Abstract
Tuberculosis (TB) affects around 10 million people worldwide in 2019. Approximately 3.4 % of new TB cases are multidrug-resistant. The gold standard method for detecting Mycobacterium tuberculosis , which is the aetiological agent of TB, is still based on microbiological culture procedures, followed by species identification and drug sensitivity testing. Sputum is the most commonly obtained clinical specimen from patients with pulmonary TB. Although smear microscopy is a low-cost and widely used method, its sensitivity is 50–60 %. Thus, owing to the need to improve the performance of current microbiological tests to provide prompt treatment, different methods with varied sensitivity and specificity for TB diagnosis have been developed. Here we discuss the existing methods developed over the past 20 years, including their strengths and weaknesses. In-house and commercial methods have been shown to be promising to achieve rapid diagnosis. Combining methods for mycobacterial detection systems demonstrates a correlation of 100 %. Other assays are useful for the simultaneous detection of M. tuberculosis species and drug-related mutations. Novel approaches have also been employed to rapidly identify and quantify total mycobacteria RNA, including assessments of global gene expression measured in whole blood to identify the risk of TB. Spoligotyping, mass spectrometry and next-generation sequencing are also promising technologies; however, their cost needs to be reduced so that low- and middle-income countries can access them. Because of the large impact of M. tuberculosis infection on public health, the development of new methods in the context of well-designed and -controlled clinical trials might contribute to the improvement of TB infection control.
Keywords: diagnosis, drug-resistant tuberculosis, nucleic acid amplification techniques, tuberculosis
Introduction
State of the art
Methods for detecting Mycobacterium tuberculosis , the aetiological agent of tuberculosis (TB), in a given sample might have respectable sensitivity (i.e. the ability to detect the presence of bacillus in the specimen) and specificity (i.e. the ability to detect only the target bacillus). There are numerous commercial serological tests for the diagnosis of TB in many settings; however, they are not recommended because of their poor performance. Although we have acknowledged them, we have not revisited in depth any current immunological test for detecting M. tuberculosis infection. Moreover, serological tests were found to be inconsistent and inaccurate, with widely varying values for sensitivity and specificity and high proportions of discordance. Hence, the World Health Organization (WHO) issued a guideline not recommending the use of such tests for the diagnosis of TB [1]. On the one hand, the only in vivo test available to evaluate M. tuberculosis infection is the tuberculin skin test, which has fair sensitivity but poor specificity [2]. On the other hand, the new interferon-gamma release assays are specific ex vivo tests. Both methods are based on the measurement of adaptive host immune response. However, none of these tests can accurately distinguish between latent and active TB [2–4]. Other diagnostic tools have been developed for the detection of M. tuberculosis , as well as drug susceptibility and viability, which can be evaluated by metabolic activity responsiveness (detection of respiration or mRNA synthesis), cell membrane integrity, or nucleic acid detection [5]. Along with these tests, conventional solid and new liquid media-based methods, which can obtain rapid results, have been developed; however, these tests are quite expensive [6]. Other methods have also been described for the detection of pathogenic mycobacteria (Table 1).
Table 1.
Commonly used methods for the detection of pathogenic mycobacteria
|
Assay |
Reference |
|---|---|
|
Polymerase chain reaction (PCR)* |
[149] |
|
Reverse transcription (RT)-PCR |
[88, 150] |
|
Enzyme-linked immunosorbent assay (ELISA) |
[151] |
|
Potentiometric biosensors |
[152] |
|
Surface plasmon resonance |
[153] |
|
Bioluminescence |
[154] |
|
Fluorescent labelling |
[155] |
|
Flow cytometry (FCM) |
[156] |
|
Transcriptomic |
[115] |
*Including TB-LAMP [84].
The aetiological agent of TB
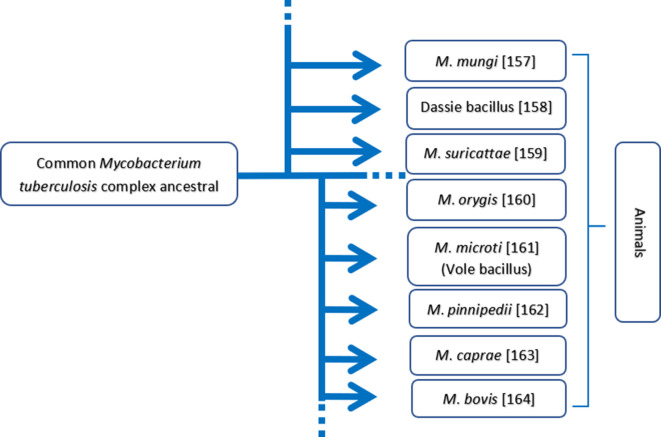
Approximately 50 mycobacterial species cause human diseases. M. tuberculosis belongs to the family Mycobacteriaceae and is a member of the M. tuberculosis complex that includes M. tuberculosis , M. canettii and M. africanum , as well as other members that cause disease in other animal species (Fig. 1). All bacilli belonging to he family Mycobacteriaceae have a lipid-rich cell wall that confers resistance against chemotherapeutic agents but not against physical agents such as ultraviolet radiation and heat [7–10].
Fig. 1.
Genealogical tree assembling a few members of the Mycobacterium tuberculosis complex causing disease to various animal species. The relevant references are shown alongside [157–164] .
The gold standard for diagnosing TB is a positive M. tuberculosis culture [11]. M. tuberculosis is a rod-shaped, non-spore-forming and strictly aerobic bacterium. As a facultative intracellular pathogen, it is capable of living in human phagocytic cells. Because of its low growth rate, cell division occurs every 18–20 h, and it takes several weeks for the bacterium to be detected as visible colonies in solid culture media [12]. Importantly, this could be considered to be a major obstacle for rapid diagnosis.
The method of Mycobacterium sp. characterization from culture is more sensitive than ordinary bacilloscopy, such as the acid-fast bacilli (AFB) microscopy, which allows for the detection of 10–100 bacilli ml−1 of a concentrated clinical sample [13]. The two most widely used culture media are the egg-based Löwenstein–Jensen (LJ) slopes and the Middlebrook series of agars (7H10 and 7H11), which are both solid-phase broths [14]. Among the two, LJ is more efficient for the detection of growth rate, whereas Middlebrook promotes faster bacterial growth [15]. Liquid culture media are used rationally to both increase the number of cells and store strains. The use of biphasic cultures in the same bottle enables more accurate analysis of the colony aspect [16]. In the case of samples obtained from co-infected TB/HIV patients, which usually have a high incidence of nontuberculous mycobacteria (NTM), species must be identified via biochemical tests or the use of specific genetic probes [17].
Following accurate diagnosis, patients must immediately begin TB chemotherapy with specific agents; however, as indicated earlier, drug resistance and compliance at this stage are a major concern. The inefficacy of treatment has been strongly related to the selection of mutant bacilli that are resistant to standard TB therapy as resistance to every anti-mycobacterial agent used is often caused by spontaneous mutations of M. tuberculosis in the target genes [18]. The two resistance mechanisms observed in M. tuberculosis are (i) overexpression of the drug target [19] and (ii) alteration of the drug target structure [20]. As indicated, a quicker diagnostic process, including the identification of resistant strains, is needed to initiate the most efficient therapeutic regimen. There are numerous target genes that can be potentially used to detect the resistant forms, such as katG, inhA, mabA and ahpC for isoniazid (INH), rpoB for rifampicin (RMP), rpsL and rrs for streptomycin (SM), embA and embB for ethambutol (EMB), and girA for fluoroquinolones. These genes can also be used to detect M. tuberculosis in clinical samples using molecular approaches [21].
Since the increase in the number of cases of treatment failure in active TB cases, the development of fast, reliable, simple and accurate methods for identifying the Mycobacterium sp. and its drug resistance has become paramount. Therefore, the detection of M. tuberculosis using a myriad of approaches has succeeded in the past 20 years. Because of a lack of review papers on this subject, we aimed to introduce the main tests developed in the current millennium and currently used for the detection of this pathogen in clinical samples and discuss their weaknesses and strengths.
Detection of M. tuberculosis
Microscopic analysis
With regard to bacteriological analysis, the detection of AFB in fresh, stained smears of sputum of suspected patients examined microscopically provides initial evidence of the presence of mycobacteria in clinical specimens. In low- and middle-income (LMI) countries, diagnosis is made primarily via microscopic examination, but this method only has a sensitivity of 50–60 % in cases of confirmed (bacillary) pulmonary TB and even lower sensitivity (< 30 %) in HIV-positive or immunosuppressed patients and in children [22]. However, it is inexpensive, easy to perform and analyse, has a short time frame (1 day) and is correlated with the infectiousness of the case [23, 24]. In short, the traditional AFB is a method referred to for bacteria that are resistant to acid discolouration after staining procedures such as the Ziehl–Neelsen (ZN) and Kinyoun techniques. This property is due to the lipid component, which represents approximately 60 % of the dry weight of the cell wall. The same lipid-rich structure is responsible for both the slow growth and the resistance of bacteria to acids [25].
Among several factors related to the virulence of M. tuberculosis , the lipid profile of the cell wall has drawn much interest owing to its unique composition, which is implicated in giving the pathogen an advantage over the host [26]. This lipid-rich cell wall is a dynamic structure that is also involved in the regulation of the transport of anti-tuberculosis drugs [27]. In fact, M. tuberculosis also alters its fatty acid metabolism to survive the host conditions; this is reflected in a different cell wall composition in terms of lipids, thereby increasing its virulence. Furthermore, this profile has been shown to modulate the immune responses launched by the host [26]. Upregulated expression of the isocitrate lyase gene, indicative of a shift in the central carbon metabolism, has been observed in M. tuberculosis cultured on long-chain fatty acids in lipid-loaded macrophages [28]. Upregulation of lipid storage in this bacillus as a means to recover from reductive stress-induced damage was shown to result in a slower growth rate and a drug-tolerant phenotype in the lipid-rich in vivo milieu in a macrophage environment [28]. Two recent broad reviews on this fascinating topic have been published [26, 27].
Before the development of new diagnostic methods, mycobacteria were detected in morning sputum samples via direct microscopy using the ZN technique [21]. However, this method has serious drawbacks (Table 2). First, the time required for adequate detection is crucial; expert technicians take approximately 5 min to observe at least 200–300 microscopic fields in only 1 smear. This leads to exhaustion under the microscope (reading fatigue) and consequently to false-negative results; moreover, in overworked laboratories, the recommended time for analysis might not be followed [29, 30]. Second, this method has poor sensitivity: in 45 % of patients with pulmonary TB and 75 % of patients with EP-TB, mycobacteria are mostly not detected because of the minimum number of bacilli per sputum sample (104 ml−1) required [31]. Lastly, specimens should be transported rapidly to the facility to avoid overgrowth of other contaminants. The cetylpyridinium chloride (CPC) method is widely used for the transport of sputum specimens; however, the detection of AFB via ZN staining can be significantly reduced in specimens preserved by CPC [32]. In addition, specimens treated with CPC should be preferentially inoculated in egg-based media as agar-based media have insufficient neutralizing activity for this quaternary ammonium compound. Sodium carbonate has been found to be a better preservative for sputum specimens for AFB smear microscopy and culture [33]. Nevertheless, aside from the weakness related to this method, a recent study has documented that DNA can be extracted from ZN smears and RMP resistance markers can be evaluated by a single polymerase chain reaction (PCR), namely nested PCR [34], thereby allowing more precise analysis of the sample, which is relevant for multidrug therapy. In a systematic review, the sensitivity of microscopy compared with that of culture ranged from 0–100 % in induced sputum samples; only 8 of 23 studies reported on the species of mycobacteria isolated in culture [35]. Notably, in settings with a high level of M. tuberculosis infection, the ZN technique is the cheapest method [36].
Table 2.
Overview of the most used methods to detect M. tuberculosis and its variants including cost-effectiveness
|
Assay |
Accessibility/cost |
Sensitivity |
Quantification |
Turnaround time* |
Resistance identification |
|---|---|---|---|---|---|
|
Bacilloscopy |
High |
Low |
Intermediate |
2–3 days |
No |
|
Solid culture |
Cheap |
Low |
Intermediate |
30–60 days |
No |
|
Liquid culture |
Intermediate |
Intermediate |
Intermediate |
15–30 days |
No |
|
Flow cytometry |
Low |
High |
High |
2–3 days |
Yes |
|
Nested PCR/RT-PCR |
Low |
Intermediate |
Low |
2–4 days |
Yes |
|
qRT-PCR |
Low |
Low |
Intermediate |
2–4 days |
No† |
|
GeneXpert MTB/RIF |
Low |
High |
High |
90 min |
Yes |
|
Fluorescence microscopy |
Intermediate |
High |
High |
1–2 days |
Yes |
*Time to detect the presence or absence of M. tuberculosis, according to the Foundation for Innovative New Diagnostics (FIND).
†Except for EP-TB [92].
