Abstract
Prions, proteins that can convert between structurally and functionally distinct states and serve as non-Mendelian mechanisms of inheritance, were initially discovered and only known in eukaryotes, and consequently considered to likely be a relatively late evolutionary acquisition. However, the recent discovery of prions in bacteria and viruses has intimated a potentially more ancient evolutionary origin. Here, we provide evidence that prion-forming domains exist in the domain archaea, the last domain of life left unexplored with regard to prions. We searched for archaeal candidate prion-forming protein sequences computationally, described their taxonomic distribution and phylogeny, and analyzed their associated functional annotations. Using biophysical in vitro assays, cell-based and microscopic approaches, and dye-binding analyses, we tested select candidate prion-forming domains for prionogenic characteristics. Out of the 16 tested, eight formed amyloids, and six acted as protein-based elements of information transfer driving non-Mendelian patterns of inheritance. We also identified short peptides from our archaeal prion candidates that can form amyloid fibrils independently. Lastly, candidates that tested positively in our assays had significantly higher tyrosine and phenylalanine content than candidates that tested negatively, an observation that may help future archaeal prion predictions. Taken together, our discovery of functional prion-forming domains in archaea provides evidence that multiple archaeal proteins are capable of acting as prions—thus expanding our knowledge of this epigenetic phenomenon to the third and final domain of life and bolstering the possibility that they were present at the time of the last universal common ancestor.
Keywords: prion, amyloid, evolution, archaea, LUCA
Significance
This work establishes that amyloid-forming, prion-like domains exist in Archaea and are capable of vertically transmitting their prion phenotype—allowing them to function as protein-based elements of inheritance. These observations, coupled with prior discoveries in Eukarya and Bacteria, suggest that prion-based self-assembly was potentially present in life’s last universal common ancestor, and therefore may be one of the most ancient epigenetic mechanisms.
Introduction
One of the most notable and puzzling disease outbreaks of the last 50 years was bovine spongiform encephalopathy. Its baffling patterns of transmission arose because it was caused by a mysterious agent devoid of nucleic acids (Prusiner 1998). The disease left 177 people and over four million cattle dead, harmed the economy of the United Kingdom, and, for a time, left the scientific community with no mechanism to explain the phenomenon (Murdoch and Murdoch 2015). The cause was eventually determined to be a misfolded form of an endogenous protein, designated the Prion Protein (PrP) (Prusiner 1998).
Although initially named for “proteinaceous infectious particles” (Prusiner 1998), ascribing a stable, concise definition to the term “prion” is not trivial. As our scientific understanding of prion-like entities continues to develop alongside the concept’s integration into society’s lexicon, the term itself is under relatively rapid linguistic evolutionary pressure. One generalizable and useful definition is: “prions are proteins that convert between structurally and functionally distinct states, at least one of which is self-propagating and self-perpetuating” (slightly modified from Alberti et al. [2009] and Garcia and Jarosz [2014]). And although certainly useful, this does not adequately convey the whole story because it assigns the property of prion to the protein itself as an individual unit, whereas some of the key hallmarks of prion activity (e.g., self-propagation and non-Mendelian inheritance) only manifest through the interaction of multiple copies of a protein under specific conditions. Conceptually this is not entirely dissimilar from how quorum sensing can be thought of as an emergent property, rather than being ascribed to a single unit. It can therefore be useful to also think of the term “prion” as a state that emerges when a specific suite of characteristics is met for multiple copies of a protein that is capable of facilitating that state. That said, unless noted otherwise, herein the term “prion” will be utilized in the former, generalized form, referring to an individual protein.
Although prions were being studied in mammals, puzzling nonchromosomal genetic elements were concurrently being discovered in yeast: [URE3] (Lacroute 1971) and [PSI+] (Young and Cox 1971). As the prion story unfolded, it soon became clear that [URE3] and [PSI+] were prions of the Ure2p and Sup35p proteins, respectively (Chernoff et al. 1993; Ter-Avanesyan et al. 1994; Wickner 1994; Lindquist et al. 1998; Liebman and Derkatch 1999). All known mammalian prion diseases are the result of the misfolding of the endogenous prion protein (PrP). By contrast, fungal prions are more diverse, sparking phenotypes produced by the alternative folds of many different proteins (Wickner 1994; Derkatch et al. 1997; Du et al. 2008; Rogoza et al. 2010; Suzuki et al. 2012). Prion conversion of PrP is pathogenic, whereas acquisition of many yeast prions is not. As more work has been done to elucidate nonpathogenic prion-forming proteins, it has been shown that prion conversion can serve as a molecular switch for multiple biological functions, often exerting a strong influence on survival. They can play a role in adaptive responses to environmental fluctuations (Halfmann and Lindquist 2010; Rogoza et al. 2010; Itakura et al. 2020), contribute to evolvability by acting as epigenetic elements (True and Lindquist 2000; True et al. 2004; Holmes et al. 2013; Du et al. 2015), and act as evolutionary capacitors (Shorter and Lindquist 2005; Masel and Siegal 2009) and bet-hedging devices (King and Masel 2007; Namy et al. 2008; Halfmann et al. 2010). For example, Saccharomyces cerevisiae employs the [GAR+] prion to switch between specialist and generalist carbon-source utilization strategies, a switch that is heritable (Brown and Lindquist 2009; Jarosz et al. 2014). The [ESI+] prion drives the emergence and transgenerational inheritance of an activated chromatin state that can result in broad resistance to environmental stress, including antifungal drugs (Harvey et al. 2020). The [SMAUG+] prion allows yeast to anticipate nutrient repletion after periods of starvation, providing a strong selective advantage (Itakura et al. 2020). Other nondetrimental prions were found in many distinct species. In Podospora anserina, prion [Het-s] carries out normal cell functions in the process of self/nonself recognition (Dalstra et al. 2005; Riek and Saupe 2016). In the angiosperm Arabidopsis thaliana, the Luminidependens protein can form a prion and is responsible for signaling flowering as a result of detecting the temperature change in the environment (Chakrabortee, Kayatekin, et al. 2016). In baculoviruses, the prion-forming LEF-10 protein was found to be responsible for efficient viral replication and expression (Nan et al. 2019). The discovery of functional prions across a broad phylogenetic diversity of organisms raises the question of their evolutionary origins.
The first prions to be discovered form highly ordered aggregates called amyloids (Sabate, Rousseau, Schymkowitz, Batlle, et al. 2015) and many, but not all, currently known prions adopt an amyloid conformation. Amyloids are long, unbranched protein aggregates characterized by a fibrillar morphology and cross β-sheet quaternary protein structure (Sipe et al. 2016). Just like prions, amyloids were first discovered in association with neurodegeneration. The most well-known amyloids form protein deposits in the human brain as in the cases of Aβ peptide and Tau protein in Alzheimer’s disease, huntingtin in Huntington’s disease, α-synuclein in Parkinson’s disease, and SOD1 in Amyotrophic lateral sclerosis (Chiti and Dobson 2006). Similar to prions, amyloids exist in myriad organisms, including microbes. Although their presence correlates with neurodegeneration, some have argued that amyloid itself might be overall more beneficial relative to other oligomeric forms of aggregation-prone proteins (Walsh et al. 2002; Benilova et al. 2012). Amyloids have functional and nonpathogenic roles in species as phylogenetically distinct as Escherichia coli and humans (Chapman et al. 2002; Biesecker et al. 2018). The term “functional amyloids” was created to describe nondetrimental amyloids (Fowler et al. 2005). In microbes, functional amyloids are often found in the extracellular matrix contributing to cellular protection and biofilm formation (Chapman et al. 2002; Cegelski et al. 2009). The first functional amyloid identified in humans was the PMEL 17 protein (Fowler et al. 2005; Bissig et al. 2016). Important in melanogenesis, amyloids of PMEL 17 create a scaffold for melanin distribution (Fowler et al. 2005; Bissig et al. 2016). Amyloid-forming proteins have been detected and experimentally verified in all domains of life: Eukaryota, Bacteria, and Archaea (Gebbink et al. 2005; Greenwald and Riek 2010; Shanmugam et al. 2019). Prions, on the other hand, have to date only been discovered and verified in Eukarya and Bacteria (Prusiner 1998; Yuan and Hochschild 2017).
The propagation of known prions relies on a protein fragment called the prion domain (PrD). These domains are often enriched in glutamine and asparagine residues and are usually found in the disordered fragments of proteins. Leveraging these principles, bioinformatic algorithms can be used to predict novel PrDs (Li and Lindquist 2000; Michelitsch and Weissman 2000; Alberti et al. 2009). Here, we use computational prediction methods on 1,262 archaeal proteomes to assess the distribution of prion proteins across the archaeal domain. We identify candidate PrDs (cPrDs) and investigate the functions associated with the proteins harboring them. We additionally perform a suite of biochemical and genetic experiments on a subset of our identified cPrDs to validate their capacity to form infectious aggregates. Our data reveal that multiple archaeal proteins can act as prions, forming heritable protein aggregates that are capable of acting as protein-based epigenetic elements. These observations, coupled with prior discoveries in eukarya and bacteria, suggest that prion-based self-assembly is an ancient form of epigenetics that may have been present in life’s last universal common ancestor (LUCA).
