Abstract
Autoimmune hepatitis (AIH) is a chronic non-resolving liver disease characterized by diffuse hypergammaglobulinemia, the presence of autoantibodies and characteristic histological findings. The disease can have catastrophic outcome with the development of end-stage liver disease if misdiagnosed/undiagnosed and left untreated. AIH pathogenesis remains obscure and the main hypothesis supports its development in genetically predisposed individuals after being exposed to certain environmental triggers. Genetic predisposition is linked to the presence of certain HLA alleles, mainly HLA-DR3 and HLA-DR4. However, a wide number of non-HLA epitopes have also been associated with the disease although data vary significantly among different ethnic groups. Therefore, it is likely that epigenetic alterations may also play a crucial role in disease's pathogenesis, although not yet extensively studied. The aim of this review was to summarize the genetic and environmental factors that have been associated with AIH, but also to open new insights towards the role of epigenetic modifications in the etiology of the disease.
Keywords: Autoimmune hepatitis, HLA-DR, Single nucleotide polymorphisms, Epigenetics
Graphical abstract
Highlights
-
•
Autoimmune hepatitis (AIH) pathogenesis remains unknown.
-
•
AIH genetic predisposition is associated mainly with HLADR3 and HLADR4 alleles but also several non-HLA epitopes.
-
•
Environmental factors such as viruses, drugs, xenobiotics have been implicated.
-
•
Recently, epigenetic factors have also been found to confer to AIH pathogenesis.
-
•
These findings may contribute to the development of novel epigenetic-based therapeutic strategies.
Abbreviations
- AIH
Autoimmune hepatitis
- ALD
Alcoholic Liver Disease
- ANA
antinuclear antibodies
- anti-LKM1
anti-liver kidney microsomal type 1
- anti-LC1
anti-liver cytosol type 1
- CARD10
Caspase Recruitment Domain Family Member 10
- CTLA-4
Cytotoxic T lymphocyte antigen-4
- DI-AIH
Drug Induced Autoimmune Hepatitis
- DILI
Drug Induced Liver Injury
- DNMTs
DNA methyl-transferases
- GWAS
Genome Wide Association Studies
- HLA
Human Leucocyte Antigen
- IgG
Immunoglobulin G
- IFNγ
Interferon γ
- meQTL
methylation Quantitative Trait Loci
- MHC
Major Histocompatibility Complex
- miRNAs
micro-RNAs
- NAFLD
Non Alcoholic Liver Disease
- PBC
Primary Biliary Cholangitis
- SH2B3
Scr homology 2 adaptor protein 3
- SMA
anti-smooth muscle antibodies
- SNPs
Single Nucleotide Polymorphisms
- TETs
Ten-eleven translocation enzymes
- TGFB1
Transforming Growth Factor Beta 1
- TH
T Helper
- TNF
Tumor necrosis factor
- Tregs
T regulatory cells
- VDR
Vitamin D receptor
- 5mC
5-methyl cytosine
- 5hmC
5-hydroxy-methyl cytosine
1. Introduction
Autoimmune hepatitis (AIH) is a chronic liver disease of unknown etiology characterized by interface hepatitis on liver histology, presence of non-organ specific autoantibodies and selective elevation of serum immunoglobulin G (IgG) levels [1]. Although AIH was first described mainly in young women, it is now clear that affects all ages of both sexes and all ethnic groups. Indeed, approximately 30% of patients are males and about 30% are older than 60 years at first diagnosis [[2], [3], [4], [5], [6], [7], [8]] [[2], [3], [4], [5], [6], [7], [8]] [[2], [3], [4], [5], [6], [7], [8]]. It is considered a relatively rare disease with prevalence rates from 10 to 24 per 100,000 inhabitants in Europe [[4], [5], [6], [7], [8]].
Due to the absence of a single diagnostic marker, AIH diagnosis is based on diagnostic criteria comprising of a combination of four parameters namely, IgG, the presence of autoantibodies, liver histology and absence of viral hepatitis markers [9]. The disease is further sub-typed in AIH-type 1, characterized by the presence of antinuclear antibodies (ANA) and anti-smooth muscle antibodies (SMA) and AIH-type 2, which predominantly affects children and adolescents and is characterized by the presence of anti-liver kidney microsomal type 1 (anti-LKM1) and/or anti-LKM type 3 (anti-LKM3)antibodies, with or without anti-liver cytosol type 1 (anti-LC1) antibodies [[10], [11], [12], [13], [14]].
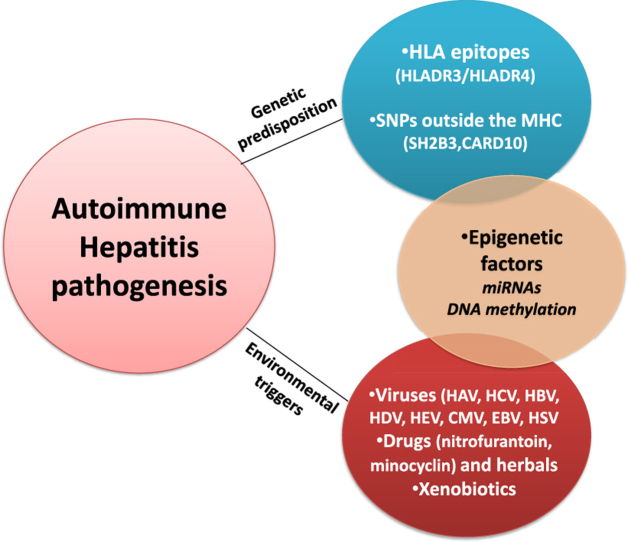
The precise etiology of AIH remains unknown, however a complex interplay between genetic, environmental and yet epigenetic factors has been considered to confer to the disease occurrence and outcome. The dominant hypothesis supports that AIH develops in a genetically predisposed individual after being exposed to one or certain environmental factors resulting to loss of self-tolerance. Thereafter, the autoimmune attack is perpetuated via: 1) T helper 1 (TH1) response exerting cytotoxicity upon recognition of antigen-major histocompatibility complex (MHC) class I complex, 2) TH2 response linked to antibody-mediated cellular cytotoxicity and complement activation, 3) TH17 cell response producing pro-inflammatory cytokines [IL17, IL22 and tumor necrosis factor a (TNFa)] and 4) CD4+CD25+FOXP3+ T regulatory-cells (Tregs) dysfunction [1,11,13,15,16].
Genetic predisposition in AIH is mainly associated with human leucocyte antigen (HLA) genes [1,17]. Moreover, a significant number of single nucleotide polymorphisms (SNPs) outside the MHC, has been identified and linked to the clinical phenotype of AIH [17]. On the other hand, several environmental factors such as xenobiotics, drugs, herbals and viruses have been associated with loss of self-tolerance through neoantigens production, as well as initiation and perpetuation of the immune response [11,18,19]. Finally, epigenetic modifications triggered by environmental factors influence genes expression and confer to immune dysregulation and disease pathogenesis [20].
Herein, we summarize the genetic, environmental and epigenetic factors that seem to contribute to AIH pathogenesis.
2. Genetic risk factors
2.1. HLA alleles
The association of AIH with the presence of specific HLA alleles dates back more than 40 years, when for the first time Mackay et al. reported an association between AIH and HLA-DR3 [21]. In the following years a considerable number of studies confirmed the role of HLA antigens, mainly HLA-DR3 and HLA-DR4, in disease occurrence and outcome [[22], [23], [24], [25], [26], [27]].
The prominent predisposing role of HLA alleles, especially HLA-DR3 and HLA-DR4, in AIH was further described by Boer et al. [28] in the largest genome wide association study (GWAS) conducted so far, in The Netherlands and Germany/Switzerland. The latter study demonstrated a strong association between DRB1*0301 and DRB1*0401 with AIH-1susceptibility, as was previously shown in diverse Caucasian populations from Europe and North America [[27], [28], [29], [30], [31]]. Furthermore, another recent study from The Netherlands, revealed that HLADRB1*0301/HLADRB1*0401 positive patients had higher IAIHG scores compared to the respective HLA-DRB1*0301/DRB1*0401 negative patients suggesting that HLA haplotypes may modify the presentation and outcome in patients with AIH-1 [32].
However, predisposing HLA genes vary among different ethnicities and geographic regions [[33], [34], [35], [36], [37], [38], [39], [40]] as shown in Table 1. Indeed, in populations of Latin ancestry, susceptibility to AIH-1 was linked to the presence of DRB1*0405, DRB1*0404 as well as DRB1*1301, while DRB1*1302 and DQB1*0301 were considered protective alleles [[33], [34], [35], [36], [37], [38], [39]]. In addition, in Japan, although HLA-DR3 is scarce, HLADRB1*0405 and HLADRB1*0404 also confer susceptibility to the disease, DRB1*0802 and DRB1*0803 predispose to disease development when DRB1*0405 is also present, while DRB1*1501 is considered a protective allele [Table 1; 25,41–43]. Moreover, studies in Asian populations have shown an association between DRB1*13 and DRB1*14 with AIH susceptibility [Table 1; 44,45].
Table 1.
Human Leucocyte Antigen (HLA)alleles associated with autoimmune hepatitis (AIH).
| HLA allele | Association | Population | Study |
|---|---|---|---|
| DRB1*0301 and DRB1*0401 | AIH-1 | European, North Americans | Mackay et al., 1980 [21] Krawitt et al., 1987 [22] Donaldson et al., 1991 [23] Seki et al., 1990 [24] Doherty et al., 1994 [26] Strettel et al., 1997 [27] Boer et al., 2014 [28] Donaldson et al., 2002 [29] |
| DRB1*0404, DRB1*0405 and DRB1*1301 | AIH-1 | Latin Americans Japanese |
Umemura et al., 2014 [25] Pando et al., 1999 [34] Czaja et al., 2002 [35] Czaja et al., 2008 [36] Goldberg et al., 2001 [37] Fortes et al., 2007 [38] Fainboin et al., 1994 [39] Furumoto et al., 2015 [42] Seki et al., 1992 [41] |
| DRB1*0802 and DRB1*0803 | AIH-1 | Japanese | Oka et al., 2017 [43] |
| DRB1*13 and DRB1*14 | AIH-1 | Indian, Iranian | Amarapurkar et al., 2003 [44] Baharlu et al., 2016 [45] |
| DRB1*07 and DRB1*03 | AIH-2 | European | Ma et al., 2006 [43] Underhill et al., 2002 [46] |
| DRB1*15 | AIH-2 | Egyptian | Elfaramawy et al., 2010 [48] |
Abbreviations: HLA, human leucocyte antigen; AIH, autoimmune hepatitis.
Both DRB1*0301and DRB1*0401 are heterodimers that contain a lysine residue at position 71 of the DRB1 peptide and the hexamer amino acid sequence LLEKQR at positions 67–72 [29,30,33]. On the other hand, DRB1*0405 and DRB1*0404 encode arginine rather than lysine at position 71 of DRB1 polypeptide, sharing the motif LLEQR [33]. Therefore, these two amino acids (lysine and arginine) in the sequence of LLEQR may be crucial for susceptibility to AIH.
As far as AIH-2 is concerned, a limited number of studies have been conducted due to the low prevalence of this subtype of the disease (about 10% of all AIH cases in Europe and North America) [[10], [11], [12]]. However, HLADRB1*07 and HLADRB1*03 have been identified as predisposing alleles [46,47], while in Egypt, AIH-2 was associated with HLADRB1*15 [48].
Although susceptibility to AIH was strongly associated with the presence of certain HLA alleles, the same alleles have been linked to different clinical phenotypes of the disease [25,26]. Since’90s,DRB1*0401 haplotype was associated with older age at disease onset, female predominance and better response to corticosteroids [31], as well as more frequent concurrence of extrahepatic autoimmune diseases [49]. Furthermore, the presence of DRB1*0401 allele was correlated with milder disease and less frequent relapses during treatment compared to DRB1*0301 [26]. On the other hand, DRB1*0301 was associated with more severe disease sinceDRB1*0301 positive patients were more likely to receive immunosuppressive treatment and liver transplant, while it was correlated with higher IgG levels [32]. Accordingly, in Japan the HLADRB1*0405 haplotype was correlated with elevated serum IgG levels and SMA positivity [25].
