Abstract
Tandem repeats of the pentanucleotide 5′-CCGNN (where N indicates any base) were previously shown to exclude nucleosomes in vitro (Y.-H. Wang and J. D. Griffith, Proc. Natl. Acad. Sci. USA 93:8863–8867, 1996). To determine the in vivo effects of these sequences, we replaced the upstream regulatory sequences of the HIS4 gene of Saccharomyces cerevisiae with either 12 or 48 tandem copies of CCGNN. Both tracts activated HIS4 transcription. We found that (CCGNN)12 tracts elevated meiotic recombination (hot spot activity), whereas the (CCGNN)48 tract repressed recombination (cold spot activity). In addition, a “pure” tract of (CCGAT)12 activated both transcription and meiotic recombination. We suggest that the cold spot activity of the (CCGNN)48 tract is related to the phenomenon of the suppressive interactions of adjacent hot spots previously described in yeast (Q.-Q. Fan, F. Xu, and T. D. Petes, Mol. Cell. Biol. 15:1679–1688, 1995; Q.-Q. Fan, F. Xu, M. A. White, and T. D. Petes, Genetics 145:661–670, 1997; T.-C. Wu and M. Lichten, Genetics 140:55–66, 1995; L. Xu and N. Kleckner, EMBO J. 16:5115–5128, 1995).
The basic unit of DNA compaction in the assembly of the eukaryotic chromosome is the nucleosome, two superhelical turns of DNA wrapped around an octamer of histones (12, 20). Gene expression is associated with destabilization of nucleosomes at the promoter region to allow for access of sequence-specific transcription factors and the general transcription machinery to DNA (reviewed in references 14 and 45). The promoter regions of most active genes, therefore, represent regions of nucleosome-free “open” chromatin (reviewed in reference 21). Nucleosome formation also appears to inhibit the initiation of meiotic recombination in yeast, since all recombination hot spots thus far characterized are located in regions of open chromatin (reviewed in reference 24). The formation and positioning of nucleosomes are sensitive to DNA sequence. For example, tandem arrays of the sequence 5′ (G/C)3NN(A/T)3NN favor nucleosome formation (36), whereas tracts of 5′ (CCG)n (associated with the fragile X syndrome), poly(A), and poly(G) exclude nucleosomes (16, 31, 47).
Tandem arrays of the repeat 5′ CCGNN are also a poor substrate for nucleosome formation in vitro. Wang and Griffith (46) designed this repeat (i) to incorporate the triplet repeat 5′ (CCG)n which is the basis for five folate-sensitive fragile sites (including fragile X) that strongly exclude nucleosomes (47) and (ii) to place major groove wedges, such as (G/C)3 in an arrangement that should inhibit bending around the histone octamer (36). Both CCG and CCGNN are members of the motif [(C/G)3NN]n and, in a search of the GenBank database, 75 examples were found in which there was at least 85% homology to a tract of [(C/G)3NN]48 (46). In 31 of these examples, this motif was in the promoter region, and 20 of these 31 genes lacked a TATA box, suggesting a role of these sequences in the activation of transcription for TATA-less genes. In addition, in two of these genes, the human dihydrofolate reductase and ETS-2 genes, the [(C/G)3NN] tracts were in nuclease-hypersensitive regions (25, 35), indicating that these sequences might exclude nucleosomes in vivo.
Most eukaryotic promoters consist of upstream regulatory sequences, bound by sequence-specific transcriptional activators, and the TATA element, the binding site for TFIID (reviewed in reference 39). Deletion of the upstream regulatory sequences or elimination of the transcription factors that bind to this region reduces or eliminates transcription. Iyer and Struhl (15) showed that insertion of either poly(A) or poly(G) sequences upstream of the yeast HIS3 gene stimulated transcription. These authors suggested these simple repetitive elements generated a nucleosome-free region that allowed increased accessibility of transcription factors to their binding sites.
The upstream regulatory sequences and the transcription factors required for expression of the yeast HIS4 gene have been extensively characterized by Fink and coworkers. Four sequence-specific transcription factors, Bas1p, Bas2p, Rap1p, and Gcn4p, are involved in stimulating HIS4 expression (2, 5, 44), and deletion of all of the binding sites for these transcription factors reduces HIS4 expression to a very low level (27).
The promoter region of HIS4 also represents a strong meiotic recombination hot spot (4). Hot spot activity at HIS4, as at other hot spots (24), is associated with local recombination-initiating double-stranded DNA breaks (DSBs) (8). Although there is an approximately linear relationship between the level of DSBs and the frequency of recombination at the HIS4 locus, there is a basal level of recombination that is not associated with an observable DSB (8). It is not clear whether these basal recombination events reflect a different type of DNA lesion involved in initiating recombination or a diffuse scattering of DSBs difficult to visualize by Southern analysis.
Formation of DSBs at the wild-type HIS4 recombination hot spot requires binding of the Bas1p, Bas2p, and Rap1p transcription factors (49, 50). This requirement for transcription factors for hot spot activity, however, does not represent a requirement for high levels of transcription, since deletion of the HIS4 TATA sequence, which reduces expression at least 20-fold, has no effect on hot spot activity (48). These results indicate that the binding of transcription factors stimulates recombination either by creating open chromatin, allowing access of the recombination machinery (49), or by directly tethering the recombination machinery to the chromosome (9). Transcription factor-dependent meiotic recombination hot spot activity has also been observed in Schizosaccharomyces pombe (19).
All recombination hot spots analyzed thus far in Saccharomyces cerevisiae are in regions of chromatin that are sensitive to DNase I and/or micrococcal nuclease (24). Mutations that reduce HIS4 recombination hot spot activity (mutations of the promoter or mutations of transcription factors that bind to the HIS4 promoter) reduce the size of the DNase I-sensitive region at the 5′ end of HIS4 (9). Even mutations that eliminate HIS4 transcription and hot spot activity, however, do not result in complete loss of DNase I sensitivity in the HIS4 upstream region (9). One interpretation of this result is that regions of open chromatin are necessary, but not sufficient, for hot spot activity and for the activation of transcription (9, 52).
Since tandemly repeated CCGNN sequences are poor substrates for nucleosome formation and since nucleosome-depleted regions are associated with high levels of transcription and recombination (as described above), we examined the effects of these sequences on gene expression and meiotic recombination in vivo by replacing the upstream regulatory HIS4 sequences with 12 or 48 copies of CCGNN. Although both (CCGNN)12 and (CCGNN)48 arrays stimulated transcription, only the (CCGNN)12 arrays resulted in hot spot activity. The longer CCGNN array actively suppressed local recombination.
MATERIALS AND METHODS
Plasmid constructions.
