Abstract
Long noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is upregulated in various cancers, and its overexpression is associated with tumor growth and metastasis. MALAT1 has been recognized as a key player in the regulation of RNA splicing and transcription; however, the landscape of gene expression regulated by MALAT1 remains unclear. In this study, we employed an integrated transcriptomics and proteomics strategy to characterize the alterations in gene expression induced by MALAT1 knockdown in hepatocellular carcinoma (HCC) cells and identified 2662 differentially expressed transcripts and 1149 differentially expressed proteins. Interestingly, downregulation of MALAT1 reduced the abundances of multiple genes in the AMP-activated protein kinase (AMPK) signaling and biosynthesis of unsaturated fatty acids pathways. Further investigation showed that MALAT1 knockdown inhibited glucose uptake and lipogenesis by reducing the expression levels of these lipid metabolism related genes, which contributes to the oncogenic role of MALAT1 in tumor cell proliferation and invasion. This study uncovers the function of MALAT1 in the modulation of cancer lipid metabolism, reveals the underlying molecular mechanism, and further supports the potential therapeutic opportunities for targeting MALAT1 in HCC treatment.
Keywords: hepatocellular carcinoma, lipid metabolism, long noncoding RNA, proteomics, transcriptomics
Abbreviations: 2-NBDG, 2-[N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy-D-glucose; AA, arachidonic acid; ACC, acetyl-CoA carboxylase; AMPK, AMP-activated protein kinase; ChIP, chromatin immunoprecipitation; DHA, docosahexaenoic acid; FA, formic acid; FASN, fatty acid synthase; FRAP, fluorescence recovery after photobleaching; HCC, hepatocellular carcinoma; KEGG, Kyoto Encyclopedia of Genes and Genomes; LC–MS/MS, liquid chromatography–tandem mass spectrometry; lncRNA, long noncoding RNA; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; MRM, multiple reaction monitoring; MUFAs, monounsaturated fatty acids; PAs, phosphatidic acids; PCA, principal component analysis; PGs, phosphatidylglycerols; PIs, phosphatidylinositols; PSs, phosphatidylserines; PUFAs, polyunsaturated fatty acids; qRT-PCR, quantitative real-time polymerase chain reaction; RAB14, Ras-related protein Rab-14; RAP, RNA antisense purification; SCD, stearoyl-CoA desaturase; SFAs, saturated fatty acids; siRNA, small interfering RNA; SREBF1, sterol regulatory element-binding protein 1; TG, triglyceride
Graphical Abstract
Highlights
-
•
Multiomic analysis characterizes MALAT1-regulated gene expression in HCC cells.
-
•
MALAT1 knockdown reduces the expression of genes involving in lipid metabolism.
-
•
MALAT1 knockdown inhibits glucose uptake and lipogenesis.
-
•
MALAT1 promotes HCC cell proliferation and migration through regulating lipid metabolism.
In Brief
Here we investigate how long noncoding RNA MALAT1 regulates gene expression in liver cancer cell line using an integrated transcriptomics and proteomics strategy. We show that MALAT1 modulates the expression of multiple genes in the AMP-activated protein kinase signaling and lipid metabolism pathways. Additionally, MALAT1 knockdown inhibits lipogenesis and thereby reduces cancer cell proliferation and invasion. This study uncovers the role of MALAT1 in regulating lipid metabolism and further supports the potential therapeutic opportunities by targeting MALAT1.
Aberrant lipid metabolism is emerging as a critical hallmark of cancer (1). The multiple functions of lipids in cell signaling, membrane components, and energy storage are essential for cancer cells (2, 3). Substantial evidence has implicated that the dysregulation of cellular lipid metabolism is highly associated with the development and progression of cancer, including hepatocellular carcinoma (HCC) (4, 5). HCC is typically characterized by elevated lipogenesis, accompanied by increased de novo synthesized saturated and monounsaturated fatty acids (SFAs and MUFAs), and reduced polyunsaturated FAs (PUFAs) obtained from circulating lipids (6, 7). Lipid dysregulation has also been described in viral hepatitis and other liver diseases that are closely associated with the carcinogenesis of HCC (8, 9). Although the high lipogenic phenotype of cancer cells is now widely recognized, the underlying mechanism remains unclear.
Multiple lipogenesis-related genes are found to be upregulated in various cancers, such as acetyl-CoA carboxylase (ACC), fatty acid synthase (FASN), and stearoyl-CoA desaturase (SCD). SCD catalyzes the desaturation of long chain fatty acids, especially stearoyl-CoA, to form 18:1 MUFAs, which are then converted to triglycerides (TG) for energy storage or phospholipids for constructing cell membrane. The expression of SCD is controlled by transcription factor sterol regulatory element-binding protein 1 (SREBF1) (10). SCD has emerged as a key player in lipogenesis, and its overexpression is associated with poor prognosis in various cancer types, such as prostate, breast, kidney, and liver cancer (11, 12, 13, 14). Studies have shown that SCD inhibitors could reduce growth of xenografts in mice, suggesting the potential therapeutic benefits of targeting SCD in cancer treatment (15).
Long noncoding RNAs (lncRNAs) are a class of mRNA-like transcripts, longer than 200 nucleotides without protein coding capability (16). LncRNAs play key roles in the regulation of gene expression at multiple levels, such as chromatin remodeling, transcriptional regulation, posttranscriptional processing, RNA translation, and protein stability (17). The dysregulation of lncRNA has been associated with tumorigenesis and development of malignant tumors by regulating various biological processes in cancer cells, such as cell growth, invasion, differentiation, proliferation, apoptosis, and cell cycle (18, 19).
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is one of the first lncRNAs discovered with designated roles in cancers (20, 21). The MALAT1 transcript is approximately 8700 nt in length, and unlike many other lncRNAs, MALAT1 exhibits a high level of evolutionary conservation (22). Accumulating evidence has shown that MALAT1 is upregulated in many cancers, including HCC (23, 24). MALAT1 overexpression promotes tumor cell progression and metastasis (20, 25, 26, 27, 28). MALAT1 may exert its biological roles by regulating gene expression via multiple mechanisms. For example, MALAT1 has been implicated in regulating pre-mRNA splicing through interacting with various splicing factors, such as serine/arginine-rich splicing factor 1 and serine/arginine-rich splicing factor 3 (29, 30). Our previous study also showed that MALAT1 could bind with various spliceosome components and RNA splicing related proteins (31). Furthermore, MALAT1 can also interact with transcription factors and epigenetic regulators, e.g., polycomb 2 protein and cell cycle and apoptosis regulator protein 2, to modulate gene transcription (31, 32). However, a comprehensive understanding of the full spectrum of gene expression regulated by MALAT1 is still missing.
In this study, we first systematically investigated the impact of MALAT1 knockdown on gene expression in HCC cells by integrating transcriptomics and proteomics. We found that downregulation of MALAT1 reduced the abundances of multiple molecules from the fatty acid metabolism pathway and the upstream AMP-activated protein kinase (AMPK) signaling pathway, such as SCD, SREBF1, two subunits of AMPK, and Ras-related protein Rab-14 (RAB14). Further investigation suggested that MALAT1 may regulate the expression of these genes through RNA splicing or transcription. This study uncovers a previously unknown role of MALAT1 in the modulation of cellular lipid metabolism and reveals the underlying molecular mechanism.
Experimental Procedures
Cell Culture and Transfection
Hep3B, HepG2, and PLC HCCcell lines were obtained from the American Type Culture Collection (ATCC). Huh7 HCC cell line was purchased from Japanese Collection of Research Bioresources (JCRB). MHCC97L, MHCC97H, and HCCLM3 HCC cell lines were purchased from the Cell Bank of Type Culture Collection (Chinese Academy of Sciences). Cells were cultured in Dulbecco’s modified Eagle’s high glucose medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (100 μg/ml) at 37 °C in a humidified incubator in the presence of 5% CO2. For siRNA-mediated gene knockdown, cells were transfected with siRNA targeting MALAT1 (siMALAT1) or a negative control siRNA (siNC) using the X-tremeGENE siRNA Transfection Reagent (Roche Diagnostics), according to the manufacturer’s instructions. siRNAs were synthesized by RiboBio Co Ltd, and the sequences were listed in supplemental Table S1. For overexpression, cDNA of the fragment 6918 to 8441 nt of MALAT1 was amplified via PCR using the Premix Taq DNA Polymerase (TaKaRa) and cloned into the pcDNA3.1(+) vector (Invitrogen). The MALAT1 and empty vector plasmids were then transfected into HCCLM3 cells using Lipofectamine 2000 (Invitrogen).
RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from the cells using TRIzol reagent (Invitrogen), and complementary DNA (cDNA) was synthesized using the FastQuant RT kit (TianGen) according to the manufacturer’s instructions. Quantitative RNA expression analysis was performed on a 7500 Fast Real-Time PCR System (ABI) using the SuperReal SYBR Green PreMix (TianGen) following the manufacturer’s protocol. Each sample was analyzed in triplicate, and the relative RNA expression fold changes were calculated with 2−ΔΔCT and normalized to a housekeeping gene, GAPDH. The primer sequences used in this study were listed in supplemental Table S2.
Protein Extraction and Western Blotting
Adherent cells were washed with cold PBS and lysed with SDS lysis buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerinum) supplemented with 1× protease inhibitor cocktail (Roche Diagnostics). Protein concentration was quantitated using the BCA Protein Assay Kit (Thermo Fisher Scientific). Proteins were resolved by SDS-PAGE and transferred to the Immobilon-P membrane (0.25 μm pore size, Millipore). Rabbit anti-SCD, rabbit anti-PRKAB1, and rabbit anti-PRKAG1 primary antibodies were purchased from Abcam. Rabbit anti-SREBF1 and mouse anti-GAPDH primary antibodies were purchased from Proteintech. Mouse anti-RAB14 primary antibody was purchased from Santa Cruz Biotechnology. Primary antibodies were incubated with the membranes at 4 °C overnight in 5% skim milk (BD Biosciences). Blots were later developed by enhanced chemiluminescence by using horseradish-peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology).