Results
Computational Identification and Distribution of cPrDs in Archaea
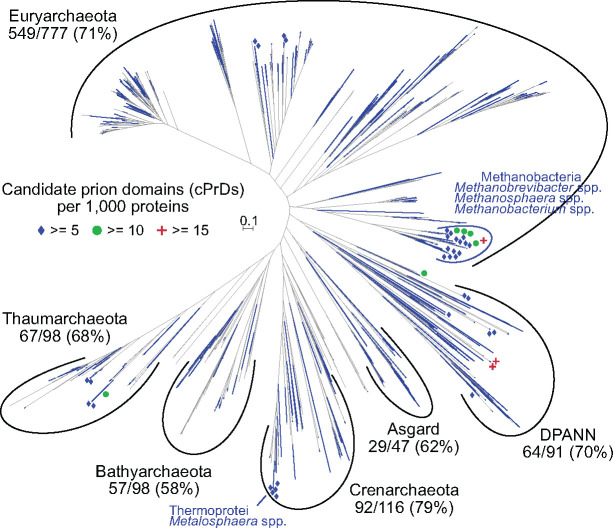
Leveraging the strong enrichment of glutamine and asparagine in prion domains, we utilized the program PLAAC (Lancaster et al. 2014) to scan for cPrDs in archaeal proteomes sourced from the UniProt (Bateman 2019) database (see fig. 1 for overview). Of the 1,262 archaeal proteomes scanned, at least one cPrD was identified in 873 proteomes (∼69%). Of the 2,805,234 proteins scanned, 2,797 were identified as containing cPrDs (∼0.10%). The distribution of proteomes with at least one cPrD identified was largely homogenous across the archaeal domain (fig. 2). There were modestly lower proportions within the Bathyarchaeota (58%) and the Asgard group (62%), and a higher proportion within the Crenarchaeota (79%). We note however that these taxonomic summary delineations are somewhat arbitrary, and as with all databases there can be underlying confounding factors about which lineages may be over- or under-represented as compared with others. For example, a commonly sequenced organism will likely be overrepresented, as compared with those less commonly sequenced.
Fig. 1.
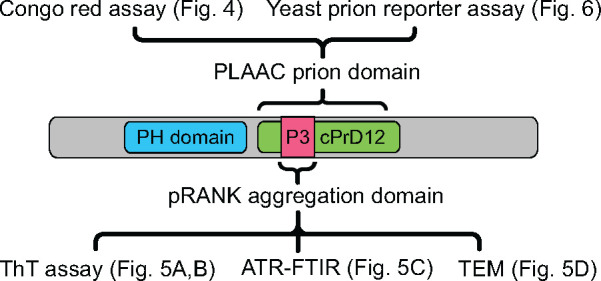
Fragments of protein and methods used to analyze them. Schematic representation of PH domain-containing protein from Sulfolobus sp. Candidate prion domain identified by PLAAC (cPrD12) and aggregation domain identified by pRANK (P3) are marked. Methods applied to analyze the designated fragments are listed with references to other figures.
Fig. 2.
A phylogenomic tree of incorporated Archaea with distribution of cPrDs overlain. Tips with at least 1 cPrD are colored blue. Blue diamonds are places near those with ≥ to five cPrDs per 1,000 genes; green circles near those with ≥10; and red pluses near those with ≥15. Major taxa summary values include the number within that group with identified cPrDs out of the total in that group followed by that percentage. The Asgard group here includes: Thorarchaeota, Odinarchaeota, and Heimdallarchaeota. DPANN group here includes: Woesearchaeota, Pacearchaeota, Nanoarchaeota, Aenigmarchaeota, Diapherotrites, Micrarchaeota, and Altiarchaeota. The unrooted tree is an estimated maximum-likelihood of 76 single-copy orthologs forming an alignment of 12,330 amino acids (see Materials and Methods).
The frequency of identified cPrDs within each proteome was ∼2.21 ± 3.38 (mean ± 1 SD) with a median of one: 30% of the scanned proteomes contained 0; ∼25% had the median of 1: ∼35% had between 2 and 5; and ∼10% had >5 (supplementary data set S1, Supplementary Material online). Normalizing these values to the total number of proteins in each individual proteome (done as cPrDs per 1,000 proteins) returns ∼1.14 ± 1.89 (mean ± 1 SD) with a median of 0.6 (supplementary data set S1, Supplementary Material online; histogram in supplementary fig. S1, Supplementary Material online). Based on these normalized values, three proteomes were found to have >15 identified cPrDs per 1,000 genes (placing them in percentile ranks of >99.8) including two closely related Nanoarchaeota (GCA_003086475.1 and GCA_003086415.1) and a Euryarchaeota of the class Methanobacteria (GCA_001639265.1; demarcated with red plus signs in fig. 2). Other proteomes with relatively high-densities of identified cPrDs included 10 with ≥10 cPrDs per 1,000 proteins (99.2 percentile rank) and 46 with ≥5 (96.4th percentile). Many of these were concentrated within the class Methanobacteria (across the genera Methanobrevibacter, Methanosphaera, and Methanobacterium) and within representatives of Metallosphaera sedula of the Thermoprotei class (fig. 2).
Enriched Functions of Proteins Containing cPrDs
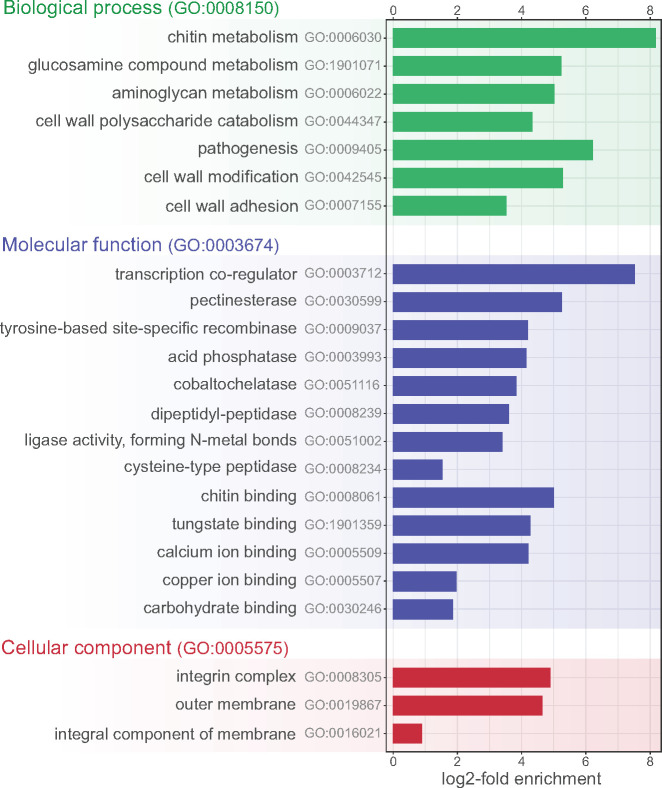
We utilized Gene Ontology (GO) (Ashburner et al. 2000) annotations (GO “terms”) and goatools (Klopfenstein et al. 2018) to test for enrichment or purification of specific GO terms in our identified cPrD-containing proteins (2,797) as compared with all of the proteins scanned (2,805,234). The identified cPrD-containing proteins were associated with 706 GO terms, 523 of which were found to be significantly enriched or purified in the cPrD group relative to all proteins (based on Benjamini–Hochberg false-discovery rates ≤0.05; supplementary data set S2, Supplementary Material online). The vast majority of these (469/523; ∼90%) were found to be purified in proteins harboring cPrDs relative to all proteins (underrepresented), and 54 were found to be enriched—a subset of which are presented in figure 3.
Fig. 3.
Select enriched GO terms of proteins harboring cPrDs as compared with all scanned archaeal proteins. Displayed are log2-fold enrichments of the given GO term’s frequency among total cPrD-containing proteins (2,797) as compared its frequency among all computationally scanned archaeal proteins (2,805,234). Considered enrichments were those with Benjamini–Hochberg false-discovery rates of ≤0.05 (see Materials and Methods). Full results in supplementary data set S3, Supplementary Material online.
GO breaks its terms down into three namespaces: Biological Process, Molecular Function, and Cellular Component. Significantly enriched GO terms included proteins involved in: chitin metabolism (GO:0006030), cell wall polysaccharide catabolism (GO:0044347), and cell wall adhesion (GO:0007155) within the Biological Process namespace; transcription coregulator (GO:0003712), tyrosine-based site-specific recombinase (GO:0009037), and calcium (GO:0005509) and copper ion binding (GO:0005507) within the Molecular Function namespace; and outer membrane (GO:0019867) within the Cellular Component namespace (fig. 3; full results in supplementary data set S2, Supplementary Material online). Significantly purified (underrepresented) GO terms included: cellular amino acid (GO:0008652) and nucleotide biosynthesis (GO:0009165) within Biological Process; nucleotide (GO:0000166) and metal ion binding (GO:0046872) within Molecular Function; and cytoplasm (GO:0005737) and ribosome (GO:0005840) within Cellular Component (supplementary data set S2, Supplementary Material online).
Multiple Archaeal cPrDs Bind Congo Red
Selective binding to dyes, including Congo red, is a diagnostic characteristic of amyloid. We investigated this property in the cPrDs of the highest scoring (COREscore ≥30) putative prions from archaeal proteomes. Many cPrDs consisted of sequences with long stretches of only Q or N. Because of the low complexity of these sequences, the synthesis of coding DNA in many cases proved impossible. Thus, we chose sequences complex enough to allow their synthesis. From this group, we focused on genes with functional annotations, and synthesized 16 coding sequences (table 1). We cloned these sequences into a commercially available vector allowing synthesis and export of investigated cPrDs (Sivanathan and Hochschild 2013).
Table 1.