The plasmids pY(CCGNN)48-9, pMD55, pMD60, and pMD65 (which were used to construct yeast strains in which the wild-type upstream regulatory sequences were replaced by CCGNN repeats) were derived from plasmid pPD5 (49). The pPD5 plasmid contains a Sau3AI fragment from the upstream HIS4 region with a 171-bp deletion removing the transcription factor binding sites (his4-Δ52 [27]) and an XhoI linker replacing the deleted sequences. To construct pY(CCGNN)48-9, we treated p(CCGNN)48 (46) with SalI and EcoRI and ligated the resulting fragments to XhoI-treated pPD5. The DNA samples were then used as a substrate for the Klenow fragment of DNA polymerase to fill in the end resulting from EcoRI cleavage, as well as the unligated XhoI-generated end. The mixture was extracted with phenol-chloroform and then ethanol precipitated. After the pellet was resuspended, we treated the sample with DNA ligase and transformed the products into Escherichia coli. The resulting transformants were examined by DNA sequence analysis. The plasmid pMD55 was constructed by the same procedure, except that p(CCGNN)12 was used (46) instead of p(CCGNN)48.
The pY(CCGNN)48-9 and pMD55 plasmids contained polylinker sequences in addition to the CCGNN repeats (see Fig. 1). The plasmid pMD60 is identical to pMD55 except that it lacks the polylinker sequences. This plasmid was constructed by annealing two oligonucleotides: 5′ CTAGATCGTCGACCGTACCGATCCGAACCGGACCGCTCCGAGCCGTCCCGTACCGCACCGCCCCG TTCCGAGTCGACATGGTAC and 5′ GTACCATGTCGACTCGGAACGGGGCGGTGCGGTACGGGACGGCTCGGAGCGGTCCGGTTCGGATCGGT ACGGTCGACGATCTAG. The double-stranded oligonucleotide was treated with SalI, and the resulting fragment was ligated with XhoI-treated pPD5. Due to the method of construction, the last repeat is CCGAG in pMD60, compared to CCGAT in pMD55. The plasmid pMD65 is identical to pMD60, except that the two oligonucleotides that were annealed were 5′ CTAGATCGTCGA(CCGAT)12GTCGACATGGTAC and 5′ GTACCATGTCGAC(ATCGG)12TCGACGATCTAG. The resulting double-stranded oligonucleotide was treated with SalI and then ligated with XhoI-treated pPD5. For all plasmids, the orientation of the CCGNN repeats is the same (CCGNN sequences adjacent to the non-transcribed sequences of HIS4).
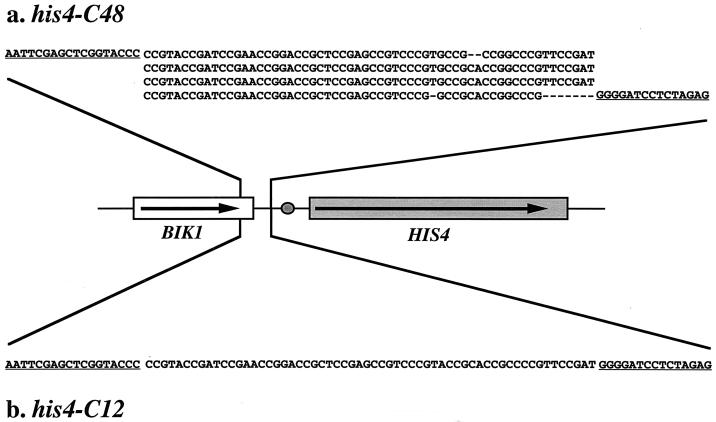
FIG. 1.
Sequences of his4-C48 and his4-C12 insertions and position of these insertions relative to the HIS4 coding sequence. Although the his4-C48 insertion has only 47 copies of CCGNN, we use the his4-C48 term to keep the nomenclature consistent with the previous study (46). The HIS4 and BIK1 genes are boxed, with the direction of transcription indicated by arrows. The oval shows the position of the HIS4 TATA sequence. The CCGNN repeats replace the upstream regulatory sequences of HIS4 and a portion of the neighboring BIK1 gene as shown. The polylinker sequences are underlined. Sequences: a, (CCGNN)48 insertion (his4-C48); b, (CCGNN)12 insertion (his4-C12).
Strain constructions.
All strains were isogenic except for changes introduced by transformation; the relevant genotypes for all haploids are given in Table 1. The haploid strains were derived from AS4 (MATα trp1 arg4 tyr7 ade6 ura3) and AS13 (MATa leu2 ade6 ura3 rme1) (38) or from published derivatives of these strains. Tracts of (CCGNN)48 were inserted upstream of HIS4 by performing a two-step transplacement of strains PD80, PD57, and PD63 with MfeI-treated pY(CCGNN)48-9, generating strains DTK255, DTK292, and DTK227, respectively. Several strains were constructed by two-step transplacements of PD80 and PD63 with various plasmids, including DTK344 (PD80 with MfeI-treated pMD55), DTK345 (PD63 with MfeI-treated pMD55), DTK361 (PD80 with MfeI-treated pMD60), DTK362 (PD63 with MfeI-treated pMD60), DTK458 (PD80 with MfeI-treated pMD65), and DTK457 (PD63 with MfeI-treated pMD65). Strains DTK322, DTK350, DTK321, and DTK351 are rad50S derivatives of DTK255, DTK344, DTK227, and DTK345, respectively; these strains were constructed by one-step transplacements by using BamHI/EcoRI-treated pNKY349 (1). The strain DTK303 was made by a two-step transplacement of DTK292 with BsrGI-treated pSH17 (10).
TABLE 1.
Relevant genotypes of haploid strains used in this study
| Straina | HIS4 upstream alterationb | HIS4 coding sequence alterationc | Other relevant alterationsd | Referencee |
|---|---|---|---|---|
| AS4 | WT | WT | WT | 38 |
| PD63 | his4-Δ52 | WT | WT | 4 |
| MW74 | his4-Δ52 | his4-203 | WT | 10 |
| HF6 | his4-202 | his4-lopc | rad50S | 8 |
| DNY107 | WT | WT | rad50S | 8 |
| DTK227 | his4-C48 | WT | WT | — |
| DTK321 | his4-C48 | WT | rad50S | — |
| DTK345 | his4-C12 | WT | WT | — |
| DTK351 | his4-C12 | WT | rad50S | — |
| DTK362 | his4-C12d | WT | WT | — |
| DTK457 | his4-CCGAT12 | WT | WT | — |
| AS13 | WT | WT | WT | 38 |
| PD57 | his4-Δ52 | WT | WT | 4 |
| PD80 | his4-Δ52 | his4-lopc | WT | 4 |
| DNY25 | WT | his4-lopc | WT | 26 |
| HF4 | WT | his4-ATC | rad50S | 8 |
| HF5 | his4-202 | WT | rad50S | 8 |
| DTK255 | his4-C48 | his4-lopc | WT | — |
| DTK292 | his4-C48 | WT | WT | — |
| DTK303 | his4-C48 | his4-203 | WT | — |
| DTK322 | his4-C48 | his4-lopc | rad50S | — |
| DTK344 | his4-C12 | his4-lopc | WT | — |
| DTK350 | his4-C12 | his4-lopc | rad50S | — |
| DTK361 | his4-C12d | his4-lopc | WT | — |
| DTK458 | his4-CCGAT12 | his4-lopc | WT | — |
The first 11 strains listed were derived by transformation from the haploid AS4, and the remainder were from the haploid AS13.
