Summary
The gut microbiome has been implicated as a key regulator of brain function in health and disease. But the impact of gut microbiota on functional brain connectivity is unknown. We used resting-state functional magnetic resonance imaging in germ-free and normally colonized mice under naive conditions and after ischemic stroke. We observed a strong, brain-wide increase of functional connectivity in germ-free animals. Graph theoretical analysis revealed significant higher values in germ-free animals, indicating a stronger and denser global network but with less structural organization. Breakdown of network function after stroke equally affected germ-free and colonized mice. Results from histological analyses showed changes in dendritic spine densities, as well as an immature microglial phenotype, indicating impaired microglia-neuron interaction in germ-free mice as potential cause of this phenomenon. These results demonstrate the substantial impact of bacterial colonization on brain-wide function and extend our so far mainly (sub) cellular understanding of the gut-brain axis.
Subject areas: microbiome, Neuroscience
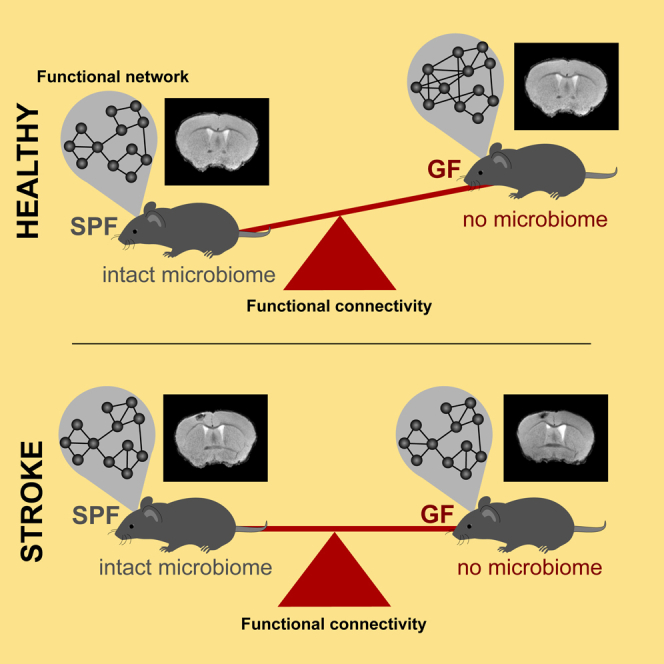
Graphical abstract
Highlights
-
•
Germ-free mice show strong brain-wide increase of functional connectivity
-
•
Global network of germ-free mice is denser but less structurally organized
-
•
Stroke leads to equal network breakdown in germ-free and normally colonized mice
-
•
Results show: bacterial colonization has substantial impact on brain-wide function
Microbiome; Neuroscience
Introduction
Over the past decade, the importance of the gut microbiota (the trillions of microorganisms in the intestinal compartment) for brain development, its physiological function, and role in disease has become increasingly appreciated. Indeed, microbiota have now been implicated as key regulators of the gut-brain axis (Cryan et al., 2019). The microbiota and the brain communicate with each other via various routes including immunological, nervous, and metabolic pathways (Cryan et al., 2019).
Key information on the intricate relationship between gut microbiota and brain function has been gained from germ-free (GF) animal models – animals that have been raised and housed under sterile (axenic) conditions. GF mice have been shown to have an impaired blood-brain barrier, alterations in synaptic plasticity, and learning deficits (Diaz Heijtz et al., 2011; Sharon et al., 2016; Arentsen et al., 2015). More specifically, a previous landmark study demonstrated that GF status of mice substantially affected neuronal dendrite and dendritic spine morphology, the morphological correlates of neuronal communication, thus establishing the concept that developmental gut microbiota colonization is required for normal brain development (Luczynski et al., 2016).
Besides its function in this critical aspect of brain development and maturation, a functional role of the gut microbiota has also been implicated in various neurodevelopmental, autoimmune, and acute disorders of the brain (Sharon et al., 2016; Delgado Jimenez and Benakis, 2020). We and others have previously identified that the gut microbiota is a key modulator of stroke outcome in rodent stroke models (Delgado Jimenez and Benakis, 2020; Sadler et al., 2017). We have shown that stroke changes the bacterial composition in the gut, which was causally linked to a pro-inflammatory T cell polarization and worse stroke outcome (Singh et al., 2016). Similarly, antibiotic treatment induced dysbiosis and affected stroke outcome (Benakis et al., 2016). Following up on these first studies which demonstrated a previously unrecognized key role of changes in microbiota composition on stroke outcome, we then demonstrated in a proof-of-concept study using GF mice that bacterial colonization per se affects stroke outcome and post-stroke neuroinflammation (Singh et al., 2018). Although these earlier studies linking a functional role of the gut microbiota to stroke pathophysiology have focused on acute stroke outcome, we recently observed a pro-regenerative function of key microbial metabolites, short-chain fatty acids, on long-term stroke outcome (Sadler et al., 2020). These results consistently demonstrated the relevance of gut microbiota as bona fide modulators of secondary neuroinflammation and regeneration after experimental stroke. Specifically, we identified that short-chain fatty acids — key bacterial metabolites that readily cross the blood-brain barrier (Frost et al., 2014; Morrison and Preston, 2016) — modulate cortical network plasticity as well as markers of synaptic plasticity in the chronic phase after experimental stroke in mice (Sadler et al., 2020).
However, a critical limitation of this previous work was the lack of in-depth analysis of the connection between gut microbiota colonization and functional brain connectivity in health and disease beyond morphological parameters (e.g., dendritic spines) or only cortical interhemispheric connections as previously studied by us and others (Luczynski et al., 2016; Sadler et al., 2020). Moreover, the previous studies analyzing the impact of gut microbiota colonization on brain network function, its morphological correlate and implications for recovery after brain injury using optical imaging or histological assessment were limited to analyses of excitatory neurons (Sundman et al., 2017; Luczynski et al., 2016; Sadler et al., 2020). Therefore, a more comprehensive and unbiased analysis of the role of the gut microbiota on functional brain network connectivity is required to advance our understanding of physiological and pathological brain function under the control of the microbiota.
In order to achieve this, we used resting-state functional magnetic resonance imaging (fMRI) together with graph theoretical network analysis to analyze brain-wide network function, which has so far not been established for analyzing the gut-brain axis. Resting-state fMRI (rsfMRI) has gained wide attention for its noninvasive information on brain-wide neuronal networks, both in clinical and animal experimental studies (Damoiseaux, 2012; Grandjean et al., 2016). With such functional connectivity maps, we have characterized in previous studies the age dependence of whole-brain networks (Egimendia et al., 2019), described functional connectivity disturbances in brain disease (Green et al., 2019a; Minassian et al., 2020), and unraveled intrinsic regeneration-driven plasticity or treatment-induced functional improvement after stroke (Minassian et al., 2020; Green et al., 2019a; Aswendt et al., 2021a). We further adopted graph theoretical analysis of human brain networks (Rubinov and Sporns, 2010) to quantitatively compare global and local network changes related to experimental stroke (Pallast et al., 2020). In the present investigation, we applied rsfMRI and graph theory to study brain connectivity patterns in germ-free and in normally colonized (specified pathogen free) mice under naive conditions and at the chronic phase after experimental stroke.
Results
Functional connectivity of naive animals
Animals were scanned with T2-weighted MRI for anatomical characterization and with resting-state fMRI (rsfMRI) for functional brain network analysis. No apparent anatomical anomalies were noted for the naive brains of germ-free (GF) animals relative to standard SPF brain images (Figure S1).
Global brain analysis
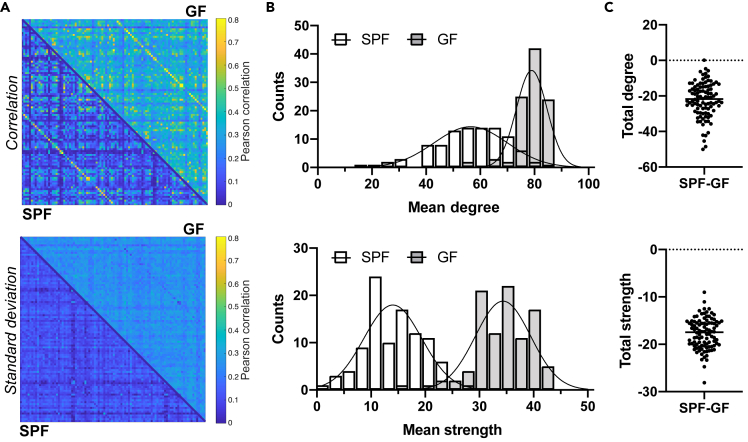
The Pearson correlation values expressing functional connectivity strength between pairs of anatomically defined nodes were averaged across animals per group and expressed in an adjacency matrix of 98 anatomically defined brain regions, so-called seeds (cf Table S1), showing an apparently higher and more variable functional connectivity in GF compared to SPF animals (Figure 1A). The qualitative differences in whole brain matrices directly relate to the frequency plots of the graph theoretical parameters degree (the number of connections of a node) and node strength (the sum of all edge weights connected to a node). The distributions (Figure 1B) show a clear shift to higher values in the GF group and a different Poisson regression curve (p < 0.001), e.g., with 2.4-fold larger amplitude compared to the SPF group degree values. Calculating the difference for degree and node strength between both groups for each of the 98 seeds (Figure 1C) confirmed the substantially weaker degree and node strength in the SPF group (p < 0.001).