To recap, it seems that certain HLA alleles and especially HLA-DR4 and HLA-DR3 confer susceptibility to AIH in some populations, however, these associations are not universal suggesting that factors beyond MHC complex may confer to AIH susceptibility.
2.2. Single nucleotide polymorphisms (SNPs) outside the MHC complex
Besides the critical role of HLA alleles in AIH occurrence and outcome, several SNPs outside the MHC complex have also been linked to disease susceptibility. However, as shown in Table 2, the results are often controversial among different studies and in many cases are not verified in replication studies in different ethnic groups.
Table 2.
Single nucleotide polymorphisms (SNPs) outside the Major Histocompatibility Complex (MHC) associated with autoimmune hepatitis (AIH).
| SNP | Gene | Association | Population | Study |
|---|---|---|---|---|
| (-308) | TNF | AIH-1 | European | Czaja et al., 1999 [50] Cookson et al., 1999 [51] |
| (AT)8 (dinucleotide repeat) and A (exon 1) | CTLA-4/CD28 | AIH-1 | European, American | Agarwal et al., 2000 [52] Djilali-Saiah et al., 2001 [53] |
| exon 2 restriction fragment length polymorphism with Fok1 | VDR | AIH-1 | European Chinese |
Vogel et al., 2002 [56] Fan et al., 2005 [57] |
| (-670) | FAS | AIH-1 | Japanese | Hiraide et al., 2005 [59] |
| (+869) and (+915) | TGFB1 | AIH-1 | Latin Americas | Paladino et al., 2010 [60] |
| (-1993) | TBX21 | AIH-1 | Chinese | Chen et al., 2011 [61] |
| rs7574865 | STAT4 | AIH-1 | Japanese | Migita et al., 2013 [62] |
| rs3184504*A | SH2B3 | AIH-1 | European | Boer et al., 2014 [28] |
| rs6000782 | CARD10 | AIH-1 | European | Boer et al., 2014 [28] |
| rs755622 | MIF | AIH-1 | Japanese North American |
Assis et al., 2016 [66] |
| rs121741, rs1217388, rs1217407 and rs2488458 | PTPN22 | AIH-1 | Japanese | Umemura et al., 2016 [67] |
| rs4325730G | ICOS | AIH-1 | Japanese | Higuchi et al., 2017 [64] |
| rs1106594 | SH2B3 | AIH-1 | Japanese | Umemura et al., 2017 [65] |
| rs7708392 | TNIP1 | AIH-1 | Japanese | Oka et al., 2018 [69] |
| rs7708392 | TNIP1 | AIH-1 | Japanese | Oka et al., 2018 [69] |
| (-590) | IL-4 | AIH-1 | Egyptian | Mansour et al., 2018 [70] |
| deleterious variants (c.116 A > G, c.305 A > G, c.1897G > C) | TNFAIP3 | AIH-1 | Japanese | Higuchi et al., 2019 [68] |
Abbreviations: AIH, autoimmune hepatitis; SH2BP3, SH2B Adapter Protein 3; CARD10, Caspase Recruitment Domain Family Member 10; CTLA4, Cytotoxic T-Lymphocyte Antigen 4; TNF, tumor necrosis factor; VDR, vitamin D receptor; TBX21, T-Box Transcription Factor 21; ICOS, Inductible T-cell Costimulator; FAS, Fas Cell Surface Death Receptor; MIF, Macrophage Migration Inhibitory Factor; PTPN22, Protein Tyrosine Phosphatase Non-Receptor Type 22; STAT4, Signal Transducer and Activator of Transcription 4; TNFAIP3, TNF Alpha Induced Protein 3; TNIP1, TNFAIP3 Interacting Protein 1; IL-4, interleukin-4, TGFB1, transforming growth factor beta 1.
Given that polymorphisms can affect immunoregulation by controlling cytokine production, the primary candidate genes predisposing to AIH were those encoding proinflammatory and immunoregulatory cytokines. Indeed, during late ‘90s, a genetic variant of TNF gene (−308) was reported to occur more commonly in AIH patients, associate with poor response to treatment and present a strong linkage disequilibrium with the HLAA1-B8-DR3 haplotype [50,51].
Further to these findings, several genes outside the MHC have been investigated as candidates for AIH susceptibility, but in small study cohorts. Interestingly, most of the identified genetic polymorphisms concerned immunological pathways of T cell activation and response, as well as, the production and action of proinflammatory cytokines.
SNPs of the Cytotoxic T-Lymphocyte Antigen 4 (CTLA-4/CD28) region have been correlated with susceptibility to AIH both in adult and pediatric European populations [52,53]. However, there is large heterogeneity between different studies, as for example, the absence of CTLA-4/CD28SNPsassociation with AIH development in Japanese patients [54,55]. Furthermore, a SNP in the vitamin D receptor (VDR) gene (exon 2 restriction fragment length polymorphism with Fok1) was associated with AIH in European and Chinese populations [56,57], a correlation, which has also been reported in other autoimmune diseases [58]. Accordingly, a SNP (−670) upstream of FAS gene (Fas Cell Surface Death Receptor)that regulates the programmed cell death [59] as well as two genetic risk loci (+869) and (+915) in the exon 1 of the TGFB1 gene(60) have been associated with AIH in Japan and Latin America, respectively. Of note, SNPs in the TGFB1 gene have been correlated with disease severity, by affecting the production of TGF-β and therefore, the development of fibrosis [60]. In addition, two independent groups, from Europe [61] and Japan [62], reported an association of SNPs in genes which regulate activation and function of T-cells [a SNP in the promoter of T-Box Transcription Factor 21 (TBX21) gene (−1993) [61] and a SNP in Signal Transducer And Activator Of Transcription 4 (STAT4) (rs7574865) [62] with AIH.
In 2014, the largest so far, GWAS study by Boer et al. [28], identified an association of the rs3184504*A allele of the Scr homology 2 adaptor protein 3 (SH2B3) gene with AIH. SH2B3 is a negative regulator of T-cell activation, TNF and Janus kinase (JAK) 2 and 3 signaling and is associated with higher expression levels of several genes involved in interferon-gamma production (IFNγ), suggesting that the risk allele leads to an increased inflammatory response [63]. Moreover, the same genetic risk allele was associated with concomitant autoimmune diseases such as primary biliary cholangitis (PBC), primary sclerosing cholangitis and celiac disease indicating a genetic overlap between AIH and other immune mediated diseases. In the same study [28], the variant rs6000782 of Caspase Recruitment Domain Family Member 10 (CARD10) gene -a scaffold protein that activates the pro-inflammatory NF-kB pathway, inducing the expression of pro-inflammatory and fibrogenic cytokines-was also highlighted as a predisposing genetic risk variant.
In contrast, one of the largest Japanese studies [64], failed to confirm the associations between rs3784504 of SH2B3 and rs6000782 of CARD10 genes with AIH-1. Instead, the authors detected another SNP, the rs4325730G upstream of the Inductible T-cell Co-stimulator (ICOS), to be linked with AIH-1 susceptibility. This risk locus was in strong linkage disequilibrium with the rs4675374 of ICOS, a SNP also associated with celiac disease. Of interest, although the reported rs3784504 of SH2B3 gene was not polymorphic in Japanese patients, another SNP of the same gene, rs1106594, was associated with AIH in this patient cohort [65].
During the latest years a number of independent studies highlighted several genetic risk variants that confer susceptibility to AIH in the Japanese population. A SNP (rs755622) in the Macrophage Migration Inhibitory Factor (MIF) promoter that serves as a proinflammatory cytokine, has been found to implicate with AIH-1 development [66]. Accordingly, diverse risk loci in the Protein Tyrosine Phosphatase Non-Receptor Type 22 (PTPN22) gene (rs121741, rs1217388, rs1217407 and rs2488458) affecting the activation of T-cells [67], were found to associate with AIH in Japanese, while various SNPs of the TNFAIP3 (TNF Alpha Induced Protein 3) gene have been correlated with the development of AIH-related cirrhosis [68]. In addition, the rs7708392 SNP in the TNIP1 (TNFAIP3 Interacting Protein) gene, which is implicated to the NF-κB pathway activation, has also been found to predispose to AIH in Japanese [69].
In North Africa and more precisely in Egypt, a SNP (−590) at IL-4 promoter has been associated with AIH in children [70], while pediatric AIH-1 has been correlated with increased functional form of KIR2DS4 (killer cell immunoglobulin-like receptors) in Latin America [71].
Taken together, the above-mentioned data indicate that up to the present, the detected SNPs in the non-HLA loci, which have been associated with AIH, present ethnic and geographic heterogeneity, suggesting that they cannot explain with confidence AIH predisposition.
2.3. Sex and hormones
As in most autoimmune diseases, AIH is characterized by a strong female predominance (female/male ratio: 3–4/1). Given that many genes involved in immunological tolerance are located on X-chromosome [72], abnormalities on X-chromosome could partly explain this predominance. In female patients with PBC, a higher frequency of monosomy X in B and T cells [73] and random X inactivation in somatic cells have been reported [74]. Furthermore, females express higher immunoglobulin serum levels and a more intense cell-mediated immune response upon immunization, while sex hormones play a critical role in this process promoting a TH2 rather than a TH1 inflammatory response [[75], [76], [77]]. However, these mechanisms have not been so far investigated in AIH, so the role of X-chromosome linked abnormalities and the effect of sex hormones is yet unclear in AIH pathogenesis.
3. Environmental triggers
A number of triggering factors have been proposed including viruses, xenobiotics and drugs, but none has been conclusively shown to be involved in AIH pathogenesis. The main hypothesis comprises the development of AIH, in genetically predisposed individuals, when they are exposed to environmental triggers, mainly via mechanisms of molecular mimicry or non-specific activation of T lymphocytes, which leads to dysregulation of immunoregulatory networks and emergence of autoreactive T cells and/or alteration in gene expression [19].
3.1. Viruses
Hepatitis A, B, C, D and E viruses (HAV, HBV, HCV, HDV, HEV), but also Cytomegalovirus (CMV), Epstein-Barr virus (EBV) and Herpes Simplex virus-1 (HSV-1) have been associated with AIH development since early 90s (Table 3). Indeed, Vento et al. [78], described an association between HAV and AIH in a cohort of 58 first degree relatives of AIH patients. Interestingly, AIH development was associated with a preexisting defect in suppressor-inducer T lymphocytes specifically controlling immune responses towards the asialo-glycoprotein receptor (ASGPR) antigen on the surface of hepatocytes. Afterwards, a number of case reports followed supporting the role of HAV as a potential AIH trigger [[79], [80], [81], [82], [83], [84], [85], [86], [87], [88]].
Table 3.
Viral triggers associated with autoimmune hepatitis (AIH).
| Viral agent | Potential mechanism | Autoantibodies detected |
|---|---|---|
| HAV [[78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88]] | defect in suppressor-inducer T lymphocytes controlling immune responses towards the asialo-glycoprotein receptor (ASGPR) | anti-ASGPR |
| HBV/HDV [[101], [102], [103], [104], [105], [106], [107]] | development of non-organ specific autoantibodies | ANA, SMA, anti-LKM3 |
| HCV [[89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99]] | sequence homology with CYP2D6 | anti-LKM1 |
| HEV [[108], [109], [110], [111], [112]] | cross-reactivity between viral and liver antigens | ANA, SMA |
| EBV [[113], [114], [115], [116], [117], [118]] | trigger of immunological response | ANA, SMA, anti-LKM1 |
| CMV [97,98] | sequence homology with CYP2D6 | anti-LKM1 |
| HSV-1 [97,98] | sequence homology with CYP2D6 | anti-LKM1 |
Abbreviations: HAV, hepatitis A; HBV/HDV, hepatitis B/hepatitis D; HCV, hepatitis C; HEV, hepatitis E; CMV, cytomegalovirus; EBV, Ebstein Bar virus; HSV-1, herpes simplex 1; ASGPR, asialo-glycoprotein receptor; ANA, antinuclear antibodies; SMA, anti-smooth muscle antibodies; anti-LKM1, anti-liver kidney microsomal type 1; CYP2D6, cytochrome P450 2D6.