Fluorescence microscopy (FM) is another option to detect M. tuberculosis in a given sample. The use of auramine as a fluorescent marker was introduced during the 1940s [37], and the sensitivity of direct microscopy can be improved by concentrating the sputum in a sediment and applying auramine-O fluorescence staining, although this is not sufficient to distinguish M. tuberculosis from other mycobacteria [21]. In 2003, Kivihya-Ndugga and colleagues [38] compared the efficiency and cost-effectiveness of FM with those of the ZN method for the analysis of the sputum of patients with pulmonary TB. When considering cost-effectiveness, FM has been shown to have better sensitivity (78 % vs 60 % for ZN), a key factor leading to savings for both the healthcare system and the patient [38]. A summary of these findings is presented in Table 2. In fact, the use of FM is promising when compared with the poor sensitivity of light microscopy for children with TB [39]. It has been found to have poor performance for the identification of smear-negative TB in HIV-positive patients [40–43]. The increased sensitivity of FM compared with that of traditional light microscopy for the detection of pulmonary TB has recently been supported in a systematic review [44]. Additionally, a meta-analysis has found that FM might increase the sensitivity of sputum smears by 10 % compared with the conventional method [45]. However, the equipment required for FM is expensive; thus, its use has been limited to regions that can afford it. In addition, the fluorescence fades with time. For this reason, the slides must be read within 24 h after development [39].
Fluorescent markers are particularly useful for in vitro and in vivo studies. They may be critical for understanding M. tuberculosis biology and disease progression as well as the development of new technologies for diagnosis and treatment, such as dyes used for flow cytometry analysis (as discussed below). A decade ago, a new near-infrared fluorogenic substrate for an endogenous mycobacteria enzyme was developed. This method using mycobacterial β-lactamase-sensitive compounds is capable of faster detection in vitro, with a limit of 6×102 colony-forming units, as well as in the murine lung, with a limit of approximately 1×104 colony-forming units [46]. However, sputum may interfere during the detection of this enzyme, increasing the probability of false-positive results. Hence, it has been suggested that unknown β-lactamase present in the clinical samples might be cleaved by the fluorogenic substrate because the hypothetical enzyme could have a similar active site to that of the M. tuberculosis enzyme [47].
Solid and liquid media for culture
The gold standard recommended by the WHO for the diagnosis of TB is the use of the culture method and the identification of species based on their physiological and biochemical features as well as the time of culture growth [11]. In biological specimens, sputum has invariably been used for that purpose.
The introduction of prokaryotic cell culture techniques has enabled both clinical and research laboratories to identify the Mycobacterium sp. and its susceptibility to antibiotics, leading to more efficacious treatment for patients with TB (Table 2). Culture is much more sensitive than the prior microscopic examination: 50 % of the pulmonary TB cases and an even greater proportion of documented EP-TB cases are negative under microscopy and are therefore only diagnosed via culture, as this method is capable of detecting few bacteria per millilitre [48, 49]. As stated previously, the traditional methods to determine the viability and growth of the Mycobacterium sp. are employed using solid agar media such as Middlebrook 7H10 or LJ; the latter has been employed for the culture of M. tuberculosis in LMI countries. Ogawa medium is another egg-based medium that is comparable to LJ in terms of its composition. It is more affordable because of the replacement of asparagine with sodium glutamate, an amino acid that is more readily available and much cheaper [50]. Nevertheless, in these solid culture media, it takes 6 weeks for M. tuberculosis to be readily detected by culture [51, 52].
In 2007, the WHO endorsed the use of liquid culture media as a gold standard for TB diagnosis based on the recommendations of international experts and studies that demonstrated that this method can be implemented in LMI settings to improve multidrug-resistant (MDR) TB diagnosis and AFB smear-negative pulmonary TB detection. The WHO’s recommendation is consistent with a large body of scientific literature regarding liquid culture media. Studies have reported a higher rate of mycobacteria isolation in a short time frame when compared with the same specimens from solid culture media [53]. Hence, the latter have been ruled out from the clinical laboratory routine [54].
Currently, numerous liquid-based media for culture with automated incubation and reading methods are available, and several studies with regard to their use for antimicrobial susceptibility testing (AST) have been revised [6, 55]. Among methods involving liquid culture media, there are radiometric-based methods, such as the Becton Dickinson BACTEC 460 system [56], and colorimetric methods based on bacterial gas production, such as the bioMérieux MB/BacT System [57]. The BACTEC 460 procedure is based on the production of radioactive carbon dioxide from palmitic acid [58]. It is well established and extensively used for AST; it is now considered to be a standard method [59, 60]. Unfortunately, this method has drawbacks, such as logistics regarding radioactive waste disposal and the requirement for needle inoculation of specimens into the liquid culture medium. To overcome these problems, a nonradiometric colorimetric system was developed, namely, the BACTEC Mycobacteria Growth Indicator Tube (MGIT) 960, which seems to be more reliable than the BACTEC 460 for AST [61–64]. This method has the advantage of being fully automated. An oxygen-quenching fluorescent sensor is used for detection, and it eliminates the need for needles to be used. Another colorimetric system is the MB/BacT, which has been compared with the BACTEC 460 [59]. Strikingly, the results for AST produced good agreement and concordance with the values for INH (96.3 %), RMP (98.8 %) and EMB (98.8 %) [65].
A study by Sorlozano and colleagues [66] comparing BACTEC MGIT 960 vs MB/BacT and the LJ solid culture media revealed 86.5 % mycobacterial recovery from BACTEC MGIT 960 in clinical samples compared with approximately 79.5 % mycobacterial recovery from the two others. The study also revealed that the combination of liquid-based and solid-based media exhibited better performance (95.5 % recovery). The period taken to isolate mycobacteria is quite important to achieve rapid diagnosis [67]. Isolation in the BACTEC MGIT 960 method (15.3±6.1 days) was shorter than that in the other two methods (20.1±8.6 and 32.6±11.8 days for MB/BacT and LJ, respectively). Detection with liquid-based media is faster and slightly more sensitive than with solid-based media, although the high sensitivity of that method is prone to contamination with environmental mycobacteria and other micro-organisms [68, 69]. This contamination may be associated with NTM because liquid-based media were found to be more effective than solid media for the detection of NTM [69].
Collectively, liquid-based media better support the growth of the M. tuberculosis complex than solid-based media [50]. Although both liquid- and solid-based media have fair sensitivity and specificity, their diagnostic values restrict them to qualitative analysis. Conversely, quantitative methods such as direct microscopic counting of AFB account for the exact number of mycobacteria present in the sample.
PCR and other molecular methods
Molecular identification has emerged as an alternative or a complement to traditional microbiological identification. It is now regarded as a promising approach [70]. Nucleic acid amplification tests (NAATs) such as based PCR, nested PCR, reverse-transcription (RT)-PCR and TB loop-mediated isothermal amplification (LAMP), use molecular probes that hybridize specifically with M. tuberculosis complex, M. avium complex, M. kansasii , or M. gordonae [71]. For a comprehensive review see [5, 72]. These assays have sensitivities and specificities of almost 100 % in the presence of at least 1×105 organisms, offering better accuracy than AFB microscopy and faster results than culture [73]. Accordingly, a comparative study found a 100 % correlation between the IS6110-based PCR and the BACTEC MGIT 960 system, and the authors suggested both methods to be used as a combined protocol for routine clinical diagnosis [74]. In paucibacillary, smear-negative TB, the sensitivity of NAAT is invariably approximately 50–60 %, but the specificity is almost 99 % [75]. In general, PCR is less sensitive than culture, and its use is limited by costs and the need for laboratory expertise and infrastructure [76].
In the early 1990s, a pioneer NAAT study was conducted by Eisenach and colleagues [77] to detect M. tuberculosis using PCR, in which sensitivity, specificity and concluding results were available within 48 h. This method can also be employed to identify drug-resistant bacilli (Table 2). In fact, a broad review on the topic of drug sensitivity testing was performed in 2014 [78], and thus we intended to update this subject (Table 3). Accordingly, we conducted a phenotypic and genetic analysis of a drug-resistant phenotype and resistance-conferring mutations in patients at a referral hospital in Fortaleza, Brazil [79]. Primary resistance was high in the participants (50.9 %), and analysis via multiplex allele-specific PCR and sequencing detected and identified mutations in katG, rpoB, inhA promoter and gyrA (Table 3). Spatial analysis revealed distinct isolates distributed in areas with low socioeconomic status in the city. Our results emphasized the importance of detecting resistance to TB drugs [79]. Previously, multiplex-PCR technology has been used to recognize INH-resistant M. tuberculosis from isolates in India [80]. Although PCR is unable to recognize viable vs non-viable bacilli (as discussed below), which is related to the inherent deficiency in quantifying the number of mycobacteria, NAAT approaches as a whole help to achieve a diagnosis of TB quickly and expedite the decision-making process for the best drug treatment protocol (Tables 2 and 3).
Table 3.
Overview of the most recent methods (2014 onwards) for detecting M. tuberculosis drug resistance, highlighting the respective target and drug
|
Assay |
Target gene |
Referring drug |
Reference |
|---|---|---|---|
|
GenoType MTBDRplus* |
rpoB, katG and inhA |
INH and RMP |
[165–170] |
|
GenoType MTBDRsl* |
embB, gyrA and rrs |
EMB, fluoroquinolones, aminoglycosides and cyclic peptide |
[171–173] |
|
RT-PCR* |
katG, rpoB, MPB64 and IS6110 |
RMP and INH |
|
|
Abbott Real-Time MTB RIF/INH† |
katG, inhA and rpoB |
RMP and INH |
[181–183] |
|
Multiplex allele-specific PCR*,‡ |
rpoB, katG, inhA, pncA and embB |
RMP, INH, pyrazinamide, fluoroquinolones and aminoglycosides |
|
|
PCR-RFLP* |
katG, embB, rpsL and gyrA |
INH, streptomycin and fluoroquinolones |
[193–195] |
|
Genedrive* |
rpoB |
RMP |
[196] |
|
Anyplex Plus MTB/NTM* |
katG and inhA |
RMP and INH |
[197–199] |
|
AuNP-based lateral flow* |
katG |
INH |
[200] |
|
Electrochemical DNA sensors†,§ |
rpoB |
RMP |
[201] |
|
Binary deoxyribozyme sensors† |
rpoB, katG, inhA and gyrA |
RMP, INH and fluoroquinolones |
[202] |
|
Luminex MicroPlex microsphere† |
rpoB, GyrA and inhA |
RMP and fluoroquinolones |
[203] |
|
Nipro Genoscholar† |
pncA |
Pyrazinamide |
[204, 205] |
|
Sequencing* |
rpoB, katG, embB, gyrA, gyrB, inhA, rpsL and rrs |
RMP, INH, fluoroquinolone and streptomycin |
[206–214] |
*Sputum samples.
†M. tuberculosis strain samples.
‡Distinguishes M. tuberculosis and M. avium complexes from other mycobacteria directly from clinical specimens [215].
§Based on polypyrrole/Fe3O4 nanocomposite-bearing redox naphthoquinone tag on PAMAM (spaNQ/PAMAM/PPy/Fe3O4).
In addition to PCR, there are three commercially available tests to directly identify the TB bacillus by 16S ribosomal transcripts, namely, (i) the M. tuberculosis direct test (MTD) using an amplicon detected using a DNA probe [81, 82]; (ii) the Amplicor using genus-specific primers by means of a colorimetric reaction [83]; and (iii) the TB-LAMP single-tube technique for the isothermal amplification of DNA or RNA [84]. The first two tests have been compared with the culture and clinical parameters, with both exhibiting high sensitivity and specificity in smear-positive specimens; however, low values were obtained in smear-negative specimens [85]. In addition, TB-LAMP is another low-cost alternative test for AFB ((84; [86]. TB-LAMP is simple, self-contained and efficacious for the early diagnosis of suspected cases of TB, with the advantages of having high throughput and no requirements for sophisticated equipment and complex biosafety facilities [84]. A meta-analysis found that additional factors, such as cost, feasibility and acceptability in settings that remain reliant on AFB, should be considered when deciding to implement this approach [86].
RT-PCR has been employed in cultured and clinical specimens. It is a scale-up technology employed to overcome the limitation of PCR in the quantification of samples. Its sensitivity ranges from 71–98 % and its specificity is close to 100 % [87]. The results are usually obtained 1.5 to 2 h after DNA extraction, and there is a low risk of contamination (reaction and detection occur in a single tube). In 2006, Ortu and colleagues [88] employed RT-PCR to detect and identify M. tuberculosis . The authors suggested that this method can be useful for assessing the treatment response in patients with TB, as it is capable of quantifying the exact number of DNA copies of the pathogen, indicating the degree of infection in the patient. They also highlighted the risk of sensitivity reduction when samples contain small amounts of M. tuberculosis DNA contaminants.
