Abstract
The FUS (TLS)-ERG chimeric protein associated with t(16;21)(p11;q22) acute myeloid leukemia is structurally similar to the Ewing’s sarcoma chimeric transcription factor EWS-ERG. We found that both FUS-ERG and EWS-ERG could induce anchorage-independent proliferation of the mouse fibroblast cell line NIH 3T3. However, only FUS-ERG was able to inhibit the differentiation into neutrophils of a mouse myeloid precursor cell line L-G and induce its granulocyte colony-stimulating factor-dependent growth. We constructed several deletion mutants of FUS-ERG lacking a part of the N-terminal FUS region. A deletion mutant lacking the region between amino acids 1 and 173 (exons 1 to 5) lost the NIH 3T3-transforming activity but retained the L-G-transforming activity. On the other hand, a mutant lacking the region between amino acids 174 and 265 (exons 6 and 7) lost the L-G-transforming activity but retained the NIH 3T3-transforming activity. These results indicate that the N-terminal region of FUS contains two independent functional domains required for the NIH 3T3 and L-G transformation, which we named TR1 and TR2, respectively. Although EWS intrinsically possessed the TR2 domain, the EWS-ERG construct employed lacked the EWS sequence containing this domain. Since the TR2 domain is always found in chimeric proteins identified from t(16;21) leukemia patients but not in chimeric proteins from Ewing’s sarcoma patients, it seems that the TR2 function is required only for the leukemogenic potential. In addition, we identified three cellular genes whose expression was altered by ectopic expression of FUS-ERG and found that these are regulated in either a TR1-dependent or a TR2-dependent manner. These results suggest that FUS-ERG may activate two independent oncogenic pathways during the leukemogenic process by modulating the expression of two different groups of genes simultaneously.
Specific chromosomal translocations are frequently found in hematopoietic malignancies and certain types of solid tumors (37). The t(16;21)(p11.2;q22.2) translocation is a recurrent chromosomal abnormality found in acute myeloid leukemia. This translocation juxtaposes the FUS (TLS) gene on chromosome 16 and the ERG gene on chromosome 21 and forms the FUS-ERG fusion gene (11, 40). The FUS gene was first discovered as a translocated gene in myxoid liposarcoma (7, 36) and encodes an RNA-binding protein (7). The N-terminal region of this protein is Ser, Tyr, Gly, and Gln rich and consists of degenerative Ser-Tyr-Gly-Gln-Gln-Ser repeats (SYGQQS repeat region), and the central and C-terminal regions consist of three Arg-Gly-Gly triplet-rich regions (RGG repeat region), an RNA-recognition motif (RRM), and a Zn finger motif. The RGG repeat regions and RRM are involved in the RNA-binding activity of this protein (35). Its heterogeneous nuclear ribonucleoprotein-like behavior and association with a basic transcription factor TFIID were reported (2, 4), but the biological function of this protein is still unclear. On the other hand, the ERG gene encodes an external transcribed spacer (ETS) family transcription factor (39). A transcriptional activation domain (ETA domain) is located in the N-terminal region, and a DNA-binding domain (ETS domain) is located in the C-terminal region (41) (see Fig. 1A). In the chimeric protein produced by the FUS-ERG fusion gene, the ETA domain of ERG is replaced by the FUS N-terminal region, containing the SYGQQS repeat region and the first RGG repeat region (11, 17) (see Fig. 1A). Because the ETS DNA-binding domain is retained, this chimeric protein is thought to function as a transcription factor.
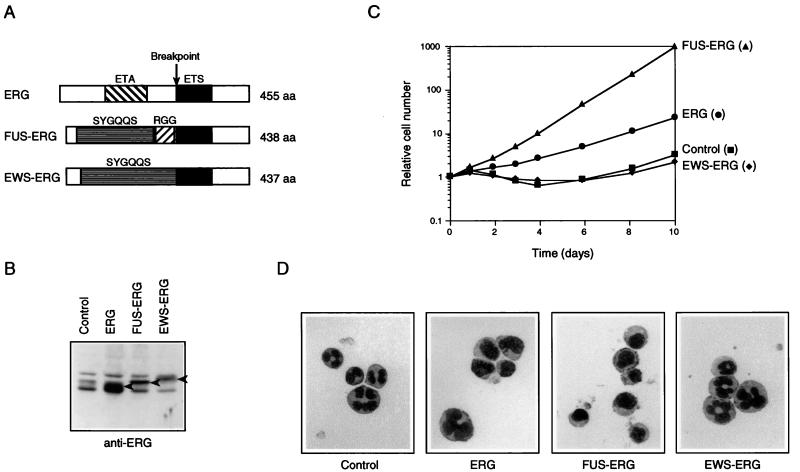
FIG. 1.
FUS-ERG but not EWS-ERG inhibits the differentiation of L-G cells into neutrophils and induces their G-CSF-dependent proliferation. (A) Structures of ERG, FUS-ERG, and EWS-ERG. The ETA domain, ETS domain, SYGQQS repeat region, and RGG repeat region are indicated. The N-terminal 282-amino-acid region of ERG was replaced by the N-terminal 265-amino-acid region of FUS and the N-terminal 264-amino-acid region of EWS in FUS-ERG and EWS-ERG, respectively. (B) Immunoblot analysis of ERG, FUS-ERG, and EWS-ERG expression in infected L-G cells. Whole-cell extracts were fractionated by SDS-PAGE, and the bands of ERG, FUS-ERG, and EWS-ERG proteins were detected with anti-ERG antibody (arrowheads). The relatively slower migration of FUS-ERG and EWS-ERG, which have lower molecular weights than ERG, may be due to high Gly, Ser, and Gln contents of their N-terminal regions (35). (C) Growth curves of the infected L-G cells in the presence of 10 ng of G-CSF per ml. The relative numbers of viable cells are indicated. (D) Nuclear morphology of the infected cells. The cells cultured in the presence of G-CSF for 6 days were stained with May-Gruenwald’s and Giemsa’s solutions.
The FUS gene is highly related to the EWS gene (9). The EWS protein also contains the N-terminal SYGQQS repeat region, three RGG repeat regions, RRM, and a Zn finger motif and shows overall amino acid sequence similarity to FUS. In addition to this structural similarity, both of these genes are translocated and fused to transcription factor genes in several malignant tumors. FUS is fused to CHOP in myxoid liposarcoma (7, 36) and to ERG in acute myeloid leukemia (11, 40), as described above. EWS is fused to FLI1, ERG, and other ETS family genes in Ewing’s sarcoma (9, 13, 14, 32, 42, 46), to ATF1 in malignant melanoma of soft tissues (45), to WT1 in desmoplastic round cell tumor (19), to TEC in extraskeletal myxoid chondrosarcoma (18), and to CHOP in myxoid liposarcoma (28). In all of the products of these fusion genes, the N-terminal region of FUS or EWS is fused to the DNA-binding domain of the relevant transcription factors. Thus, it is believed that these chimeric proteins alter the expression of cellular genes, which is regulated by their original transcription factors, resulting in characteristic tumors. In addition, since FUS and EWS are highly homologous and since both of the FUS-CHOP and EWS-CHOP fusion genes were found in the same myxoid liposarcoma, FUS and EWS are expected to play the same role in the oncogenic potential of the chimeric proteins.
Thus, we expected that FUS-ERG associated with acute myeloid leukemia and EWS-ERG associated with Ewing’s sarcoma would have the same oncogenic potentials. However, we found that FUS-ERG differed from EWS-ERG in its ability to inhibit the differentiation into neutrophils of a mouse myeloid precursor cell line L-G and to induce its granulocyte colony-stimulating factor (G-CSF)-dependent growth. Here, we report that the N-terminal regions of FUS and EWS have two potential domains, TR1 and TR2, which are required for NIH 3T3 and L-G transformation, respectively, but that the TR2 domain is not contained in the EWS-ERG chimeric protein in most cases of Ewing’s sarcoma. In addition, the TR1 and TR2 domains would appear to function as transcriptional regulation domains which determine the specificity of target genes of FUS-ERG and EWS-ERG.
MATERIALS AND METHODS
Construction of retroviral expression vectors for ERG, FUS-ERG, EWS-ERG, and FUS-ERG derivatives.
ERG and FUS-ERG cDNAs were produced by reverse transcription-PCR from total RNA of a t(16;21) acute myeloid leukemia cell line, UTP-L12 (11). For ERG cDNA, we used primers ERGGF-HindIII and ERGDR-HindIII, which are capable of amplifying from nucleotides −30 to +53 of open reading frames (ORFs) of p55 and p49 isoforms (10) and which introduce HindIII sites at both ends. The amplified product was neither p55 nor p49 and was a 455-amino-acid isoform which has the A81 exon but not the A72 exon defined by Duterque-Coquillaud et al. (10). At this time, no products were obtained with primers which could amplify erg-1, erg-2, and erg-3 isoforms (34, 38). For FUS-ERG cDNA, we used primers FUS6F-HindIII and ERGDR-HindIII, which are capable of amplifying nucleotides −9 to +53 of the FUS-ERG ORF and which introduce HindIII sites at both ends. The amplified product was a 438-amino-acid isoform, which is the smaller of the two isoforms expected as the result of the difference in the splicing acceptor site of FUS exon 3.