Summary of Results from Congo Red and Yeast Prion Reporter Assay Together with the Explanation of Prion Candidates’ IDs Used in the Text.
| ID | Congo Red Assay, Results | Yeast Prion Reporter Assay, Results | NCBI Accession | NCBI Taxonomy | NCBI Annotation |
|---|---|---|---|---|---|
| cPrD1 | CR+ | YPR+ | AEH61342.1 | Methanosalsum zhilinae | Transcriptional repressor, CopY family |
| cPrD2 | CR− | YPR− | AGN16545.1 | Methanobrevibacter sp. | Adhesin-like protein |
| cPrD3 | CR− | YPR+ | ARM76690.1 | Acidianus manzaensis | Zinc ribbon domain-containing protein |
| cPrD4 | CR+ | YPR− | KZX15367.1 | Methanobrevibacter filiformis | tRNA biosynthesis protein MnmC |
| cPrD5 | CR+ | YPR− | OQD58440.1 | Methanobrevibacter arboriphilus | Putative peptidase |
| cPrD6 | CR− | YPR+ | WP_009487818.1 | Halobacterium sp. | DNA starvation/stationary phase protection Dps |
| cPrD7 | CR− | YPR+ | WP_013774943.1 | Acidianus hospitalis | Zinc ribbon domain-containing protein |
| cPrD8 | CR+ | YPR+ | WP_014027445.1 | Pyrolobus fumarii | Tyrosine-type recombinase/integrase |
| cPrD9 | CR+ | YPR+ | WP_042706453.1 | Methanomicrobium mobile | YIP1 family protein |
| cPrD10 | CR− | YPR+ | WP_054838334.1 | Sulfolobus metallicus | Zinc ribbon domain-containing protein |
| cPrD11 | CR+ | YPR+ | WP_066970924.1 | Methanobrevibacter filiformis | NADAR family protein |
| cPrD12 | CR+ | YPR+ | WP_066972153.1 | Methanobrevibacter filiformis | PH domain-containing protein |
| cPrD13 | CR+ | YPR+ | WP_069282784.1 | Sulfolobus sp. | Zinc ribbon domain-containing protein |
| cPrD14 | CR− | YPR− | WP_074791546.1 | Haloferax larsenii | Ribonuclease BN |
| cPrD15 | CR− | YPR− | WP_082223394.1 | Halosimplex carsbadense | CBS domain-containing protein |
| cPrD16 | CR− | YPR− | WP_084269446.1 | Methanobrevibacter curvatus | ATP-dependent protease LonB |
To test if the putative prions were capable of forming amyloid fibrils, we used a bacterial export system for generating extracellular amyloid aggregates (Sivanathan and Hochschild 2013). The use of this system enables rapid screening for the ability of the aggregates to bind to the Congo red dye. A full-length prion protein is not always necessary, nor optimal for prion formation (King et al. 1997; Taylor et al. 1999). For example, in most yeast prions proteins, overproduction of only the amyloid-forming portion of the protein is sufficient for prion induction (Ter-Avanesyan et al. 1994; Masison et al. 1997). This induction is sometimes more efficient than with overproduction of the full-length protein (Masison and Wickner 1995; Kochneva-Pervukhova et al. 1998). Another characteristic of known PrDs is their modularity, which allows them to be cloned into other proteins thus transferring the ability to form prion aggregates (Li and Lindquist 2000). Preliminary tests also demonstrated that the use of a cPrD increases the sensitivity of the Congo red assay compared with full-length proteins (supplementary fig. S2, Supplementary Material online). For these reasons, and synthesis considerations, we tested cPrDs instead of full-length proteins.
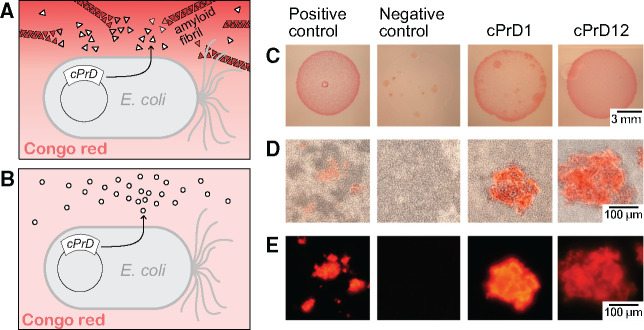
Bacteria transformed with plasmids encoding the cPrDs were plated on an induction medium containing Congo red. Because in the C-DAG system the expressed cPrDs are exported outside of the cell they can interact with the dye in the culture medium. Bacterial colonies that produce cPrDs capable of forming amyloids bind Congo red, which then changes the color of the colony to red. A clear change in color was observed in eight out of 16 colonies (fig. 4 and see supplementary fig. S3, Supplementary Material online, for results of all 16).
Fig. 4.
Comparison of colonies and corresponding protein aggregates produced by bacteria transformed with plasmids encoding cPrDs. (A and B) Two variants of possible results described in a cartoon. (A) Candidate cPrD forms amyloid aggregates that bind Congo red. (B) cPrD does not form amyloid aggregates therefore does not bind Congo red. (C) Colonies of Escherichia coli grew on the induction medium containing 0.1% Congo red. The red color of the colony indicates that the exported protein binds to Congo red. Binding of Congo red is typical for amyloid fibrils. (D) Light microscopy of preparations made of the same bacterial colonies show aggregates binding Congo red and bacterial cells concentrating around them. (E) Fluorescent microscopy of protein aggregates from panel (B) is shown. The amyloidogenic fragment of Sup35 (residues 2–253) was used as a positive control. Fragment of Sup35 not able to form amyloids (residues 125–253) was used as a negative control. Protein names and organisms from which candidate prion domains (cPrDs) were selected are listed in table 1.
Microscopy samples were prepared from the colonies that obtained the strongest color (fig. 4D). Using light microscopy, we confirmed the presence of Congo red binding aggregates in eight samples as well as in positive controls, but saw no binding to negative controls. We observed that E. coli cells tend to concentrate around the aggregates, consistent with the known role of amyloid aggregates in cell adhesion in biofilms (Larsen et al. 2007). Detection of Congo red fluorescence in some cases made aggregates more noticeable (fig. 4E). In summary, we showed that several cPrDs from Archaea can bind amyloid specific dye. This is the first step in verification of possible prion nature of the proteins.
Archaeal cPrDs Form Amyloids In Vitro
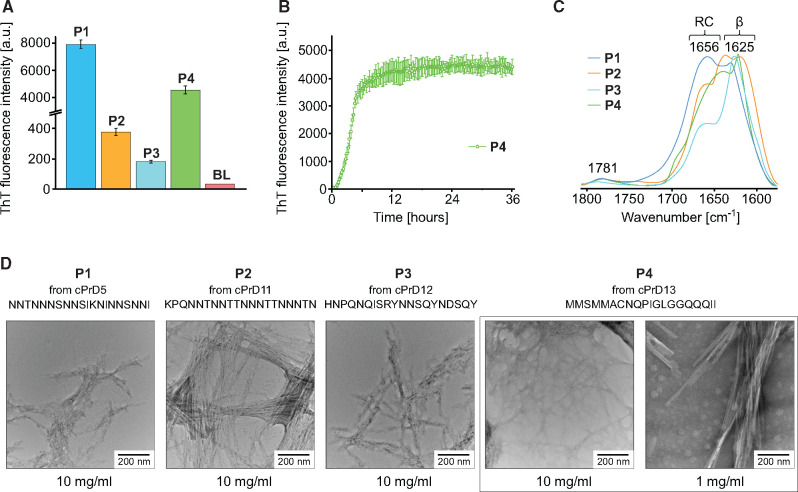
Congo red staining alone is insufficient for confirmation of the amyloid nature of protein aggregates (Yakupova et al. 2019). Thus, to further evaluate the ability of archaeal prion candidates to form amyloids, we employed an additional set of tests. Following the hypothesis that small regions of a protein are often both necessary and sufficient to drive prion behavior (Kochneva-Pervukhova et al. 1998), we analyzed even further shortened segments of the cPrDs. The length of cPrDs that yielded positive results in the Congo red assay varied from 62 to 179 amino acids. To choose fragments of putative archaeal prions most likely to drive aggregation, we used pRANK that assigns 20 amino acid long sequence window. As a principle, we selected for further analysis only these peptides which were initially soluble in water. We observed a very significant increase in thioflavin T (ThT) fluorescence for aggregated forms of peptides P1, P4 and moderate increase of ThT emission for P2 and P3 (fig. 5). Binding of ThT to amyloids increases the quantum yield of fluorescence of the dye (Biancalana and Koide 2010). Varying enhancement of ThT fluorescence is often observed for amyloid fibrils, depending on structural features that may reduce ThT binding affinity (Cloe et al. 2011). Furthermore, we found that dissolving some peptides in water will trigger a spontaneous increase in ThT emission intensity until a plateau is reached (e.g., P4—see fig. 5B), a hallmark of amyloid formation. To examine the secondary structure of the aggregates, we used attenuated total reflectance Fourier transform infrared spectroscopy (ATR−FTIR) (fig. 5C). The major spectral components of the vibrational amide I band were below 1,630 cm−1 for all four peptides we tested, implying the presence of parallel β-sheet structure characteristic of amyloid fibrils (Zandomeneghi et al. 2009). For P1, the β-sheet signal at ∼1,629 cm−1 flanks the dominant spectral component at 1,656 cm−1 indicating possible coexistence of tightly packed amyloidal β-core with abundant and less ordered conformations. A similar observation has been made for amyloid fibrils of Tau protein (Wegmann et al. 2013). We also performed a molecular dynamics simulation of peptide P2 showing that presence of a preformed β-sheet promotes the formation of a new filament of β-sheet out of unfolded P2 peptide (supplementary fig. S4, Supplementary Material online). Finally, to further examine the morphology of the aggregates, we performed transmission electron microscopy. These analyses confirmed the presence of amyloid fibrils in all analyzed preparations (fig. 5D). Collectively these data establish that amyloid conversion is promoted by specific sequences within the cPrDs.
Fig. 5.
Analysis of aggregates formed by pRANK-peptides. (A) Histogram showing ThT fluorescence intensity levels of samples containing peptides derived from prion candidates OQD58440.1 (P1); WP_066970924.1 (P2); WP_066972153.1 (P3); WP_069282784.1(P4). Prior to measurement, samples with the composition: 2.5 mg ml−1 peptide, 0.05 M NaCl, 20 µM Th, H2O, pH around 3, were incubated for 24 h at 37°. Three independent measurements were made for each sample, which was the basis for calculating the mean and error bar. For comparison, the result for the blank is also presented (BL). (B) ThT fluorescence-monitored reassociation of WP_069282784.1 (P4) peptide. Sample composition and measurement conditions are the same as in the case of (A). (C) ATR−FTIR spectra of the amide I band region of samples containing pRANK-peptides. Before measurement, small portions of lyophilized peptides were suspended in water, which was then gently evaporated. (D) Micrographs of all four peptides tested obtained using a transmission electron microscope show the fibrillar morphology of the aggregates. Peptide WP_069282784.1 (P4) is shown in two different concentrations.