Figure 1.
Germ-free mice with increased functional connectivity at the whole-brain level
(A) Top: The Pearson correlation values expressing functional connectivity strength between pairs of anatomically defined nodes were expressed in an adjacency matrix for all 98 × 98 connections, with the SPF group in the lower triangle of the matrix and in the GF group in upper triangle of the matrix. Bottom: the corresponding standard deviation values of the group-wise Pearson correlation values are given. Color bar represents the correlation range from 0 (dark blue) to 0.8 (yellow).
(B) Degree and node strength distribution for SPF and GF group (Poisson regression with likelihood ratio test: different curve for each dataset, p < 0.001).
(C) Difference in total degree and node strength between SPF and GF mice (Student's t test, p < 0.001).
Data are represented as a split adjacency matrix (average Pearson correlation coefficient across animals per connection and related SD), histogram (counts of mean degree or mean node strength values), and individual data points. SPF n = 8, GF n = 15.
To investigate whether the functional connectivity increase in the GF group is similar for all seeds and subnetworks, we applied a novel approach to the rsfMRI network analysis. We randomly selected three nodes (Olfactory areas - OLF, Auditory areas - AUD, and Primary somatosensory area, barrel field - SSp-bfd) among the list of regions with highest degree in the GF group. For these nodes, we focused on their 20 strongest connections (i.e., highest edge weights), independently for each group. Connections appearing only in one of the groups are marked in red (Figure S2). High agreement between groups was found varying between 80% (OLF; AUD) and 90% (SSp-bfd), pointing to a rather homogeneous global upscale of functional connectivity strength in the GF group relative to the SPF group.
Analysis of distinct subnetworks
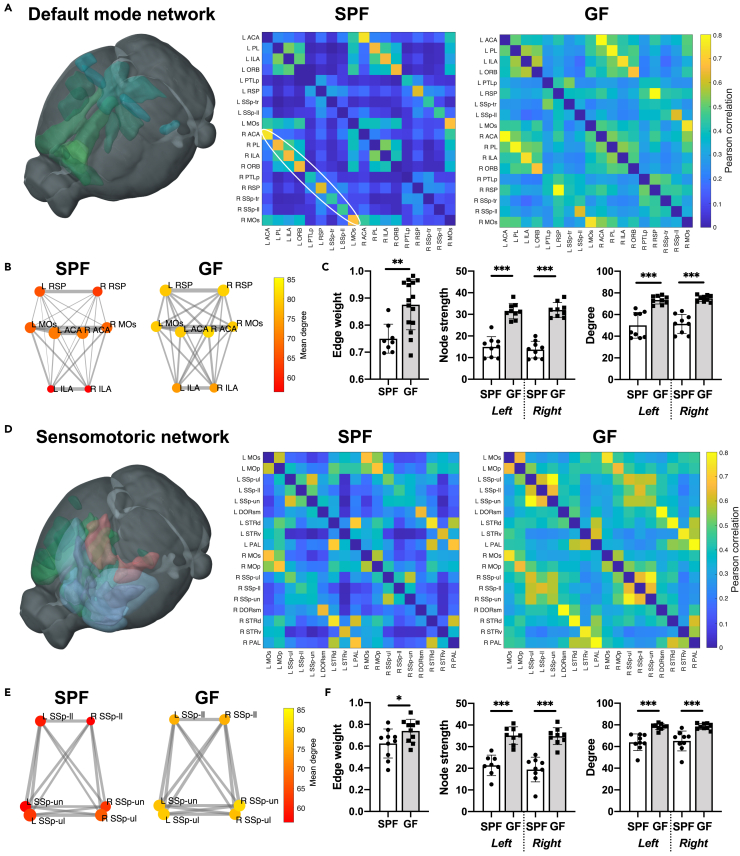
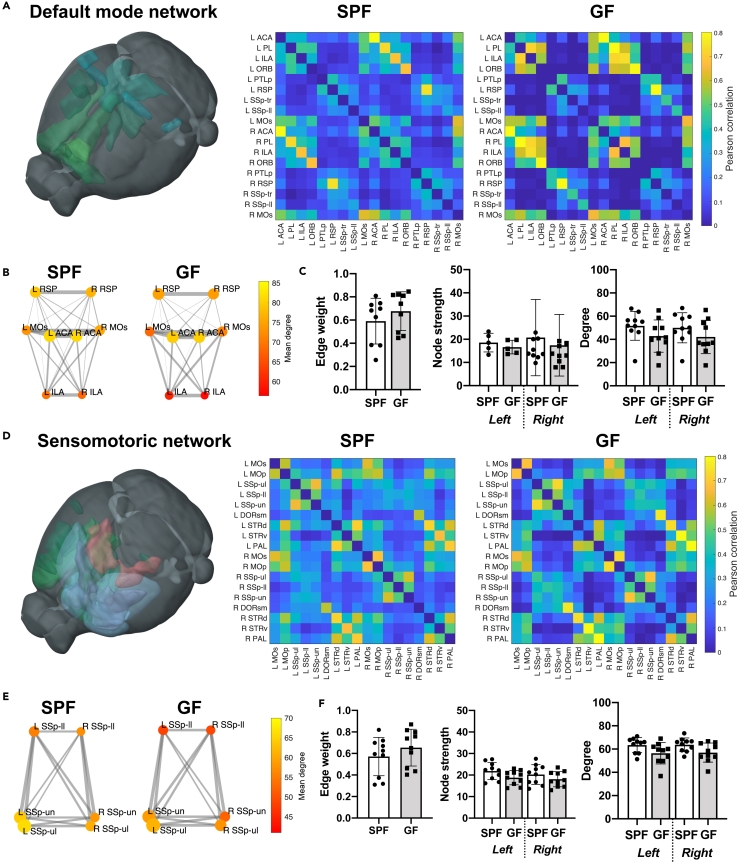
We selected the default mode network (DMN) as previously defined for the mouse (Whitesell et al., 2021) and representative regions of the sensorimotor network (SMN) as two exemplary subnets within the whole-brain functional network (Figure 2). In both, the DMN and the SMN, the adjacency matrix comparison shows a rather homogeneous upscaling from the SPF to the GFP group (Figures 2A and 2D). In the quantitative graph theoretical analysis, edge weight, node degree, and node strength (representing the seed-to-seed connection strength and the network density), are highly significantly increased in the GF group relative to the SPF group. Interestingly, the interhemispheric homotopic connections, represented by the off-diagonal in the adjacency matrix (marked by an ellipsoid in Figure 2A), show consistently higher values than all other intra- and inter-hemispheric connections.
Figure 2.
Germ-free mice with increased functional connectivity in the default mode and sensorimotor network
(A) 3D-illustration of brain regions related to the default mode network (DMN) (represented by the regions ACA, PL, ILA, ORB, PTLp, RSP, SSp-tr, SSp-ll, and MOs). The adjacency matrices present the Pearson correlation values for the related regions in the SPF and GF group.
(B) Graph representation of intra- and interhemispheric connectivity between a subset of DMN regions. Circle color represents mean degree. The edge weight line thickness was scaled 0-1 and the node size represents the node strength.
(C) Quantitative comparison of edge weight, node strength and degree for DMN regions (Student's t test and two-way ANOVA with FDR correction for multiple comparisons). Edge weight calculated for homotopic connections only (e.g., Left ACA – Right ACA), node strength and degree reported for left and right hemisphere separately.
(D) 3D-illustration of brain regions related to the sensorimotor network (SMN) (represented by the regions MOp, MOs, SSp-ul, -ll, -un, DORsm, DORpm, STRd, STRv, and PAL). The adjacency matrices present the Pearson correlation values for the related regions in the SPF and GF group.
(E) Graph representation of intra- and interhemispheric connectivity between a subset of SMN regions SSp-ll/-ul/-un. Circle color represents mean degree. The edge weight line thickness was scaled 0-1 and the node size represents the node strength.
(F) Quantitative comparison of edge weight, node strength, and degree for SMN regions (Student's t test and two-way ANOVA with FDR correction for multiple comparisons). Edge weight calculated for homotopic connections only (e.g., Left SSp-ul – Right SSp-ul), node strength and degree reported for left and right hemisphere separately.
Data are represented as an adjacency matrix with mean values across groups, and bar plots with individual data points, mean and SD. SPF n = 8, GF n = 15.
Stroke lesion characterization
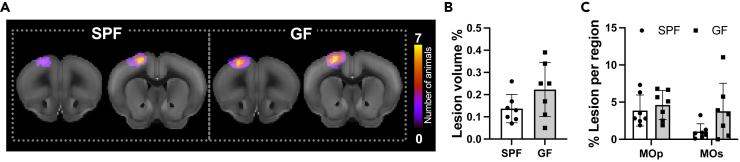
The T2-weighted MR images show a clearly demarcated hyperintense region in the left motor cortex at four weeks after induction of the photothrombotic stroke (Figure 3A). Location of the lesion is equivalent for both the SPF and the GF group, and lesion volume is not significantly different between groups (Figure 3B). In addition, the lesion distribution between primary and secondary motor cortex is not significantly different, excluding a direct impact of differences in lesion size or location on the functional network analysis between these two groups.
Figure 3.
Stroke lesion size and extent are not dependent on the microbiome
(A) Incidence maps of coronal T2-weighted images with the individual lesion extent outlined show the high reproducibility of the photothrombotic lesion in each group and present the lesion position agreeing well between the two groups.