HA-FUS-ERG, HFEΔFUS, HFEΔERG, HFEΔ1–64, HFEΔ1–110, and HFEΔ1–173 cDNAs were produced by PCR with FUS-ERG cDNA as a template. We used primers HAFUS9F-HindIII and ERGDR-HindIII for HA-FUS-ERG, HAERGKF-HindIII and ERGDR-HindIII for HFEΔFUS, HAFUS9F-HindIII and FUS16Rstop-HindIII for HFEΔERG, HAFUS10F-HindIII and ERGDR-HindIII for HFEΔ1–64, HAFUS11F-HindIII and ERGDR-HindIII for HFEΔ1–110, and HAFUS12F-HindIII and ERGDR-HindIII for HFEΔ1–173; these primers introduce HindIII sites at both ends and a hemagglutinin (HA) tag (Tyr-Pro-Tyr-Asp-Val-Pro-Asp-Tyr-Ala) at the N terminus.
HFEΔETS cDNA was produced by ligation between the N-terminal fragment of the HA-FUS-ERG cDNA, which has the upstream end digested with HindIII and the downstream end digested with BamHI and treated with the Klenow fragment of DNA polymerase I, and the C-terminal fragment, which was digested with Eco47III and HindIII.
For EWS-ERG, HFEΔ174–265, HFEΔ111–265, HFEΔ67–265, and HFEΔFUS+W265–333 cDNAs, the respective N-terminal parts and the common ERG C-terminal part were separately produced by reverse transcription-PCR from total RNA of UTP-L12 or by PCR with FUS-ERG cDNA as a template and joined later. The common C-terminal part was amplified from UTP-L12 RNA with primers ERGFF-FspI and ERGDR-HindIII, which are capable of amplifying from ERG exon 9 to nucleotide +53 of the ERG ORF and which introduce an FspI site at the upstream end and a HindIII site at the downstream end. The N-terminal part of EWS-ERG was amplified from UTP-L12 RNA with primers EWS1F-HindIII and EWS1R-ScaI, which are capable of amplifying from nucleotide −26 of the EWS ORF to EWS exon 7 and which introduce an HindIII site at the upstream end and a ScaI site at the downstream end. The N-terminal parts for HFEΔ174–265, HFEΔ111–265, and HFEΔ67–265 were amplified from FUS-ERG cDNA with primers HAFUS9F-HindIII and FUS12R-BsrBI, HAFUS9F-HindIII and FUS11R-BsrBI, and HAFUS9F-HindIII and FUS10R-BsrBI, respectively, all of which introduce a HindIII site at the upstream end and a BsrBI site at the downstream end as well as a HA tag at the N terminus. The N-terminal part of HFEΔFUS+W265–333 was amplified from UTP-L12 RNA with primers HAEWS4F-HindIII and EWS3R-BsrBI, which could introduce a HindIII site at the upstream end and a BsrBI site at the downstream end as well as a HA tag at the N terminus. The N-terminal part and the C-terminal part were digested with appropriate restriction enzymes and ligated.
We used a retroviral expression vector, pLNCX (25) for preparation of retroviruses. The cDNAs of ERG, FUS-ERG, EWS-ERG, and FUS-ERG derivatives were digested with HindIII and cloned into a HindIII site of pLNCX. These cDNA inserts were all confirmed by nucleotide sequencing.
The sequences of primers we used are as follows (restriction sites which we introduced are underlined): ERGDR-HindIII (AAGCTTTGTGGCGATGGGCTGGTG), ERGGF-HindIII (AAGCTTGATTGCATTATGGCCAGC), ERGFF-FspI (TGCGCAGTGGCCAGATCCAGCTT), EWS1F-HindIII (AAGCTTGAGAGAACGAGGAGGAAG), EWS1R-ScaI (AGTACTGCTGCTGCCCGTAGCTGCTGC), EWS3R-BsrBI (CCGCTCCAGGCTTATTGAGCCACCT), FUS6F-HindIII (AAGCTTGCTTGCTTGCCTGTGCGC), FUS7R-BsrBI (CCGCTCCAAATTTATTGAAGCCACCAC), FUS10R-BsrBI (CCGCTCCATAGCTGTTCTGGCTCTG), FUS11R-BsrBI (CCGCTCCCGAGGTGCTGCTGGGA), FUS12R-BsrBI (CCGCTCCACCTCCACCTCCACCT), FUS16Rstop-HindIII (AAGCTTTTAGCCACCAAATTTATTGAAGCCAC), HAERGKF-HindIII (AAGCTTAGGCCTCTAGACCATGGCATACCCATACGACGTGCCTGACTACGCCTCCGGCAGTGGCCAGATCCAGCTT), HAEWS4F- HindIII (AAGCTTAGGCCTCTAGACCATGGCATACCCATACGACGTGCCTGACTACGCCTCCAGTTCATTCCGACAGGACCAC), HAFUS9F- HindIII (AAGCTTAGGCCTCTAGACCATGGCATACCCATACGACGTGCCTGACTACGCCTCCGCCTCAAACGATTATACCCAAC), HAFUS10F- HindIII (AAGCTTAGGCCTCTAGACCATGGCATACCCATACGACGTGCCTGACTACGCCTCCTATGGAACTCAGTCAACTCCCC), HAFUS11F- HindIII (AAGCTTAGGCCTCTAGACCATGGCATACCCATACGACGTGCCTGACTACGCCTCCAGTTACGGTAGCAGTTCTCAGA), and HAFUS12F- HindIII (AAGCTTAGGCCTCTAGACCATGGCATACCCATACGACGTGCCTGACTACGCCTCCGGTAACTATGGCCAAGATCAATC).
Retrovirus production and cell culture.
For production of retroviruses, we used BOSC23 cells (29). Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and were transfected with pLNCX-derived expression vectors by the calcium phosphate precipitation method. After a 48- to 72-h culture, supernatants were saved as retrovirus solutions. L-G cells (15) were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 0.1 ng of recombinant mouse interleukin-3 (IL-3) (a generous gift from Kirin Brewery Co.) per ml, and 50 μM β-mercaptoethanol. Infection was carried out by adding the retrovirus solutions to L-G cell cultures, and the infected cells were selected with 1 mg of G418 per ml. When L-G cells were exposed to G-CSF, the cells maintained in the presence of IL-3 were washed twice with phosphate-buffered saline (PBS) and incubated in medium containing 10 ng of recombinant human G-CSF (a generous gift from Chugai Pharmaceutical Co.) per ml in place of IL-3. Viable cells were counted with a Coulter counter. Nuclear morphologies were observed after staining with May-Gruenwald’s solution and Giemsa’s solution (Merck). NIH 3T3 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% calf serum. Infection was carried out by adding the retrovirus solutions to NIH 3T3 cell cultures, and the infected cells were selected with 0.4 mg of G418 per ml. For the colony formation assay, cells were trypsinized and plated into soft agar medium containing 0.3% agarose (5 × 103 cells/60-mm plate). After 2 weeks of incubation, the macroscopically visible colonies were counted.
Immunoblotting analysis.
Cells were harvested and suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 5% β-mercaptoethanol, 10% glycerol) at 2 × 107 cells/ml. After boiling and centrifugation, 10 μl of cleared lysates (2 × 105 cells equivalent) was electrophoresed in SDS–10% polyacrylamide gels, and transferred to nitrocellulose membranes (Hybond ECL; Amersham). The membranes were blocked at 4°C overnight with 5% skim milk dissolved in PBS containing 0.1% Tween-20 (PBS-T), incubated at room temperature for 2 h with 1 μg of anti-ERG antibody (C-20; Santa Cruz) per ml or 0.1 μg of anti-HA antibody (3F10; Boehringer Mannheim) per ml dissolved in PBS-T, and then incubated at room temperature for 1 h with appropriate horseradish peroxidase-conjugated second antibodies dissolved in PBS-T. The antibody-bound proteins were detected with ECL Western blotting detection reagents (Amersham).
mRNA differential display.
Total RNAs were prepared by the acid guanidinium thiocyanate-phenol-chloroform method (6) from L-G cells cultured in the presence of IL-3 and purified by phenol-chloroform extraction and ethanol precipitation. mRNA differential display screening was performed by the method of Ito et al. (12). cDNAs were synthesized by using four oligo(dT) primers (GT15MN; M = A + C + G; N = A, C, G, or T) and SuperScript II reverse transcriptase (Gibco BRL). PCR-amplification was carried out between the same oligo(dT) primers and arbitrary 10-mers (Operon Technologies) by using Taq DNA polymerase (Boehringer Mannheim) with 1 cycle of denaturation at 94°C for 3 min, annealing at 40°C for 5 min, and extension at 72°C for 5 min followed by 24 cycles of denaturation at 95°C for 15 s, annealing at 40°C for 2 min, and extension at 72°C for 1 min. The PCR products were separated by gel electrophoresis in 6% polyacrylamide gels, stained with SYBR green I (Molecular Probes), and detected with FluorImager SI (Molecular Dynamics). The bands whose intensities were altered were cut out of the gel, reamplified by PCR, cloned into pGEM-T vector (Promega), and sequenced.