The possible link between HCV infection and AIH has been extensively studied in the past years [[89], [90], [91], [92], [93], [94]]. The most characteristic case was in a girl who contracted HCV infection after liver transplantation for alpha 1 anti-trypsin deficiency and developed anti-LKM1 autoantibodies 2 weeks after transplantation, resulting in overt de novo AIH-2 8 years later [95]. Indeed, a number of studies have found that about 10% of HCV infected patients develop anti-LKM1 autoantibodies and autoimmune features [92,93,96,97]. In addition, specific humoral or T cell epitopes cross-reacting between sequencies of HCV and cytochrome P450 2D6 (CYP2D6, the main target autoantigen of anti-LKM 1 in AIH-2) have been described in AIH [98]. Of note, it has been shown in animal models that DNA vaccination of mice with a plasmid encoding CYP2D6 could lead to a peak serum aminotransferases level after 4–7 months and the development of periportal, portal and lobular liver inflammatory infiltrates, suggesting that CYP2D6 plays a crucial role in AIH-2 pathogenesis [99,100]. These data led to the hypothesis that HCV induce anti-LKM1 through the mechanism of molecular mimicry leading to the development of AIH-2 [98]. Interestingly, sequence homologies have also been detected between CMV and HSV-1 and CYP2D6 [97,98].
Furthermore, HBV, HDV and HEV have also been proposed as AIH triggers. Non-organ specific autoantibodies particularly ANA and SMA have been reported in up to 7% of patients with chronic HBV infection [101], but they are actually considered as part of the natural course of the disease, as they have not been linked with AIH development [[101], [102], [103]]. However, cases with concurrent chronic hepatitis B and AIH have been reported bearing similar outcome rates to that of AIH alone [104].
Historically, HBV/HDV co-infection has been associated withanti-LKM3antibodies directed against family 1 uridine 5'-diphosphate-glucuronosyl-transferases (UGT-1), as they are detected in about 13–24% of patients [105,106]. In addition, we have reported that almost two thirds of HBV/HDV patients have at least one detectable autoantibody before treatment initiation with interferon (ANA 33%, SMA 27%, anti-LKM3 24%) [106]. However, very few patients developed AIH after interferon treatment, while the presence of autoantibodies did not affect the response to treatment [106]. Of note, patients with chronic HBV/HDV infection and anti-LKM3 (anti-UGT) autoantibodies were more prone to develop a severe course of HDV infection, albeit they had lower titers of these antibodies compared to AIH-2 patients [107].
Regarding HEV, acute infection has also been reported to associate with the detection of non-specific autoantibodies, suggesting either a cross-reactivity between HEV and liver antigens or even HEV itself acting as a possible AIH trigger [108,109]. Of interest, several studies have reported significantly higher prevalence of anti-HEV antibodies in patients with AIH than in individuals with other chronic liver diseases or patients with other autoimmune diseases and healthy controls, possibly implying that HEV infection could elicit immune events [[110], [111], [112]].
Another viral candidate, the EBV, has been associated with the development of various autoimmune diseases including AIH-1, in several case reports [[113], [114], [115]] and in a clinical follow-up of 13 patients [116]. Recently, the first case in children ever reported in the English literature considering the development of AIH-2 after well-established EBV infection was described [117]. Although no molecular mimicry could be established by sequence analysis, EBV could also be a trigger for AIH-related immunological reactions [118].
3.2. Drugs and alcohol
Many drugs have been associated with the development of a syndrome similar to “true” AIH. Historically, nitrofurantoin and minocycline have been associated with induction of AIH, implicated in 90% of drug-induced AIH (DI-AIH)cases [119]. Other drugs and herbals, such as oxyphenisatin, ornidazole, methyldopa, diclofenac, interferon, atorvastatin, highly active antiretroviral treatment and biologic agents such as infliximab, natalizumab and adalimumab, have also been reported occasionally to induce AIH [11,[119], [120], [121]].
DI-AIH is a specific phenotype of drug induced liver injury (DILI) [122]. DI-AIH has been well described in the past for tienilic acid and dihydralazine, which are no longer in use [123,124]. Hepatic metabolism of the drugs lead to reactive metabolites, which bind to cellular proteins such as components of CYP450 (CYP2C9 in the case of tienilic acid and CYP1A2 in the case of dihydralazine), and are subsequently recognized by the immune system as neoantigens [123,124]. In a genetically predisposed individual, this may lead to a chronic process, with a permanent need for immunosuppression [125]. This entity has to be differentiated from immune mediated DILI (IM-DILI), which refers to acute or chronic liver injury that may or may not resolve with drug withdrawal [125]. When the injury does not resolve after drug cessation, the patients usually need corticosteroids administration followed by rapid tapering schedule aiming to achieve complete biochemical response and sustained remission without relapse after corticosteroids discontinuation. However, these patients need long term follow up (6 monthly for at least 3 years) in order not to miss a late relapse of AIH [10].
On the other hand, alcohol could be another potential trigger of liver autoimmunity. Products of alcohol metabolism such as acetaldehyde and malondialdehyde induce the production of autoantibodies both in human and in animal models [126,127]. Moreover, despite the results of a case-control study in New Zealand [128] where low to moderate alcohol consumption decreased the risk for AIH diagnosis, results from a recent study from our group showed a significant overlap between AIH and alcoholic liver disease (ALD) [129]. Diagnosis of AIH in patients with significant alcohol consumption is often difficult, however it is of great importance as patients with AIH/ALD variant are more frequently autoantibodies positive and have worse outcome despite the appropriate administration of immunosuppression [129].
3.3. Xenobiotics
Xenobiotics are foreign chemicals or toxins that may accumulate in the body, usually due to exposure to pollutants and deactivated and/or secreted primary by the liver. Xenobiotics could induce an immune response either by non-specific activation of lymphocytes, as observed in a murine experimental model of Concanavalin A (ConA) immune-mediated liver injury [130] or by exerting hepatotoxic effect, which leads to autoantigen formation or acting as haptens, modifying hepatic proteins and rendering them immunogenic [19]. In PBC, there are clear indications of case clustering near toxic waste sites and/or other environmental pollutants [131,132]. Additionally, it has been shown that anti-mitochrondrial antibodies (AMA) positive sera from PBC patients, but not from controls, react to a number of xenobiotic-modified E2 component of pyruvate dehydrogenase (PDC-E2) structures -the major target autoantigen of PBC [133,134]. Up to the present, similar data do not exist for AIH.
As exposure to xenobiotics is difficult to be recognized, and in most cases more than one agent is implicated, the evidence of toxin-induced AIH relies basically on the results from animal studies. As an example, carbon tetrachloride (CCl4), an organic compound once used as a cleaning fluid, is known to generate toxic hepatitis and fibrosis in animals [135]. Accordingly, trichloroethylene (TCE), an organic solvent, has been linked to a variety of autoimmune diseases including AIH, a fact that was also confirmed in animal models [135]. In both cases an increase of IFNγ and the development of a chronic T-cell mediated AIH are implicated [135].
4. Epigenetic factors
Epigenetic modifications that influence the expression of genes along with their functions without affecting the sequence of nucleotides have been implicated in the development of several autoimmune diseases. These changes are reversible, cell-type specific and are impacted by age, sex and environmental factors. Epigenetic changes not only confer to the etiopathogenesis of autoimmune diseases, but they also represent a potential target for therapeutic interventions. Epigenetic studies in AIH are scarce and they were mainly engaged to the investigation of the role of micro-RNAs (miRNAs) in disease pathogenesis, while only one study up to date has addressed the implication of DNA methylation modifications (Table 4).
Table 4.
Epigenetic studies in autoimmune hepatitis (AIH).
| Study | Design | Epigenetic Mechanism | Findings |
|---|---|---|---|
| Migita et al., 2015 [137] | 46 AIH patients vs. 40 CHCV and 13 HC | miRNAs | •Increased levels of miR-21 associated with liver inflammation •Decreased levels of miR-21 and miR-122 in cirrhosis |
| Yamaura et al., 2017 [138] | 28 patients (CHBV, AIH, PBC,NASH, CHCV) vs. 4 HC | miRNAs | •Reduced levels of miR-218, miR-363, miR-518f, miR-628–5p, miR-888, miR-523, miR-141, miR-302 b, miR-643, and miR-57-not capable to distinguish AIH from other liver diseases |
| Chen et al., 2018 [139] | animal model | miRNAs | •miR-223: protective effect on liver, regulating the nucleotide binding oligonucleotide domain like receptor 3 and caspase 1 |
| Su et al., 2016 [140] | animal model | miRNAs | •miR-674–5p acted as a negative regulator of 5-lipoxygenase mediated liver injury |
| Xia et al., 2018 [141] | animal model | miRNAs | •miR-155 attenuated concanavalin A induced AIH by inhibiting TH17 cell function |
| Zachou et al., 2021 [148] | 10 AIH at diagnosis vs. 8 AIH at remission, vs 9 PBC and 10HC | •DNMTs/TETs •total 5mC/5hmC •IHC •cell specific DNA methylation |
•No difference in 5mC/5hmC •Altered expression of DNMT3A and TET1 in AIH at diagnosis •Association of DNMT3A with disease activity and treatment •Increased 5hmC staining of the periportal infiltrating lymphocytes •Gene specific epigenetic alterations in pathways of immune response |
Abbreviations: AIH, autoimmune hepatitis; PBC, primary biliary cholangitis; CHBV, chronic HBV infection; CHCV, chronic HCV infection; NASH, non-alcoholic steatohepatitis; HC, healthy controls; miRNAs, micro-RNAs; DNMTs, DNA methyl-transferases; TETs, ten eleven-translocation enzymes; 5mC/5hmC: 5-methylcytosine/5-hydroxymethyl cytosine; IHC, immunohistochemistry.
4.1. miRNAs
miRNAs are small non-coding RNA molecules with conserved properties, which function by base-pairing with complementary sequences in mRNAs. After binding to the mRNA target they induce its degradation leading to the suppression of gene expression at a post-transcriptional level [136].
Unlike other liver diseases such as non-alcoholic fatty liver disease (NAFLD) and PBC, AIH is just entering the epigenetic era. Up to now epigenetic studies in AIH have focused on the role of circulating miRNAs in disease occurrence. Migita et al. [137], found that miR-21 was increased and significantly associated with the degree of liver inflammation in AIH patients. In contrast, circulating levels of miR-21 and miR-122 were reduced in patients with cirrhosis and were inversely correlated with increased stage of fibrosis. Accordingly, Yamaura et al. [138], studied the serum miRNA profiles in patients with diverse liver diseases compared to healthy controls and observed reduced levels of several miRNAs (miR-218, miR-363, miR-518f, miR-628–5p, miR-888, miR-523, miR-141, miR-302 b, miR-643, and miR-573) in the patients cohort, which unfortunately were not able to distinguish AIH from other liver diseases.
miRNAs have also been studied in experimental animal models in AIH. miR-223 derived from exosomes of bone marrow-derived mesenchymal stem cells was found to induce a liver protective effect, regulating the nucleotide binding oligonucleotide domain like receptor 3and caspase 1 [139]. Moreover, in a ConA-induced AIH animal model, it was shown that miR-674–5p acted as a negative regulator of 5-lipoxygenase mediated liver injury [140], while miR-155 was found to attenuate the liver injury by inhibiting TH17 cell function [141].
4.2. DNA methylation
DNA methylation is the main and most well studied epigenetic mechanism in eucaryotic cells and consists of the addition of a methyl-group at the position 5’ of the pyrimidine ring of cytosine, catalyzed by DNA methyl-transferases (DNMTs) [142]. An active demethylation process is also taking place, catalyzed by Ten-eleven translocation enzymes (TETs) which oxidize 5-methyl cytosine (5mC) to 5-hydroxy-methyl cytosine (5hmC) and finally the methyl-groups are removed by DNA repairing enzymes [143].