RNA Sequencing
A total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using UltraTM RNA Library Prep Kit for Illumina (New England Biolabs) following manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the libraries were sequenced on an Illumina HiSeq X 10 platform (Illumina), and 150-bp paired ends were generated.
Protein Extraction and Digestion
HCCLM3 cells were disrupted using 8 M urea solution in 50 mM NH4HCO3 supplemented with 1 mM NaF, 1 mM Na3VO4, and protease inhibitor cocktail, and the cell debris was removed by centrifugation at 16,000g for 30 min at 4 °C. Cellular proteins were quantified using BCA assay. The proteins were reduced by 10 mM DTT (Promega) for 30 min at 37 °C and alkylated by 40 mM IAA (Sigma-Aldrich) for 45 min at room temperature in the dark, then the 8 M urea was removed by filter-aided sample preparation (FASP) with 10 Kd filter (Millipore). The proteins were digested by incubating with trypsin at a protease: protein ratio of 1:50 at 37 °C overnight. The digested peptides were further quantified using Pierce Quantitative Colorimetric Peptide Assay (Thermo Fisher Scientific). For proteomics analysis, 100 ug peptides of each sample were fractionated into eight fractions by a high pH reversed-phase peptide fractionation kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Then the peptides were dried with Savant SpeedVac (Thermo Fisher Scientific) and resuspended in 0.1% formic acid (FA) before mass spectrometric analyses.
Liquid Chromatography–Tandem Mass Spectrometry (LC–MS/MS)
Peptides from each fraction were analyzed by nano-LC-MS/MS using an Easy-nLC 1000 system (Thermo Fisher Scientific) coupled to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) with a nanoelectrospray ion source (Thermo Fisher Scientific). In brief, peptides were loaded onto a 2-cm self-packed trap column (150 μm inner diameter, ReproSil-Pur C18-AQ, 3 μm; Dr Maisch) using buffer A and separated on a self-packed 150-μm-inner-diameter column with a length of 30 cm (ReproSil-Pur C18-AQ, 1.9 μm; Dr Maisch) over a 120-min gradient (buffer A, 0.1% FA in water; buffer B, 0.1% FA in 80% ACN) at a flow rate of 500 nl/min (0–10 min, 8–12% B; 10–79 min, 12–27% B;79–107 min, 27–45% B; 107–110 min, 45–95% B; and 110–120 min, 95% B). The Orbitrap Fusion Lumos was set to OT-IT mode. For a full mass spectrometry survey scan, the target value was 5E5 and the scan ranged from 350 to 1800 m/z at a resolution of 120,000 and a maximum injection time of 50 ms. For the MS2 scan, a duty cycle of 3 s was set with the top-speed mode. Ions with a charge state of 2 to 8 were selected for fragmentation by higher-energy collision dissociation with a normalized collision energy of 35%. The MS2 spectra were acquired in the ion trap in rapid mode with an AGC target of 5E3 and a maximum injection time of 50 ms.
Quantification of Gene Expression
Raw data (raw reads) of fastq format were first processed through the in-house perl scripts to remove reads containing adaptor, reads containing ploy-N, and low-quality reads. The clean reads per sample were mapped to the reference using STAR (version: 2.7.3a) aligner (33). The reference consists of the human reference genome and the Ensembl annotated human transcriptome (GRCh38 assembly, Ensembl release 94). FeatureCounts (version: 1.6.4) was used to calculate the number of aligned reads per gene (34). RSEM (version 1.3.3) was used to quantify genes and transcripts expression levels (35). The genes with total count >10 in all samples were considered as stably expressed.
Protein Identification and Quantification
The tandem mass spectra were searched against the reviewed human UniProt database (version: 20180820, 20,325 sequences) using MaxQuant (version: 1.6.3.4) (36). Trypsin was selected as the proteolytic enzyme, and two missed cleavages sites were allowed. Cysteine carbamidomethylation was set as a fixed modification. The oxidation of M and acetylation of the protein N-terminal were set as variable modifications. Peptides with the minimum of seven amino acid length were considered and the required FDR was set to 1% at the peptide and protein level. Match between run was applied with default parameters. The first search mass tolerance was 20 ppm, and the main search peptide tolerance was 4.5 ppm. Quantification in MaxQuant was performed using the Label-Free Quantification algorithm. Proteins with at least two identifications were used for subsequent quantitative analysis. Missing values were imputed with the minimum value across the proteome data set for algorithms that could not handle missing values.
Bioinformatics Analysis
For the transcriptomics data, differential gene expression was performed using the R/Bioconductor package DESeq2 (version: 1.28.1) (37). Gene symbols and transcript biotype were annotated using R/bioconductor package biomaRt (version: 2.44.4). To identify differently expressed proteins between siNC and siMALAT1 groups, statistical significance was calculated by a simple linear model and moderated t-statistics using an empirical Bayes shrinkage method implemented in the R/Bioconductor package limma (version: 3.44.3) (38). Functional enrichment analysis was performed using PANTHER (39) (http://www.pantherdb.org/) for protein class classification and R/bioconductor package clusterProfiler (version: 3.16.1) for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment (40). Cytoscape (version: 3.8.0) application KEGGscape (version: 0.9.0) was used to visualize the KEGG pathway map. For the transcription factor (TF) network construction, the differentially expressed TFs and mRNAs were analyzed using the Chromatin immunoprecipitation (ChIP) Enrichment Analysis (ChEA, version: 3) by searching against three ChIP-seq databases, including ENCODE_ChIP-seq, Literature_ChIP-seq, and ReMap_ChIP-seq (https://maayanlab.cloud/chea3/) (41). Then, the gene expression network was constructed and visualized in Cytoscape (version: 3.8.0).
Measurement of Glucose Uptake
The fluorescent glucose analog 2-[N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy-D-glucose (2-NBDG, Sigma-Aldrich) was used to measure glucose uptake. Prestarved cells were incubated with 2-NBDG (10 μM) for 15 min, washed twice by PBS, and analyzed by flow cytometry (BD Biosciences).
Unsaturated Fatty Acid and Triacylglycerol Measurement
Cells were disrupted in methanol supplemented with 2% FA and 0.01 M butylated hydroxytoluene (Sigma-Aldrich) by repeated freeze–thaw, and then the lipid content was extracted by ethyl acetate. The concentration of unsaturated fatty acids was determined by the Lipid Assay Kit (unsaturated fatty acids) (Abcam) following the manufacturer’s protocol. For the analysis of triacylglycerol, cells were disrupted and the concentration of triacylglycerol was assayed using a triglyceride assay kit according to the manufacturer's protocol (Applygen Technologies Inc).
The Measurement of Lipids
The lipids of interest were analyzed by targeted LC-MS/MS using an established protocol as reported previously (42). Briefly, the harvested cells were resuspended in 50 μl of water and then mixed with 450 μl of methanol containing 10 pmol of 14:0 lysophosphatidic acid, 100 pmol 12:0 lysophosphatidylcholine, and 50 pmol of 17:0 ceramide, which were used for quality control. The lipid standards were purchased from Avanti Polar Lipids. After vortexing and centrifugation at 10,000g for 5 min under room temperature, the supernatant was examined by the negative ion multiple reaction monitoring (MRM) mode for the analysis of negatively charged lipids, such as phosphatidic acids (PAs) and phosphatidylserines (PSs). The positive ion MRM mode analyses were performed for the other lipids, such as phosphatidylglycerols (PGs) and phosphatidylinositols (PIs).
MS analysis was performed using the API 4500 QTRAP mass spectrometer (SCIEX) with the Analyst data acquisition system. Nitrogen was used as the nebulizer and desolvation gas. The operating parameters were set as follows: curtain gas, 25; collision gas, medium; ion source gas 1, 45; ion source gas 2, 50; electrospray voltage 5500 V with positive ion MRM mode or 4500 V with negative ion MRM mode. Information-dependent acquisition threshold was set at 200 counts per second. The collision energy in the Enhanced Production Ion mode was set at 35, and the collision energy spread was set at 15. The dynamic fill time function was used to prevent the space charges effect. The peak areas of lipids were used for relative quantification. Two-sided Student’s t test was performed for statistical analysis, and p value <0.05 was considered to be statistically significant.
Fluorescence Recovery after Photobleaching (FRAP)
5 × 104 HCCLM3 cells transfected with siNC or siMALAT1 were cultured in confocal dishes for 24 h. Then the cells were washed three times with PBS and incubated for 10 min at 37 °C with 1 ml of 10 mM Dil perchlorate (Beyotime). Olympus FV1000 confocal microscope system was used to acquire images with the following parameters: excitation wavelength, 549 nm; receive wavelength, 565 nm; photobleaching wavelength, 488 nm; photobleaching time, 15 s.
