Abstract
Divergence of gene function and expression during development can give rise to phenotypic differences at the level of cells, tissues, organs, and ultimately whole organisms. To gain insights into the evolution of gene expression and novel genes at spatial resolution, we compared the spatially resolved transcriptomes of two distantly related nematodes, Caenorhabditis elegans and Pristionchus pacificus, that diverged 60–90 Ma. The spatial transcriptomes of adult worms show little evidence for strong conservation at the level of single genes. Instead, regional expression is largely driven by recent duplication and emergence of novel genes. Estimation of gene ages across anatomical structures revealed an enrichment of novel genes in sperm-related regions. This provides first evidence in nematodes for the “out of testis” hypothesis that has been previously postulated based on studies in Drosophila and mammals. “Out of testis” genes represent a mix of products of pervasive transcription as well as fast evolving members of ancient gene families. Strikingly, numerous novel genes have known functions during meiosis in Caenorhabditis elegans indicating that even universal processes such as meiosis may be targets of rapid evolution. Our study highlights the importance of novel genes in generating phenotypic diversity and explicitly characterizes gene origination in sperm-related regions. Furthermore, it proposes new functions for previously uncharacterized genes and establishes the spatial transcriptome of Pristionchus pacificus as a catalog for future studies on the evolution of gene expression and function.
Keywords: comparative genomics, orphan genes, de novo gene, duplication, Pristionchus pacificus, Caenorhabditis elegans, meiosis
Introduction
The immense morphological diversity across all living organisms has raised the fundamental question, how novelty arises in the first place. Genetic screens in multiple model systems have dissected the underlying architecture of many traits and have highlighted the role of cis-regulatory evolution, as well as the emergence of novel genes in shaping phenotypic diversity (Wray 2007; Khalturin et al. 2009). Novel genes can arise from a spectrum of processes ranging from duplication of existing genes to de novo formation from noncoding sequences (McLysaght and Hurst 2016; Rödelsperger, Prabh, et al. 2019; Van Oss and Carvunis 2019). In addition, sequence divergence can contribute to the formation of orphan genes without detectable homologs in other lineages. Such orphan genes make up to one-third of all genes in a given genome (Tautz and Domazet-Lošo 2011; Rödelsperger, Prabh, et al. 2019). Their high abundance combined with the paucity of homologs raises the fundamental question, where orphan genes are coming from and what their biological function might be. Although the origin of orphan genes has been extensively studied in yeast (Carvunis et al. 2012; Vakirlis et al. 2018), vertebrates (Toll-Riera et al. 2008; Knowles and McLysaght 2009), and insects (Levine et al. 2006; Wissler et al. 2013), recent studies in plants and other animal phyla such as nematodes have broadened our knowledge of novel gene formation (Prabh et al. 2018; Prabh and Rödelsperger 2019; Zhang et al. 2019). In particular, nematodes with their small genome sizes and species richness are ideal model systems to study the evolution of novel genes by means of phylogenomic approaches relying on deep taxon sampling (Prabh et al. 2018; Rödelsperger 2018; Rödelsperger, Prabh, et al. 2019). Specifically, each of the two nematode genera Caenorhabditis and Pristionchus has around 40 described species and continuous sampling efforts discover new species almost every year (Rödelsperger et al. 2018; Herrmann et al. 2019; Stevens et al. 2019). The combination of deep taxon sampling with genome sequencing allowed us to characterize the evolutionary dynamics and regulation of novel genes in Pristionchus nematodes as well as to elucidate several mechanisms including de novo formation that give rise to orphan genes (Prabh et al. 2018; Werner et al. 2018; Prabh and Rödelsperger 2019). Moreover, the two nematode species Caenorhabditis elegans and Pristionchus pacificus have the additional advantage to be established model systems with genetic toolkits including transgenes and genome editing that allow for comparative studies of gene function (Witte et al. 2015; Markov et al. 2016; Moreno et al. 2017, 2018).
In this study, we focus on the spatial expression of novel genes, which can provide further insights into their birth processes and enables to generate hypotheses about their functions. The P. pacificus genome has around 8,000 orphan genes without homologs in other nematode families (Rödelsperger 2018; Prabh and Rödelsperger 2019). However, only two of these orphan genes have been functionally characterized in unbiased genetic investigations. First, the orphan gene dauerless was found to control the entry into the dauer stage, which represents a highly conserved dispersal and long-term survival strategy in nematodes (Mayer et al. 2015). The second orphan gene self-1 encodes a small peptide responsible for self-recognition in natural populations of P. pacificus. Nematodes of the P. pacificus lineage developed tooth-like structures that allow them to predate on other nematodes (Bento et al. 2010; Wilecki et al. 2015). Simultaneously, they evolved a self-recognition system to prevent cannibalism. Notably, a change of a single amino acid in the SELF-1 peptide was sufficient for a mutant line to be killed by its parent (Lightfoot et al. 2019). Although these examples indicate that orphan genes can be identified in unbiased genetic studies, developing rationales for large-scale targeting of novel genes remains difficult.
Therefore, in this study, we focus on the spatial expression of novel genes using RNA tomography (tomo-seq), which can provide insight into their birth processes and allows to generate hypotheses about their functions. Previously, the combination of spatial transcriptomics with knockdown experiments in C. elegans provided strong evidence that regional expression was indeed highly indicative for function (Ebbing et al. 2018). To acquire information about regionalized gene expression in P. pacificus on a genome-wide scale, we performed spatial transcriptomics of young adult hermaphrodites. First, this allows us to investigate in which anatomical structure a particular gene is expressed. This is of particular importance for the Pristionchus research community as spatial expression based on reporter lines is only known for a few P. pacificus genes (Ragsdale et al. 2013; Kieninger et al. 2016; Sieriebriennikov et al. 2020) and only a single tissue-specific RNA-seq data set is currently available (Lightfoot et al. 2016). Second, this data set can be used for a comparative analysis of gene expression between P. pacificus and C. elegans, which have been estimated to have diverged 60–90 Ma (Prabh et al. 2018; Werner et al. 2018). Finally, we can compare the distribution and evolutionary dynamics of genes of different age classes including novel genes across different anatomical regions. This analysis revealed a striking enrichment of novel genes in sperm-related regions, providing first evidence for the “out of testis” hypothesis in nematodes (Levine et al. 2006; Kaessmann 2010; Soumillon et al. 2013; Witt et al. 2019). “Out of testis” genes represent a mix of products of pervasive transcription as well as fast evolving members of ancient gene families. Together, the spatially resolved transcriptome of P. pacificus extends our understanding of novel gene formation in nematodes and will be an important resource for future studies of gene function.
Results
Spatial Transcriptomics Identifies Thousands of Genes with Regional Expression
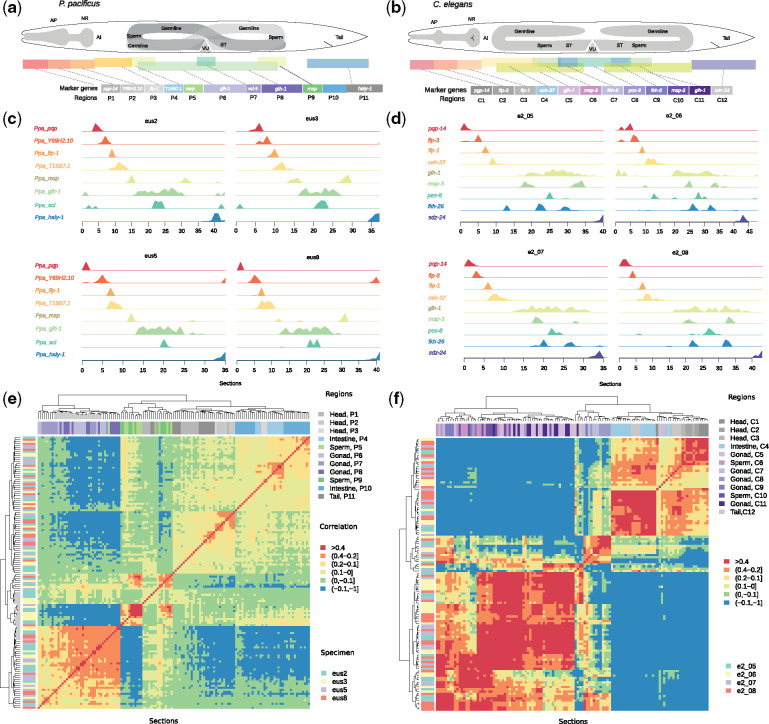
In order to acquire genome-wide gene expression data with spatial resolution, we performed RNA tomography (Junker et al. 2014; Ebbing et al. 2018) which combines cryosectioning of individual P. pacificus specimens with RNA sequencing. This approach has been applied previously to compare the spatial transcriptomes of C. elegans hermaphrodites and males (Ebbing et al. 2018). In total, four young adult P. pacificus hermaphrodites were frozen and cut into around 40 sections per animal (20 µm per section) along the anterior–posterior (A–P) axis. RNA-seq library preparation and sequencing resulted in the detection of 21,046–32,930 (median of all sections per animal) transcripts per section. In total, we detected 12,265 P. pacificus genes with expression in all four specimens (supplementary table S1, Supplementary Material online). Reanalysis of the spatial transcriptomics data from C. elegans hermaphrodites with the same filtering criteria yielded 6,890 robustly expressed genes (Ebbing et al. 2018). We used orthologs of previously established C. elegans marker genes with punctuated spatial expression to define distinct regions (P1–P11 in P. pacificus and C1–C12 in C. elegans) that would correspond to different anatomical structures (fig. 1a and b) (Ebbing et al. 2018). For example, the neuropeptide encoding gene flp-1 (P3, C3), which in C. elegans is expressed in the nerve ring (Nelson 1998), defines regions P3, C3, and the anterior regions (P1, P2, C1, and C2) as different parts of the head (fig. 1a and b). The gonads including the germline are defined by expression of the helicase glh-1 (P5–P9 and C5–C11) (Gruidl et al. 1996). Within the gonads, the expression of major sperm protein (MSP) genes marks sperm-related regions (P5, P9 and C6, C10) (Ebbing et al. 2018; Tzur et al. 2018). In contrast to males, spermatogenesis in hermaphrodites occurs in a finite duration during development. In P. pacificus hermaphrodites, spermatogenesis is delayed relative to C. elegans hermaphrodites and starts only at the last larval molt, with the transition to oogenesis occurring 4–6 h into adulthood (Rudel et al. 2005). As mature sperm, which are stored in the spermatheca (fig. 1a and b), are transcriptionally and translationally silent, the detected MSP expression rather represents residual mRNA levels than de novo transcription. Regions P4 and C4 are enriched for signals from the intestine that are not intermingled with either neuronal or germline expression. The comparison of the spatial transcriptome with previously generated P. pacificus reporter lines showed a high level of agreement (supplementary fig. S1, Supplementary Material online) (Ragsdale et al. 2013; Kieninger et al. 2016; Sieriebriennikov et al. 2020). For example, eud-1, the master regulator of the mouth form dimorphism in P. pacificus (Ragsdale et al. 2013; Kieninger et al. 2016), which is expressed in a few head neurons, is exclusively enriched in region P2 (supplementary fig. S1, Supplementary Material online). Importantly, the expression profiles of many of these marker genes are highly similar across species and individuals (fig. 1c and d). Taken together, these first analyses indicate that our data set represents a high-quality, spatially resolved transcriptome of P. pacificus that is suited for comparative analysis with C. elegans.
