Abstract
Summary
Single-cell RNA-Seq (scRNA-Seq) data is useful in discovering cell heterogeneity and signature genes in specific cell populations in cancer and other complex diseases. Specifically, the investigation of condition-specific functional gene modules (FGM) can help to understand interactive gene networks and complex biological processes in different cell clusters. QUBIC2 is recognized as one of the most efficient and effective biclustering tools for condition-specific FGM identification from scRNA-Seq data. However, its limited availability to a C implementation restricted its application to only a few downstream analysis functionalities. We developed an R package named IRIS-FGM (Integrative scRNA-Seq Interpretation System for Functional Gene Module analysis) to support the investigation of FGMs and cell clustering using scRNA-Seq data. Empowered by QUBIC2, IRIS-FGM can effectively identify condition-specific FGMs, predict cell types/clusters, uncover differentially expressed genes and perform pathway enrichment analysis. It is noteworthy that IRIS-FGM can also take Seurat objects as input, facilitating easy integration with the existing analysis pipeline.
Availability and implementation
IRIS-FGM is implemented in the R environment (as of version 3.6) with the source code freely available at https://github.com/BMEngineeR/IRISFGM.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Single-cell RNA-Seq (scRNA-Seq) data characterizes the cell heterogeneity in complex tissues and diseases that can reveal cell subpopulations and their unique gene expression patterns. A condition-specific FGM is a highly structured expression pattern of a gene set, which tend to be functionally related or co-regulated in a particular cell cluster or cell type. Biclustering is a widely accepted approach for identifying such FGMs (i.e. biclusters) in a gene expression dataset. Our previously developed tool, QUBIC2 (Xie et al., 2020), outperformed existing methods, such as FABIA (Hochreiter et al., 2010), ISA (Bergmann et al., 2003), Plaid (Lazzeroni et al., 2002) and Bimax (Prelic et al., 2006), in identifying biologically meaningful biclusters on 10X scRNA-Seq data. These condition-specific FGMs were successfully applied to revealed regulatory signals and their targeted gene in a specific cell type (Ma et al., 2020). Furthermore, the investigation of FGM can help to understand gene-gene interaction networks and complex biological processes from scRNA-Seq data(Xie et al., 2020). However, previously QUBIC2 was only available as a C implementation, and its applicative power was also restricted to only a few downstream analysis functionalities. On the other hand, its usability and interpretability will be further improved by being integrated with a powerful and functional interpretation system, e.g. our comprehensive web server-based RNA-Seq interpretation system (Monier et al., 2019). To this end, we developed an R package named IRIS-FGM (Integrative scRNA-Seq Interpretation System for Functional Gene Module analysis) to support the investigation of FGMs and cell clustering using scRNA-Seq data. Empowered by QUBIC2, IRIS-FGM can effectively identify co-expressed and co-regulated FGMs, predict cell types/clusters, uncover differentially expressed gene (DEG) patterns and perform functional enrichment analysis.