As an inherent concern already highlighted, the detection of pathogen DNA via PCR does not distinguish viable from non-viable bacilli, and some NAAT studies have described the occurrence of false positivity due to contamination [89–91]. To minimize this possibility, real-time mRNA-based quantitative (q)RT-PCR, instead of DNA, might be useful to detect viable M. tuberculosis bacilli and for the diagnosis of active TB [92]. RNA has a short half-life [93] and it is predicted to be found in only viable cells. In addition, M. tuberculosis mRNA is quite stable, with a half-life of roughly 9 min [94]. In fact, the mRNA of Ag85B has only been detected in viable bacteria [95–97]. However, the sensitivity of the assay is low, and it is burdensome to work with RNA on a routine basis (Table 2). Thus, more effort is needed to develop simpler and more cost-effective PCR tests that can be used regularly in LMI countries to achieve efficient TB diagnosis [92].
Although PCR restriction fragment length polymorphism (RFLP) was developed in the last century, PCR-RFLP analysis of the genetic code has been employed to rapidly identify mycobacteria, including M. tuberculosis , in the current millennium [98]. In some cases, PCR-RFLP has been compared with conventional biochemical tests for diagnostic use [99, 100]. In addition, DNA microarrays have been used for the identification of M. avium , M. chelonae , M. fortuitum , M. gordonae , M. intracellulare , M. kansasii , M. scrofulaceum , M. smegmatis , M. tuberculosis and M. xenopi isolates [101]. We employed spoligotyping and georeferencing systems in an attempt to elucidate the genetic diversity of M. tuberculosis isolates circulating in patients with pulmonary TB from Fortaleza, Brazil [102]. Drug susceptibility testing and spoligotyping assay were both conducted, and the residences of the patients were georeferenced. Approximately 44.3 % of isolates were resistant to at least one drug, whereas 55.7 % were sensitive to all the drugs tested. A high frequency of resistance was observed in previously treated TB cases and among new cases. It was observed that the spoligopattern family distribution paralleled that reported for South America. A high case rate occurred among the resistant TB group because of transmitted and acquired resistance.
An automated method based on PCR technology has been described as a promising tool for fast and specific detection of M. tuberculosis . The Cepheid GeneXpert MTB/RIF system and its next-generation Xpert Ultra [103] uses a plastic cartridge containing all of the reagents required for DNA extraction to amplify the rpoB gene. GeneXpert MTB/RIF detects the amplicons in association with a point-of-care device, with results obtained in 2 h and with minimal hands-on technical time [104]. Recent studies have indicated the use of this approach to suppress the risk of bioaerosol infection as well as its use in point-of-care settings [105]. Accordingly, a single test using GeneXpert MTB/RIF might detect TB in 99 % of patients with smear-positive and >80 % of patients with smear-negative pulmonary TB [76]. Furthermore, GeneXpert MTB/RIF can detect RMP resistance with a sensitivity of 95.1 % and a specificity of 98.4 % [105], and a meta-analysis has suggested that this approach should be preferred in settings where resource and infrastructure requirements are adequate and where co-infection TB/HIV or drug resistance is a concern [86]. In addition, the updated Xpert Ultra test might detect HIV-associated TB with high sensitivity, thereby reducing TB-related mortality in co-infected TB/HIV patients [106]. The WHO initially recommended this technology in early 2011 [67]; the organization is monitoring the global rollout of this method to promote coordination [107]. In the past 3 years, one systematic and four literature reviews of the performance of GeneXpert MTB/RIF for the diagnosis of TB in several settings have been published [108–112]. Because comprehensive reviews on the diagnostic accuracy of the GeneXpert MTB/RIF system were published in 2013 [113], 2014 [114] and 2019 [103], we have not focused on this topic here. Accordingly, those reviews were made available for the detection of pulmonary TB and RMP resistance as part of a WHO process to develop structured guidelines on the use of the test.
Finally, the assessments of global gene expression measured in whole blood recently allowed for the diagnosis of TB. Accordingly, a pioneer study prospectively identified host-derived, blood-based 16-gene expression biomarkers in people from both South Africa and The Gambia who are at risk of developing active TB, thereby indicating the possibility of preventing the disease via a targeted intervention using non-sputum-based tests [115]. In a single year, the 16-gene signature predicted TB progression with a sensitivity of 66.1 % and a specificity of 80.6 %. In the same period, the risk signature in untouched groups exhibited a sensitivity of 53.7 % and a specificity of 82.8 %. Subsequently, another robust and simple PCR-based host blood transcriptomic signature, the so-called RISK6, was developed to identify individuals at risk of incident disease, to screen for subclinical or clinical TB and to monitor TB treatment [116]. Its performance in the diagnosis of subclinical and clinical diseases in HIV-uninfected and co-infected TB/HIV patients exceeded 85 %. As a screening test for TB, RISK6 fulfilled the benchmarks established in the WHO target product profiles for those tests. The RISK6 scores were correlated with lung immunopathology activity and tracked treatment response. RISK6 predicted treatment failure prior to chemotherapy initiation. To obtain further details, two systematic reviews of the performance of the host blood global gene expression for diagnosing and predicting the progression of TB disease in different cohorts have recently been published [117, 118]. Collectively, these results indicate that both the 16-gene signature and RISK6 hold promise for worldwide applicability as field-friendly, point-of-care triage, diagnostic and predictive tests for TB based on the detection of biomarker profiles.
All the NAAT techniques described above have advantages over conventional techniques, such as their rapid detection and identification of TB, fast turnaround times for results, reliability and reproducibility. However, when using these techniques, additional equipment and trained personnel are needed. To date, these factors limit the implementation of these methodologies in LMI countries.
Flow cytometry
The evolution of methods to detect M. tuberculosis as described earlier has drastically reduced the time required for susceptibility testing from weeks to hours. Among these methods, FCM is a promising and potential tool that has become one of the best options for the rapid detection and quantification of various bacteria from the environment, food and clinical samples [119, 120]. An overview of its features is presented in Table 2.
In 1995, Norden and colleagues [121] described the use of FCM as a rapid test for drug susceptibility in a pioneer study. Using fluorescein diacetate (FDA), the authors tested M. tuberculosis strain H37Ra, which is susceptible to anti-mycobacterial agents, within 24 h and obtained similar results to those reported by others [122, 123]. Another study confirmed an agreement of approximately 94 % between the agar proportion method and FCM (by detection of FDA hydrolysis) for the INH-resistant strain, as well as total agreement for the EMB and RMP tests [124]. Although results can be obtained quickly using FCM, its biosafety remains an important drawback to its large-scale application owing to the production of infectious aerosols. Accordingly, Moore and colleagues [125] incubated specimens with paraformaldehyde before FCM analysis. However, it was observed that the tubercle bacilli trapped in the container could have escaped from the later treatment.
Another method to test the susceptibility of M. tuberculosis to drugs using FCM has been described to be fast and safe [126]. In this protocol, mycobacteria are heat-killed and probed with SYTO 16 stain, a non-symmetric cyanine with three positive charges that, when linked to pathogen nucleic acids, increases the intensity of the fluorescence signal and thus marks dead cells as bright green. After incubation of M. tuberculosis with SM, INH, RMP and EMB, the cells are stained and analysed using FCM. An excellent correlation between BACTEC MGIT 960 and FCM has been observed when compared with 12 to 15 days or 72 h of incubations, respectively, indicating that SYTO 16 enables clear distinction for drug susceptibility tests [126].
Recently, Qin and colleagues [127] discussed the importance of brighter fluorescent labels to improve the sensitivity of FCM and suggested the use of luminescent nanoparticles, as their superiority over conventional fluorophores in terms of fluorescence intensity and photostability was remarkable. In their study, an improved two-colour FCM approach was developed using a combination of Rubpy dye-doped silica nanoparticles with SYBR Green I dye to detect M. tuberculosis , thereby avoiding false positivity. However, neither drug susceptibility testing nor clinical sample detection was available in that setting. Another study recommended the use of FCM for the discrimination of live, drug-injured and dead M. tuberculosis [128]. Using SYTO 9, propidium iodide and ethidium monoazide (a compound that irreversibly binds to the DNA of dead cells), the authors could discriminate the effectiveness of anti-tuberculosis agents as targets to induce killing.
As FCM is a powerful, fast and safe tool that is essential for M. tuberculosis drug susceptibility testing in TB diagnostics that must remain simple and cost-effective. Janossy [129] claimed that there is a requirement to introduce this method in settings that have remained virtually untouched by contemporary laboratory technologies. Further, issues related to maintenance and special operational training restrict the wide use of FCM, particularly with regard to implementing the method in LMI countries.
Other nonconventional methods
Rapid detection of mycobacteria can be performed manually on the basis of the reduction of a tetrazolium salt indicator in a liquid-based medium [130]. This technique was compared with the use of the BACTEC 460 system and LJ solid culture media; it is a field-friendly device in which the antibiotic supplement is already incorporated, and easy and immediate reading of the results is guaranteed [131]. However, no additional parameters, such as specificity and sensitivity, were reported in that study.
The ESP Culture System II (ESP II) is based on the detection of pressure changes in the culture medium [64]. This method has been evaluated by comparing its performance with that of the BACTEC 460 and Middlebrook 7H11 systems. ESP II is a less labour-intensive alternative to BACTEC 460 for detection of mycobacteria [64]. Similarly to BACTEC 460 or other liquid culture media, ESP II is recommended to be used in combination with another culture method rather than as a stand-alone system. ESP II plus BACTEC 460 yielded the highest mycobacterial recovery rate; however, in most laboratories, such a combination would probably be expensive [64].
In recent years, mass spectrometry (MS) has exhibited high efficacy in the identification of bacteria in routine clinical work [132, 133]. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS was introduced during the 1980s [134], and in 2018 Cao and colleagues [135] conducted a systematic review and meta-analysis to confirm its accuracy in the identification of mycobacteria. The authors reported that the method might precisely identify 92 % of M. tuberculosis isolates. In addition, several other studies corroborated that MALDI-TOF MS exhibits good sensitivity in the identification of M. tuberculosis [136–138]. However, this technique struggles to differentiate mycobacterial species with high genetic similarity [139]. In any case, MALDI-TOF MS is considered to be a promising diagnostic method that potentially accelerates the identification of slow-growing mycobacterial species and might also identify drug-resistant M. tuberculosis strains [140]. Besides the above-mentioned advantages, the drawback of MS for wide use in TB diagnosis is its high cost, especially when it is implemented in LMI countries.
The use of DNA sequencing began in the 1970s when Frederick Sanger developed the chain termination method [141]. However, despite its development, the Sanger technique presented limitations at that time, which made it impossible to generate a large amount of data at a low cost. Since then, scientific advances in the sequencing technique have emerged, which has led to the advent of new-generation sequencers [142]. Whole-genome sequencing (WGS) is becoming central in epidemiological investigations of TB because of its better resolution and cost-effectiveness compared with traditional typing approaches. Numerous systematic reviews on the performance of this technique were published in 2016 and 2017 in several settings, indicating the speed with which this predicted diagnostic tool has taken off in recent times [143–145]. In one systematic review, the authors revealed that WGS has an average sensitivity and specificity for detecting drug-resistant forms of M. tuberculosis strains of 98 and 97 % for RMP and 97 and 96 % for INH, respectively [145]. However, Witney and colleagues [146] reported that WGS may yield false-positive results when polymorphism occurs in regions correlated with RMP resistance. In another study, the authors concluded that there is still much to learn about the origins of the growing genetic diversity that influence the interpretation from the understanding of M. tuberculosis transmission in each setting, and public health teams and researchers should combine epidemiological, clinical and WGS data to strengthen investigations of TB transmission [144]. Finally, besides the absence of a head-to-head comparison in M. tuberculosis isolates, a study found that WGS had greater discriminatory power than conventional genotyping and detected transmission events missed by epidemiological investigations [143].
Although this method exhibits high sensitivity, its implementation in the clinical laboratory routine is hindered by two factors: (i) the requirement for bacterial growth to obtain a certain amount of DNA necessary for analysis and (ii) the high cost of system maintenance. One of the major challenges of WGS in the diagnosis of M. tuberculosis is direct sequencing of sputum specimens (Table 3). In 2015, a study employed the biotinylated RNA technique specifically designed to detect M. tuberculosis genomes directly from the sputum of TB patients [147]. In 2018, the WHO published guidelines for the use of this technology for M. tuberculosis complex diagnosis and for the detection of drug-related mutations [148].
Conclusions
In this review, we demonstrated that the current microbiological tests for the diagnosis of TB in clinical samples are challenging. The most common and cost-effective and oldest test (developed over a century ago) for TB diagnosis is AFB sputum smear microscopy.
Currently, TB detection in LMI countries is performed using different methods, which involve clinical history, physical examination and complementary tests, such as sputum smears, M. tuberculosis culture, radiological examination, and histopathological and immunological approaches, all of which are supervised by well-trained healthcare workers who analyse the results of the complementary tests. On the other hand, in the modern laboratories of developed countries, suspected TB cases may be diagnosed by means of newer methods of cultivation using quicker molecular approaches, and this leads to accurate diagnosis in point-of-care settings [67].