Fig. 1.
Comparative spatial transcriptomics of nematodes. (a, b) The schematics represent anatomical overviews of Pristionchus pacificus (a) and Caenorhabditis elegans (b) hermaphrodites together with expression domains of selected marker genes which were used to define regions across the A–P axis. For example, P5 was defined as the first peak of Ppa_msp, P9 as the second peak, and P10 was defined indirectly as sections between P9 and P11. AP, anterior pharynx region; NR, nerve ring; AI, anterior intestine region; ST, spermatheca; VU, vulva and uterus region. (c, d) The plots show the normalized expression of marker genes across all sections for four specimens of P. pacificus (c) and C. elegans (d). (e, f) Hierarchical clustering of P. pacificus (e) and C. elegans (f) sections based on Pearson correlation indicates that sections within a region are generally highly similar on the whole transcriptome level.
Based on the partitioning into regions, we summarized expression values per gene by taking the median z-score for all sections in a given region. We further defined regional genes based on a relative enrichment of expression in a given region (median z-score >1). This identified 3,502 regional genes in P. pacificus (supplementary table S3, Supplementary Material online) and 3,656 for the reanalyzed C. elegans data (supplementary table S4, Supplementary Material online) (Ebbing et al. 2018). Next, we performed a clustering approach to assess whether regions that reflect similar anatomical structures (as defined using only a few marker genes) show similar transcriptomic profiles on a global scale. Despite the variable degrees of cellular heterogeneity across sections, hierarchical clustering based on Pearson correlation grouped individual sections by anatomical region and not by specimen (fig. 1e and f). Thus, anatomical regions identified in individual animals are reproducible across different animals, and they are not affected by batch effects or individual-to-individual variation. Together, our data set of 3,502 genes with regional expression profiles represents an enormous extension to the knowledge about spatial expression in P. pacificus.
Regional Expression Is Not Highly Conserved between P. pacificus and C. elegans
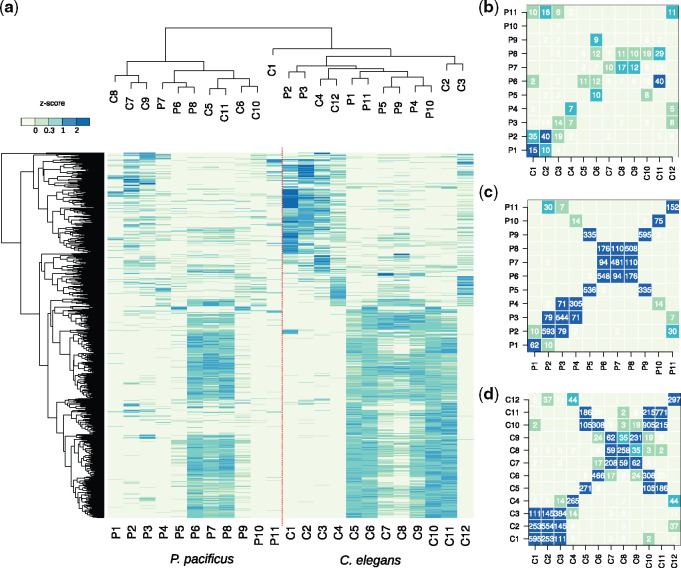
The availability of spatially resolved and highly comparable transcriptomes of C. elegans and P. pacificus provides the opportunity to study the evolution of regional expression in nematodes. Despite the high conservation of expression patterns of individual marker genes such as flp-3, glh-1, and MSP genes and their orthologs in P. pacificus (fig. 1c and d), it remains elusive to what extent regional expression is generally conserved between species. In addition, since partially nonorthologous marker genes were selected to segment the spatial transcriptomes of P. pacificus and C. elegans (fig. 1a–d), it is unclear how the different regions of both species correspond to each other. To assess the overall degree of similarity between different regions across species, we compared expression profiles of regional one-to-one orthologs. We combined z-score normalized expression data of 1,559 one-to-one orthologous genes with regional expression in at least one of the species and performed a clustering analysis (fig. 2a). The most dominant signal is the separation between the gonadal regions (P6–P8, C5–C11) and the rest of the worms. Specifically, the majority of genes is highly expressed in the gonadal regions of both species resulting in a joint clustering of data from both species (fig. 2a). In contrast, outside the gonadal regions, the clustering appears to be predominantly driven by species-specific signals, such as varying levels of intestinal or cuticular expression and similarities between head and tail regions (fig. 2a). Interestingly, the most anterior regions of C. elegans (C1–C4) show hundreds of genes with highly specific expression that are absent in P. pacificus suggesting substantial divergence between the two species. These genes are mostly involved in muscle-related functions, such as mitochondrion (P < 10−15), striated muscle (P < 10−11), and locomotion (P < 10−3) (Huang et al. 2009). This difference is consistent with a recent morphological study showing a pronounced shift in the P. pacificus lineage from muscular to glandular tissues in the mouth regions (Riebesell and Sommer 2017). To quantify the amounts of regional genes with highly conserved expression more explicitly, we counted the number of shared one-to-one orthologs in every pairwise comparison (fig. 2b and supplementary table S5, Supplementary Material online). Surprisingly, only a few dozens of one-to-one orthologs are shared between regions. This finding suggests substantial expression divergence, because overall, 25–30% of the P. pacificus genes have one-to-one orthologs in C. elegans (Rödelsperger, Athanasouli, et al. 2019). Thus, regionally expressed genes seem to be largely composed of one-to-one orthologs with divergent expression and novel genes.
Fig. 2.
Limited conservation of highly spatially restricted expression. (a) The heatmap shows the median z-score-normalized expression values for one-to-one orthologs of regional genes in both species. Joint clustering of regions separates the germline from the rest of the worms. (b–d) The heatmaps show the numbers of shared one-to-one orthologs (b) or shared genes in Pristionchus pacificus (c) and Caenorhabditis elegans (d) between different regions. The color code scales with the significance of the overlap (Fisher’s exact test).
Conserved and Symmetric Expression Patterns Support the Homology of Anatomical Structures between P. pacificus and C. elegans
Although the overall level of conserved regional expression is low, the comparison of regional expression based on shared orthologs also provides some indication how the different regions in both nematodes correspond to each other. In particular, the head and tail regions exhibit a high similarity across the data sets of both species. For example, the P. pacificus region P2 shares 40 regional one-to-one orthologs with C. elegans region C2, but also 35 with C. elegans region C1 (fig. 2b). This shows that there is no unambiguous mapping between individual regions in the anterior part of P. pacificus to their counterparts in C. elegans. However, as a single unit, the three most anterior P. pacificus regions together (P1–P3) share dozens of one-to-one orthologs specifically with the most anterior regions in C. elegans (C1–C3) and thus, represent distinct segments of the heads (fig. 2b). Also, the intestinal regions P4 and C4 from both species seem to correspond to one another, and the most posterior regions (P11 and C12) show highly significant sharing of one-to-one orthologs (fig. 2b). Within regions representing the gonads (consisting of both the somatic and germline gonad), the signal is obscured partially by the symmetry of gonad structures and the anatomical differences between the species (fig. 1a and b). To illustrate this, we tested for gene sharing between regions of the same species. This perfectly recapitulated the symmetry of the gonads, as many of these regions exhibit highly significant gene sharing with themselves and one additional region. For example, both MSP expressing regions (P5 and P9 in P. pacificus and C6 and C10 in C. elegans) have hundreds of genes in common (fig. 2c and d). In the case of C. elegans, sperm-related signals seem to be stronger mixed with general gonadal expression than in P. pacificus because the global transcriptomic profiles in C. elegans are more similar to other gonadal regions and the amount of shared regional genes is higher (figs. 1e, 1f, and 2d). Thus, even though the overall level of conserved regional expression is rather low, it is still sufficient to identify homologous anatomical regions across species as follows: head, intestine, general gonad, sperm-related, and tail regions.