Northern hybridization analysis.
Total RNAs (5 μg) were electrophoresed in a formaldehyde–1% agarose gel and transferred to a nylon membrane (Hybond N+; Amersham) by standard methods. Hybridization was carried out at 42°C overnight in hybridization mixture (6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 50% formamide, 1% SDS, 1× Denhardt’s solution, 10% dextran sulfate, 100 μg of denatured herring testis DNA per ml). The membranes were washed three times at 65°C in washing buffer (0.1× SSC, 0.1% SDS), and the hybridized transcripts were observed with a BAS2000 image analyzer (Fuji Film).
In vitro transcription-translation and electrophoretic mobility shift assay.
For synthesis of HA-FUS-ERG, HFEΔ1–173, HFEΔFUS, HFEΔ174–265, and HFEΔETS proteins, the corresponding cDNAs, which were cloned in the pLNCX expression vectors, were digested with HindIII and recloned into a HindIII site of pSP64poly(A). By using these vectors, an in vitro transcription-translation reaction was carried out with the TnT SP6 quick-coupled transcription-translation system (Promega). The in vitro-translated extracts were diluted with mock extract to equalize the concentrations of the synthesized proteins as judged by the intensity of the bands in immunoblotting analysis with anti-ERG. E74 oligonucleotide probe was prepared by annealing of synthetic oligonucleotides E74F (AATAACCGGAAGTAACTC) and E74R (GAGTTACTTCCGGTTATT) and by 32P labelling with T4 polynucleotide kinase and [γ-32P]ATP. Mutant E74 oligonucleotide competitor was prepared by annealing of synthetic oligonucleotides E74Fm (AATAACCCCAAGTAACTC) and E74Rm (GAGTTACTTGGGGTTATT). 32P-labelled E74 oligonucleotide (50 pmol) and 0.5 to 2 μl of the diluted extracts were incubated with 10 nmol of mutant E74 oligonucleotide competitor in 20 μl of binding buffer [20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 100 μg of bovine serum albumin per ml, 50 μg of poly(dI-dC) per ml] at room temperature for 20 min, and complexes were separated by electrophoresis in 4% polyacrylamide gels. The shifted bands were observed with a BAS2000 image analyzer.
Nucleotide sequence accession number. The sequence of C14G220 has been assigned accession no. AB028209.
RESULTS
FUS-ERG transforms L-G myeloid precursor cells.
The t(16;21) leukemia-associated chimeric protein FUS-ERG structurally resembles the Ewing’s sarcoma-associated chimeric protein EWS-ERG. To investigate the role of FUS-ERG in leukemogenesis by comparison with ERG and EWS-ERG, we constructed recombinant retroviruses to express these proteins (Fig. 1A). Concerning FUS-ERG and EWS-ERG, some variants attributable to differences in their translocational breakpoints have been described (17, 46). We used the smallest forms of these proteins containing FUS exons 1 to 5 or EWS exons 1 to 7 fused to ERG exon 9. In addition, ERG has several splicing isoforms, as reported by Rao et al. (38), Duterque-Coquillaud et al. (10), and Prasad et al. (34). We used a 455-amino-acid isoform (see Materials and Methods), since this form was mainly expressed in the t(16;21) leukemia cells which we examined.
A mouse myeloid precursor cell line, L-G, has been used successfully to investigate the leukemogenic function of the fusion gene associated with acute myeloid leukemia (16). L-G cells proliferate in the presence of IL-3 and differentiate into mature neutrophils when G-CSF is added to medium in place of IL-3 (15). We used this cell line to analyze the effects of ERG, FUS-ERG, and EWS-ERG on myeloid-cell differentiation. L-G cells were infected with retroviruses to express these proteins, and the infected cells were selected by G418 resistance. The expression of the ERG, FUS-ERG, and EWS-ERG proteins in the infected cells was confirmed by immunoblotting analysis with anti-ERG antibody (Fig. 1B). The cells expressing ERG, FUS-ERG, and EWS-ERG proliferated in the presence of IL-3 and died in the absence of cytokines, like control cells infected with a mock retrovirus (data not shown). In the presence of G-CSF, the control and EWS-ERG-expressing cells did not proliferate (Fig. 1C) but morphologically differentiated into mature neutrophils (Fig. 1D). In contrast, the FUS-ERG-expressing cells proliferated exponentially in response to G-CSF (Fig. 1C) without differentiating into neutrophils (Fig. 1D). These results indicated that FUS-ERG possessed a transformation activity that inhibited the differentiation of L-G cells into neutrophils and induced their G-CSF-dependent proliferation. This was in contrast to EWS-ERG, which possessed no such activity. The ERG-expressing cells displayed an intermediate phenotype, as shown by their weak proliferation and sporadic differentiation (Fig. 1C and D).
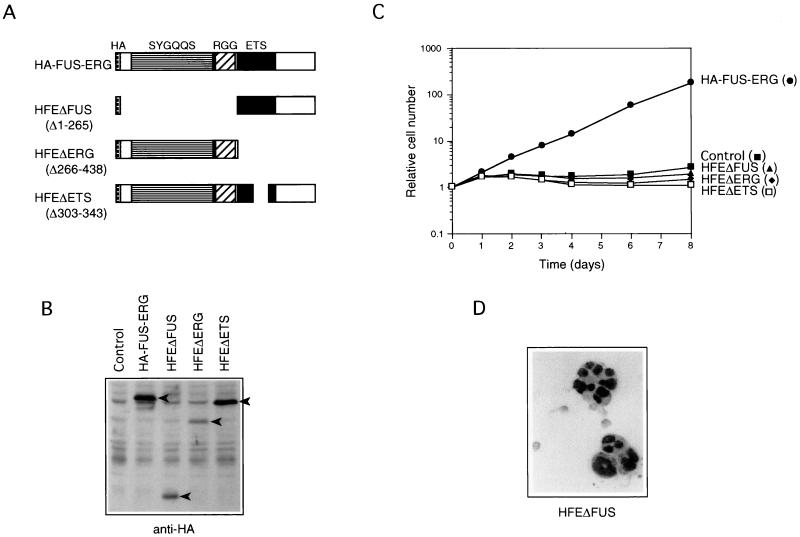
To determine the contribution of the N-terminal FUS and C-terminal ERG regions of FUS-ERG in the L-G-transforming activity, we constructed HA-tagged deletion mutants lacking parts of the chimeric protein (Fig. 2A) and introduced them into L-G cells by infecting the cells with recombinant retroviruses. The expression of these mutant proteins in the infected cells was confirmed by immunoblot analysis with anti-HA antibody (Fig. 2B). The cells expressing HA-tagged FUS-ERG proliferated without differentiating in the presence of G-CSF (Fig. 2C and D), like the cells expressing FUS-ERG without the tag, indicating that HA tagging did not inhibit the L-G-transforming activity. On the other hand, the cells expressing the FUS (amino acids 1 to 265)-deleted mutant HFEΔFUS or the ERG (amino acids 266 to 438)-deleted mutant HFEΔERG did not proliferate (Fig. 2C) and morphologically differentiated into mature neutrophils (Fig. 2D and data not shown), like the control cells. These results indicated that both of the FUS and ERG regions are required for the transformation activity of FUS-ERG to inhibit the differentiation and stimulate the G-CSF-dependent proliferation of L-G cells. In addition, we constructed a mutant lacking amino acids 303 to 343 within the ETS DNA-binding domain (Fig. 2A). The cells expressing this mutant HFEΔETS also did not proliferate (Fig. 2C) and morphologically differentiated into mature neutrophils in the presence of G-CSF (data not shown). Although this mutant lacks a part of the ETS domain which contains the nuclear localization signal, this protein was still localized to the nucleus (data not shown). Thus, these results suggested that the DNA-binding activity of the ETS domain is required and FUS-ERG functions as a chimeric transcription factor in the transformation of L-G cells.
FIG. 2.
The FUS N-terminal region and the ERG ETS domain are necessary for transformation of L-G cells. (A) Structures of HA-FUS-ERG, HFEΔFUS, HFEΔERG, and HFEΔETS. Amino acids 1 to 265, 266 to 438, and 303 to 343 of FUS-ERG are deleted in HFEΔFUS, HFEΔERG, and HFEΔETS, respectively. (B) Immunoblot analysis of HA-FUS-ERG, HFEΔFUS, HFEΔERG, and HFEΔETS expression in the infected L-G cells. Whole-cell extracts were fractionated, and the bands of HA-FUS-ERG, HFEΔFUS, HFEΔERG, and HFEΔETS proteins were detected with anti-HA antibody (arrowheads). (C) Growth curves of the infected L-G cells in the presence of 10 ng of G-CSF per ml. (D) Nuclear morphology of the HFEΔFUS-expressing L-G cells. The cells cultured in the presence of G-CSF for 6 days were stained with May-Gruenwald’s and Giemsa’s solutions.