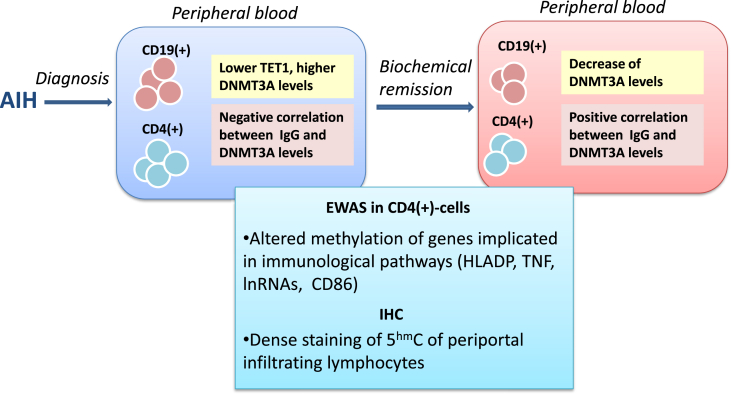
Although studies on DNA methylation alterations have been conducted in other chronic liver diseases such as NAFLD and PBC [[144], [145], [146], [147]], to the best of our knowledge no methylation studies have been conducted in AIH. However, a recent study from our group [148] investigated the DNA methylation alterations in peripheral CD19(+) and CD4(+) lymphocytes as well as in liver sections from AIH patients before and after achieving complete remission compared to healthy controls and PBC patients. The results highlighted the increased expression of DNMT3A and reduced expression of TET1 compared to PBC patients and controls, respectively. Moreover, DNMT3A expression was negatively associated with circulating IgG levels indicating an association with disease activity, while immunosuppressive treatment reduced DNMT3A levels. Alterations in DNMT3A and TET1 transcriptional levels were not associated with differences in total 5mC/5hmC levels indicating gene specific epigenetic alterations. The latter was confirmed by whole genome methylation analysis in CD4(+) T cells, which revealed hypomethylation of the majority of genes in AIH before treatment administration in contrast to the hypermethylation detected at disease remission and the hypermethylation of almost all the implicated genes in PBC. Interestingly, the hypomethylation of immune cells in AIH before treatment was also confirmed in the histological level, since infiltrating periportal lymphocytes from AIH patients’ liver sections presented increased 5hmC staining in immunohistochemistry. These findings hint towards an extensive methylation alteration of many genes of the immune cells in AIH, as well as a methylation “shift” between the active and the remission stage of the disease. Furthermore, they suggest that different epigenetic profiles exist between distinct autoimmune liver diseases. Of note, the majority of the genes found hypomethylated in AIH at diagnosis were those implicated in immune response pathways further supports the notion that methylation alterations may contribute to AIH pathogenesis (Fig. 1) [148].
Fig. 1.
DNA methylation alterations in peripheral immune cells and liver sections from AIH patients. Increased DNMT3A and reduced TET1 expression is observed in CD4(+) and CD19(+) lymphocytes from AIH patients at diagnosis compared to PBC patients and healthy controls, respectively. DNMT3A expression is negatively associated with circulating IgG levels, while Immunosuppressive treatment reduces DNMT3A levels in AIH at remission. Alterations in DNMT3A and TET1 transcriptional levels are associated with altered methylation status of several genes implicated in immune response pathways. Immunohistochemistry on liver sections from AIH patients at diagnosis reveals a dense 5hmC staining of periportal infiltrating lymphocytes indicating hypomethylation.
Abbreviations: AIH: autoimmune hepatitis, EWAS: epigenome wide association studies, DNMT3A: DNA methyl-transferase 3A, TET1: ten-eleven-translocation enzyme 1, IHC: immunohistochemistry, IgG: immunoglobulin G, HLADP: human leucocyte antigen DP, lnRNA: long non-coding RNA, TNF: tumor necrosis factor, 5 hmC: 5-hydroxymethyl cytosine.
5. Overview and future perspectives
Taking into consideration the abovementioned data, it is obvious that complex mechanisms based on the interaction between genetic, environmental and epigenetic factors comprises the backbone of AIH pathogenesis. Genetic impact alone is usually modest with the exception of certain HLA alleles, while environmental factors are, in most cases, difficult to be identified and linked directly to the etiopathogenesis of AIH. Therefore, epigenetic mechanisms seem to play a critical role in AIH pathogenesis and further investigation in this field could possibly open new insights not only towards to the better understanding of the disease, but also for the development of novel epigenetic-based treatment interventions.
Previous studies in other autoimmune diseases and now in AIH showed that regulatory genes are in the majority of cases hypomethylated, transcriptional activity is increased and circulating miRNAs intervene to genes expression at the post-transcriptional level [149,150]. Therefore, interventions that alter methylation pattern such as targeting methyl-binding domains (MBDs) or modulating miRNAs could affect the function of implicated genes. However, these epigenetic-based strategies in autoimmune diseases are still in their infancy [151,152].
Linking genetic variants with epigenetic marks is a novel and promising approach for better understanding autoimmune diseases pathogenesis. As we have recently described [153] in Sjӧgren's syndrome, when predisposing SNPs are located within the proximal gene regulatory regions close to transcription start sites (TSS), they affect transcription by controlling the binding of transcriptional factors. However, when conferring SNPs are located in distal regulatory elements, they can affect transcription through an association with altered DNA methylation leading to the description of methylation quantitative trait loci (meQTL) [153,154]. Thereby, adequate control of meQTLs could be a valuable tool for unveiling pathogenetic mechanisms in AIH and guide future therapeutic interventions.
6. Conclusion
In conclusion, the precise mechanisms that lead to the development of AIH and affect the outcome of the disease remain obscure. The impact of genetic and environmental factors in AIH pathogenesis is already supported by several studies, but merits further exploration, in order to be fully clarified. Since up to know neither genetic nor environmental factors alone are sufficient to explain AIH pathogenesis, epigenetics seem a promising tool to provide better understanding of disease etiopathogenesis. In fact, epigenetic modifications that alter genes transcription and, their simultaneous analysis and association with genetic variants of the disease represent the field towards which future investigation needs to be oriented not only for establishing a better pathogenetic basis of the disease, but also for developing novel epigenetic-based therapeutic strategies.
Author contributions
Study concept and design: George N Dalekos and Kalliopi Zachou. Acquisition of research data: Kalliopi Zachou, Aggeliki Lyberopoulou and Pinelopi Arvaniti. Analysis, interpretation of data and drafting of the article: Kalliopi Zachou, Aggeliki Lyberopoulou and Pinelopi Arvaniti. Critical revision and editing of the manuscript: George N Dalekos.
Funding
This project was supported in part by a research grant from the Hellenic Association for the Study of the Liver (HASL; No ID number is applicable) and the Research Committee of the University of Thessaly (No. 2466). P. Arvaniti was also supported by an award for rare diseases by the Federation for the Development of Internal Medicine in Europe (FDIME; No ID number is applicable).
Declaration of competing interest
The authors declare no conflict of interest.
Contributor Information
Kalliopi Zachou, Email: zachoukalliopi@gmail.com.
Pinelopi Arvaniti, Email: peni.arvaniti@gmail.com.
Aggeliki Lyberopoulou, Email: aglyber@gmail.com.
George N. Dalekos, Email: georgedalekos@gmail.com.
References
- 1.Gatselis N.K., Zachou K., Koukoulis G.K., Dalekos G.N. Autoimmune hepatitis, one disease with many faces: etiopathogenetic, clinico-laboratory and histological characteristics. World J. Gastroenterol. 2015;21:60–83. doi: 10.3748/wjg.v21.i1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manns M.P., Lohse A.W., Vergani D. Autoimmune hepatitis – update 2015. J. Hepatol. 2015;62:S100–S111. doi: 10.1016/j.jhep.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Dalekos G.N., Azariadis K., Lygoura V., Arvaniti P., Gampeta S., Gatselis N.K. Autoimmune hepatitis in patients aged 70 years or older: disease characteristics, treatment response and outcome. Liver Int. 2021 doi: 10.1111/liv.14900. [DOI] [PubMed] [Google Scholar]
- 4.Grønbæk L., Otete H., Ban L., Crooks C., Card T., Jepsen P., West J. Incidence, prevalence and mortality of autoimmune hepatitis in England 1997-2015. A population-based cohort study. Liver Int. 2020;40:1634–1644. doi: 10.1111/liv.14480. [DOI] [PubMed] [Google Scholar]
- 5.Grønbæk L., Vilstrup H., Jepsen P. Autoimmune hepatitis in Denmark: incidence, prevalence, prognosis, and causes of death. A nationwide registry-based cohort study. J. Hepatol. 2014;60:612–617. doi: 10.1016/j.jhep.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Valgeirsson K.B., Hreinsson J.P., Björnsson E.S. Increased incidence of autoimmune hepatitis is associated with wider use of biological drugs. Liver Int. 2019;39:2341–2349. doi: 10.1111/liv.14224. [DOI] [PubMed] [Google Scholar]
- 7.Puustinen L., Barner-Rasmussen N., Pukkala E., Färkkilä M. Incidence, prevalence, and causes of death of patients with autoimmune hepatitis: a nationwide register-based cohort study in Finland. Dig. Liver Dis. 2019;51:1294–1299. doi: 10.1016/j.dld.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 8.van Gerven Ν.Μ.F., Verwer B.J., Witte B.I., van Erpecum K.J., R van Buuren H., Maijers I. Scand J Gastroenterol .; 2014. Epidemiology and Clinical Characteristics of Autoimmune Hepatitis in the Netherlands; pp. 1245–1254. [DOI] [PubMed] [Google Scholar]
- 9.Hennes E.M., Zeniya M., Czaja A.J., Parés A., Dalekos G.N., Krawitt E.L., Bittencourt P.L., Porta G., Boberg K.M., Hofer H., Bianchi F.B., Shibata M., Schramm C., Eisenmann de Torres B., Galle P.R., McFarlane I., Dienes H.-P., Lohse A.W. International autoimmune hepatitis group, simplified criteria for the diagnosis of autoimmune hepatitis. Hepatology. 2008;48:169–176. doi: 10.1002/hep.22322. [DOI] [PubMed] [Google Scholar]
- 10.European association for the study of the liver, EASL clinical practice guidelines: autoimmune hepatitis. J. Hepatol. 2015;63:971–1004. doi: 10.1016/j.jhep.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 11.Zachou K., Muratori P., Koukoulis G.K., Granito A., Gatselis N., Fabbri A., Dalekos G.N., Muratori L. Review article: autoimmune hepatitis -- current management and challenges. Aliment. Pharmacol. Ther. 2013;38:887–913. doi: 10.1111/apt.12470. [DOI] [PubMed] [Google Scholar]