2 Framework design
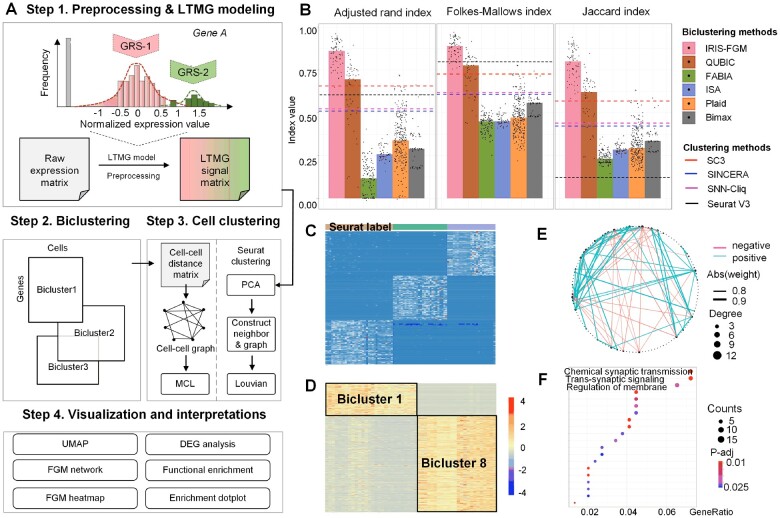
The IRIS-FGM framework consists of four key steps (Fig. 1A). In the first step, the raw expression matrix (with rows presenting genes and columns representing cells) is imported into the R environment as an IRIS-FGM object, and it is further preprocessed by removing low-quality cells based on numbers of features and normalizing expression values (Supplementary Method S2). We employed a left-truncated mixture Gaussian (LTMG) model (Wan et al., 2019), a robust statistical model that effectively detects regulatory signals for each gene while being robust against the high level of zero inflation and low signal-to-noise ratio. The LTMG model identifies regulatory signal components using the normalized read counts and generates the discretized gene-cell matrix. In the second step, QUBIC2 is applied to identify biclusters (i.e. FGMs) from the LTMG discretized matrix. In the third step, we first include the widely used cell clustering package Seurat (Butler et al., 2018). Meanwhile, the Markov clustering algorithm (MCL) is implemented to identify cell clusters on a cell-cell graph constructed by integrating all the biclusters from the second step. Specifically, in such a graph, each node represents a cell, and an edge indicates that the two connected cells belong to the same bicluster (Xie et al., 2020). In a comparison study of cell clustering approaches on the test dataset, IRIS-FGM outperforms other five popular biclustering tools [QUBIC (Li et al., 2009), FABIA, ISA, Plaid, Bimax] and four clustering tools [i.e. SC3 (Kiselev et al., 2017), SINCERA (Guo et al., 2015), SNN-Cliq (Shi et al., 2017) and Seurat (Butler et al., 2018)] (Fig. 1B, Supplementary Method S1). In the fourth step, IRIS-FGM provides six functions that allow users to visualize the analytical results of condition-specific FGMs and cell clustering results: UMAP plot, DEGs across all clusters (Fig. 1C), FGM heatmap (Fig. 1D), co-expression network (Fig. 1E) and pathway enrichment results (Fig. 1F). Detailed information on the above four steps can be found in the following section and Supplementary Method S2. In support of the connection of IRIS-FGM and the Seurat package for downstream analyses, IRIS-FGM can take and generate the Seurat object, including but not limited to dimension reduction results and clustering results from raw expression matrix from Seurat.
Fig. 1.
The overview of IRIS-FGM workflow and data interpretations. (A) The IRIS-FGM workflow includes three main steps: preprocessing and LTMG modeling, biclustering and cell clustering and downstream interpretations. GRS-1 and GRS-2 represent gene regulatory signals 1 and 2. (B) Cell clustering evaluation of IRIS-FGM against the five popular biclustering methods (bars) and four clustering methods (dashed lines) on Yan’s data in terms of the Adjusted Rand index, Folkes-Mallows index and Jaccard index. Dots represent the results of different parameters used for each biclustering methods. (C) Global marker heatmap shows the top 50 differentially expressed genes for three labels predicted by the Seurat framework. (D) FGM heatmap visualization of biclusters 1 and 8 identified from Yan’s data. (E) FGM networks of Bicluster 1 based on Figure 1D. The size of the nodes (black) indicates the degree of a node. The thickness of edges indicates the correlation coefficient estimates. The edge color shows the positive (green) and the negative (red) relationships between the two genes. (F) Dot plot shows the pathway enrichment result derived from Bicluster 8 based on Figure 1D. Dot color indicates statistical significance and dot size indicates the number of genes both in the bicluster and pathway gene list
3 Functions and examples
IRIS-FGM contains 27 functions (Supplementary Method S2), and the main functions of IRIS-FGM are summarized below as four categories corresponding to the four steps in Figure 1A. We further demonstrate the applicative power of IRIS-FGM using the 90 human embryonic cells (Yan et al., 2013), 2700 normal human peripheral blood mononuclear cells (PBMCs) from 10X official website, 1955 CD8+ T cells from human non-small-cell lung cancer (Guo et al., 2018) and 6454 cells from mouse melanoma (Davidson et al., 2020). More details of coding demonstration and corresponding results can be found in Supplementary Example S1–S4. In the following sections, we only list the names of our package’s main functions, and the full list can be found in Supplementary Table S1.
