Abstract
Direct comparisons between historical and contemporary populations allow for detecting changes in genetic diversity through time and assessment of the impact of habitat fragmentation. Here, we determined the genetic architecture of both historical and modern lions to document changes in genetic diversity over the last century. We surveyed microsatellite and mitochondrial genome variation from 143 high-quality museum specimens of known provenance, allowing us to directly compare this information with data from several recently published nuclear and mitochondrial studies. Our results provide evidence for male-mediated gene flow and recent isolation of local subpopulations, likely due to habitat fragmentation. Nuclear markers showed a significant decrease in genetic diversity from the historical (HE = 0.833) to the modern (HE = 0.796) populations, whereas mitochondrial genetic diversity was maintained (Hd = 0.98 for both). Although the historical population appears to have been panmictic based on nDNA data, hierarchical structure analysis identified four tiers of genetic structure in modern populations and was able to detect most sampling locations. Mitogenome analyses identified four clusters: Southern, Mixed, Eastern, and Western and were consistent between modern and historically sampled haplotypes. Within the last century, habitat fragmentation caused lion subpopulations to become more geographically isolated as human expansion changed the African landscape. This resulted in an increase in fine-scale nuclear genetic structure and loss of genetic diversity as lion subpopulations became more differentiated, whereas mitochondrial structure and diversity were maintained over time.
Keywords: Keywords: lion, Panthera leo, mitogenome, nDNA, mtDNA, aDNA, spatiotemporal, historical DNA, habitat fragmentation, male-mediated gene flow, genetic diversity
Introduction
Effective, long-term conservation of wildlife populations benefits from knowledge of a species’ genetic history and the genetic variation that exists across its range. Coupled with an understanding of human-mediated landscape changes, knowledge of spatiotemporal trends in genetic diversity can help inform the management of wildlife populations (Martínez-Cruz et al. 2007; Spurgin et al. 2014; Casas-Marce et al. 2017).
Multiple published investigations document the genetic consequences of large-scale landscape changes over short periods of time (Martínez-Cruz et al. 2007; Tracy and Jamieson 2011; Spurgin et al. 2014; Borrell et al. 2018) (i.e., 100 years). Levels of genetic diversity are directly related to a species ability to adapt, survive, and thrive, and loss of genetic diversity can be detrimental to overall population health and long-term survival (Reed and Frankham 2003; Allendorf et al. 2013; Yoder et al. 2018; Leigh et al. 2019). However, without a baseline for comparison, it is difficult to assess what effect landscape changes in recent history may have on a species. With the recent advances in sequencing technology and methodology for isolating genetic material (DNA) from archived museum specimens, genetic information can now be accessed from both historical individuals and their contemporary counterparts. Combining these data sets provide a quantitative measure of changes in genetic diversity.
The exponential increase in the human population, urban development, and rural expansion (Cameron 1990) since the late-1800s has resulted in changes to the African landscape and fragmentation to once contiguous wildlife ranges (Newmark 2008; Riggio et al. 2015; Crooks et al. 2017). Many species across Africa are declining due to human-induced threats (Craigie et al. 2010; Ceballos et al. 2015, 2017), even in protected areas (Craigie et al. 2010). The adverse effects of low genetic diversity have been observed in small feline populations that exist in heavily managed fenced reserves (Trinkel et al. 2008; Johnson et al. 2010; Bertola et al. 2012; Creel et al. 2013; Kerk et al. 2019). The lion population has changed dramatically over the past 100 years (Chardonnet 2002; Barnett et al. 2006a; Riggio et al. 2015), particularly in terms of size and distribution, in response to habitat availability and anthropogenic pressures related to a growing human population (Wittemyer et al. 2008; Riggio et al. 2012; Blackburn et al. 2016; United Nations 2017).
Around the turn of the 19th century, explorers, naturalists, and hunters went on expeditions to collect biological specimens for preservation in natural history museums. These expeditions resulted in hundreds of lion specimens, being deposited in museums across the world, which predate the exponential human population growth across Africa (Cameron 1990; David 2011). With the continued development of techniques for improved isolation and sequencing of degraded genetic material (ancient DNA, aDNA), these collections now provide access to genetic information from historical as well as contemporary individuals.
Previous studies that sampled nuclear genetic diversity reported both high (Lyke et al. 2013; Morandin et al. 2014; Smitz et al. 2018) and low (Tende et al. 2014b; VanHooft et al. 2018; Curry et al. 2019) levels of gene flow, but this was largely dependent on the amount of connectivity present between sampling locations, for example, isolated populations such as those in the Kainji Lake National Park and Yankari Game Reserve in Nigeria (Tende et al. 2014b) and Kafue National Park and the Luangwa Valley Ecosystem in Zambia (Curry et al. 2019). Genetic differentiation can even be seen between populations within national parks (VanHooft et al. 2018). However, where there are no geographic or man-made barriers to limit movement, there is only weak evidence of population structure and high levels of gene flow (Morandin et al. 2014; Smitz et al. 2018).
Studies including historical and ancient lion samples have been primarily restricted to mtDNA analyses incorporated within a modern lion data set (Barnett et al. 2006a, 2006b, 2009, 2014, 2016). A recent study including historical individuals focused on assessing changes in the recent past but was confined to a local analysis of the Kavango–Zambezi (KAZA) transfrontier conservation area (Dures et al. 2019). Here, we report the first range-wide study assessing changes in genetic diversity of the lion, based on both historical and modern samples collected throughout Africa and India. By comparing diversity estimates from samples from different time periods, we can detect and evaluate changes in genetic diversity that have occurred during this time of landscape and anthropogenic change.
Results
Nuclear DNA
The modern data set (MD) contained 135 lions from 14 sampling locations collected from the 1990 to 2012 and the historical data set (HD) consisted of 143 lions dating prior to 1949 (table 1 and supplementary appendix S1, Supplementary Material online). Nine microsatellite loci (Leo006, Leo008, Leo085, Leo098, Leo126, Leo224, Leo230, Leo247, and Leo281) were shared between the two data sets. The MD had >75% allele calls reported, and the HD had an average of 90% amplification success across the nine loci. Sampling was similarly distributed across the lion range for both the MD and HD (fig. 1).