New methods may be tested in well-designed and -controlled clinical trials, in addition to being used in high-incidence LMI settings. In this direction, both the 16-gene signature and RISK6 hold promise for worldwide applicability as field-friendly, point-of-care triage, diagnostic and predictive tests for TB based on the detection of biomarker profiles [117]. It is important to bear in mind that the time between the onset of TB disease and correct TB diagnosis and initiation of the correct anti-mycobacterial regimen is often protracted. In short, despite progress in the development of methods for mycobacterial detection helping to improve TB infection control, major gaps persist, and the diagnosis of TB requires more rigorous practices.
Funding information
This work received no specific grant from any funding agency. PRZA was supported by FIOCRUZ and CNPq PQ-2 fellowships. This work is part of both PhD (TAC) and master’s theses (PRCS) in Pathology (UFC, Ceara, Brazil), which are currently supported by Funcap (Ceara State grant agency) and CAPES (Ministry of Education). The members of the PRZA team are in debt to INCT-TB for kindly granting access to their scholarships.
Author contributions
Conceptualization – L.L.N., C.C.F., P.R.Z. A. Data curation – T.A.C., P.R.C.S., P.R.Z.A. Investigation – T.A.C., P.R.C.S., P.R.Z.A. Project administration – P.R.Z.A. Supervision – L.L.N., C.C.F., P.R.Z.A. Visualization – T.A.C., P.R.C.S. Writing (original draft) – T.A.C., P.R.C.S., L.L.N., C.C.F., P.R.Z.A. Writing (review and editing) – T.A.C., P.R.C.S., L.L.N., C.C.F., P.R.Z.A.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AFB, acid-fast bacilli; AST, antimicrobial susceptibility testing; CPC, cetylpyridinium chloride; EMB, ethambutol; FCM, flow cytometry; FDA, fluorescein diacetate; FM, fluorescence microscopy; INH, isoniazid; LAMP, loop-mediated isothermal amplification; LJ, Löwenstein-Jensen; LMI, low- and middle-income; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; MS, mass spectrometry; MTD, M. tuberculosis direct test; NAAT, nucleic acid amplification tests; NTM, nontuberculous mycobacteria; qRT-PCR, Real Time Quantitative PCR reverse transcription; RFLP, restriction fragment length polymorphism; RMP, rifampicin; RT-PCR, reverse transcription polymerase chain reaction; SM, streptomycin; TB, tuberculosis; WGS, whole-genome sequencing; WHO, World Health Organization; ZN, Ziehl-Neelsen.
References
- 1.Morris K. WHO recommends against inaccurate tuberculosis tests. Lancet. 2011;377:113–114. doi: 10.1016/s0140-6736(11)60005-6. [DOI] [PubMed] [Google Scholar]
- 2.Andersen P, Munk ME, Pollock JM, Doherty TM. Specific immune-based diagnosis of tuberculosis. Lancet. 2000;356:1099–1104. doi: 10.1016/s0140-6736(00)02742-2. [DOI] [PubMed] [Google Scholar]
- 3.Dockrell HM, Weir RE. Whole blood cytokine assays-a new generation of diagnostic tests for tuberculosis? Int J Tuberc Lung Dis. 1998;2:441–442. [PubMed] [Google Scholar]
- 4.WHO Global tuberculosis report 2019. Geneva. 2019 [Google Scholar]
- 5.Keer JT, Birch L. Molecular methods for the assessment of bacterial viability. J Microbiol Methods. 2003;53:175–183. doi: 10.1016/s0167-7012(03)00025-3. [DOI] [PubMed] [Google Scholar]
- 6.Kwak M, Lee WK, Lim YJ, Lee SH, Ryoo S. Systematic review and meta-analysis of the nitrate reductase assay for drug susceptibility testing of Mycobacterium tuberculosis and the detection limits in liquid medium. J Microbiol Methods. 2017;141:1–9. doi: 10.1016/j.mimet.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Warner DF, Mizrahi V. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin Microbiol Rev. 2006;19:558–570. doi: 10.1128/CMR.00060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Argyrou A, Vetting MW, Blanchard JS. New insight into the mechanism of action of and resistance to isoniazid: interaction of Mycobacterium tuberculosis enoyl-ACP reductase with INH-NADP. J Am Chem Soc. 2007;129:9582–9583. doi: 10.1021/ja073160k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeevan A, Sharma AK, McMurray DN. Ultraviolet radiation reduces resistance to Mycobacterium tuberculosis infection in BCG-vaccinated guinea pigs. Tuberculosis (Edinb) 2009;89:431–438. doi: 10.1016/j.tube.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Donaghy J, Keyser M, Johnston J, Cilliers FP, Gouws PA, et al. Inactivation of Mycobacterium avium ssp. paratuberculosis in milk by UV treatment. Lett Appl Microbiol. 2009;49:217–221. doi: 10.1111/j.1472-765X.2009.02644.x. [DOI] [PubMed] [Google Scholar]
- 11.Gholoobi A, Masoudi-Kazemabad A, Meshkat M, Meshkat Z. Comparison of culture and PCR methods for diagnosis of Mycobacterium tuberculosis in different clinical specimens. Jundishapur J Microbiol. 2014;7:e8939. doi: 10.5812/jjm.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rageade F, Picot N, Blanc-Michaud A, Chatellier S, Mirande C, et al. Performance of solid and liquid culture media for the detection of Mycobacterium tuberculosis in clinical materials: meta-analysis of recent studies. Eur J Clin Microbiol Infect Dis. 2014;33:867–870. doi: 10.1007/s10096-014-2105-z. [DOI] [PubMed] [Google Scholar]
- 13.Hobby GL, Holman AP, Iseman MD, Jones JM. Enumeration of tubercle bacilli in sputum of patients with pulmonary tuberculosis. Antimicrob Agents Chemother. 1973;4:94–104. doi: 10.1128/AAC.4.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.American Thoracic Society Diagnostic standards and classification of Tuberculosis. Am Rev Respir Dis. 1990;142:725–735. doi: 10.1164/ajrccm/142.3.725. [DOI] [PubMed] [Google Scholar]
- 15.Naveen G, Peerapur BV. Comparison of the Lowenstein-jensen medium, the middlebrook 7H10 medium and MB/BacT for the isolation of Mycobacterium tuberculosis (MTB) from clinical specimens. J Clin Diagn Res. 2012;6:1704–1709. doi: 10.7860/JCDR/2012/4603.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelhay A, Magnin JP, Gondrexon N, Baup S, Willison J. Adaptation of a Mycobacterium strain to phenanthrene degradation in a biphasic culture system: influence on interfacial area and droplet size. Biotechnol Lett. 2009;31:57–63. doi: 10.1007/s10529-008-9832-0. [DOI] [PubMed] [Google Scholar]
- 17.Bammann RH, Zamarioli LA, Pinto VS, Vázquez CM, Litvoc MN, et al. High prevalence of drug-resistant tuberculosis and other mycobacteria among HIV-infected patients in Brazil: a systematic review. Mem Inst Oswaldo Cruz. 2010;105:838–841. doi: 10.1590/s0074-02762010000600019. [DOI] [PubMed] [Google Scholar]
- 18.Rodrigues L, Aínsa JA, Amaral L, Viveiros M. Inhibition of drug efflux in mycobacteria with phenothiazines and other putative efflux inhibitors. Recent Pat Antiinfect Drug Discov. 2011;6:118–127. doi: 10.2174/157489111796064579. [DOI] [PubMed] [Google Scholar]
- 19.Larsen MH, Vilchèze C, Kremer L, Besra GS, Parsons L, et al. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis . Mol Microbiol. 2002;46:453–466. doi: 10.1046/j.1365-2958.2002.03162.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Yew WW. Mechanisms of drug resistance in Mycobacterium tuberculosis: update 2015. Int J Tuberc Lung Dis. 2015;19:1276–1289. doi: 10.5588/ijtld.15.0389. [DOI] [PubMed] [Google Scholar]
- 21.Cheng VC, Yew WW, Yuen KY. Molecular diagnostics in tuberculosis. Eur J Clin Microbiol Infect Dis. 2005;24:711–720. doi: 10.1007/s10096-005-0039-1. [DOI] [PubMed] [Google Scholar]
- 22.Sester M, Giehl C, McNerney R, Kampmann B, Walzl G, et al. Challenges and perspectives for improved management of HIV/Mycobacterium tuberculosis co-infection. Eur Respir J. 2010;36:1242–1247. doi: 10.1183/09031936.00040910. [DOI] [PubMed] [Google Scholar]
- 23.ATS Diagnostic standards and classification of tuberculosis in adults and children. Am J Respir Crit Care Med. 2000;161:1376–1395. doi: 10.1164/ajrccm.161.4.16141. [DOI] [PubMed] [Google Scholar]
- 24.WHO Systematic screening for active tuberculosis: principles and recommendations. WHO/HTM/TB/2013.04. Geneva. 2013;1 [PubMed] [Google Scholar]
- 25.Abrahams KA, Besra GS. Mycobacterial cell wall biosynthesis: a multifaceted antibiotic target. Parasitology. 2018;145:116–133. doi: 10.1017/S0031182016002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghazaei C. Mycobacterium tuberculosis and lipids: Insights into molecular mechanisms from persistence to virulence. J Res Med Sci. 2018;23:63. doi: 10.4103/jrms.JRMS_904_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Queiroz A, Riley LW. Bacterial immunostat: Mycobacterium tuberculosis lipids and their role in the host immune response. Rev Soc Bras Med Trop. 2017;50:9–18. doi: 10.1590/0037-8682-0230-2016. [DOI] [PubMed] [Google Scholar]
- 28.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 2011;7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cambanis A, Ramsay A, Wirkom V, Tata E, Cuevas LE. Investing time in microscopy: an opportunity to optimise smear-based case detection of tuberculosis. Int J Tuberc Lung Dis. 2007;11:40–45. [PubMed] [Google Scholar]
- 30.Poeta P, Silva V, Guedes A, Eduardo Pereira J, Cláudia Coelho A, et al. Tuberculosis in the 21th century: Current status of diagnostic methods. Exp Lung Res. 2018;44:352–360. doi: 10.1080/01902148.2018.1545880. [DOI] [PubMed] [Google Scholar]
- 31.Garg SK, Tiwari RP, Tiwari D, Singh R, Malhotra D, et al. Diagnosis of tuberculosis: available technologies, limitations, and possibilities. J Clin Lab Anal. 2003;17:155–163. doi: 10.1002/jcla.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smithwick RW, Stratigos CB, David HL. Use of cetylpyridinium chloride and sodium chloride for the decontamination of sputum specimens that are transported to the laboratory for the isolation of Mycobacterium tuberculosis . J Clin Microbiol. 1975;1:411–413. doi: 10.1128/jcm.1.5.411-413.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bobadilla-del-Valle M, Ponce-de-León A, Kato-Maeda M, Hernández-Cruz A, Calva-Mercado JJ, et al. Comparison of sodium carbonate, cetyl-pyridinium chloride, and sodium borate for preservation of sputa for culture of Mycobacterium tuberculosis . J Clin Microbiol. 2003;41:4487–4488. doi: 10.1128/JCM.41.9.4487-4488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suresh N, Singh UB, Arora J, Pande JN, Seth P, et al. Rapid detection of rifampicin-resistant Mycobacterium tuberculosis by in-house, reverse line blot assay. Diagn Microbiol Infect Dis. 2006;56:133–140. doi: 10.1016/j.diagmicrobio.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 35.Hepple P, Ford N, McNerney R. Microscopy compared to culture for the diagnosis of tuberculosis in induced sputum samples: a systematic review. Int J Tuberc Lung Dis. 2012;16:579–588. doi: 10.5588/ijtld.11.0617. [DOI] [PubMed] [Google Scholar]
- 36.Sohn H, Minion J, Albert H, Dheda K, Pai M. TB diagnostic tests: how do we figure out their costs? Expert Rev Anti Infect Ther. 2009;7:723–733. doi: 10.1586/eri.09.52. [DOI] [PubMed] [Google Scholar]
- 37.Levinson L, White RL. The detection of tubercle bacilli by fluorescence microscopy. N Engl J Med. 1947;237:186. doi: 10.1056/NEJM194708072370604. [DOI] [PubMed] [Google Scholar]
- 38.Kivihya-Ndugga LE, van Cleeff MR, Githui WA, Nganga LW, Kibuga DK, et al. A comprehensive comparison of Ziehl-Neelsen and fluorescence microscopy for the diagnosis of tuberculosis in a resource-poor urban setting. Int J Tuberc Lung Dis. 2003;7:1163–1171. [PubMed] [Google Scholar]
- 39.Steingart KR, Henry M, Ng V, Hopewell PC, Ramsay A, et al. Fluorescence versus conventional sputum smear microscopy for tuberculosis: A systematic review. Lancet Infect Dis. 2006;6:570–581. doi: 10.1016/S1473-3099(06)70578-3. [DOI] [PubMed] [Google Scholar]