Archaeal cPrDs Can Promote Protein-Based Inheritance
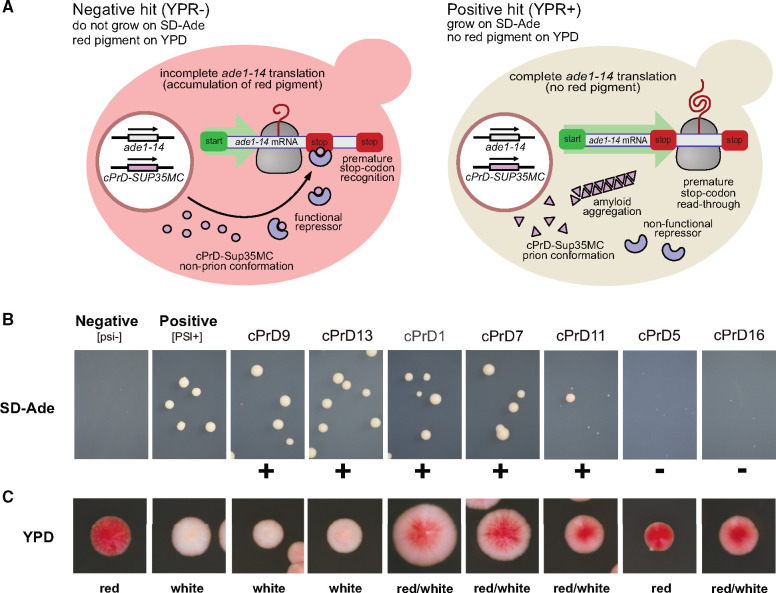
The unusual folding landscapes of prion proteins allow them to act as epigenetic mechanisms of inheritance (Alberti et al. 2009). To test whether this was true for our identified archaeal cPrDs, we employed an assay based on the well-characterized prion phenotypes of S. cerevisiae that depend on the prion state of Sup35 protein (Alberti et al. 2009). Normally, the protein Sup35 is a part of a translation-termination complex responsible for recognition of the stop codon and termination of translation. The yeast strain that we used in these experiments has a premature stop codon in ADE1 gene coding for SAICAR synthetase. Premature translation termination in ADE1 causes accumulation of red pigment—the product of polymerization of aminoimidazole ribotide (AIR) (Smirnov et al. 1967; Nevzglyadova et al. 2011)—and the ensuing lack of adenine production precludes the growth of naive, [psi−] cells on adenine-deficient medium (SD-Ade). Occasionally, Sup35 undergoes conformational conversion to a prion state in which it forms amyloid fibrils. In this state, the protein is sequestered into aggregates, inhibiting translation termination—the [PSI+] phenotype—and promoting readthrough of the premature stop codon in ADE1. Colonies with this phenotype can grow on SD-Ade medium, and appear white on nutrient-rich media like Yeast Peptone Dextrose (YPD) because accumulation of the red pigment is reduced.
Sup35 protein consists of an N-terminal PrD, a middle region (M), and a C-terminal functional domain (C). The PrD of Sup35 and other prions are modular and can be transferred to nonprion proteins creating new protein-based elements of inheritance (Li and Lindquist 2000). In our experiment, we tested if the original PrD of Sup35 can be substituted by cPrDs from archaea while preserving the ability to undergo prion-dependent phenotypic change. If a cPrD can function as a bona fide prion domain, the yeast colony bearing the chimeric protein should have a phenotype resembling [PSI+]. In contrast, a cPrD that is unable to facilitate the prion-like conversion would have a [psi−] phenotype.
We substituted the original PrD of Sup35 with 16 different archaeal cPrDs, generating a series of cPrD-Sup35MC chimeric proteins. In this yeast-based prion reporter assay (YPR), we found that 10 chimeric strains grew colonies of a significant size on SD-Ade media. To distinguish quantitatively between positive (YPR+) and negative (YPR−) results of the assay, images of the Petri dishes were analyzed using ImageJ (see Materials and Methods for details). Most chimeric strains when plated on YPD medium grew colonies much whiter than the negative control, suggesting aberrant synthesis of red pigment (fig. 6; see supplementary figs. S5, S7, S8.1, and S8.2, Supplementary Material online, for all positive and negative results; results are also summarized in table 1 and supplementary data set S3, Supplementary Material online). The reduction of red pigment is characteristic for [PSI+] phenotype and indicates the presence of prion form of Sup35. We restreaked the colonies that generated a phenotype resembling [PSI+] (YPR+) on YPD media. Some colonies showed reversion to [psi−] phenotype suggesting that cPrD-Sup35MC is in a metastable state allowing reversion of prion phenotype (supplementary fig. S6, Supplementary Material online).
Fig. 6.
cPrD-Sup35MC strains display different ability to grow on media lacking adenine and a variety of colony colors. (A) Cartoon explaining the mechanism of accumulation of red pigment, which depends on the formation of functional repressor. (B and C) Representative images of the colonies of cPrD-Sup35MC-expressing strains growing on SD-Ade and YPD plates respectively (see supplementary fig. S5, Supplementary Material online, for all 16 tested and supplementary fig. S7, Supplementary Material online, for images of the whole area of Petri dishes). Colonies phenotypes [psi−] and [PSI+] are shown for comparison. Positive results are marked with “+” and are considered positive results of the yeast prion reporter assay (YPR+). Negative results are marked with “−” and are considered negative results of the yeast prion.
Based on this set of experiments, we conclude that archaeal cPrDs can functionally substitute PrD of Sup35 and facilitate the formation of prion-based elements of inheritance.
Tyrosine and/or Phenylalanine Content Distinguishes Functional cPrDs
In the experiments described above, we found a total of six cPrDs that tested positive both in Congo red and yeast prion reporter assays (CR+/YPR+), and four that tested negative in both assays (CR−/YPR−). We investigated the differences in sequence composition of cPrDs from these two groups. The distribution of glutamine and asparagine content, a key driver of prion behavior (Michelitsch and Weissman 2000), was similar in each (fig. 7A and supplementary fig. S10A, Supplementary Material online). Yet in searching for motifs in the sequences of the proteins that tested positive in both assays, we observed an additional pattern of consecutively occurring proline and glutamine (P and Q) with nearby aromatic residues such as tyrosine and phenylalanine (Y and F; supplementary fig. S10B and C, Supplementary Material online)—which was absent in those that tested negative in both assays (supplementary fig. S10D, Supplementary Material online). Tyrosine content was generally higher in the CR+/YPR+ group (Kruskal–Wallis H-test P = 0.036; Wilcoxon rank-sum test P = 0.042; fig. 7A). The only CR+/YPR+ cPrD that contained no tyrosine, harbored the highest amount of phenylalanine (∼5%) among all cPrDs. Tyrosine and phenylalanine content was significantly higher in the CR+/YPR+ group, and also in Q/N-rich PrDs of known prion proteins in yeast, than in the CR−/YPR− group (Kruskal–Wallis H-test P = 0.01, 0.005; Wilcoxon rank-sum test P = 0.01, 0.005, respectively; fig. 7B). Moreover, the frequency of tyrosine and phenylalanine in PrDs of the only two currently known bacterial prion proteins—transcription termination factor Rho from Clostridium botulinum and single-stranded DNA-binding protein SSB from Campylobacter hominis—also revealed elevated levels as compared with our CR−/YPR− group (fig. 7B). The difference between CR−/YPR− and other groups was enhanced further when we compared the content of all amino acids with aromatic side chains (supplementary fig. S10E, Supplementary Material online). Thus, content of aromatic residues, especially of tyrosine and phenylalanine, in Q/N-rich PrDs may play a key role in determining the formation of prions across the evolutionary spectrum.
Fig. 7.
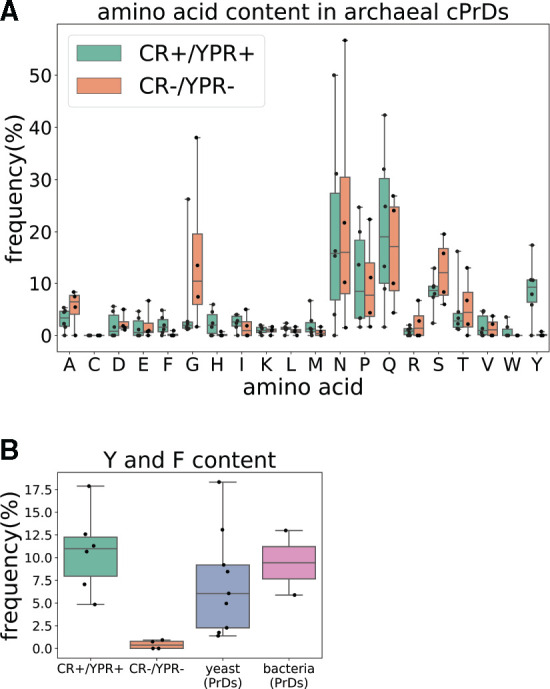
Tyrosine and/or phenylalanine distinguish positive-testing cPrDs from negative-testing cPrDs. (A) Comparison of amino acid frequency in cPrDs that tested positive in both Congo red and Sup35-based yeast-assay (CR+/YPR+), and ones that tested negative in both experimental assays (CR−/YPR−). Tyrosine (Y) content of the CR+/YPR+ cPrDs were higher than the CR−/YPR− cPrDs in general. The CR+/YPR+ cPrD with no tyrosine (lowest datapoint in distribution of Y) is the same with highest phenylalanine (F) frequency (highest data point in distribution of F). (B) Frequencies of tyrosine and phenylalanine in archaeal cPrDs, yeast, and bacterial PrDs. CR+/YPR+ cPrDs as well as yeast PrDs contain significantly higher Y/F than CR−/YPR− cPrDs. The Y/F frequency distribution in bacterial PrDs is also similar to that in CR+/YPR+ cPrDs and yeast PrDs. Since the bacterial group contained PrDs of only the two experimentally confirmed prions (Rho and SSB), no statistics were performed to compare the group with other ones.