Table 1.
Historical Versus Modern Genetic Diversity for nDNA and mtDNA.
| Historical | Modern | Significance | Trend | |||
|---|---|---|---|---|---|---|
| nDNA (nine msat loci) | N | 143 | N | 135 | ↓ | |
| H E | 0.833 | H E | 0.796 | *** | ||
| A | 15 | A | 11.6 | *** | ||
| PA | 6.2 | PA | 1.2 | *** | ||
| M | 0.41 | M | 0.32 | *** | ||
| mtDNA (280 SNPs) | N | 102 | N | 19 | → | |
| s | 280 | s | 258 | |||
| π | 0.222 | π | 0.258 | n.s. | ||
| H d | 0.98 | H d | 0.98 | n.s. | ||
| H | 74 | H | 17 | |||
| PM | 22 | PM | 1 | |||
Note.—N, sample size; HE, expected heterozygosity; A, allelic richness; PA, private alleles; M, Garza–Williamson index; s, segregating sites; π, nucleotide diversity; Hd, haplotype diversity; H, number of haplotypes; PM, private mutations; SNP, single nucleotide polymorphism. Trend is based on statistical significance from a comparison of means. HE, A, PA, and M had a P value <0.005 (***) indicating a downward trend (↓) from historical to modern. The P value for π and Hd was >0.05 (n.s.) indicating maintained diversity (→) from historical to modern.
Fig. 1.
Map of lion sample locations. Dot size coincides with sample size for each location.
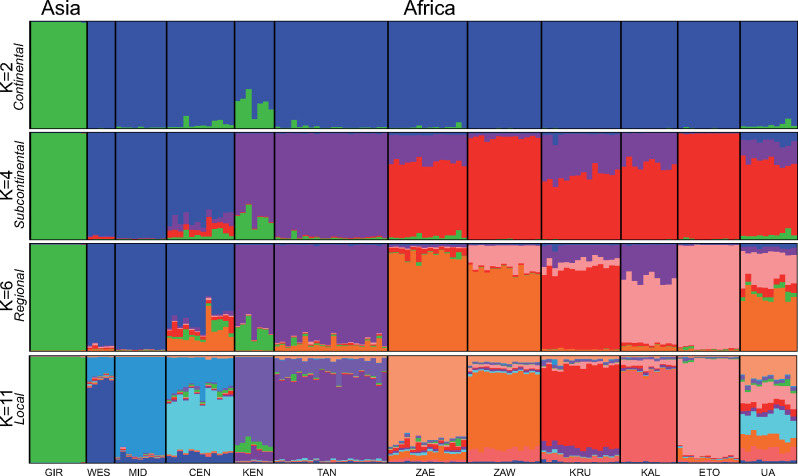
Fine-scale structure was observed in the MD but not the HD. The MD hierarchical structure analysis resulted in four tiers of structure: continental (K = 2), subcontinental (K = 4), regional (K = 6), and local (K = 11), as seen in the final analysis with location priors (fig. 2 and supplementary appendix S2.a, Supplementary Material online, shows a graphical display of the step-by-step hierarchical population structure). The initial run had a ΔK of two separating Asia (GIR) from Africa. The African population could then be further broken down into a Western, Eastern, and Southern population with a ΔK of three from structural analysis of only the African population. Analysis of the Western population also resulted a ΔK of three separating a West African (WES), Central African (CEN), and a population between the two (MID). The Eastern population had a ΔK of two separating lions in Kenya (KEN) from all lions sampled in Tanzania (TAN). The Southern population is separated into five local populations that can be grouped into three regional populations as ΔK was 5, however, there was a sizable peak also seen at K = 3. Eastern and western Zambia (ZAE and ZAW) make up a Southeast population, whereas Etosha and Kalahari (ETO and KAL) make up a Southwest population. Kruger was identified as a single population at both K = 3 and K = 5. Population clustering in the MD principal coordinate analysis (PCoA) follows the subcontinental tier (fig. 3). Mean heterozygosity across polymorphic loci (HE) is lowest in Gir and highest in CEN although only 44% of loci are polymorphic in this population (supplementary appendix S3, Supplementary Material online).
Fig. 2.
The four tiers of modern lion population structure as determined by hierarchical structure analysis based on nine microsatellites. Groups are colored based on figure 1.
Fig. 3.
Results of a PCoA of nine microsatellite loci analyzed in historical and modern lion samples.
Bayesian clustering in STRUCTURE did not identify any population structure in the HD. Although ΔK was also 2 for the initial run, individuals could not be assigned to meaningful populations (supplementary appendix S2.b, Supplementary Material online). Further evidence of this lack of structure was observed in the PCoA results (fig. 3). PCoA did not reveal any population clusters but did show weak evidence of isolation-by-distance (IBD; supplementary appendix S5, Supplementary Material online), indicating an admixed population.
A significant decrease (P value <0.005) from HD to MD was evident across diversity indices (table 1). Correcting for sample size through rarefaction, the HD has an allelic richness of 14.2 and private allelic richness of 4.6, higher than the MD at 11.3 and 1.7, respectively. The Garza–Williamson index (M) of the HD is 0.41 and the MD is 0.32.
Mitochondrial DNA
Results from mitochondrial analyses can be found in table 1. There was no significant difference found between mtDNA nucleotide diversity (π) and haplotype diversity (Hd). The historical population had 74 haplotypes with 22 private mutations, whereas the modern population had 17 haplotypes with only 1 private mutation. Two haplotypes (Hap_33 and Hap_66) are in both data sets. The low number of private mutations in the modern population was likely a result of the small number of mitogenomes compared with the historical population. Although we were able to obtain a large number of historical mitogenomes from museum samples, the number of modern mitogenomes was restricted to published data.