(B) Quantitative analysis of the lesion volume, derived from the T2-weighted RARE images (Unpaired Student's t test, n.s.).
(C) Detailed analysis of lesion fraction within primary and secondary motor cortex (two-way ANOVA, n.s.).
Functional connectivity after stroke
Global brain analysis
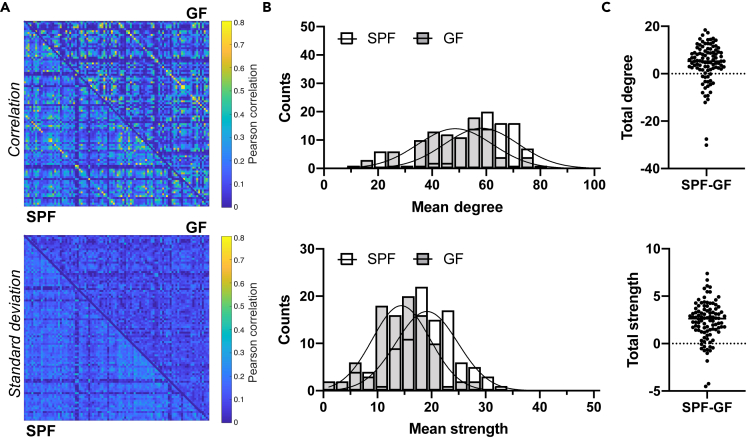
Induction of stroke led to a minor global average increase of connectivity strength across the whole brain at the chronic ischemic phase in SPF mice, encompassing also the contralateral healthy hemisphere. This global increase, calculated as the mean edge weight relative to the naive state, is only 17%. In contrast, in the ischemic GF group, stroke resulted in a massive global average reduction of connectivity strength by 53%. These opposite changes of the functional connectivity strength upon stroke lead to connectivity matrices with closely similar correlation values in the SPF and GF group (Figure 4A).
Figure 4.
Stroke-induced changes in functional connectivity reverse germ-free effects
(A) Whole-brain matrices of Pearson correlation values and standard deviation for all 98 × 98 connections in the SPF and GF group, respectively.
(B) Distribution of degree and node strength (Poisson regression with likelihood ratio test: different curve for each dataset, p < 0.001).
(C) Difference in total degree or node strength between SPF and GF (Student's t test, p < 0.05 for degree; p < 0.01 for node strength).
Data are represented as split adjacency matrix (average Pearson correlation coefficient across animals per connection and related SD), histogram (counts of mean degree or mean strength values), and individual data points. SPF n = 8, GF n = 15
This close similarity between the functional networks of SPF and GF animals is further reflected in the histogram distribution of the graph theoretical variables, mean degree and mean node strength (Figure 4B). Although still statistically significantly different, their two Poisson distribution curves almost completely fall together for the two groups with similar amplitude (14.01 vs. 14.17 for the degree values in GF and SPF, respectively). Interestingly, quantitative analyses of the difference of degree and strength between both groups for each of the 98 seeds showed that both, total degree and total node strength of the GF group have decreased even below the values of the SPF mice (p < 0.05 for total degree; p < 0.01 for total strength) (Figure 4C).
Analysis of distinct subnetworks
The global similarity of functional connectivity values after stroke in both groups is found again in the adjacency matrices of the SMN (Figure 5D). In the DMN, a cluster of transhemispheric connections (including Anterior cingulate area - ACA, Prelimbic area - PL, Infralimbic area - ILA, and Orbital area - ORB) is clearly raised above the otherwise close similarity of the DMN network in both matrices (Figure 5A). Quantitative analysis of edge weight, node strength, and node degree (Figures 5C and 5F) show that for both subnetworks (DMN and SMN), the edge weights of the GF animals remain somewhat higher than for the SPF mice, similar as in the naive brain situation, although no longer statistically significant here. Interestingly, both, degree and strength, show a non-significant trend to lower values for the GF group (Figures 5C and 5F), different to the naive brain situation where edge weights, degree, and strength all were significantly higher in GF animals.
Figure 5.
Functional connectivity in the default mode and sensorimotor network after stroke
(A) 3D-illustration of brain regions related to the default mode network (DMN) (represented by the regions ACA, PL, ILA, ORB, PTLp, RSP, SSp-tr, SSp-ll, and MOs). The adjacency matrices present the Pearson correlation values for the related regions in the SPF and GF groups.
(B) Graph representation of intra- and interhemispheric connectivity between a subset of DMN regions. Circle color represents mean degree. The edge weight line thickness was scaled 0-1 and the node size represents the node strength.
(C) Quantitative comparison of edge weight, node strength and degree for DMN regions (Student's t test and two-way ANOVA with FDR correction for multiple comparisons). Edge weight calculated for homotopic connections only (e.g. Left ACA – Right ACA), node strength and degree reported for left and right hemisphere separately.
(D) 3D-illustration of brain regions related to the sensorimotor network (SMN) (represented by the regions MOp, MOs, SSp-ul, -ll, -un, DORsm, DORpm, STRd, STRv, PAL). The adjacency matrices present the Pearson correlation values for the related regions in the SPF and GF group.
(E) Graph representation of intra- and interhemispheric connectivity between a subset of SMN regions SSp-ll/-ul/-un. Circle color represents mean degree. The edge weight line thickness was scaled 0-1 and the node size represents the node strength.
(F) Quantitative comparison of edge weight, node strength and degree for SMN regions (Student's t test and two-way ANOVA with FDR correction for multiple comparisons). Edge weight calculated for homotopic connections only (e.g. Left SSp-ul – Right SSp-ul), node strength and degree reported for left and right hemisphere separately.
Data are represented as an adjacency matrix with mean values across groups, and bar plots with individual data points, mean and SD. SPF n = 7, GF n = 7.
Comparison between naive and ischemic condition
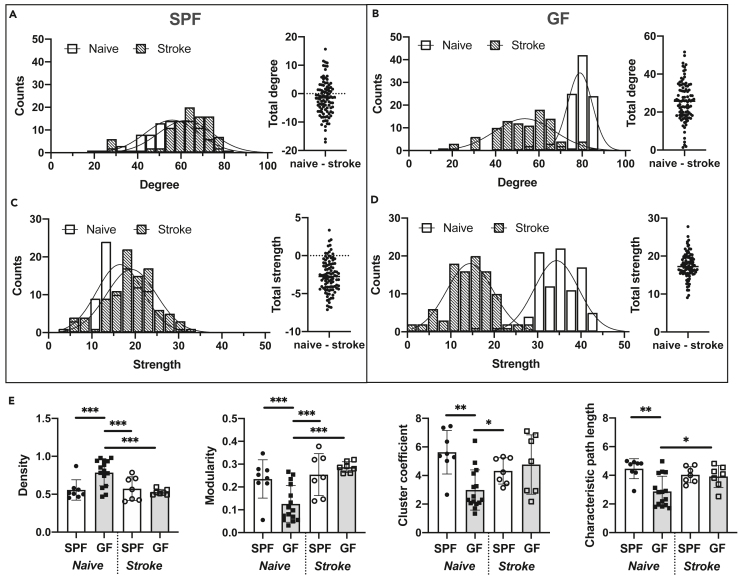
The study design required separate animal groups for healthy, naive, and for ischemic conditions, independent of SPF or GF status. Although the earlier analysis above focused on the comparison between SPF and GF animals, separately for naive or for ischemic conditions, we here compare the functional network conditions between naive and ischemic conditions, separately for SPF and GF groups. The histograms for degree and node strength of the SPF groups (Figures 6A and 6C) both show a close overlay between naive and ischemic status for the Poisson distribution curves (p < 0.01), with a slight shift to higher values after stroke, in agreement with expected hyperconnectivity in mild strokes with small ischemic lesions (Blaschke et al., 2021; Minassian et al., 2020). In the quantitative differences for all 98 nodes across the brain, total degree was no longer statistically significant, whereas total strength showed a significant increase (p < 0.001) following stroke (Figures 6A and 6C).
Figure 6.
Global network alterations in germ-free mice after stroke
(A) Degree distribution (Poisson regression with likelihood ratio test: different curve for each dataset, p < 0.01) and difference in total degree between naive and stroke SPF mice (Student's t test, n.s.).
(B) Degree distribution (Poisson regression with likelihood ratio test: different curve for each dataset, p < 0.001) and difference in total degree (naive – stroke) for GF mice (Student's t test, p < 0.001).
(C) Node strength distribution (Poisson regression with likelihood ratio test: different curve for each dataset, p < 0.01) and difference in total strength (naive – stroke) for SPF mice (Student's t test, p < 0.001).
(D) Node strength distribution (Poisson regression with likelihood ratio test: different curve for each dataset, p < 0.001) and difference in total strength (naive – stroke) for GF mice (Student's t test, p < 0.001).
(E) Global graph theoretical measures for naive and stroke SPF and GF mice, respectively (two-way ANOVA with FDR correction for multiple comparisons).
Data are represented as histogram (counts of mean degree/node strength values) and bar plots (mean and SD with individual data points per animal). naive/stroke: SPF n = 8/7, GF n = 15/7.
In contrast, the histograms of the GF groups showed a significant separation of the Poisson distribution curves (p < 0.001) between naive and ischemic groups, but with the naive status remaining at clearly higher values (Figures 6B and 6D). This was further confirmed by the quantitative differences for all 98 nodes across the brain (p < 0.001 for both total degree and total strength) (Figures 6B and 6D). Thus, relative to the naive status, stroke resulted in a significantly reduced connectivity for the GF group.