FUS amino acids 174 to 265 corresponding to exons 6 and 7 are required to transform L-G cells.
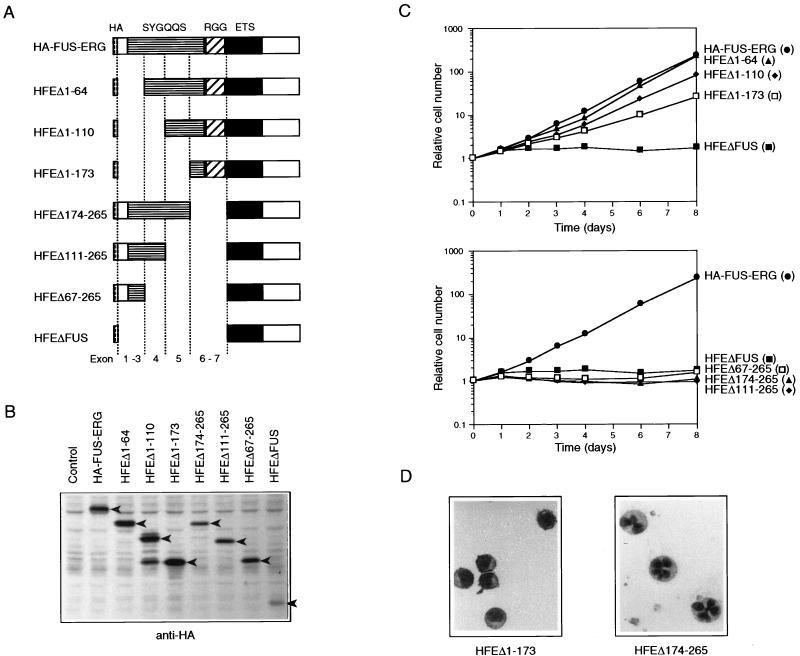
FUS-ERG transformed L-G cells, whereas EWS-ERG did not. To identify a critical region within the N-terminal FUS region for the transformation of L-G cells, we constructed several mutants with deletions from the N-terminal or C-terminal end of the FUS region (Fig. 3A). The deletions in these mutants corresponded approximately to the exon-intron structures of the FUS gene, as shown in Fig. 3A. These mutants were introduced into L-G cells by infection with retroviruses, and the transformation phenotypes of the infected cells were examined. The expression of these mutant proteins in the infected cells was confirmed by immunoblot analysis with anti-HA antibody (Fig. 3B). The N-terminal deletion mutants HFEΔ1–64, HFEΔ1–110, and HFEΔ1–173 stimulated the G-CSF-dependent proliferation (Fig. 3C) and inhibited the differentiation (Fig. 3D and data not shown), while the growth rates of the cells expressing these mutants were decreased to some extent by these deletions. The HFEΔ1–173-expressing cells proliferated at almost the same growth rate as the ERG-expressing cells did (Fig. 1C and 3C), but their differentiation was likely to be inhibited more strongly (see Fig. 1D and 3D). On the other hand, the C-terminal deletion mutants HFEΔ174–265, HFEΔ111–265, and HFEΔ67–265 neither stimulated the G-CSF-dependent proliferation (Fig. 3C) nor inhibited differentiation (Fig. 3D and data not shown), like HFEΔFUS, a mutant lacking all of the FUS region. These results indicated that the region between amino acids 174 and 265 is critical for the L-G-transforming activity. This region corresponds to FUS exons 6 and 7 and contains a part of the SYGQQS repeat region and all of the first RGG repeat region (Fig. 3A).
FIG. 3.
FUS amino acids 174 to 265 are necessary for transformation of L-G cells. (A) Structures of HA-FUS-ERG deletion mutants lacking parts of the N-terminal FUS portion. The deletions of these mutants nearly correspond to the exon-intron structure of the FUS gene, as indicated. (B) Immunoblot analysis of the expression of HA-FUS-ERG N-terminal deletion mutants in the infected L-G cells. Whole-cell extracts were fractionated, and the bands of these deletion mutants were detected with anti-HA antibody (arrowheads). HFEΔ1–110-expressing cells generated two major bands. It is likely that the smaller one was a C-terminus-truncated protein, since it could not be detected with anti-ERG antibody. (C) Growth curves of the infected L-G cells in the presence of 10 ng of G-CSF per ml. (D) Nuclear morphology of the HFEΔ1–173- and HFEΔ174–265-expressing L-G cells. The cells cultured in the presence of G-CSF for 6 days were stained with May-Gruenwald’s and Giemsa’s solutions.
FUS amino acids 1 to 173, corresponding to exons 1 to 5, are required to transform NIH 3T3 cells.
The Ewing’s sarcoma-associated chimeric transcription factor EWS-FLI1 and its artificial derivative FUS-FLI1 were reported to induce anchorage-independent growth of fibroblast cells in agar medium when a mouse fibroblast cell line, NIH 3T3, was used (20, 23, 24, 44). We examined such an activity of ERG, FUS-ERG, EWS-ERG, and some FUS-ERG mutants. NIH 3T3 cells were infected with recombinant retroviruses, and the infected cells were selected by their G418 resistance. The expression of these proteins was confirmed by immunoblot analysis with anti-ERG or anti-HA antibody (data not shown). The infected cells were plated in soft agar medium, and macroscopically visible colonies were counted after 2 weeks of culture. The cells expressing FUS-ERG and EWS-ERG efficiently formed colonies, while the cells expressing ERG did not, like control cells infected with a mock retrovirus (Table 1). The cells expressing HFEΔFUS, HFEΔERG, and HFEΔETS also did not form colonies (Table 1). These results indicated that, like EWS-FLI1 and FUS-FLI1, both FUS-ERG and EWS-ERG have the transformation potential to induce colony formation of NIH 3T3 cells in soft agar medium and that this NIH 3T3-transforming activity requires both the N-terminal FUS region and the ETS DNA-binding domain.
TABLE 1.
Transformation of NIH 3T3 cells by ERG, FUS-ERG, EWS-ERG, and FUS-ERG mutants
| Construct | No. of colonies in soft agara |
|---|---|
| Vector | 0 |
| ERG | 0 |
| FUS-ERG | 89.0 ± 18.4 |
| EWS-ERG | 92.3 ± 14.6 |
| HA-FUS-ERG | 82.7 ± 21.4 |
| HFEΔFUS | 0 |
| HFEΔERG | 0 |
| HFEΔETS | 0 |
| HFEΔ1–173 | 0.7 ± 1.2 |
| HFEΔ174–265 | 92.7 ± 8.5 |
A total of 5 × 103 cells of NIH 3T3 infectants were plated, and macroscopically visible colonies were counted after a 2-week culture. Values represent means ± standard deviations of counts from three plates.
HFEΔ1–173 and HFEΔ174–265 were also introduced into NIH 3T3 cells by using the retrovirus vector. The cells expressing HFEΔ174–265 formed colonies as efficiently as wild-type FUS-ERG did, while the cells expressing HFEΔ1–173 did not (Table 1). Accordingly, HFEΔ1–173, which transformed L-G cells, did not transform NIH 3T3 cells, and HFEΔ174–265, which did not transform L-G cells, transformed NIH 3T3 cells. These results indicated that the region between amino acids 1 and 173, which corresponds to FUS exons 1 to 5, contains a functional domain required for the NIH 3T3-transforming activity but not for the L-G-transforming activity, and that the region between amino acids 174 and 265, which corresponds to FUS exons 6 and 7, contains another functional domain required for the L-G-transforming activity but not for the NIH 3T3-transforming activity. Because the ETS DNA-binding domain is always required for these transformation activities, these two domains may function as transcriptional regulation domains. We have named these domains TR1 and TR2 (transforming regulation domain 1 and 2).
EWS amino acids 266 to 337, corresponding to exons 8 and 9, possess the TR2 function.