Spatial Expression Patterns Reveal Regional Clustering of Large Gene Families
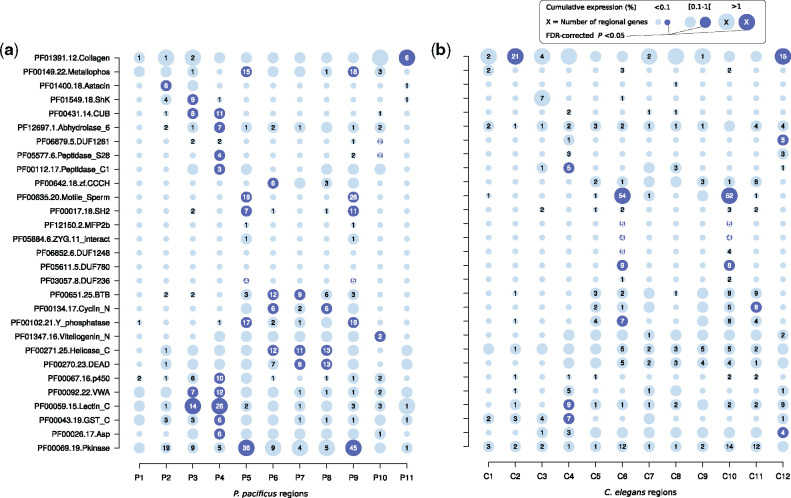
The limited degree of conservation at the level of one-to-one orthologous genes (fig. 2b) is indirect evidence that certain novel gene families are enriched among regional genes. This would be consistent with a previous studies showing that large portions of developmentally regulated genes in C. elegans and P. pacificus originated from lineage-specific duplication events and therefore, do not have one-to-one orthologs (Castillo-Davis and Hartl 2002; Baskaran et al. 2015). Thus, we tested regional genes for an overrepresentation of gene families as defined by the presence of certain protein domains (fig. 3a and b). Consistent with previous analysis based on reporter lines, we found a significant enrichment (false discovery rate-corrected P value <0.05) of astacin genes in the head region P2 of P. pacificus (Sieriebriennikov et al. 2020), which is not observed in the C. elegans data (fig. 3a and b). This example of expression divergence might be an indirect effect caused by cellular heterogeneity across species as the corresponding gland cell expressing these astacins is greatly expanded in P. pacificus (Riebesell and Sommer 2017; Sieriebriennikov et al. 2020). Overall, this analysis revealed that most regions are strongly biased toward the expression of members of a particular gene family. The numbers of such lineage-specific duplicates can by far exceed the amount of one-to-one orthologs in a given region (fig. 3a and b). For example, although regions P4 and C4 only share seven one-to-one orthologs (fig. 2b), P4 and C4 contain 26 and 9 regionally expressed C-type lectins. C-type lectins have been implicated in the innate immunity of C. elegans and were previously found to be expressed in the intestines of both nematodes (Schulenburg et al. 2008; Lightfoot et al. 2016). Similarly, MSP expressing regions share almost no one-to-one orthologs, yet both nematode species express between 19 and 54 different MSPs in a regional manner (fig. 3a and b). Thus, gene families that have undergone recent duplications show strong regional clustering in the spatial transcriptomes of P. pacificus and C. elegans.
Fig. 3.
Overrepresentation of protein domains among regional genes. (a, b) We identified 29 protein domains that are significantly enriched in regional genes in at least one of the species. The plots show the number of regional genes, the cumulative expression, and an indication of significance for a given protein domain in Pristionchus pacificus (a) and Caenorhabditis elegans (b).
Previously Uncharacterized Gene Families Are Expressed in Sperm-Related Regions
Although many of the enriched gene families such as C-type lectins, MSPs, vitellogenins, and kinases have well-characterized molecular or biological functions, we observed multiple domains of unknown function (DUF) with regional expression as well (fig. 3a and b). DUFs mostly represent evolutionary constrained sequence stretches in lineage-specific clusters of orthologous and paralogous genes (El-Gebali et al. 2019). On a functional level, these novel gene families are still poorly characterized, probably because different paralogs are functionally redundant or because large-scale RNAi screens did not yield obvious developmental or morphological phenotypes (Kamath et al. 2003). For example, the spatial expression profile of DUF1261-containing genes is suggestive of intestinal expression (fig. 3a and b) and indeed five of them were previously detected in the intestinal transcriptome of C. elegans (Lightfoot et al. 2016). Interestingly, DUF780, DUF236, and DUF1248 show the highest signal in MSP expressing sperm-related regions in at least one of the species (fig. 3a and b). It is important to note that all these gene families are largely restricted to nematodes and thus, homologous sequences are unknown from vertebrates and most insects (El-Gebali et al. 2019). Following the idea that regional expression could be an indicator of potential biological function (Ebbing et al. 2018), we would hypothesize that the coexpression of the three gene families DUF780, DUF236, and DUF1248 together with MSPs possibly indicates a role in sperm cells (fig. 3a and b).
The Phylogenies of Sperm-Related Gene Families Are Consistent with Recent Expansions
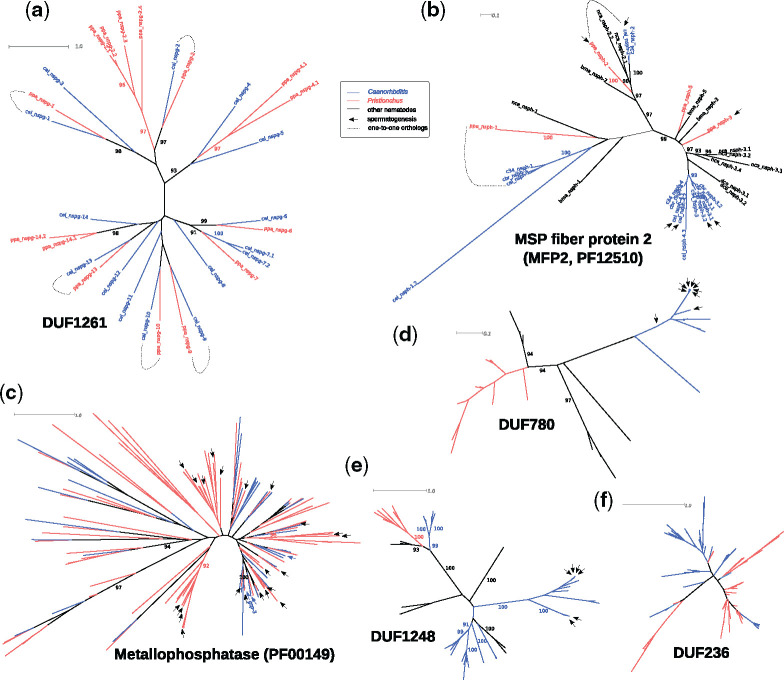
Given that reproductive genes are frequently associated with rapid evolution (Swanson and Vacquier 2002; Kasimatis and Phillips 2018), we tested whether the phylogenies of the corresponding gene families are indeed consistent with rapid evolution and exhibit signatures of recent lineage-specific gene expansions. To this end, we reconstructed phylogenies of members of all three families and compared these with the phylogeny of DUF1261-containing proteins (as an example for a DUF family without sperm-related expression profile, fig. 4a). In addition, we generated phylogenies for two other gene families that are also coexpressed with MSPs, metallophosphatases including C. elegans gsp-3 and homologs of MSP fiber protein 2 (MFP2), which are involved in sperm motility (Buttery et al. 2003; Wu et al. 2012) (fig. 4b and c). This analysis revealed that individual members of the DUF1261 gene family are perfectly preserved as one-to-one orthologs between P. pacificus and C. elegans (fig. 4a). In contrast, the two sperm-related gene families are consistent with recent gene expansions (fig. 4b and c). For example, four out of five of the sperm-related MFP2 genes from C. elegans arose very recently from duplication after the split from its sister species Caenorhabditis inopinata (C. sp34, fig. 4b) (Kanzaki et al. 2018). Similarly, sperm-related metallophoshophatases in P. pacificus tend to cluster in lineage-specific groups of paralogs which is a common sign of recent duplications. Also, in the case of the three uncharacterized but putatively sperm-related gene families DUF780, DUF236, and DUF1248, clusters of paralogous genes most-likely point toward recent lineage-specific duplication (fig. 4d–f). We conclude from these observations that the phylogenies of all five sperm-related gene families show signatures of recent expansions that are consistent with rapid evolution.
Fig. 4.
Rapid gene expansion of sperm-related gene families. (a) DUF1261 represents an uncharacterized gene family that is expressed in the nematode intestine. A number of genes within this gene family are preserved as one-to-one orthologs between Pristionchus pacificus and Caenorhabditis elegans. (b, c) Members of the MFP2 (b) and metallophosphatase (c) gene families are known to be involved in spermatogenesis and sperm motility. Multiple sperm-related genes in both families are found in lineage-specific clusters and likely arose by recent gene duplication. (d–f) The phylogenies of other uncharacterized gene families also exhibit strong signatures of recent gene expansions and sperm-related genes are found in Caenorhabditis-specific subtrees of DUF780 (d) and DUF1248 (e). DUF236 containing genes usually have multiple repeated DUF236 domains; the phylogenetic tree (f) represents the relationship of individual repeat units. Thus, individual sperm-related genes cannot be indicated.