Unlike yeast (or similar prokaryotic) prions which essentially harbor modular, transferable Q/N-rich PrDs, mammalian prion protein (PrP) does not have such a region. In addition, PrP and yeast prion proteins do not share significant sequence similarity. Interestingly, a segment of the human prion protein (PrP) 169-YSNQNNF-175 containing glutamine, asparagine, tyrosine, and phenylalanine, is required for its self-assembly (Huang and Caflisch 2015b). Moreover, the phenylalanine F175 is highly conserved, and the tyrosine Y169 is strictly conserved in mammalian PrPs and may function as an aggregation gatekeeper (Huang and Caflisch 2015a) (supplementary fig. S11, Supplementary Material online). During self-assembly mediated by this segment, stacks of tyrosine and phenylalanine stabilize the amyloid core formed by the stacks of Q/Ns (Gallagher-Jones et al. 2018). In addition, tyrosine Y218 which is strictly conserved in mammalian PrPs (supplementary fig. S11, Supplementary Material online), stabilizes the amyloid fibril formed by full-length human PrP (Wang et al. 2020). This suggests a potentially generalizable role of tyrosine/phenylalanine in prion formation, not only limited to Q/N-rich PrDs.
Discussion
Long viewed as enigmatic drivers of disease, prions have emerged as a form of epigenetics beyond the chromosome that can be adaptive in eukaryotes ranging from fungi (Namy et al. 2008; Brown and Lindquist 2009; Li and Kowal 2012; Jarosz et al. 2014; Harvey et al. 2018) to humans (Hou et al. 2011). Early analyses (Michelitsch and Weissman 2000) suggested prions were absent in other domains of life, consistent with a prevailing view that they are evolutionarily young elements. The recent discovery of prions in bacteria (Yuan and Hochschild 2017) challenged this dogma, yet prions have not yet been identified in archaea. In this study, we show that multiple archaeal cPrDs can form amyloids and act as bona fide elements of information transfer in living cells, fulfilling fundamental tenets of the prion characterization. To the best of our knowledge, neither prions nor intracellular amyloids have been reported in the domain Archaea to date. Confirming their presence would indicate their existence in all three domains of life, suggesting either that prions were present during the time of LUCA, or that they are convergently evolved.
Though many prions do not form amyloid (Wickner et al. 2006; Chakrabortee, Byers, et al. 2016; Chakravarty et al. 2020), in the current study, we focused on amyloid-forming cPrDs because recently identified bacterial prions have been shown to form amyloids (Yuan and Hochschild 2017), and amyloid fibril formation can also be detected with a relatively high-throughput experimental survey like the Congo red assay used here. We utilized the computational program PLAAC (Lancaster et al. 2014) to identify novel cPrDs. Although PLAAC’s underlying algorithm was trained on yeast PrDs, it has recently proven successful in facilitating the identification of prions in bacteria (Yuan and Hochschild 2017; Fleming et al. 2019). Taken with our work here, in which PLAAC also enabled the identification of likely PrDs in Archaea, this speaks to the chemically conserved nature of at least a subset of (c)PrDs (amyloid-forming) across all three domains of life. We stress, however, that in this initial exploration of potential prions within the domain Archaea, we have performed a somewhat limited rather than exhaustive search (being guided by current information largely sourced from Q/N-rich, amyloid-forming, yeast-derived prions). Bacteria and archaeal proteomes appear to have fewer Q/N-rich regions in general as compared with eukaryotes (Michelitsch and Weissman 2000), but it is as yet still unclear if this translates to them having fewer prions overall, or if it is the case that they are encoded with a different chemical signature to which we currently remain naive.
To investigate our computationally identified cPrDs experimentally, we utilized biochemical and genetic approaches (listed in regard to protein fragment, see fig. 1). This included staining with amyloid specific dye (fig. 4), infrared spectroscopy (fig. 5A–C), transmission electron microscopy (fig. 5D), and the yeast prion reporter assay (fig. 6). We utilized the model organisms E. coli and S. cerevisiae in our experiments. Future work will require working with the source organism of the cPrD and testing the entire protein.
Looking across the archaeal domain, many species harbor at least 1 cPrD. However, Methanobacteria stand out as especially rich in cPrDs (fig. 2, right side). Several members of the genera Methanobrevibacter, Methanosphaera, and Methanobacterium have five or more cPrDs per 1,000 proteins (supplementary data set S1, Supplementary Material online). Out of 16 candidates tested, six were from the Class Methanobacteriaceae, two of which tested positive under both experimental assays (table 1 and supplementary data set S3, Supplementary Material online). By far the dominant Gene Ontology (GO) term associated with the cPrDs identified in this group was integral component of membrane from the Cellular Component namespace (GO:0016021; ∼70% of all assigned GO terms; supplementary data set S4, Supplementary Material online), but this was also the case when looking exclusively outside of the Methanobacteria—with ∼58% of all GO terms associated with cPrDs being GO:0016021, which was enriched overall (fig. 3). The majority of the remaining cPrDs from the Methanobacteria were not annotated (supplementary data set S4, Supplementary Material online). Possibly contributing to their relatively higher cPrD frequencies is that they had on average fewer proteins overall. The average number of proteins from the incorporated Methanobacteria was 1,915 ± 324 (mean ± 1 SD), as compared with 2,238 ± 1,028 for all others. Likely further contributing is that the Q/N-rich regions PLAAC is identifying inherently tend to be higher in AT-content due to the possible codons encoding them (N = AAT or AAC; Q = CAA or CAG; supplementary data set S5, Supplementary Material online), and Methanobacteria have relatively higher AT-content compared with all others (65 ± 7.32% vs. 51 ± 11.9%; Welch t-test P = 7.64e-23). In general, those with higher AT content have higher normalized frequencies of cPrDs (supplementary fig. S9, Supplementary Material online).
Our GO enrichment analysis revealed that of the annotations that were significantly enriched or purified in proteins containing our cPrDs, the vast majority (∼90%) were purified (e.g., depleted). As such, there is a high level of functional constraint with regard to which functional domains cPrDs tend to stably coexist with (meaning within the same protein). It is notable that overall metal ion binding (GO:0046872) is purified in the proteins harboring cPrDs relative to all proteins (supplementary data set S2, Supplementary Material online), yet specific “child” GO terms nested underneath it such as calcium (GO:0005509) and copper ion binding (GO:0005507) are enriched (fig. 3). This suggests that there is something chemically consistent with Q/N-rich amyloids within these specific ion-binding proteins that is distinct from other metal ion-binding proteins. Although we observed an enrichment in outer membrane cellular components and in several cell wall-related biological processes such as chitin binding, chitin metabolism, and cell wall adhesion (fig. 3 and supplementary data set S2, Supplementary Material online), we note that it may simply be the case that cell-wall related proteins like these might tend to harbor Q/N-rich amyloids. This may place them disproportionately in the category of being detected by PLAAC, but not likely to present prion-like characteristics upon further experimental scrutiny. In this case, the only cell-wall related cPrD we tested was found to be negative in both the Congo red and yeast-prion reporter assays (cPrD2; adhesin-like protein; table 1 and supplementary data set S3, Supplementary Material online).
We observed that tyrosine and phenylalanine content, as well as total content of aromatic residues, was significantly higher in the CR+/YPR+ group. Tyrosine was also the most abundant hydrophobic residue in PrDs identified in yeast (Alberti et al. 2009; Sabate et al. 2015). MacLea et al. (2015) showed that tyrosine is necessary for stable prion variants of Sup35. Later it was proposed that aromatic residues are favored in PrDs, because they promote prion formation and chaperone-dependent prion propagation while avoiding detection by the degradation machinery (Gonzalez Nelson et al. 2014; Cascarina et al. 2018). Recently prion formation has been associated with phase behavior (Franzmann et al. 2018). Interestingly it was shown that phase behavior is determined by the number of aromatic residues like tyrosine and phenylalanine, and their patterning (Martin et al. 2020). Our observation adds to the above showing that the role of these aromatic residues in prion formation is universal across domains of life.
The first prions discovered in the Domain Eukaryota (yeast translation release factor Sup35) (Ter-Avanesyan et al. 1994) and the Domain Bacteria (transcription termination factor Rho) (Yuan and Hochschild 2017) were both related to cellular regulation. Further, amyloids in particular have been noted for their roles in signal transduction such as those involved with programmed cell death (Daskalov et al. 2015). In the current study, one of our positive-testing prion candidates was transcriptional repressor CopY (cPrD1; table 1 and supplementary data set S3, Supplementary Material online), and one of our most highly enriched GO terms is involved with transcription regulation (GO:0003712; fig. 3). As amyloid-based prions themselves can orchestrate a regulatory network (Gilks 2004; Hou et al. 2011; Yuan and Hochschild 2017; Jakobson and Jarosz 2018), this suggests that additional regulatory factors may be controlled by aggregation if they harbor a cPrD. This would provide a scenario in which the aggregation state of the protein (as dictated by intracellular conditions) might be a mechanism for controlling its activity. As in the case with the eukaryotic Sup35 (upon which aggregation results in less efficient translation termination) (True and Lindquist 2000), and with the bacterial transcriptional terminator Rho (in which aggregation results in transcriptional read-through) (Yuan and Hochschild 2017), these types of “interferences” ultimately may provide a selective benefit by enabling a population under stress to rapidly explore a broader phenotypic landscape. Indeed, it has been demonstrated that prions in S. cerevisiae can confer a fitness advantage (Halfmann et al. 2010; Suzuki et al. 2012; Itakura et al. 2020). The observation that cPrDs and PrDs are commonly found within genes involved in cellular regulation across all three domains of life suggests either that: 1) PrDs within these gene families are convergently evolved; or 2) PrDs may be a relatively early-evolved manifestation of cellular regulation that was present at the time of LUCA. For now, which of these scenarios unfolded throughout Earth’s biological history is a question that remains outstanding.
Conclusion
With this work, we show that archaeal cPrDs can facilitate the acquisition of the prion phenotype, allowing them to function as protein-based elements of inheritance—thus expanding our knowledge of this epigenetic phenomenon to the third and final domain of life. This adds support to the hypothesis that amyloid-based prions were present at the earliest stages of life’s evolution. The current study represents an example of a top-down approach that pushes the story of amyloid-based functions further back in evolutionary time. This is complementary to work investigating the “amyloid world” theory of the origin of life (Chernoff 2004; Maury 2018) that proposes that amyloids may have been forming from very short peptides before the emergence of what we might consider the first “living” system (Rode et al. 1999; Greenwald and Riek 2012; Rufo et al. 2014; Greenwald et al. 2016; Rout et al. 2018). These two approaches are heading toward each other, slowly closing the gap between them, with many interesting questions remaining to be answered and asked. Does a continuity exist between prebiotic amyloids and modern amyloid-based prions (Lupi et al. 2006)? Are extant amyloid-based prions the biochemically evolved descendants of a prebiotic system capable of self-assembly and self-aggregation? Did the self-assembly and self-aggregation of these short peptides facilitate an expansion in range of potential interactions and functionalities? This work demonstrating protein-based, epigenetic inheritance via PrDs derived from the archaeal domain of life is a valuable step toward bridging this gap, but clearly there are many more to be taken.