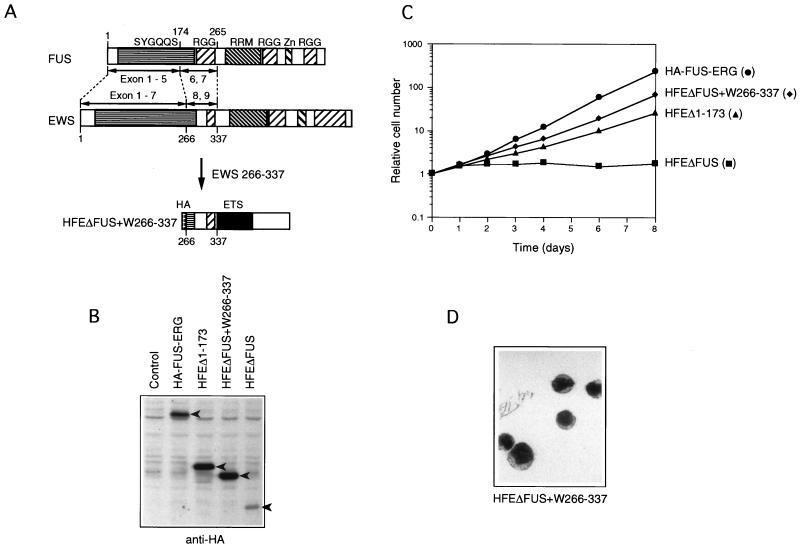
EWS-ERG transformed NIH 3T3 cells but not L-G cells, while FUS-ERG transformed both types of cells. FUS and EWS exhibit similarity not only at the amino acid sequence level but also in their exon-intron structures (1, 26, 33). FUS exons 6 and 7 corresponded to EWS exons 8 and 9 (Fig. 4A), and these two regions contain the first RGG repeat region. The EWS-ERG construct which we used contained only EWS exons 1 to 7. Thus, to examine whether EWS has a domain required for the L-G transformation, we constructed a mutant, HFEΔFUS+W266–337 (Fig. 4A), in which EWS amino acids 266 to 337, corresponding to EWS exons 8 and 9, were fused to the C-terminal ERG portion, and examined its transformation potential in L-G cells. The cells expressing HFEΔFUS+W266–337 proliferated exponentially in response to G-CSF (Fig. 4C) without differentiating into mature neutrophils (Fig. 4D). This result indicated that EWS possesses the TR2 function between amino acids 266 and 337. It is likely that EWS-ERG failed to transform L-G cells in the above experiment because it lacked this region.
FIG. 4.
EWS amino acids 266 to 337 are functional for transformation of L-G cells. (A) Structures of FUS, EWS, and HFEΔFUS+W266–337. The SYGQQS repeat region, RGG repeat region, RRM, and Zn finger motif are indicated. FUS exons 6 and 7 and EWS exons 8 and 9 are structurally conserved (1, 26, 33). In HFEΔFUS+W266–337, EWS amino acids 266 to 337, which correspond to exons 8 and 9, were joined to the C-terminal ERG portion. (B) Immunoblot analysis of HA-FUS-ERG, HFEΔ1–173, HFEΔFUS+W266–337, and HFEΔFUS expression in the infected L-G cells. Whole-cell extracts were fractionated, and the bands of HA-FUS-ERG, HFEΔ1–173, HFEΔFUS+W266–337, and HFEΔFUS proteins were detected with anti-HA antibody (arrowheads). (C) Growth curves of the infected L-G cells in the presence of 10 ng of G-CSF per ml. (D) Nuclear morphology of the HFEΔFUS+W266–337-expressing L-G cells. The cells cultured in the presence of G-CSF for 6 days were stained with May-Gruenwald’s and Giemsa’s solutions.
Alteration of cellular gene expression induced by FUS-ERG.
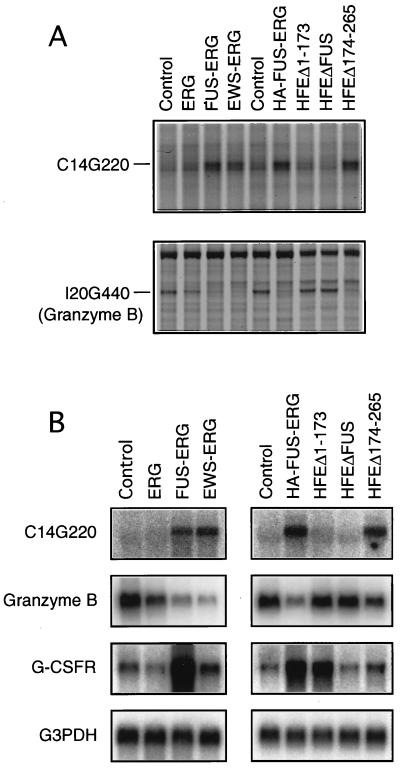
Because the ETS DNA-binding domain is required for the transformation of both L-G and NIH 3T3 cells by FUS-ERG, it seems likely that FUS-ERG functions as a transcription factor to change the expression of cellular genes. To investigate any changes in gene expression induced by FUS-ERG, we used the mRNA differential display method (12, 21). First, total RNAs were prepared from FUS-ERG-expressing L-G cells and control cells. Next, cDNAs were synthesized by using four oligo(dT) primers (GT15MN, where M = A + C + G and N = A, C, G, or T) and PCR amplified between the same four oligo(dT) primers and 160 arbitrary 10-mers (in total, 640 primer pairs). Then the resulting PCR products were separated by gel electrophoresis and their intensities were compared between FUS-ERG-expressing cells and control cells. We screened about 20,000 bands with 640 primer pairs and identified 2 bands whose intensities were enhanced or repressed by FUS-ERG expression. Cloning and sequencing analysis of the enhanced band C14G220 and the repressed band I20G440 revealed that I20G440 was derived from the granzyme B gene but that C14G220 did not match any known genes in the database. We examined the expression of these genes in L-G cells expressing ERG, FUS-ERG, EWS-ERG, HA-FUS-ERG, HFEΔ1–173, HFEΔFUS, and HFEΔ174–265 (Fig. 5A). The expression of C14G220 was up-regulated and the expression of granzyme B was down-regulated in cells expressing FUS-ERG, EWS-ERG, or HFEΔ174–265, but their levels remained unchanged in HFEΔ1–173-expressing cells (Fig. 5A). These alterations of expression were confirmed by Northern hybridization analysis (Fig. 5B).
FIG. 5.
Alteration of cellular gene expression by FUS-ERG and FUS-ERG mutants. (A) mRNA differential-display patterns of C14G220 and I20G440 (granzyme B) expression in control and ERG, FUS-ERG, EWS-ERG, and FUS-ERG mutant-expressing L-G cells. (B) Northern analysis of C14G220, granzyme B, and G-CSF receptor expression in control and ERG, FUS-ERG, EWS-ERG, and FUS-ERG mutant-expressing L-G cells. Glyceraldehyde 3-phosphate dehydrogenase (G3PDH) was used as a control. The C14G220 expression was enhanced and the granzyme B expression was repressed in cells expressing TR1-containing constructs, FUS-ERG, EWS-ERG, HA-FUS-ERG, and HFEΔ174–265. The G-CSF receptor (G-CSFR) expression was enhanced in cells expressing TR2-containing constructs, FUS-ERG, HA-FUS-ERG, and HFEΔ1–173.
In addition to mRNA differential-display screening, the expression of some of the genes known to be involved in myeloid-cell proliferation and differentiation was compared between the FUS-ERG-expressing and control L-G cells by Northern hybridization analysis. We found that the expression of the G-CSF receptor gene was enhanced approximately five- to eightfold in FUS-ERG-expressing cells. This enhancement of expression was also observed in HFEΔ1–173-expressing cells but not in cells expressing EWS-ERG or HFEΔ174–265 (Fig. 5B).
The above data is summarized in Table 2. FUS-ERG, EWS-ERG, and HFEΔ174–265, which contain TR1, changed the expression of two genes identified by the mRNA differential display, and FUS-ERG and HFEΔ1–173, which contain TR2, enhanced the expression of the G-CSF receptor gene. Thus, among three genes studied, the expression of two genes seems to be regulated in a TR1-dependent manner and the expression of the G-CSF receptor gene seems to be regulated in a TR2-dependent manner.
TABLE 2.
Alteration of cellular gene expression by FUS-ERG, EWS-ERG, and FUS-ERG mutants
| Construct | N-terminal domain(s) | Regulation of expression of:
|
||
|---|---|---|---|---|
| C14G220 | Granzyme B | G-CSF receptor | ||
| FUS-ERG | TR1 + TR2 | Up | Down | Up |
| EWS-ERG | TR1 | Up | Down | |
| HA-FUS-ERG | TR1 + TR2 | Up | Down | Up |
| HFEΔFUS | None | |||
| HFEΔ1–173 | TR2 | Up | ||
| HFEΔ174–265 | TR1 | Up | Down | |
DNA-binding properties of FUS-ERG mutants.
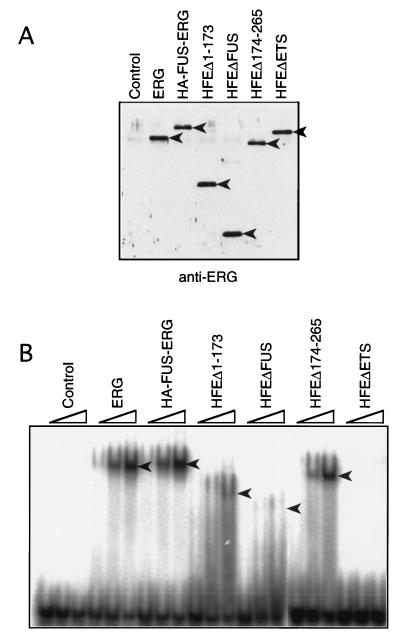
Prasad et al. (35) showed that ERG and FUS-ERG bind to the E74 sequence in a sequence-specific manner in an electrophoretic mobility shift assay. We examined the binding to the E74 sequence of some FUS-ERG mutants with a similar experiment involving in vitro-translated proteins (Fig. 6). When a mutant E74 oligonucleotide (with a GGAA-to-CCAA change in the ETS core-binding site) was added as a competitor, ERG and HA-FUS-ERG specifically bound to the E74 oligonucleotide, as in the previous work, while an ETS DNA-binding domain-deleted mutant HFEΔETS did not (Fig. 6B). All of the FUS region-deleted mutants which we examined, HFEΔ1–173 lacking TR1, HFEΔ174–265 lacking TR2, and HFEΔFUS lacking both TR1 and TR2, bound to the E74 sequence specifically but showed different band intensities (Fig. 6B). HFEΔ174–265 bound as efficiently as ERG and HA-FUS-ERG, but HFEΔ1–173 and HFEΔFUS bound more weakly to the E74 oligonucleotide. It seems likely that the N-terminal region of ERG and TR1 of FUS stabilize the ETS domain-E74 complex.