Spatially Resolved Phylostratigraphy Supports the Out of Testis Hypothesis in Nematodes
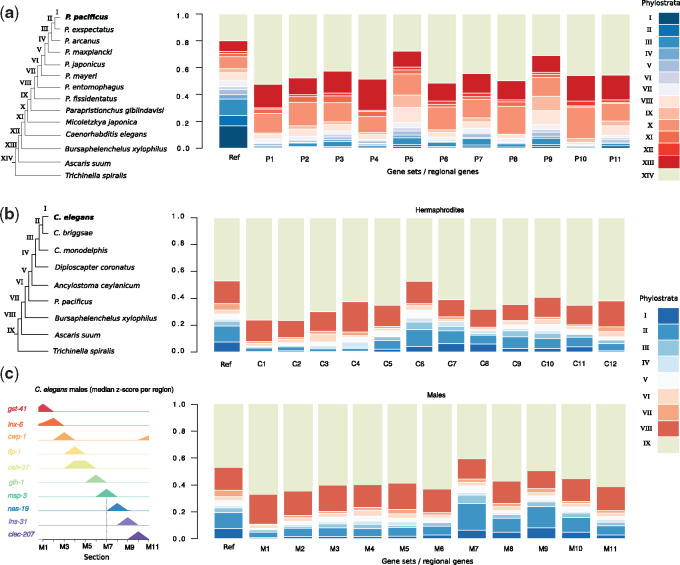
Genomic studies in Drosophila and mammals revealed that novel genes frequently exhibit testis-biased expression, resulting in the “out of testis” hypothesis, which postulates that testis might represent a special environment facilitating the formation of novel genes (Levine et al. 2006; Soumillon et al. 2013; Witt et al. 2019). The above-mentioned examples of the novel gene families (DUF780, DUF236, and DUF1248) that are expressed in sperm-related regions provide first evidence that the “out of testis” hypothesis may also hold true in nematodes. To provide additional support for the “out of testis” hypothesis in nematodes, we systematically assessed the amount of novel genes that are expressed in different anatomical structures, by performing a phylostratigraphic analysis of all regionally expressed genes (Domazet-Loso et al. 2007). To this end, we combined comparative genomic data from multiple non-Pristionchus species with the genomes of seven additional Pristionchus species that were recently sequenced on the same platform with similar protocols to study the evolutionary dynamics of novel gene families (Prabh et al. 2018) (fig. 5a). The underlying ladder-like phylogeny enables us to date the emergence of a gene based on the presence/absence patterns of homologous sequences in other taxa. This defined phylostrata (age classes), which can be quantified across anatomical regions (fig. 5a). For example, phylostratum I denotes genes that are specific to P. pacificus, whereas genes of other phylostrata are shared with more distantly related species and are therefore to be considered older. The P. pacificus data set supports the “out of testis” hypothesis as regions P5 and P9 show the overall highest ratio of young phylostrata (I–XIII, fig. 5a). We repeated the phylostratigraphic analysis for C. elegans using a different phylogenomic context, which included the high-quality Caenorhabditis briggsae genome and the most basal Caenorhabditis species C. monodelphis (Stein et al. 2003; Slos et al. 2017). Also, in this data set, the sperm-related regions (C6 and C10) exhibit the highest fraction of young phylostrata (I–VIII, fig. 5b). To further support this observation, we reanalyzed tomo-seq data of C. elegans males (Ebbing et al. 2018). Consistent with the fact that C. elegans males have only one gonadal arm, we observe a single peak of young phylostrata in region M7 which also expresses MSP genes (fig. 5c). Interestingly, region M9 that overlaps the seminal vesicle and vas deferens (Ebbing et al. 2018) also exhibits high levels of young phylostrata suggesting a role of novel genes in sperm maturation and seminal fluid function (fig. 5c). To further narrow down the location of highest expression of novel genes, we repeated this analysis with a previously published C. elegans tomo-seq data set consisting of ten gonadal regions (Tzur et al. 2018). Although expression of MSP was not detectable in hermaphrodite data of the investigated developmental stage (L4 + 24 h), we observed a pronounced peak of young phylostrata in regions of crossover formation and desynapsis (supplementary fig. S2, Supplementary Material online). These findings indicate that meiotic spermatocytes exhibit the highest levels of young phylostrata and they are consistent across data sets generated from different laboratories, nematode sexes, and species. Together, our observations provide strong support for the “out of testis” hypothesis in nematodes.
Fig. 5.
Expression of novel genes in sperm-related regions. (a, b) We classified Pristionchus pacificus genes into gene ages by means of phylostratigraphy and plotted the fraction of phylostrata across the different regional gene sets. The stacked bars labeled as “Ref” show the genome-wide, phylostratigraphic distribution which is dependent on the phylogenetic resolution around the focal species. Sperm-related regions in P. pacificus (a, P5, P9) and Caenorhabditis elegans (b, C6, C10) showed the highest fraction of young age classes which is consistent with the “out of testis” hypothesis. (c) Reanalysis of tomo-seq data of C. elegans males identifies 11 regions (M1–M11) and confirms that the MSP expressing region (M7) indeed shows the highest fraction of young phylostrata.
Young Out of Testis Genes May Result from Pervasive Transcription and Evolve Rapidly
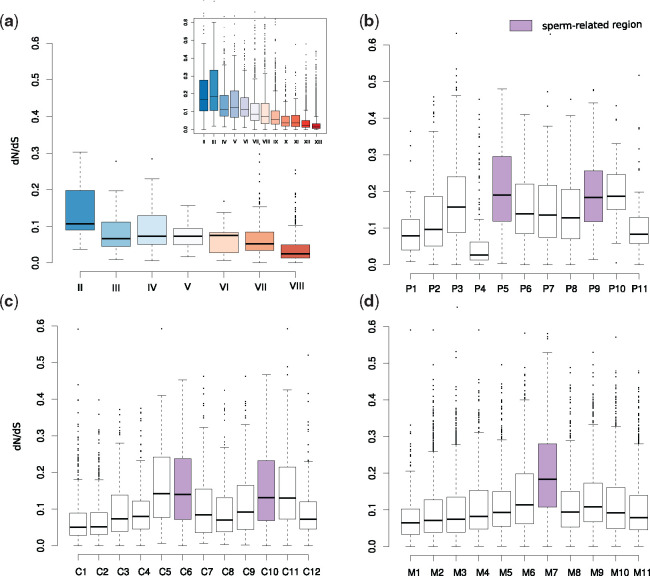
Despite widespread support for the “out of testis” hypothesis in insects and mammals, it is still not entirely clear to what extent novel gene formation is driven by sexual selection or by an overall permissive chromatin state. A comprehensive, phylotranscriptomic analysis in mammals supported the idea that the observed pattern is largely a product of the permissive chromatin state resulting in pervasive transcription (Soumillon et al. 2013). In agreement with this, we found that not only are more novel genes expressed in sperm-related regions, but that sperm-related regions also tend to express the overall highest numbers of genes irrespective of the gene’s age (supplementary fig. S3, Supplementary Material online). This observation suggests that the expression of novel genes in nematodes is also, at least partially, a byproduct of the higher transcriptomic complexity in sperm-related regions. To test if sperm-related genes in C. elegans are actually functional, we used signatures of purifying selection as indirect evidence of function (Prabh and Rödelsperger 2016). Comparison of nonsynonymous versus synonymous substitutions (dN/dS) of orthologous pairs between C. elegans and C. briggsae indicated that basically all C. elegans genes with C. briggsae orthologs exhibit strong purifying selection, which is indirect evidence of biological function (Prabh and Rödelsperger 2016) (fig. 6). Contrasting different phylostrata, we found that younger genes are generally less constrained than genes of more ancient phylostrata, which is consistent with previous phylogenomic analysis of P. pacificus and Pristionchus exspectatus orthologs (Prabh et al. 2018) (fig. 6a). When estimating selective constraint across the sets of regional genes, we found in all three data sets that sperm-related regions exhibit high ratios of genes under relaxed evolutionary constraint (fig. 6b–d). Thus, pervasive transcription but also rapid evolution, seem to be inherent features of sperm-related regions.
Fig. 6.
Relaxed evolutionary constraint in sperm-related regions. (a) The boxplots show median and interquartile ranges of dN/dS ratios inferred from orthologs between Caenorhabditis elegans and C. briggsae for different phylostrata. The inset shows equivalent data for Pristionchus pacificus and P. exspectatus orthologs. Generally, older genes underlie stronger purifying selection than more recent gene classes. (b–d) The distribution of dN/dS values across sets of regional genes in P. pacificus hermaphrodites (b), C. elegans hermaphrodites (c), and C. elegans males (d) indicates that sperm-related regions have high ratios of genes underlying relaxed evolutionary constraint.