Materials and Methods
Computational Identification and Classification of Prion Candidates
Archaeal reference proteomes and annotation information, including Gene Ontology (GO) annotations (Ashburner et al. 2000), were downloaded from the UniProt database (Bateman 2019). To systematically exclude low-quality proteomes, only those ranked as “standard” based on UniProt’s “Complete Proteome Detector” algorithm were incorporated, resulting in 1,262 proteomes holding 2,805,234 proteins as accessed on March 27, 2020.
The command-line version of PLAAC (Lancaster et al. 2014) (installed from https://github.com/whitehead/plaac in April of 2020) was used to identify proteins with putative prion domains in each proteome individually with default settings other than “-a 0” — which tells PLAAC to calculate and use the current proteome’s background amino acid frequencies entirely, rather than those of S. cerevisiae. From those for which a core length of at least 60 was identified, several candidates that also possessed corresponding annotations were selected for experimental validation.
GO enrichment analyses were performed to identify enrichment/purification of frequencies of GO annotations in the proteins holding identified cPrDs (2,797) as compared with all scanned proteins (2,805,234) using the goatools v0.6.10 (Klopfenstein et al. 2018) “find_enrichment.py” script with default settings. Statistical significance was defined as those with Benjamini-Hochberg false-discovery rates of ≤0.05. Many related GO terms were statistically significant at different depth levels and in those cases the term with the lowest depth was represented.
The phylogenomic tree was produced with GToTree v1.4.11 (Lee and Ponty 2019), within which 76 single-copy orthologs specific to the archaeal domain (using the “Archaea.hmm” included with GToTree) were identified with HMMER3 v3.2.1 (Eddy 2011), individually aligned with muscle v3.8.1551 (Edgar 2004), trimmed with trimal (Capella-Gutiérrez et al. 2009), and concatenated prior to phylogenetic estimation with FastTree2 v2.1.10 (Price et al. 2010). Tree was initially visualized and edited with the Interactive Tree of Life interface (Letunic and Bork 2007).
Aggregation domains (20-amino acid long) from prion candidates were selected for synthesis with pRANK (http://faculty.pieas.edu.pk/fayyaz/prank.html, last accessed February 2021) (Afsar Minhas et al. 2017) using default settings.
Colony Color Phenotype—Congo Red Assay
Curli-dependent amyloid generator (C-DAG) was used to direct the export of heterologous cPrDs to the cell surface of E. coli strain VS45 (Sivanathan and Hochschild 2013). Both bacterial strain and expression vector enabling protein production under the control of the arabinose-inducible PBAD promoter was part of commercially available C-DAG amylyloidogenicity kit https://www.kerafast.com/productgroup/540/c-dag-amyloidogenicity-kit (last accessed February 2021). Export-directed fusion proteins contained the first 42 residues of CsgA signal sequence at the N terminus as described in the kit protocol. Sec translocon signal sequence is removed during translocation across the inner membrane, the 22-residue targeting sequence is retained at the N-terminus.
Lumio tag (CCPGCCGAGG) and His6 tag were added to the cPrD sequence at the C terminus during gene synthesis (www.idtdna.com, last accessed February 2021).
For assessment of colony color phenotype, overnight cultures of VS45 were transformed with plasmids containing cPrDs. Bacteria were diluted to OD600 0.01 in LB supplemented with the appropriate antibiotics (100 mg ml−1 carbenicillin, 25 mg ml−1 chloramphenicol). After 30 min of growth at 37 °C, 5 ml of the culture was spotted on LB agar plates supplemented with inducers (0.2% [w/v] L-arabinose, 1 mM IPTG), antibiotics (100 mg ml−1 carbenicillin; 25 mg ml−1 chloramphenicol), and, where indicated, on LB plates supplemented with Congo red (5 mg ml−1). Plates were then incubated for 4 days at room temperature. Photos of colonies were acquired using Canon EOS 6D SLR camera and the EOS Utility 3 software (https://www.usa.canon.com/internet/portal/us/home/support/self-help-center/eos-utility/, last accessed February 2021).
Light and Fluorescent Microscopy
Extracellular aggregates of heterologous candidate prion proteins stained with Congo Red were visualized using Zeiss Axioimager Z.1 with a Canon EOS SLR camera and 6D and EOS Utility 3 software.
Transmission Electron Microscopy
Peptide solutions were adsorbed onto formvar/carbon-coated nickel grids (TED PELLA, CA) in water, blotted dry, negatively stained with 1% uranyl acetate (TED PELLA, CA), for 1 min, blotted dry, and then viewed on transmission electron microscope Hitachi H-9500 TEM (Hitachi High Technologies America) at 300 kV with an Advanced Microscopy Techniques 4-megapixel digital camera (AMT XR41B, Low-Dose CCD).
Peptide Synthesis
Peptides corresponding to the 20 amino acid long fragments of putative PrDs were synthesized by ELIM Biopharm (Hayward, CA). The peptides were of 95% purity as determined by HPLC and mass spectrometry analyses. Peptides were dissolved in deionized water and stored at −80 °C. In our experiments, we used only the peptides that were initially soluble in water. pRANK-peptide sequences named P1, P2, P3, and P4 are presented in figure 5 and in the supplementary data set S3, Supplementary Material online.
Measurements of ThT Fluorescence
For ThT fluorescence-based measurements, (λ ex. 440 nm/λ em. 485 nm) of fibrillization kinetics, a CLARIOstar plate reader from BMG LABTECH (Offenburg, Germany), and 96-well black microplates were used. All samples were slightly acidified to pH approximately 3. Typically, wells were filled with 150 μl volumes of diluted peptide samples containing ThT. Measurements were carried out at 37 °C with agitation for 70 h; Each kinetic trace was calculated as an average from three independently collected trajectories (the error bars correspond to the standard deviations).
Attenuated Total Reflectance Fourier Transform Infrared Spectroscopy
Measurements were carried out using single-reflection diamond ATR (attenuated total reflectance) accessory of the Nicolet iS50 FTIR spectrometer. The liquid suspensions of peptides were gently dried in situ until they formed films. Infrared spectra of the films were collected. Typically, for a single spectrum, 32 interferograms of 2 cm−1 resolution were coadded. Due to ambiguity in determining real values of refractive indices of amyloid aggregates, uncorrected ATR-FTIR data is shown. The spectra were corrected for water vapor, baseline, and normalized. Data processing was performed using GRAMS software (Thermo Nicolet).
Yeast Vector Preparation
Fragments of genes corresponding to predicted prion domains were cloned into a yeast vector PDJ1776 (pAG415-ADH1-ccdB-SUP35C) using the Gateway Cloning enzymes (Invitrogen Clonase Gateway BP 11789020 and LR 11791020 Clonase II Enzyme Mix). Gateway Cloning procedures were based on methods described in Alberti et al. (2009). Primers used to amplify cPrDs and to attach the recombinogenic attB1 and attB2 sites for Gateway cloning are listed in supplementary data set S6, Supplementary Material online.
Yeast Strains
Saccharomyces cerevisiae strain YDJ1420 was transformed with plasmid pAG415-ADH1-cPrD#-SUP35C and subsequently cured of plasmid pAG426-GPD-SUP35C (plasmid shuffle). The resulting strains are labeled YDJ6455-6472. Example: strain YDJ6455-pAG415-ADH1-cPrD1-SUP35C (Mata, leu2-3,112; his3-11,-15; trp1-1; ura3-1; ade1-14; can1-100; sup35::HygB; pAG415-ADH1-cPrD1-SUP35C [RNQ+], [psi−]). Saccharomyces cerevisiae strain YDJ1528 was used as positive control (Mata, leu2-3,112; his3-11,-15; trp1-1; ura3-1; ade1-14; can1-100; sup35::HygB; AG415-ADH1-Sup35NM-Sup35C, [RNQ+], [psi−]). Saccharomyces cerevisiae strain YDJ1420 was used as negative control (Mata, leu2-3,112; his3-11,-15; trp1-1; ura3-1; ade1-14; can1-100; sup35::HygB; pAG426-GPD-SUP35C, [RNQ+], [psi−]). Standard rich (YPD) and Synthetic Dropout (SD) Media were used to culture yeast cells at 30 °C.
Plasmid Shuffling and Phenotypic Analysis
Plasmids containing archaeal prion domains fused with the carboxy-terminal domain of Sup35 (cPrD#-Sup35C) were, respectively, transformed into strain YDJ1420 by the standard transformation method (Gietz et al. 1995). The resulting transformants were incubated on leucine-deficient synthetic medium (SD-Leu) for 3 days at 30 °C and then plated on SD-Leu medium supplemented with 1 mg ml−1 of 5-fluoroorotic acid (RPI Research Products F10501) to eliminate the Sup35 maintainer plasmid (pAG426-GPD-SUP35C) generating strains expressing an cPrD#-Sup35C fusion protein as the only source of functional Sup35. Phenotypes were assessed by the growth of colonies for 3 days at 30 °C in SD-Leu medium and plating of different dilutions of these cultures on YPD agar.
Postshuffled yeast strains were plated on adenine-deficient synthetic medium (SD-Ade) medium to verify the readthrough status of the premature UGA stop codon in the ade1-14 allele. In [PSI+] cells, the majority of Sup35 is aggregated unavailable for translation termination, which results in the read-through of the ade1-14 premature stop codon. The synthesis of full-length Ade1 protein (N-succinyl-5-aminoimidazole-4-carboxamide ribotide synthetase) results in white colonies on rich medium and growth on SD-Ade medium. [psi−] cells showed no read-through of the ade1-14 premature stop codon and did not grow on SD-Ade medium. Images of SD-Ade plates with colonies were analyzed with the ImageJ program (https://imagej.nih.gov/ij/, last accessed February 2021) to quantitatively determine the outcome of the experiment. To be counted as a colony, the minimum area for a particle had to be 10% more than the largest particle area found in the negative control (area ≥0.3 mm2) that has a circularity ≥0.75 (see supplementary figs. S8.1 and S8.2, Supplementary Material online). The dish with at least one colony meeting these criteria is categorized as positive results of the yeast prion reporter assay (YPR+). If a dish has zero colonies, then it is categorized as a negative result (YPR−).