FIG. 6.
Binding to the E74 sequence of FUS-ERG mutants. (A) Immunoblot analysis of in vitro-translated ERG, HA-FUS-ERG, HFEΔ1–173, HFEΔFUS, HFEΔ174–265, and HFEΔETS. The in vitro-translated extracts were diluted to equalize the concentrations of the synthesized proteins (see Materials and Methods), and the same amounts of the diluted extracts were fractionated by SDS-PAGE. The bands of ERG, HA-FUS-ERG, HFEΔ1–173, HFEΔ174–265, and HFEΔETS proteins were detected with anti-ERG antibody (arrowhead). (B) Electrophoretic mobility shift assay with in vitro-translated proteins. 32P-labelled E74 oligonucleotide and different amounts (0.5, 1, and 2 μl) of the diluted extracts were incubated with cold mutant E74 oligonucleotide. The protein-bound E74 oligonucleotides were fractionated in a 4% polyacrylamide gel (arrowheads).
DISCUSSION
We have found that the t(16;21) leukemia-associated chimeric protein FUS-ERG inhibits the differentiation of L-G cells into neutrophils and stimulates their G-CSF-dependent proliferation and, in addition, induces the anchorage-independent growth of NIH 3T3 cells. These two transformation activities depend on two different domains, TR1 and TR2, located in the N-terminal FUS region of FUS-ERG. TR1, which is located between FUS amino acids 1 and 173 (exons 1 to 5), is required for NIH 3T3 transformation but not for L-G transformation, and TR2, which is located between FUS amino acids 174 and 265 (exons 6 and 7), is required for L-G transformation but not for NIH 3T3 transformation.
TR1 and TR2 domains in FUS and EWS.
FUS and EWS belong to a distinct family of RNA-binding proteins that exhibit a high degree of similarity in their amino acid sequences and exon-intron structures. Unlike FUS-ERG, EWS-ERG was unable to transform L-G cells in early experiments. However, FUS-ERG and EWS-ERG had the same transformation activities when appropriate regions of FUS and EWS were fused to the C-terminal region of ERG. FUS exons 1 to 5 (amino acids 1 to 173) containing TR1 correspond to EWS exons 1 to 7 (amino acids 1 to 264). Our EWS-ERG construct containing EWS amino acids 1 to 264 was able to transform NIH 3T3 cells, indicating the presence of the TR1 function in EWS. FUS exons 6 and 7 (amino acids 174 to 265) containing TR2 correspond to EWS exons 8 and 9 (amino acids 265 to 335). An EWS-ERG derivative, HFEΔFUS+W265–335, containing EWS amino acids 265 to 335, was able to transform L-G cells, indicating the presence of the TR2 function in EWS. The presence of the TR1 and TR2 functions in EWS suggests that TR1 and TR2 are conserved functional domains between FUS and EWS, although the normal functions of these domains are still unknown.
Involvement of TR1 and TR2 domains in other malignant tumors.
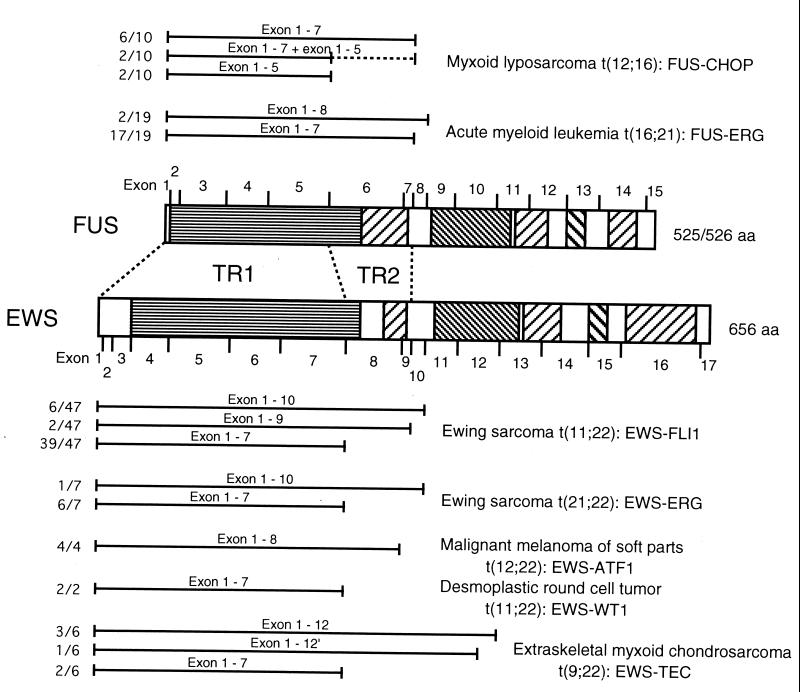
Translocations of FUS and EWS are involved in many malignant tumors including t(16;21) acute myeloid leukemia, myxoid liposarcoma, and Ewing’s sarcoma. The N-terminal regions of FUS and EWS, which we analyzed in the chimeric proteins with ERG, are expected to function through common or similar mechanisms when fused to other transcription factors. We summarize the translocated regions of FUS and EWS in these tumors in Fig. 7. In most of these tumors, the translocated regions included in the chimeric proteins varied among patients, depending on the location of the translocation breakpoint. FUS exons 1 to 7 or 1 to 8 were translocated and fused to ERG in t(16;21) acute myeloid leukemia (17). Thus, the resulting FUS-ERG chimeric proteins always contained the TR2 domain, confirming the importance of this domain for the leukemogenic potential of FUS-ERG. On the other hand, FUS exons 1 to 5 or 1 to 7 were translocated and fused to CHOP in myxoid liposarcoma (28). EWS exons 1 to 7, 1 to 9, or 1 to 10 were translocated and fused to FLI1 or ERG in Ewing’s sarcoma (46). In all of these tumors except t(16;21) leukemia and malignant melanoma of soft tissue, exons 1 to 5 of FUS or exons 1 to 7 of EWS were the minimal common regions included in the relevant chimeric proteins, which contain the TR1 domain but not the TR2 domain. In malignant melanoma of soft tissue, EWS exons 1 to 8 were always translocated and fused to ATF1, but this is probably due to formation of an in-frame junction, because fusion of EWS exon 7 to ATF1 results in the formation of an out-of-frame junction (45). Accordingly, the TR1 domain is always involved in all tumor-associated FUS or EWS chimeric proteins. In contrast, the TR2 domain is involved in only FUS-ERG of t(16;21) leukemia. Thus, it is likely that TR1 is an effector domain that functions in a broad spectrum of malignant tumors and that TR2 plays a specific role in acute myeloid leukemia.
FIG. 7.
The TR2 domain is always present in FUS-ERG chimeric protein of t(16;21) acute myeloid leukemia but not in chimeric proteins of other malignant tumors. Translocated regions of FUS and EWS found in myxoid liposarcoma with t(12;16) (27), acute myeloid leukemia with t(16;21) (17), Ewing’s sarcoma with t(11;22) and t(21;22) (46), malignant melanoma of soft tissue with t(12;22) (45), desmoplastic round-cell tumor with t(11;22) (19), and extraskeletal myxoid chondrosarcoma with t(9;22) (18) are summarized. Regions which are translocated and present in chimeric proteins are indicated by bars. Ratios of the number of relevant cases to the total number of cases are also shown on the left. In two cases of myxoid liposarcoma, two types of FUS-CHOP chimeric proteins containing exons 1 to 5 and exons 1 to 7 were present at the same time. In one case of extraskeletal myxoid chondrosarcoma, translocation occurred within exon 12 of EWS. The SYGQQS repeat region, RGG repeat region, RRM, and Zn finger motif are indicated by hatched boxes, as in Fig. 4. aa, amino acids.
TR1 and TR2 may function as transcriptional regulation domains to determine target gene specificity.
