Abstract
Summary
The need for an efficient and cost-effective method is compelling in biomolecular NMR. To tackle this problem, we have developed the Poky suite, the revolutionized platform with boundless possibilities for advancing research and technology development in signal detection, resonance assignment, structure calculation and relaxation studies with the help of many automation and user interface tools. This software is extensible and scalable by scripting and batching as well as providing modern graphical user interfaces and a diverse range of modules right out of the box.
Availability and implementation
Poky is freely available to non-commercial users at https://poky.clas.ucdenver.edu.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Computer science-related interdisciplinarity creates great synergies in a variety of subjects, including the biological sciences. NMR spectroscopy, one of the major techniques for obtaining 3D structures of proteins at the near-atomistic resolution, is suitable to make use of this new trend of technology because all the tasks are performed on computers, other than the sample preparation step. We introduced the CHESPA/CHESCA-SPARKY software package that performs the principal component analysis (PCA) and the hierarchical agglomerative clustering for detecting long-range allosteric networks (Shao et al., 2020). D.F. Hansen introduced how a deep neural network (DNN) can be used to reconstruct sparsely acquired data (Hansen, 2019). Klukowski et al. introduced NMRNet which detects NMR signals by deep learning approach (Klukowski et al., 2018). We have strived to promote new technologies by increasing usability through establishing an integrative NMR platform that compromises NMRFAM-SPARKY, PONDEROSA-C/S and I-PINE webserver (Lee et al., 2014, 2016, 2019), which would then allow us to attain a new robust foundation to keep up with all the rapid technological breakthroughs and the sunset of Python 2.
Here, we introduce the Poky suite for multidimensional NMR spectroscopy and three-dimensional structure calculation of biomolecules. The Poky suite is a superset and enhanced version of popular integrated software packages supported by new integrated programs. It is not only a significant advance in the research platform but also a new software platform for convenient technology development. Its new extensible and scalable features by scripting and batching will enable users to freely use the offered automated analysis templates and build their own library sets to keep up with the rapid pace of technology.
2 Implementation
The Poky Suite has been written in different computer languages and library kits exhibiting the maximum efficiency and performance in each program. All the programs in the Poky suite are cross-platform supporting Windows, Mac and Linux.