Sequence Analysis of Experimentally Tested Archaeal cPrDs and Known PrDs of Yeast, and Bacteria
We used glam2 from MEME suite version 5.1.1 (Frith et al. 2008; Bailey et al. 2009) for finding motifs via gapped alignment of CR+/YPR+ and CR−/YPR− cPrDs separately. For each group of cPrDs, keeping all other parameters set to default values, we programmatically performed runs of glam2 with varying values of the parameter “-n” which controls the number of iterations after which glam2 ends each run without improvement. For each glam2 run corresponding to a value of “n,” the number of alignment runs (“-r,” replicates) was set to 10 as a default. Initially the values of “n” were chosen from [4,000, 6,000, 8,000, 10,000, 20,000, 40,000, 60,000, 80,000, 100,000]. At n = 60,000, we found the single optimal motif for the CR−/YPR− group to be reproduced by all 10 replicates. The score of this 30 amino acid long CR−/YPR− motif was 39.0692 (as shown in supplementary fig. S10D, Supplementary Material online). On the other hand, although we found similar motifs across replicates for CR+/YPR+ cPrDs within the above-mentioned range of values for “n,” we strived to obtain identical motifs supported by multiple replicates. We then increased the number of iterations and chose the following values: [200,000, 400,000, 800,000, 1,000,000, 2,000,000, 4,000,000]. At n = 4 million, we obtained the two optimal motifs with score 84.0115, and of length 42 and 40 amino acids, respectively (as shown in supplementary fig. S10B and C, Supplementary Material online). Each of these motifs was supported by 5/10 replicates. They also showed similarity between themselves.
The coordinates of Q/N-rich prion domains (PrDs) of known yeast (S. cerevisiae) prion proteins were obtained from Uniprot. The list of these proteins included: SWI1, SUP35, NEW1, RNQ1, MOT3, URE2, CYC8, SFP1, and NUP100. The PrD coordinates of NUP100 were obtained from Halfmann et al. (2012). The coordinates of bacterial PrD from transcription termination factor Rho (Clostridium botulinum) and single-stranded DNA-binding protein SSB (Campylobacter hominis) were obtained from Yuan and Hochschild (2017) and Fleming et al. (2019), respectively (Yuan and Hochschild 2017; Fleming et al. 2019). The PrD sequences were programmatically extracted from the full-length fasta sequences of these proteins obtained from Uniprot.
The stats module from SciPy was used for the statistical tests (https://docs.scipy.org/doc/scipy/reference/stats.html, last accessed February 2021).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
T.Z. was funded by Ministerstwo Nauki i Szkolnictwa Wyższego under the Mobilność Plus program (1656/MOB/V/2017/0). A.C. was funded under the Initiative for Maximizing Student Development program at University of California, Santa Cruz National Institutes of Health (2R25GM058903-20 NIH/NIGMS). D.F.J. was funded under the National Science Foundation (MCB1453762), the Vallee Foundation, and the David and Lucile Packard Foundation (2015-63121). Transmission electron microscopy image acquisition made possible by the support of the Universities Space Research Association Science & Technology Innovation Labs Program managed under National Aeronautics and Space Administration contract NNA16BD14C. We thank Thomas Lozanoski for his help in the yeast prion reporter assays. We thank Anupam Chakravarty for his advice on the experimental approach. We thank Stanisław Dunin-Horkawicz for his intellectual support at the early stages of the project. We acknowledge the support of the Origins Research Group of Exobiology Laboratory (ORGEL) at NASA Ames Research Center.
Author Contributions
T.Z., M.D.L., S.S.M., and L.J.R. designed research. T.Z., M.D.L., S.S.M., A.C., and R.D. performed research. T.Z., M.D.L., S.S.M., A.C., R.D., P.D.B., R.W.P., J.E.S., N.B.B., and W.D. analyzed data. T.Z., M.D.L., S.S.M., A.C., R.D., P.D.B., W.D., D.F.J., and L.J.R. wrote the paper.
Data Availability
References
- Afsar Minhas FUA, Ross ED, Ben-Hur A, Cotter A, Li W, Schmitt-Ulms G.. 2017. Amino acid composition predicts prion activity. PLoS Comput Biol. 13(4):e1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S.. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137(1):146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. Nat Genet. 25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS.. 2009. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server):W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A.2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47(D1):D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benilova I, Karran E, De Strooper B.. 2012. The toxic Aβ oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat Neurosci. 15(3):349–357. [DOI] [PubMed] [Google Scholar]
- Biancalana M, Koide S.. 2010. Molecular mechanism of thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta. 1804(7):1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker SG, Nicastro LK, Paul Wilson R, Tükel Ç.. 2018. The functional amyloid curli protects Escherichia coli against complement-mediated bactericidal activity. Biomolecules 8(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig C, Rochin L, van Niel G.. 2016. PMEL amyloid fibril formation: the bright steps of pigmentation. Int J Mol Sci. 17(9):1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JCS, Lindquist S.. 2009. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 23(19):2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascarina SM, Paul KR, Machihara S, Ross ED.. 2018. Sequence features governing aggregation or degradation of prion-like proteins. PLoS Genet. 14(7):e1007517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Åberg V, Walker JN, Seed PC, Almqvist F, et al. 2009. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol. 5(12):913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, Byers JS, Jones S, Garcia DM, Bhullar B, Chang A, She R, et al. 2016. Intrinsically disordered proteins drive emergence and inheritance of biological traits. Cell 167(2):369–381.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, Kayatekin C, Newby GA, Mendillo ML, Lancaster A, Lindquist S.. 2016. Luminidependens (LD) is an Arabidopsis protein with prion behavior. Proc Natl Acad Sci U S A. 113(21):6065–6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty AK, Smejkal T, Itakura AK, Garcia DM, Jarosz DF.. 2020. A non-amyloid prion particle that activates a heritable gene expression program. Mol Cell. 77(2):251–265.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ.. 2002. Role of Escherichia coli Curli operons in directing amyloid fiber formation. Science 295(5556):851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO.2004. Amyloidogenic domains, prions and structural inheritance: rudiments of early life or recent acquisition? Curr Opin Chem Biol. 8(6):665–671. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Derkach IL, Inge-Vechtomov SG.. 1993. Multicopy SUP35 gene induces de-novo appearance of Psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 24(3):268–270. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM.. 2006. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 75(1):333–366. [DOI] [PubMed] [Google Scholar]
- Cloe AL, Orgel JPRO, Sachleben JR, Tycko R, Meredith SC.. 2011. The Japanese mutant Aβ (ΔE22-Aβ 1-39) forms fibrils instantaneously, with low-thioflavin T fluorescence: seeding of wild-type Aβ 1-40 into atypical fibrils by Δe22-Aβ 1-39. Biochemistry 50(12):2026–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalstra HJP, Van Der Zee R, Swart K, Hoekstra RF, Saupe SJ, Debets AJM.. 2005. Non-Mendelian inheritance of the HET-s prion or HET-s prion domains determines the Het-S spore killing system in Podospora anserina. Fungal Genet Biol. 42(10):836–847. [DOI] [PubMed] [Google Scholar]
- Daskalov A, Habenstein B, Martinez D, Debets AJM, Sabaté R, Loquet A, Saupe SJ.. 2015. Signal transduction by a fungal NOD-like receptor based on propagation of a prion amyloid fold. PLoS Biol. 13(2):e1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW.. 1997. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147(2):507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Park KW, Yu H, Fan Q, Li L.. 2008. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 40(4):460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhang Y, Li L.. 2015. The yeast prion [SWI+] abolishes multicellular growth by triggering conformational changes of multiple regulators required for flocculin gene expression. Cell Rep. 13(12):2865–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR.2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC.2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming E, Yuan AH, Heller DM, Hochschild A.. 2019. A bacteria-based genetic assay detects prion formation. Proc Natl Acad Sci U S A. 116(10):4605–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW.. 2005. Functional amyloid formation within mammalian tissue. PLoS Biol. 4(1):e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nüske E, Richter D, et al. 2018. Phase separation of a yeast prion protein promotes cellular fitness. Science 359(6371):eaao5654. [DOI] [PubMed] [Google Scholar]
- Frith MC, Saunders NFW, Kobe B, Bailey TL.. 2008. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput Biol. 4(5):e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher-Jones M, Glynn C, Boyer DR, Martynowycz MW, Hernandez E, Miao J, Zee CT, et al. 2018. Sub-ångström cryo-EM structure of a prion protofibril reveals a polar clasp. Nat Struct Mol Biol. 25(2):131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DM, Jarosz DF.. 2014. Rebels with a cause: molecular features and physiological consequences of yeast prions. FEMS Yeast Res. 14(1):136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebbink MFBG, Claessen D, Bouma B, Dijkhuizen L, Wösten HAB.. 2005. Amyloids – a functional coat for microorganisms. Nat Rev Microbiol. 3(4):333–341. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA.. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS‐DNA/PEG procedure. Yeast 11(4):355–360. [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P.. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 15(12):5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Nelson AC, Paul KR, Petri M, Flores N, Rogge RA, Cascarina SM, Ross ED.. 2014. Increasing prion propensity by hydrophobic insertion. PLoS One 9(2):e89286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald J, Friedmann MP, Riek R.. 2016. Amyloid aggregates arise from amino acid condensations under prebiotic conditions. Angew Chem. 128(38):11781–11785. [DOI] [PubMed] [Google Scholar]
- Greenwald J, Riek R.. 2010. Biology of amyloid: structure, function, and regulation. Structure 18(10):1244–1260. [DOI] [PubMed] [Google Scholar]
- Greenwald J, Riek R.. 2012. On the possible amyloid origin of protein folds. J Mol Biol. 421(4–5):417–426. [DOI] [PubMed] [Google Scholar]
- Halfmann R, Alberti S, Lindquist S.. 2010. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 20(3):125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S.. 2010. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science 330(6004):629–632. [DOI] [PubMed] [Google Scholar]