Finally, further global graph theoretical parameters, density (fraction of existing edges relative to the theoretical maximum number of edges), modularity (level of network segregation), cluster coefficient (node connections with the nearest nodes proportionally to the maximum number of connections), and characteristic path length (average shortest path length in the whole network) were calculated for naive and stroke condition in SPF and GF groups (Figure 6E). Naive GF mice showed a highly significantly increased density, which, however, reduced to SPF level after stroke induction. Modularity as well as cluster coefficient and characteristic path length were significantly lowered in naive GF group; again, after stroke, all three parameters became indistinguishable between the GF and SPF groups.
Histological analysis of dendritic spines and microglial morphology
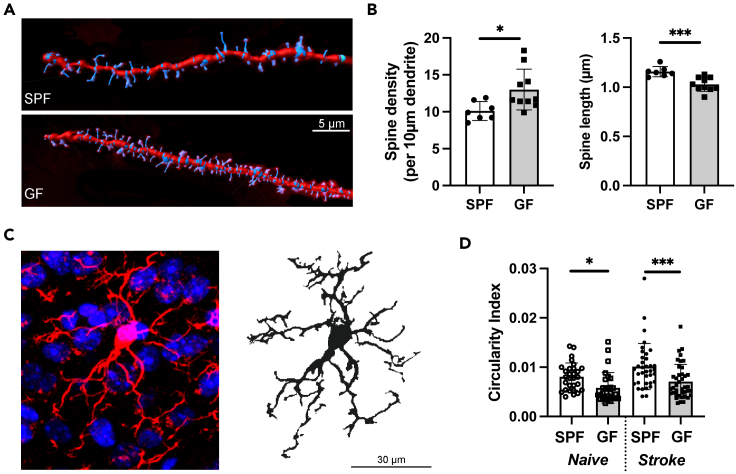
After stroke, there is a highly intricate reorganization of the neuronal network. Microglia eliminates synaptic elements in an activity dependent manner; additionally neurons undergo dynamic structural changes (Wake et al., 2009). Therefore, in order to study a potential mechanistic cause of the observed phenomena in the rsfMRI analysis, we histologically assessed neuronal synapse pruning and microglial activity. We quantified the spines on dendrites of cortical pyramidal neurons found in the layer 2/3, 400 μm away from the lesion. We then reconstructed the dendrites in Imaris for quantification of spine densities per dendrite and analysis of spine lengths (Figure 7A). We found that the spine density was significantly increased (p < 0.05) in the ipsilateral cortex of GF mice compared to SPF mice at 28 days post lesion induction; however, the average dendritic spine length was significantly shorter in GF mice (p < 0.001, Figure 7B). These results indicated an increase of overall spines most likely due to an abundance of immature or dysfunctional spines which were not pruned (Segal et al., 2000).
Figure 7.
Differences in dendritic spines and microglial morphology between GF and SPF mice
(A) Representative images of Imaris 3D reconstruction for pyramidal neuronal spine density analysis from cortex. White line is 5 μm. SPF (above) and GF (below).
(B) Spine density per 10 μm of dendrite and spine length in the ipsilateral hemisphere of GF and SPF mice 28 d after ischemic stroke (Student's t test, p < 0.05 and p < 0.001; n(SPF) = 7; n(GF):10).
(C) Representative image of Iba1 immunostaining (red) with counterstained cell nuclei (blue) (left) and the binary mask for microglia staining with Iba1 (right).
(D) Circularity index for microglia in naive GF and SPF mice as well as 28 d post-stroke. n(naive) = 5 mice per group; n(stroke) = 7 mice per group, Student's t test, p < 0.05 and p < 0.001).
Recent studies suggest a key role for microglial activation in synapse pruning during physiological brain maturation and under disease conditions (Trapp et al., 2007). Moreover, impaired microglial activation in GF compared to SPF mice has previously already been reported by us and others (Mezo et al., 2020; Erny et al., 2015; Sadler et al., 2020). To histologically investigate microglial activation status in naive mice and at 28 d post-stroke, we analyzed microglial circularity which has been shown to correlate with cellular activation (Heindl et al., 2018) (Figure 7C). We observed a significantly reduced circularity index in both naive and post-stroke microglia of the ipsilateral hemisphere in GF compared to SPF mice (Figure 7D), indicating a less activated or less mature microglial phenotype in GF mice, which is in accordance with previous reports (Thion et al., 2018; Singh et al., 2018).
Taken together, we observed an association of increased functional network strength with morphological markers of increased synapse density and reduced microglial activation. These findings suggest that the changes observed in GF mice could be a result of altered (microglia-driven) synaptic plasticity.
Discussion
Resting-state functional MRI (rsfMRI) has become of increasing interest as it allows to simultaneously monitor the status of functional neuronal networks across the whole brain without external stimuli. In clinical studies, rsfMRI has been widely used to characterize brain states during varying pathological conditions (Damoiseaux, 2012; Silasi and Murphy, 2014). Furthermore, it has successfully been used to diagnose progression of various brain disorders (Badhwar et al., 2017). Next to the focus on interregional correlations of the low frequency alterations of the BOLD signal, graph theory is becoming a valuable approach to study connectivity patterns and organization at multiple scales of the brain network (Rubinov and Sporns, 2010; Farahani et al., 2019). rsfMRI has also become an important tool in experimental studies using mouse models for neurodegenerative diseases (Grandjean et al., 2016; Green et al., 2019b; Shah et al., 2016; Zerbi et al., 2014) and stroke (Green et al., 2018; Minassian et al., 2020; van Meer et al., 2012). For the first time, we here show that the modulation of functional network strength by gut microbiota in the naive mouse brain leads to profound hyperconnectivity in GF mice. We also demonstrate that stroke induced a severe drop in the functional neuronal network in GF animals, in contrast to SPF animals, in which stroke had only a minor effect on network connectivity in the chronic phase.
Naive brain situation
The absence of gut microbiota was found to result in a strong increase of the functional networks throughout the whole mouse brain to twice the connectivity strength of mice housed under conventional SPF conditions. The increase of functional network strength was rather homogeneous in GF mice, as demonstrated with the closely equal networks of examples of the top connections. This homogeneous upscale from SPF to GF connectivity strength was confirmed for the individual subnetworks DMN and SMN. The graph analytical parameters edge weights, node degree, and node strength, all showed significantly higher values in the GF group, indicating a stronger and denser global network but with less structural organization, as seen by the significantly lower modularity and cluster coefficient. Both, lower modularity and cluster coefficient point toward a more random functional network, whereas the opposite, i.e. higher modularity and cluster coefficient indicate a functional network consisting of more segregated functional units in SPF mice (Rubinov and Sporns, 2010; Paldino et al., 2017).
Changes in network connectivity after ischemic stroke
The photothrombotic induction of stroke led to a small, circumscribed cortical lesion. In the SPF group, this resulted in a slight increase of functional connectivity strength at four weeks after stroke induction. This hyperconnectivity in the chronic ischemic phase is in good agreement with earlier reports by Minassian and coworkers (Minassian et al., 2020) in a mouse cortical lesion model and by Blaschke et al. (Blaschke et al., 2021) in a recent comparison of small focal stroke lesions in mice and humans. Both these studies found a clear increase in functional connectivity strength. This hyperconnectivity was reflected in our graph theoretical analysis by a small but significant increase in both node degree and node strength upon stroke induction, further supported by the reported increase in node strength in different mouse models of cortical stroke and in human patients with mild stroke (Blaschke et al., 2021).
In contrast, the ischemic germ-free mice showed a strong and global decrease in network connectivity. This shift to lower values in node degree and strength resulted in close alignment of the mean degree and mean strength distribution between the GF and SPF groups, with the GF values even slightly below those of the SPF group. Interestingly, the global network characterizing values - density, modularity, cluster coefficient, and characteristic path length - having been significantly different from the SPF group under naive, healthy condition, becomes indistinguishable between both groups under ischemic condition. These findings of substantially different functional network conditions in the naive state and closely similar network conditions in the ischemic state indicate that the strong influence of the gut microbiome absence on the healthy functional network is overridden by the dominant influence of the ischemic lesion.
Interpretation of the hyperconnectivity
Under neuropathological conditions, hyperconnectivity has been described as a hallmark of neurological disturbance (Hillary et al., 2014) and has been reported for mild cases of stroke in mice and men (Minassian et al., 2020; Blaschke et al., 2021). In a hypothetical model of functional connectivity alterations, Gorges and colleagues reported that connectivity strength increases as “a potentially compensatory response to ongoing cell degeneration in order to maintain normal behavioral performance” (Gorges et al., 2017). This dichotomy of true regeneration—in the sense of restoration of physiological function—versus compensation by an aberrant function can also be observed in the behavioral recovery of rodents after stroke (Murphy and Corbett, 2009; Levin et al., 2009; Whishaw, 2000). However, the functional basis of increased connectivity for recovery or compensation after acute injuries to the brain is still largely unknown and requires a more detailed mapping of the neuronal connectivity-behavior axis.