Poky main window. The Poky main window is the main hub for accessing all programs while providing the current status of loaded data (Fig. 1). Main menu. Menu buttons access the multidimensional NMR data visualization and analysis functions (Fig. 1A). Pointer mode. Buttons with mouse cursor icons change the operation mode on the spectrum (Fig. 1B). Tab widgets. Three tabbed widgets in the middle are changed by clicking the tab title (Fig. 1C). ‘Experiments’ shows currently loaded spectra, associated views and buttons to operate them (Supplementary Fig. S1A). ‘Resonances’ shows resonance chemical shifts for selected molecules and conditions and their operation (Supplementary Fig. S1B). ‘Poky Automation Guide (PAG)’ shows available end-to-end workflows or protocols to get the specific job done in Poky (Supplementary Fig. S1C). Protein NMR workflows for both solution and solid-state NMR have been made. Automation tools for nucleic acids, small molecules and metabolomics are on our agenda. 3D structure calculation. Buttons to launch Poky Structure Builder and Poky Structure Analyzer programs for automated structure calculation are located below the tab widgets (Fig. 1D; Supplementary Fig. S2AB). Compared to PONDEROSA-C/S (Lee et al., 2014), the workflow is much simpler. Simply by clicking a few buttons and doing a couple of drag-and-drops can calculate structures and visualize results. Spectral conversion. The single spectrum conversion button asks the user to select raw data to convert to the UCSF format from Bruker TopSpin, NMRPipe (Delaglio et al., 1995) and NMRView (Johnson, 2004) when many data files can be converted by Easy NMRPIPE2UCSF, Easy BRUKER2UCSF and Easy NMRVIEW2UCSF programs by a few clicks (Fig. 1E; Supplementary Fig. S3). Converted UCSF data can be further edited by the Easy UCSFDATA program (Supplementary Fig. S4). Visualization. Poky supports a few plotting programs. Spectral views inherited from NMRFAM-SPARKY are useful for visualizing and analyzing contours from NMR data (Supplementary Fig. S5A). Various plots generated by MatPlotLib are suitable for static visualization and high-quality figure making (Supplementary Fig. S5B). NDP-Plot is an interactive 2D chart plotting program (Supplementary Fig. S5C). Text editor for scripting. Poky Notepad is a powerful text editor with many customizable ‘Poky Scripts’ templates (Supplementary Fig. S6). Users can make their script library by saving customized versions of them or writing from scratch in the local machine. The user can use the file browsers to navigate and drag-and-drop from one’s library folder. Batching. Automated batching jobs without human operations are also supported. Adding the ‘-b’ parameter followed by a batch script file when running Poky will let the program know it is a batch and process the file. Our group plans to maintain our GitHub repository for community-wide development of these script files and incorporate them in the following versions of the Poky suite. User support. User support buttons are accessible from the main window directly (Supplementary Fig. S7A). Whenever there is a question or a curiosity, the user can ask developers and other users using the ‘Question?’ button or browse previous questions and answers in the user forum using the ‘User Forum’ button. The ‘Commands’ button shows a dialog with searchable functions (Supplementary Fig. S7B), and the ‘Cheat Sheet’ shows useful keyboard shortcuts (Supplementary Fig. S7C).
Fig. 1.
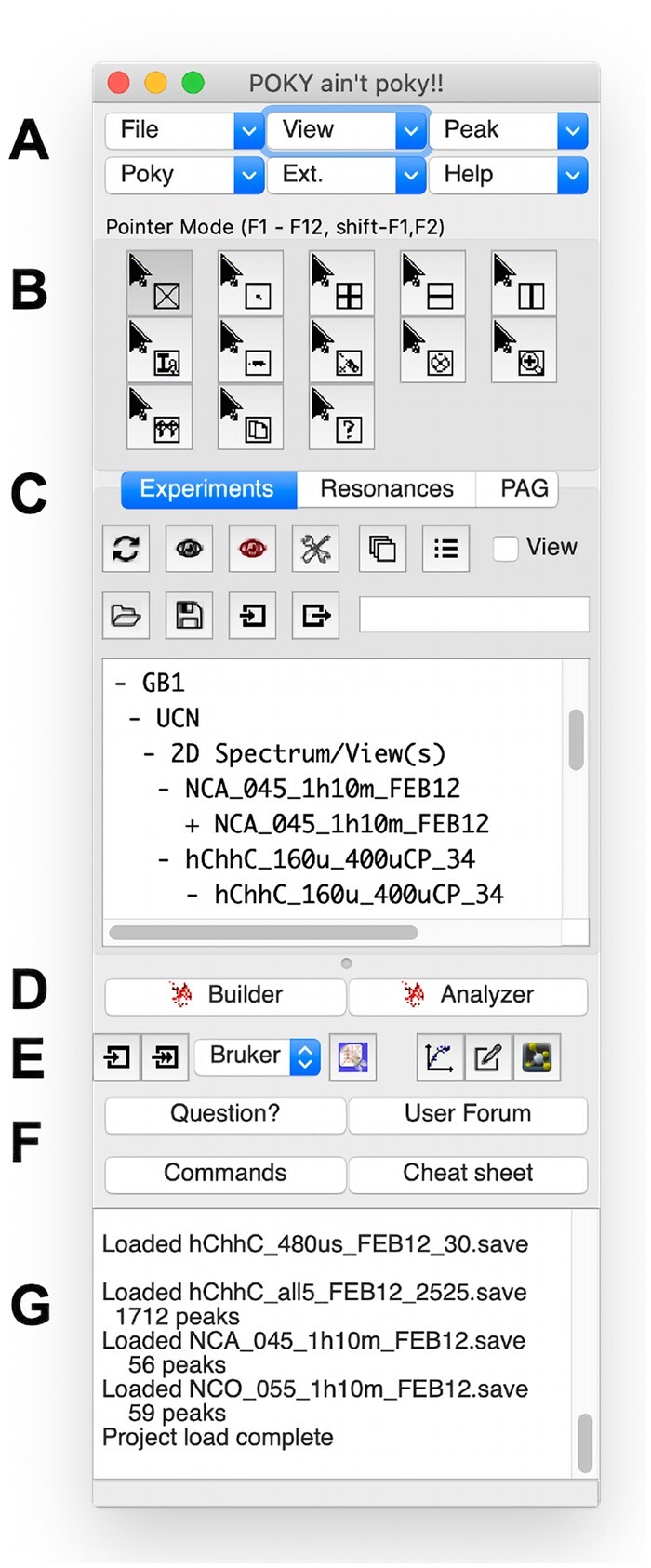