Mitochondrial genome analyses identified four major clades: Southern, Mixed, Eastern, and Western. Each clade was represented by at least one of the 19 modern lions. A Northern subclade was nested within the Western clade. Bootstrap values in the unrooted maximum likelihood (ML) tree (fig. 4) indicating strong support for these four clades. The four clusters in the principal component analysis (PCA, supplementary appendix S6, Supplementary Material online) and the four main branches of the haplotype network (supplementary appendix S7, Supplementary Material online) also support these same four clades.
Fig. 4.
ML tree based on 280 variable sites in 121 lion mitogenomes showing nodes with >70% bootstrap support. Black dots denote the nodes of the four major clades. Arcs indicate clade boundaries. The gray dot denotes the nested Gir Forest clade. Color corresponds to sampling location according to conventionally recognized regions in figure 1.
There are five conventionally recognized regions of Africa according to the United Nations geoscheme for Africa: Southern, Eastern, Western, Central, and Northern (https://unstats.un.org/unsd/methodology/m49/). The Southern clade includes the conventionally recognized regions of Southern Africa incorporating Botswana, South Africa, Namibia, and Zimbabwe. Haplotypes from Botswana and South Africa were present in both the Southern and the Mixed clades. The Mixed clade consists of haplotypes from the Southern, Eastern, and Western subcontinental groups.
The Western clade included countries in the conventionally recognized regions of Central and Western Africa including present-day Democratic Republic of Congo, Benin, Central African Republic, and Cameroon. The Eastern clade consists primarily of historical lions from British East Africa, specifically present-day Kenya, Tanzania and Uganda, and a modern lion from Somalia. Eastern haplotypes within the Western clade are from bordering countries suggesting gene flow between neighboring regions, which is in line with previously published patterns (Bertola et al. 2016).
The Mixed clade is intermediary, consisting of lions from Southern and Eastern Africa. Zambia and Malawi were found exclusively in the Mixed clade, whereas all other countries found in the Mixed clade are also within the Southern and/or Western clades. The historical samples from the Congo Region (present day Republic of the Congo and Gabon) were assigned to the Western subcontinental group by convention (fig. 1), but mitochondrial analyses consistently clustered them within the Southern and Mixed clades. The Congo Region is below the Congolese rainforests, a geographical barrier isolating it from West and Central Africa to the north. For the lion, the Congo Region is, therefore, closer to East and Southern Africa.
Discussion
A century ago, the lion population consisted of close-proximity prides with enough overlapping movement to appear panmictic. Our comparison of nDNA and mtDNA data between historical and modern samples indicates the presence of substantial historical male-mediated gene flow and evidence of recent isolation of local subpopulations likely due to habitat fragmentation.
Although lions currently exhibit fine-scale population structure, the historical population lacked any apparent structure, suggesting lions acted as a panmictic population only a century ago. This result was supported by structure analysis and PCoA. The modern populations cluster into four subcontinental groups, which are not recovered in the historical population, even between Asia and Africa.
Four tiers of modern nuclear genetic structure were identified through hierarchical structure analysis. Continental structure separated Asia from Africa. The subcontinental tier identified three main groups in Africa: Western, Eastern, and Southern. This is consistent with previous studies that also observed strong differentiation between Africa and Asia as well as subcontinental structure within Africa (Bertola et al. 2015, 2019; Manuel et al. 2020). Smitz et al. (2018) identified only two groups at the subcontinental scale; lack of identification of a Southern group was likely due to low sampling. The regional tier divides the Southern group into a Southwest, South, and Southeast group. The highest level of population structure was able to detect most sampling locations as in Antunes et al. (2008). Only sampling locations in Tanzania were unable to be individually identified, similar to findings of Smitz et al. (2018). Admixture was evident within local groups (fig. 2, K = 11) indicating recent gene flow. The UA group comprised individuals sampled within local groups that could not be assigned to a particular tier due to admixture. Other range-wide studies of lions have shown a similar localized structure pattern with individuals assigned to sampling populations with evidence of isolation-by-distance (Antunes et al. 2008; Dubach et al. 2013). Although several previous studies on lion phylogeography did include historical samples (Barnett et al. 2006a, 2014; Bertola et al. 2016; Manuel et al. 2020), they did not study the effect of sample age on derived population structure.
Habitat fragmentation restricts gene flow and often leads to loss of genetic diversity (Fahrig 2003; Delaney et al. 2010). The KA–ZA transfrontier conservation area has seen a decrease in genetic and allelic diversity over the past century (Dures et al. 2019). Our study showed that, range-wide, there has been a significant decrease in nDNA diversity from the historical to the modern lion population (table 1). Expected heterozygosity, allelic richness, and number of private alleles have all significantly decreased (P value <0.0005). Although there is evidence of a recent reduction in population size in both the historical and modern populations, both displaying an M value <0.67 (Garza and Williamson 2001; table 1), M is significantly lower in the modern population. This indicates that the reduction predates Africa’s exponential human population growth, but the reduction has increased in the past century.
Given the strong signal differentiating the Asiatic and African lions in the analysis of modern populations, we predicted the historical lions from the Gir Forest National Park (NP) in India would cluster independently of the historical African lions. The PCoA, however, showed that the Gir Forest lions cluster together in the center of a single historical lion cluster (fig. 3). Lions were at the brink of extinction in Asia at the beginning of the 20th century (Singh and Gibson 2011) when these samples were collected (1906–1929). Today there are over 500 lions in the Gir Forest NP (Jhala et al. 2019). The historical and modern samples from the Gir Forest NP were collected before the peak of a recent bottleneck and its subsequent population restoration. Our comparisons document this severe bottleneck that resulted in low genetic diversity in Asia compared with Africa (supplementary appendix S3, Supplementary Material online) and demonstrate how diversity shared with the rest of the population is lost to a bottleneck. Habitat fragmentation leading to the isolation of subpopulations within Africa appears to be following the same trend as the Asiatic lion a century ago.