Young Out of Testis Genes Affect Spermatogenesis and Meiosis
Finally, to further support the idea that recently evolved “out of testis” genes are not just a byproduct of pervasive transcription but actually got integrated into the biology of their host (Rödelsperger, Prabh, et al. 2019), we exploited the enormous genetic knowledge of C. elegans to screen for novel genes with well-characterized biological functions. Although the largest portion (∼90%) of novel genes (Phylostrata I–III) has not even been characterized in C. elegans, we found a number of recently evolved genes with functions in spermatogenesis and meiosis (table 1). Notably, we found regional expression of several meiosis-related genes such as him-5 (Meneely et al. 2012), lab-1 (Tzur et al. 2012), syp-2 and syp-3 (Schild-Prüfert et al. 2011), kca-1 (McNally et al. 2012), mesp-1 (Wolff et al. 2016), brc-2 (Checchi et al. 2014), and ify-1 (Kitagawa et al. 2002) in sperm-related regions C6 and C10 of C. elegans hermaphrodites. The same regions also express other spermatogenesis-related genes such as spe-11 (L’Hernault et al. 1988) and fog-2 (Nayak et al. 2005). In region CM7 of C. elegans males, we found expression of multiple novel spermatogenesis and meiosis-related genes such as spe-12 (Nance et al. 1999), spe-27 (Minniti et al. 1996), spe-11 (L’Hernault and Singson 2000), spe-19 (Geldziler et al. 2005), spe-38 (Chatterjee 2005), lab-1 (Tzur et al. 2012), and ify-1 (Kitagawa et al. 2002) as well as nine members of another uncharacterized nematode-specific protein family (NSP group A, table 1). These results demonstrate that multiple seemingly newly arisen genes have been integrated into the biology of their host and modulate universal processes including spermatogenesis and meiosis.
Table 1.
Candidate List of Novel, Sperm-Related Caenorhabditis elegans Genes.
| Category | C. elegans Genes |
|---|---|
| Meiosis | him-5, cdc-26, lab-1, kca-1, syp-2, syp-3, mesp-1, brc-2, ify-1, hcp-2, and rsa-2 |
| Spermatogenesis | fog-2, spe-27, spe-19, spe-11, spe-38, and sss-1 |
| Other F-box proteins | fbxb-38, fbxb-36, fbxc-20, fbxa-201, fbxa-196, fbxa-218, fbxa-125, fbxc-44, and fbxb-52 |
| Nematode-specific protein group A | nspa-1, nspa-2, nspa-3, nspa-4, nspa-5, nspa-6, nspa-7, nspa-8, and nspa-10 |
| Germ granules | pgl-1, pgl-2, and pgl-3 |
| Others | sdz-27, hil-8, cec-2, numr-2 egl-1, dlc-6, kbp-1, mut-16, hmg-12, hpo-40, gpr-1, gpr-2, clec-260, and clec-142 |
Note.—Caenorhabditis elegans genes of phylostrata I–III that have been characterized previously.
Discussion
Nematodes are one of the most species-rich animal phyla and with C. elegans, they harbor one of the best studied model organisms (Lambshead 1993; Blaxter 1998). Many of the free-living nematodes, including C. elegans, are characterized by their small body, which was one the key features for the selection and success of C. elegans as a model system. However, for a long time, their small body size has also hindered the generation of large-scale tissue-specific expression catalogs that are comparable to mammals (Dupuy et al. 2007; Spencer et al. 2011; GTEx Consortium 2013; Smith et al. 2019). With the advent of spatial transcriptomics and single-cell sequencing, the nematode research community can now overcome this hindrance (Cao et al. 2017; Ebbing et al. 2018). Although single-cell RNA-seq still has obvious advantages in terms of resolution, spatial transcriptomics provides additional positional information that is very useful to disentangle signals from symmetric anatomical structures such as the hermaphroditic gonads. In this study, we combined cryosectioning of individual P. pacificus worms with RNA-seq in order to study the evolution of novel genes at spatial resolution and to establish a resource for comparative studies of gene function between C. elegans and P. pacificus (Markov et al. 2016; Moreno et al. 2017, 2018). Since individual sections represent a mix of heterogeneous tissues and cells (e.g., cuticle, neurons, intestine, somatic gonad, and germline), we focused on the analysis of regional genes with enriched expression in a given region relative to the whole worm in order to minimize contributions from other tissues. However, further experimental analysis would be needed to confirm the expression of these regional transcripts in the proposed tissue. By comparison with the spatial transcriptome of C. elegans hermaphrodites, we found that only a small number of one-to-one orthologs show conserved regional expression. This result resembles recent findings of expression divergence between the developmental transcriptomes of C. elegans and C. briggsae (Lu et al. 2020) and together, these results suggest that nematode transcriptomes harbor considerable species-specific signals at a spatial and temporal level. However, further analysis will be needed to disentangle to what extent the limited spatial conservation is caused by expression divergence, heterogeneous composition of cells and tissues across sections, lineage-specific duplications, or low expression levels that are below the current limits of detection (individual sections of our spatial transcriptome are sequenced at a substantially lower depth than samples in previous RNA-seq studies [Baskaran et al. 2015; Lightfoot et al. 2016]). At the same time, the limited level of conserved regional expression highlighted important differences between species, including the gain of spatially restricted expression of complete gene families. Focusing on sperm-related regions, we identified some previously uncharacterized gene families that might play roles in spermatogenesis or sperm cell biology (fig. 4) and we found broad support for the “out of testis” hypothesis in nematodes (fig. 5). This is particularly interesting as unlike Drosophila and most mammals, C. elegans and P. pacificus are androdioecious species where most individuals are hermaphrodites that are capable of self-fertilization and only a small percentage of a population is composed of males. Hermaphrodites of both species produce spermatids during larval development which are activated upon contact with oocytes that are formed after transitioning to the adult stage (L’Hernault 2006). Although these spermatids are transcriptionally and translationally quiescent (no de novo transcription and translation), transcripts are still detectable which allowed us to investigate the “out of testis” hypothesis in young adult hermaphrodites of both species. Note however, that the enrichment of novel genes in sperm-related regions was strongest in the spatial transcriptomic data of C. elegans males with active spermatogenesis (fig. 5c and supplementary fig. S2, Supplementary Material online). Interestingly, the evolution of selfing in nematodes led to genome contractions and gene losses that preferentially affect male-biased and sperm-related genes (Fierst et al. 2015; Rödelsperger et al. 2018; Yin et al. 2018). Thus, the analysis of gonochoristic sister species could reveal sperm-related expression of further novel genes that have been lost in the P. pacificus and C. elegans lineages.
Gene duplication is a major source for generating novel genes and functions (Ohno 1970), but other processes such as divergence and de novo formation can also generate novel orphan genes (Schlötterer 2015; McLysaght and Hurst 2016; Van Oss and Carvunis 2019) and first cases of these processes have been recently characterized in nematodes (Prabh and Rödelsperger 2019; Rödelsperger, Prabh, et al. 2019; Zhang et al. 2019). The spermatogenesis-related gene fog-2 is a good example of sequence divergence. Mutations in fog-2 transform hermaphrodites into females that are not capable of self-fertilization (Schedl and Kimble 1988). fog-2 arose recently via gene duplication in C. elegans and has no one-to-one ortholog in C. briggsae (Nayak et al. 2005). According to our phylostratigraphic analysis, it is classified as Caenorhabditis specific. However, the FOG-2 protein and also other sperm-related genes (table 1) contain a widely distributed F-box motif that is also recognized by searches using profile hidden Markov models (El-Gebali et al. 2019), supporting the notion that the absence of sequence similarity is due to divergence. Similarly, BRC-2 has been shown to be homologous to the mammalian tumor suppressor BRCA2 (Bork et al. 1996), suggesting that the actual gene age of highly divergent members of ancient gene families can be underestimated in phylostratigraphic analysis (Moyers and Zhang 2015). However, even if more sensitive homology searches can still detect some level of sequence similarity, we would argue that such a sequence has diverged enough from its ancestor to be called novel (Rödelsperger, Prabh, et al. 2019). On top of the lack of strong evolutionary constraint, positive selection can additionally drive sequence divergence, especially in the context of reproductive genes (Swanson and Vacquier 2002; Kasimatis and Phillips 2018). Consistent with this, previous analysis has found evidence for positive selection acting on fog-2 (Nayak et al. 2005). Interestingly, fog-2 is not the only F-box gene and many of its paralogs are also expressed in sperm-related regions (table 1). Thus, following the argument that spatial expression is indicative for biological function (Ebbing et al. 2018), we would also hypothesize that these F-box genes might play some previously undescribed roles during spermatogenesis. Nevertheless, not every gene that is found to be coexpressed with MSPs is likely to perform biological functions in sperm cells. The high transcriptional complexity in sperm-related regions as a result of a permissive chromatin state may lead to the expression of putative de novo genes without any specific function. Thus, the transcriptome of sperm-related regions might harbor a mix of many classes of novel genes some of which are simply byproducts of pervasive transcription. Even if such sequences do not have any biological function, they form the raw material out of which new genes may evolve. Some of these potential de novo genes may start to interact with other cellular components and gain biological functions (Keeling et al. 2019). At the same time, recent gene duplicates and rapidly evolving members of ancient gene families also contribute to this heterogenous pool of novel genes that is highly expressed in sperm-related regions. This is further supported by the recent finding in mammals that even though error correction processes such as transcriptional scanning exist, male reproductive genes exhibit an unusually high level of sequence divergence (Xia et al. 2020). In future, more detailed analysis and functional studies of individual genes and gene families are needed to comprehensively characterize the roles of novel genes in sperm cell biology.
Conclusion
By comparison of the spatial transcriptomes between P. pacificus and C. elegans, we have shed light on the evolution of novel genes in a tissue-specific context. We present highly spatially restricted expression profiles for many previously uncharacterized gene families in C. elegans and P. pacificus, which might help future work to study their function in a more targeted context. Focusing on sperm-related genes, we provide first evidence in nematodes supporting the previously postulated “out of testis” hypothesis and demonstrate that the sequences expressed during spermatogenesis represent a mix of biologically relevant genes as well as potential byproducts of pervasive transcription. Furthermore, we find evidence for rapid evolution of sperm-related genes at the level of sequence and copy number, which reflects typical patterns of the evolution of reproductive genes. Exploiting the wealth of genetic knowledge in C. elegans, we could show that even universal cellular processes such as meiosis are affected by the birth of novel genes. Finally, our data set may be taken as a starting point to investigate how gene expression changes and genomic novelty coincide with morphological and behavioral diversity.