The screenshot of the POKY main window. (A) Main menu. (B) Pointer mode. (C) Tab widgets. (D) 3D structure calculation. (E) Spectral conversion, editing, visualization and text editing tools. (F) User support buttons. (G) Notification area
3 Results and conclusions
We have developed the Poky suite as a complete research and technology development platform for automated NMR processing and analysis, non-uniform sampling (NUS) reconstruction, structure calculation and visualization. All the plugins in NMRFAM-Sparky (∼200) have been upgraded including recently developed PINE-SPARKY.2 (Lee and Markley, 2018), PISA-SPARKY (Weber et al., 2020) and CHESCA/CHESPA-SPARKY. Furthermore, data science and machine learning packages are included and we provide script templates in Poky. We plan to update versions with new functions and tutorial videos routinely. Although the objective of this development is futuristic, the fundamental workflow is similar to NMRFAM-Sparky/PONDEROSA-C/S, so the users can effortlessly master the operation of POKY.
Funding
This work was supported by the National Science Foundation [DBI-2051595 and DBI-1902076 to W.L.] and the startup support from the University of Colorado Denver.
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Woonghee Lee, Department of Chemistry, University of Colorado Denver, Denver, CO 80204, USA.
Mehdi Rahimi, Department of Chemistry, University of Colorado Denver, Denver, CO 80204, USA.
Yeongjoon Lee, Department of Chemistry, University of Colorado Denver, Denver, CO 80204, USA.
Abigail Chiu, Department of Chemistry, University of Colorado Denver, Denver, CO 80204, USA.
References
- Delaglio F. et al. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Hansen D.F. (2019) Using deep neural networks to reconstruct non-uniformly sampled NMR spectra. J. Biomol. NMR, 73, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A. (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. Clifton NJ, 278, 313–352. [DOI] [PubMed] [Google Scholar]
- Klukowski P. et al. (2018) NMRNet: a deep learning approach to automated peak picking of protein NMR spectra. Bioinformatics, 34, 2590–2597. [DOI] [PubMed] [Google Scholar]
- Lee W. et al. (2016) Integrative NMR for biomolecular research. J. Biomol. NMR, 64, 307–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. et al. (2019) I-PINE web server: an integrative probabilistic NMR assignment system for proteins. J. Biomol. NMR, 73, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Markley J.L. (2018) PINE-SPARKY.2 for automated NMR-based protein structure research. Bioinformatics, 34, 1586–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. et al. (2014) PONDEROSA-C/S: client-server based software package for automated protein 3D structure determination. J. Biomol. NMR, 60, 73–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H. et al. (2020) CHESPA/CHESCA-SPARKY: automated NMR data analysis plugins for SPARKY to map protein allostery. Bioinformatics, btaa781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber D.K. et al. (2020) PISA-SPARKY: an interactive SPARKY plugin to analyze oriented solid-state NMR spectra of helical membrane proteins. Bioinformatics, 36, 2915–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