- 40.Harries AD, Maher D, Nunn P. An approach to the problems of diagnosing and treating adult smear-negative pulmonary tuberculosis in high-HIV-prevalence settings in sub-Saharan Africa. Bull World Health Organ. 1998;76:651–662. [PMC free article] [PubMed] [Google Scholar]
- 41.Foulds J, O’Brien R. New tools for the diagnosis of tuberculosis: the perspective of developing countries. Int J Tuberc Lung Dis. 1998;2:778–783. [PubMed] [Google Scholar]
- 42.Shingadia D, Novelli V. Diagnosis and treatment of tuberculosis in children. Lancet Infect Dis. 2003;3:624–632. doi: 10.1016/s1473-3099(03)00771-0. [DOI] [PubMed] [Google Scholar]
- 43.Siddiqi K, Lambert ML, Walley J. Clinical diagnosis of smear-negative pulmonary tuberculosis in low-income countries: the current evidence. Lancet Infect Dis. 2003;3:288–296. doi: 10.1016/s1473-3099(03)00609-1. [DOI] [PubMed] [Google Scholar]
- 44.Steingart KR, Henry M, Laal S, Hopewell PC, Ramsay A, et al. Commercial serological antibody detection tests for the diagnosis of pulmonary tuberculosis: a systematic review. PLoS Med. 2007;4:e202. doi: 10.1371/journal.pmed.0040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steingart KR, Ng V, Henry M, Hopewell PC, Ramsay A, et al. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. 2006;6:664–674. doi: 10.1016/S1473-3099(06)70602-8. [DOI] [PubMed] [Google Scholar]
- 46.Kong Y, Yao H, Ren H, Subbian S, Cirillo SLG, et al. Imaging tuberculosis with endogenous β-lactamase reporter enzyme fluorescence in live mice. Proc Natl Acad Sci U S A. 2010;107:12239–12244. doi: 10.1073/pnas.1000643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sule P, Tilvawala R, Mustapha T, Hassounah H, Noormohamed A, et al. Rapid tuberculosis diagnosis using reporter enzyme fluorescence. J Clin Microbiol. 2019;57 doi: 10.1128/JCM.01462-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeager H, Lacy J, Smith LR, LeMaistre CA. Quantitative studies of mycobacterial populations in sputum and saliva. Am Rev Respir Dis. 1967;95:998–1004. doi: 10.1164/arrd.1967.95.6.998. [DOI] [PubMed] [Google Scholar]
- 49.Truffot-Pernot C, Veziris N. Bacteriological tests for tuberculosis. Rev Mal Respir. 2011;28:1034–1047. doi: 10.1016/j.rmr.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Antas PRZ, Santos DO, Pinheiro RO, Barbosa T. Current Diagnosis of Infant Tuberculosis Infection. Rio de Janeiro: Bentham Science Publishers; 2012. [Google Scholar]
- 51.Sharp SE, Lemes M, Sierra SG, Poniecka A, Poppiti RJ. Löwenstein-jensen Media. No longer necessary for mycobacterial isolation. Am J Clin Pathol. 2000;113:770–773. doi: 10.1309/JHDD-1HF4-2KCN-7ANP. [DOI] [PubMed] [Google Scholar]
- 52.Fair E, Hopewell PC, Pai M. International Standards for Tuberculosis Care: revisiting the cornerstones of tuberculosis care and control. Expert Rev Anti Infect Ther. 2007;5:61–65. doi: 10.1586/14787210.5.1.61. [DOI] [PubMed] [Google Scholar]
- 53.WHO The Use of Liquid Medium for Culture and DST. Geneva: World Health Organization; 2007. [Google Scholar]
- 54.Procop GW, Church DL, Hall GS, Janda WM. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. Jones & Bartlett Publishers: 2020. [Google Scholar]
- 55.Parsons LM, Somoskövi A, Urbanczik R, Salfinger M. Laboratory diagnostic aspects of drug resistant tuberculosis. Front Biosci. 2004;9:2086–2105. doi: 10.2741/1290. [DOI] [PubMed] [Google Scholar]
- 56.Aggarwal P, Singal A, Bhattacharya SN, Mishra K. Comparison of the radiometric BACTEC 460 TB culture system and Löwenstein-Jensen medium for the isolation of mycobacteria in cutaneous tuberculosis and their drug susceptibility pattern. Int J Dermatol. 2008;47:681–687. doi: 10.1111/j.1365-4632.2008.03675.x. [DOI] [PubMed] [Google Scholar]
- 57.Piersimoni C, Scarparo C, Callegaro A, Tosi CP, Nista D, et al. Comparison of MB/Bact alert 3D system with radiometric BACTEC system and Löwenstein-Jensen medium for recovery and identification of mycobacteria from clinical specimens: a multicenter study. J Clin Microbiol. 2001;39:651–657. doi: 10.1128/JCM.39.2.651-657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Venkataraman P, Herbert D, Paramasivan CN. Evaluation of the BACTEC radiometric method in the early diagnosis of tuberculosis. Indian J Med Res. 1998;108:120–127. [PubMed] [Google Scholar]
- 59.Garrigó M, Aragón LM, Alcaide F, Borrell S, Cardeñosa E, et al. Multicenter laboratory evaluation of the MB/BacT Mycobacterium detection system and the BACTEC MGIT 960 system in comparison with the BACTEC 460TB system for susceptibility testing of Mycobacterium tuberculosis . J Clin Microbiol. 2007;45:1766–1770. doi: 10.1128/JCM.02162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodrigues CS, Shenai SV, Almeida D, Sadani MA, Goyal N, et al. Use of bactec 460 TB system in the diagnosis of tuberculosis. Indian J Med Microbiol. 2007;25:32–36. doi: 10.4103/0255-0857.31059. [DOI] [PubMed] [Google Scholar]
- 61.Ardito F, Posteraro B, Sanguinetti M, Zanetti S, Fadda G. Evaluation of BACTEC Mycobacteria Growth Indicator Tube (MGIT 960) automated system for drug susceptibility testing of Mycobacterium tuberculosis . J Clin Microbiol. 2001;39:4440–4444. doi: 10.1128/JCM.39.12.4440-4444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang TS, HZ T, Lee SS, Huang WK, Liu YC. Antimicrobial susceptibility testing of Mycobacterium tuberculosis to first-line drugs: comparisons of the MGIT 960 and BACTEC 460 systems. Ann Clin Lab Sci. 2002;32:142–147. [PubMed] [Google Scholar]
- 63.Ganeswrie R, Chui CS, Balan S, Puthucheary SD. Comparison of BACTEC MGIT 960 system and BACTEC 460 TB system for growth and detection of Mycobacteria from clinical specimens. Malays J Pathol. 2004;26:99–103. [PubMed] [Google Scholar]
- 64.Williams-Bouyer N, Yorke R, Lee HI, Woods GL. Comparison of the BACTEC MGIT 960 and ESP culture system II for growth and detection of mycobacteria. J Clin Microbiol. 2000;38:4167–4170. doi: 10.1128/JCM.38.11.4167-4170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nishiyama H, Aono A, Sugamoto T, Mizuno K, Chikamatsu K, et al. Optimization of the microscopic observation drug susceptibility assay for four first-line drugs using Mycobacterium tuberculosis reference strains and clinical isolates. J Microbiol Methods. 2014;101:44–48. doi: 10.1016/j.mimet.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 66.Sorlozano A, Soria I, Roman J, Huertas P, Soto MJ, et al. Comparative evaluation of three culture methods for the isolation of mycobacteria from clinical samples. J Microbiol Biotechnol. 2009;19:1259–1264. doi: 10.4014/jmb.0901.0059. [DOI] [PubMed] [Google Scholar]
- 67.WHO Roadmap for rolling out Xpert MTB/RIF for rapid diagnosis of TB and MDR-TB. Geneva; 2011. Contract No. 2010;6 [Google Scholar]
- 68.Hanna BA, Ebrahimzadeh A, Elliott LB, Morgan MA, Novak SM, et al. Multicenter evaluation of the BACTEC MGIT 960 system for recovery of mycobacteria. J Clin Microbiol. 1999;37:748–752. doi: 10.1128/JCM.37.3.748-752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dowdy DW, Lourenço MC, Cavalcante SC, Saraceni V, King B, et al. Impact and cost-effectiveness of culture for diagnosis of tuberculosis in HIV-infected Brazilian adults. PloS One. 2008;3:e4057. doi: 10.1371/journal.pone.0004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chin KL, Sarmiento ME, Norazmi MN, Acosta A. DNA markers for tuberculosis diagnosis. Tuberculosis (Edinb) 2018;113:139–152. doi: 10.1016/j.tube.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 71.Shinnick TM, Good RC. Diagnostic mycobacteriology laboratory practices. Clin Infect Dis. 1995;21:291–299. doi: 10.1093/clinids/21.2.291. [DOI] [PubMed] [Google Scholar]
- 72.Machado D, Couto I, Viveiros M. Advances in the molecular diagnosis of tuberculosis: From probes to genomes. Infect Genet Evol. 2019;72:93–112. doi: 10.1016/j.meegid.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 73.Peralta G, Barry P, Pascopella L. Use of nucleic acid amplification tests in tuberculosis patients in California, 2010-2013. Open Forum Infect Dis. 2016;3:ofw230. doi: 10.1093/ofid/ofw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun JR, Lee SY, Perng CL, JJ L. Detecting Mycobacterium tuberculosis in Bactec MGIT 960 cultures by inhouse IS6110-based PCR assay in routine clinical practice. J Formos Med Assoc. 2009;108:119–125. doi: 10.1016/S0929-6646(09)60042-5. [DOI] [PubMed] [Google Scholar]
- 75.Das S, Mangold KA, Shah NS, Peterson LR, Thomson RB, et al. Performance and utilization of a laboratory-developed nucleic acid amplification test (NAAT) for the diagnosis of pulmonary and extrapulmonary tuberculosis in a low-prevalence area. Am J Clin Pathol. 2020;154:115–123. doi: 10.1093/ajcp/aqaa031. [DOI] [PubMed] [Google Scholar]
- 76.Zar HJ. Diagnosis of pulmonary tuberculosis in children-what’s new? S Afr Med J. 2007;97:983–985. [PubMed] [Google Scholar]
- 77.Eisenach KD, Sifford MD, Cave MD, Bates JH, Crawford JT. Detection of Mycobacterium tuberculosis in sputum samples using a polymerase chain reaction. Am Rev Respir Dis. 1991;144:1160–1163. doi: 10.1164/ajrccm/144.5.1160. [DOI] [PubMed] [Google Scholar]
- 78.Palomino JC, Vandamme P, Martin A. Classical and new assays for detecting drug resistance in tuberculosis. Biomark Med. 2014;8:1105–1114. doi: 10.2217/bmm.14.73. [DOI] [PubMed] [Google Scholar]
- 79.Campelo TA, Lima LNC, Lima KVB, Silva CS, Conceição MLD, et al. Molecular characterization of pre-extensive drug resistant Mycobacterium tuberculosis in Northeast Brazil. Rev Inst Med Trop Sao Paulo. 2020;62:e4. doi: 10.1590/S1678-9946202062004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mathuria JP, Nath G, Samaria JK, Anupurba S. Molecular characterization of INH-resistant Mycobacterium tuberculosis isolates by PCR-RFLP and multiplex-PCR in North India. Infect Genet Evol. 2009;9:1352–1355. doi: 10.1016/j.meegid.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 81.Abe C, Hirano K, Wada M, Kazumi Y, Takahashi M, et al. Detection of Mycobacterium tuberculosis in clinical specimens by polymerase chain reaction and Gen-probe amplified Mycobacterium tuberculosis direct test. J Clin Microbiol. 1993;31:3270–3274. doi: 10.1128/jcm.31.12.3270-3274.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Causse M, Ruiz P, Gutiérrez-Aroca JB, Casal M. Comparison of two molecular methods for rapid diagnosis of extrapulmonary tuberculosis. J Clin Microbiol. 2011;49:3065–3067. doi: 10.1128/JCM.00491-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miller N, Infante S, Cleary T. Evaluation of the LiPA MYCOBACTERIA assay for identification of mycobacterial species from BACTEC 12B bottles. J Clin Microbiol. 2000;38:1915–1919. doi: 10.1128/JCM.38.5.1915-1919.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gray CM, Katamba A, Narang P, Giraldo J, Zamudio C, et al. Feasibility and operational performance of tuberculosis detection by loop-mediated isothermal amplification platform in decentralized settings: Results from a multicenter study. J Clin Microbiol. 2016;54:1984–1991. doi: 10.1128/JCM.03036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rajalahti I, Vuorinen P, Liippo K, Nieminen MM, Miettinen A. Evaluation of commercial DNA and rRNA amplification assays for assessment of treatment outcome in pulmonary tuberculosis patients. Eur J Clin Microbiol Infect Dis. 2001;20:746–750. doi: 10.1007/s100960100593. [DOI] [PubMed] [Google Scholar]