Materials and Methods
Worm Culturing and Sample Preparation
Pristionchus pacificus strain PS312 was grown on OP50 Escherichia coli strain on S-medium-agar petri plates (Stiernagle 2006; Werner et al. 2017). Bleached egg populations were seeded on to plates to obtain synchronized worm cultures. Worms were screened ∼64 h post bleaching, to determine their developmental stage, under a high-magnification stereomicroscope. Young adults were sought out for, by looking out for the invaginated vulval shape and absence of embryos in the germline (Sommer 1997; Mok et al. 2015), and were selectively picked from the plates for subsequent cryosectioning and sample preparation steps.
Cryosectioning and Preparation of Sequencing Libraries
Young adult worms were straightened in cryosolution (OCT). The head and the tail were marked by a single blue bead (Affigel Blue Gel agarose beads). The worms were then frozen, within 10 min of preparation, on dry ice and stored at −80 °C prior to cryosectioning. The embedded worms were sectioned using a cooled histology cryotome (−20 °C). Sections of 20 μm width were made starting from the first blue bead until the last blue bead. Typically, a worm, in-between two blue beads, would comprise about 40 sections. The sections of one individual worm were placed in a cooled pre-prepared 96-well plate, previously stored at −80 °C. The 96-well plates contain barcoded primers used for the CELseq protocol; consisting of a PolyT stretch, a unique 8-nt section-specific barcode, a 4-nt random barcode, the 5′ Illumina adaptor, and a T7 promotor for in vitro transcription. 96-Well plates containing sectioned worms were sealed, spinned down at 300 rpm at 4 °C, and stored at −80 °C. mRNA processing, reverse transcription, in vitro transcription, and illumina sequencing library preparation were performed according to the robotized version of the CELseq2 protocol (Hashimshony et al. 2012, 2016; Muraro et al. 2016), by the Single Cell Facility at the Hubrecht Institute. Sample processing was performed within a week of cryosectioning.
Preparation of a Reference Transcriptome
As current gene annotations for P. pacificus do not include 3′UTRs (Rödelsperger, Athanasouli, et al. 2019), we generated a customized P. pacificus reference annotation based on a previously established strand-specific transcriptome assembly (Rödelsperger et al. 2016, 2018). To reduce isoform redundancy, we applied the following set of rules to pick one representative isoform per gene: 1) pick the isoform that had a poly-A tail which was defined as the last six nucleotides or more being adenine, 2) in case of multiple isoforms with a poly-A tail, the isoform that had the most 3′ alignment to the best protein hit in C. elegans was chosen, 3) if neither rule 1 nor rule 2 could resolve the ambiguity, we picked the longest isoform. Open reading frames were then extracted from these sequences by taking the longest partial or complete translatable peptide sequence that was not interfered by a stop codon (Rödelsperger et al. 2018). We then applied an Inparanoid-like approach of detection of one-to-one orthologs with C. elegans (Remm et al. 2001). In summary, BlastP searches against combined protein data from both species were performed (e-value <0.001) and only best-reciprocal pairs with lowest cross-species e-value (self-hits were excluded) were reported to avoid calling one-to-one orthologs in the presence of inparalogs. Finally, to annotate protein domains in the reference annotation, we ran the hmmsearch program of the HMMER3.0 package (-E 0.001) (Mistry et al. 2013).
Analysis of Tomo-Seq Data Expression Data
Raw reads were demultiplexed, aligned to the reference annotation, and transcript counts were quantified as described in Ebbing et al. (2018). Transcript counts were normalized by the total number of transcripts per section. Subsequently, for each gene in a single worm, z-scores were calculated using the scale function as implemented in R on the transcript normalized expression values across all sections. To make sure that the P. pacificus and C. elegans data sets are maximally comparable, we reanalyzed the previously generated spatial transcriptomics data of C. elegans males and hermaphrodites (Ebbing et al. 2018) using the same normalization procedures and rules for defining the anatomical regions. We then used the expression pattern of selected marker genes to classify the sections into regions. Data from multiple individuals were combined by pooling all sections corresponding to a certain region and taking the median z-score from all these sections. Clustering of sections based on Pearson correlation and clustering of regions based on z-scores of one-to-one orthologous genes were done using complete linkage clustering as implemented in the hclust and heatmap functions of R. The tomo-seq data of C. elegans hermaphrodites and males were analyzed in the same way. Similarly, spatial transcriptomic data from the study of Tzur et al. (2018) were extracted from supplementary tables S2 and S10, Supplementary Material online, and were z-score normalized. For all data sets, expressed genes were defined as genes with a positive median normalized transcript count of all sections corresponding to a region and regional genes were defined as genes with a z-score >1. Overrepresentation analysis of protein domains (results of hmmsearch) was done by calculating the intersection between a set of regional genes and a given protein domain and quantifying the significance by performing a Fisher’s exact test against a background of all expressed genes. To correct for multiple comparisons, P values were adjusted by the false discovery rate method.
Phylostratigraphic, Phylogenetic, and Selection Analysis
Phylostratigraphic analysis for C. elegans was performed by obtaining annotated protein sets for C. elegans (WS260), C. briggsae (WS260), C. monodelphis (JU1667 v1), Diploscapter coronatus (WBPS10), Ancylostoma ceylanicum (WS248), P. pacificus (WS260), Bursaphelenchelus xylophilus (WS248), Ascaris suum (WS248), and Trichinella spiralis (WS248). The set of C. elegans proteins (longest isoform per gene) were taken as query sequences and phylostrata were defined from the pairwise BlastP (version 2.6) searches against all other species (e-value <0.001) (Domazet-Loso et al. 2007). Phylostratigraphy for P. pacificus was done analogously using the predicted proteins from recently sequenced diplogastrid genomes (Prabh et al. 2018) and proteins from C. elegans, Bursaphelenchelus xylophilus, Ascaris suum, and Trichinella spiralis as target BlastP databases (e-value <0.001). To account for potentially incorrectly annotated open reading frames (ORFs) in the P. pacificus reference annotation, additional BlastN searches against the corresponding diplogastrid transcriptome assemblies (Rödelsperger et al. 2018) were performed (e-value <10−5). Phylogenetic analysis of selected gene families was done by extracting C. elegans and P. pacificus protein sequences based on the protein domain annotation created by hmmsearch. Further sequences were included after manual Blast searches on pristionchus.org, wormbase.org, and parasite.wormbase.org. A multiple sequence alignment was then generated by the MUSCLE program (version 3.8.31) and maximum likelihood trees were reconstructed using the phangorn R package (pml and optim.pml function with the following parameters, model = “LG,” optNni = TRUE, optBf = TRUE, and optInv = TRUE). One hundred bootstrap pseudoreplicates were calculated and edges with support values above 90 were labeled in the resulting trees using the Dendroscope and Inkscape softwares. In order to quantify the strength of selection, we first computed orthologous clusters between C. elegans and C. briggsae proteins, P. pacificus and P. exspectatus proteins, and P. pacificus and Pristionchus fissidentatus proteins with the help of orthAgogue (Ekseth et al. 2014). Subsequently, pairs of orthologous proteins were aligned by MUSCLE (version 3.8.31) (Edgar 2004) and translated into a codon alignment by the program pal2nal (version 14) (Suyama et al. 2006). We then ran the codeml program of the paml package (version 4) to estimate one dN/dS value for each orthologous pair (codeml model 0) (Yang 2007). For each query gene for C. elegans and P. pacificus, we selected the ortholog with lowest estimated dS as one-to-one ortholog for further selection analysis. dN/dS values for subsets of genes, for example, different phylostrata or sperm-related genes, were then visualized as boxplots in R. For better comparison of dN/dS values across anatomical regions and nematode genera, we chose a pair of Pristionchus species that is separated by similar evolutionary distance (measured as median dS) as C. elegans and C. briggsae. As median dS between C. elegans and C. briggsae was 2.1, the matching Pristionchus species pair corresponded to P. pacificus and P. fissidentatus (median dS = 1.9) which was considerably more diverged than P. pacificus and P. exspectatus (median dS = 0.3).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank all members of the Sommer lab for helpful discussions. Moreover, we are grateful to Marco C. Betist for help with freezing the samples and to Judith Vivié and Mauro Muraro from Single Cell Discoveries for processing of the samples and library preparation. This work was funded by the research program (14NOISE01) of the Foundation for Fundamental Research on Matter (FOM) of the Netherlands Organization for Scientific Research (NWO) (A.E. and H.C.K.) and the Max Planck Society (C.R., D.R.S., M.O., and R.J.S).
Author Contributions
H.C.K., C.R., and R.J.S. performed conceptualization; A.E., D.R.S., M.O., R.J.S., and H.C.K. provided resources; C.R., A.E., D.R.S., and M.O. contributed to investigation; C.R. and A.E. performed data curation, formal analysis, and visualization; C.R. was responsible for the writing original draft; C.R., A.E., R.J.S., H.C.K., D.R.S., and M.O. were responsible for the writing review and editing; C.R. was responsible for project administration; C.R., H.C.K., and R.J.S. were responsible for supervision; and H.C.K. and R.J.S. were responsible for funding acquisition.
Sequencing data have been submitted to the European Nucleotide Archive under the study accession PRJEB37439.
