Abstract
During a 13-month period, 11 equine patients visiting a veterinary teaching hospital for various diagnostic and surgical procedures developed postprocedural infections from which methicillin (oxacillin)-resistant Staphylococcus aureus (MRSA) strains were isolated. The S. aureus isolates were identified by conventional methods that included Gram staining, tests for colonial morphology, tests for clumping factor, and tests for coagulase and urease activities and were also tested with the API STAPH IDENT system. Antimicrobial susceptibility tests were performed by the disk diffusion method. The biochemical profile and antibiogram of each isolate suggested that the isolates may have come from a common source. Because MRSA strains are very uncommon animal isolates but are rather common human isolates, a nasal swab specimen for culture was collected voluntarily from five persons associated with equine surgery and recovery in an attempt to identify a possible source of the organisms. MRSA strains were isolated from three of the five people, with one person found to be colonized with two biotypes of MRSA. The MRSA isolates from the people appeared to be identical to the isolates from horses. Further study of the isolates included SmaI and EagI macrorestriction analysis by pulsed-field gel electrophoresis conducted in two different laboratories. The results indicated that both the equine and human isolates were members of a very closely related group which appear to have originated from a common source. On the basis of the pattern associated with the infection, it is speculated that the members of the Veterinary Teaching Hospital staff were the primary source of the infection, although the specific mode of transmission is unclear.
Staphylococcus aureus is a pathogen for numerous animal species and humans. Human isolates of S. aureus, unlike animal isolates, are frequently resistant to the penicillinase-resistant penicillins. Organisms exhibiting this type of resistance are referred to as methicillin (oxacillin)-resistant S. aureus (MRSA). In the 1980s, MRSA emerged as a major clinical and epidemiological pathogen in human hospitals (16). The seriousness of this problem has been compounded by the fact that these organisms are frequently resistant to most of the commonly used antimicrobial agents, including the aminoglycosides, macrolides, chloramphenicol, and tetracycline. Although initially susceptible to the fluoroquinolones, MRSA strains have rapidly developed widespread resistance to this class of antimicrobial agent (20). In addition, in accordance with the National Committee for Clinical Laboratory Standards (NCCLS), MRSA strains should be considered to be resistant to all cephalosporins, cephems, and other β-lactams, such as ampicillin-sulbactam, amoxicillin-clavulanic acid, ticarcillin-clavulanic acid, piperacillin-tazobactam, and the carbapenems, regardless of the in vitro test results obtained with those agents (22). The justification for this is the poor clinical response to those antimicrobial agents by MRSA.
While there are numerous publications on outbreaks of nosocomial infections in human hospitals (1, 4, 6, 7, 10, 11, 14, 15, 18, 19, 24), there are limited publications on the epidemiological aspects of nosocomial infections in the animal hospital and laboratory setting (3, 13, 17, 28). Although no reports of a nosocomial spread of an MRSA infection in a veterinary hospital could be found, there have been veterinary reports of MRSA infections in dairy herds with mastitis (8, 9) and in companion animals (5, 17, 28) and of an isolated incident in a horse (13). This report presents data on the isolation of MRSA from 11 equine patients seen at an active midwestern veterinary teaching hospital over a 13-month period for various diagnostic and surgical procedures. The facts that the horses presented are from different farms and were seen over a long period of time and that MRSA is a very uncommon equine isolate suggest that the probable source of this organism was the human caregivers. However, the exact mode of transmission is unknown. To our knowledge, this paper is the first publication on MRSA strains as a cause of a nosocomial epidemic in an animal hospital.