Interestingly, besides external factors—such as microbial colonization, microglial activity, and others—also the lesion location and severity itself has a critical impact on the observed changes in brain connectivity after stroke. Mild strokes have been shown to lead to hyperconnectivity with slow but continuously increasing network strength over 12 weeks (Minassian et al., 2020). On the other hand, severe tissue damage, such as induced by the commonly used middle cerebral artery occlusion mouse model (Llovera et al., 2021), was shown to result in early and persistent hypoconnectivity (Green et al., 2018). Again, in Gorges' hypothetical model of connectivity alterations, it is assumed that “when a critical cell loss is reached, the functional reserves are exhausted, […] and functional connectivity decreases” (Gorges et al., 2017). In the present study, the hyperconnectivity, although weak, of functional neuronal networks at 4 weeks post stroke induction points to the category of mild ischemic lesion in our stroke model of cortical infarct, in full accordance with earlier reports (Minassian et al., 2020; Blaschke et al., 2021). Interestingly, the level of functional network strength reached after stroke in both SPF and GF groups of mice, is very similar. This is remarkable as the GF group shows a massive hyperconnectivity under naive conditions. This hyperconnectivity (relative to SPF naive state); however, is completely antagonized by the stroke induced reduction in network connectivity. This suggests that stroke is the dominant parameter of functional network modulation, whereas in the naive state, the absence of gut microbiota has a predominant influence on neuronal network function.
In this study, we observed that changes in network connectivity strength were associated with alteration of dendritic spine density, which is commonly perceived as the morphological correlate of synaptic inter-neuronal communication (Arnsten et al., 2010). Indeed, correlative changes in network connectivity as well as spine density or morphology have been demonstrated in a number of brain disorders including Alzheimer's disease, epilepsy, and psychiatric diseases (Palmer, 2014; Penzes et al., 2011). Besides these primary brain disorders, changes in spine density and morphology have also been observed in previous reports investigating GF mice in comparison to conventionally colonized animals (Luczynski et al., 2016). In accordance with this previous report, which has focused mainly on dendrites in the amygdala and hippocampus, we confirm here a significant impact of gut colonization on spine densities as well as their morphology also for excitatory neurons in the neocortex. An increased spine density has previously been suggested to reflect an increased number of excitatory neuronal input (Moser et al., 1994). This would suggest that GF mice have higher levels of excitatory input into the cortex near the lesion after stroke, which is potentially reflected in the increased connectivity strength and density observed in naive GF mice.
Increased spine densities might be a result of cell-autonomous, neuronal mechanisms or because of a reduction of synapse pruning, the mechanism of spine removal by microglial cells. It is a limitation of this study that we did not characterize the impact of bacterial colonization on neuronal activity, endogenous plasticity, and expression of pro-regenerative factors. Yet, we have observed a microglial morphology resembling a less activated or immature microglial phenotype (Heindl et al., 2018; Singh et al., 2018). Activation of microglia has previously been shown to be associated with the active pruning of synapses in the same brain areas as investigated in this study (i.e., layer 2/3 of the mouse neocortex) (Wilton et al., 2019). Interestingly, it was recently demonstrated that physiological microglial function is critically required - via maintenance of synapse turnover - to prevent a hyperexcitation of the neuronal network, as potentially reflected by the increased connectivity strength in our dataset. In accordance with our results on microglial morphology, previous studies have also shown that microglia in GF mice display an immature phenotype with impaired innate immune responses (Erny et al., 2015). Hence, our results align well with the concept of altered brain network function in GF mice owing to impaired synapse turnover by immature microglia. Yet, further studies are required to test the causality along such a supposed multicellular process of microbiota-microglia-neuronal alterations under the control of microbiota and bacterial metabolites.
In conclusion, we demonstrate here the previously unrecognized vast impact of gut colonization on global functional brain networks. The increase in brain connectivity in microbiota-deficient animals resulted in a denser but less structured network, pointing toward a less efficient system during compensation efforts of the brain. The hyperconnectivity is masked after acute injuries, most likely due to the overall breakdown of network integrity after ischemic lesion. Histological data from this study, in conjunction with results of previous reports, suggest a potential role of microglia to mediate the microbiome's effect on network connectivity strength.
Limitations of the study
The study did not allow a longitudinal examination of the functional neuronal network data on the same individuals as the animals could not continuously be kept under GF conditions during the rsfMRI experiments, thus requiring comparison of separate groups between healthy naive and ischemic status. Thus, it was also not possible within the scope of the project to include further time points after stroke in order to observe the apparently competing effects of gut microbiome deficiency and stroke on the functional neuronal network conditions. For future experiments, it will be of particular interest to investigate the effect of microbiome deficiency within the first days after stroke induction as earlier reports have shown a continuous rise in hyperconnectivity over several weeks following stroke induction (Aswendt et al., 2021b; Minassian et al., 2020). Finally, the differences in functional network patterns described in the present investigation remain without correlation with appropriate behavior studies. Here, again as with a longitudinal rsfMRI study design, it would be extremely challenging and beyond the scope of the present study to realize comprehensive behavior analyses under GF condition.
In addition, our study was not able to clarify the mechanistic cause of the hyperconnectivity in GF mice at naive state and the difference in the response of connectivity strength after stroke between the two study groups. The confirmation of a microglial morphology which suggests an impaired microglial activity in GF mice as well as the differences in dendritic spine densities which are in line with previous reports studying GF mice, suggest the possibility that microglial dysfunction and microglia-dependent synaptic plasticity could be involved in the observed effects on functional network connectivity. However, a causal validation of the mechanisms along the microbiota-microglia-neuronal axis is beyond the scope of this study and warrants further in-depth analyses.
STAR★Methods
Key recources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse Iba1 (rabbit) | WAKO | Cat# 019-19,741; RRID: AB_839504 |
| Anti-rabbit AF594 (goat) | Thermo Fisher Scientific | Cat# A32740; RRID: AB_2762824 |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI (4′,6-Diamidino-2-Phenylindole) | Thermo Fisher Scientific | D1306; RRID: AB_2629482 |
| Fluoromount aqueous mounting medium | Merck | F4680 |
| Critical commercial assays | ||
| Slice golgi Kit | Bioenno Lifesciences | 003760 |
| Deposited data | ||
| Raw and analyzed data | This paper | https://dx.doi.org/10.17617/3.5p |
| Experimental models: organisms/strains | ||
| Mouse: C57BL6/J | Charles River | 000,664 |
| Software and algorithms | ||
| MATLAB_R2019b | The MathWorks, Inc | |
| Microsoft Excel | Microsoft Corporation | |
| GraphPad Prism 8/9 | Graphpad SoftwareInc | |
| AIDAmri_v1.1 | Own development | https://github.com/aswendtlab/AIDAmri |
| AIDAconnect_v1.0 | Own development | https://github.com/aswendtlab/AIDAconnect |
| FSL 5.0.1 | https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/ | |
| ITK-SNAP 3.8.0 | http://www.itksnap.org/pmwiki/pmwiki.php | |
| Brain connectivity Toolbox | https://sites.google.com/site/bctnet/ | |
| Python 3.6 | https://www.python.org/downloads/release/python-360/ | |
| ImageJ 2.1.0 | https://imagej.net/ | |
| Fiji 2.0 | https://imagej.net/software/fiji/ | |
| Imaris x64 version 8.4.0 | BITPLANE | |
| ParaVision (PV) version 6.0.1 and 7.0 | Bruker Biospin | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Markus Aswendt (markus.aswendt@uk-koeln.de).
Material availability
This study did not generate new unique reagents.
Experimental model and subject details
Animal experiments
A total of 48 male C57BL/6J mice were used for the study. 20 SPF C57BL/6J male mice were purchased from Charles River (Sandhofer Weg 7, 97,633 Sulzfeld, Germany). 28 germ-free (GF) C57BL/6J mice were obtained from the Clean Mouse Facility, University of Bern, Switzerland and the Institute of Systems Immunology, Julius-Maximilians University Würzburg. On the day of arrival, mice were aged 9-10 weeks. GF mice were housed in sterile HAN-gnotocages and received the same sterile water and pelleted food as conventional (SPF) mice. All surgical procedures and cage changes were performed in a disinfected laminar flow microbiological safety cabinet. All experimental protocols were performed in accordance with the guidelines for the use of experimental animals and were approved by the governmental committee (Regierung von Oberbayern, Munich, Germany). Detailed exclusion criteria are described below. Animals were randomly assigned to treatment groups and all analyses were performed by investigators blinded to group allocation. All animal experiments were performed and reported in accordance with the ARRIVE guidelines (Kilkenny et al., 2010).
Method details
Axenic status assessment
GF status was routinely monitored by culture-dependent and -independent methods and confirmed to be pathogen-free. Briefly, samples from each material imported in the isolator (such as food, bedding, and water) were collected and extensive aerobic and anaerobic cultures were performed. Serology and PCR analysis for exclusion of pathogens following FELASA annual recommendations and guidelines was performed in collaboration with IDEXX.
Experimental protocol
Animals were shipped in HAN-gnotocages directly to the Max Planck Institute for Metabolism Research in Cologne for the in vivo MRI investigations. In the first phase of the study, naive GF (n = 18) and normally colonized, specific pathogen free (SPF) animals (n = 10) were shipped in separate HAN-gnotocages (4-5 mice per cage) in order to maintain the microbiological status. The Cologne lab was blinded to the assignment of animals to SPF and GF group. 24 h after acceptance in Cologne, one cage was opened, and the corresponding mice were directly scanned in the 9.4T MRI system. Directly following the MRI scan, the animals were sacrificed by perfusion fixation. Then, the procedure was repeated with the next cage. Unblinding was done after the completion of image data post-processing. During the analysis of the rsfMRI data, five animals needed to be excluded (n = 2, SPF group; n = 3; GF group) because of measurement artifacts prohibiting reliable functional connectivity analysis, resulting in n = 8 and n = 15 datasets for the SPF and GF group, respectively, entering the functional connectivity analysis.