RNA Antisense Purification (RAP)
Cultured adherent cells were first cross-linked with 2% formaldehyde solution for 10 min at 37 °C, and the cross-linking reaction was terminated by adding 500 mM glycine solution. Cross-linked cells were disrupted using the lysis buffer (10 mM Tris-HCl pH 7.5, 500 mM LiCl, 0.5% DDM (n-Dodecyl-beta-D-maltoside), 0.2% SDS, 0.1% sodium deoxycholate, protease inhibitor cocktail, RNase Inhibitor) for 30 s on ice by the Ultrasonic Cell Disruptor (Sonics). The cell lysate was treated with DNase for 10 min at 37 °C, followed by termination with a stop buffer (10 mM EDTA, 5 mM EGTA, and 2.5 mM TCEP). Then the cell lysate was incubated with 2 × volume of the hybrid buffer (1.5 mM Tris-HCl pH 7.5, 7.5 mM EDTA, 750 mM LiCl, 0.75% DDM, 0.3% SDS, 0.15% sodium deoxycholate, 6 M urea, and 3.75 mM TCEP) for 10 min on the ice, followed by 10 min centrifugation at 16,000g at 4 °C. Next, the supernatant was used for hybridization reaction with biotin-labeled antisense probes against MALAT1 at 37 °C for 2 h, and the probes against lacZ were used as control. The designed antisense probes were synthesized by GENEWIZ, and the sequences were listed in supplemental Table S3. Streptavidin-coated magnetic beads (Thermo Fisher Scientific) were added to bind the probes by incubating for 30 min and separated by a magnetic stand. To elute the probe-binding RNAs, the beads were resuspended by the elution buffer (20 mM Tris-HCl pH 7.5, 10 M EDTA, 2% N-lauroylsarcosine and 2.5 mM TCEP) and heated to 95 °C for 10 min, then a final concentration of 1 mg/ml Proteinase K (Beyotime) was added to digest the protein for 1 h at 55 °C. Finally, TRIzol was used to isolate RNAs from the digested mixture.
Cell Proliferation Assay
Transfected cells were plated at 2000 cells per well in 96-well plates and incubated with or without 10 μM oleic acid (Sigma-Aldrich) for 48 h at 37 °C. Then the cells were measured with the CCK8 assay (Dojindo) according to the manufacturer’s protocol. Briefly, 10 μl of CCK8 solution was added to each well, and the samples were incubated at 37 °C for 2 h before the absorbance was measured at 450 nm.
Wound Healing and Invasion Assays
Wound healing and invasion assays were carried out as previously described (43). In brief, for the wound healing assay, cells were seeds in 6-well plates and grown to 80 to 90% confluence. The cells were scratched with a pipet tip in the middle of the plate and then washed with PBS to remove the detached cells. Then, the plates were incubated in DMEM containing 1% FBS and 10 μM oleic acid (Sigma-Aldrich) for the rescue experiments. The wound closure was monitored microscopically at different time points (0, 3, 6, 9, 12, 24 h) and photographed at 0 and 24 h, respectively.
The invasion assay was performed using Matrigel-coated transwell inserts (Millipore) containing polycarbonate membranes with 8 μm pores. Serum-starved cells were trypsinized and counted. Then 5 × 104 cells in serum-free DMEM medium were added to the upper chamber, and 600 μl of DMEM supplemented with 10% FBS was added to the lower chamber. After incubation for 24 h at 37 °C, nonmigrating cells on the upper membrane surface were removed with a cotton swab. The migrating cells on the under surface were fixed and stained with a Richard-Allan Scientific Three-Step stain kit (Thermo Fisher Scientific). The migrating cells were counted in six random regions at 200× under microscope.
Experimental Design and Statistical Rationale
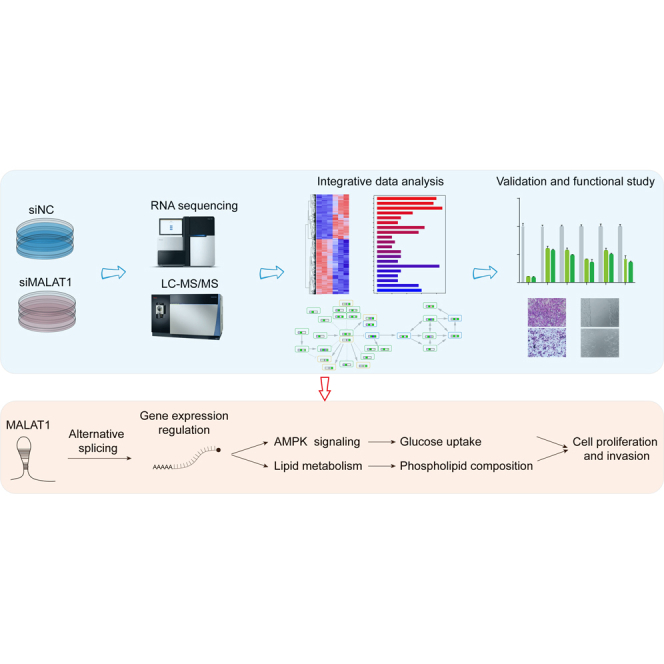
The proteomics and transcriptomics experiments were performed as described in Figure 1, and three biological replicates for each condition were investigated for both analyses. To get a deep coverage of the proteome, the tryptic peptides were separated using high-pH reversed-phase chromatography before LC-MS/MS analysis, and proteins observed in at least two biological replicates were considered for the label-free quantification of the proteome. The statistical significance was evaluated by a simple linear model and moderated t-statistics using an empirical Bayes shrinkage method implemented in the R/Bioconductor package limma (version: 3.44.3) (38). Proteins with a fold change larger than 1.5 and a p value <0.05 were defined as significantly differentially expressed. Cell assays were performed in triplicates and analyzed by Student′s t test for comparison between two groups. Wound healing results were analyzed by repeated measures analysis of variance (ANOVA). All statistical analyses were performed using R (version: 3.6.3) or GraphPad Prism (version: 8.4.3). Data were presented as means ± sd, and p < 0.05 was considered as a significant difference.
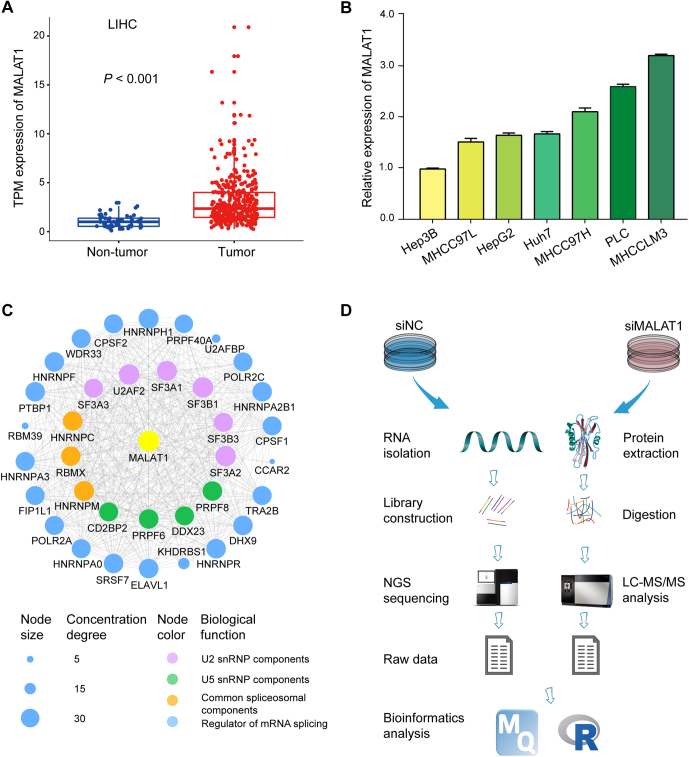
Fig. 1.
The expression of MALAT1 in HCC tumor tissues and cell lines.A, comparison of the expression of MALAT1 between tumor and nontumor samples from the TCGA-LIHC (Liver Hepatocellular Carcinoma) transcriptomics data. Transcripts Per Million (TPM) values were used for quantification. Boxplot center lines = median, lower bound = 25% quantile, upper bound = 75% quantile, lower whisker = the smallest observation greater than or equal to the lower hinge −1.5 × interquartile range (IQR), upper whisker = the largest observation less than or equal to the upper hinge +1.5 × IQR. Wilcoxon rank-sum test was used for statistical analysis. B, relative levels of MALAT1 across seven HCC cell lines. Data represent mean ± s.d. C, interacting proteome network of MALAT1 involving in RNA splicing. The network was constructed by Cytoscape (version: 3.8.0). D, the workflow of the experimental approach employed in this study.
Results
MALAT1 Is Overexpressed in HCC
To investigate the level of MALAT1 in HCC, we obtained RNA sequencing data of liver tumors and normal tissues from The Cancer Genome Atlas (TCGA) database, including 50 normal tissues and 371 liver tumors. The results showed that the level of MALAT1 was significantly increased in HCC tumors compared with normal tissues (fold change = 2.85, p < 0.001) (Fig. 1A). Next, we analyzed the level of MALAT1 in seven different HCC cell lines, including Hep3B, MHCC97L, HepG2, Huh7, MHCC97H, PLC, and HCCLM3. The results suggested that the level of MALAT1 elevated with the increasing invasiveness of the investigated HCC cells (Fig. 1B). In the three HCC cells with lower invasion capability, such as Hep3B and MHCC97L, MALAT1 was detected with relatively lower abundance. The level of MALAT1 was markedly increased in HCC cells with higher metastatic ability, such as HCCLM3 (Fig. 1B).
Earlier studies have shown that one of the major mechanisms by which MALAT1 exerts its biological function is to regulate mRNA splicing (29, 30). Our previous study also demonstrated that MALAT1 could pull down multiple subunits of the RNA spliceosome and other splicing-related proteins (31). We reanalyzed the data and constructed an interactome network for MALAT1 associated with RNA splicing, showing MALAT1 as an important factor in this critical step of gene expression (Fig. 1C). However, the target genes regulated by MALAT1 are still largely unknown. Therefore, in order to systematically understand the regulatory roles of MALAT1 in HCC, an integrated transcriptomics and proteomics analysis was performed in this study to investigate the alterations in the landscape of gene expression in HCCLM3 cells at both mRNA and protein levels induced by MALAT1 knockdown (Fig. 1D).