Historical and modern mtDNA show strikingly different patterns than nuclear data. Although nuclear diversity has decreased significantly, mtDNA diversity has remained constant over time (table 1). mtDNA is matrilineally inherited, and localized studies show that there is little or no female-mediated gene flow between subpopulations across Africa (Tende et al. 2014a; Curry et al. 2015, 2019). Female lions primarily remain with their natal pride, whereas males disperse (Pusey et al. 1987; Spong and Creel 2001). Therefore, pride structure can dictate mtDNA population structure. With females remaining close to their natal prides, habitat fragmentation will not greatly alter pride structure, thereby keeping mtDNA diversity constant over time. Accordingly, the distribution of modern haplotypes within clades (N = 17) is geographically consistent with historical haplotypes (N = 74). The four major clades (Southern, Mixed, Eastern, and Western) geographically follow the subcontinental groups identified by nDNA analysis (Southern, Eastern, Western, and Northern).
The Western clade includes lions from West and Central Africa as well as Asiatic lions. Previous studies have suggested that the Asiatic, West, and Central African lions should be grouped taxonomically (Bauer et al. 2015; Bertola et al. 2016). United States Fish and Wildlife Services recently updated the lion taxonomy under the Endangered Species Act to recognize these populations as the same subspecies, Panthera leo leo, and East and Southern Africa populations as Panthera leo melanochaita (USFWS 2015). Our mitogenome results support this dichotomy, placing the Gir Forest NP lions within the Western clade in all analyses.
If unobstructed by geographic or artificial barriers, a male lion’s home range can be hundreds to thousands of kilometer square (Stander 2006; Ngwenya et al. 2013; Gatta 2016) and can span different habitats (Bauer et al. 2003; Lehmann et al. 2008; Loveridge et al. 2009). However, the original range of the lion has been severely reduced as a direct result of the growing human population (Riggio et al. 2012) and changes in land-use, such as expansion of large-scale cultivation and increased movement of livestock into protected areas (Tumenta et al. 2013; Masanja 2014; Sogbohossou et al. 2014). As lion habitat has become more fragmented and groups of prides become more isolated, gene flow is restricted, and subpopulations become more genetically distinguishable. The dichotomy between the historic nuclear and mitochondrial structure is indicative of male-mediated gene flow and female philopatry. This pattern is not as evident in the modern population because fragmentation has hindered the ability of males to migrate between increasingly isolated subpopulations.
The differences evident between the historical and modern lion populations have several important conservation implications. If left unattended, these subpopulations could become completely isolated leading to further differentiation and reduction in genetic diversity. Managing a species as a continuous population without a continuous habitat requires considerable resources. Lions currently reside in 28 countries whose different approaches to wildlife policy could complicate range-wide management (Nyhus 2016) and act as additional artificial barriers. Cooperative international management would be needed to restore historical levels of connectivity. Currently, the African Lion Working Group recommends using regional guidelines for sourcing lions for translocations (ALWG 2016). Although mtDNA structure should still be considered, strict guidelines dictated by nDNA genetic similarities within regional populations may not be as critical for maintaining the population’s genetic diversity if the goal is to reflect historical levels of gene flow. Rather, natural dispersal capability should be used as a guideline for selecting suitable source and target populations, although long-distance translocations, especially transcontinental translocations, are not recommended.
Connectivity is critical to enable gene flow between subpopulations to avoid the erosion of genetic diversity (Keyghobadi 2007). As an iconic flagship species, these results expose the influence of habitat fragmentation, potentially affecting hundreds of other species. We already are observing the initial effects of this fragmentation on lions through increased nDNA structure and decreased nDNA diversity. Science-based management policies and informed stewardship can help mitigate the loss of nDNA diversity and continued preservation of mtDNA diversity. Intervention, ideally in the form of restoring and protecting natural wildlife corridors, is needed to increase gene flow between lion subpopulations and reduce the effects of habitat fragmentation.
Materials and Methods
Nuclear Analysis
Biological material from 162 lions dating prior to 1949 was collected from museums (fig. 1, supplementary appendix S9, Supplementary Material online) in the form of bone fragments, whole teeth or tooth fragments, nasal turbinate bones, and/or dried tissue. Microsatellite amplification was performed following protocols and procedures described in Curry and Derr (2019) using their panel of microsatellite loci designed using historical samples having minimal linkage disequilibrium. Nine microsatellite loci (Leo006, Leo008, Leo085, Leo098, Leo126, Leo224, Leo230, Leo247, and Leo281) that had >75% allele call coverage across both the MD and the HD for all loci were employed in the final analyses. Only lions with known sampling date and location and >70% amplification success were used in downstream analyses. Sample preparation, DNA extraction and storage, PCR amplification, allele calling, and call verification followed protocols described in Curry and Derr (2019). Further details are found in supplementary appendix S11, Supplementary Material online.
The MD consists of microsatellite allele calls from Bertola et al. (2015) (MD-1), Driscoll et al. (2002) (MD-2), and Curry et al. (2019) (MD-3). Six additional lions were included from the African Wildlife Genomics collection at Texas A&M University (MD-4). These data sets were combined to expand sample size and range for structure analysis and population statistics as well as for direct comparison with the HD. Data calibration is needed when combining microsatellite allele calls from different studies (Ellis et al. 2011). Details on calibration of allele calls can be found in supplementary appendix S10, Supplementary Material online.
Nuclear diversity calculations were done using Arlequin v3.5 (Excoffier et al. 2005), GenePop (Rousset 2008), HPRare (Kalinowski 2005), and GenAlEx v6.5 (Peakall and Smouse 2012). MD and HD were analyzed separately, and results were then compared. A comparison of means was used to determine statistical significance of differences between historical to modern metrics.