- 86.Shete PB, Farr K, Strnad L, Gray CM, Cattamanchi A. Diagnostic accuracy of TB-LAMP for pulmonary tuberculosis: a systematic review and meta-analysis. BMC Infect Dis. 2019;19:268. doi: 10.1186/s12879-019-3881-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Broccolo F, Scarpellini P, Locatelli G, Zingale A, Brambilla AM, et al. Rapid diagnosis of mycobacterial infections and quantitation of Mycobacterium tuberculosis load by two real-time calibrated PCR assays. J Clin Microbiol. 2003;41:4565–4572. doi: 10.1128/JCM.41.10.4565-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ortu S, Molicotti P, Sechi LA, Pirina P, Saba F, et al. Rapid detection and identification of Mycobacterium tuberculosis by Real Time PCR and Bactec 960 MIGT. New Microbiol. 2006;29:75–80. [PubMed] [Google Scholar]
- 89.Rajalahti I, Ruokonen EL, Kotomäki T, Sintonen H, Nieminen MM. Economic evaluation of the use of PCR assay in diagnosing pulmonary TB in a low-incidence area. Eur Respir J. 2004;23:446–451. doi: 10.1183/09031936.04.00009704. [DOI] [PubMed] [Google Scholar]
- 90.Dundar D, Sayan M, Arslan Z, Tamer GS, Dundar V. Routine using pattern and performance of diagnostic tests for tuberculosis on a university hospital. Am J Med Sci. 2010;339:244–248. doi: 10.1097/MAJ.0b013e3181cbfe40. [DOI] [PubMed] [Google Scholar]
- 91.Noordhoek GT, Mulder S, Wallace P, van Loon AM. Multicentre quality control study for detection of Mycobacterium tuberculosis in clinical samples by nucleic amplification methods. Clin Microbiol Infect. 2004;10:295–301. doi: 10.1111/j.1198-743X.2004.00825.x. [DOI] [PubMed] [Google Scholar]
- 92.Mehta PK, Raj A, Singh N, Khuller GK. Diagnosis of extrapulmonary tuberculosis by PCR. FEMS Immunol Med Microbiol. 2012;66:20–36. doi: 10.1111/j.1574-695X.2012.00987.x. [DOI] [PubMed] [Google Scholar]
- 93.Sahmi F, Sahmi M, Gévry N, Sahadevan P, Allen BG, et al. A putative protein-RNA complex regulates posttranscriptional processing of cytochrome P450 aromatase (CYP19A1) in bovine granulosa cells. Mol Reprod Dev. 2019;86:1901–1908. doi: 10.1002/mrd.23289. [DOI] [PubMed] [Google Scholar]
- 94.Rustad TR, Minch KJ, Brabant W, Winkler JK, Reiss DJ, et al. Global analysis of mRNA stability in Mycobacterium tuberculosis . Nucleic Acids Res. 2013;41:509–517. doi: 10.1093/nar/gks1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hellyer TJ, DesJardin LE, Hehman GL, Cave MD, Eisenach KD. Quantitative analysis of mRNA as a marker for viability of Mycobacterium tuberculosis . J Clin Microbiol. 1999;37:290–295. doi: 10.1128/JCM.37.2.290-295.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pai SR, Actor JK, Sepulveda E, Hunter RL, Jagannath C. Identification of viable and non-viable Mycobacterium tuberculosis in mouse organs by directed RT-PCR for antigen 85B mRNA. Microb Pathog. 2000;28:335–342. doi: 10.1006/mpat.2000.0353. [DOI] [PubMed] [Google Scholar]
- 97.Jou NT, Yoshimori RB, Mason GR, Louie JS, Liebling MR. Single-tube, nested, reverse transcriptase PCR for detection of viable Mycobacterium tuberculosis . J Clin Microbiol. 1997;35:1161–1165. doi: 10.1128/jcm.35.5.1161-1165.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sankar S, Ramamurthy M, Nandagopal B, Sridharan G. An appraisal of PCR-based technology in the detection of Mycobacterium tuberculosis . Mol Diagn Ther. 2011;15:1–11. doi: 10.1007/BF03257188. [DOI] [PubMed] [Google Scholar]
- 99.da Silva Rocha A, da Costa Leite C, Torres HM, de Miranda AB, Pires Lopes MQ, et al. Use of PCR-restriction fragment length polymorphism analysis of the hsp65 gene for rapid identification of mycobacteria in Brazil. J Microbiol Methods. 1999;37:223–229. doi: 10.1016/s0167-7012(99)00062-7. [DOI] [PubMed] [Google Scholar]
- 100.Ergin A, Kocagöz T, Us D. Evaluation of 120 mycobacterial strains isolated from clinical specimens to the species level by polymerase chain reaction-restriction enzyme analysis. Scand J Infect Dis. 2000;32:657–662. doi: 10.1080/003655400459586. [DOI] [PubMed] [Google Scholar]
- 101.Gingeras TR, Ghandour G, Wang E, Berno A, Small PM, et al. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 102.RoS L, Suffys P, Barroso EC, Kerr LR, Duarte CR, et al. Genotyping and drug resistance patterns of Mycobacterium tuberculosis strains observed in a tuberculosis high-burden municipality in Northeast, Brazil. Braz J Infect Dis. 2013;17:338–345. doi: 10.1016/j.bjid.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Horne DJ, Kohli M, Zifodya JS, Schiller I, Dendukuri N, et al. Xpert MTB/RIF and Xpert MTB/RIF Ultra for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2019;6:CD009593. doi: 10.1002/14651858.CD009593.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williamson DA, Roberts SA, Bower JE, Vaughan R, Newton S, et al. Clinical failures associated with rpoB mutations in phenotypically occult multidrug-resistant Mycobacterium tuberculosis . Int J Tuberc Lung Dis. 2012;16:216–220. doi: 10.5588/ijtld.11.0178. [DOI] [PubMed] [Google Scholar]
- 105.Banada PP, Sivasubramani SK, Blakemore R, Boehme C, Perkins MD, et al. Containment of bioaerosol infection risk by the Xpert MTB/RIF assay and its applicability to point-of-care settings. J Clin Microbiol. 2010;48:3551–3557. doi: 10.1128/JCM.01053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.MacLean E, Saravu K, Pai M. Diagnosing active tuberculosis in people living with HIV: an ongoing challenge. Curr Opin HIV AIDS. 2019;14:46–54. doi: 10.1097/COH.0000000000000512. [DOI] [PubMed] [Google Scholar]
- 107.Evans D, Sineke T, Schnippel K, Berhanu R, Govathson C, et al. Impact of Xpert MTB/RIF and decentralized care on linkage to care and drug-resistant tuberculosis treatment outcomes in Johannesburg, South Africa. BMC Health Serv Res. 2018;18:973. doi: 10.1186/s12913-018-3762-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sagili KD, Muniyandi M, Nilgiriwala KS, Shringarpure KS, Satyanarayana S, et al. Cost-effectiveness of GeneXpert and LED-FM for diagnosis of pulmonary tuberculosis: A systematic review. PLoS One. 2018;13:e0205233. doi: 10.1371/journal.pone.0205233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Méchaï F, Bouchaud O. Tuberculous meningitis: Challenges in diagnosis and management. Rev Neurol (Paris) 2019;175:451–457. doi: 10.1016/j.neurol.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 110.Tiberi S, Zumla A, Migliori GB. Multidrug and extensively drug-resistant tuberculosis: Epidemiology, clinical features, management and treatment. Infect Dis Clin North Am. 2019;33:1063–1085. doi: 10.1016/j.idc.2019.09.002. [DOI] [PubMed] [Google Scholar]
- 111.Yong YK, Tan HY, Saeidi A, Wong WF, Vignesh R, et al. Immune biomarkers for diagnosis and treatment monitoring of tuberculosis: current developments and future poospects. Front Microbiol. 2019;10:2789. doi: 10.3389/fmicb.2019.02789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liang CN, Zhao HW, Kang J, Hou G, Yin Y. Acute mediastinitis associated with tracheobronchial tuberculosis and aspergillosis: a case report and literature review. J Int Med Res. 2020;48:300060520918469. doi: 10.1177/0300060520918469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Steingart KR, Sohn H, Schiller I, Kloda LA, Boehme CC, et al. XPERT MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2013;1:CD009593. doi: 10.1002/14651858.CD009593.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Steingart KR, Schiller I, Horne DJ, Pai M, Boehme CC, et al. XPERT MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2014;1:CD009593. doi: 10.1002/14651858.CD009593.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Penn-Nicholson A, Mbandi SK, Thompson E, Mendelsohn SC, Suliman S, et al. RISK6, a 6-gene transcriptomic signature of TB disease risk, diagnosis and treatment response. Sci Rep. 2020;10:8629. doi: 10.1038/s41598-020-65043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mulenga H, Zauchenberger CZ, Bunyasi EW, Mbandi SK, Mendelsohn SC, et al. Performance of diagnostic and predictive host blood transcriptomic signatures for Tuberculosis disease: A systematic review and meta-analysis. PLoS One. 2020;15:e0237574. doi: 10.1371/journal.pone.0237574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Warsinske H, Vashisht R, Khatri P. Host-response-based gene signatures for tuberculosis diagnosis: A systematic comparison of 16 signatures. PLoS Med. 2019;16:e1002786. doi: 10.1371/journal.pmed.1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yamaguchi N, Sasada M, Yamanaka M, Nasu M. Rapid detection of respiring Escherichia coli O157:H7 in apple juice, milk, and ground beef by flow cytometry. Cytometry A. 2003;54:27–35. doi: 10.1002/cyto.a.10045. [DOI] [PubMed] [Google Scholar]
- 120.Lenaerts J, Lappin-Scott HM, Porter J. Improved fluorescent in situ hybridization method for detection of bacteria from activated sludge and river water by using DNA molecular beacons and flow cytometry. Appl Environ Microbiol. 2007;73:2020–2023. doi: 10.1128/AEM.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Norden MA, Kurzynski TA, Bownds SE, Callister SM, Schell RF. Rapid susceptibility testing of Mycobacterium tuberculosis (H37Ra) by flow cytometry. J Clin Microbiol. 1995;33:1231–1237. doi: 10.1128/jcm.33.5.1231-1237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bownds SE, Kurzynski TA, Norden MA, Dufek JL, Schell RF. Rapid susceptibility testing for nontuberculosis mycobacteria using flow cytometry. J Clin Microbiol. 1996;34:1386–1390. doi: 10.1128/jcm.34.6.1386-1390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kirk SM, Schell RF, Moore AV, Callister SM, Mazurek GH. Flow cytometric testing of susceptibilities of Mycobacterium tuberculosis isolates to ethambutol, isoniazid, and rifampin in 24 hours. J Clin Microbiol. 1998;36:1568–1573. doi: 10.1128/JCM.36.6.1568-1573.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.DeCoster DJ, Vena RM, Callister SM, Schell RF. Susceptibility testing of Mycobacterium tuberculosis: comparison of the BACTEC TB-460 method and flow cytometric assay with the proportion method. Clin Microbiol Infect. 2005;11:372–378. doi: 10.1111/j.1469-0691.2005.01127.x. [DOI] [PubMed] [Google Scholar]
- 125.Moore AV, Kirk SM, Callister SM, Mazurek GH, Schell RF. Safe determination of susceptibility of Mycobacterium tuberculosis to antimycobacterial agents by flow cytometry. J Clin Microbiol. 1999;37:479–483. doi: 10.1128/JCM.37.3.479-483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pina-Vaz C, Costa-de-Oliveira S, Rodrigues AG. Safe susceptibility testing of Mycobacterium tuberculosis by flow cytometry with the fluorescent nucleic acid stain SYTO 16. J Med Microbiol. 2005;54:77–81. doi: 10.1099/jmm.0.45627-0. [DOI] [PubMed] [Google Scholar]
- 127.Qin D, He X, Wang K, Tan W. Using fluorescent nanoparticles and SYBR Green I based two-color flow cytometry to determine Mycobacterium tuberculosis avoiding false positives. Biosens Bioelectron. 2008;24:626–631. doi: 10.1016/j.bios.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 128.Soejima T, Iida K, Qin T, Taniai H, Yoshida S, et al. anti-tuberculosis agent-injured, and dead Mycobacterium tuberculosis using flow cytometry. FEMS Microbiol Lett. 2009;294:74–81. doi: 10.1111/j.1574-6968.2009.01549.x. [DOI] [PubMed] [Google Scholar]
- 129.Janossy G. The changing pattern of “smart” flow cytometry (S-FC) to assist the cost-effective diagnosis of HIV, tuberculosis, and leukemias in resource-restricted conditions. Biotechnol J. 2008;3:32–42. doi: 10.1002/biot.200700200. [DOI] [PubMed] [Google Scholar]
- 130.Cambau E, Wichlacz C, Truffot-Pernot C, Jarlier V. Evaluation of the new MB redox system for detection of growth of mycobacteria. J Clin Microbiol. 1999;37:2013–2015. doi: 10.1128/JCM.37.6.2013-2015.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Samra Z, Kaufman L, Bechor J, Bahar J. Comparative study of three culture systems for optimal recovery of mycobacteria from different clinical specimens. Eur J Clin Microbiol Infect Dis. 2000;19:750–754. doi: 10.1007/s100960000369. [DOI] [PubMed] [Google Scholar]