MALAT1 Modulates Gene Expression in HCC Cells
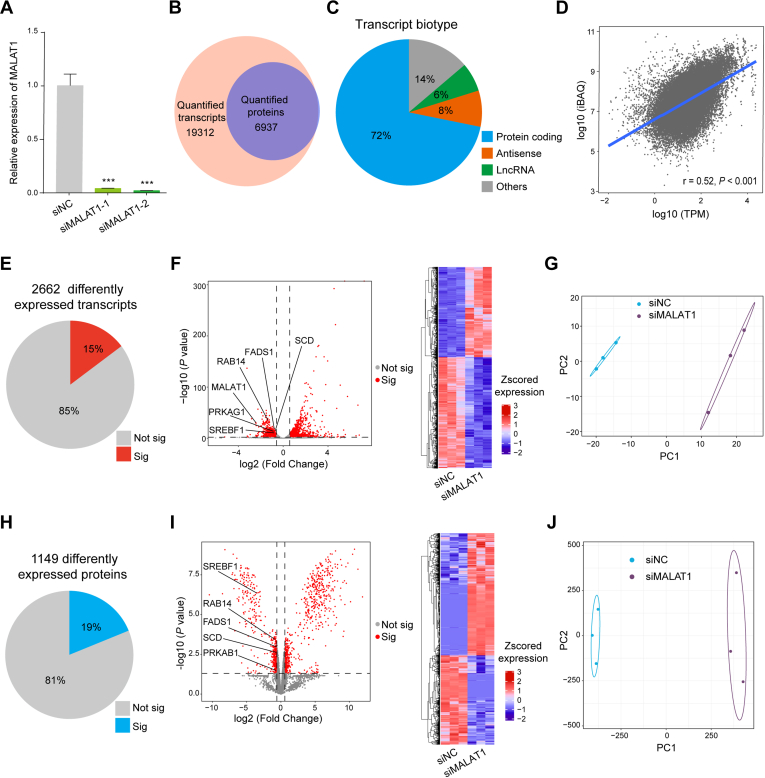
The transcriptome analysis quantified 19,312 transcripts, the majority of which were protein-coding RNAs (72%), while lncRNAs and antisense RNAs accounted for 6% and 8% of the total identified transcripts, respectively (Fig. 2, A–C and supplemental Table S4). A total of 6937 proteins were detected by the proteomic analysis, and 6836 proteins (98.5%) were also detected at the RNA level in the transcriptome data (Fig. 2B). Correlation analysis showed decent reproducibility between the replicates (supplemental Fig. S1), and the correlation analysis between the transcriptome and proteome data showed a moderate degree of correlation between the two datasets (r = 0.52, p < 0.001) (Fig. 2D).
Fig. 2.
MALAT1 knockdown induced perturbation of gene expression at the transcriptome and proteome level.A, the relative expression of MALAT1 in the negative control (siNC) and siMALAT1-treated HCCLM3 cells as measured by qRT-PCR. Data represent mean ± s. d. of triplicate independent experiments (∗∗∗p < 0.001, by two-sided Student’s t test). B, the overlap of genes quantified at the RNA and protein levels. C, percentages of the indicated biotypes of the quantified transcripts. D, comparison of the protein intensities as in intensity-based absolute quantification (iBAQ) values versus mRNA intensities (TPM). Pearson correlation analysis was conducted. E and H, the percentages of the differently expressed transcripts (E) and proteins (H). F and I, volcano plots and heatmaps showing the differently expressed transcripts (F) and proteins (I). G and J, principal component analysis of all quantified RNAs (G) and proteins (J).
For the transcriptomics analysis, a total of 2662 differentially expressed transcripts (15%) were detected induced by MALAT1 knockdown (fold change > 1.5, p < 0.01) (Fig. 2E and supplemental Table S4), of which 1195 were upregulated in MALAT1 knockdown cells and 1467 were downregulated, and MALAT1 itself was decreased by 5.4-fold (Fig. 2F). Principal component analysis (PCA) results showed that the cells in the siNC group and the siMALAT1 group exhibited significant differences at the transcriptome level (Fig. 2G).
For the quantitative proteomics analysis, a total of 1149 differentially expressed proteins (19%) were detected after MALAT1 knockdown (fold change > 1.5, p < 0.05) (Fig. 2H and supplemental Table S4), of which 664 were upregulated and 485 were downregulated (Fig. 2I). PCA analysis showed significant difference between the siNC cells and the siMALAT1 cells in their proteomes (Fig. 2J). The integrated transcriptomics and proteomics analysis indicated that MALAT1 knockdown posed profound effect on the global landscape of gene expression in HCC cells.
Genes Regulated by MALAT1 Exert Diverse Biological Functions
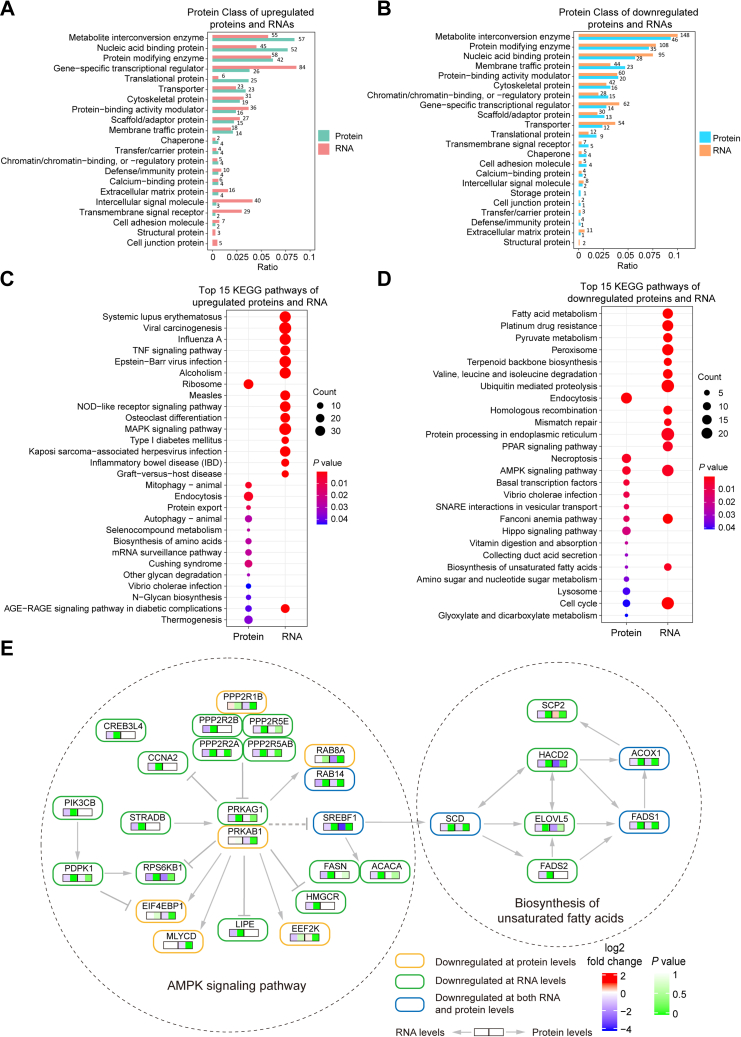
To better understand the biological functions of the differentially expressed genes potentially regulated by MALAT1, we classified them by the Panther online annotation tool Protein Class (39). The most enriched protein classes of the differentially expressed proteins and RNAs included metabolite interconversion enzyme, nucleic acid binding protein, protein modifying enzyme, gene-specific transcriptional regulator, membrane traffic protein, etc. (Fig. 3, A and B and supplemental Table S5). Of note, we observed that many TFs were differentially expressed in MALAT1 knockdown cells. In order to further reveal the roles of MALAT1 in gene regulation, we constructed a MALAT1-TFs-Targets network by mapping the TFs with changes at the protein level and the differentially expressed transcripts based on information acquired from the CHIP-seq databases (supplemental Fig. S2). The results show that TFs, such as myc proto-oncogene protein (MYC), transcription factor jun-B (JUNB), and CCAAT/enhancer-binding protein β (CEBPB), may act as mediators for MALAT1 to regulate the expressions of a large spectrum of genes in HCC cells.
Fig. 3.
Functional analysis of the differently expressed mRNAs and proteins induced by MALAT1 knockdown.A and B, protein class classification of the upregulated (A) and downregulated (B) proteins and mRNAs. Pantherdb was used for the analysis (http://www.pantherdb.org/). C and D, KEGG pathway overrepresentation analysis of the upregulated (C) and downregulated (D) proteins and mRNAs. The analysis was conducted by clusterProfiler (version: 3.16.0) R/Bioconductor package. E, differently expressed genes detected in the AMPK signaling and biosynthesis of unsaturated fatty acids pathways.