Knowing that population structure has been found regionally (Antunes et al. 2008; Morandin et al. 2014; Miller et al. 2015; Bertola et al. 2016; Tensen 2016; VanHooft et al. 2018; Smitz et al. 2018), we implemented a hierarchical strategy to uncover any hidden structure that may be lost when subpopulations are analyzed together (Coulon et al. 2008; Noss 1990; Balkenhol et al. 2014). STRUCTURE runs were performed on each of the full MD and HD data sets without priors for 15 iterations of K 1–15 for 100,000 Markov chain Monte Carlo replications with 10% burn-in. STRUCTURE was rerun for each population as determined through ΔK values from STRUCTURE HARVESTER (Earl and vonHoldt 2012) with individuals assigned to populations based on Q scores from runs combined in CLUMPP (Jakobsson and Rosenberg 2007). To determine structural tiers, this was continued until no additional population structure was found. Samples were assigned to the finest level of structure then run as a full population with location priors for 15 iterations of K 1–12. Runs were combined using CLUMPP (Jakobsson and Rosenberg 2007) and visualized at each tier using DISTRUCT (Rosenberg 2004). To further look at structure patterns, a mantel test for IBD and PCoA were performed in GenAlEx v6.5 (Peakall and Smouse 2012).
Mitochondrial Analysis
Only polymorphic sites found in the sequences generated in this study were used for downstream analyses. Conservative filtering was implemented to accommodate the higher error rate associated with DNA damage possible in older samples (Shapiro and Hofreiter 2012; Templeton et al. 2013; Gorden et al. 2018). However, conservative filtering in the historical mitogenomes may increase false negative variation present in the published, modern mitogenomes. Therefore, polymorphic sites found only in the published mitogenomes were excluded to reduce potential biases produced by differences in sequencing between studies.
Whole mitogenomes were assembled based on whole-genome sequencing of 155 samples (152 historical and 3 modern). Details on whole-genome sequencing, quality filtering, and single nucleotide polymorphism identification can be found in supplementary appendix S11, Supplementary Material online. After filtering, 102 historical lions and 3 modern lions were of sufficient quality for downstream analyses (NCBI Bioproject: PRJNA602714). Sixteen additional modern lion sequences from GenBank (KP001493–KP001506 [Bertola et al. 2016], KP202262 [Davis et al. 2010], KC834784 [Bagatharia et al. 2013]) were added for a total of 19 modern lions.
Mitochondrial diversity analyses of the multiple sequence alignment of 280 polymorphic sites were performed using PLINK v1.9 (Purcell et al. 2007), Arlequin v3.5 (Excoffier et al. 2005), and DnaSP v6 (Rozas et al. 2017). PCA was performed using R package SNPRelate (Zheng et al. 2012) through calculation of eigenvectors and visualized using the plot3D function in the rgl R package. A median-joining haplotype network was created using POPArt (Bandelt et al. 1999) and an unrooted ML tree was inferred in RAxML using a rapid bootstrap with 1000 replicates evaluated under the GTR + GAMMA + I substitution model (Stamatakis 2014).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We extend our deepest appreciation to the natural history museums that allowed us to sample their collections: American Museum of Natural History, Carnegie Museum of Natural History, Field Museum of Natural History, Kansas University Natural History Museum, Natural History Museum of Los Angeles County, Naturalis Biodiversity Center, Royal Belgian Institute of Natural Sciences, Swedish Royal Museum of Natural History, The Museum of Vertebrate Zoology at Berkeley, Yale Peabody Museum, and Zoological Museum Amsterdam. Additionally, we are grateful to Dr Hans de Iongh and Dr Klaas Vrieling for their assistance in the procurement of data and samples. A special thank you goes to the laboratory personnel at the DNA Technologies Core Laboratory at Texas A&M University for their assistance during laboratory procedures and analysis. This project was made possible in part by the Texas A&M University College of Veterinary Medicine Trainee Grant, Dallas Safari Club, Dallas Safari Club Foundation, Safari Club International Foundation, Houston Safari Club Foundation Dan L. Duncan Scholarship Award Program, the Explorer’s Club Exploration Fund, the Boore Family Foundation, Curry Family donations, and all the generous backers of the Experiment.com Cat Challenge.
References
- Allendorf FW, Luikart G, Aitken SN.. 2013. Conservation and the genetics of populations. 2nd ed. West Sussex (UK: ): John Wiley & Sons, Ltd. [Google Scholar]
- ALWG. 2016. Proposed ALWG statement on genetic considerations for translocations involving African lions. Available from: https://www.africanliongroup.org/resources. Accessed July 20, 2020.