- Halfmann R, Wright JR, Alberti S, Lindquist S, Rexach M.. 2012. Prion formation by a yeast GLFG nucleoporin. Prion 6(4):391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey ZH, Chakravarty AK, Futia RA, Jarosz DF.. 2020. A prion epigenetic switch establishes an active chromatin state. Cell 180(5):928–940.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey ZH, Chen Y, Jarosz DF.. 2018. Protein-based inheritance: epigenetics beyond the chromosome. Mol Cell. 69(2):195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DL, Lancaster AK, Lindquist S, Halfmann R.. 2013. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell 153(1):153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang Q-X, Chen ZJ.. 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146(3):448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Caflisch A.. 2015a. Evolutionary conserved Tyr169 stabilizes the Β2-Α2 loop of the prion protein. J Am Chem Soc. 137(8):2948–2957. [DOI] [PubMed] [Google Scholar]
- Huang D, Caflisch A.. 2015b. The roles of the conserved tyrosine in the Β2-Α2 loop of the prion protein. Prion 9(6):412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura AK, Chakravarty AK, Jakobson CM, Jarosz DF.. 2020. Widespread prion-based control of growth and differentiation strategies in Saccharomyces cerevisiae. Mol Cell. 77(2):266–278.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobson CM, Jarosz DF.. 2018. Organizing biochemistry in space and time using prion-like self-assembly. Curr Opin Syst Biol. 8:16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Lancaster AK, Brown JCS, Lindquist S.. 2014. An evolutionarily conserved prion-like element converts wild fungi from metabolic specialists to generalists. Cell 158(5):1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C-Y, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K.. 1997. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci. 94(13):6618–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Masel J.. 2007. The evolution of Bet-Hedging adaptations to rare scenarios. Theor Popul Biol. 72(4):560–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DV, Zhang L, Pedersen BS, Ramírez F, Warwick Vesztrocy A, Naldi A, Mungall CJ, Yunes JM, Botvinnik O, Weigel M, et al. 2018. GOATOOLS: a python library for gene ontology analyses. Sci Rep. 8(1):10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva-Pervukhova NV, Poznyakovski AI, Smirnov VN, Ter-Avanesyan MD.. 1998. C-terminal truncation of the Sup35 protein increases the frequency of de novo generation of a prion-based [PSI+] determinant in Saccharomyces cerevisiae. Curr Genet. 34(2):146–151. [DOI] [PubMed] [Google Scholar]
- Lacroute F.1971. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol. 106(2):519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AK, Nutter-Upham A, Lindquist S, King OD.. 2014. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30(17):2501–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH.. 2007. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 9(12):3077–3090. [DOI] [PubMed] [Google Scholar]
- Lee MD, Ponty Y.. 2019. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35(20):4162–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2007. Interactive tree of life (ITOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23(1):127–128. [DOI] [PubMed] [Google Scholar]
- Li L, Kowal AS.. 2012. Environmental regulation of prions in yeast. PLoS Pathog. 8(11):e1002973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lindquist S.. 2000. Creating a protein-based element of inheritance. Science 287(5453):661–664. [DOI] [PubMed] [Google Scholar]
- Liebman SW, Derkatch IL.. 1999. The yeast [PSI+] prion: making sense of nonsense. J Biol Chem. 274(3):1181–1184. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Debburman SK, Glover JR, Kowal AS, Liu JJ, Schirmer EC, Serio TR.. 1998. Amyloid fibres of Sup35 support a prion-like mechanism of inheritance in yeast. Biochem Soc Trans. 26(3):486–490. [DOI] [PubMed] [Google Scholar]
- Lupi O, Dadalti P, Cruz E, Sanberg PR.. 2006. Are prions related to the emergence of early life? Med Hypotheses. 67(5):1027–1033. [DOI] [PubMed] [Google Scholar]
- MacLea KS, Paul KR, Ben-Musa Z, Waechter A, Shattuck JE, Gruca M, Ross ED.. 2015. Distinct amino acid compositional requirements for formation and maintenance of the [PSI +] prion in yeast. Mol Cell Biol. 35(5):899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, Mittag T.. 2020. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J, Siegal ML.. 2009. Robustness: mechanisms and consequences. Trends Genet. 25(9):395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC, Maddelein LM, Wickner RB.. 1997. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc Natl Acad Sci U S A. 94(23):12503–12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC, Wickner RB.. 1995. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270(5233):93–95. [DOI] [PubMed] [Google Scholar]
- Maury CPJ.2018. Amyloid and the origin of life: self-replicating catalytic amyloids as prebiotic informational and protometabolic entities. Cell Mol Life Sci. 75(9):1499–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS.. 2000. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci. 97(22):11910–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch BM, Murdoch GK.. 2015. Genetics of prion disease in cattle. Bioinform Biol Insights. 9S4:BBI.S29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Galopier A, Martini C, Matsufuji S, Fabret C, Rousset JP.. 2008. Epigenetic control of polyamines by the prion [PSI+]. Nat Cell Biol. 10(9):1069–1075. [DOI] [PubMed] [Google Scholar]
- Nan H, Chen H, Tuite MF, Xu X.. 2019. A viral expression factor behaves as a prion. Nat Commun. 10(1):359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevzglyadova OV, Kuznetsova IM, Mikhailova EV, Artamonova TO, Artemov AV, Mittenberg AG, Kostyleva EI, Turoverov KK, Khodorkovskii MA, Soidla TR.. 2011. The effect of red pigment on the amyloidization of yeast proteins. Yeast 28(7):505–526. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB.1998. Nobel lecture: prions. Proc Natl Acad Sci U S A. 95(23):13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riek R, Saupe SJ.. 2016. The HET-S/s prion motif in the control of programmed cell death. Cold Spring Harb Perspect Biol. 8(9):a023515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode BM, Flader W, Sotriffer C, Righi A.. 1999. Are prions a relic of an early stage of peptide evolution? Peptides 20(12):1513–1511. [DOI] [PubMed] [Google Scholar]
- Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, Volkov K, Mironova L.. 2010. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc Natl Acad Sci. 107(23):10573–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout SK, Friedmann MP, Riek R, Greenwald J.. 2018. A prebiotic template-directed peptide synthesis based on amyloids. Nat Commun. 9(1):234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufo CM, Moroz YS, Moroz OV, Stöhr J, Smith TA, Hu X, DeGrado WF, Korendovych IV.. 2014. Short peptides self-assemble to produce catalytic amyloids. Nat Chem. 6(4):303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabate R, Rousseau F, Schymkowitz J, Batlle C, Ventura S.. 2015. Amyloids or prions? That is the question. Prion 9(3):200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabate R, Rousseau F, Schymkowitz J, Ventura S.. 2015. What makes a protein sequence a prion? PLoS Comput Biol. 11(1):e1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam N, Baker MODG, Ball SR, Steain M, Pham CLL, Sunde M.. 2019. Microbial functional amyloids serve diverse purposes for structure, adhesion and defence. Biophys Rev. 11(3):287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S.. 2005. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 6(6):435–450. [DOI] [PubMed] [Google Scholar]
- Sipe JD, Benson MD, Buxbaum JN, Ikeda S-I, Merlini G, Saraiva MJM, Westermark P.. 2016. Amyloid fibril proteins and amyloidosis: chemical identification and clinical classification International Society of Amyloidosis 2016 Nomenclature guidelines. Amyloid 23(4):209–213. [DOI] [PubMed] [Google Scholar]
- Sivanathan V, Hochschild A.. 2013. A bacterial export system for generating extracellular amyloid aggregates. Nat Protoc. 8(7):1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov MN, Smirnov VN, Budowsky EI, Inge-Vechtomov SG, Serebrjakov NG.. 1967. Red pigment of adenine-deficient yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 27(3):299–292. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M.. 2012. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 336(6079):355–359. [DOI] [PubMed] [Google Scholar]
- Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB.. 1999. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283(5406):1339–1343. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN.. 1994. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [Psi+] in the yeast Saccharomyces cerevisiae. Genetics 137(3):671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Berlin I, Lindquist SL.. 2004. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431(7005):184–187. [DOI] [PubMed] [Google Scholar]
- True HL, Lindquist SL.. 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407(6803):477–483. [DOI] [PubMed] [Google Scholar]
- Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ.. 2002. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416(6880):535–539. [DOI] [PubMed] [Google Scholar]
- Wang L-Q, Zhao K, Yuan H-Y, Wang Q, Guan Z, Tao J, Li X-N, Sun Y, Yi C-W, Chen J, et al. 2020. Cryo-EM structure of an amyloid fibril formed by full-length human prion protein. Nat Struct Mol Biol. 27(6):598–602. [DOI] [PubMed] [Google Scholar]
- Wegmann S, Medalsy ID, Mandelkow E, Müller DJ.. 2013. The fuzzy coat of pathological human tau fibrils is a two-layered polyelectrolyte brush. Proc Natl Acad Sci U S A. 110(4):E313–E321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB.1994. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264(5158):566–569. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Shewmaker F.. 2006. How to find a prion: [URE3], [PSI+] and [β]. Methods 39(1):3–8. [DOI] [PubMed] [Google Scholar]
- Yakupova EI, Bobyleva LG, Vikhlyantsev IM, Bobylev AG.. 2019. Congo red and amyloids: history and relationship. Biosci Rep. 39(1):BSR20181415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young CSH, Cox BS.. 1971. Extrachromosomal elements in a super–suppression system of yeast: I. A nuclear gene controlling the inheritance of the extrachromosomal elements. Heredity 26(3):413–422. [Google Scholar]
- Yuan AH, Hochschild A.. 2017. A bacterial global regulator forms a prion. Science 355(6321):198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandomeneghi G, Krebs MRH, McCammon MG, Fändrich M.. 2009. FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci. 13(12):3314–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.