- 12.Dalekos G., Koskinas J., Papatheodoridis G.V. Hellenic association for the study of the liver clinical practice guidelines: autoimmune hepatitis. Ann. Gastroenterol. 2019:1–23. doi: 10.20524/aog.2018.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mack C.L., Adams D., Assis D.N., Kerkar N., Manns M.P., Mayo M.J., Vierling J.M., Alsawas M., Murad M.H., Czaja A.J. 2020. Diagnosis and Management of Autoimmune Hepatitis in Adults and Children: 2019 Practice Guidance and Guidelines from the American Association for the Study of Liver Diseases Hepatology; pp. 671–722. [DOI] [PubMed] [Google Scholar]
- 14.Wang G., Tanaka A., Zhao H., Jia J., Ma X., Harada K., Wang F.-S., Wei L., Wang Q., Sun Y., Hong Y., Rao H., Efe C., Lau G., Payawal D., Gani R., Lindor K., Jafri W., Omata M., Sarin S.K. The Asian Pacific Association for the Study of the Liver clinical practice guidance: the diagnosis and management of patients with autoimmune hepatitis. Hepatol Int. 2021;15:223–257. doi: 10.1007/s12072-021-10170-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Longhi M.S., Mieli-Vergani G., Vergani D. Regulatory T cells in autoimmune hepatitis: an updated overview. J. Autoimmun. 2021;119:102619. doi: 10.1016/j.jaut.2021.102619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longhi M.S., Hussain M.J., Mitry R.R., Arora S.K., Mieli-Vergani G., Vergani D., Ma Y. Functional study of CD4 + CD25 + regulatory T cells in health and autoimmune hepatitis. J. Immunol. 2006;176:4484–4491. doi: 10.4049/jimmunol.176.7.4484. [DOI] [PubMed] [Google Scholar]
- 17.Higuchi T., Oka S., Furukawa H., Tohma S., Yatsuhashi H., Migita K. Genetic risk factors for autoimmune hepatitis: implications for phenotypic heterogeneity and biomarkers for drug response. Hum. Genom. 2021;15:6. doi: 10.1186/s40246-020-00301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Floreani A., Restrepo-Jiménez P., Secchi M.F., De Martin S., Leung P.S.C., Krawitt E., Bowlus C.L., Gershwin M.E., Anaya J.-M. Etiopathogenesis of autoimmune hepatitis. J. Autoimmun. 2018;95:133–143. doi: 10.1016/j.jaut.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Béland K., Lapierre P., Alvarez F. Influence of genes, sex, age and environment on the onset of autoimmune hepatitis. WJG. 2009;15:1025. doi: 10.3748/wjg.15.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czaja A.J. Epigenetic changes and their implications in autoimmune hepatitis. Eur. J. Clin. Invest. 2018;48 doi: 10.1111/eci.12899. [DOI] [PubMed] [Google Scholar]
- 21.Mackay I.R., Tait B.D. HLA associations with autoimmune-type chronic active hepatitis: identification of B8-DRw3 haplotype by family studies. Gastroenterology. 1980;79:95–98. doi: 10.1016/0016-5085(80)90080-3. [DOI] [PubMed] [Google Scholar]
- 22.Krawitt E.L., Kilby A.E., Albertini R.J., Schanfield M.S., Chastenay B.F., Harper P.C., Mickey R.M., McAuliffe T.L. Immunogenetic studies of autoimmune chronic active hepatitis: HLA, immunoglobulin allotypes and autoantibodies. Hepatology. 1987;7:1305–1310. doi: 10.1002/hep.1840070621. [DOI] [PubMed] [Google Scholar]
- 23.Donaldson P.T., Doherty D.G., Hayllar K.M., McFarlane I.G., Johnson P.J., Williams R. Susceptibility to autoimmune chronic active hepatitis: human leukocyte antigens DR4 and A1-B8-DR3 are independent risk factors. Hepatology. 1991;13:701–706. doi: 10.1002/hep.1840130415. [DOI] [PubMed] [Google Scholar]
- 24.Seki T., Kiyosawa K., Inoko H., Ota M. Association of autoimmune hepatitis with HLA-Bw54 and DR4 in Japanese patients. Hepatology. 1990;12:1300–1304. doi: 10.1002/hep.1840120609. [DOI] [PubMed] [Google Scholar]
- 25.Umemura T., Katsuyama Y., Yoshizawa K., Kimura T., Joshita S., Komatsu M., Matsumoto A., Tanaka E., Ota M. Human leukocyte antigen class II haplotypes affect clinical characteristics and progression of type 1 autoimmune hepatitis in Japan. PloS One. 2014;9 doi: 10.1371/journal.pone.0100565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doherty D.G., Donaldson P.T., Underhill J.A., Farrant J.M., Duthie A., Mieli-Vergani G., McFarlane I.G., Johnson P.J., Eddleston A.L., Mowat A.P. Allelic sequence variation in the HLA class II genes and proteins in patients with autoimmune hepatitis. Hepatology. 1994;19:609–615. doi: 10.1002/hep.1840190311. [DOI] [PubMed] [Google Scholar]
- 27.Strettell M.D., Donaldson P.T., Thomson L.J., Santrach P.J., Moore S.B., Czaja A.J., Williams R. Allelic basis for HLA-encoded susceptibility to type 1 autoimmune hepatitis. Gastroenterology. 1997;112:2028–2035. doi: 10.1053/gast.1997.v112.pm9178696. [DOI] [PubMed] [Google Scholar]
- 28.de Boer Y.S., van Gerven N.M.F., Zwiers A., Verwer B.J., van Hoek B., van Erpecum K.J., Beuers U., van Buuren H.R., Drenth J.P.H., den Ouden J.W., Verdonk R.C., Koek G.H., Brouwer J.T., Guichelaar M.M.J., Vrolijk J.M., Kraal G., Mulder C.J.J., van Nieuwkerk C.M.J., Fischer J., Berg T., Stickel F., Sarrazin C., Schramm C., Lohse A.W., Weiler-Normann C., Lerch M.M., Nauck M., Völzke H., Homuth G., Bloemena E., Verspaget H.W., Kumar V., Zhernakova A., Wijmenga C., Franke L., Bouma G. Genome-wide association study identifies variants associated with autoimmune hepatitis type 1. Gastroenterology. 2014;147:443–452. doi: 10.1053/j.gastro.2014.04.022. e5. [DOI] [PubMed] [Google Scholar]
- 29.Donaldson P.T. Genetics in autoimmune hepatitis. Semin. Liver Dis. 2002;22:353–364. doi: 10.1055/s-2002-35705. [DOI] [PubMed] [Google Scholar]
- 30.Donaldson P.T. Genetics of liver disease: immunogenetics and disease pathogenesis. Gut. 2004;53:599–608. doi: 10.1136/gut.2003.031732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Czaja A.J., Carpenter H.A., Santrach P.J., Moore S.B. Significance of HLA DR4 in type 1 autoimmune hepatitis. Gastroenterology. 1993;105:1502–1507. doi: 10.1016/0016-5085(93)90157-8. [DOI] [PubMed] [Google Scholar]
- 32.van Gerven N.M.F., de Boer Y.S., Zwiers A., Verwer B.J., Drenth J.P.H., van Hoek B., van Erpecum K.J., Beuers U., van Buuren H.R., den Ouden J.W., Verdonk R.C., Koek G.H., Brouwer J.T., Guichelaar M.M.J., Vrolijk J.M., Coenraad M.J., Kraal G., Mulder C.J.J., van Nieuwkerk C.M.J., Bloemena E., Verspaget H.W., Kumar V., Zhernakova A., Wijmenga C., Franke L., Bouma G. Dutch Autoimmune Hepatitis Study Group, HLA-DRB1*03:01 and HLA-DRB1*04:01 modify the presentation and outcome in autoimmune hepatitis type-1. Gene Immun. 2015;16:247–252. doi: 10.1038/gene.2014.82. [DOI] [PubMed] [Google Scholar]
- 33.Czaja A.J., Donaldson P.T. Genetic susceptibilities for immune expression and liver cell injury in autoimmune hepatitis. Immunol. Rev. 2000;174:250–259. doi: 10.1034/j.1600-0528.2002.017401.x. [DOI] [PubMed] [Google Scholar]
- 34.Pando M., Larriba J., Fernandez G.C., Fainboim H., Ciocca M., Ramonet M., Badia I., Daruich J., Findor J., Tanno H., Cañero-Velasco C., Fainboim L. Pediatric and adult forms of type I autoimmune hepatitis in Argentina: evidence for differential genetic predisposition. Hepatology. 1999;30:1374–1380. doi: 10.1002/hep.510300611. [DOI] [PubMed] [Google Scholar]
- 35.Czaja A.J., Souto E.O., Bittencourt P.L., Cancado E.L.R., Porta G., Goldberg A.C., Donaldson P.T. Clinical distinctions and pathogenic implications of type 1 autoimmune hepatitis in Brazil and the United States. J. Hepatol. 2002;37:302–308. doi: 10.1016/s0168-8278(02)00182-4. [DOI] [PubMed] [Google Scholar]
- 36.Czaja A.J., Carpenter H.A., Moore S.B. HLA DRB1*13 as a risk factor for type 1 autoimmune hepatitis in North American patients. Dig. Dis. Sci. 2008;53:522–528. doi: 10.1007/s10620-007-9859-4. [DOI] [PubMed] [Google Scholar]
- 37.Goldberg A.C., Bittencourt P.L., Mougin B., Cançado E.L., Porta G., Carrilho F., Kalil J. Analysis of HLA haplotypes in autoimmune hepatitis type 1: identifying the major susceptibility locus. Hum. Immunol. 2001;62:165–169. doi: 10.1016/s0198-8859(00)00234-2. [DOI] [PubMed] [Google Scholar]
- 38.Fortes M. del P., Machado I.V., Gil G., Fernández-Mestre M., Dagher L., León R.V., Bianco N.E., Tassinari P. Genetic contribution of major histocompatibility complex class II region to type 1 autoimmune hepatitis susceptibility in Venezuela. Liver Int. 2007;27:1409–1416. doi: 10.1111/j.1478-3231.2007.01581.x. [DOI] [PubMed] [Google Scholar]
- 39.Fainboim L., Marcos Y., Pando M., Capucchio M., Reyes G.B., Galoppo C., Badía I., Remondino G., Ciocca M., Ramonet M. Chronic active autoimmune hepatitis in children. Strong association with a particular HLA-DR6 (DRB1*1301) haplotype. Hum. Immunol. 1994;41:146–150. doi: 10.1016/0198-8859(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 40.Umemura T., Ota M. Genetic factors affect the etiology, clinical characteristics and outcome of autoimmune hepatitis. Clin J Gastroenterol. 2015;8:360–366. doi: 10.1007/s12328-015-0620-9. [DOI] [PubMed] [Google Scholar]
- 41.Seki T., Ota M., Furuta S., Fukushima H., Kondo T., Hino K., Mizuki N., Ando A., Tsuji K., Inoko H. HLA class II molecules and autoimmune hepatitis susceptibility in Japanese patients. Gastroenterology. 1992;103:1041–1047. doi: 10.1016/0016-5085(92)90041-v. [DOI] [PubMed] [Google Scholar]
- 42.Furumoto Y., Asano T., Sugita T., Abe H., Chuganji Y., Fujiki K., Sakata A., Aizawa Y. Evaluation of the role of HLA-DR antigens in Japanese type 1 autoimmune hepatitis. BMC Gastroenterol. 2015;15:144. doi: 10.1186/s12876-015-0360-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oka S., Furukawa H., Yasunami M., Kawasaki A., Nakamura H., Nakamura M., Komori A., Abiru S., Nagaoka S., Hashimoto S., Naganuma A., Naeshiro N., Yoshizawa K., Yamashita H., Ario K., Ohta H., Sakai H., Yabuuchi I., Takahashi A., Abe K., Yatsuhashi H., Tohma S., Ohira H., Tsuchiya N., Migita K. HLA-DRB1 and DQB1 alleles in Japanese type 1 autoimmune hepatitis: the predisposing role of the DR4/DR8 heterozygous genotype. PloS One. 2017;12 doi: 10.1371/journal.pone.0187325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amarapurkar D.N., Patel N.D., Amarapurkar A.D., Kankonkar S.R. HLA genotyping in type-I autoimmune hepatitis in Western India. J. Assoc. Phys. India. 2003;51:967–969. [PubMed] [Google Scholar]
- 45.Baharlou R., Faghihi-Kashani A., Faraji F., Najafi-Samei M., Setareh M., Zamani F., Tajik N. HLA-DRB1 alleles of susceptibility and protection in Iranians with autoimmune hepatitis. Hum. Immunol. 2016;77:330–335. doi: 10.1016/j.humimm.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Ma Y., Bogdanos D.P., Hussain M.J., Underhill J., Bansal S., Longhi M.S., Cheeseman P., Mieli-Vergani G., Vergani D. Polyclonal T-cell responses to cytochrome P450IID6 are associated with disease activity in autoimmune hepatitis type 2. Gastroenterology. 2006;130:868–882. doi: 10.1053/j.gastro.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 47.Underhill J., Ma Y., Bogdanos D., Cheeseman P., Mieli-Vergani G., Vergani D. Different immunogenetic background in autoimmune hepatitis type 1, type 2 and autoimmune sclerosing cholangitis. Journal of Hepatology - J HEPATOL. 2002;36 doi: 10.1016/S0168-8278(02)80564-5. 156–156. [DOI] [Google Scholar]