- Antunes A, Troyer JL, Roelke ME, Pecon-Slattery J, Packer C, Winterbach C, Winterbach H, Hemson G, Frank L, Stander P, et al. 2008. The evolutionary dynamics of the lion Panthera leo revealed by host and viral population genomics. PLoS Genet. 4(11):e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagatharia SB, Joshi MN, Pandya RV, Pandit AS, Patel RP, Desai SM, Sharma A, Panchal O, Jasmani FP, Saxena AK.. 2013. Complete mitogenome of Asiatic lion resolves phylogenetic status within Panthera. BMC Genomics. 14(1):572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkenhol N, Holbrook JD, Onorato D, Zager P, White C, Waits LP.. 2014. A multi-method approach for analyzing hierarchical genetic structures: a case study with cougars Puma concolor. Ecography (Cop.) 37(6):552–563. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A.. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16(1):37–48. [DOI] [PubMed] [Google Scholar]
- Barnett R, Shapiro B, Barnes I, Ho SYW, Burger J, Yamaguchi N, Higham TFG, Wheeler HT, Rosendahl W, Sher AV, et al. 2009. Phylogeography of lions (Panthera leo ssp.) reveals three distinct taxa and a late Pleistocene reduction in genetic diversity. Mol Ecol. 18(8):1668–1677. [DOI] [PubMed] [Google Scholar]
- Barnett R, Yamaguchi N, Barnes I, Cooper A.. 2006. a. The origin, current diversity and future conservation of the modern lion (Panthera leo). Proc R Soc B. 273(1598):2119–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett R, Yamaguchi N, Barnes I, Cooper A.. 2006. b. Lost populations and preserving genetic diversity in the lion Panthera leo: implications for its ex situ conservation. Conserv Genet. 7(4):507–514. [Google Scholar]
- Barnett R, Yamaguchi N, Shapiro B, Ho SYW, Barnes I, Sabin R, Werdelin L, Cuisin J, Larson G.. 2014. Revealing the maternal demographic history of Panthera leo using ancient DNA and a spatially explicit genealogical analysis. BMC Evol Biol. 14(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Chapron G, Nowell K, Henschel P, Funston P, Hunter LTB, Macdonald DW, Packer C.. 2015. Lion (Panthera leo) populations are declining rapidly across Africa, except in intensively managed areas. Proc Natl Acad Sci U S A. 112(48):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, de Iongh HH, Di Silvestre I.. 2003. Lion (Panthera leo) social behaviour in the West and Central African savannah belt. Mamm Biol. 68(4):239–243. [Google Scholar]
- Bertola LD, Jongbloed H, Gaag KJ, Van Der Knijff PD, Yamaguchi N, Hooghiemstra H, Henschel P, White PA, Driscoll CA, Tende T, et al. 2016. Phylogeographic patterns in Africa and high resolution delineation of genetic clades in the lion (Panthera leo). Sci Rep. 6(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertola LD, Tensen L, van Hooft P, White PA, Driscoll CA, Henschel P, Caragiulo A, Dias-Freedman I, Sogbohossou EA, Tumenta PN, et al. 2015. Autosomal and mtDNA markers affirm the distinctiveness of lions in west and central Africa. PLoS One. 10(10):e0137975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertola LD, Vermaat M, Lesilau F, Chege M, Tumenta P, Sogbohossou E, Bauer H, Patterson B, White P, de Iongh H, et al. 2019. Whole genome sequencing and the application of a SNP panel reveal primary evolutionary lineages and genomic diversity in the lion (Panthera leo). BioRxiv 1–28. 10.1101/814103. [DOI] [PMC free article] [PubMed]
- Bertola LD, Vrieling K, de Iongh HH.. 2012. Conservation genetics of the lion: new approaches to species conservation. Genetic diversity: new research. United Kingdom: Nova Biomedical. p. 61–82.
- Blackburn S, Hopcraft JGC, Ogutu JO, Matthiopoulos J, Frank L, Singh N.. 2016. Human-wildlife conflict, benefit sharing and the survival of lions in pastoralist community-based conservancies. J Appl Ecol. 53(4):1195–1205. [Google Scholar]
- Borrell JS, Wang N, Nichols RA, Buggs R.. 2018. Genetic diversity maintained among fragmented populations of a tree undergoing range contraction. Heredity (Edinburgh) 121(4):304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron R.1990. A concise economic history of the world: from paleolithic times to the present. New York: Oxford University Press. [Google Scholar]
- Casas-Marce M, Marmesat E, Soriano L, Martínez-Cruz B, Lucena-Perez M, Nocete F, Rodríguez-Hidalgo A, Canals A, Nadal J, Detry C, et al. 2017. Spatiotemporal dynamics of genetic variation in the Iberian lynx along its path to extinction reconstructed with ancient DNA. Mol Biol Evol. 34(11):2893–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM.. 2015. Accelerated modern human-induced species losses: entering the sixth mass extinction. Sci Adv. 1(5):e1400253.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos G, Ehrlich PR, Dirzo R.. 2017. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proc Natl Acad Sci U S A. 114(30):E6089–E6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardonnet P.2002. Conservation of the African lion: contribution to a status survey. International Foundation for the Conservation of Wildlife. Fr Conserv Force, USA (September). p. 102–171. [Google Scholar]
- Coulon A, Fitzpatrick JW, Bowman R, Stith BM, Makarewich CA, Stenzler LM, Lovette IJ.. 2008. Congruent population structure inferred from dispersal behaviour and intensive genetic surveys of the threatened Florida scrub-jay (Aphelocoma cœrulescens). Mol Ecol. 17(7):1685–1701. [DOI] [PubMed] [Google Scholar]
- Craigie ID, Baillie JEM, Balmford A, Carbone C, Collen B, Green RE, Hutton JM.. 2010. Large mammal population declines in Africa’s protected areas. Biol Conserv. 143(9):2221–2228. [Google Scholar]
- Creel S, Becker MS, Durant SM, M’Soka J, Matandiko W, Dickman AJ, Christianson D, Dröge E, Mweetwa T, Pettorelli N, et al. 2013. Conserving large populations of lions—the argument for fences has holes. Ecol Lett. 16(11):1413. [DOI] [PubMed] [Google Scholar]
- Crooks KR, Burdett CL, Theobald DM, King SRB, Di Marco M, Rondinini C, Boitani L.. 2017. Quantification of habitat fragmentation reveals extinction risk in terrestrial mammals. Proc Natl Acad Sci U S A. 114(29):7635–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry CJ, Derr JN.. 2019. Development of lion MiniSTRs for use with modern and historical DNA samples. African J Wildl Res. 49(1):64–74. [Google Scholar]