MATERIALS AND METHODS
The data presented here are for 11 equine patients admitted to Michigan State University’s Veterinary Teaching Hospital for various medical and surgical procedures between September 1993 and October 1994. The animals were discharged with no signs of infection; however, within 2 to 3 weeks following discharge, the horses were readmitted to the Veterinary Teaching Hospital with wound infections originating at the site of the therapeutic procedure. The therapeutic procedures included colic surgery (n = 6), joint invasion (n = 2), laryngeal hemiplegia (n = 1), and soft-tissue invasion, i.e., vaccination (n = 2). Samples were collected from the affected wounds and were submitted to the Bacteriology/Mycology Laboratory at the Michigan State University Animal Health Diagnostic Laboratory for bacterial isolation and susceptibility testing. The isolate common to all of the samples submitted was oxacillin (methicillin)-resistant S. aureus. These organisms were identified by conventional methods including Gram staining, tests for colonial morphology, tests for clumping factor, and tests for coagulase and urease activities and were also tested with the API STAPH IDENT system. Disk diffusion susceptibility testing was performed in accordance with NCCLS guidelines (23). Oxacillin MICs were determined by a broth microdilution test method in accordance with NCCLS guidelines (22).
Because of the possibility that the equine isolates were of human origin, a request was made to the members of the equine medicine and surgery faculty and staff that they provide nasal swab specimens for culture in the hope of identifying a potential source. Only 5 of the more than 20 potential candidates consented to providing nasal swab specimens for culture. From these cultures, four MRSA isolates were isolated from specimens from three individuals. To determine the commonality of the isolates, the human and equine isolates were sent to both the University of Iowa and North Carolina State University for phenotypic (biotypes and antibiograms) and genomic studies in a blind, coded format.
At the University of Iowa, genomic DNA was prepared for restriction fragment analysis by modifications of previously published techniques (25) and was then digested with SmaI (New England Biolabs, Beverly, Mass.). Pulsed-field gel electrophoresis (PFGE) was performed on the CHEF-DR II apparatus (Bio-Rad, Richmond, Calif.) with the following: 0.5× TBE (Tris-borate-EDTA), 1% agarose, a temperature of 13°C, and 200 V for 24 h with a switch interval ramped from 10 to 90 s. The investigators at North Carolina State University compared colony morphologies on four different medium types (Trypicase soy agar [TSA], P agar, 5% sheep blood plus TSA, and 5% horse blood plus TSA). The genomic DNA was prepared for PFGE by modifications of previously published techniques (12) and was then analyzed with the restriction enzymes SmaI and EagI (New England Biolabs). The fragments from the digest were separated in the agarose gel slab by using the CHEF-DR II (Bio-Rad) PFGE unit with the following: 0.5× TBE, 1% agarose, a temperature of 13°C, and 200 V for 22 h with a switch interval ramped from 15 to 55 s.
RESULTS
A total of 15 MRSA isolates were collected and analyzed by both laboratories. Eleven of these were from equine patients and four were from the surgical and technical staff of the equine hospital. Of the 15 isolates analyzed, 12 were beta-hemolytic and three were gamma-hemolytic (Table 1). One of the people who tested positive for MRSA possessed two phenotypically different isolates. One of these was beta-hemolytic and the other was gamma-hemolytic. Two other people possessed an MRSA isolate. One person had a beta-hemolytic strain, whereas the other person possessed a gamma-hemolytic strain. One of the 11 equine isolates was gamma-hemolytic, while the other 10 isolates exhibited beta-hemolysis. The results from the susceptibility profiles (Table 2) indicated that the beta-hemolytic isolates had the same antibiogram, as did the gamma-hemolytic isolates. By the disk diffusion test, all of the isolates were categorized as being susceptible in vitro to amikacin, cephalothin, ciprofloxacin, clindamycin, vancomycin, and imipenem. All isolates were considered to be resistant to ampicillin, cefoxitin, ceftiofur, erythromycin, gentamicin, kanamycin, penicillin, tetracycline, and trimethoprim-sulfamethoaxzole, with four exceptions. One equine isolate had an intermediate zone size when it was tested against ceftiofur, and all three gamma-hemolytic isolates had intermediate zone sizes when they were tested against oxacillin. Oxacillin resistance was confirmed by determining the MICs for all 15 isolates. For the 12 beta-hemolytic isolates, MICs were 4 to 16 μg/ml, and thus, their resistance was confirmed. The MICs for the three gamma-hemolytic isolates ranged from 1.0 to 2.0 μg/ml, and they would thus be considered susceptible by the broth microdilution test method.