- 48.Elfaramawy A.A.M., Elhossiny R.M., Abbas A.A., Aziz H.M.A. HLA-DRB1 as a risk factor in children with autoimmune hepatitis and its relation to hepatitis A infection. Ital. J. Pediatr. 2010;36:73. doi: 10.1186/1824-7288-36-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czaja A.J., Strettell M.D., Thomson L.J., Santrach P.J., Moore S.B., Donaldson P.T., Williams R. Associations between alleles of the major histocompatibility complex and type 1 autoimmune hepatitis. Hepatology. 1997;25:317–323. doi: 10.1002/hep.510250211. [DOI] [PubMed] [Google Scholar]
- 50.Czaja A.J., Cookson S., Constantini P.K., Clare M., Underhill J.A., Donaldson P.T. Cytokine polymorphisms associated with clinical features and treatment outcome in type 1 autoimmune hepatitis. Gastroenterology. 1999;117:645–652. doi: 10.1016/s0016-5085(99)70458-0. [DOI] [PubMed] [Google Scholar]
- 51.Cookson S., Constantini P.K., Clare M., Underhill J.A., Bernal W., Czaja A.J., Donaldson P.T. Frequency and nature of cytokine gene polymorphisms in type 1 autoimmune hepatitis. Hepatology. 1999;30:851–856. doi: 10.1002/hep.510300412. [DOI] [PubMed] [Google Scholar]
- 52.Agarwal K., Czaja A.J., Jones D.E., Donaldson P.T. Cytotoxic T lymphocyte antigen-4 (CTLA-4) gene polymorphisms and susceptibility to type 1 autoimmune hepatitis. Hepatology. 2000;31:49–53. doi: 10.1002/hep.510310110. [DOI] [PubMed] [Google Scholar]
- 53.Djilali-Saiah I., Ouellette P., Caillat-Zucman S., Debray D., Kohn J.I., Alvarez F. CTLA-4/CD 28 region polymorphisms in children from families with autoimmune hepatitis. Hum. Immunol. 2001;62:1356–1362. doi: 10.1016/s0198-8859(01)00344-5. [DOI] [PubMed] [Google Scholar]
- 54.Bittencourt P.L., Palácios S.A., Cançado E.L.R., Porta G., Carrilho F.J., Laudanna A.A., Kalil J., Goldberg A.C. Cytotoxic T lymphocyte antigen-4 gene polymorphisms do not confer susceptibility to autoimmune hepatitis types 1 and 2 in Brazil. Am. J. Gastroenterol. 2003;98:1616–1620. doi: 10.1111/j.1572-0241.2003.07525.x. [DOI] [PubMed] [Google Scholar]
- 55.Umemura T., Ota M., Yoshizawa K., Katsuyama Y., Ichijo T., Tanaka E., Kiyosawa K. Association of cytotoxic T-lymphocyte antigen 4 gene polymorphisms with type 1 autoimmune hepatitis in Japanese. Hepatol. Res. 2008;38:689–695. doi: 10.1111/j.1872-034X.2008.00337.x. [DOI] [PubMed] [Google Scholar]
- 56.Vogel A., Strassburg C.P., Manns M.P. Genetic association of vitamin D receptor polymorphisms with primary biliary cirrhosis and autoimmune hepatitis. Hepatology. 2002;35:126–131. doi: 10.1053/jhep.2002.30084. [DOI] [PubMed] [Google Scholar]
- 57.Fan L., Tu X., Zhu Y., Zhou L., Pfeiffer T., Feltens R., Stoecker W., Zhong R. Genetic association of vitamin D receptor polymorphisms with autoimmune hepatitis and primary biliary cirrhosis in the Chinese. J. Gastroenterol. Hepatol. 2005;20:249–255. doi: 10.1111/j.1440-1746.2005.03532.x. [DOI] [PubMed] [Google Scholar]
- 58.Panierakis C., Goulielmos G., Mamoulakis D., Petraki E., Papavasiliou E., Galanakis E. Vitamin D receptor gene polymorphisms and susceptibility to type 1 diabetes in Crete, Greece. Clin. Immunol. 2009;133:276–281. doi: 10.1016/j.clim.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 59.Hiraide A., Imazeki F., Yokosuka O., Kanda T., Kojima H., Fukai K., Suzuki Y., Hata A., Saisho H. Fas polymorphisms influence susceptibility to autoimmune hepatitis. Am. J. Gastroenterol. 2005;100:1322–1329. doi: 10.1111/j.1572-0241.2005.41053.x. [DOI] [PubMed] [Google Scholar]
- 60.Paladino N., Flores A.C., Fainboim H., Schroder T., Cuarterolo M., Lezama C., Ballerga E.G., Levi D., Tanno H., Costanzo G., Arruvito L., Fainboim L. The most severe forms of type I autoimmune hepatitis are associated with genetically determined levels of TGF-beta1. Clin. Immunol. 2010;134:305–312. doi: 10.1016/j.clim.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 61.Chen S., Zhao W., Tan W., Luo X., Dan Y., You Z., Kuang X., Wang Y., Deng G. Association of TBX21 promoter polymorphisms with type 1 autoimmune hepatitis in a Chinese population. Hum. Immunol. 2011;72:69–73. doi: 10.1016/j.humimm.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 62.Migita K., Nakamura M., Abiru S., Jiuchi Y., Nagaoka S., Komori A., Hashimoto S., Bekki S., Yamasaki K., Komatsu T., Shimada M., Kouno H., Hijioka T., Kohjima M., Nakamuta M., Kato M., Yoshizawa K., Ohta H., Nakamura Y., Takezaki E., Nishimura H., Sato T., Ario K., Hirashima N., Oohara Y., Naganuma A., Muro T., Sakai H., Mita E., Sugi K., Yamashita H., Makita F., Yatsuhashi H., Ishibashi H., Yasunami M. Association of STAT4 polymorphisms with susceptibility to type-1 autoimmune hepatitis in the Japanese population. PloS One. 2013;8 doi: 10.1371/journal.pone.0071382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y., He X., Schembri-King J., Jakes S., Hayashi J. Cloning and characterization of human Lnk, an adaptor protein with pleckstrin homology and Src homology 2 domains that can inhibit T cell activation. J. Immunol. 2000;164:5199–5206. doi: 10.4049/jimmunol.164.10.5199. [DOI] [PubMed] [Google Scholar]
- 64.Higuchi T., Oka S., Furukawa H., Nakamura M., Komori A., Abiru S., Nagaoka S., Hashimoto S., Naganuma A., Naeshiro N., Yoshizawa K., Shimada M., Nishimura H., Tomizawa M., Kikuchi M., Makita F., Yamashita H., Ario K., Yatsuhashi H., Tohma S., Kawasaki A., Ohira H., Tsuchiya N., Migita K. Association of a single nucleotide polymorphism upstream of ICOS with Japanese autoimmune hepatitis type 1. J. Hum. Genet. 2017;62:481–484. doi: 10.1038/jhg.2016.155. [DOI] [PubMed] [Google Scholar]
- 65.Umemura T., Joshita S., Hamano H., Yoshizawa K., Kawa S., Tanaka E., Ota M. Association of autoimmune hepatitis with Src homology 2 adaptor protein 3 gene polymorphisms in Japanese patients. J. Hum. Genet. 2017;62:963–967. doi: 10.1038/jhg.2017.74. [DOI] [PubMed] [Google Scholar]
- 66.Assis D.N., Takahashi H., Leng L., Zeniya M., Boyer J.L., Bucala R. A macrophage migration inhibitory factor polymorphism is associated with autoimmune hepatitis severity in US and Japanese patients. Dig. Dis. Sci. 2016;61:3506–3512. doi: 10.1007/s10620-016-4322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Umemura T., Joshita S., Yamazaki T., Komatsu M., Katsuyama Y., Yoshizawa K., Tanaka E., Ota M. Genetic association of PTPN22 polymorphisms with autoimmune hepatitis and primary biliary cholangitis in Japan. Sci. Rep. 2016;6:29770. doi: 10.1038/srep29770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Higuchi T., Oka S., Furukawa H., Nakamura M., Komori A., Abiru S., Hashimoto S., Shimada M., Yoshizawa K., Kouno H., Naganuma A., Ario K., Kaneyoshi T., Yamashita H., Takahashi H., Makita F., Yatsuhashi H., Ohira H., Migita K. Role of deleterious single nucleotide variants in the coding regions of TNFAIP3 for Japanese autoimmune hepatitis with cirrhosis. Sci. Rep. 2019;9:7925. doi: 10.1038/s41598-019-44524-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oka S., Higuchi T., Furukawa H., Nakamura M., Komori A., Abiru S., Nagaoka S., Hashimoto S., Naganuma A., Naeshiro N., Yoshizawa K., Shimada M., Nishimura H., Tomizawa M., Kikuchi M., Makita F., Yamashita H., Ario K., Yatsuhashi H., Tohma S., Kawasaki A., Tsuchiya N., Migita K. Association of a single nucleotide polymorphism in TNIP1 with type-1 autoimmune hepatitis in the Japanese population. J. Hum. Genet. 2018;63:739–744. doi: 10.1038/s10038-018-0440-0. [DOI] [PubMed] [Google Scholar]
- 70.Mansour A.I., Behairy O.G., Abd Almonaem E.R., Abd-Rabuh R.M., Ahmed I.A.E. Association of interleukin (IL)-4 variable number of tandem repeats (VNTRs) and IL-4-590 promoter polymorphisms with susceptibility to pediatric autoimmune hepatitis type 1. Cytokine. 2018;110:243–247. doi: 10.1016/j.cyto.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 71.Podhorzer A., Paladino N., Cuarterolo M.L., Fainboim H.A., Paz S., Theiler G., Capucchio M., López S.I., Machicote A., Montal S., Podesta G., Fainboim L. The early onset of type 1 autoimmune hepatitis has a strong genetic influence: role of HLA and KIR genes. Gene Immun. 2016;17:187–192. doi: 10.1038/gene.2016.7. [DOI] [PubMed] [Google Scholar]
- 72.Invernizzi P., Pasini S., Selmi C., Gershwin M.E., Podda M. Female predominance and X chromosome defects in autoimmune diseases. J. Autoimmun. 2009;33:12–16. doi: 10.1016/j.jaut.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 73.Invernizzi P., Miozzo M., Battezzati P.M., Bianchi I., Grati F.R., Simoni G., Selmi C., Watnik M., Gershwin M.E., Podda M. Frequency of monosomy X in women with primary biliary cirrhosis. Lancet. 2004;363:533–535. doi: 10.1016/S0140-6736(04)15541-4. [DOI] [PubMed] [Google Scholar]
- 74.Miozzo M., Selmi C., Gentilin B., Grati F.R., Sirchia S., Oertelt S., Zuin M., Gershwin M.E., Podda M., Invernizzi P. Preferential X chromosome loss but random inactivation characterize primary biliary cirrhosis. Hepatology. 2007;46:456–462. doi: 10.1002/hep.21696. [DOI] [PubMed] [Google Scholar]
- 75.Czaja A.J., dos Santos R.M., Porto A., Santrach P.J., Moore S.B. Immune phenotype of chronic liver disease. Dig. Dis. Sci. 1998;43:2149–2155. doi: 10.1023/a:1018836004279. [DOI] [PubMed] [Google Scholar]
- 76.McFarlane I.G., Heneghan M.A. Autoimmunity and the female liver. Hepatol. Res. 2004;28:171–176. doi: 10.1016/j.hepres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 77.Whitacre C.C., Reingold S.C., O'Looney P.A. A gender gap in autoimmunity. Science. 1999;283:1277–1278. doi: 10.1126/science.283.5406.1277. [DOI] [PubMed] [Google Scholar]
- 78.Vento S., Garofano T., Dolci L., Di Perri G., Concia E., Bassetti D. Identification of hepatitis A virus as a trigger for autoimmune chronic hepatitis type 1 in susceptible individuals. Lancet. 1991;337:1183–1187. doi: 10.1016/0140-6736(91)92858-Y. [DOI] [PubMed] [Google Scholar]
- 79.Huppertz H.-I., Triechel U., Gassel A.M., Jeschke R., Meyer K.-H. Zum Büschenfelde, Autoimmune hepatitis following hepatitis A virus infection. J. Hepatol. 1995;23:204–208. doi: 10.1016/0168-8278(95)80336-X. [DOI] [PubMed] [Google Scholar]