- Curry CJ, White PA, Derr JN.. 2015. Mitochondrial haplotype diversity in Zambian lions: bridging a gap in the biogeography of an iconic species. PLoS One. 10(12):e0143827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry CJ, White PA, Derr JN.. 2019. Genetic analysis of African lions (Panthera leo) in Zambia support movement across anthropogenic and geographical barriers. PLoS One. 14(5):e0217179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S.2011. Slavery and the “Scramble for Africa.” BBC Hist. Available from: http://www.bbc.co.uk/history/british/abolition/scramble_for_africa_article_01.shtml
- Davis BW, Li G, Murphy WJ.. 2010. Supermatrix and species tree methods resolve phylogenetic relationships within the big cats, Panthera (Carnivora: Felidae). Mol Phylogenet Evol. 56(1):64–76. [DOI] [PubMed] [Google Scholar]
- Delaney KS, Riley SPD, Fisher RN.. 2010. A rapid, strong, and convergent genetic response to urban habitat fragmentation in four divergent and widespread vertebrates. PLoS One. 5(9):e12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll CA, Menotti-Raymond M, Nelson G, Goldstein D, O’Brien SJ.. 2002. Genomic microsatellites as evolutionary chronometers: a test in wild cats. Genome Res. 12(3):414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubach JM, Briggs MB, White PA, Ament BA, Patterson BD.. 2013. Genetic perspectives on “Lion Conservation Units” in Eastern and Southern Africa. Conserv Genet. 14(4):741–755. [Google Scholar]
- Dures SG, Carbone C, Loveridge AJ, Maude G, Midlane N, Aschenborn O, Gottelli D.. 2019. A century of decline: loss of genetic diversity in a southern African lion-conservation stronghold. Divers Distrib. 25(6):870–810. [Google Scholar]
- Earl DA, vonHoldt BM.. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 4(2):359–361. [Google Scholar]
- Ellis JS, Gilbey J, Armstrong A, Balstad T, Cauwelier E, Cherbonnel C, Consuegra S, Coughlan J, Cross TF, Crozier W, et al. 2011. Microsatellite standardization and evaluation of genotyping error in a large multi-partner research programme for conservation of Atlantic salmon (Salmo salar L.). Genetica 139(3):353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S.. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 1:117693430500100–117693430500150. [PMC free article] [PubMed] [Google Scholar]
- Fahrig L.2003. Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst. 34(1):487–515. [Google Scholar]
- Garza JC, Williamson EG.. 2001. Detection of reduction in population size using data from microsatellite loci. Mol Ecol. 10(2):305–318. [DOI] [PubMed] [Google Scholar]
- Gatta M.2016. Population structure, home ranges and movement of Nairobi National Park lions (Panthera leo melanochaita) in relation to livestock depredation [MSc Thesis]. Leiden, Netherlands: Leiden University.
- Gorden EM, Sturk-Andreaggi K, Marshall C.. 2018. Repair of DNA damage caused by cytosine deamination in mitochondrial DNA of forensic case samples. Forensic Sci Int Genet. 34(February):257–264. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA.. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(14):1801–1806. [DOI] [PubMed] [Google Scholar]
- Jhala YV, Banerjee K, Chakrabarti S, Basu P, Singh K, Dave C, Gogoi K.. 2019. Asiatic lion: ecology, economics, and politics of conservation. Front Ecol Evol. 7(Aug):1–21. [Google Scholar]
- Johnson WE, Onorato DP, Roelke ME, Land ED, Cunningham M, Belden RC, McBride R, Jansen D, Lotz M, Shindle D, et al. 2010. Genetic restoration of the Florida panther. Science 329(5999):1641–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST.2005. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes. 5(1):187–189. [Google Scholar]
- Kerk M, Onorato DP, Hostetler JA, Bolker BM, Oli MK.. 2019. Dynamics, persistence, and genetic management of the endangered Florida panther population. Wildl Monogr. 203(1):3–35. [Google Scholar]
- Keyghobadi N.2007. The genetic implications of habitat fragmentation for animals. Can J Zool. 85(10):1049–1064. [Google Scholar]
- Lehmann MB, Funston PJ, Owen CR, Slotow R.. 2008. Home range utilisation and territorial behaviour of lions (Panthera leo) on Karongwe Game Reserve, South Africa. PLoS One. 3(12):e3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh DM, Hendry AP, Vázquez-Domínguez E, Friesen VL.. 2019. Estimated six per cent loss of genetic variation in wild populations since the industrial revolution. Evol Appl. 12(8):1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveridge AJ, Valeix M, Davidson Z, Murindagomo F, Macdonald DW.. 2009. Changes in home range size of African lions in relation to pride size and prey biomass in a semi-arid savanna. Ecography 32(6):953–962. [Google Scholar]
- Lyke MM, Dubach J, Briggs MB.. 2013. A molecular analysis of African lion (Panthera leo) mating structure and extra-group paternity in Etosha National Park. Mol Ecol. 22(10):2787–2796. [DOI] [PubMed] [Google Scholar]
- Manuel MD, Barnett R, Sandoval-Velasco M, Yamaguchi N, Vieira FG, Mendoza LZ, Shiping L, Martin MD, Sinding M-H, Mak SST, et al. 2020. The evolutionary history of extinct and living lions. Proc Natl Acad Sci U S A. 117(20):10927–10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cruz B, Godoy JA, Negro JJ.. 2007. Population fragmentation leads to spatial and temporal genetic structure in the endangered Spanish imperial eagle. Mol Ecol. 16(3):477–486. [DOI] [PubMed] [Google Scholar]
- Masanja GF.2014. Human population growth and wildlife extinction in Ugalla ecosystem, Western Tanzania. J Sustain Dev Stud. 5(2):192–217. [Google Scholar]
- Miller SM, Harper CK, Bloomer P, Hofmeyr J, Funston PJ.. 2015. Fenced and fragmented: conservation value of managed metapopulations. PLoS One. 10(12):e0144605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandin C, Loveridge AJ, Segelbacher G, Elliot N, Madzikanda H, Macdonald DW, Höglund J.. 2014. Gene flow and immigration: genetic diversity and population structure of lions (Panthera leo) in Hwange National Park, Zimbabwe. Conserv Genet. 15(3):697–706. [Google Scholar]