The upstream regulatory region of HIS4 in wild-type (WT) strains has binding sites for the transcriptional activators Rap1p, Bas1p, Bas2p, and Gcn4p, and strains with the his4-Δ52 allele have deletions at these sites. In his4-202 strains, the sequences deleted in his4-Δ52 were replaced with about 50 bp of telomeric sequences (49). In his4-C48 and his4-C12 strains, the wild-type upstream regulatory region was replaced with 48 or 12 copies, respectively, of CCGNN repeats (Fig. 1). Strains with the his4-C12d insertion are identical to those with his4-C12, except that the polylinker flanking the CCGNN repeats has been deleted. In strains with the his4-CCGAT12 allele, the wild-type upstream regulatory region of HIS4 was replaced with 12 copies of CCGAT.
The his4-lopc allele is a 26-bp palindromic insertion at position 467 of the coding sequence. When strains heterozygous for this insertion are sporulated and dissected, high levels of postmeiotic segregation are observed, indicating that this marker results in a poorly repaired mismatch when located within a heteroduplex (26). The his4-203 allele is an insertion of about 50 bp of telomeric DNA at position 96 of the coding sequence (10). The his4-ATC allele is a single-base-pair change at position 3 of the coding sequence (3).
When diploid strains homozygous for the rad50S mutation are sporulated, meiosis-specific DSBs associated with hot spots are formed, but the broken ends are not processed (1).
—, strain constructed in this study.
Isogenic diploids were constructed by mating the following haploids (AS4 strains listed before AS13 strains): DNY26 (AS4 × DNY25), FX3 (DNY107 × HF4), FX4 (HF6 × HF5), MW155 (MW74 × PD57), PD81 (PD63 × PD80), DTK269 (DTK227 × DNY25), DTK270 (AS4 × DTK255), DTK273 (DTK227 × DTK255), DTK312 (DTK227 × DTK303), DTK324 (DTK321 × DTK322), DTK347 (DTK345 × DTK344), DTK352 (DTK351 × DTK350), DTK364 (DTK362 × DTK361), and DTK460 (DTK457 × DTK458).
Media and genetic procedures.
With the exceptions noted below, standard protocols and media were used (13). Sporulation plates contained 1% potassium acetate, 0.1% yeast extract, 0.05% glucose, 6 μg of adenine per ml, and 2% agar. As in previous experiments involving this genetic background, diploids were sporulated at 18°C and dissected onto plates containing rich growth medium. After colonies had formed, they were replica plated to various omission media. Postmeiotic segregation events at the HIS4 locus were detected as sectored His+/His− colonies (4). In strains heterozygous for two his4 mutant alleles, allelism tests were performed as described previously (10).
In some tetrads derived from DTK329, we examined the pattern of aberrant segregation of the heterozygous cold spot insertion. Since the cold spot does not alter the ability of cells to grow in the absence of histidine, this analysis was done by PCR with primers specific for the cold spot sequences or the wild-type hot spot. DNA was isolated from spore colonies in which the his4-lopc mutation had an aberrant segregation pattern. For all four spore colonies of each tetrad, PCR was performed by using two pairs of primers. PCR done with primers 23201 (5′ CAGTTGGAACAGGCTCAAGCAC) and 23202 (5′ AGTCACTGTGCATGGGTTTAGC) generate a DNA fragment of about 130 bp in DNA samples containing the wild-type hot spot; PCR performed with primers 23201 and 23203 (5′ GGGGGTACCGAGCTCGAATTTC) result in a DNA fragment of about 240 bp in DNA samples containing the cold spot.
DNA and RNA isolation and analysis.
To examine meiosis-specific DSBs at the HIS4 locus, we sporulated cells in 1% potassium acetate at 25°C (8). The method of DNA isolation was that of Goyon and Lichten (11). DNA from meiotic cells was treated with BglII, and the resulting fragments were separated on an 0.8% agarose gel. Standard Southern analysis was done by using an XbaI-XhoI fragment of plasmid pDN42 (containing sequences from the 5′ end of HIS4 [8]). The analysis of the DNase I-sensitive sites in HIS4 chromatin was done exactly as described previously (9). DSBs at the ARG4 locus were detected with an EcoRV-BglII fragment derived from plasmid pAK1 (8).
Total RNA was prepared (37) from premeiotic or meiotic cells grown in liquid cultures. Standard Northern analysis was done (33). The nylon filter with the transferred RNA was hybridized to a mixture of two probes: the same HIS4 probe used in the Southern analysis (described above) and an actin gene probe. The ACT1 probe was a 1-kb gel-purified HindIII-XhoI fragment of plasmid pGAL1-ACT1 (provided by R. Sia, Duke University). Levels of hybridization (for both Northern and Southern analyses) were quantitated with a PhosphorImager (Molecular Dynamics).
Statistical analysis.
Statistical comparisons were done using the Instat 1.12 program for Macintosh. Results were considered statistically significant if P was <0.05.
RESULTS
Nuclease sensitivity of the HIS4 upstream regulatory region in strains with (CCGNN)48 and (CCGNN)12 repeats.
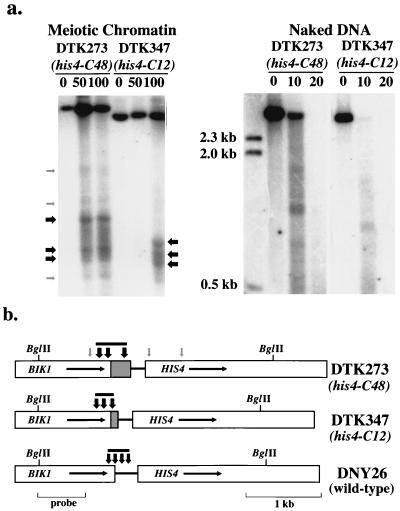
If the CCGNN repeats are a poor substrate for nucleosome formation in vivo, the chromatin containing the upstream regulatory sequences of HIS4 in strains with insertions of this repeat should be sensitive to DNase I. We constructed diploid strains (DTK273 and DTK347) in which the wild-type HIS4 upstream regulatory sequences were replaced with (CCGNN)48 or (CCGNN)12 repeats on both homologous chromosomes; the mutant alleles are his4-C48 or his4-C12, respectively. The sequences and insertion positions of these alleles are shown in Fig. 1. We examined the DNase I sensitivity of chromatin in cells from these strains after the cells were incubated for 6 h in sporulation medium (Fig. 2). We found previously that meiosis-specific double-strand DNA breaks appear after the cells have been incubated 4 to 6 h (8).
FIG. 2.