Since the ETS DNA-binding domain is absolutely required for both L-G and NIH 3T3 transformation by FUS-ERG, it is suggested that FUS-ERG functions as a transcription factor to alter the transcription pattern of cellular genes, which leads to cell transformation. Actually, it was reported that both FUS-ERG and ERG function as transcriptional activators on an artificial promoter containing the E74 ETS-binding sequence (35). However, FUS-ERG was unable to activate transcription from natural promoters known to be regulated by ERG, such as those of stromelysin 1 and vimentin (3, 5), in our reporter assay system with NIH 3T3 cells, although ERG activated these promoters (11a). In the present study, we identified three cellular genes whose expression was altered by ectopic expression of FUS-ERG in L-G cells, although it is unknown at present whether these genes were regulated directly or indirectly. The expression of these genes was not affected or was only slightly influenced by ERG (Fig. 5). In particular, the expression of the G-CSF receptor, which was up-regulated by FUS-ERG, was instead down-regulated by ERG. Although ERG stimulated the G-CSF-dependent proliferation of L-G cells, it seems likely that the underlying mechanisms and downstream target genes of ERG and FUS-ERG are different. Concerning the downstream genes, it is more important that the expression of granzyme B and C14G220 was not changed by a FUS-ERG mutant lacking TR1 and that the expression of G-CSF receptor was not up-regulated by a mutant lacking TR2. These results indicated that FUS-ERG regulates the different sets of genes in TR1- and TR2-dependent manners (Fig. 8). TR1 and TR2 may function as transcriptional regulation domains to determine target gene specificity.
FIG. 8.
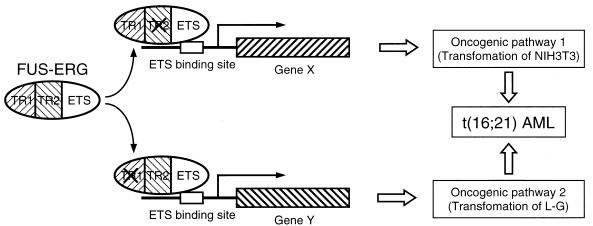
Model for the simultaneous activation of two oncogenic pathways for leukemogenesis by FUS-ERG. FUS-ERG enhances (or represses) the expression of a group of genes (represented by gene X) in a TR1-dependent and TR2-independent manner, and these genes activate an oncogenic pathway which causes the transformation of NIH 3T3 cells. At the same time, FUS-ERG enhances (or represses) the expression of another group of genes (represented by gene Y) in a TR1-independent and TR2-dependent manner, and these genes activate another oncogenic pathway, which causes the transformation of L-G cells. The simultaneous regulation of different groups of genes by FUS-ERG may cause the simultaneous activation of two different oncogenic pathways, resulting in acute myeloid leukemia.
How do TR1 and TR2 determine the target gene specificity? As shown in Fig. 6, all of FUS-ERG, a TR1 deletion mutant, HFEΔ1–173, and a TR2 deletion mutant, HFEΔ174–265 specifically bound to the E74 sequence. It is known that ERG regulates genes synergistically with other transcription factors like AP-1 (3). Thus, it is possible that protein-protein interaction mediated by the TR1 and TR2 domains determines the specificity of target genes regulated by FUS-ERG. On the other hand, HFEΔ1–173 bound weakly to the E74 sequence compared to HFEΔ174–265. Thus, it is also conceivable that HFEΔ1–173 may bind to other target sequences better than HFEΔ174–265 does. The alteration of binding specificity can explain the difference in target gene specificity between the FUS-ERG mutants lacking TR1 and TR2. Prasad et al. (35) reported that FUS fusion to ERG causes a slight reduction of the binding-sequence specificity of the ETS domain of ERG. This reduction may reflect TR2-dependent modulation of the binding specificity. Recently, Perrotti et al. (31) reported that FUS functions as a downstream effector of the BCR-ABL chimeric tyrosine kinase associated with chronic myeloid leukemia and represses the expression of the G-CSF receptor gene. Thus, it is possible that FUS-ERG dominant-negatively represses the function of FUS, resulting in up-regulation of the G-CSF receptor gene. However, this seems unlikely, because the ETS DNA-binding domain is required for the G-CSF receptor up-regulation (data not shown).
Enhanced expression of the G-CSF receptor.
In this study, we found that the expression of the G-CSF receptor is enhanced by FUS-ERG. The AML1-MTG8 chimeric protein, which is associated with t(8;21) acute myeloid leukemia and has a similar L-G-transforming activity (16), also enhances the G-CSF receptor expression (40a). G-CSF is a cytokine known to stimulate the proliferation of myeloid precursor cells. The enhanced expression of the G-CSF receptor in L-G cells induces their G-CSF-dependent proliferation like FUS-ERG and AML1-MTG8, although it does not inhibit the differentiation (40a). In addition, proliferation of a t(16;21) leukemia cell line, YNH-1, is stimulated by G-CSF (43). These findings suggest that the G-CSF receptor is one of target genes responsible for the oncogenic activity of FUS-ERG and that overexpression of the G-CSF receptor and resulting enhanced G-CSF signalling may contribute to the leukemogenesis of the t(16;21) leukemia.
Simultaneous activation of two independent oncogenic pathways by FUS-ERG.
FUS-ERG can inhibit the differentiation and stimulate the G-CSF-dependent proliferation of L-G cells as well as induce the anchorage-independent growth of NIH 3T3 cells. Recently, Pereira et al. (30) reported that FUS-ERG has the potential to enhance the proliferative and self-renewal capacity of myeloid progenitor cells. The relation of this potential to the transforming activities that we identified is not known, because its structural requirement has not been reported. On the other hand, the requirement of distinct domains for L-G- and NIH 3T3-transforming activities suggests that these activities reflect independent oncogenic potentials of FUS-ERG and that FUS-ERG simultaneously activates two independent oncogenic pathways in t(16;21) leukemia cells (Fig. 8).
As described above, AML1-MTG8 has a similar L-G-transforming activity. The BCR-ABL chimeric protein associated with chronic myelogenous leukemia transforms a mouse fibroblast cell line, Rat-1 (22). It is still unknown whether the L-G transformation by FUS-ERG and AML1-MTG8 occurs through the same mechanism. In addition, it is likely that the fibroblast cell transformation activities of FUS-ERG and BCR-ABL reflect the different oncogenic potentials, since BCR-ABL does not transform NIH 3T3 cells (8). However, the similarity in the transformation activities between FUS-ERG and other leukemia chimeric proteins suggests the importance of these two transformation activities of FUS-ERG in leukemogenesis. Kong et al. (17) reported that t(16;21) acute myeloid leukemia has a very poor prognosis compared to other types of leukemia. The simultaneous activation of two pathways may explain this poor prognosis.
ACKNOWLEDGMENTS
We thank A. D. Miller for providing pLNCX vector, D. Baltimore for providing BOSC23 cells, T. Honjo for providing L-G cells, Y. Sakamoto for providing NIH 3T3 cells, and T. Ito for making suggestions about the mRNA differential-display technique.
This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, and Culture; by a grant from the Special Coordination Funds for the Promotion of Science and Technology from the Science and Technology Agency; by a Grant-in-Aid for the 2nd Term Comprehensive 10-year Strategy for Cancer Control and a Research Grant on Human Genome and Gene Therapy from the Ministry of Health and Welfare; and by the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Drug ADR Relief, R&D Promotion, and Product Review of Japan.
REFERENCES
- 1.Åman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, Höglund M, Forster A, Rabbitts T H, Ron D, Mandahl N, Mitelman F. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics. 1996;37:1–8. doi: 10.1006/geno.1996.0513. [DOI] [PubMed] [Google Scholar]
- 2.Bertolotti A, Lutz Y, Heard D J, Chambon P, Tora L. hTAFII68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 3.Butticè G, Duterque-Coquillaud M, Basuyaux J P, Carrère S, Kurkinen M, Stéhelin D. Erg, an Ets-family member, differentially regulates human collagenase1 (MMP1) and stromelysin1 (MMP3) gene expression by physically interacting with the Fos/Jun complex. Oncogene. 1996;13:2297–2306. [PubMed] [Google Scholar]
- 4.Calvio C, Neubauer G, Mann M, Lamond A I. Identification of hnRNP P2 as TLS/FUS using electrospray mass spectrometry. RNA. 1995;1:724–733. [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J H, Vercamer C, Li Z, Paulin D, Vandenbunder B, Stehelin D. PEA3 transactivates vimentin promoter in mammary epithelial and tumor cells. Oncogene. 1996;13:1667–1675. [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Crozat A, Åman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 8.Daley G Q, McLaughlin J, Witte O N, Baltimore D. The CML-specific P210 bcr/abl protein, unlike v-abl, does not transform NIH/3T3 fibroblasts. Science. 1987;237:532–535. doi: 10.1126/science.2440107. [DOI] [PubMed] [Google Scholar]
- 9.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, Aurias A, Thomas G. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumors. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 10.Duterque-Coquillaud M, Niel C, Plaza S, Stehelin D. New human erg isoforms generated by alternative splicing are transcriptional activators. Oncogene. 1993;8:1865–1873. [PubMed] [Google Scholar]
- 11.Ichikawa H, Shimizu K, Hayashi Y, Ohki M. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 1994;54:2865–2868. [PubMed] [Google Scholar]
- 11a.Ichikawa, H. Unpublished data.