References
- Baskaran P, Rödelsperger C, Prabh N, Serobyan V, Markov GV, Hirsekorn A, Dieterich C.. 2015. Ancient gene duplications have shaped developmental stage-specific expression in Pristionchus pacificus. BMC Evol Biol. 15:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento G, Ogawa A, Sommer RJ.. 2010. Co-option of the hormone-signalling module dafachronic acid-DAF-12 in nematode evolution. Nature 466(7305):494–497. [DOI] [PubMed] [Google Scholar]
- Blaxter M.1998. Caenorhabditis elegans is a nematode. Science 282(5396):2041–2046. [DOI] [PubMed] [Google Scholar]
- Bork P, Blomberg N, Nilges M.. 1996. Internal repeats in the BRCA2 protein sequence. Nat Genet. 13(1):22–23. [DOI] [PubMed] [Google Scholar]
- Buttery SM, Ekman GC, Seavy M, Stewart M, Roberts TM.. 2003. Dissection of the Ascaris sperm motility machinery identifies key proteins involved in major sperm protein-based amoeboid locomotion. Mol Biol Cell 14(12):5082–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. 2017. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357(6352):661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis A-R, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. 2012. Proto-genes and de novo gene birth. Nature 487(7407):370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Hartl DL.. 2002. Genome evolution and developmental constraint in Caenorhabditis elegans. Mol Biol Evol. 19(5):728–735. [DOI] [PubMed] [Google Scholar]
- Chatterjee I.2005. The Caenorhabditis elegans spe-38 gene encodes a novel four-pass integral membrane protein required for sperm function at fertilization. Development 132(12):2795–2808. [DOI] [PubMed] [Google Scholar]
- Checchi PM, Lawrence KS, Van MV, Larson BJ, Engebrecht J.. 2014. Pseudosynapsis and decreased stringency of meiotic repair pathway choice on the hemizygous sex chromosome of Caenorhabditis elegans males. Genetics 197(2):543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Brajković J, Tautz D.. 2007. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 23(11):533–539. [DOI] [PubMed] [Google Scholar]
- Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al. 2007. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol. 25(6):663–668. [DOI] [PubMed] [Google Scholar]
- Ebbing A, Vértesy Á, Betist MC, Spanjaard B, Junker JP, Berezikov E, van Oudenaarden A, Korswagen HC.. 2018. Spatial transcriptomics of C. elegans males and hermaphrodites identifies sex-specific differences in gene expression patterns. Dev Cell 47(6):801–813.e6. [DOI] [PubMed] [Google Scholar]
- Edgar RC.2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekseth OK, Kuiper M, Mironov V.. 2014. orthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics 30(5):734–736. [DOI] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47(D1):D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierst JL, Willis JH, Thomas CG, Wang W, Reynolds RM, Ahearne TE, Cutter AD, Phillips PC.. 2015. Reproductive mode and the evolution of genome size and structure in Caenorhabditis nematodes. PLoS Genet. 11(6):e1005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldziler B, Chatterjee I, Singson A.. 2005. The genetic and molecular analysis of spe-19, a gene required for sperm activation in Caenorhabditis elegans. Dev Biol. 283(2):424–436. [DOI] [PubMed] [Google Scholar]
- Gruidl ME, Smith PA, Kuznicki KA, McCrone JS, Kirchner J, Roussell DL, Strome S, Bennett KL.. 1996. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 93(24):13837–13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, et al. 2016. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, Yanai I.. 2012. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2(3):666–673. [DOI] [PubMed] [Google Scholar]
- Herrmann M, Kanzaki N, Weiler C, Yoshida K, Rödelsperger C, Sommer RJ.. 2019. Two new species of Pristionchus (Nematoda: Diplogastridae) include the gonochoristic sister species of P. fissidentatus. J Nematol. 51:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- Junker JP, Noël ES, Guryev V, Peterson KA, Shah G, Huisken J, McMahon AP, Berezikov E, Bakkers J, van Oudenaarden A.. 2014. Genome-wide RNA Tomography in the zebrafish embryo. Cell 159(3):662–675. [DOI] [PubMed] [Google Scholar]
- Kaessmann H.2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20(10):1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421(6920):231–237. [DOI] [PubMed] [Google Scholar]
- Kanzaki N, Tsai IJ, Tanaka R, Hunt VL, Liu D, Tsuyama K, Maeda Y, Namai S, Kumagai R, Tracey A, et al. 2018. Biology and genome of a newly discovered sibling species of Caenorhabditis elegans. Nat Commun. 9(1):3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimatis KR, Phillips PC.. 2018. Rapid gene family evolution of a nematode sperm protein despite sequence hyper-conservation. G3 (Bethesda) 8:353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling DM, Garza P, Nartey CM, Carvunis A-R.. 2019. The meanings of “function” in biology and the problematic case of de novo gene emergence. eLife 8:e47014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG.. 2009. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 25(9):404–413. [DOI] [PubMed] [Google Scholar]
- Kieninger MR, Ivers NA, Rödelsperger C, Markov GV, Sommer RJ, Ragsdale EJ.. 2016. The nuclear hormone receptor NHR-40 acts downstream of the sulfatase EUD-1 as part of a developmental plasticity switch in Pristionchus. Curr Biol. 26(16):2174–2179. [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Law E, Tang L, Rose AM.. 2002. The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr Biol. 12(24):2118–2123. [DOI] [PubMed] [Google Scholar]
- Knowles DG, McLysaght A.. 2009. Recent de novo origin of human protein-coding genes. Genome Res. 19(10):1752–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambshead PJD.1993. Recent developments in marine benthic biodiversity research. Oceanis 19:5–24. [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ.. 2006. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci U S A. 103(26):9935–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hernault SW.2006. Spermatogenesis. WormBook, ed. The C. elegans Research Community, WormBook. Available from: 10.1895/wormbook.1.85.1. [DOI] [Google Scholar]
- L’Hernault SW, Shakes DC, Ward S.. 1988. Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Genetics 120:435–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hernault SW, Singson AW.. 2000. Developmental genetics of spermatogenesis in the nematode Caenorhabditis elegans. In: The testis. New York: Springer. p. 109–119. [Google Scholar]
- Lightfoot JW, Chauhan VM, Aylott JW, Rödelsperger C.. 2016. Comparative transcriptomics of the nematode gut identifies global shifts in feeding mode and pathogen susceptibility. BMC Res Notes 9:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot JW, Wilecki M, Rödelsperger C, Moreno E, Susoy V, Witte H, Sommer RJ.. 2019. Small peptide–mediated self-recognition prevents cannibalism in predatory nematodes. Science 364(6435):86–89. [DOI] [PubMed] [Google Scholar]
- Lu MR, Lai C-K, Liao B-Y, Tsai IJ.. 2020. Comparative transcriptomics across nematode life cycles reveal gene expression conservation and correlated evolution in adjacent developmental stages. Genome Biol Evol. 12(7):1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov GV, Meyer JM, Panda O, Artyukhin AB, Claaßen M, Witte H, Schroeder FC, Sommer RJ.. 2016. Functional conservation and divergence of daf-22 paralogs in Pristionchus pacificus dauer development. Mol Biol Evol. 33(10):2506–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MG, Rödelsperger C, Witte H, Riebesell M, Sommer RJ.. 2015. The orphan gene dauerless regulates dauer development and intraspecific competition in nematodes by copy number variation. PLoS Genet. 11(6):e1005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Hurst LD.. 2016. Open questions in the study of de novo genes: what, how and why. Nat Rev Genet. 17(9):567–578. [DOI] [PubMed] [Google Scholar]
- McNally KLP, Fabritius AS, Ellefson ML, Flynn JR, Milan JA, McNally FJ.. 2012. Kinesin-1 prevents capture of the oocyte meiotic spindle by the sperm aster. Dev Cell 22(4):788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely PM, McGovern OL, Heinis FI, Yanowitz JL.. 2012. Crossover distribution and frequency are regulated by him-5 in Caenorhabditis elegans. Genetics 190(4):1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minniti AN, Sadler C, Ward S.. 1996. Genetic and molecular analysis of spe-27, a gene required for spermiogenesis in Caenorhabditis elegans hermaphrodites. Genetics 143(1):213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Finn RD, Eddy SR, Bateman A, Punta M.. 2013. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41(12):e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DZL, Sternberg PW, Inoue T.. 2015. Morphologically defined sub-stages of C. elegans vulval development in the fourth larval stage. BMC Dev Biol. 15:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E, Lenuzzi M, Rödelsperger C, Prabh N, Witte H, Roeseler W, Riebesell M, Sommer RJ.. 2018. DAF-19/RFX controls ciliogenesis and influences oxygen-induced social behaviors in Pristionchus pacificus. Evol Dev. 20(6):233–243. [DOI] [PubMed] [Google Scholar]