- Newmark WD.2008. Isolation of African protected areas. Front Ecol Environ. 6(6):321–328. [Google Scholar]
- Ngwenya MM, Mkandhla NM, Muzvondiwa JV.. 2013. A comparison of lion home range sizes in two management blocks in a semi-arid savannah National Park of Zimbabwe. Int J Sci Res. 2(2):388–396. [Google Scholar]
- Noss RF.1990. Indicators for monitoring biodiversity : a hierarchical approach. Conserv Biol. 4(4):355–364. [Google Scholar]
- Nyhus PJ.2016. Human–wildlife conflict and coexistence. Annu Rev Environ Resour. 41(1):143–171. [Google Scholar]
- Peakall R, Smouse PE.. 2012. GenALEx 6.5: genetic analysis in excel. Population genetic software for teaching and research: an update. Bioinformatics 28(19):2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusey AE, Packer C, Erhoff-Mulder MB.. 1987. The evolution of sex-biased dispersal in lions. Behaviour 101(4):275–310. [Google Scholar]
- Reed DH, Frankham R.. 2003. Correlation between fitness and genetic diversity. Conserv Biol. 17(1):230–237. [Google Scholar]
- Riggio J, Caro T, Dollar L, Durant SM, Jacobson AP, Kiffner C, Pimm SL, van Aarde RJ.. 2015. Lion populations may be declining in Africa but not as Bauer et al. suggest. Proc Natl Acad Sci U S A. 113(2):201521506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggio J, Jacobson A, Dollar L, Bauer H, Becker M, Dickman A, Funston P, Groom R, Henschel P, Iongh HD, et al. 2012. The size of savannah Africa: a lion’s (Panthera leo) view. Biodivers Conserv. 22(1):17–35. [Google Scholar]
- Rosenberg NA.2004. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 4(1):137–138. [Google Scholar]
- Rousset F.2008. GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 8(1):103–106. [DOI] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sanchez-Gracia A.. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 34(12):3299–3302. [DOI] [PubMed] [Google Scholar]
- Shapiro B, Hofreiter M.. 2012. Ancient DNA. New York: Springer Science + Business Media, LLC. [Google Scholar]
- Singh HS, Gibson L.. 2011. A conservation success story in the otherwise dire megafauna extinction crisis: the Asiatic lion (Panthera leo persica) of Gir forest. Biol Conserv. 144(5):1753–1757. [Google Scholar]
- Smitz N, Jouvenet O, Ligate FA, Crosmary W-G, Ikanda D, Chardonnet P, Fusari A, Meganck K, Gillet F, Melletti M, et al. 2018. A genome-wide data assessment of the African lion (Panthera leo) population genetic structure in Tanzania. PLoS One. 13(11):e0205395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogbohossou EA, Bauer H, Loveridge A, Funston PJ, De Snoo GR, Sinsin B, De Iongh HH.. 2014. Social structure of lions (Panthera leo) is affected by management in Pendjari Biosphere Reserve, Benin. PLoS One. 9(1):e84674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spong G, Creel S.. 2001. Deriving dispersal distances from genetic data. Proc R Soc Lond B. 268(1485):2571–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgin LG, Wright DJ, van der Velde M, Collar NJ, Komdeur J, Burke T, Richardson DS.. 2014. Museum DNA reveals the demographic history of the endangered Seychelles warbler. Evol Appl. 7(9):1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stander PE.2006. Population ecology and demography of Kunene Lions January 2006. Research Paper: 2006/1.
- Templeton JEL, Brotherton PM, Llamas B, Soubrier J, Haak W, Cooper A, Austin JJ.. 2013. DNA capture and next-generation sequencing can recover whole mitochondrial genomes from highly degraded samples for human identification. Invest Genet. 4(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tende T, Bensch S, Ottosson U, Hansson B.. 2014. a. Dual phylogenetic origins of Nigerian lions (Panthera leo). Ecol Evol. 4(13):2668–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tende T, Hansson B, Ottosson U, Akesson M, Bensch S.. 2014. b. Individual identification and genetic variation of lions (Panthera leo) from two protected areas in Nigeria. PLoS One. 9(1):e84288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tensen L.2016. Under what circumstances can wildlife farming benefit species conservation? Glob Ecol Conserv. 6:286–298. [Google Scholar]
- Tracy LN, Jamieson IG.. 2011. Historic DNA reveals contemporary population structure results from anthropogenic effects, not pre-fragmentation patterns. Conserv Genet. 12(2):517–526. [Google Scholar]
- Trinkel M, Ferguson N, Reid A, Reid C, Somers M, Turelli L, Graf J, Szykman M, Cooper DJ, Haverman P, et al. 2008. Translocating lions into an inbred lion population in the Hluhluwe-iMfolozi Park, South Africa. Anim Conserv. 11(2):138–143. [Google Scholar]
- Tumenta PN, van’t Zelfde M, Croes BM, Buij R, Funston PJ, Udo de Haes HA, De Iongh HH.. 2013. Changes in lion (Panthera leo) home range size in Waza National Park, Cameroon. Mamm Biol. 78(6):461–469. [Google Scholar]
- United Nations. 2017. World population prospects: the 2017 revision, key findings and advance tables. Report No.: ESA/P/WP/248. New York: United Nations.
- USFWS. 2015. 50 CFR Part 17. Endangered and Threatened Wildlifeand Plants; Listing Two Lion Subspecies. Available from: https://www.govinfo.gov/content/pkg/FR-2015-12-23/pdf/2015-31958.pdf. Accessed July 20, 2020.
- VanHooft P, Keet DF, Brebner DK, Bastos ADS, Hooft PV, Keet DF, Brebner DK, Bastos ADS, van Hooft P, Keet DF, et al. 2018. Genetic insights into dispersal distance and disperser fitness of African lions (Panthera leo) from the latitudinal extremes of the Kruger National Park, South Africa. BMC Genet. 19(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittemyer G, Elsen P, Bean WT, Burton AC, Brashares JS.. 2008. Growth at protected area edges. Science 321(5885):123–126. [DOI] [PubMed] [Google Scholar]
- Yoder AD, Poelstra JW, Tiley GP, Williams RC, Kumar S.. 2018. Neutral theory is the foundation of conservation genetics. Mol Biol Evol. 35(6):1322–1326. [DOI] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS.. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28(24):3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.