Mapping of the DNase I-hypersensitive sites in HIS4 chromatin isolated from strains DTK273 (homozygous for his4-C48) and DTK347 (homozygous for his4-C12). (a) Chromatin was prepared from cells incubated for 6 h at 25°C in sporulation medium (9). Chromatin was digested with 0, 50, and 100 U of DNase I, and DNA was then extracted. Naked DNA was incubated with 0, 10, or 20 U of DNase I. Samples were then treated with BglII and examined by Southern analysis; a BglII-PvuII fragment of BIK1 was used as a hybridization probe. Numbers on the left indicate the sizes of DNA markers. Arrows mark the positions of DNase I-hypersensitive sites; black and gray arrows indicate strong and weak sites, respectively. (b) The positions of the DNase I-hypersensitive sites relative to restriction maps for the strains are shown by vertical arrows. As above, black and gray arrows indicate strong and weak hypersensitive sites, respectively. The gray bars above the arrows indicate the extent of the major open chromatin region in each strain. The promoter region in DNY26 (wild type) is replaced by the (CCGNN)48 sequence (hatched region) in DTK273 and the (CCGNN)12 sequence in DTK347. Data for DNY26 is derived from Fan and Petes (9).
Although there were some differences in the location of DNase I-hypersensitive sites between the strains, both strains with CCGNN repeats had DNase I-sensitive promoter regions (Fig. 2a). The strain with 48 repeats (DTK273) had the largest open region, with three strong hypersensitive sites within this region. The strain with 12 repeats (DTK347) also had three strong hypersensitive sites, but the open region was compressed. The hypersensitive sites were located within and upstream of the CCGNN repeats but did not extend into the coding region of HIS4 (Fig. 2b). DNase I-hypersensitive sites were found previously (9) at similar positions in the isogenic wild-type strain DNY26 (Fig. 2b). The DNase I-hypersensitive sites observed in chromatin were not present in naked DNA samples (Fig. 2a). In summary, these results suggest that the CCGNN repeats represent open chromatin, a result in agreement with our previous observations (46). The difference in the sizes of open chromatin associated with the (CCGNN)48 and (CCGNN)12 tracts was greater than expected from the difference in sizes of the two tracts. In addition, the regions of open chromatin overlapped with, but were not identical to, the positions of the tracts (Fig. 2b). These results suggest that the CCGNN repeats can affect nucleosome formation in regions directly adjacent to the tracts.
We also compared the DNase I sensitivity of the HIS4 chromatin in DTK273 and DTK347 relative to ARG4 chromatin in the same strains. Blots containing DNase I-treated samples were first examined by using a HIS4-specific probe and then stripped and rehybridized to an ARG4-specific probe. We measured the level of radioactivity in the intact fragments (undigested by DNase I) for each probe. The ratio of these values (ARG4/HIS4) was determined for the samples of 0, 50, and 100 U (DNase I). These ratios were then normalized by dividing by the ARG4/HIS4 ratio for the 0-U samples. Values greater than one indicate that the HIS4 chromatin is more sensitive to DNase I treatment than the ARG4 chromatin. These normalized ratios for DTK273 (values from two experiments) were 0 U (normalized to 1), 50 U (1.3, 1.42), and 100 U (1.3, 1.83). The normalized ratios for DTK347 were 0 U (normalized to 1), 50 U (1.0 and 1.1), and 100 U (1.1 and 1.1). These results suggest that the DNase I sensitivity of the HIS4 and ARG4 chromatin is approximately the same in strain DTK347. The HIS4 chromatin in DTK273 is slightly, but significantly (P = 0.02 by the Mann-Whitney nonparametric test), more DNase I sensitive than the HIS4 chromatin in DTK347.
Insertions of (CCGNN)48 and (CCGNN)12 stimulate HIS4 transcription.
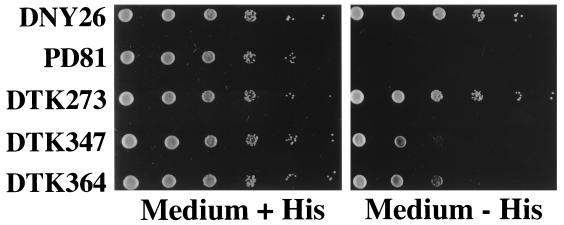
Strains with the his4-Δ52 mutation are phenotypically His− (PD81; Fig. 3). Although the TATA sequence and the HIS4 coding sequences are intact, elimination of the binding sites for the transcription factors Bas1p, Bas2p, Rap1p, and Gcn4p results in very low levels of HIS4 transcription (27). When we replaced the deleted sequences with (CCGNN)48 repeats (DTK273), the strain became His+ (Fig. 3), indicating that these sequences stimulated transcription. Similarly, replacement of the wild-type upstream sequences with (CCGNN)12 repeats (his4-C12) resulted in a His+ phenotype (DTK347). The strain DTK364 has the same (CCGNN)12 repeats as DTK347 but lacks polylinker sequences flanking the repetitive tracts. The (CCGNN)12 tracts stimulated HIS4 gene expression less than the (CCGNN)48 tracts.
FIG. 3.
Stimulation of HIS4 transcription by insertions of CCGNN repeats in the upstream regulatory region. Single yeast colonies were suspended in water, and 1:10 serial dilutions were performed. A 10-μl portion of each dilution was placed on solid medium, with the most concentrated suspension on the left side of the plates. Each row represents a different strain as follows: DNY26, wild-type HIS4 upstream regulatory sequences; PD81, homozygous for his4-Δ52 mutation which removes binding sites for all known HIS4 transcription activators; DTK273, homozygous for his4-C48; DTK347, homozygous for his4-C12; and DTK364, homozygous for his4-C12d.
We also examined the level of HIS4 mRNA by Northern analysis in both vegetative and meiotic cells (data not shown). The levels of mRNA in vegetative cells of DTK273 and DTK347 were roughly 50 and 25%, respectively, of that found in vegetative cells of the wild-type strain (DNY26). After 6 h in sporulation medium, the level of HIS4 RNA in DTK273 was reduced about 10-fold, whereas HIS4 expression was reduced in the wild-type strain about 5-fold. In summary, insertion of CCGNN repeats upstream of HIS4 stimulates gene expression; possible mechanisms for this stimulation will be discussed below.
Insertion of (CCGNN)48 suppresses local meiotic recombination.
The frequency of meiotic recombination at particular sites in the yeast genome can be assayed either genetically or physically. Genetic analysis requires tetrad analysis of a diploid heterozygous for an alteration in the region of interest (30). If a high frequency of non-Mendelian segregation is observed at the heterozygous site, the marker is located near a recombination hot spot. To describe aberrant segregation tetrads, we will use the nomenclature derived for eight-spored fungi. By this nomenclature, standard Mendelian segregation is 4:4 and gene conversion events are either 6:2 (three wild-type spore colonies:one mutant spore colony) or 2:6 (one wild-type spore colony:three mutant spore colonies). Tetrads with a single postmeiotic segregation (PMS) event are detected as spore colonies that are sectored for the wild-type and mutant alleles and will be described as 5:3 (two wild-type spore colonies:one mutant spore colony:one sectored colony) or 3:5 (one wild-type spore colony:two mutant spore colonies:one sectored colony) segregations (30).