- Moreno E, Sieriebriennikov B, Witte H, Rödelsperger C, Lightfoot JW, Sommer RJ.. 2017. Regulation of hyperoxia-induced social behaviour in Pristionchus pacificus nematodes requires a novel cilia-mediated environmental input. Sci Rep. 7(1):17550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyers BA, Zhang J.. 2015. Phylostratigraphic bias creates spurious patterns of genome evolution. Mol Biol Evol. 32(1):258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, D, Koning EJP, et al. 2016. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3(4):385–394.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J, Minniti AN, Sadler C, Ward S.. 1999. spe-12 encodes a sperm cell surface protein that promotes spermiogenesis in Caenorhabditis elegans. Genetics 152(1):209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S, Goree J, Schedl T.. 2005. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 3(1):e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LS.1998. Disruption of a neuropeptide gene, flp-1, causes multiple behavioral defects in Caenorhabditis elegans. Science 281(5383):1686–1690. [DOI] [PubMed] [Google Scholar]
- Ohno S.1970. Evolution by gene duplication. New York: Springer. [Google Scholar]
- Prabh N, Rödelsperger C.. 2016. Are orphan genes protein-coding, prediction artifacts, or non-coding RNAs? BMC Bioinf. 17(1):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabh N, Rödelsperger C.. 2019. De novo, divergence, and mixed origin contribute to the emergence of orphan genes in Pristionchus nematodes. G3 (Bethesda) 9:2277–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabh N, Roeseler W, Witte H, Eberhardt G, Sommer RJ, Rödelsperger C.. 2018. Deep taxon sampling reveals the evolutionary dynamics of novel gene families in Pristionchus nematodes. Genome Res. 28(11):1664–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale EJ, Müller MR, Rödelsperger C, Sommer RJ.. 2013. A developmental switch coupled to the evolution of plasticity acts through a sulfatase. Cell 155(4):922–933. [DOI] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL.. 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 314(5):1041–1052. [DOI] [PubMed] [Google Scholar]
- Riebesell M, Sommer RJ.. 2017. Three-dimensional reconstruction of the pharyngeal gland cells in the predatory nematode Pristionchus pacificus. J Morphol. 278(12):1656–1666. [DOI] [PubMed] [Google Scholar]
- Rödelsperger C.2018. Comparative genomics of gene loss and gain in Caenorhabditis and other nematodes. Comp Genomics. 419–432. [DOI] [PubMed] [Google Scholar]
- Rödelsperger C, Athanasouli M, Lenuzzi M, Theska T, Sun S, Dardiry M, Wighard S, Hu W, Sharma DR, Han Z.. 2019. Crowdsourcing and the feasibility of manual gene annotation: a pilot study in the nematode Pristionchus pacificus. Sci Rep. 9(1):18789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C, Menden K, Serobyan V, Witte H, Baskaran P.. 2016. First insights into the nature and evolution of antisense transcription in nematodes. BMC Evol Biol. 16(1):165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C, Prabh N, Sommer RJ.. 2019. New gene origin and deep taxon phylogenomics: opportunities and challenges. Trends Genet. 35(12):914–922. [DOI] [PubMed] [Google Scholar]
- Rödelsperger C, Röseler W, Prabh N, Yoshida K, Weiler C, Herrmann M, Sommer RJ.. 2018. Phylotranscriptomics of Pristionchus nematodes reveals parallel gene loss in six hermaphroditic lineages. Curr Biol. 28(19):3123–3127.e5. [DOI] [PubMed] [Google Scholar]
- Rudel D, Riebesell M, Sommer RJ.. 2005. Gonadogenesis in Pristionchus pacificus and organ evolution: development, adult morphology and cell–cell interactions in the hermaphrodite gonad. Dev Biol. 277(1):200–221. [DOI] [PubMed] [Google Scholar]
- Schedl T, Kimble J.. 1988. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119(1):43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild-Prüfert K, Saito TT, Smolikov S, Gu Y, Hincapie M, Hill DE, Vidal M, McDonald K, Colaiácovo MP.. 2011. Organization of the synaptonemal complex during meiosis in Caenorhabditis elegans. Genetics 189(2):411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C.2015. Genes from scratch—the evolutionary fate of de novo genes. Trends Genet. 31(4):215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H, Hoeppner MP, Weiner J, Bornberg-Bauer E.. 2008. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology 213(3–4):237–250. [DOI] [PubMed] [Google Scholar]
- Sieriebriennikov B, Sun S, Lightfoot JW, Witte H, Moreno E, Rödelsperger C, Sommer RJ.. 2020. Conserved nuclear hormone receptors controlling a novel plastic trait target fast-evolving genes expressed in a single cell. PLoS Genet. 16(4):e1008687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slos D, Sudhaus W, Stevens L, Bert W, Blaxter M.. 2017. Caenorhabditis monodelphis sp. n.: defining the stem morphology and genomics of the genus Caenorhabditis. BMC Zool. 2:4. [Google Scholar]
- Smith CM, Hayamizu TF, Finger JH, Bello SM, McCright IJ, Xu J, Baldarelli RM, Beal JS, Campbell J, Corbani LE, et al. 2019. The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Res. 47(D1):D774–D779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RJ.1997. Evolution and development–the nematode vulva as a case study. BioEssays 19(3):225–231. [DOI] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthès P, Kokkinaki M, Nef S, Gnirke A, et al. 2013. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 3(6):2179–2190. [DOI] [PubMed] [Google Scholar]
- Spencer WC, Zeller G, Watson JD, Henz SR, Watkins KL, McWhirter RD, Petersen S, Sreedharan VT, Widmer C, Jo J, et al. 2011. A spatial and temporal map of C. elegans gene expression. Genome Res. 21(2):325–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1(2):E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L, Félix M-A, Beltran T, Braendle C, Caurcel C, Fausett S, Fitch D, Frézal L, Gosse C, Kaur T, et al. 2019. Comparative genomics of 10 new Caenorhabditis species. Evol Lett. 3(2):217–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T.2006. Maintenance of C. elegans. WormBook, ed. The C. elegans Research Community, WormBook. Available from: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD.. 2002. The rapid evolution of reproductive proteins. Nat Rev Genet. 3(2):137–144. [DOI] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T.. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12(10):692–702. [DOI] [PubMed] [Google Scholar]
- Toll-Riera M, Bosch N, Bellora N, Castelo R, Armengol L, Estivill X, Mar Alba M.. 2008. Origin of primate orphan genes: a comparative genomics approach. Mol Biol Evol. 26(3):603–612. [DOI] [PubMed] [Google Scholar]
- Tzur YB, Egydio de Carvalho C, Nadarajan S, Van Bostelen I, Gu Y, Chu DS, Cheeseman IM, Colaiácovo MP.. 2012. LAB-1 targets PP1 and restricts Aurora B kinase upon entrance into meiosis to promote sister chromatid cohesion. PLoS Biol. 10(8):e1001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur YB, Winter E, Gao J, Hashimshony T, Yanai I, Colaiácovo MP.. 2018. Spatiotemporal gene expression analysis of the germline uncovers a syncytial expression switch. Genetics 210(2):587–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakirlis N, Hebert AS, Opulente DA, Achaz G, Hittinger CT, Fischer G, Coon JJ, Lafontaine I.. 2018. A molecular portrait of de novo genes in yeasts. Mol Biol Evol. 35(3):631–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oss SB, Carvunis A-R.. 2019. De novo gene birth. PLoS Genet. 15(5):e1008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MS, Sieriebriennikov B, Loschko T, Namdeo S, Lenuzzi M, Dardiry M, Renahan T, Sharma DR, Sommer RJ.. 2017. Environmental influence on Pristionchus pacificus mouth form through different culture methods. Sci Rep. 7(1):7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MS, Sieriebriennikov B, Prabh N, Loschko T, Lanz C, Sommer RJ.. 2018. Young genes have distinct gene structure, epigenetic profiles, and transcriptional regulation. Genome Res. 28(11):1675–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilecki M, Lightfoot JW, Susoy V, Sommer RJ.. 2015. Predatory feeding behaviour in Pristionchus nematodes is dependent on phenotypic plasticity and induced by serotonin. J Exp Biol. 218(9):1306–1313. [DOI] [PubMed] [Google Scholar]
- Wissler L, Gadau J, Simola DF, Helmkampf M, Bornberg-Bauer E.. 2013. Mechanisms and dynamics of orphan gene emergence in insect genomes. Genome Biol Evol. 5(2):439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt E, Benjamin S, Svetec N, Zhao L.. 2019. Testis single-cell RNA-seq reveals the dynamics of de novo gene transcription and germline mutational bias in Drosophila. eLife 8:e47138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte H, Moreno E, Rödelsperger C, Kim J, Kim J-S, Streit A, Sommer RJ.. 2015. Gene inactivation using the CRISPR/Cas9 system in the nematode Pristionchus pacificus. Dev Genes Evol. 225(1):55–62. [DOI] [PubMed] [Google Scholar]
- Wolff ID, Tran MV, Mullen TJ, Villeneuve AM, Wignall SM.. 2016. Assembly of Caenorhabditis elegans acentrosomal spindles occurs without evident microtubule-organizing centers and requires microtubule sorting by KLP-18/kinesin-12 and MESP-1. Mol Biol Cell 27(20):3122–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA.2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8(3):206–216. [DOI] [PubMed] [Google Scholar]
- Wu J-C, Go AC, Samson M, Cintra T, Mirsoian S, Wu TF, Jow MM, Routman EJ, Chu DS.. 2012. Sperm development and motility are regulated by PP1 phosphatases in Caenorhabditis elegans. Genetics 190(1):143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Yan Y, Baron M, Wagner F, Barkley D, Chiodin M, Kim SY, Keefe DL, Alukal JP, Boeke JD, et al. 2020. Widespread transcriptional scanning in the testis modulates gene evolution rates. Cell 180(2):248–262.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yin D, Schwarz EM, Thomas CG, Felde RL, Korf IF, Cutter AD, Schartner CM, Ralston EJ, Meyer BJ, Haag ES.. 2018. Rapid genome shrinkage in a self-fertile nematode reveals sperm competition proteins. Science 359(6371):55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gao Y, Long M, Shen B.. 2019. Origination and evolution of orphan genes and de novo genes in the genome of Caenorhabditis elegans. Sci China Life Sci. 62(4):579–593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.