In the second phase, photothrombotic stroke was induced in GF (n = 12) and SPF (n = 8) animals in Munich. Four weeks later, mice were sent to Cologne in HAN-gnotocages. Four GF animals were excluded based on T2-weighted MRI: two had no stroke, and a further two had undefined cortical anomalities.
After MRI experiments, mice were sacrificed under deep anesthesia (4-5% isoflurane in breathing gas (70/30% N2/O2), perfused first with saline, and then with aldehyde golgi-fixative solution (Bioenno). Brains were carefully removed and placed in fixative solution at 4°C for 6 days and sent back to Munich for further analysis (see below).
Stroke induction
At 9-10 weeks of age, focal cerebral ischemia was induced by photothrombosis of the primary motor cortex as previously described (Cramer et al., 2019; Llovera et al., 2021). All surgical procedures including stroke induction, mouse handling and cage changes were performed under sterile conditions in a sterile microbiological laminar flow cabinet as previously described in detail (Singh et al., 2018). In brief, mice were anesthetized with an intraperitoneal injection of fentanyl (0.05 mg/kg), midazolam (5 mg/kg), and medetomidine (0.5 mg/kg) in a disinfected laminar flow microbiological safety cabinet. Mice were transferred into a stereotaxic apparatus, and body temperature was maintained at 37°C. Dexpanthenol eye ointment was applied to both eyes. Following a skin incision, bregma was first located and then, using the laser, the lesion location was marked in the left hemisphere, −1.5 mm lateral and 1.0 mm rostral. Mice received intraperitoneally 10 μL/g body weight of 1% Rose Bengal (sigma 198,250-5g) in saline and were left for 10 min incubation to allow circulation. Shielding was placed on skull allowing a 2 mm diameter circular exposure over the anticipated lesion area. The laser was applied to the lesion area for 17 min, Cobolt Jive 50, 561 nm wavelength, and power at 25 mW. Fiber Collimation Package: 543 nm, f = 7.66 mm, FC/APC. Immediately after surgery, mice were recovered via intraperitoneal antagonistic injection of a combination of naloxone (1.2 mg/kg), flumazenil (0.5 mg/kg), and atipamezole (2.5 mg/kg).
MRI data acquisition
MRI measurements were performed with a 9.4 Tesla animal scanner (Bruker Biospin, Ettlingen, Germany) with 20 cm horizontal bore diameter, equipped with actively shielded gradient coils. Radio frequency transmission and signal reception were performed with a helium-cooled mouse 1H quadrature cryogenic surface coil (CryoProbe, Bruker Biospin). Paravision 6.0.1 and 7.0 (Bruker Biospin) was used to execute the MRI acquisition protocols. The animal was placed in an animal holder equipped with a mask supplying the breathing gases (70/30% N2/O2). The mouse head was fixed with teeth bar and ear bars. The animal's body temperature was measured via a fiber optic rectal probe (1025T System, SA Instruments, Stony Brook, NY, USA) and kept constant at 37 ± 1.0°C by an adjustable water circulating system (medres, Cologne, Germany). Additionally, breathing rate was continuously monitored (1025T System), recorded (DASYlab Software, National Instruments, Austin, TX, USA), and synchronized with the MRI sequence.
Anatomical brain imaging
3-plane pilot reference scans using FLASH were acquired to confirm correct positioning of the mouse head, followed by a FieldMap with consecutive local shim to optimize magnetic field homogeneity. Next, a high-resolution anatomical T2-weighted TurboRARE dataset was acquired with a RARE factor of 8 and two averages. The field-of-view (FOV) was set to 17.5 × 17.5 mm2, and 35 or 48 coronal slices of 0.3 mm thickness without inter-slice gap were placed to cover the whole mouse brain. The matrix dimension was set to 256 x 256, repetition time (TR) to 5,500 ms, and echo spacing to 10.8 ms, resulting in an effective echo time (TE) of 32.5 ms. In-plane resolution was (68 μm)2 and a bandwidth of 39.062 Hz.
Resting-state fMRI (rsfMRI)
A bolus of 0.1 mg/kg medetomidine (Domitor, Elanco), suspended in 250 μL NaCl, was administered subcutaneously 15 min prior to the functional imaging scan, with subsequent reduction of isoflurane level to 0.5%, following an earlier reported protocol (Green et al., 2018, 2019a). A gradient-echo echo-planar imaging (GE-EPI) sequence was used for rsfMRI with the following parameters: FOV: 17.5 × 17.5 mm2, matrix size: 96 × 96, in-plane resolution: 182 × 182 μm2, TR = 1,420 ms, and TE = 18 ms. 335 image sets were acquired with 20 slices each, with slice thickness of 0.4 mm and inter-slice gap of 0.1 mm, recorded non-interleaved and covering the whole forebrain, starting only after a minimal time of 10 min on reduced isoflurane levels.
Tissue preparation
At 28 days post-surgery, mice were sacrificed in deep anesthesia (4-5% isoflurane in breathing gas (70/30% N2/O2), perfused first with saline and then with aldehyde fixative solution (Bioenno). Brains were removed and placed in fixative solution at 4°C for 6 days. 100 μm coronal thick vibratome sections were consecutively collected with alternative sections in either 10x PBS for Golgi-Cox staining, or 1x phosphate-buffered solution (PBS) for microglia analysis.
Spine density analysis
Vibratome slices were removed from 10x PBS and incubated in impregnation solution sliceGolgi Kit (Bioenno) for 5 days in the dark. Staining and post-staining was performed as per manufacturer's protocol. The analysis was performed on cortical pyramidal neurons as found in layer 2/3 400 μM around the perimeter of the lesion. In total, per animal 25 dendrites (5 dendrites from 5 neurons) in both hemispheres were recorded using an Axio Imager.M2 and a 100× objective (EC Plan-Neofluar, NA = 1.3, immersion: oil, acquisition at 18-20°C) using the AxioCam MRc and AxioVision 4.8.2 software. Dendrites from the images were then 3D-reconstructed and the spine density evaluated on the reconstructed 3D surface using Imaris x64 (8.4.0, Bitplane).
Microglia analysis
Vibratome slices were removed from 1x PBS and washed for a further 3 times, exchanging PBS every 5 min. Free-floating sections were blocked for 1 h at room temperature with 5% normal goat blocking buffer. Sections were then stained with rabbit anti-Iba1 (1:200, Wako) and placed on a gentle shaker overnight at 4°C. Next day, after washing, sections were stained with secondary antibody, goat anti-rabbit AF594 (1:200, Thermo Fisher Scientific) for 2 h in the dark at room temperature. Nuclei were stained with DAPI (1:5000, Invitrogen). Sections were mounted onto microscope slides, and a coverslip affixed with Fluoromount aqueous mounting medium (Sigma). Confocal z stack images were acquired at 40× objective 300 μm lateral to the lesion in cortical pyramidal layer 2/3.
Morphological analysis was performed using FIJI software (Version 2.0). Microglia were chosen for analysis if the entire cell was located in the z stack. In brief, the maximum intensity projection of the cell was created only selecting images in the stack which contained the cell. From the maximum intensity projections, threshold steps were performed; first brightness and contrast were adjusted, and then a binary mask was created. For each microglia, the following formula was used to calculate the circularity index: (4 x 3.1415926 x Area)/(Perimeter2). For each mouse, at least 5 cells were analyzed.
MRI postprocessing
The raw data of Bruker file format was converted to NifTI format, pre-processed and registered with the Allen Brain Reference atlas (ccf v3) (Lein et al., 2007) – in the text referred to as the atlas - using the in-house developed Python pipeline AIDAmri as described elsewhere (Pallast et al., 2019). Briefly, the data was converted to NifTI, bias-field corrected, and brain-extracted, followed by a two-step registration process. The raw data remained untouched and the transformation was applied to the atlas with a high-resolution MRI template as intermediate step. We used a modified version of the atlas, comprising 98 regions split between both hemispheres and selected to match parental regions with the MR image resolution. A list of all the abbreviations of all 98 regions used here is provided in the Table S1. The AIDAmri code and the modified atlas are available online (https://github.com/aswendtlab/AIDAmri).
The pre-processing of the rsfMRI data included skull stripping, spatial filtering with a Gaussian kernel sigma of 0.1 mm, slice time correction (for the repetition time), slice-wise motion correction, and regression of the respiration, bandpass filtered between 0.01 and 0.1 Hz. Regressors for breathing, motion correction, and drift variations were generated from the physiological monitoring data (see above), from the results from MCFLIRT and calculated drift functions (first, second, and third order), respectively. Fluctuations correlating with respiration, motion correction, and drift were regressed out of the functional data slice-wise using FSL Regfilt.