TABLE 1.
PFGE typing results
| Isolate no. | Hemolytic pattern | PFGE pattern
|
|
|---|---|---|---|
| Univ. of Iowaa | N.C. State Univ.b | ||
| 1 | Beta | B1 | A1 |
| 2 | Beta | B1 | A1 |
| 3 | Beta | B1 | A1 |
| 4 | Beta | B1 | A1 |
| 5 | Beta | B1 | A1 |
| 6 | Gamma | B1 | A1 |
| 7 | Beta | B1 | A1 |
| 8 | Beta | B2 | A2 |
| 9 | Beta | B1 | A1 |
| 10 | Beta | B1 | A1 |
| 11 | Beta | B1 | A1 |
| 12 | Beta | B1 | A1 |
| 13 | Gamma | B1 | A1 |
| 14 | Beta | B1 | A1 |
| 15 | Gamma | B1 | A1 |
Univ. of Iowa, University of Iowa. All isolates had the same digest pattern except isolate 8, which had a minor variation.
N.C. State Univ., North Carolina State University. All isolates had the same digest pattern except isolate 8, which had a minor variation. The colonial morphology and hemolysis of isolates 6, 13, and 15 were different from the rest.
TABLE 2.
Susceptibility profiles for equine and human S. aureus isolates
| Study no. | Source of isolate | Hemolytic pattern | Susceptibility (MIC [μg/ml])a
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AN | AMP | FOX | CFb | CIP | XNL | CC | E | GM | K | OXc | P | TE | SXT | VA | IMP | |||
| 1 | Equine | Beta | S | R | R | S | S | R | S | R | R | R | R (4.0) | R | R | R | S | S |
| 2 | Equine | Beta | S | R | R | S | S | R | S | R | R | R | R (8.0) | R | R | R | S | S |
| 3 | Equine | Beta | S | R | R | S | S | R | S | R | R | R | R (8.0) | R | R | R | S | S |
| 4 | Equine | Beta | S | R | R | S | S | R | S | R | R | R | R (8.0) | R | R | R | S | S |
| 5 | Equine | Beta | S | R | R | S | S | Id | S | R | R | R | R (8.0) | R | R | R | S | S |
| 6 | Equine | Gamma | S | R | R | S | S | R | S | R | R | R | I (2.0) | R | R | R | S | S |
| 7 | Equine | Beta | S | R | R | S | S | R | S | R | R | R | R (4.0) | R | R | R | S | S |
| 8 | Equine | Beta | S | R | R | S | S | R | S | R | R | R | R (8.0) | R | R | R | S | S |
| 9 | Equine | Beta | S | R | R | S | S | R | S | R | R | R | R (8.0) | R | R | R | S | S |
| 10 | Equine | Beta | S | R | R | S | S | R | S | R | R | R | R (16.0) | R | R | R | S | S |
| 11 | Equine | Beta | S | R | R | S | S | R | S | R | R | R | R (8.0) | R | R | R | S | S |
| 12 | Human 1 | Beta | S | R | R | S | S | R | S | R | R | R | R (8.0) | R | R | R | S | S |
| 13 | Human 1 | Gamma | S | R | R | S | S | R | S | R | R | R | I (1.0) | R | R | R | S | S |
| 14 | Human 2 | Beta | S | R | R | S | S | R | S | R | R | R | R (8.0) | R | R | R | S | S |
| 15 | Human 3 | Gamma | S | R | R | S | S | R | S | R | R | R | I (1.0) | R | R | R | S | S |
AN, amikacin; AMP, ampicillin; FOX, cefoxitin; CF, cephalothin; CIP, ciprofloxacin; XNL, ceftiofur; CC, clindamycin; E, erythromycin; GM, gentamicin; K, kanamycin; OX, oxacillin; P, penicillin; TE, tetracycline; SXT, trimethoprim-sulfamethoxazole; VA, vancomycin; IMP, imipenem; S, susceptible; R, resistant.