- 80.Hilzenrat N., Zilberman D., Klein T., Zur B., Sikuler E. Autoimmune hepatitis in a genetically susceptible patient: is it triggered by acute viral hepatitis A? Dig. Dis. Sci. 1999;44:1950–1952. doi: 10.1023/a:1026645629103. [DOI] [PubMed] [Google Scholar]
- 81.Skoog S.M., Rivard R.E., Batts K.P., Smith C.I. Autoimmune hepatitis preceded by acute hepatitis A infection. Am. J. Gastroenterol. 2002;97:1568–1569. doi: 10.1016/S0002-9270(02)04107-2. [DOI] [PubMed] [Google Scholar]
- 82.Grünhage F., Spengler U., Fischer H.P., Sauerbruch T. Autoimmune hepatitis – sequel of a relapsing hepatitis A in a 75-year-old woman. DIG. 2004;70:187–191. doi: 10.1159/000082253. [DOI] [PubMed] [Google Scholar]
- 83.Tanaka H., Tujioka H., Ueda H., Hamagami H., Kida Y., Ichinose M. Autoimmune hepatitis triggered by acute hepatitis A. World J. Gastroenterol. 2005;11:6069–6071. doi: 10.3748/wjg.v11.i38.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tabak F., Özdemir F., Tabak O., Erer B., Tahan V., Ozaras R. Autoimmune hepatitis induced by the prolonged hepatitis A virus infection. Ann. Hepatol. 2008;7:177–179. doi: 10.1016/S1665-2681(19)31878-2. [DOI] [PubMed] [Google Scholar]
- 85.Kim Y.D., Kim K.-A., Rou W.S., Lee J.S., Song T.J., Bae W.K., Kim N.-H. A case of autoimmune hepatitis following acute hepatitis A. Korean J. Gastroenterol. 2011;57:315–318. doi: 10.4166/kjg.2011.57.5.315. [DOI] [PubMed] [Google Scholar]
- 86.Singh G., Palaniappan S., Rotimi O., Hamlin P.J. Autoimmune hepatitis triggered by hepatitis A. Gut. 2007;56:304. doi: 10.1136/gut.2006.111864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grave A., Juel J., Vyberg M., Olesen S.S., Hansen J.B. Autoimmune hepatitis preceded by hepatitis A, Ugeskr Laeger. 2015;177:V12140669. [PubMed] [Google Scholar]
- 88.Subramanian S.K., Patel J.M., Younes M., Nevah Rubin M.I. Postinfectious autoimmune hepatitis-induced liver failure: a consequence of hepatitis A virus infection. ACG Case Rep J. 2020;7 doi: 10.14309/crj.0000000000000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lenzi M., Ballardini G., Fusconi M., Cassani F., Selleri L., Volta U., Zauli D., Bianchi F.B. Type 2 autoimmune hepatitis and hepatitis C virus infection. Lancet. 1990;335:258–259. doi: 10.1016/0140-6736(90)90070-l. [DOI] [PubMed] [Google Scholar]
- 90.Michel G., Ritter A., Gerken G., Meyer zum Büschenfelde K.H., Decker R., Manns M.P. Anti-GOR and hepatitis C virus in autoimmune liver diseases. Lancet. 1992;339:267–269. doi: 10.1016/0140-6736(92)91332-3. [DOI] [PubMed] [Google Scholar]
- 91.Lunel F., Abuaf N., Frangeul L., Grippon P., Perrin M., Le Coz Y., Valla D., Borotto E., Yamamoto A.M., Huraux J.M. Liver/kidney microsome antibody type 1 and hepatitis C virus infection. Hepatology. 1992;16:630–636. doi: 10.1002/hep.1840160304. [DOI] [PubMed] [Google Scholar]
- 92.Dalekos G.N., Wedemeyer H., Obermayer-Straub P., Kayser A., Barut A., Frank H., Manns M.P. Epitope mapping of cytochrome P4502D6 autoantigen in patients with chronic hepatitis C during alpha-interferon treatment. J. Hepatol. 1999;30:366–375. doi: 10.1016/s0168-8278(99)80092-0. [DOI] [PubMed] [Google Scholar]
- 93.Dalekos G.N., Makri E., Loges S., Obermayer-Straub P., Zachou K., Tsikrikas T., Schmidt E., Papadamou G., Manns M.P. Increased incidence of anti-LKM autoantibodies in a consecutive cohort of hepatitis C patients from central Greece. Eur. J. Gastroenterol. Hepatol. 2002;14:35–42. doi: 10.1097/00042737-200201000-00007. [DOI] [PubMed] [Google Scholar]
- 94.Dalekos G.N., Obermayer-Straub P., Bartels M., Maeda T., Kayser A., Braun S., Loges S., Schmidt E., Gershwin M.E., Manns M.P. Cytochrome P450 2A6: a new hepatic autoantigen in patients with chronic hepatitis C virus infection. J. Hepatol. 2003;39:800–806. doi: 10.1016/s0168-8278(03)00356-8. [DOI] [PubMed] [Google Scholar]
- 95.Mackie F.D., Peakman M., Yun M., Sallie R., Smith H., Davies E.T., Mieli-Vergani G., Vergani D. Primary and secondary liver/kidney microsomal autoantibody response following infection with hepatitis C virus. Gastroenterology. 1994;106:1672–1675. doi: 10.1016/0016-5085(94)90426-x. [DOI] [PubMed] [Google Scholar]
- 96.Vergani D., Mieli-Vergani G. Autoimmune manifestations in viral hepatitis. Semin. Immunopathol. 2013;35:73–85. doi: 10.1007/s00281-012-0328-6. [DOI] [PubMed] [Google Scholar]
- 97.Kerkar N., Choudhuri K., Ma Y., Mahmoud A., Bogdanos D.P., Muratori L., Bianchi F., Williams R., Mieli-Vergani G., Vergani D. Cytochrome P4502D6(193-212): a new immunodominant epitope and target of virus/self cross-reactivity in liver kidney microsomal autoantibody type 1-positive liver disease. J. Immunol. 2003;170:1481–1489. doi: 10.4049/jimmunol.170.3.1481. [DOI] [PubMed] [Google Scholar]
- 98.Bogdanos D.P., Dalekos G.N. Enzymes as target antigens of liver-specific autoimmunity: the case of cytochromes P450s. Curr. Med. Chem. 2008;15:2285–2292. doi: 10.2174/092986708785747508. [DOI] [PubMed] [Google Scholar]
- 99.Lapierre P., Djilali-Saiah I., Vitozzi S., Alvarez F. A murine model of type 2 autoimmune hepatitis: xenoimmunization with human antigens. Hepatology. 2004;39:1066–1074. doi: 10.1002/hep.20109. [DOI] [PubMed] [Google Scholar]
- 100.Lapierre P., Béland K., Martin C., Alvarez F., Alvarez F. Forkhead box p3+ regulatory T cell underlies male resistance to experimental type 2 autoimmune hepatitis. Hepatology. 2010;51:1789–1798. doi: 10.1002/hep.23536. [DOI] [PubMed] [Google Scholar]
- 101.Gregorio G.V., Jones H., Choudhuri K., Vegnente A., Bortolotti F., Mieli-Vergani G., Vergani D. Autoantibody prevalence in chronic hepatitis B virus infection: effect in interferon alfa. Hepatology. 1996;24:520–523. doi: 10.1002/hep.510240309. [DOI] [PubMed] [Google Scholar]
- 102.Dalekos G.N., Zachou K., Liaskos C., Gatselis N. Autoantibodies and defined target autoantigens in autoimmune hepatitis: an overview. Eur. J. Intern. Med. 2002;13:293–303. doi: 10.1016/s0953-6205(02)00089-4. [DOI] [PubMed] [Google Scholar]
- 103.Gatselis N.K., Georgiadou S.P., Koukoulis G.K., Tassopoulos N., Zachou K., Liaskos C., Hatzakis A., Dalekos G.N. Clinical significance of organ- and non-organ-specific autoantibodies on the response to anti-viral treatment of patients with chronic hepatitis C. Aliment. Pharmacol. Ther. 2006;24:1563–1573. doi: 10.1111/j.1365-2036.2006.03165.x. [DOI] [PubMed] [Google Scholar]
- 104.Rigopoulou E.I., Zachou K., Gatselis N., Koukoulis G.K., Dalekos G.N. Autoimmune hepatitis in patients with chronic HBV and HCV infections: patterns of clinical characteristics, disease progression and outcome. Ann. Hepatol. 2013;13:127–135. doi: 10.1016/S1665-2681(19)30914-7. [DOI] [PubMed] [Google Scholar]
- 105.Crivelli O., Lavarini C., Chiaberge E., Amoroso A., Farci P., Negro F., Rizzetto M. Microsomal autoantibodies in chronic infection with the HBsAg associated delta (delta) agent. Clin. Exp. Immunol. 1983;54:232–238. [PMC free article] [PubMed] [Google Scholar]
- 106.Zachou K., Yurdaydìn C., Drebber U., Schlaphoff V., Dienes H.-P., Manns M., Wedemeyer H., Dalekos G.N. P0576 : impact of organ and non-organ-specific autoantibodies on the treatment outcome of patients with hepatitis D virus infection. J. Hepatol. 2015;62:S531–S532. doi: 10.1016/S0168-8278(15)30782-0. [DOI] [Google Scholar]
- 107.Strassburg C.P., Obermayer-Straub P., Alex B., Durazzo M., Rizzetto M., Tukey R.H., Manns M.P. Autoantibodies against glucuronosyltransferases differ between viral hepatitis and autoimmune hepatitis. Gastroenterology. 1996;111:1576–1586. doi: 10.1016/s0016-5085(96)70020-3. [DOI] [PubMed] [Google Scholar]
- 108.Terziroli Beretta-Piccoli B., Ripellino P., Gobbi C., Cerny A., Baserga A., Di Bartolomeo C., Bihl F., Deleonardi G., Melidona L., Grondona A.G., Mieli-Vergani G., Vergani D., Muratori L. Autoimmune liver disease serology in acute hepatitis E virus infection. J. Autoimmun. 2018;94:1–6. doi: 10.1016/j.jaut.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 109.Elfert K.A., Qasim H.M., Faisal M.M., Elghazali A., Siddiqui M.Y.A., Petkar M., Sadik N. Hepatitis E viral association with autoimmune hepatitis: a viral trigger or cross-reactivity. CRG. 2021;15:115–122. doi: 10.1159/000509494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pischke S., Gisa A., Suneetha P.V., Wiegand S.B., Taubert R., Schlue J., Wursthorn K., Bantel H., Raupach R., Bremer B., Zacher B.J., Schmidt R.E., Manns M.P., Rifai K., Witte T., Wedemeyer H. Increased HEV seroprevalence in patients with autoimmune hepatitis. PloS One. 2014;9 doi: 10.1371/journal.pone.0085330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Taubert R., Diestelhorst J., Junge N., Kirstein M.M., Pischke S., Vogel A., Bantel H., Baumann U., Manns M.P., Wedemeyer H., Jaeckel E. Increased seroprevalence of HAV and parvovirus B19 in children and of HEV in adults at diagnosis of autoimmune hepatitis. Sci. Rep. 2018;8:17452. doi: 10.1038/s41598-018-35882-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eder M., Strassl R., Beinhardt S., Stättermayer A., Kozbial K., Lagler H., Holzmann H., Trauner M., Hofer H. High seroprevalence of anti‐hepatitis E antibodies in Austrian patients with autoimmune hepatitis. Liver Int. 2018;39 doi: 10.1111/liv.14005. [DOI] [PubMed] [Google Scholar]
- 113.Aceti A., Mura M.S., Babudieri S., Bacciu S.A. A young woman with hepatitis after a sore throat. Lancet. 1995;346:1603. doi: 10.1016/s0140-6736(95)91932-5. [DOI] [PubMed] [Google Scholar]
- 114.Kojima K., Nagayama R., Hirama S., Maeda T., Takikawa H., Miyake K., Yamanaka M., Shiga J. Epstein-Barr virus infection resembling autoimmune hepatitis with lactate dehydrogenase and alkaline phosphatase anomaly. J. Gastroenterol. 1999;34:706–712. doi: 10.1007/s005350050324. [DOI] [PubMed] [Google Scholar]
- 115.Nobili V., Comparcola D., Sartorelli M.R., Devito R., Marcellini M. Autoimmune Hepatitis type 1 after Epstein-Barr virus infection. Pediatr. Infect. Dis. J. 2003;22:387. doi: 10.1097/01.inf.0000060825.68086.9c. [DOI] [PubMed] [Google Scholar]
- 116.Vento S., Guella L., Mirandola F., Cainelli F., Di Perri G., Solbiati M., Ferraro T., Concia E. Epstein-Barr virus as a trigger for autoimmune hepatitis in susceptible individuals. Lancet. 1995;346:608–609. doi: 10.1016/s0140-6736(95)91438-2. [DOI] [PubMed] [Google Scholar]