The time-series of the regions were calculated as the mean time series of all voxels belonging to the specific region. The correlation matrix was calculated for all 98 regions with Pearson's correlation on the full dataset. Graphs were constructed by applying a lower threshold of 0.1. Functional connectivity data was presented using network science and graph theory, in which a brain region relates to a node and the connection between two nodes to an edge. We calculated the global graph properties density, modularity, cluster coefficient, and characteristic path length as well as the local measures degree, node strength, and edge weight. The weighted, undirected graphs were analyzed using a custom-version of the Brain Connectivity Toolbox (Rubinov and Sporns, 2010). The custom software AIDAconnect and a detailed step-by-step guide to replicate the analysis is available online (https://github.com/aswendtlab/Project_Microbiome/). Atlas renderings were generated with ©Allen Institute for Brain Science. Brain Explorer 2. Available from: https://mouse.brain-map.org/static/brainexplorer.
Lesion volume determination
Three-dimensional (3D) lesion masks were drawn semi-automatically using ITKsnap (www.itksnap.org (Yushkevich et al., 2006)) as described previously (Ito et al., 2018). The resulting lesion masks were averaged for the experimental groups per time point and presented as incidence maps, where each pixel reflects in color code, in how many mice that pixel was inside the stroke mask. Furthermore, we recorded the frequency and the fraction of a given brain region that was affected by stroke. From the individual stroke masks, the lesion volume (relation to whole brain volume in %) and the lesion area per brain region were extracted. For a qualitative comparison, pixels inside the stroke masks were added across mice and displayed as incidence maps superimposed on the atlas template.
Quantification and statistical analysis
Graphpad Prism software (Graphpad Software, San Diego, CA, USA) was used for statistical analysis. Significance was considered if p < 0.05. Two-way analysis of variance (ANOVA) with false discovery rate (FDR) correction for multiple comparisons was used to compare global and local measures in naive GF/SPF and stroke GF/SPF mice, respectively. Unpaired t test was applied for total degree and strength comparisons and for analysis of the histological data. The histogram plot with frequency of degree and node strength per group was fitted with a Poisson regression. The likelihood ratio test was applied to test whether the distributions differ from each other. Details of statistical analysis for each experiment is reported in the respective figure legend.
Acknowledgments
We gratefully acknowledge the professional graphical work of Mrs Ulla Uhlenküken. This study was funded by the Friebe Foundation (T0498/28960/16) to M.A. and the European Research Council (ERC-StGs 802305) and the German Research Foundation (DFG) under Germany ´s Excellence Strategy (EXC 2145 SyNergy – ID 390857198) to A.L.
Author contributions
M.A., A.L., M.H. designed and supervised the study, analyzed the data, wrote the manuscript.
M.A., C.G., R.S., G.L. L.D., S.V., F.W., performed MRI and histological experiments.
M.D., D.W. performed image data post processing.
M.GdA., A.L. provided the germ-free animals.
Declaration of interests
All authors declare no competing interests.
Published: October 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103095.
Contributor Information
Markus Aswendt, Email: markus.aswendt@uk-koeln.de.
Arthur Liesz, Email: arthur.liesz@med.uni-muenchen.de.
Mathias Hoehn, Email: nc-hoehnma4@netcologne.de.
Supplemental information
Data and code availability
All original MRI raw and processed data are publicly available in the Edmond open data repository of the Max Planck Society under https://edmond.mpdl.mpg.de/imeji/collection/Ce5QnWU4SktzEPN.
The software used to process the MRI data is available on GitHub: https://github.com/aswendtlab/Project_Microbiome.
References
- Arentsen T., Raith H., Qian Y., Forssberg H., Diaz Heijtz R. Host microbiota modulates development of social preference in mice. Microb. Ecol. Health Dis. 2015;26:29719. doi: 10.3402/mehd.v26.29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnsten A.F., Paspalas C.D., Gamo N.J., Yang Y., Wang M. Dynamic Network Connectivity: a new form of neuroplasticity. Trends Cogn. Sci. 2010;14:365–375. doi: 10.1016/j.tics.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswendt M., Pallast N., Wieters F., Baues M., Hoehn M., Fink G.R. Lesion size- and location-dependent recruitment of contralesional thalamus and motor cortex facilitates recovery after stroke in mice. Transl. Stroke Res. 2021;12:87–97. doi: 10.1007/s12975-020-00802-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswendt M., Wilhelmsson U., Wieters F., Stokowska A., Schmitt F.J., Pallast N., De Pablo Y., Mohammed L., Hoehn M., Pekna M., Pekny M. Reactive astrocytes prevent maladaptive plasticity after ischemic stroke. bioRxiv. 2021 doi: 10.1101/2021.06.16.448657. [DOI] [PubMed] [Google Scholar]
- Badhwar A., Tam A., Dansereau C., Orban P., Hoffstaedter F., Bellec P. Resting-state network dysfunction in Alzheimer's disease: a systematic review and meta-analysis. Alzheimers Dement. 2017;8:73–85. doi: 10.1016/j.dadm.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benakis C., Brea D., Caballero S., Faraco G., Moore J., Murphy M., Sita G., Racchumi G., Ling L., Pamer E.G. Commensal microbiota affects ischemic stroke outcome by regulating intestinal gammadelta T cells. Nat. Med. 2016;22:516–523. doi: 10.1038/nm.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke S., Hensel L., Minassian A., Vlachakis S., Tscherpel C., Vay S.U., Rabenstein M., Schroeter M., Fink G.R., Hoehn M. Translating functional connectivity after stroke: fMRI detects comparable network changes in mice and humans. Stroke. 2021;52:9. doi: 10.1161/STROKEAHA.120.032511. [DOI] [PubMed] [Google Scholar]
- Cramer J.V., Gesierich B., Roth S., Dichgans M., During M., Liesz A. In vivo widefield calcium imaging of the mouse cortex for analysis of network connectivity in health and brain disease. Neuroimage. 2019;199:570–584. doi: 10.1016/j.neuroimage.2019.06.014. [DOI] [PubMed] [Google Scholar]
- Cryan J.F., O'riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M., Codagnone M.G., Cussotto S., Fulling C., Golubeva A.V. The microbiota-gut-brain Axis. Physiol. Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- Damoiseaux J.S. Resting-state fMRI as a biomarker for Alzheimer's disease? Alzheimers Res. Ther. 2012;4:8. doi: 10.1186/alzrt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado Jimenez R., Benakis C. The gut ecosystem: a critical player in stroke. Neuromol. Med. 2020;23:236–241. doi: 10.1007/s12017-020-08633-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz R., Wang S., Anuar F., Qian Y., Bjorkholm B., Samuelsson A., Hibberd M.L., Forssberg H., Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U S A. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egimendia A., Minassian A., Diedenhofen M., Wiedermann D., Ramos-Cabrer P., Hoehn M. Aging reduces the functional brain networks strength-a resting state fMRI study of healthy mouse brain. Front. Aging Neurosci. 2019;11:277. doi: 10.3389/fnagi.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D., Hrabe De Angelis A.L., Jaitin D., Wieghofer P., Staszewski O., David E., Keren-Shaul H., Mahlakoiv T., Jakobshagen K., Buch T. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farahani F.V., Karwowski W., Lighthall N.R. Application of graph theory for identifying connectivity patterns in human brain networks: a systematic review. Front. Neurosci. 2019;13:585. doi: 10.3389/fnins.2019.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost G., Sleeth M.L., Sahuri-Arisoylu M., Lizarbe B., Cerdan S., Brody L., Anastasovska J., Ghourab S., Hankir M., Zhang S. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat. Commun. 2014;5:3611. doi: 10.1038/ncomms4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorges M., Roselli F., Müller H.-P., Ludolph A.C., Rasche V., Kasubek J. Functional connectivity mapping in the animal model: principles and applications of resting-state fMRI. Front. Neurol. 2017;8:200. doi: 10.3389/fneur.2017.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean J., Derungs R., Kulic L., Welt T., Henkelman M., Nitsch R.M., Rudin M. Complex interplay between brain function and structure during cerebral amyloidosis in APP transgenic mouse strains revealed by multi-parametric MRI comparison. Neuroimage. 2016;134:1–11. doi: 10.1016/j.neuroimage.2016.03.042. [DOI] [PubMed] [Google Scholar]
- Green C., Minassian A., Vogel S., Diedenhofen M., Beyrau A., Wiedermann D., Hoehn M. Sensorimotor functional and structural networks after intracerebral stem cell grafts in the ischemic mouse brain. J. Neurosci. 2018;38:1648–1661. doi: 10.1523/JNEUROSCI.2715-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C., Minassian A., Vogel S., Diedenhofen M., Wiedermann D., Hoehn M. Persistent quantitative vitality of stem cell graft is necessary for stabilization of functional brain networks after stroke. Front. Neurol. 2019;10:335. doi: 10.3389/fneur.2019.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C., Sydow A., Vogel S., Anglada-Huguet M., Wiedermann D., Mandelkow E., Mandelkow E.-M., Hoehn M. Functional networks are impaired by elevated tau-protein but reversible in a regulatable Alzheimer's disease mouse model. Mol. Neurodegen. 2019;14:13. doi: 10.1186/s13024-019-0316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindl S., Gesierich B., Benakis C., Llovera G., Duering M., Liesz A. Automated morphological analysis of microglia after stroke. Front. Cell. Neurosci. 2018;12:106. doi: 10.3389/fncel.2018.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary F.G., Roman C.A., Bajo R., Castellanos N.D., Venkatesan U., Rajtmajer S.M. Hyperconnectivity is a fundamental response to neurological disruption. Neuropsychology. 2014;29:59–75. doi: 10.1037/neu0000110. [DOI] [PubMed] [Google Scholar]