By performing tetrad analysis with a large number of diploid strains heterozygous for markers at different positions within the HIS4 gene and the neighboring BIK1 gene, we previously found a gradient of aberrant segregation (polarity gradient) with the highest level of recombination in the promoter region of HIS4 (4); these experiments define this region as a recombination hot spot. As previously mentioned, subsequent analysis showed that both transcription factor binding sites (4, 50) and transcription factors (49, 50) were required for hot spot activity. It should be emphasized that loss of HIS4 hot spot activity does not completely eliminate meiotic recombination in the HIS4 region (4).
Meiotic recombination hot spots can also be defined as sites with high levels of meiosis-specific DSBs. As first shown at the ARG4 locus (40), meiotic recombination events in yeast are initiated by DSBs. At the HIS4 locus, a meiosis-specific DSB occurs in the HIS4 promoter region in wild-type strains, and this DSB is eliminated by the loss of transcription factors or transcription factor binding sites (8).
In previous studies, we analyzed the aberrant segregation frequency of his4-lopc (a 26-bp palindromic insertion in the HIS4 coding sequence) in DNY26 (wild-type upstream sequences) and PD81 (homozygous for a deletion of the transcription factor binding sites, his4-Δ52) (4, 8, 26). These data are reproduced in Table 2. In DNY26, 51% of the tetrads underwent non-Mendelian segregation of his4-lopc. This rate is very high relative to most other loci in yeast (30), indicating the strength of the wild-type HIS4 hot spot. In PD81, the rate of aberrant segregation was reduced two- to threefold, and the rate of HIS4-LEU2 crossovers was reduced by 40%. The LEU2 gene is about 20 kb centromere proximal to HIS4.
TABLE 2.
Meiotic recombination segregation patterns of HIS4 in strains with various upstream regulatory sequences
| Straina | HIS4 upstream regulatory sequencesb | Coding sequence mutationc | Total tetrads | No. of tetrads with various segregation patternsd
|
HIS4-LEU2 dist (cM)e | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4:4 | 6:2 | 2:6 | 5:3 | 3:5 | Ab 4:4 | Other Ab Seg | % Ab Seg | |||||
| DNY26 | HIS4/HIS4 | HIS4/his4-lopc | 493 | 244 | 29 | 17 | 88 | 67 | 15 | 33 | 51 | 31 |
| PD81 | his4-Δ52/his4-Δ52 | HIS4/his4-lopc | 398 | 313 | 14 | 6 | 26 | 32 | 5 | 2 | 21 | 19 |
| DTK273 | his4-C48/his4-C48 | HIS4/his4-lopc | 317 | 282 | 2 | 2 | 10 | 19 | 1 | 1 | 11 | 10 |
| DTK269 | his4-C48/HIS4 | HIS4/his4-lopc | 340 | 286 | 14 | 2 | 27 | 7 | 1 | 3 | 16 | 13 |
| DTK270 | HIS4/his4-C48 | HIS4/his4-lopc | 260 | 218 | 3 | 6 | 11 | 20 | 0 | 2 | 16 | 14 |
| MW155 | his4-Δ52/his4-Δ52 | HIS4/his4-203 | 234 | 144 | 54 | 18 | 1 | 2 | 0 | 15 | 39 | 28 |
| DTK312 | his4-C48/his4-C48 | HIS4/his4-203 | 250 | 232 | 8 | 7 | 1 | 0 | 0 | 2 | 7 | 11 |
| DTK347 | his4-C12/his4-C12 | HIS4/his4-lopc | 327 | 130 | 10 | 12 | 58 | 78 | 13 | 26 | 60 | 36 |
| DTK364 | his4-C12d/his4-C12d | HIS4/his4-lopc | 233 | 92 | 8 | 6 | 41 | 50 | 12 | 24 | 61 | 38 |
| DTK460 | his4-CCGAT12/his4-CCGAT12 | HIS4/his4-lopc | 269 | 142 | 14 | 8 | 41 | 46 | 8 | 10 | 47 | 26 |
The genotypes for the upstream regulatory sequences for each diploid are listed (allele contributed by the AS4 parent followed by allele contributed by the AS13 parent). The descriptions of the alleles are as follows: HIS4 (wild-type upstream regulatory sequences), his4-Δ52 (deletion removing all transcription factor binding sites), his4-C48 [replacement of wild-type transcription factor binding sites with (CCGNN)48 repeats], his4-C12 [replacement of wild-type transcription factor binding sites with (CCGNN)12 repeats], his4-C12d [replacement of wild-type transcription factor binding sites with (CCGNN)12 repeats with the polylinker deleted], and his4-CCGAT12 [replacement of wild-type transcription factor binding sites with (CCGAT)12 repeats].
The his4-lopc allele is a palindromic insertion of 26 bp (26). The his4-203 allele is an insertion of about 50 bp of telomeric DNA (10). Both alleles result in a His− phenotype. The allele contributed by the AS4 parent is followed by the allele contributed by the AS13 parent.
The segregation patterns indicate segregation of the HIS4 coding sequence alleles. For all patterns, the first number indicates the wild-type allele and the second is the mutant allele. The segregation patterns are as follows: 4:4 (normal Mendelian segregation), 6:2 and 2:6 (gene conversion events), 5:3 and 3:5 (single-PMS tetrads), Ab 4:4 (aberrant 4:4; one wild-type spore colony, one mutant spore colony, and two sectored colonies), and Other Ab Seg (tetrads with more than one PMS and/or gene conversion event).
All strains were heterozygous for HIS4 and LEU2 markers. We classified tetrads as parental ditype, nonparental ditype, and tetratype and then calculated the recombination distance between the two markers in centimorgans (cM) by standard methods (29).
In DTK273, the (CCGNN)48 insertion replaces the normal transcription factor binding sites of HIS4 on both homologues. This insertion reduced the level of aberrant segregation of his4-lopc to 11%, significantly (P < 0.001, by the Fisher exact test) below the frequency observed in PD81 (21%). In addition, the frequency of HIS4-LEU2 crossovers relative to that observed in PD81 was significantly (P = 0.001, by the chi-square contingency test) reduced by the insertion. Since we have previously shown that the his4-Δ52 mutation in PD81 eliminates hot spot activity as measured genetically and physically, the stronger effect of the (CCGNN)48 insertion suggests that these sequences actively repress recombination; additional experimental support for this conclusion will be provided below.
It should be pointed out that the suppression of recombination observed in DTK273 appears to be specific for the region near HIS4. The rate of aberrant segregation for a heterozygous marker located in the ARG4 gene located on a different chromosome was 9%, which is not significantly different from the rates observed in DNY26 (7%) or PD81 (10%). In addition, recombination distances between LEU2 and the centromere were approximately equal in DNY26, PD81, and DTK273 (distances of 8, 9, and 9 centimorgans, respectively).
The strains DTK269 and DTK270 were heterozygous for the (CCGNN)48 insertion and his4-lopc. In strain DTK270, the insertion was on the same chromosome as the his4-lopc allele and, in DTK269, the insertion was on the opposite chromosome. For both strains, the rates of aberrant segregation and the HIS4-LEU2 recombination distances were similar and were intermediate between the rates observed in PD81 and DTK273 (Table 2).