Next, we performed KEGG pathway overrepresentation test on the genes with altered expression induced by MALAT1 knockdown (Fig. 3, C and D and supplemental Table S6). The analysis results showed that the differentially expressed mRNAs and proteins were enriched in distinct pathways. For example, the upregulated proteins were most significantly enriched in the ribosome, mitophagy and endocytosis pathways, while the upregulated mRNAs were highly associated with immune related pathways (Fig. 3C). Intriguingly, the downregulated mRNAs and proteins were both significantly enriched in the AMPK signaling and biosynthesis of unsaturated fatty acids pathways (Fig. 3D and supplemental Fig. S3). AMPK signaling promotes the uptake of glucose that is converted to citrate in the tricarboxylic acid (TCA) cycle, and citrate is then transported back to the cytosol to produce acetyl-CoA for fatty acid biosynthesis (44, 45). We observed 18 differentially expressed mRNAs and nine differentially expressed proteins enriched in the AMPK signaling pathway (supplemental Table S6). SREBF1and RAB14 were detected to be downregulated at both mRNA and protein levels (Figs. 2, F and I and 3E). Moreover, seven mRNAs and three proteins were enriched in the biosynthesis of unsaturated fatty acids pathway (supplemental Table S6). For example, SCD, fatty acid desaturase 1 (FADS1), and acyl-CoA oxidase 1 (ACOX1) were downregulated at both mRNA and protein levels (Figs. 2, F and I and 3E). These results suggest that MALAT1 may play an important regulatory role in lipid metabolism by targeting multiples genes in the AMPK and unsaturated fatty biosynthesis pathways.
MALAT1 Knockdown Inhibited the Expression of Genes Regulating Lipid Metabolism
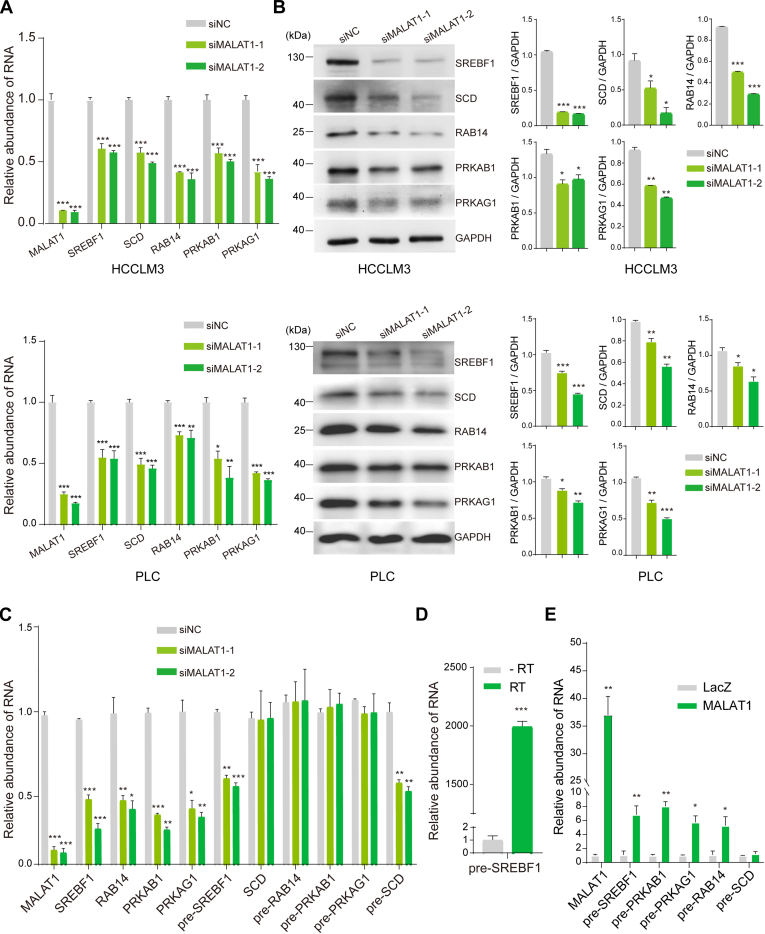
Accumulating evidence has shown the multifaceted functions of MALAT1 in tumor progression by regulating cell processes such as cell proliferation, apoptosis, migration, and angiogenesis (20, 21, 24). A recent study showed that MALAT1 may promote hepatic steatosis and insulin resistance by modulating lipid accumulation through SREBP-1c (46). However, its complete role in lipid metabolism is still largely unclear. Intriguingly, our data showed that MALAT1 knockdown reduced the expression level of multiple signaling molecules and key enzymes involved in lipogenesis. For example, SCD is a crucial enzyme that catalyzes the biosynthesis of monounsaturated fatty acids (47). The expression of SCD is known to be regulated by SREBF1, a membrane-bound transcription factor that controls the rate of lipid synthesis in animal cells by activating fatty acid and triglyceride synthesis genes (48). The RNA sequencing and mass spectrometry data showed significantly downregulation of SCD and SREBF1 at both of the mRNA and protein levels (Figs. 2, F and I, and 3E). Therefore, we further validated this observation by qRT-PCR and immunoblotting using two different types of HCC cells, including PLC and HCCLM3. The results showed MALAT1 knockdown decreased the abundances of both SCD and SREBF1 (Fig. 4, A and B).
Fig. 4.
MALAT1 knockdown inhibited the expression of genes in the AMPK signaling and lipid metabolism pathways through pre-mRNA splicing or transcription.A, the relative RNA levels of MALAT1, SREBF1, SCD, RAB14, PRKAB1, and PRKAG1 were measured by qRT-PCR in HCCLM3 (upper panel) and PLC (lower panel) cell lines. B, the expression of the proteins of interest in HCCLM3 (upper panel) and PLC (lower panel) cells was examined by western blotting. The densities of the corresponding protein bands were measured by Image J and displayed on the right. C, relative quantification of the mRNAs and the corresponding pre-mRNAs by qRT-PCR. D, measurement of the pre-mRNA of SREBF1 by qRT-PCR with or without reverse transcriptase (RT). E, qRT-PCR analysis of the pre-mRNAs copurified with MALAT1 by biotinylated anti-MALAT1 probes. The anti-LacZ probes were used as negative control. Data represent mean ± s.d. of triplicate independent experiments (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, by two-sided Student’s t test).
AMPK is a key regulator of cellular energy homeostasis, comprised of a catalytic α subunits and the regulatory β and γ subunits. AMPK increases the activity of Rab family G proteins, such as RAB14, and induces membrane translocation of GLUT4 to stimulate glucose uptake (49). From the omics data, we found that the expression levels of AMPK subunit β1 (PRKAB1), subunit γ1 (PRKAG1), and RAB14 were downregulated at mRNA or protein levels (Figs. 2, F and I, and 3E). Next, we examined their expression alterations by qRT-PCR and western blotting, and the results showed that the levels of all these three genes were decreased at both mRNA and protein levels in siMALAT1 cells (Fig. 4, A and B).
Furthermore, to understand how MALAT1 regulates the expression of the genes of interest, we examined the pre-mRNA levels of these genes using primers targeting the exon–intron junctions. The results showed that MALAT1 downregulation had no significant effect on the pre-mRNA levels of SREBF1, PRKAB1, PRKAG1, and RAB14; however, pre-SCD was reduced to a similar level compared with its mRNA (Fig. 4C). Notably, pre-SREBF1 was not amplified in samples treated without reverse transcriptase, excluding the possibility of DNA contamination (Fig. 4D). To further validate the role of MALAT1 in regulating the expression of selected genes. We overexpressed the 6918 to 8441 nt fragment of MALAT1 in HCCLM3 cells and examined the mRNA and pre-mRNA levels of SREBF1, RAB14, PRKAB1, PRKAG1, and SCD (supplemental Fig. S4). In agreement with the results from the knockdown experiment, the mRNA levels of these genes were significantly increased by the overexpression of MALAT1, and no significant change of the pre-mRNAs was observed, except for pre-SCD. The data indicated that the investigated genes were likely regulated by MALAT1 at the posttranscriptional level, except for SCD.
Engreitz et al. (30) reported that MALAT1 could indirectly bind many pre-mRNAs through protein mediation in spliceosome and regulate their splicing. Therefore, we hypothesized that MALAT1 may modulate the expression of SREBF1, PRKAB1, PRKAG1, and RAB14 via RNA splicing. To test this hypothesis, we investigated their interactions with MALAT1 by cross-linking assisted RAP. HCCLM3 cells were first cross-linked with formaldehyde, and then MALAT1 and the associated pre-mRNAs were purified with biotin-labeled antisense probes. The results showed that MALAT1 was efficiently enriched in the pull-down sample (fold enrichment = 36.6, p = 0.002), and pre-SREBF1(fold enrichment = 6.3, p = 0.007), pre-PRKAB1(fold enrichment = 8.0, p = 0.002), pre-PRKAG1(fold enrichment = 5.7, p = 0.01), and pre-RAB14 (fold enrichment = 4.8, p = 0.02) were copurified with MALAT1 (Fig. 4E). Interestingly, pre-SCD was not isolated together with MALAT1 (Fig. 4E), suggesting that it did not bind with MALAT1 as the other investigated RNAs.
To further explore the effect of MALAT1 on regulating RNA splicing, we investigated the alternative splicing events using the transcriptomics data by rMATS (50). The analysis identified a total of 3475 differential alternative splicing events, including 2394 skipped exon (SE) events, 453 mutually exclusive exons (MXE) events, 192 alternative 5′ splice site (A5SS) events, 153 alternative 3′ splice site (A3SS) events, and 283 retained intron (RI) events (supplemental Fig. S5A). The results revealed the broad effect of MALAT1 on the process of alternative splicing. Interestingly, we detected differential SE events of SREBF1 and PRKAB1 (supplemental Fig. S5B). The inclusion levels (ψ) of SREBF1 exon 2 and PRKAB1 exon 5 were significantly lower in siMALAT1 cells compared with the control cells. The results further demonstrated the regulatory role of MALAT1 in the expression of lipid metabolism-related genes by modulating RNA splicing.
In summary, these results indicated that MALAT1 could regulate the expression of multiple genes related to lipogenesis in HCC cells via distinct mechanisms. MALAT1 may modulate the splicing of the pre-mRNAs of SREBF1, PRKAB1, PRKAG1 and RAB14, but regulate the expression of SCD at transcriptional level likely through SREBF1, a transcription factor well-known for targeting SCD.