In accordance with NCCLS guidelines, an S. aureus isolate resistant to oxacillin should also be considered resistant to all other β-lactams.
For the three gamma-hemolytic isolates, the zone size is intermediate and the MIC range is 1.0 to 2.0 μg/ml.
The zone size for this organism was 20 mm. NCCLS M31-T document-proposed guidelines indicate that resistant organisms have zone sizes of ≤17 mm.
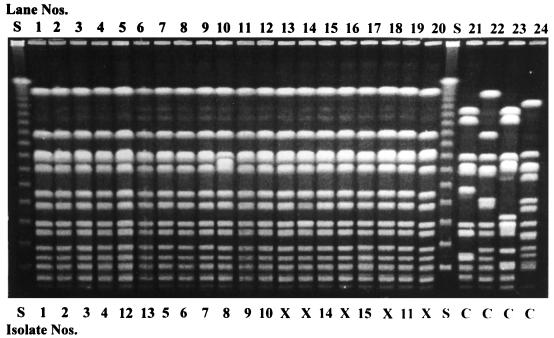
Results from the University of Iowa concluded that, on the basis of restriction enzyme analysis of genomic DNA by PFGE, one major PFGE type (type B) was identified in all 15 isolates (Fig. 1, lanes 1 to 12, 15, 17, and 19). Isolates were considered to be the same strain if all bands matched, subtypes of the same strain if the patterns differed by one to three bands, and different strains if the patterns differed by more than three bands (25). Two subgroups of type B were noticed. Isolate 8 (Fig. 1, lane 10) was labeled type B2, whereas all other isolates were considered type B1 (Table 1). Although the subtypes were not identical, they should be considered the same strain for epidemiological purposes because of the variation in only one to three lanes. The isolates in lanes 13, 14, 16, 18, and 20 in Fig. 1 represent additional equine isolates collected during the earlier part of the same time period and under the same circumstances as those for the isolates in the other lanes (Fig. 1, lanes 1 to 12, 15, 17, and 19). However, these isolates were not submitted to North Carolina State University for comparison, and thus, data for these isolates are not included in this report other than as illustrated in these lanes. Lanes 21 to 24 of Fig. 1 represent unrelated MRSA isolates that served as controls.
FIG. 1.
Fingerprints of MRSA isolates obtained by PFGE with SmaI digestion. Lane S, 48.5-kb bacteriophage lambda ladder; lanes 13, 14, 16, and 20, MRSA isolates (X) from equine patients that were not analyzed in both laboratories; lanes 21 to 24, unrelated MRSA isolates that served as controls (C).
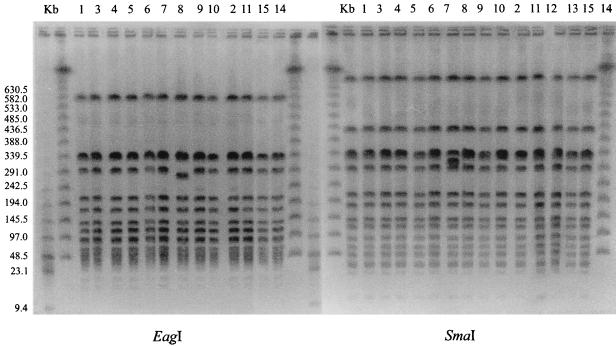
At North Carolina State University, a comparison of colony morphology and hemolysis on the four different kinds of media indicated that there were two groups of isolates. Twelve of the 15 isolates were in group 1, while 3 isolates made up group 2. Group 1 isolates were beta-hemolytic. The colony morphology of group 2 isolates (isolates 6, 13, and 15) was different from that of group 1 isolates, and group 1 isolates were gamma-hemolytic (Table 1). However, results from the SmaI digestion indicated that all 15 strains were very closely related. These isolates were considered members of the same strain because their digestion patterns differed by no more than one band. Because of some minor variations in patterns (isolate 8) and colony morphology (isolates 6, 13, and 15), digestion with EagI was done for greater delineation of the isolates and to confirm their similarities. As with the results from the University of Iowa, the pattern for isolate 8 varied compared to the patterns for the other strains (Fig. 2).