3.1 Preprocessing and LTMG modeling of scRNA-seq (9 functions)
In this step, IRIS-FGM will perform cell filtering (SubsetData), normalization (ProcessData) and LTMG modeling based on scRNA-seq data. We implement the LTMG model using function RunLTMG, and it takes the IRIS-FGM object as input and returns a regulatory signal matrix. The signal matrix can also be integrated into the Seurat object, which can be called by object@LTMG@ Tmp.seurat. The Seurat object with the signal matrix can be further analyzed with the Seurat analysis pipeline, such as cell clustering and DEG analysis (full function list can be found in Supplementary Table S1).
3.2 Biclustering for condition-specific FGM identification (6 functions)
IRIS-FGM provides a biclustering algorithm to predict condition-specific FGMs from the gene expression matrix by implementing QUBIC 2.0 in the function RunBicluster. Specifically, it is equipped with two discretization methods: (i) a quantile discretization way for raw expression matrix (RunDiscretization) and (ii) a binarization method (CalBinaryMultiSignal) for the preprocessed signal matrix from Step 1.
3.3 Cell clustering (3 functions)
To identify cell clusters, IRIS-FGM implements the MCL clustering algorithm in FindClassBasedOnMC while also employing cell clustering methods from Seurat by a dimension reduction RunDimensionReduction and the Louvain clustering RunClassification. Considering the computational complexity, Seurat is recommended when the number of cells is large.
3.4 Visualization and interpretations (9 functions)
To further elucidate cell heterogeneity, IRIS-FGM integrated the efficient DEG identification method from Seurat (FindGlobalMarkers) and a highly accurate DEG detection methods from DEsingle (FindMarker) (Miao et al., 2018; Wang et al., 2019). Pathway enrichment analysis of a given gene list is performed by clusterProfiler R package (RunPathway) (Yu et al., 2012). Moreover, IRIS-FGM also provides six visualization functions to facilitate an intuitive understanding of cell cluster distribution (PlotDimension), gene regulatory network (PlotModuleNetwork), etc.
4 Conclusion and discussion
We developed a robust and multifunctional R package, IRIS-FGM, for scRNA-Seq data analysis that enables identifying condition-specific FGMs and cell clusters, visualizing a cell-cell network and implementing functional enrichment analysis of gene signatures. Furthermore, the intermediate product (Seurat object with LTMG discretized matrix) of IRIS-FGM can be used in and integrated with the Seurat analysis pipeline. The elucidation of condition-specific FGMs has far-reaching impacts on how differentially activated transcriptional regulatory signals affect cell states and evolutionary cell trajectories, among other phenotypic characteristics. In the long run, the novel knowledge derived using IRIS-FGM will shed light on the gene regulatory network among various cell types within a complex tissue or disease microenvironment.
Funding
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) [R01-GM131399 to Q.M.; R01-GM122078 to D.C.], the National Cancer Institute of the NIH [R01-CA188419 to Z.L.; R21-CA209848 to D.C.] and the National Institute on Drug Abuse of the NIH [U01-DA045300 to D.C.].
Conflict of Interest: none declared.
Data availability
The data underlying this article are available in GEO, 10x Genomics, and ArrayExpress. All datasets can be derived from sources in the public domain: the human embryo cells data were from GSE36552; the PBMCs data were from https://support.10xgenomics.com/single-cell-gene-expression/datasets/1.1.0/pbmc3k; the human lung cancer data were from GSE99254; and the mouse melanoma data were downloaded from https://www.ebi.ac.uk/gxa/sc/experiments/E-EHCA-2/Results.