- 117.Zellos A., Spoulou V., Roma-Giannikou E., Karentzou O., Dalekos G.N., Theodoridou M. Autoimmune hepatitis type-2 and Epstein-Barr virus infection in a toddler: art of facts or an artifact? Ann. Hepatol. 2013;12:147–151. [PubMed] [Google Scholar]
- 118.Cabibi D. Autoimmune hepatitis following Epstein-Barr virus infection. BMJ Case Rep. 2008;2008 doi: 10.1136/bcr.06.2008.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Björnsson E., Talwalkar J., Treeprasertsuk S., Kamath P.S., Takahashi N., Sanderson S., Neuhauser M., Lindor K. Drug-induced autoimmune hepatitis: clinical characteristics and prognosis. Hepatology. 2010;51:2040–2048. doi: 10.1002/hep.23588. [DOI] [PubMed] [Google Scholar]
- 120.Czaja A.J. Drug-induced autoimmune-like hepatitis. Dig. Dis. Sci. 2011;56:958–976. doi: 10.1007/s10620-011-1611-4. [DOI] [PubMed] [Google Scholar]
- 121.Saitis A., Gatselis N., Zachou K., Dalekos G.N. Use of TNFα antagonists in refractory AIH: revealing the unforeseen. J. Hepatol. 2013;59:197–198. doi: 10.1016/j.jhep.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 122.European Association for the Study of the Liver Electronic address: easloffice@easloffice.eu, clinical practice guideline panel: chair:, panel members, EASL governing board representative:, EASL clinical practice guidelines: drug-induced liver injury. J. Hepatol. 2019;70:1222–1261. doi: 10.1016/j.jhep.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 123.Beaune P., Dansette P.M., Mansuy D., Kiffel L., Finck M., Amar C., Leroux J.P., Homberg J.C. Human anti-endoplasmic reticulum autoantibodies appearing in a drug-induced hepatitis are directed against a human liver cytochrome P-450 that hydroxylates the drug. Proc. Natl. Acad. Sci. U. S. A. 1987;84:551–555. doi: 10.1073/pnas.84.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bourdi M., Larrey D., Nataf J., Bernuau J., Pessayre D., Iwasaki M., Guengerich F.P., Beaune P.H. Anti-liver endoplasmic reticulum autoantibodies are directed against human cytochrome P-450IA2. A specific marker of dihydralazine-induced hepatitis. J. Clin. Invest. 1990;85:1967–1973. doi: 10.1172/JCI114660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weiler-Normann C., Schramm C. Drug induced liver injury and its relationship to autoimmune hepatitis. J. Hepatol. 2011;55:747–749. doi: 10.1016/j.jhep.2011.02.024. [DOI] [PubMed] [Google Scholar]
- 126.Paronetto F. Immunologic reactions in alcoholic liver disease. Semin. Liver Dis. 1993;13:183–195. doi: 10.1055/s-2007-1007348. [DOI] [PubMed] [Google Scholar]
- 127.Xu D., Thiele G.M., Beckenhauer J.L., Klassen L.W., Sorrell M.F., Tuma D.J. Detection of circulating antibodies to malondialdehyde-acetaldehyde adducts in ethanol-fed rats. Gastroenterology. 1998;115:686–692. doi: 10.1016/S0016-5085(98)70148-9. [DOI] [PubMed] [Google Scholar]
- 128.Ngu J.H., Gearry R.B., Frampton C.M., Stedman C.A.M. Autoimmune hepatitis: the role of environmental risk factors: a population-based study. Hepatol Int. 2013;7:869–875. doi: 10.1007/s12072-013-9448-x. [DOI] [PubMed] [Google Scholar]
- 129.Rigopoulou E.I., Gatselis N., Arvaniti P., Koukoulis G.K., Dalekos G.N. Alcoholic liver disease and autoimmune hepatitis: sometimes a closer look under the surface is needed. Eur. J. Intern. Med. 2021;85:86–91. doi: 10.1016/j.ejim.2020.12.024. [DOI] [PubMed] [Google Scholar]
- 130.Tiegs G., Hentschel J., Wendel A. A T cell-dependent experimental liver injury in mice inducible by concanavalin A. J. Clin. Invest. 1992;90:196–203. doi: 10.1172/JCI115836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ala A., Stanca C.M., Bu-Ghanim M., Ahmado I., Branch A.D., Schiano T.D., Odin J.A., Bach N. Increased prevalence of primary biliary cirrhosis near Superfund toxic waste sites. Hepatology. 2006;43:525–531. doi: 10.1002/hep.21076. [DOI] [PubMed] [Google Scholar]
- 132.Gatselis N.K., Zachou K., Lygoura V., Azariadis K., Arvaniti P., Spyrou E., Papadamou G., Koukoulis G.K., Dalekos G.N., Rigopoulou E.I. Geoepidemiology, clinical manifestations and outcome of primary biliary cholangitis in Greece. Eur. J. Intern. Med. 2017;42:81–88. doi: 10.1016/j.ejim.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 133.Gatselis N.K., Dalekos G.N. Molecular diagnostic testing for primary biliary cholangitis. Expert Rev. Mol. Diagn. 2016;16:1001–1010. doi: 10.1080/14737159.2016.1217159. [DOI] [PubMed] [Google Scholar]
- 134.Amano K., Leung P.S.C., Rieger R., Quan C., Wang X., Marik J., Suen Y.F., Kurth M.J., Nantz M.H., Ansari A.A., Lam K.S., Zeniya M., Matsuura E., Coppel R.L., Gershwin M.E. Chemical xenobiotics and mitochondrial autoantigens in primary biliary cirrhosis: identification of antibodies against a common environmental, cosmetic, and food additive, 2-octynoic acid. J. Immunol. 2005;174:5874–5883. doi: 10.4049/jimmunol.174.9.5874. [DOI] [PubMed] [Google Scholar]
- 135.Gilbert K.M. Xenobiotic exposure and autoimmune hepatitis. Hepat Res Treat. 2010;2010:248157. doi: 10.1155/2010/248157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zare-Shahabadi A., Renaudineau Y., Rezaei N. MicroRNAs and multiple sclerosis: from physiopathology toward therapy. Expert Opin. Ther. Targets. 2013;17:1497–1507. doi: 10.1517/14728222.2013.838219. [DOI] [PubMed] [Google Scholar]
- 137.Migita K., Komori A., Kozuru H., Jiuchi Y., Nakamura M., Yasunami M., Furukawa H., Abiru S., Yamasaki K., Nagaoka S., Hashimoto S., Bekki S., Kamitsukasa H., Nakamura Y., Ohta H., Shimada M., Takahashi H., Mita E., Hijioka T., Yamashita H., Kouno H., Nakamuta M., Ario K., Muro T., Sakai H., Sugi K., Nishimura H., Yoshizawa K., Sato T., Naganuma A., Komatsu T., Oohara Y., Makita F., Tomizawa M., Yatsuhashi H. Circulating microRNA profiles in patients with type-1 autoimmune hepatitis. PloS One. 2015;10 doi: 10.1371/journal.pone.0136908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yamaura Y., Tatsumi N., Takagi S., Tokumitsu S., Fukami T., Tajiri K., Minemura M., Yokoi T., Nakajima M. Serum microRNA profiles in patients with chronic hepatitis B, chronic hepatitis C, primary biliary cirrhosis, autoimmune hepatitis, nonalcoholic steatohepatitis, or drug-induced liver injury. Clin. Biochem. 2017;50:1034–1039. doi: 10.1016/j.clinbiochem.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 139.Chen L., Lu F., Chen D., Wu J., Hu E., Xu L., Zheng M., Li H., Huang Y., Jin X., Gong Y., Lin Z., Wang X., Chen Y. BMSCs-derived miR-223-containing exosomes contribute to liver protection in experimental autoimmune hepatitis. Mol. Immunol. 2018;93:38–46. doi: 10.1016/j.molimm.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 140.Su K., Wang Q., Qi L., Hua D., Tao J., Mangan C.J., Lou Y., Li L. MicroRNA-674-5p/5-LO axis involved in autoimmune reaction of Concanavalin A-induced acute mouse liver injury. Toxicol. Lett. 2016;258:101–107. doi: 10.1016/j.toxlet.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 141.Xia G., Wu S., Wang X., Fu M. Inhibition of microRNA-155 attenuates concanavalin-A-induced autoimmune hepatitis by regulating Treg/Th17 cell differentiation. Can. J. Physiol. Pharmacol. 2018;96:1293–1300. doi: 10.1139/cjpp-2018-0467. [DOI] [PubMed] [Google Scholar]
- 142.Renaudineau Y., Youinou P. Epigenetics and autoimmunity, with special emphasis on methylation. Keio J. Med. 2011;60:10–16. doi: 10.2302/kjm.60.10. [DOI] [PubMed] [Google Scholar]
- 143.D Rasmussen K., Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pirola C.J., Gianotti T.F., Burgueño A.L., Rey-Funes M., Loidl C.F., Mallardi P., Martino J.S., Castaño G.O., Sookoian S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62:1356–1363. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 145.Pirola C.J., Scian R., Gianotti T.F., Dopazo H., Rohr C., Martino J.S., Castaño G.O., Sookoian S. Epigenetic modifications in the biology of nonalcoholic fatty liver disease: the role of DNA hydroxymethylation and TET proteins. Medicine (Baltim.) 2015;94 doi: 10.1097/MD.0000000000001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lleo A., Zhang W., Zhao M., Tan Y., Bernuzzi F., Zhu B., Liu Q., Tan Q., Malinverno F., Valenti L., Jiang T., Tan L., Liao W., Coppel R., Invernizzi P., Lu Q., Adams D.H., Gershwin M.E. DNA methylation profiling of the X chromosome reveals an aberrant demethylation on CXCR3 promoter in primary biliary cirrhosis. Clin. Epigenet. 2015;7 doi: 10.1186/s13148-015-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Selmi C., Cavaciocchi F., Lleo A., Cheroni C., De Francesco R., Lombardi S.A., De Santis M., Meda F., Raimondo M.G., Crotti C., Folci M., Zammataro L., Mayo M.J., Bach N., Shimoda S., Gordon S.C., Miozzo M., Invernizzi P., Podda M., Scavelli R., Martin M.R., Seldin M.F., Lasalle J.M., Gershwin M.E. Genome-wide analysis of DNA methylation, copy number variation, and gene expression in monozygotic twins discordant for primary biliary cirrhosis. Front. Immunol. 2014;5:128. doi: 10.3389/fimmu.2014.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zachou K, Arvaniti P, Lyberopoulou A, Sevdali E, Speletas M, Ioannou M, Koukoulis G.K, Renaudineau Y, Dalekos G.N. Altered DNA Methylation Pattern Characterizes the Peripheral Immune Cells of Patients with Autoimmune Hepatitis. medRxivhttps://doi.org/10.1101/2021.07.01.21259836. [DOI] [PubMed]
- 149.Meda F., Folci M., Baccarelli A., Selmi C. The epigenetics of autoimmunity. Cell. Mol. Immunol. 2011;8:226–236. doi: 10.1038/cmi.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jeffries M.A., Sawalha A.H. Autoimmune disease in the epigenetic era: how has epigenetics changed our understanding of disease and how can we expect the field to evolve? Expet Rev. Clin. Immunol. 2015;11:45–58. doi: 10.1586/1744666X.2015.994507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Baubec T., Ivánek R., Lienert F., Schübeler D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell. 2013;153:480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 152.Szyf M. Epigenetic therapeutics in autoimmune disease. Clin. Rev. Allergy Immunol. 2010;39:62–77. doi: 10.1007/s12016-009-8172-8. [DOI] [PubMed] [Google Scholar]
- 153.Arvaniti P., Le Dantec C., Charras A., Arleevskaya M.A., Hedrich C.M., Zachou K., Dalekos G.N., Renaudineau Y. Linking genetic variation with epigenetic profiles in Sjögren’s syndrome. Clin. Immunol. 2020;210:108314. doi: 10.1016/j.clim.2019.108314. [DOI] [PubMed] [Google Scholar]
- 154.Banovich N.E., Lan X., McVicker G., van de Geijn B., Degner J.F., Blischak J.D., Roux J., Pritchard J.K., Gilad Y. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]