FIG. 2.
Fingerprints of MRSA obtained by PFGE with SmaI and EagI digestion. Lane Kb, a 48.5-kb bacteriophage lambda ladder. For the SmaI digestion, lanes 1 to 15 represent both equine and human isolates associated with the MRSA outbreak. For the EagI digestion, the isolates associated with the MRSA outbreak are represented in lanes 1 to 11, 14, and 15.
DISCUSSION
The animals in this study were originally brought from a variety of locations throughout the upper Midwest to the Michigan State University Veterinary Teaching Hospital for evaluation of a variety of conditions. The most common complaint upon arrival was acute abdominal crisis requiring surgery (55% of the horses). Two patients had surgery involving joints, and the remaining three horses were treated for medical conditions that involved soft-tissue invasion with needles. None of the patients had signs of infection on initial presentation. Within 2 to 3 weeks of initial treatment, they were readmitted for evaluation of surgical or therapeutic site infections. From routine diagnostic cultures, an MRSA isolate was identified in all of the samples submitted.
Because MRSA had rarely been isolated in the veterinary diagnostic bacteriology laboratory and never from a horse, the isolation of this organism from several horses over a period of a few months prompted an investigation as to a potential source. In previous reports on the isolation of MRSA in a veterinary environment (8, 9, 13), it was concluded that the isolates were not of animal origin and were most likely from humans. Attempts were therefore made to identify a possible human source of the organisms. In humans, S. aureus has frequently been isolated from the anterior nares and the vaginal, rectal, and perineal regions (20). None of the equine staff members were required to submit to a sample for culture; however, five did consent to provide a nasal swab specimen for culture. Two of the five cultures were negative for MRSA. Four MRSA strains were isolated from three of five individuals who consented to provide samples for culture. The PFGE comparison performed in two laboratories indicated that the equine isolates and the human isolates were most likely from the same source.
It is known that S. aureus strains, as a species, may show considerable polymorphism in pulsed-field patterns (2, 26). However, although MRSA isolates do not have identical pulsed-field patterns, they show more similarity to each other than S. aureus isolates as a whole show to each other. In viewing the pulsed-field patterns for the isolates in this study, it was concluded that all of the isolates were from a common group or lineage and that, for epidemiological purposes, all of the isolates should be considered representatives of the same strain. The similarities of the equine and human isolates described in this report are clear compared to the similarities of the control MRSA strains.
Although PFGE cannot always distinguish among isolates with differences in colony type and antibiograms, we can speculate, when looking at all of the compiled data, that among these isolates there is a subdivision which is indicative of a separate clonal status. Alternatively, the results presented above may be suggestive of a clonal variation within a population. This can also be seen when viewing the oxacillin MIC results for the 15 isolates included in this study. The beta-hemolytic strains are resistant to oxacillin (MICs ≥ 4 μg/ml), whereas the gamma-hemolytic strains appear to be susceptible (MIC range, between 1 and 2 μg/ml). However, the PFGE results indicate the similarities of all of the isolates, and, thus, it is concluded that they are the same strain from a single source. In other words, while there was some limited phenotypic variation among the 15 isolates, by macrorestriction patterns they appeared to be the same.
On the basis of our data and the temporal patterns associated with the infections, it is concluded that all of the isolates were members of a very closely related group which appears to have originated from a common source. The initial source of this outbreak is probably of human origin, since the horses originally arrived at the hospital over a 13-month period from a variety of locations and with no apparent staphylococcal infection at the time of arrival. It is difficult to know how many other horses were seen at the teaching hospital during this time interval. It is also difficult to know how many, if any, of those horses developed MRSA infections but were treated by the referring veterinarian without any further contact with the Veterinary Teaching Hospital. A number of people potentially had access to the horses during the horses’ initial visits. Among the three individuals from whom four MRSA strains were isolated, there was no common relationship between the affected patients and these staff members, except that the staff members all worked in the same area in which the patients were seen. The transmission could have spread from human to horse during any of the invasive procedures or during care throughout the animal’s hospital stay. In humans, staphylococci are usually transmitted from person to person via contaminated hands (21). All of the horses involved in this study received hands-on treatment. However, since samples from all personnel involved with the horses were not cultured, the specific mode of transmission is unclear since many of the patients were examined and treated by different members of the hospital staff and samples from only a small percentage of the Veterinary Teaching Hospital equine staff were cultured.
