERBB2-), MDA-MB-231 (PR-, ER-, ERBB2-) and MDA-MB-468 (also triple-negative). The inclusion of multiple cell lines, with diverse mutations, is part of validation since the morphology of the 3D colony formation depends on mutation. In addition, colonies were fixed on days 2, 5 or 7 to make training invariant to colony morphogenesis. Each cultured cell line was imaged with a resolution of 0.25 microns in the XY-plane and a step-size of 1 micron in the Z-direction and was represented isomorphically. Additionally, each image was divided into patches using a sliding window of size 128×128×32 with no and 50% overlap for training and testing, respectively. This is a validated approach (Ronneberger, 2015) in segmentation for eliminating the boundary effects during reconstruction. Finally, the training patches were extended through data augmentation with a 60–40% split for training and testing, respectively.
All comparisons were performed against an extension of the UNet (Ronneberger et al., 2015) for delineating cells imaged by phase-contrast microscopy, which we extended to 3D for comparative analysis as shown in Supplementary Figure S2. We refer to this method as the UNet-Cell in this manuscript, which outperforms the traditional UNet for cell-based segmentation. For each experimental condition, we report the F1-score and Aggregated Jaccard Index (AJI). The model was trained with an Adam optimizer for ten epochs and a learning rate of 1e-5. To correct for the class imbalance, foreground and background pixels were weighted by one and five, respectively. Additionally, an early stopping criterion was implemented for any epoch when the cost function failed to improve. Finally, indices associated with colony morphologies (e.g. roundness, number of cells) were computed as outlined in Supplementary SB.
3 Results
We report the system performance following tuning of hyper parameters and visual inspection of the segmentation results as function of the mutation landscape. Hyperparameter tuning includes both the (i) regularization parameter λ and (ii) the threshold for probability map that resulted in an optimum performance with λ = 10 and per detailed simulations shown in Supplementary SC and Supplementary Figures S3–S5. Having completed the tuning, Table 1 summarizes an improved performance of the proposed method in terms of the mean AJI, pixel- and object-level F1-scores with 95% confidence intervals. Visual inspections of model predictions, for each mutation, with their respective ground truths, are provided in Figure 1 with additional examples provided in Supplementary SD and Supplementary Figures S6–S8 indicating a robust performance.
Table 1.
Comparison of performance metrics between the proposed and UNet-Cell (Ronneberger et al., 2015) show an improved result for pixel- and object-level detection
| Method | AJI | Pixel F1-score | Object F1-score |
|---|---|---|---|
| UNet-cell | 0.60.01 | 0.81 0.01 | 0.79 0.02 |
| Proposed | 0.60.01 | 0.83 0.01 | 0.83 0.02 |
Fig. 1.
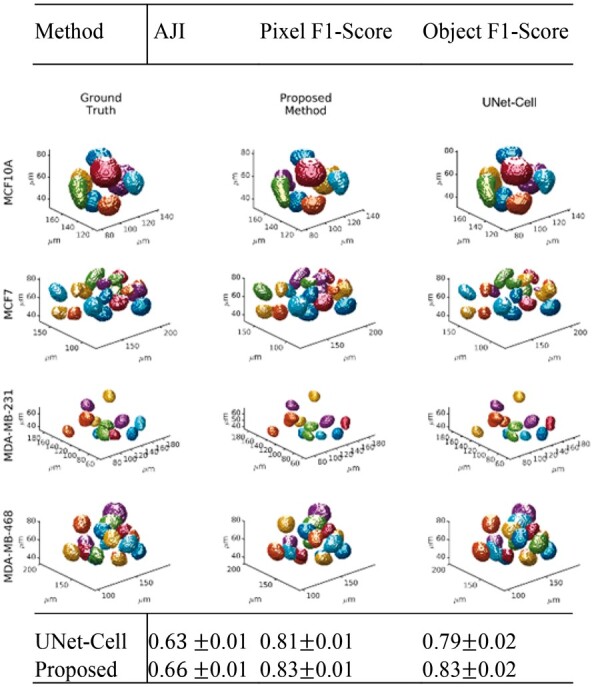
Visualizations of 3D organoid model predictions for each cell line and their distinct mutation
Funding
This work was partially supported by National Institutes of Health R15-CA235430 and National Aeronautics Space Administration 80NSSC18K1464.
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Garrett Winkelmaier, Department of Electrical and Biomedical Engineering, University of Nevada Reno, Reno, NV 89557, USA.
Bahram Parvin, Department of Electrical and Biomedical Engineering, University of Nevada Reno, Reno, NV 89557, USA; Department of Cell and Molecular Biology, University of Nevada Reno, Reno, NV 89557, USA.
References
- Bilgin C.C. et al. (2016) BioSig3D: high content screening of three-Dimensional cell culture models. PLoS One, 11, e0148379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Parvin B. (2020) Organoid model of mammographic density displays a higher frequency of aberrant colony formations with radiation exposure. Bioinformatics, 36, 1989–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshdeli M. et al. (2019) Deep fusion of contextual and object-based representations for delineation of multiple nuclear phenotypes. Bioinformatics, 35, 4860–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger O. et al. (2015) U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. NY: Springer. pp. 234–241.
- Rossi G. et al. (2018) Progress and potential in organoid research. Nat. Rev. Genet., 19, 671–687. [DOI] [PubMed] [Google Scholar]
- Simian M., Bissell M.J. (2017) Organoids: a historical perspective of thinking in three dimensions. J. Cell Biol., 216, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