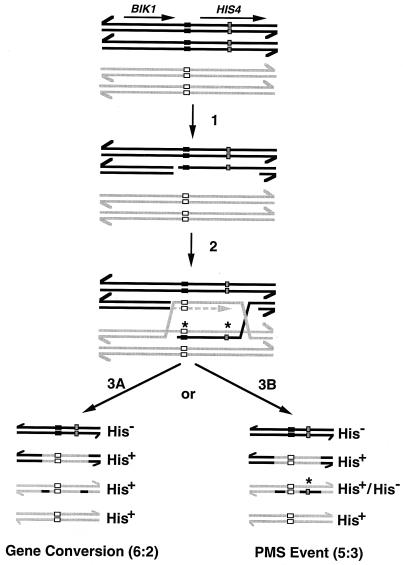
In previous studies of strains heterozygous for hot spot activity, it has been shown that classes of aberrant segregation in which the hot spot is lost are more frequent than those in which the hot spot is duplicated (reviewed in reference 30). The rationale for this bias based on the DSB repair model of recombination (42) is shown in Fig. 4. In this figure, we illustrate the configuration of markers in DTK269 in which the wild-type recombination hot spot is on the same homologue as the mutation in the HIS4 coding sequence (his4-lopc) and the opposite homologue has the cold spot sequence and the wild-type HIS4 coding sequence. If the double-strand break that initiates recombination (indicated by a vertical arrow in the figure) occurs on the chromosome with the wild-type hot spot, then the resulting recombination intermediate (shown below step 2 in Fig. 4) would have two chromatids with wild-type HIS4 genes, one chromatid with the mutant his4-lopc allele and one chromatid with a heteroduplex containing one strand with wild-type HIS4 information and one strand with mutant his4-lopc information. If this mismatch (indicated by an asterisk) is corrected to generate a fully wild-type gene, a 6:2 gene conversion event would be generated (step 3A). Failure to repair the DNA mismatch would result in 5:3 segregation (step 3B). Tetrads of the 2:6 and 3:5 classes would reflect initiation of DSB formation on the chromosome with the cold spot. Thus, for DTK269, a bias in favor of 6:2 and 5:3 tetrads over 2:6 and 3:5 tetrads suggests that the chromosome with the wild-type hot spot and his4-lopc mutation is the preferred chromosome for initiating recombination. Since the linkage relationships of the hot spot and cold spot sequences with the HIS4 coding sequence alleles in DTK270 is opposite to that in DTK269, one expects to see the opposite bias in recovery of types of aberrant segregation.
FIG. 4.
Patterns of aberrant segregation expected if recombination initiates by a DSB (42) on the chromosome with the wild-type hot spot in strain DTK269. Chromosomes are depicted as double-stranded DNA molecules, with the different homologues having either black or gray DNA strands. The coding sequences and the directions of transcription for HIS4 and the neighboring BIK1 gene are shown by horizontal arrows. The wild-type hot spot and the (CCGNN)48 cold spot are indicated by horizontal black and white rectangles, respectively. The position of the his4-lopc palindromic insertion is shown by the stippled vertical rectangle. After DSB formation, one strand of the duplex is degraded 5′ to 3′, resulting in a single-stranded “tail” (step 1). The single-stranded portion of the molecule invades the other homologue, resulting in a heteroduplex with two mismatches (indicated by asterisks) (step 2). In addition, repair synthesis occurs on the displaced DNA strand as shown by the dashed line. After resolution of the resulting junctions to retain flanking markers in the parental configuration, two alternative types of aberrant segregation could be produced. Repair of both mismatches by using the gray strand as a template (step 3A) would result in a gene conversion event of the 6:2 class (three His+ spore colonies:one His− spore colony). Failure to repair the his4-lopc mismatch (step 3B) or both mismatches would result in a PMS event of the 5:3 class (two His+ spore colonies:one His− spore colony:one sectored His+/His− spore colony). Tetrads of the 2:6 or 3:5 classes would result from DSB formation on the chromosome containing the cold spot.
The sum of the 5:3 and 6:2 tetrads in DTK269 is 41, whereas the sum of the 3:5 and 2:6 classes is 9; in DTK270, the sum of the 5:3 and 6:2 tetrads in DTK269 is 14, whereas the sum of the 3:5 and 2:6 classes is 26. The distributions of tetrads in these classes between the two strains are significantly different (P < 0.001, by the chi-square test). These results suggest that the suppression of recombination caused by (CCGNN)48 primarily occurs in cis rather than in trans. Since the frequency of aberrant segregation (16%) in the his4-C48 heterozygous strains is less than the calculated mean frequency (31%) of the wild-type (DNY26) and his4-C48 homozygous (DTK273) strains, it is possible that the heterozygous (CCGNN)48 tract may interfere with heteroduplex formation. Alternatively, although the strongest recombination-suppressing effects of (CCGNN)48 occur in cis, there may also be a small trans effect.
If the initiating DSB occurs upstream of the hot spot and cold spot sequences (as shown in Fig. 4), one would expect that the resulting heteroduplex would contain two mismatches, one representing the heterozygous his4-lopc mutation in the coding sequence and one representing the sequence differences between the hot spot and cold spot. We previously showed that mismatches resulting from heteroduplex formation involving small palindromic insertions (such as his4-lopc) were inefficiently repaired (26); as expected, most of the aberrant segregation events in DTK269 and DTK270 are PMS events. The mismatch involving the hot spot and cold spot sequences would be expected to produce a large “bubble” of single-stranded DNA, in which one strand of the bubble has the hot spot sequences and the other strand has the cold spot sequences. Since we previously found that large single-stranded DNA loops were efficiently repaired (26), we would expect that such a mismatch would be readily repaired. Since the hot spot and cold spot sequences are not within the coding sequence and since both hot spots and cold spots lead to similar levels of HIS4 expression, standard phenotypic tests did not allow us to examine the segregation of a hot spot–cold spot heterozygous marker. Using PCR methods (described in Materials and Methods), we analyzed the segregation of this marker in a limited number of tetrads derived from DTK269. We examined only those tetrads in which the his4-lopc marker underwent aberrant segregation. In six of six tetrads in which the his4-lopc marker underwent gene conversion, the hot spot–cold spot marker also underwent conversion in the same direction (both markers 2:6 or both 6:2). Of eight tetrads in which the his4-lopc marker showed 5:3 or 3:5 segregation, the hot spot–cold spot marker underwent gene conversion in four tetrads and showed 4:4 segregation in four tetrads. In summary, these results indicate that most of the recombination events that involve his4-lopc initiate upstream of the cold spot, as shown in Fig. 4.
Suppression of the activity of a closely linked hot spot by the (CCGNN)48 insertion.
In previous studies, we showed that an insertion of about 50 bp of telomeric DNA in the HIS4 coding sequence (his4-203) resulted in hot spot activity associated with DSB formation at the site of the insertion (10). Tetrad analysis of strain MW155 [heterozygous for his4-203 and homozygous for a deletion of the wild-type upstream hot spot (his4-Δ52)] revealed the bias in classes of aberrant tetrads expected for a strain heterozygous for hot spot activity (reference 10 and data reproduced in Table 2); the sum of 6:2 and 5:3 classes (55) significantly (P < 0.01) exceeded the sum of 2:6 and 3:5 classes (20).