REFERENCES
- 1.Back N A, Linnemann C C, Pfaller M A, Staneck J L, Morthland V. Recurrent epidemics caused by a single strain of erythromycin-resistant Staphylococcus aureus. JAMA. 1993;270:1329–1333. [PubMed] [Google Scholar]
- 2.Bannerman T L, Hancock G A, Tenover F C, Miller J M. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995;33:551–555. doi: 10.1128/jcm.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackmore D K, Francis R A. The apparent transmission of staphylococci of human origin to laboratory animals. J Comp Pathol. 1970;80:645–651. doi: 10.1016/0021-9975(70)90064-2. [DOI] [PubMed] [Google Scholar]
- 4.Brandenburg A H, van Belkum A, van Pelt C, Bruining H A, Mouton J W, Verburgh H A. Patient-to-patient spread of a single strain of Corynebacterium striatum causing infections in a surgical intensive care unit. J Clin Microbiol. 1996;34:2089–2094. doi: 10.1128/jcm.34.9.2089-2094.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cefai C, Ashurst S, Owens C. Human carriage of methicillin-resistant Staphylococcus aureus linked with pet dog. Lancet. 1994;344:539–540. doi: 10.1016/s0140-6736(94)91926-7. [DOI] [PubMed] [Google Scholar]
- 6.Cohen S H, Morita M M, Bradford M. A seven-year experience with methicillin-resistant Staphylococcus aureus. Am J Med. 1991;91(Suppl. 3B):233S–237S. doi: 10.1016/0002-9343(91)90374-7. [DOI] [PubMed] [Google Scholar]
- 7.De Lencastre H, De Lencastre A, Tomasz A. Methicillin-resistant Staphylococcus aureus isolates recovered from a New York City hospital: analysis by molecular fingerprinting techniques. J Clin Microbiol. 1996;34:2121–2124. doi: 10.1128/jcm.34.9.2121-2124.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devriese L A, Hommez J. Epidemiology of methicillin-resistant Staphylococcus aureus in dairy herds. Res Vet Sci. 1975;19:23–27. [PubMed] [Google Scholar]
- 9.Devriese L A, Van Damme L R, Fameree L. Methicillin (cloxacillin)-resistant Staphylococcus aureus strains isolated from bovine mastitis cases. Zentbl Veterinarmed Ser B. 1972;19:598–605. doi: 10.1111/j.1439-0450.1972.tb00439.x. [DOI] [PubMed] [Google Scholar]
- 10.Doebbeling B N, Stanley G L, Sheetz C T, Pfaller M A, Houston A K, Annis L, Li N, Wenzel R P. Comparative efficacy of alternative hand-washing agents in reducing nosocomial infections in intensive care units. N Engl J Med. 1992;327:88–92. doi: 10.1056/NEJM199207093270205. [DOI] [PubMed] [Google Scholar]
- 11.Farrington M, Ling J, Ling T, French G L. Outbreaks of infection with methicillin-resistant Staphylococcus aureus on neonatal and burns units of a new hospital. Epidemiol Infect. 1990;105:215–228. doi: 10.1017/s0950268800047828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George C, Kloos W. Comparison of the SmaI-digested chromosomes of Staphylococcus epidermidis and the closely related species Staphylococcus capitis and Staphylococcus caprae. Int J Syst Bacteriol. 1994;44:404–409. doi: 10.1099/00207713-44-3-404. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann F A, Trostle S S, Klohnen A A O. Isolation of methicillin-resistant Staphylococcus aureus from a postoperative wound infection in a horse. J Am Vet Med Assoc. 1997;221:590–592. [PubMed] [Google Scholar]