- Ito M., Aswendt M., Lee A.G., Ishizaka S., Cao Z., Wang E.H., Levy S.L., Smerin D.L., Mcnab J.A., Zelneh M. RNA-sequencing analysis revealed a distinct motor cortex transcriptome in spontaneously recovered mice after stroke. Stroke. 2018;49:2191–2199. doi: 10.1161/STROKEAHA.118.021508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C., Browne W.J., Cuthill I.C., Emerson M., Altman D.G. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A.B., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Levin M.F., Kleim J.A., Wolf S.L. What do motor "recovery" and "compensation" mean in patients following stroke? Neurorehabil. Neural Repair. 2009;23:313–319. doi: 10.1177/1545968308328727. [DOI] [PubMed] [Google Scholar]
- Llovera G., Simats A., Liesz A. Modeling stroke in mice: transient middle cerebral artery occlusion via the external carotid artery. J. Vis. Exp. 2021;171:e62573. doi: 10.3791/62573. [DOI] [PubMed] [Google Scholar]
- Luczynski P., Whelan S.O., O'sullivan C., Clarke G., Shanahan F., Dinan T.G., Cryan J.F. Adult microbiota-deficient mice have distinct dendritic morphological changes: differential effects in the amygdala and hippocampus. Eur. J. Neurosci. 2016;44:2654–2666. doi: 10.1111/ejn.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezo C., Dokalis N., Mossad O., Staszewski O., Neuber J., Yilmaz B., Schnepf D., De Aguero M.G., Ganal-Vonarburg S.C., Macpherson A.J. Different effects of constitutive and induced microbiota modulation on microglia in a mouse model of Alzheimer's disease. Acta Neuropathol. Commun. 2020;8:119. doi: 10.1186/s40478-020-00988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minassian A., Green C., Diedenhofen M., Vogel S., Hess S., Stoeber M., Dobrivojevic Radmilovic M., Wiedermann D., Kloppenburg P., Hoehn M. Human neural stem cell induced functional network stabilization after cortical stroke: a longitudinal resting-state fMRI study in mice. Front. Cell Neurosci. 2020;14:86. doi: 10.3389/fncel.2020.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D.J., Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M.B., Trommald M., Andersen P. An increase in dendritic spine density on hippocampal CA1 pyramidal cells following spatial learning in adult rats suggests the formation of new synapses. Proc. Natl. Acad. Sci. U S A. 1994;91:12673–12675. doi: 10.1073/pnas.91.26.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.H., Corbett D. Plasticity during stroke recovery: from synapse to behaviour. Nat. Rev. Neurosci. 2009;10:861–872. doi: 10.1038/nrn2735. [DOI] [PubMed] [Google Scholar]
- Paldino M.J., Zhang W., Chu Z.D., Golriz F. Metrics of brain network architecture capture the impact of disease in children with epilepsy. Neuroimage Clin. 2017;13:201–208. doi: 10.1016/j.nicl.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallast N., Diedenhofen M., Blaschke S., Wieters F., Wiedermann D., Hoehn M., Fink G., Aswendt M. Processing pipeline for atlas-based imaging data analysis of structural and fucntional mouse brain MRI (AIDAmri) Front. Neuroinform. 2019;13:e42. doi: 10.3389/fninf.2019.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallast N., Wieters F., Nill M., Fink G.R., Aswendt M. Graph theoretical quantification of white matter reorganization after cortical stroke in mice. Neuroimage. 2020;217:116873. doi: 10.1016/j.neuroimage.2020.116873. [DOI] [PubMed] [Google Scholar]
- Palmer L.M. Dendritic integration in pyramidal neurons during network activity and disease. Brain Res. Bull. 2014;103:2–10. doi: 10.1016/j.brainresbull.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Penzes P., Cahill M.E., Jones K.A., Vanleeuwen J.E., Woolfrey K.M. Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M., Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Sadler R., Cramer J.V., Heindl S., Kostidis S., Betz D., Zuurbier K.R., Northoff B.H., Heijink M., Goldberg M.P., Plautz E.J. Short-chain fatty acids improve poststroke recovery via immunological mechanisms. J. Neurosci. 2020;40:1162–1173. doi: 10.1523/JNEUROSCI.1359-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler R., Singh V., Benakis C., Garzetti D., Brea D., Stecher B., Anrather J., Liesz A. Microbiota differences between commercial breeders impacts the post-stroke immune response. Brain Behav. Immun. 2017;66:23–30. doi: 10.1016/j.bbi.2017.03.011. [DOI] [PubMed] [Google Scholar]
- Segal I., Korkotian I., Murphy D.D. Dendritic spine formation and pruning: common cellular mechanisms? Trends Neurosci. 2000;23:53–57. doi: 10.1016/s0166-2236(99)01499-x. [DOI] [PubMed] [Google Scholar]
- Shah D., Praet J., Latif Hernandez A., Hofling C., Anckaerts C., Bard F., Morawski M., Detrez J.R., Prinsen E., Villa A. Early pathologic amyloid induces hypersynchrony of BOLD resting-state networks in transgenic mice and provides an early therapeutic window before amyloid plaque deposition. Alzheimers Dement. 2016;12:964–976. doi: 10.1016/j.jalz.2016.03.010. [DOI] [PubMed] [Google Scholar]
- Sharon G., Sampson T.R., Geschwind D.H., Mazmanian S.K. The central nervous system and the gut microbiome. Cell. 2016;167:915–932. doi: 10.1016/j.cell.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silasi G., Murphy T.H. Stroke and teh connectome: how connectivity guides therapeutic intervention. Neuron. 2014;83:1354–1368. doi: 10.1016/j.neuron.2014.08.052. [DOI] [PubMed] [Google Scholar]
- Singh V., Roth S., Llovera G., Sadler R., Garzetti D., Stecher B., Dichgans M., Liesz A. Microbiota dysbiosis controls the neuroinflammatory response after stroke. J. Neurosci. 2016;36:7428–7440. doi: 10.1523/JNEUROSCI.1114-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., Sadler R., Heindl S., Llovera G., Roth S., Benakis C., Liesz A. The gut microbiome primes a cerebroprotective immune response after stroke. J. Cereb. Blood Flow Metab. 2018;38:1293–1298. doi: 10.1177/0271678X18780130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundman M.H., Chen N.K., Subbian V., Chou Y.H. The bidirectional gut-brain-microbiota axis as a potential nexus between traumatic brain injury, inflammation, and disease. Brain Behav. Immun. 2017;66:31–44. doi: 10.1016/j.bbi.2017.05.009. [DOI] [PubMed] [Google Scholar]
- Thion M.S., Low D., Silvin A., Chen J., Grisel P., Schulte-Schrepping J., Blecher R., Ulas T., Squarzoni P., Hoeffel G. Microbiome influences prenatal and adult microglia in a Sex-specific manner. Cell. 2018;172:500–516 e16. doi: 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp B.D., Wujek J.R., Criste G.A., Jalabi W., Yin X.H., Kidd G.J., Stohlman S., Ransohoff R.M. Evidence for synaptic stripping by cortical microglia. Glia. 2007;55:360–368. doi: 10.1002/glia.20462. [DOI] [PubMed] [Google Scholar]
- van Meer M.P., Otte W.M., Van Der Marel K., Nijboer C.H., Kavelaars A., Van Der Sprenkel J.W., Viergever M.A., Dijkhuizen R.M. Extent of bilateral neuronal network reorganization and functional recovery in relation to stroke severity. J. Neurosci. 2012;32:4495–4507. doi: 10.1523/JNEUROSCI.3662-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake H., Moorhouse A.J., Jinno S., Kohsaka S., Nabekura J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J. Neurosci. 2009;29:3974–3980. doi: 10.1523/JNEUROSCI.4363-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whishaw I.Q. Loss of the innate cortical engram for action patterns used in skilled reaching and the development of behavioral compensation following motor cortex lesions in the rat. Neuropharmacology. 2000;39:788–805. doi: 10.1016/s0028-3908(99)00259-2. [DOI] [PubMed] [Google Scholar]
- Whitesell J.D., Liska A., Coletta L., Hirokawa K.E., Bohn P., Williford A., Groblewski P.A., Graddis N., Kuan L., Knox J.E. Regional, layer, and cell-type-specific connectivity of the mouse default mode network. Neuron. 2021;109:545–559 e8. doi: 10.1016/j.neuron.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton D.K., Dissing-Olesen L., Stevens B. Neuron-glia signaling in synapse elimination. Annu. Rev. Neurosci. 2019;42:107–127. doi: 10.1146/annurev-neuro-070918-050306. [DOI] [PubMed] [Google Scholar]
- Yushkevich P.A., Piven J., Hazlett H.C., Smith R.G., Ho S., Gee J.C., Gerig G. User-Guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Zerbi V., Wiesmann M., Emmerzaal T.L., Jansen D., Van Beek M., Mutsaers M.P., Beckmann C.F., Heerschap A., Kiliaan A.J. Resting-state functional connectivity changes in aging apoE4 and apoE-KO mice. J. Neurosci. 2014;34:13963–13975. doi: 10.1523/JNEUROSCI.0684-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original MRI raw and processed data are publicly available in the Edmond open data repository of the Max Planck Society under https://edmond.mpdl.mpg.de/imeji/collection/Ce5QnWU4SktzEPN.
The software used to process the MRI data is available on GitHub: https://github.com/aswendtlab/Project_Microbiome.