The strain DTK312 was heterozygous for his4-203 and homozygous for the his4-C48 insertion. The insertions reduced the level of aberrant segregation of his4-203 from 39 to 7%, reflecting a significant (P < 0.001) reduction in the number of aberrant segregation tetrads. In addition, the bias in the recovery of 5:3 plus 6:2 versus 3:5 plus 2:6 classes was eliminated, as expected if the his4-203 hot spot activity was eliminated. Crossovers were also significantly reduced (P < 0.001 for comparison of DTK312 and MW155). We conclude, therefore, that the (CCGNN)48 insertion is capable of suppressing recombination in a closely linked (about 250 bp) adjacent hot spot.
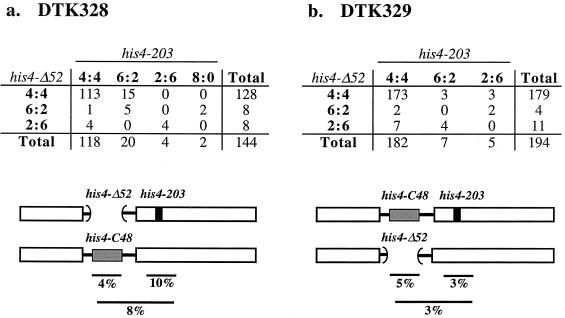
To determine whether the suppression of the activity of the adjacent hot spot occurred in cis (as predicted from our previous analysis) or in trans, we constructed diploid strains heterozygous for his4-203 and his4-C48. In strain DTK328, his4-203 and his4-C48 were on opposite chromosomes; in strain DTK329, these alterations were on the same chromosome. If his4-C48 suppressed the activity of his4-203 equivalently in cis and in trans, the frequency of aberrant segregation for his4-203 should be the same in the two strains. The tetrad data and allelic arrangements for the two strains are shown in Fig. 5. We monitored the aberrant segregation of the his4-Δ52 allele, the his4-C48 cold spot, and the his4-203 hot spot. We found that the number of aberrant segregation events involving his4-203 in DTK328 (26) significantly (P < 0.001 by Fisher exact test) exceeded those in DTK329 (12). These results demonstrate that the (CCGNN)48 tract suppresses recombination of an adjacent hot spot more effectively in cis than in trans.
FIG. 5.
Aberrant segregation patterns in yeast strains with a hot spot (his4-203) and a cold spot (his4-C48) arranged either in trans (DTK328) or in cis (DTK329). The his4-203 hot spot is shown as a black rectangle, and the his4-C48 cold spot is shown as a gray rectangle, with brackets indicating the extent of the his4-Δ52 deletion. Tetrads were dissected and allelism tests were performed in order to examine separately the segregation of his4-Δ52 and his4-203. In the top part of the figure, the column headings represent the segregation patterns for the his4-203 allele, and the row headings represent the segregation patterns for the his4-Δ52 allele. The values in the tables show the number of tetrads in each class. For example, there were 15 tetrads that segregated 6:2 for his4-203 and 4:4 for his4-Δ52 derived from DTK328. In the bottom part of the figure, lines and percentages below each allele show the frequency of recombination at the indicated location. For example, in DTK328, 4% of the tetrads reflected aberrant segregation of the his4-Δ52 allele, 10% reflected aberrant segregation of the his4-203 allele, and 8% reflected aberrant segregation for both alleles. Panels: a, numbers of aberrant segregation tetrads and arrangement of markers in DTK328; b, numbers of aberrant segregation tetrads and arrangement of markers in DTK329.
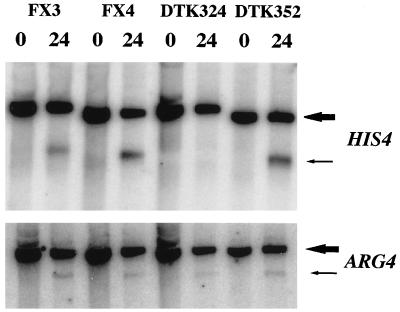
In addition to examining the recombination-suppressing activity of the (CCGNN)48 tract by tetrad analysis, we carried out Southern analysis on DTK324, a rad50S derivative of DTK273 homozygous for the his4-C48 insertion. All strains used in the Southern analysis were homozygous for the rad50S mutation, since this mutation allows DSB formation without subsequent processing of the broken DNA ends (1). As shown in Fig. 6, no meiosis-specific DSB was observed in DTK324, although DSBs were observed in control strains FX3 (rad50S strain homozygous for wild-type hot spot [8]) and FX4 (rad50S strain homozygous for his4-202 [telomeric insertion in upstream region] [8]). The levels of meiosis-specific DSBs observed at the ARG4 locus were similar in the samples for all four strains (Fig. 6), indicating that the failure to see a DSB in DTK324 at HIS4 was due to the CCGNN tract insertion and not to a global decrease in DSB formation.
FIG. 6.
Meiosis-specific DSBs at the HIS4 locus in strains with different upstream regulatory sequences. DNA was isolated from diploid strains (all homozygous for rad50S) grown vegetatively or incubated in sporulation medium for 24 h. DNA was treated with BglII and examined by Southern analysis (see Materials and Methods). The upper panel shows the Southern analysis of the HIS4 locus, and the lower panel shows a Southern analysis of the same blot after reprobing of the blot with an ARG4-specific probe. The large arrow indicates the position of the intact BglII fragment containing the 5′ end of HIS4 or ARG4, and the small arrow shows the position of meiosis-specific DSBs. The intact BglII fragments (as well as DSB-associated fragments) in different strains were not identical in size because of alterations introduced in the upstream regulatory regions. The strain FX3 (rad50S derivative of DNY26) is homozygous for the wild-type upstream regulatory sequences, and FX4 is homozygous for his4-202, an insertion of telomeric DNA with hot spot activity (8, 49). DTK324 (rad50S derivative of DTK273) is homozygous for his4-C48 and DTK352 (rad50S derivative of DTK347) is homozygous for his4-C12.
In summary, genetic and physical evidence indicates that the (CCGNN)48 insertion suppresses local meiotic recombination by inhibiting formation of the initiating DNA lesion. We failed to detect any DSB formation at the location of the normal HIS4 hot spot, although meiotic recombination at the HIS4 locus (monitored by the frequency of aberrant segregation) is not eliminated. The remaining HIS4 recombination activity could reflect DSB events initiated at sites distant from the HIS4 locus or local DSBs scattered over a large region. Alternatively, these basal recombination events could be initiated by a different DNA lesion than a DSB.
Insertions of (CCGNN)12 or (CCGAT)12 tracts result in stimulation of local meiotic recombination.