- 14.Heczko P B, Bulanda M, Hoeffler U. Nasal carriage of Staphylococcus aureus and its influence on hospital infections caused by methicillin-resistant strains. Int J Med Microbiol. 1990;274:333–341. doi: 10.1016/s0934-8840(11)80690-0. [DOI] [PubMed] [Google Scholar]
- 15.Hollis R J, Barr J L, Doebbling B N, Pfaller M A, Wenzel R P. Familial carriage of methicillin-resistant Staphylococcus aureus and subsequent infection in a premature neonate. Clin Infect Dis. 1995;21:328–332. doi: 10.1093/clinids/21.2.328. [DOI] [PubMed] [Google Scholar]
- 16.Kloos W E, Bannerman T L. Staphylococcus and Micrococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: ASM Press; 1995. pp. 282–298. [Google Scholar]
- 17.Koterba A, Torchia J, Silverthorne C, Ramphal R, Merritt A M, Manucy J. Nosocomial infections and bacterial antibiotic resistance in a university equine hospital. J Am Vet Med Assoc. 1986;189:185–191. [PubMed] [Google Scholar]
- 18.Lee S H, Yii N W, Hanifah Y A. Methicillin-resistant Staphylococcus aureus in surgical patients: Malaysian experience. J R Coll Surg Edinburgh. 1991;36:323–327. [PubMed] [Google Scholar]
- 19.Linnemann C C, Moore P, Staneck J L, Pfaller M A. Reemergence of epidemic methicillin-resistant Staphylococcus aureus in a general hospital associated with changing staphylococcal strains. Am J Med. 1991;91(Suppl. 3B):238S–244S. doi: 10.1016/0002-9343(91)90375-8. [DOI] [PubMed] [Google Scholar]
- 20.Mandell G, Douglas J, Bennett R, editors. Principles and practices of infectious diseases. 4th ed. Edinburgh, United Kingdom: Churchill Livingstone, Ltd.; 1995. [Google Scholar]
- 21.Mulligan M E, Murray-Leisure K A, Ribner B S, Standiford H C, John J F, Korvick J A, Kauffman C A, Yu V L. Methicillin-resistant Staphylococcus aureus: a consensus review of the microbiology, pathogenesis, and epidemiology with implications for prevention and management. Am J Med. 1993;94:313–328. doi: 10.1016/0002-9343(93)90063-u. [DOI] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standards. NCCLS document M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standards. NCCLS document M2-A3. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 24.Ndawula E M, Brown L. Mattress as reservoirs of epidemic methicillin-resistant Staphylococcus aureus. Lancet. 1991;337:488. doi: 10.1016/0140-6736(91)93420-e. [DOI] [PubMed] [Google Scholar]
- 25.Pfaller M A, Hollis R J, Sader H S. PFGE analysis of chromosomal restriction fragments, section 10.5.c. In: Tenover F C, editor. Clinical microbiology procedures handbook. Washington, D.C: American Society for Microbiology; 1994. [Google Scholar]
- 26.Prevost G B, Jaulhac B, Piemont Y. DNA fingerprinting by pulsed-field gel electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 1992;30:967–973. doi: 10.1128/jcm.30.4.967-973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott G M, Thomson R, Malone-Lee J, Ridgway G L. Cross-infection between animals and man: possible feline transmission of Staphylococcus aureus infection in humans? J Hosp Infect. 1988;12:29–34. doi: 10.1016/0195-6701(88)90119-3. [DOI] [PubMed] [Google Scholar]
- 28.Tomlin J, Pead M J, Lloyd D H, Howell S, Hartmann F, Jackson H A, Muir P. Methicillin-resistant Staphylococcus aureus infections in 11 dogs. Vet Rec. 1999;16:60–64. doi: 10.1136/vr.144.3.60. [DOI] [PubMed] [Google Scholar]