MALAT1 Knockdown Inhibited Lipogenesis
Since AMPK signaling is a key regulator of glucose uptake, we examined whether MALAT1 could affect glucose uptake by measuring the abundance of fluorescent glucose analog 2-NBDG in siMALAT1 cells and siNC cells using flow cytometry. We observed that the downregulation of MALAT1 reduced glucose uptake in both HCCLM3 and PLC cells (Fig. 5A). The data indicated that MALAT1 knockdown decreased glucose uptake likely by inhibiting AMPK signaling.
Fig. 5.
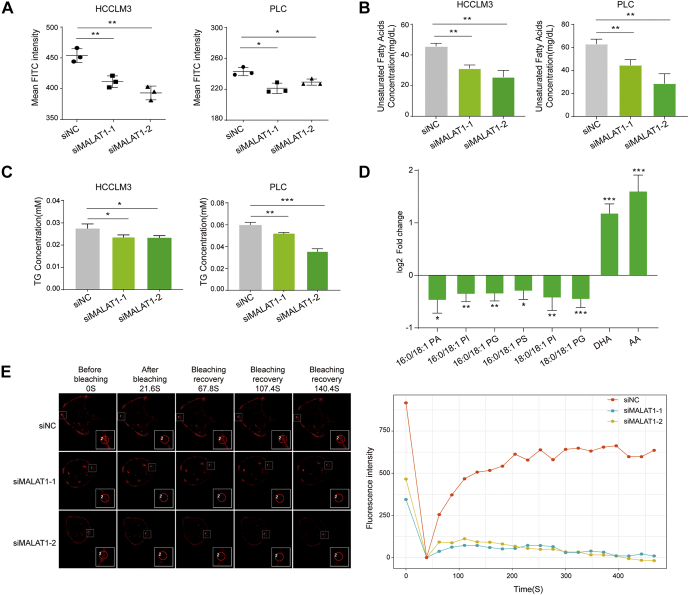
MALAT1 knockdown decreased lipogenesis in HCC cells.A, glucose uptake in HCCLM3 (left panel) and PLC (right panel) cells treated with siMALAT1. Data represent mean ± s.d. of triplicate independent experiments (∗p < 0.05, ∗∗p < 0.01, by two-sided Student’s t test). B, the concentration of unsaturated fatty acids extracted from HCCLM3 (left panel) and PLC (right panel) cells treated with siMALAT1s. Data represent mean ± s.d. of triplicate independent experiments (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, by two-sided Student’s t test). C, the concentration of TG extracted from HCCLM3 (left panel) and PLC (right panel) cells treated with siMALAT1. Data represent mean ± s.d. of triplicate independent experiments (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, by two-sided Student’s t test). D, targeted LC-MS/MS analysis of lipids with altered abundance between siNC and siMALAT1 treated HCCLM3 cells. Data represent mean ± s.d. of five biological replicates (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, by two-sided Student’s t test). E, representative cell membrane images of HCCLM3 cells at the indicated time points after photobleaching. Fluorescence intensities of the photobleached areas at indicated time points were shown on the right.
The integrated omics analysis revealed that MALAT1 knockdown reduced the expression level of SCD, an important enzyme for lipogenesis. Therefore, we examined the regulatory effect of MALAT1 on unsaturated fatty acids metabolism by measuring the abundance of total unsaturated fatty acids in MALAT1 knockdown cells and the corresponding control cells. The results showed that MALAT1 downregulation significantly reduced the total abundance of unsaturated fatty acids in two different types of HCC cells (Fig. 5B).
The de novo synthesized fatty acids can be converted to TG for energy storage. Therefore, we also examined the total amount of TG in siNC and siMALAT1 cells. The results showed that MALAT1 knockdown indeed reduced the abundance of TG in the both HCCLM3 and PLC cells (Fig. 5C).
On the other hand, fatty acids may also constitute phospholipids, one of the main components of the cell membrane. To further understand the effect of MALAT1 on lipid metabolism, we investigated several major types of lipid molecules by targeted LC-MS/MS. As a result, we observed reduced levels of multiple lipid molecules that contain 18:1 MUFAs, such as 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (16:0/18:1 PA), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol (16:0/18:1 PI) and 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (16:0/18:1 PG) (Fig. 5D and supplemental Table S7). Intriguingly, the abundance of PUFAs, such as docosahexaenoic acid (DHA) and arachidonic acid (AA), was elevated in siMALAT1 cells (Fig. 5D and supplemental Table S7). Higher levels of MUFAs have been widely observed in transformed cells and cancerous tissue as a result of enhanced lipogenesis. On the other hand, PUFAs are essential fatty acids obtained from the nutrients. The data showed that MALAT1 knockdown cells exhibited lower level of lipogenesis and higher dependence on lipid uptake from the extracellular nutrient supplies.
Furthermore, we investigated whether the alteration in phospholipid composition could affect the physical properties of the cell membrane. A fluorescence photobleaching recovery experiment was performed using Dil perchlorate, a lipophilic dye highly fluorescent when incorporated into the lipid bilayer. The results showed that knockdown of MALAT1 significantly reduced the fluidity of the cell membrane (Fig. 5E).
Taken together, our findings indicated that MALAT1 knockdown rewired the lipid metabolism in HCC cells by decreasing glucose uptake and inhibiting lipogenesis, leading to decreased TG abundance, altered phospholipid composition, and reduced membrane fluidity.
MALAT1 Knockdown Suppressed HCC Cell Proliferation, Migration, and Invasion
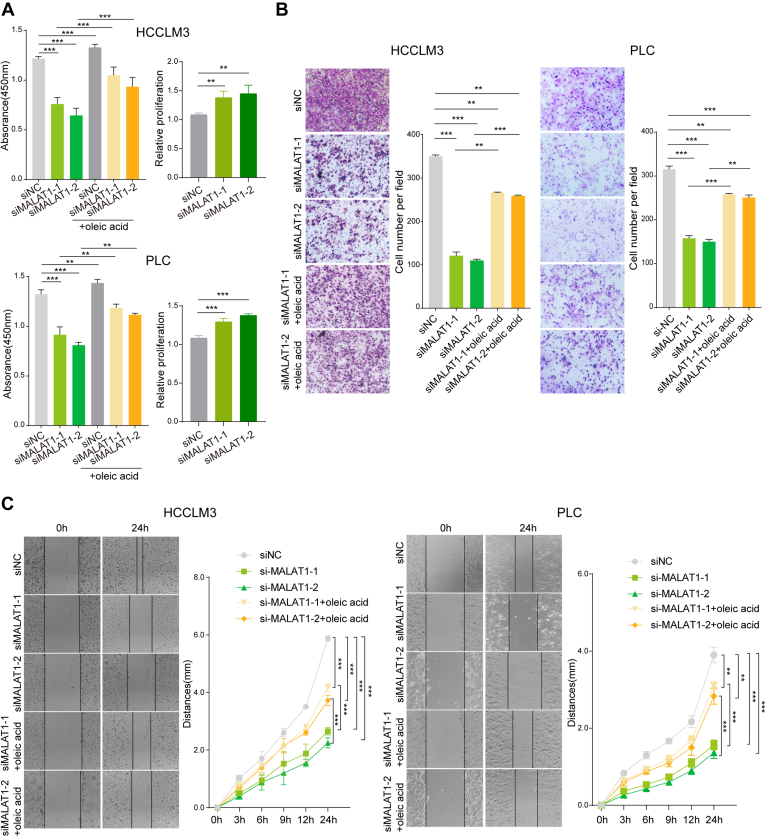
As a major component of the cell membrane, lipids are highly demanded by proliferating cells, and lipogenesis has been recognized as one of the key drivers for tumor progression. To further understand whether MALAT1 exerts its oncogenic function through regulating lipogenesis, we examined cell proliferation in HCC cells by CCK-8 experiments. The results showed that downregulation of MALAT1 inhibited the proliferation of both HCCLM3 and PLC cells, and incubation with oleic acid, the major product of SCD activity, can partially restore the inhibitory effect of siMALAT1 on cell proliferation (Fig. 6A). Additionally, siMALAT1 cells were more sensitive to oleic acid treatment compared with the control cells, suggesting that the rescue effect of oleic acid was associated with MALAT1 knockdown (Fig. 6A).
Fig. 6.
The role of MALAT1 in lipid metabolism contributes to its functions in promoting cell proliferation, migration, and invasion.A, cell proliferation of HCCLM3 (upper panel) and PLC (lower panel) cells with MALAT1 knockdown cultured with or without oleic acid (10 μM). The increase of cell proliferation delivered by oleic acid supplementation (relative proliferation) was calculated by dividing the absorbance of cells treated with oleic acid supplementation with the absorbance of the corresponding untreated cells. Data represent mean ± s.d. of triplicate independent experiments (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, by two-sided Student’s t test). B, matrigel invasion assay using HCCLM3 (left panel) and PLC (right panel) cells with MALAT1 knockdown cultured with or without oleic acid (10 μM). Representative images of the invaded cells are shown on the left in each panel. Data represent mean ± s.d. of triplicate independent experiments (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, by two-sided Student’s t test). C, migration assay using HCCLM3 (left panel) and PLC (right panel) cells with MALAT1 knockdown cultured with or without oleic acid (10 μM). Representative images of the migrated cells are shown on the left in each panel. The plots on the right show the cell migration distances at the indicated time points. Data represent means ± s.d. of triplicate independent experiments (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, by two-tailed student’s t test).