- 12.Ito T, Kito K, Adati N, Mitsui Y, Hagiwara H, Sakaki Y. Fluorescent differential display: arbitrarily primed RT-PCR fingerprinting on an automated DNA sequencer. FEBS Lett. 1994;351:231–236. doi: 10.1016/0014-5793(94)00867-1. [DOI] [PubMed] [Google Scholar]
- 13.Jeon I-S, Davis J N, Braun B S, Sublett J E, Roussel M F, Denny C T, Shapiro D N. A variant Ewing’s sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995;10:1229–1234. [PubMed] [Google Scholar]
- 14.Kaneko Y, Yoshida K, Handa M, Toyoda Y, Nishihira H, Tanaka Y, Sasaki Y, Ishida S, Higashino F, Fujinaga K. Fusion of an ETS-family gene, EIAF, to EWS by t(17;22)(q12;q12) chromosome translocation in an undifferentiated sarcoma of infancy. Genes Chromosomes Cancer. 1996;15:115–121. doi: 10.1002/(SICI)1098-2264(199602)15:2<115::AID-GCC6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Kinashi T, Lee K H, Ogawa M, Tohyama K, Tashiro K, Fukunaga R, Nagata S, Honjo T. Premature expression of the macrophage colony-stimulating factor receptor on a multipotential stem cell line does not alter differentiation lineages controlled by stromal cells used for coculture. J Exp Med. 1991;173:1267–1279. doi: 10.1084/jem.173.5.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitabayashi I, Ida K, Morohoshi F, Yokoyama A, Mitsuhashi N, Shimizu K, Nomura N, Hayashi Y, Ohki M. The AML1-MTG8 leukemic fusion protein forms a complex with a novel member of the MTG8(ETO/CDR) family, MTGR1. Mol Cell Biol. 1998;18:846–858. doi: 10.1128/mcb.18.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong X-T, Ida K, Ichikawa H, Shimizu K, Ohki M, Maseki N, Kaneko Y, Sako M, Kobayashi Y, Tojou A, Miura I, Kakuda H, Funabiki T, Horibe K, Hamaguchi H, Akiyama Y, Bessho F, Yanagisawa M, Hayashi Y. Consistent detection of TLS/FUS-ERG chimeric transcripts in acute myeloid leukemia with t(16;21)(p11;q22) and identification of a novel transcript. Blood. 1997;90:1192–1199. [PubMed] [Google Scholar]
- 18.Labelle Y, Zucman J, Stenman G, Kindblom L-G, Knight J, Turc-Carel C, Dockhorn-Dworniczak B, Mandahl N, Desmaze C, Peter M, Aurias A, Delattre O, Thomas G. Oncogenic conversion of a novel orphan nuclear receptor by chromosome translocation. Hum Mol Genet. 1995;4:2219–2226. doi: 10.1093/hmg/4.12.2219. [DOI] [PubMed] [Google Scholar]
- 19.Ladanyi M, Gerald W. Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res. 1994;54:2837–2840. [PubMed] [Google Scholar]
- 20.Lessnick S L, Braun B S, Denny C T, May W A. Multiple domains mediate transformation by the Ewing’s sarcoma EWS/FLI-1 fusion gene. Oncogene. 1995;10:423–431. [PubMed] [Google Scholar]
- 21.Liang P, Pardee A B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 22.Lugo T G, Witte O N. The BCR-ABL oncogene transforms Rat-1 cells and cooperates with v-myc. Mol Cell Biol. 1989;9:1263–1270. doi: 10.1128/mcb.9.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.May W A, Gishizky M L, Lessnick S L, Lunsford L B, Lewis B C, Delattre O, Zucman J, Thomas G, Denny C T. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci USA. 1993;90:5752–5756. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.May W A, Lessnick S L, Braun B S, Klemsz M, Lewis B C, Lunsford L B, Hromas R, Denny C T. The Ewing’s sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13:7393–7398. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller A D, Miller D G, Garcia J V, Lynch C M. Use of retroviral vectors for gene transfer and expression. Methods Enzymol. 1993;217:581–599. doi: 10.1016/0076-6879(93)17090-r. [DOI] [PubMed] [Google Scholar]
- 26.Morohoshi F, Ootsuka Y, Arai K, Ichikawa H, Mitani S, Munakata N, Ohki M. Genomic structure of the human RBP56/hTAFII68 and FUS/TLS genes. Gene. 1998;221:191–198. doi: 10.1016/s0378-1119(98)00463-6. [DOI] [PubMed] [Google Scholar]
- 27.Panagopoulos I, Mandahl N, Mitelman F, Åman P. Two distinct FUS breakpoint clusters in myxoid liposarcoma and acute myeloid leukemia with the translocations t(12;16) and t(16;21) Oncogene. 1995;11:1133–1137. [PubMed] [Google Scholar]
- 28.Panagopoulos I, Höglund M, Mertens F, Mandahl N, Mitelman F, Åman P. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene. 1996;12:489–494. [PubMed] [Google Scholar]
- 29.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1994;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereira D S, Dorrell C, Yto C Y, Gan O I, Murdoch B, Rao V N, Zou J-P, Reddy E S P, Dick J E. Retroviral transduction of TLS-ERG initiates a leukemogenic program in normal human hematopoietic cells. Proc Natl Acad Sci USA. 1998;95:8239–8244. doi: 10.1073/pnas.95.14.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrotti D, Bonatti S, Trotta R, Martinez R, Skorski T, Salomoni P, Grassilli E, Iozzo R V, Cooper D R, Calabretta B. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 1998;17:4442–4455. doi: 10.1093/emboj/17.15.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O. A new member of the ETS family fused to EWS in Ewing tumors. Oncogene. 1997;14:1159–1164. doi: 10.1038/sj.onc.1200933. [DOI] [PubMed] [Google Scholar]
- 33.Plougastel B, Zucman J, Peter M, Thomas G, Delattre O. Genomic structure of the EWS gene and its relationship to EWSR1, a site of tumor-associated chromosome translocation. Genomics. 1993;18:609–615. doi: 10.1016/s0888-7543(05)80363-5. [DOI] [PubMed] [Google Scholar]
- 34.Prasad D D K, Rao V N, Lee L, Reddy E S P. Differentially spliced erg-3 product functions as a transcriptional activator. Oncogene. 1994;9:669–673. [PubMed] [Google Scholar]
- 35.Prasad D D K, Ouchida M, Lee L, Rao V N, Reddy E S P. TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene. 1994;9:3717–3729. [PubMed] [Google Scholar]
- 36.Rabbitts T H, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 37.Rabbitts T H. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 38.Rao V N, Papas T S, Reddy E S P. erg, a human ets-related gene on chromosome 21: alternative splicing, polyadenylation, and translation. Science. 1987;237:635–639. doi: 10.1126/science.3299708. [DOI] [PubMed] [Google Scholar]
- 39.Reddy E S P, Rao V N, Papas T S. The erg gene: a human gene related to the ets oncogene. Proc Natl Acad Sci USA. 1987;84:6131–6135. doi: 10.1073/pnas.84.17.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu K, Ichikawa H, Tojo A, Kaneko Y, Maseki N, Hayashi Y, Ohira M, Asano S, Ohki M. An ets-related gene, ERG, is rearranged in human myeloid leukemia with t(16;21) chromosomal translocation. Proc Natl Acad Sci USA. 1993;90:10280–10284. doi: 10.1073/pnas.90.21.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Shimizu, K. Unpublished data.
- 41.Siddique H R, Rao V N, Lee L, Reddy E S P. Characterization of the DNA binding and transcriptional activation domains of the erg protein. Oncogene. 1993;8:1751–1755. [PubMed] [Google Scholar]
- 42.Sorensen P H B, Lessnick S L, Lopez-Terrada D, Liu X F, Triche T J, Denny C T. A second Ewing’s sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat Genet. 1994;6:146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto K, Hamaguchi H, Nagata K, Kobayashi M, Tanimoto F, Taniwaki M. Establishment of a novel human acute myeloblastic leukemia cell line (YNH-1) with t(16;21), t(1;16) and 12q13 translocations. Leukemia. 1997;11:599–608. doi: 10.1038/sj.leu.2400594. [DOI] [PubMed] [Google Scholar]
- 44.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 45.Zucman J, Delattre O, Desmaze C, Epstein A L, Stenman G, Speleman F, Fletchers C D M, Aurias A, Thomas G. EWS and ATF-1 gene fusion induced by t(12;22) translocation in malignant melanoma of soft parts. Nat Genet. 1993;4:341–345. doi: 10.1038/ng0893-341. [DOI] [PubMed] [Google Scholar]
- 46.Zucman J, Melot T, Desmaze C, Ghysdael J, Plougastel B, Peter M, Zucker J M, Triche T J, Sheer D, Turc-Carel C, Ambros P, Combaret V, Lenoir G, Aurias A, Thomas G, Delattre O. Combinatorial generation of variable fusion proteins in the Ewing family of tumors. EMBO J. 1993;12:4481–4487. doi: 10.1002/j.1460-2075.1993.tb06137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]