Supplementary Material
Contributor Information
Yuzhou Chang, Department of Biomedical Informatics, The Ohio State University, Columbus, OH 43210, USA.
Carter Allen, Department of Biomedical Informatics, The Ohio State University, Columbus, OH 43210, USA.
Changlin Wan, Center for Computational Biology and Bioinformatics and Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN 46202, USA.
Dongjun Chung, Department of Biomedical Informatics, The Ohio State University, Columbus, OH 43210, USA.
Chi Zhang, Center for Computational Biology and Bioinformatics and Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN 46202, USA.
Zihai Li, Pelotonia Institute for Immuno-Oncology, The James Comprehensive Cancer Center, The Ohio State University, Columbus, OH 43210, USA.
Qin Ma, Department of Biomedical Informatics, The Ohio State University, Columbus, OH 43210, USA.
References
- Bergmann S. et al. (2003) Iterative signature algorithm for the analysis of large-scale gene expression data. Phys. Rev. E Stat. Nonlin. Soft Matter Phys., 67, 031902. [DOI] [PubMed] [Google Scholar]
- Butler A. et al. (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol., 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S. et al. (2020) Single-cell RNA sequencing reveals a dynamic stromal niche that supports tumor growth. Cell Rep., 31, 107628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M. et al. (2015) SINCERA: a pipeline for single-cell RNA-seq profiling analysis. PLoS Comput. Biol., 11, e1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. et al. (2018) Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med., 24, 978–985. [DOI] [PubMed] [Google Scholar]
- Hochreiter S. et al. (2010) FABIA: factor analysis for bicluster acquisition. Bioinformatics, 26, 1520–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev V.Y. et al. (2017) SC3: consensus clustering of single-cell RNA-seq data. Nat. Methods, 14, 483–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzeroni L. et al. (2002) Plaid models for gene expression data. Stat. Sinica, 12, 61–86. [Google Scholar]
- Li G. et al. (2009) QUBIC: a qualitative biclustering algorithm for analyses of gene expression data. Nucleic Acids Res., 37, e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A. et al. (2020) IRIS3: integrated cell-type-specific regulon inference server from single-cell RNA-Seq. Nucleic Acids Res., 48, W275–W286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z. et al. (2018) DEsingle for detecting three types of differential expression in single-cell RNA-seq data. Bioinformatics, 34, 3223–3224. [DOI] [PubMed] [Google Scholar]
- Monier B. et al. (2019) IRIS-EDA: An integrated RNA-Seq interpretation system for gene expression data analysis. PLoS Comput. Biol., 15, e1006792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelic A. et al. (2006) A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics, 22, 1122–1129. [DOI] [PubMed] [Google Scholar]
- Shi F. et al. (2017) Identifying cell subpopulations and their genetic drivers from single-cell RNA-seq data using a biclustering approach. J. Comput. Biol., 24, 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C. et al. (2019) LTMG: a novel statistical modeling of transcriptional expression states in single-cell RNA-Seq data. Nucleic Acids Res., 47, e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. et al. (2019) Comparative analysis of differential gene expression analysis tools for single-cell RNA sequencing data. BMC Bioinformatics, 20, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J. et al. (2020) QUBIC2: a novel and robust biclustering algorithm for analyses and interpretation of large-scale RNA-Seq data. Bioinformatics, 36, 1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L. et al. (2013) Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol., 20, 1131–1139. [DOI] [PubMed] [Google Scholar]
- Yu G. et al. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS, 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in GEO, 10x Genomics, and ArrayExpress. All datasets can be derived from sources in the public domain: the human embryo cells data were from GSE36552; the PBMCs data were from https://support.10xgenomics.com/single-cell-gene-expression/datasets/1.1.0/pbmc3k; the human lung cancer data were from GSE99254; and the mouse melanoma data were downloaded from https://www.ebi.ac.uk/gxa/sc/experiments/E-EHCA-2/Results.