Next, we investigated whether MALAT1 can regulate the migration of HCC cells through lipogenesis. The results from wound-healing assay and invasion assay showed that the downregulation of MALAT1 significantly decreased the migratory and invasiveness of HCCLM3 and PLC cells. In addition, oleic acid can also partially restore the inhibitory effect of MALAT1 knockdown on cell migration and invasion (Fig. 6, B and C). Taken together, the data show that the role of MALAT1 in lipogenesis could contribute to its function in promoting the progressive phenotype of HCC cells.
Discussion
In this study, we performed an integrated transcriptomics and proteomics analysis and observed a total of 2662 transcripts and 1149 proteins differentially expressed in MALAT1 knockdown cells. Our data showed a profound impact of MALAT1 on the landscape of gene expression in HCC cells. MALAT1 has been reported to influence RNA translation as a competitive endogenous RNA (ceRNA) (51, 52). In addition, MALAT1 may regulate pre-mRNA splicing (29, 53). In this study, we found that MALAT1 can be copurified with the pre-mRNAs of SREBF1, PRKAB1, PRKAG1, and RAB14, which further confirmed its role in RNA splicing. Moreover, MALAT1 is highly involved in the process of gene transcription. A previous study has shown MALAT1 binding to Pc2 to activate gene expression by promoting transcription factor E2F1 SUMOylation (32). Two recent studies have shown that MALAT1 can extensively interact with gene sites that are actively transcribed (30, 54). In our study, we found that MALAT1 can affect the abundances of many important transcription factors, such as MYC, CEBPB, and SREBF1, and we constructed a TFs-targets network that provided a systematic view for the role of MALAT1 in gene regulation. Our data suggest that MALAT1 may regulate its target genes by coordinating various mechanisms involving gene expression regulation.
Several lncRNAs have been reported to be involved in lipid metabolism in HCC. For example, lncRNA highly upregulated in liver cancer (HULC) regulates lipid metabolism in liver cancer cells through upregulating acyl-CoA synthetase subunit ACSL1 (55). Another study reported that lncARSR promotes lipid accumulation by activating the IRS2/AKT pathway in HCC cell line (56). MALAT1 has been reported to be involved in lipid metabolism in the context of atherosclerosis (57). However, the underlying mechanism is still largely unclear, and its role in the lipid metabolism of cancer cells has not been investigated before.
In this study, we discovered that genes with altered expression induced by siMALAT1 were highly enriched in both of the AMPK signaling and fatty acid metabolism pathways. Intriguingly, our observations suggest that MALAT1 may exert its biological roles by acting at the level of pathways rather than targeting individual genes. AMPK is known to be a metabolic sensor responsible for nutrients uptake, and it plays critical roles in the regulation of both anabolism and catabolism of lipids (44, 58). Our data show that MALAT1 may rewire cellular metabolism by simultaneously promoting AMPK-mediated glucose uptake and the downstream lipogenesis to support the elevated cell proliferation.
Here we found that MALAT1 could modulate the expression of multiple lipid metabolism-related genes, including SREBF1 and SCD, at both the mRNA and protein levels. SREBF1 is a key membrane-bound transcription factor regulating lipid synthesis, and SCD is a direct target gene of SREBF1 involved in the biosynthesis of unsaturated fatty acids (48). The resultant MUFAs are then converted into TGs for energy storage or into phospholipids for membrane formation and signaling transduction. The upregulated de novo lipogenesis of cancer cells has now been widely recognized, and it may provide a survival advantage to the tumor cells to become less dependent on the circulatory lipids in the inadequately vascularized tumor microenvironment (59). Our results showed that MALAT1 knockdown significantly inhibited lipogenesis, and oleic acid partially restored the phenotype alterations in HCC cells induced by MALAT1 knockdown, suggesting that the role of MALAT1 in lipid metabolism may contribute to its oncogenic effects.
Moreover, besides regulating RNA splicing, MALAT1 may be involved in lipid metabolism through other mechanisms. For example, Zhou et al. (60) reported that MALAT1 could bind with forkhead box protein O1 (FOXO1), a transcription factor that plays an important role in both glucose and lipid metabolism. In contrast to the function of SREBF1, FOXO1 promotes lipolysis instead of lipogenesis. Additionally, our previous MALAT1 interactome study identified several enzymes involved in lipid degradation, such as propionyl-CoA carboxylase and hydroxyacyl-coenzyme A dehydrogenase, suggesting the possibility of MALAT1 to regulate lipid metabolism through directly binding to metabolic enzymes. However, further investigation is required to test such hypothesis. In addition, cross-linking-assisted enrichment may reveal other MALAT1 binding proteins and further complete the mechanisms by which MALAT1 regulates lipid metabolism.
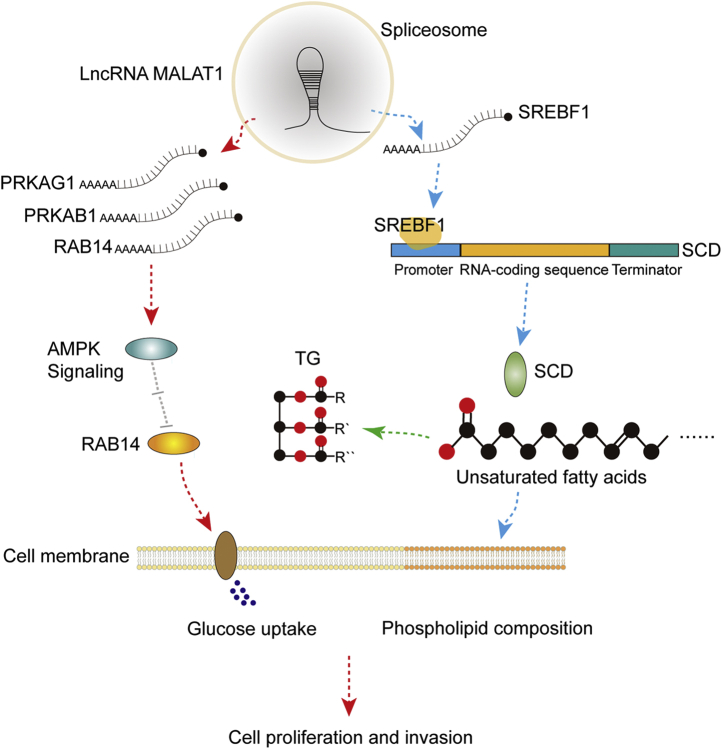
In conclusion, this study systematically describes the impact of MALAT1 on gene expression in HCC cells and reveals its role in the regulation of lipid metabolism. As shown in Figure 7, our observations suggest that MALAT1 promotes the expression of multiple genes in the AMPK signaling and unsaturated fatty acid metabolism pathways through pre-mRNA splicing and transcription, leading to upregulated glucose uptake and lipogenesis, which contributes to the role of MALAT1 in tumor cell proliferation and invasion. This study reveals the molecular mechanism by which MALAT1 involves in the dysregulation of lipid metabolism in HCC cells and further supports the potential therapeutic opportunities for targeting MALAT1 in HCC treatment.
Fig. 7.
The proposed molecular mechanism by which MALAT1 promotes cell proliferation and invasion through regulating lipogenesis. MALAT1 modulates the expression of genes in the AMPK signaling and unsaturated fatty acid biosynthesis pathways, thereby elevates glucose uptake and lipogenesis. The synthesized fatty acids are converted to TG for energy storage and phospholipids for cell membrane construction, which facilitates the cell proliferation and invasion of HCC cells.
Data Availability
The proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE (61) partner repository with the dataset identifier PXD016790. The RNA-Seq data have been deposited in NCBI’s Sequence Read Archive (SRA) and are accessible with BioProject ID PRJNA669618 (https://www.ncbi.nlm.nih.gov/bioproject).
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (21974094) and the National Key R&D Program of China (2018YFA0506900). This work was also funded by the Open Project Program of the State Key Laboratory of Proteomics (SKLP-O202004).
Author contributions
R. C. conceptualization; H. W. formal analysis; Z. Z. and R. C. funding acquisition; H. W., Y. Z., X. G., X. L., and Y. G. investigation; Y. Z. methodology; Z. Z., X. Z., and R. C. supervision; H. W., Y. Z., and X. G. visualization; H. W. and Y. Z. writing—original draft; R. C. writing—reviewing and editing.