- 132.Bizzini A, Durussel C, Bille J, Greub G, Prod’hom G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol. 2010;48:1549–1554. doi: 10.1128/JCM.01794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Carbonnelle E, Mesquita C, Bille E, Day N, Dauphin B, et al. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin Biochem. 2011;44:104–109. doi: 10.1016/j.clinbiochem.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 134.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 135.Cao Y, Wang L, Ma P, Fan W, Gu B, et al. Accuracy of matrix-assisted laser desorption Ionization-time of flight mass spectrometry for identification of Mycobacteria: a systematic review and meta-analysis. Sci Rep. 2018;8:4131. doi: 10.1038/s41598-018-22642-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Balada-Llasat JM, Kamboj K, Pancholi P. Identification of mycobacteria from solid and liquid media by matrix-assisted laser desorption ionization-time of flight mass spectrometry in the clinical laboratory. J Clin Microbiol. 2013;51:2875–2879. doi: 10.1128/JCM.00819-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang TS, Lee CC, Tu HZ, Lee SS. Rapid identification of mycobacteria from positive MGIT broths of primary cultures by MALDI-TOF mass spectrometry. PloS One. 2018;13:e0192291. doi: 10.1371/journal.pone.0192291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Quinlan P, Phelan E, Doyle M. Matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry (MS) for the identification of mycobacteria from MBBacT ALERT 3D liquid cultures and Lowenstein-Jensen (LJ) solid cultures. J Clin Pathol. 2015;68:229–235. doi: 10.1136/jclinpath-2014-202374. [DOI] [PubMed] [Google Scholar]
- 139.Kodana M, Tarumoto N, Kawamura T, Saito T, Ohno H, et al. Utility of the MALDI-TOF MS method to identify nontuberculous mycobacteria. J Infect Chemother. 2016;22:32–35. doi: 10.1016/j.jiac.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 140.Su K-Y, Yan BS, Chiu HC, Yu C-J, Chang SY, et al. Rapid sputum multiplex detection of the M. tuberculosis complex (MTBC) and resistance mutations for eight antibiotics by Nucleotide MALDI-TOF MS. Sci Rep. 2017;7:41486. doi: 10.1038/srep41486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Henson J, Tischler G, Ning Z. Next-generation sequencing and large genome assemblies. Pharmacogenomics. 2012;13:901–915. doi: 10.2217/pgs.12.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nikolayevskyy V, Kranzer K, Niemann S, Drobniewski F. Whole genome sequencing of Mycobacterium tuberculosis for detection of recent transmission and tracing outbreaks: A systematic review. Tuberculosis (Edinb) 2016;98:77–85. doi: 10.1016/j.tube.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 144.Hatherell HA, Colijn C, Stagg HR, Jackson C, Winter JR, et al. Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med. 2016;14:21. doi: 10.1186/s12916-016-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Papaventsis D, Casali N, Kontsevaya I, Drobniewski F, Cirillo DM, et al. Whole genome sequencing of Mycobacterium tuberculosis for detection of drug resistance: a systematic review. Clin Microbiol Infect. 2017;23:61–68. doi: 10.1016/j.cmi.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 146.Witney AA, Cosgrove CA, Arnold A, Hinds J, Stoker NG, et al. Clinical use of whole genome sequencing for Mycobacterium tuberculosis . BMC Med. 2016;14:46. doi: 10.1186/s12916-016-0598-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Brown AC, Bryant JM, Einer-Jensen K, Holdstock J, Houniet DT, et al. Rapid whole-genome sequencing of Mycobacterium tuberculosis isolates directly from clinical samples. J Clin Microbiol. 2015;53:2230–2237. doi: 10.1128/JCM.00486-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.WHO The Use of Next-Generation Sequencing Technologies For The Detection of Mutations Associated with Drug Resistance in Mycobacterium Tberculosis Complex: Technical Guide. Geneva: World Health Organization; 2018. [Google Scholar]
- 149.Rodríguez-Lázaro D, D’Agostino M, Herrewegh A, Pla M, Cook N, et al. Real-time PCR-based methods for detection of Mycobacterium avium subsp. paratuberculosis in water and milk. Int J Food Microbiol. 2005;101:93–104. doi: 10.1016/j.ijfoodmicro.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 150.Espasa M, González-Martín J, Alcaide F, Aragón LM, Lonca J, et al. Direct detection in clinical samples of multiple gene mutations causing resistance of Mycobacterium tuberculosis to isoniazid and rifampicin using fluorogenic probes. J Antimicrob Chemother. 2005;55:860–865. doi: 10.1093/jac/dki132. [DOI] [PubMed] [Google Scholar]
- 151.Brooks BW, Devenish J, Lutze-Wallace CL, Milnes D, Robertson RH, et al. Evaluation of a monoclonal antibody-based enzyme-linked immunosorbent assay for detection of Campylobacter fetus in bovine preputial washing and vaginal mucus samples. Vet Microbiol. 2004;103:77–84. doi: 10.1016/j.vetmic.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 152.Golichenari B, Nosrati R, Farokhi-Fard A, Faal Maleki M, Gheibi Hayat SM, et al. Electrochemical-based biosensors for detection of Mycobacterium tuberculosis and tuberculosis biomarkers. Crit Rev Biotechnol. 2019;39:1056–1077. doi: 10.1080/07388551.2019.1668348. [DOI] [PubMed] [Google Scholar]
- 153.Oh B-K, Lee W, Kim YK, Lee WH, Choi JW. Surface plasmon resonance immunosensor using self-assembled protein G for the detection of Salmonella paratyphi . J Biotechnol. 2004;111:1–8. doi: 10.1016/j.jbiotec.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 154.Gupta UD, Katoch K, Singh HB, Natrajan M, Sharma VD, et al. Assessment of viability by normal mouse foot-pad and bacillary ATP bioluminescence assay in multibacillary cases treated with an MDT regimen using conventional as well as newer drugs like minocycline and ofloxacin. Indian J Lepr. 2000;72:437–442. [PubMed] [Google Scholar]
- 155.McFeters GA, Pyle BH, Lisle JT, Broadaway SC. Rapid direct methods for enumeration of specific, active bacteria in water and biofilms. Soc Appl Microbiol symp ser. 1999;85:193S–200S. [PubMed] [Google Scholar]
- 156.Hibi K, Mitsubayashi K, Fukuda H, Ushio H, Hayashi T, et al. Rapid direct determination using combined separation by prepared immunomagnetic and flow cytometry of Flavobacterium psychrophilum . Biosensors and Bioelectronics. 2007;22:1916–1919. doi: 10.1016/j.bios.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 157.Alexander KA, Laver PN, Michel AL, Williams M, van Helden PD, et al. Novel Mycobacterium tuberculosis complex pathogen, M. mungi . Emerg Infect Dis. 2010;16:1296–1299. doi: 10.3201/eid1608.100314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cousins DV, Peet RL, Gaynor WT, Williams SN, Gow BL. Tuberculosis in imported hyrax (Procavia capensis) caused by an unusual variant belonging to the Mycobacterium tuberculosis complex. Vet Microbiol. 1994;42:135–145. doi: 10.1016/0378-1135(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 159.Parsons SD, Drewe JA, Gey van Pittius NC, Warren RM, van Helden PD. Novel cause of tuberculosis in meerkats, South Africa. Emerg Infect Dis. 2013;19:2004–2007. doi: 10.3201/eid1912.130268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.van Ingen J, Rahim Z, Mulder A, Boeree MJ, Simeone R, et al. Characterization of Mycobacterium orygis as M. tuberculosis complex subspecies. Emerg Infect Dis. 2012;18:653–655. doi: 10.3201/eid1804.110888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wells A. Tuberculosis in wild voles. Lancet. 1937;139:917. [Google Scholar]
- 162.Cousins DV, Williams SN, Reuter R, Forshaw D, Chadwick B, et al. Tuberculosis in wild seals and characterisation of the seal bacillus. Aust Vet J. 1993;70:92–97. doi: 10.1111/j.1751-0813.1993.tb03284.x. [DOI] [PubMed] [Google Scholar]
- 163.Aranaz A, Liébana E, Gómez-Mampaso E, Galán JC, Cousins D, et al. Mycobacterium tuberculosis subsp. caprae subsp. nov.: a taxonomic study of a new member of the Mycobacterium tuberculosis complex isolated from goats in Spain. Int J Syst Bacteriol. 1999;49:1263–1273. doi: 10.1099/00207713-49-3-1263. [DOI] [PubMed] [Google Scholar]
- 164.Karlson AG, Lessel EF. Mycobacterium bovis nom. nov. Int J Syst Evol Microbiol. 1970;20:273–282. [Google Scholar]
- 165.Karimi H, En-Nanai L, Oudghiri A, Chaoui I, Laglaoui A, et al. Performance of GenoType MTBDRplus assay in the diagnosis of drug-resistant tuberculosis in Tangier, Morocco. J Glob Antimicrob Resist. 2018;12:63–67. doi: 10.1016/j.jgar.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 166.Singh K, Kumari R, Tripathi R, Gupta A, Anupurba S. Mutation in MPT64 gene influencing diagnostic accuracy of SD Bioline assay (capilia. BMC Infect Dis. 2019;19:1048. doi: 10.1186/s12879-019-4671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Al-Mutairi NM, Ahmad S, Mokaddas EM. Molecular characterization of multidrug-resistant Mycobacterium tuberculosis (MDR-TB) isolates identifies local transmission of infection in Kuwait, a country with a low incidence of TB and MDR-TB. Eur J Med Res. 2019;24:38. doi: 10.1186/s40001-019-0397-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gkaravela L, Papadimitriou-Olivgeris M, Foka A, Kolonitsiou F, Spiliopoulou A, et al. Combination of commercially available molecular assays and culture based methods in diagnosis of tuberculosis and drug resistant tuberculosis. Braz J Microbiol. 2017;48:785–790. doi: 10.1016/j.bjm.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Abdel-Moein KA, Hamed O, Fouad H. Molecular detection of Mycobacterium tuberculosis in cattle and buffaloes: a cause for public health concern. Trop Anim Health Prod. 2016;48:1541–1545. doi: 10.1007/s11250-016-1125-3. [DOI] [PubMed] [Google Scholar]
- 170.Lee YS, Kang HR, Lee SH, Kim Y, Kim MY, et al. Diagnostic usefulness of the GenoType MTBDRplus assay for detecting drug-resistant tuberculosis using AFB smear-negative specimens with positive TB-PCR result. Infect Dis (Lond) 2016;48:350–355. doi: 10.3109/23744235.2015.1122831. [DOI] [PubMed] [Google Scholar]
- 171.Theron G, Peter J, Richardson M, Barnard M, Donegan S, et al. The diagnostic accuracy of the genotype MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs. Cochrane Database Syst Rev. 2014;10:CD010705. doi: 10.1002/14651858.CD010705.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mokry J, Porvaznik I, Kusnir P, Dohal M, Solovic I. Detection of resistance to anti-tuberculosis drugs in the clinical isolates of Mycobacterium tuberculosis from Slovakia through comparison between phenotypic and genetic methods and evaluation of resistance levels with clinical parameter. J Physiol Pharmacol. 2019;70 doi: 10.26402/jpp.2019.1.10. [DOI] [PubMed] [Google Scholar]
- 173.Mao X, Ke Z, Shi X, Liu S, Tang B, et al. Diagnosis of drug resistance to Fluoroquinolones, amikacin, capreomycin, kanamycin and ethambutol with genotype MTBDRsl assay: a meta-analysis. Ann Clin Lab Sci. 2015;45:533–544. [PubMed] [Google Scholar]
- 174.Sajduda A, Brzostek A, Poplawska M, Augustynowicz-Kopec E, Zwolska Z, et al. Molecular characterization of rifampin- and isoniazid-resistant Mycobacterium tuberculosis strains isolated in Poland. J Clin Microbiol. 2004;42:2425–2431. doi: 10.1128/JCM.42.6.2425-2431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Helb D, Jones M, Story E, Boehme C, Wallace E, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48:229–237. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rao P, Chawla K, Shenoy VP, Mukhopadhyay C. Role of real-time PCR for detection of tuberculosis and drug resistance directly from clinical samples. Indian J Tuberc. 2016;63:149–153. doi: 10.1016/j.ijtb.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 177.Sharma K, Modi M, Kaur H, Sharma A, Ray P, et al. rpoB gene high-resolution melt curve analysis: a rapid approach for diagnosis and screening of drug resistance in tuberculous meningitis. Diagn Microbiol Infect Dis. 2015;83:144–149. doi: 10.1016/j.diagmicrobio.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 178.Riahi F, Derakhshan M, Mosavat A, Soleimanpour S, Rezaee SA. Evaluation of point mutation detection in Mycobacterium tuberculosis with isoniazid resistance using real-time PCR and TaqMan probe assay. Appl Biochem Biotechnol. 2015;175:2447–2455. doi: 10.1007/s12010-014-1442-9. [DOI] [PubMed] [Google Scholar]
- 179.Mokrousov I, Vyazovaya A, Zhuravlev V, Otten T, Millet J, et al. Real-time PCR assay for rapid detection of epidemiologically and clinically significant Mycobacterium tuberculosis Beijing genotype isolates. J Clin Microbiol. 2014;52:1691–1693. doi: 10.1128/JCM.03193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chauhan DS, Sharma R, Parashar D, Sharma P, Das R, et al. Early detection of multidrug resistant (MDR) Mycobacterium tuberculosis in a single tube with in-house designed fluorescence resonance energy transfer (FRET) probes using real-time PCR. Indian J Exp Biol. 2016;54:229–236. [PubMed] [Google Scholar]
- 181.Wang M-G, Xue M, Wu S-Q, Zhang M-M, Wang Y, et al. Abbott RealTime MTB and MTB RIF/INH assays for the diagnosis of tuberculosis and rifampicin/isoniazid resistance. Infect Genet Evol. 2019;71:54–59. doi: 10.1016/j.meegid.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 182.Hofmann-Thiel S, Molodtsov N, Duffner C, Kadyrov A, Kalmambetova G, et al. Capacity of Abbott RealTime MTB RIF/INH to detect rifampicin- and isoniazid-resistant tuberculosis. Int J Tuberc Lung Dis. 2019;23:458–464. doi: 10.5588/ijtld.18.0615. [DOI] [PubMed] [Google Scholar]
- 183.Ruiz P, Causse M, Vaquero M, Gutierrez JB, Casal M. Evaluation of a new automated Abbott RealTime MTB RIF/INH assay for qualitative detection of rifampicin/isoniazid resistance in pulmonary and extra-pulmonary clinical samples of Mycobacterium tuberculosis . Infect Drug Resist. 2017;10:463–467. doi: 10.2147/IDR.S147272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kumari R, Banerjee T, Anupurba S. Molecular detection of drug resistance to ofloxacin and kanamycin in Mycobacterium tuberculosis by using multiplex allele-specific PCR. J Infect Public Health. 2018;11:54–58. doi: 10.1016/j.jiph.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 185.Salim S, Zaman G, Younis S, Hussain W, Khurshid U, et al. Multiplex PCR for rapid diagnosis of drug resistant Mycobacterium tuberculosis . J Coll Physicians Surg Pak. 2019;29:833–837. doi: 10.29271/jcpsp.2019.09.833. [DOI] [PubMed] [Google Scholar]
- 186.Sinha P, Banerjee T, Srivastava GN, Anupurba S. Rapid detection of drug-resistant Mycobacterium tuberculosis directly from clinical specimens using allele-specific polymerase chain reaction assay. Indian J Med Res. 2019;150:33–42. doi: 10.4103/ijmr.IJMR_374_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Munir S, Mahmood N, Shahid S, Khan MI. Molecular detection of Isoniazid, Rifampin and Ethambutol resistance to M. tuberculosis and M. bovis in multidrug resistant tuberculosis (MDR-TB) patients in Pakistan. Microb Pathog. 2017;110:262–274. doi: 10.1016/j.micpath.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 188.Tam KK, Leung KS, Siu GK, Chang KC, Wong SS, et al. Direct detection of pyrazinamide resistance in Mycobacterium tuberculosis by use of pncA PCR sequencing. J Clin Microbiol. 2019;57 doi: 10.1128/JCM.00145-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ullah I, Ahmad W, Shah AA, Shahzada A, Tahir Z, et al. Detection of rifampicin resistance of Mycobacterium tuberculosis using multiplex allele specific polymerase chain reaction (MAS-PCR) in Pakistan. Infect Genet Evol. 2019;71:42–46. doi: 10.1016/j.meegid.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 190.Mistri SK, Sultana M, Kamal SM, Alam MM, Irin F, et al. Evaluation of efficiency of nested multiplex allele-specific PCR assay for detection of multidrug resistant tuberculosis directly from sputum samples. Lett Appl Microbiol. 2016;62:411–418. doi: 10.1111/lam.12564. [DOI] [PubMed] [Google Scholar]
- 191.Majumdar T, Bhattacharya S, Barman D, Bhoumik P, Bir R. Detection of multidrug-resistant tuberculosis using MGITTM(TM) and MAS-PCR in Tripura, India. Int J Tuberc Lung Dis. 2016;20:166–169. doi: 10.5588/ijtld.14.0986. [DOI] [PubMed] [Google Scholar]
- 192.Thirumurugan R, Kathirvel M, Vallayyachari K, Surendar K, Samrot AV, et al. Molecular analysis of rpoB gene mutations in rifampicin resistant Mycobacterium tuberculosis isolates by multiple allele specific polymerase chain reaction in Puducherry, South India. J Infect Public Health. 8:619–625. doi: 10.1016/j.jiph.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 193.Riaz M, Mahmood Z, Javed MT, Javed I, Shahid M, et al. Drug resistant strains of Mycobacterium tuberculosis identified through PCR-RFLP from patients of Central Punjab, Pakistan. Int J Immunopathol Pharmacol. 2016;29:443–449. doi: 10.1177/0394632016638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Javed I, Mahmood Z, Shahid M, Khaliq T. Identification of ofloxacin-resistant Mycobacterium tuberculosis by PCR-RFLP and sequencing. Pak J Pharm Sci. 2016;29:281–286. [PubMed] [Google Scholar]
- 195.Flores-Treviño S, Morfín-Otero R, Rodríguez-Noriega E, González-Díaz E, Pérez-Gómez HR, et al. Characterization of phenotypic and genotypic drug resistance patterns of Mycobacterium tuberculosis isolates from a city in Mexico. Enferm Infecc Microbiol Clin. 2015;33:181–185. doi: 10.1016/j.eimc.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 196.Castan P, de Pablo A, Fernández-Romero N, Rubio JM, Cobb BD, et al. Point-of-care system for detection of Mycobacterium tuberculosis and rifampin resistance in sputum samples. J Clin Microbiol. 2014;52:502–507. doi: 10.1128/JCM.02209-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Luukinen BV, Vuento R, Hirvonen JJ. Evaluation of a semi-automated Seegene PCR workflow with MTB, MDR, and NTM detection for rapid screening of tuberculosis in a low-prevalence setting. APMIS. 2020;128:406–413. doi: 10.1111/apm.13040. [DOI] [PubMed] [Google Scholar]
- 198.Sali M, De Maio F, Caccuri F, Campilongo F, Sanguinetti M, et al. Multicenter evaluation of Anyplex plus MTB/NTM MDR-TB assay for rapid detection of Mycobacterium tuberculosis complex and multidrug-resistant isolates in pulmonary and extrapulmonary specimens. J Clin Microbiol. 2016;54:59–63. doi: 10.1128/JCM.01904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pérez-García F, Ruiz-Serrano MJ, López Roa P, Acosta F, Pérez-Lago L, et al. Diagnostic performance of Anyplex II MTB/MDR/XDR for detection of resistance to first and second line drugs in Mycobacterium tuberculosis . J Microbiol Methods. 2017;139:74–78. doi: 10.1016/j.mimet.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 200.Karunaratne RE, Wijenayaka LA, Wijesundera SS, De Silva KMN, Adikaram CP, et al. Use of nanotechnology for infectious disease diagnostics: application in drug resistant tuberculosis. BMC Infect Dis. 2019;19:618. doi: 10.1186/s12879-019-4259-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Haddaoui M, Sola C, Raouafi N, Korri-Youssoufi H. E-DNA detection of rpoB gene resistance in Mycobacterium tuberculosis in real samples using Fe3O4/polypyrrole nanocomposite. Biosens Bioelectron. 2019;128:76–82. doi: 10.1016/j.bios.2018.11.045. [DOI] [PubMed] [Google Scholar]
- 202.Bengtson HN, Homolka S, Niemann S, Reis AJ, da Silva PE, et al. Multiplex detection of extensively drug resistant tuberculosis using binary deoxyribozyme sensors. Biosens Bioelectron. 2017;94:176–183. doi: 10.1016/j.bios.2017.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yin F, Chan JF, Zhu Q, Fu R, Chen JH, et al. Development and in-use evaluation of a novel Luminex MicroPlex microsphere-based (TRIOL) assay for simultaneous identification of Mycobacterium tuberculosis and detection of first-line and second-line anti-tuberculous drug resistance in China. J Clin Pathol. 2017;70:342–349. doi: 10.1136/jclinpath-2016-203952. [DOI] [PubMed] [Google Scholar]
- 204.Driesen M, Kondo Y, de Jong BC, Torrea G, Asnong S, et al. Evaluation of a novel line probe assay to detect resistance to pyrazinamide, a key drug used for tuberculosis treatment. Clin Microbiol Infect. 2018;24:60–64. doi: 10.1016/j.cmi.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Willby MJ, Wijkander M, Havumaki J, Johnson K, Werngren J, et al. Detection of Mycobacterium tuberculosis pncA mutations by the Nipro genoscholar PZA-TB II assay compared to conventional sequencing. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.01871-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rowneki M, Aronson N, Du P, Sachs P, Blakemore R, et al. Detection of drug resistant Mycobacterium tuberculosis by high-throughput sequencing of DNA isolated from acid fast bacilli smears. PloS One. 2020;15:e0232343. doi: 10.1371/journal.pone.0232343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Karimi H, Oudghiri A, En-Nanei L, Mzibri ME, Laglaoui A, et al. Frequency of genomic mutations mediating resistance of Mycobacterium tuberculosis isolates to rifampicin in Northern Morocco. Rev Inst Med Trop Sao Paulo. 2020;62:e37. doi: 10.1590/S1678-9946202062037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jhanjhria S, Kashyap B, Gomber S, Gupta N, Hyanki P, et al. Phenotypic isoniazid resistance and associated mutations in pediatric tuberculosis. Indian J Tuberc. 2019;66:474–479. doi: 10.1016/j.ijtb.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 209.Yadav R, Saini A, Mankotia J, Khaneja R, Agarwal P, et al. Genetic characterization of Second-Line Drug-Resistant and extensively drug-resistant Mycobacterium tuberculosis from the Northern region of India. J Epidemiol Glob Health. 2018;8:220–224. doi: 10.2991/j.jegh.2018.02.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang Z, Xie T, Mu C, Wang C, Ju H, et al. Molecular characteristics of ofloxacin mono-resistant Mycobacterium tuberculosis isolates from new and previously treated tuberculosis patients. Journal of Clinical Laboratory Analysis. 2018;32 doi: 10.1002/jcla.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Oh TS, Kim YJ, Kang HY, Kim CK, Cho SY, et al. RNA expression analysis of efflux pump genes in clinical isolates of multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis in South Korea. Infect Genet Evol. 2017;49:111–115. doi: 10.1016/j.meegid.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 212.Jagielski T, Bakuła Z, Roeske K, Kamiński M, Napiórkowska A, et al. Mutation profiling for detection of isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J Antimicrob Chemother. 2015;70:3214–3221. doi: 10.1093/jac/dkv253. [DOI] [PubMed] [Google Scholar]
- 213.Martinez E, Holmes N, Jelfs P, Sintchenko V. Genome sequencing reveals novel deletions associated with secondary resistance to pyrazinamide in MDR Mycobacterium tuberculosis . J Antimicrob Chemother. 2015;70:2511–2514. doi: 10.1093/jac/dkv128. [DOI] [PubMed] [Google Scholar]
- 214.Jagielski T, Bakuła Z, Roeske K, Kamiński M, Napiórkowska A, et al. Detection of mutations associated with isoniazid resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates. J Antimicrob Chemother. 2014;69:2369–2375. doi: 10.1093/jac/dku161. [DOI] [PubMed] [Google Scholar]
- 215.Gopinath K, Singh S. Multiplex PCR assay for simultaneous detection and differentiation of Mycobacterium tuberculosis, Mycobacterium avium complexes and other Mycobacterial species directly from clinical specimens. J Appl Microbiol. 2009;107:425–435. doi: 10.1111/j.1365-2672.2009.04218.x. [DOI] [PubMed] [Google Scholar]