Supplemental Data
References
- 1.Robey R.B., Weisz J., Kuemmerle N.B., Salzberg A.C., Berg A., Brown D.G., Kubik L., Palorini R., Al-Mulla F., Al-Temaimi R., Colacci A., Mondello C., Raju J., Woodrick J., Scovassi A.I. Metabolic reprogramming and dysregulated metabolism: Cause, consequence and/or enabler of environmental carcinogenesis? Carcinogenesis. 2015;36:S203–S231. doi: 10.1093/carcin/bgv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer. 2018;18:33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beloribi-Djefaflia S., Vasseur S., Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5 doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budhu A., Roessler S., Zhao X., Yu Z., Forgues M., Ji J., Karoly E., Qin L.X., Ye Q.H., Jia H.L., Fan J., Sun H.C., Tang Z.Y., Wang X.W. Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes. Gastroenterology. 2013;144:1066–1075.e1. doi: 10.1053/j.gastro.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakagawa H., Hayata Y., Kawamura S., Yamada T., Fujiwara N., Koike K. Lipid metabolic reprogramming in hepatocellular carcinoma. Cancers (Basel) 2018;10:447. doi: 10.3390/cancers10110447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guri Y., Colombi M., Dazert E., Hindupur S.K., Roszik J., Moes S., Jenoe P., Heim M.H., Riezman I., Riezman H., Hall M.N. mTORC2 promotes tumorigenesis via lipid synthesis. Cancer Cell. 2017;32:807–823.e12. doi: 10.1016/j.ccell.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Wang M., Han J., Xing H., Zhang H., Li Z., Liang L., Li C., Dai S., Wu M., Shen F., Yang T. Dysregulated fatty acid metabolism in hepatocellular carcinoma. Hepat. Oncol. 2016;3:241–251. doi: 10.2217/hep-2016-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selitsky S.R., Dinh T.A., Toth C.L., Kurtz C.L., Honda M., Struck B.R., Kaneko S., Vickers K.C., Lemon S.M., Sethupathy P. Transcriptomic analysis of chronic hepatitis B and C and liver cancer reveals MicroRNA-mediated control of cholesterol synthesis programs. mBio. 2015;6:e01500–e01515. doi: 10.1128/mBio.01500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mashek D.G., Khan S.A., Sathyanarayan A., Ploeger J.M., Franklin M.P. Hepatic lipid droplet biology: Getting to the root of fatty liver. Hepatology. 2015;62:964–967. doi: 10.1002/hep.27839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabor D.E., Kim J.B., Spiegelman B.M., Edwards P.A. Identification of conserved cis-elements and transcription factors required for sterol-regulated transcription of stearoyl-CoA desaturase 1 and 2. J. Biol. Chem. 1999;274:20603–20610. doi: 10.1074/jbc.274.29.20603. [DOI] [PubMed] [Google Scholar]
- 11.Fritz V., Benfodda Z., Rodier G., Henriquet C., Iborra F., Avancès C., Allory Y., de la Taille A., Culine S., Blancou H., Cristol J.P., Michel F., Sardet C., Fajas L. Abrogation of de novo lipogenesis by stearoyl-CoA desaturase 1 inhibition interferes with oncogenic signaling and blocks prostate cancer progression in mice. Mol. Cancer Ther. 2010;9:1740–1754. doi: 10.1158/1535-7163.MCT-09-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holder A.M., Gonzalez-Angulo A.M., Chen H., Akcakanat A., Do K.A., Fraser Symmans W., Pusztai L., Hortobagyi G.N., Mills G.B., Meric-Bernstam F. High stearoyl-CoA desaturase 1 expression is associated with shorter survival in breast cancer patients. Breast Cancer Res. Treat. 2013;137:319–327. doi: 10.1007/s10549-012-2354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Roemeling C.A., Marlow L.A., Wei J.J., Cooper S.J., Caulfield T.R., Wu K., Tan W.W., Tun H.W., Copland J.A. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin. Cancer Res. 2013;19:2368–2380. doi: 10.1158/1078-0432.CCR-12-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang G.M., Jiang Q.H., Cai C., Qu M., Shen W. SCD1 negatively regulates autophagy-induced cell death in human hepatocellular carcinoma through inactivation of the AMPK signaling pathway. Cancer Lett. 2015;358:180–190. doi: 10.1016/j.canlet.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 15.Tracz-Gaszewska Z., Dobrzyn P. Stearoyl-CoA desaturase 1 as a therapeutic target for the treatment of cancer. Cancers (Basel) 2019;11:948. doi: 10.3390/cancers11070948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulitsky I., Bartel D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long Y., Wang X., Youmans D.T., Cech T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017;3 doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 20.Gutschner T., Hämmerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M., Zörnig M., MacLeod A.R., Spector D.L., Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goyal B., Yadav S.R.M., Awasthee N., Gupta S., Kunnumakkara A.B., Gupta S.C. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim. Biophys. Acta Rev. Cancer. 2021;1875:188502. doi: 10.1016/j.bbcan.2021.188502. [DOI] [PubMed] [Google Scholar]
- 22.Ji P., Diederichs S., Wang W., Böing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E., Thomas M., Berdel W.E., Serve H., Müller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 23.Lin R., Maeda S., Liu C., Karin M., Edgington T.S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimoto R., Mayeda A., Yoshida M., Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochim. Biophys. Acta. 2016;1859:192–199. doi: 10.1016/j.bbagrm.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Hu L., Wu Y., Tan D., Meng H., Wang K., Bai Y., Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okugawa Y., Toiyama Y., Hur K., Toden S., Saigusa S., Tanaka K., Inoue Y., Mohri Y., Kusunoki M., Boland C.R., Goel A. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park J.Y., Lee J.E., Park J.B., Yoo H., Lee S.H., Kim J.H. Roles of long non-coding RNAs on tumorigenesis and Glioma development. Brain Tumor Res. Treat. 2014;2:1–6. doi: 10.14791/btrt.2014.2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao H., Tang K., Liu P., Chen K., Hu J., Zeng J., Xiao W., Yu G., Yao W., Zhou H., Li H., Pan Y., Li A., Ye Z., Wang J. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–38015. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., Blencowe B.J., Prasanth S.G., Prasanth K.V. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engreitz J.M., Sirokman K., McDonel P., Shishkin A.A., Surka C., Russell P., Grossman S.R., Chow A.Y., Guttman M., Lander E.S. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen R., Liu Y., Zhuang H., Yang B., Hei K., Xiao M., Hou C., Gao H., Zhang X., Jia C., Li L., Li Y., Zhang N. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017;45:9947–9959. doi: 10.1093/nar/gkx600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L., Lin C., Liu W., Zhang J., Ohgi K.A., Grinstein J.D., Dorrestein P.C., Rosenfeld M.G. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao Y., Smyth G.K., Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 35.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 37.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keenan A.B., Torre D., Lachmann A., Leong A.K., Wojciechowicz M.L., Utti V., Jagodnik K.M., Kropiwnicki E., Wang Z., Ma'ayan A. ChEA3: Transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47:W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L., Wang L., Shangguan D., Wei Y., Han J., Xiong S., Zhao Z. Ultra-high-performance liquid chromatography electrospray ionization tandem mass spectrometry for accurate analysis of glycerophospholipids and sphingolipids in drug resistance tumor cells. J. Chromatogr. A. 2015;1381:140–148. doi: 10.1016/j.chroma.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Wang H., Shao X., He Q., Wang C., Xia L., Yue D., Qin G., Jia C., Chen R. Quantitative proteomics implicates Rictor/mTORC2 in cell adhesion. J. Proteome Res. 2018;17:3360–3369. doi: 10.1021/acs.jproteome.8b00218. [DOI] [PubMed] [Google Scholar]
- 44.Lin S.C., Hardie D.G. AMPK: Sensing glucose as well as cellular energy status. Cell Metab. 2018;27:299–313. doi: 10.1016/j.cmet.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 45.Martínez-Reyes I., Chandel N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020;11:102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan C., Chen J., Chen N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016;6:22640. doi: 10.1038/srep22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paton C.M., Ntambi J.M. Biochemical and physiological function of stearoyl-CoA desaturase. Am. J. Physiol. Endocrinol. Metab. 2009;297:E28–E37. doi: 10.1152/ajpendo.90897.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimano H., Sato R. SREBP-regulated lipid metabolism: Convergent physiology - divergent pathophysiology. Nat. Rev. Endocrinol. 2017;13:710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- 49.Jaldin-Fincati J.R., Pavarotti M., Frendo-Cumbo S., Bilan P.J., Klip A. Update on GLUT4 vesicle traffic: A cornerstone of insulin action. Trends Endocrinol. Metab. 2017;28:597–611. doi: 10.1016/j.tem.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Y., Ma L. New insights into long non-coding RNA MALAT1 in cancer and metastasis. Cancers (Basel) 2019;11:216. doi: 10.3390/cancers11020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji Q., Cai G., Liu X., Zhang Y., Wang Y., Zhou L., Sui H., Li Q. MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis. Cell Death Dis. 2019;10:378. doi: 10.1038/s41419-019-1598-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malakar P., Shilo A., Mogilevsky A., Stein I., Pikarsky E., Nevo Y., Benyamini H., Elgavish S., Zong X., Prasanth K.V., Karni R. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 2017;77:1155–1167. doi: 10.1158/0008-5472.CAN-16-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui M., Xiao Z., Wang Y., Zheng M., Song T., Cai X., Sun B., Ye L., Zhang X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 56.Chi Y., Gong Z., Xin H., Wang Z., Liu Z. Long noncoding RNA lncARSR promotes nonalcoholic fatty liver disease and hepatocellular carcinoma by promoting YAP1 and activating the IRS2/AKT pathway. J. Transl Med. 2020;18:126. doi: 10.1186/s12967-020-02225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu L., Tan L., Yao J., Yang L. Long non-coding RNA MALAT1 regulates cholesterol accumulation in ox-LDL-induced macrophages via the microRNA-17-5p/ABCA1 axis. Mol. Med. Rep. 2020;21:1761–1770. doi: 10.3892/mmr.2020.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Q., Liu S., Zhai A., Zhang B., Tian G. AMPK-mediated regulation of lipid metabolism by phosphorylation. Biol. Pharm. Bull. 2018;41:985–993. doi: 10.1248/bpb.b17-00724. [DOI] [PubMed] [Google Scholar]
- 59.Röhrig F., Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 60.Zhou L., Xu D.Y., Sha W.G., Shen L., Lu G.Y. Long non-coding RNA MALAT1 interacts with transcription factor Foxo1 to regulate SIRT1 transcription in high glucose-induced HK-2 cells injury. Biochem. Biophys. Res. Commun. 2018;503:849–855. doi: 10.1016/j.bbrc.2018.06.086. [DOI] [PubMed] [Google Scholar]
- 61.Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.W., Wang R., Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE (61) partner repository with the dataset identifier PXD016790. The RNA-Seq data have been deposited in NCBI’s Sequence Read Archive (SRA) and are accessible with BioProject ID PRJNA669618 (https://www.ncbi.nlm.nih.gov/bioproject).