The (CCGNN)48 tract was constructed by ligating together four tandem copies of (CCGNN)12 (46); during the construction of the plasmid containing this repeat [pY(CCGNN)48-9], 2 bp were lost from one repeat, 1 bp was lost from a second, and one repeat was deleted (Fig. 1a). The sequence of the 12-repeat insertion is shown in Fig. 1b. The diploid DTK347 was homozygous for the (CCGNN)12 repeat and heterozygous for his4-lopc. In contrast to the results obtained for DTK273 [homozygous for the (CCGNN)48 insertion], the 12-repeat insertion acted as a strong recombination hot spot. The rate of aberrant segregation events for his4-lopc was 60% (Table 2), significantly (P < 0.001) higher than that observed for DTK273 (11%) and for PD81 (21%). This level of recombination was even significantly (P < 0.01) higher than that observed for DNY26 (51%), which was homozygous for the wild-type recombination hot spot. Crossovers were also significantly elevated in DTK347 relative to levels observed for DTK273 and PD81 (P < 0.001 for both comparisons). In addition, in DNA isolated from meiotic cells of a rad50S derivative of DTK347 (DTK352), we found a meiosis-specific DSB near the site of the insertion (Fig. 6).
As shown in Fig. 1, the CCGNN repeats of his4-C48 and his4-C12 were flanked by polylinker sequences. We constructed a derivative of his4-C12 in which these flanking sequences were absent (his4-C12d). A diploid homozygous for his4-C12d and heterozygous for his4-lopc (DTK364) had the same frequency of aberrant segregation as observed in DTK347 (Table 2). Thus, the polylinker sequences do not appear to contribute to the hot spot activity.
The repetitive tract of his4-C12 has a mixture of different types of 5-bp sequences. To determine whether a tract of identical 5-bp repeats would also stimulate recombination, we inserted 12 copies of CCGAT into the promoter region of HIS4 (his4-CCGAT12). This insertion activated HIS4 expression to approximately the same level observed for the mixed (CCGNN) repeats (data not shown). As shown in Table 2, a diploid strain homozygous for the his4-CCGAT12 insertion (DTK460) had a level of HIS4 aberrant segregation equivalent to the level seen in a strain containing a wild-type hot spot (DNY26). Although the (CCGAT)12 repeats result in a meiotic recombination hot spot, the activity of the hot spot was reduced significantly (P = 0.002, by Fisher exact test) compared to that caused by the his4-C12 insertion (comparison of strains DTK460 and DTK347).
DISCUSSION
In this study, we examined the effects of tandem repeats of CCGNN, a sequence shown to exclude nucleosomes (46), on gene expression and meiotic recombination at the HIS4 locus of S. cerevisiae. We found that (CCGNN)12 and (CCGNN)48 tracts, replacing the normal upstream regulatory region of HIS4, generated a region of open chromatin and activated HIS4 gene expression. The insertions, however, had opposite effects on meiotic recombination, with (CCGNN)12 and (CCGNN)48 acting as recombination hot spots and cold spots, respectively. Each of these conclusions will be discussed further below.
The patterns of DNase I-sensitive sites observed in DTK273 (homozygous for his4-C48) and DTK347 (homozygous for his4-C12) were similar to those observed previously in strains with actively transcribed HIS4 genes (9). Thus, it is likely that CCGNN repeats exclude nucleosomes in vivo as expected from the in vitro studies. The pattern of DNase I-sensitive sites did not change dramatically as the cells entered meiosis (data not shown). This result is in agreement with our previous analysis of chromatin structure at the HIS4 locus. In contrast, Ohta et al. (28) showed that chromatin at the ARG4 recombination hot spot became more sensitive to micrococcal nuclease as cells entered meiosis.
In strains DTK273, DTK347, and DTK460, binding sites for all of the transcription factors known to stimulate HIS4 have been deleted and replaced with CCGNN repeats. There are about 20 bp between the border of the insertions and the TATA sequence and about 60 bp between TATA and the mRNA start site (27). None of the promoter sequences remaining in these strains are known to bind transcription factors. The demonstration that the CCGNN repeats activate transcription and exclude nucleosome formation in vivo supports the model that long tracts of [(C/G)3NN] generate an open chromatin structure, providing access of the transcriptional machinery to DNA (46); similarly, Iyer and Struhl (15) suggested that poly(A) and poly(G) sequences located upstream of the yeast HIS3 gene stimulated transcription by opening the chromatin to facilitate the binding of transcription factors.
There are several related models to explain the effects of the CCGNN repeats. First, the open chromatin may allow the direct entry of RNA polymerase to the DNA. Second, the open chromatin structure formed by the CCGNN repeats could facilitate the binding of an as-yet-undiscovered transcription factor to a site adjacent to the repeats. Third, the CCGNN repeats could be bound directly by an as-yet-undiscovered transcription factor that stimulates HIS4 expression. Although we cannot conclusively rule out any of these models because of the observation that three different nucleosome-excluding DNA sequences activate transcription in yeast, the third model is less likely than the first two.
The substitution of the wild-type upstream regulatory sequences with the (CCGNN)12 or (CCGAT)12 repeats results in a strong meiotic recombination hot spot. Since the wild-type HIS4 hot spot requires the binding of transcription factors for activity (49, 50), this observation suggests two possibilities (somewhat related to those devised to explain the effects of the insertions on gene expression). First, the (CCGNN)12 repeats may represent open chromatin, naked DNA unbound by cellular proteins; this naked DNA could be a preferred substrate for the formation of recombination-initiating complexes. Alternatively, the (CCGNN)12 insertion may bind transcription factors that result in activation of hot spot activity. This possibility is made less likely by the observation that both the CCGNN and CCGAT tracts have similar effects on transcription and recombination.
A number of other DNA sequences (including the 5′ end of the bacterial β-lactamase gene (38) and BamHI linker sequences [53]) have hot spot activity but do not bind known transcription factors. In addition, a poly(A) tract located in the 5′ region of ARG4 has been shown to be required for high levels of meiotic recombination at that locus (34). Based on these data and our observations, we favor a model in which the CCGNN tracts create an open chromatin configuration which acts as a transcription factor-independent substrate for the recombination-initiating complex. We recently suggested that yeast cells have two types of recombination hot spots: α (that require binding of a transcription factor for activity) and β (DNA sequences susceptible to DSB formation in the absence of bound proteins) (17). We suggest that the CCGNN tracts act as β hot spots. Although it is clear that 12 copies of the CCGNN tracts are sufficient to generate a hot spot, it is possible that a smaller number of the repeats would also suffice.
By any of the models proposed for the hot spot activity of the (CCGNN)12 insertion, the cold spot activity of the (CCGNN)48 repeat insertion is unexpected. We suggest this cold-spot activity is related to the phenomenon of competitive interactions between adjacent hot spots described previously in yeast (8, 10, 52, 53). If a recombination hot spot is inserted near a preexisting hot spot, the activity of both is reduced. These competitive interactions are stronger in cis than in trans (10). Although our studies at the HIS4 locus involved closely linked hot spots (<1 kb apart), Wu and Lichten (52) found competitive interactions between hot spots located as much as 17 kb apart.
A variety of mechanisms have been proposed to explain competitive hot spot interactions (10, 52). Hot spots may compete for the binding of proteins that stimulate recombination, directly or indirectly. Since the competition appears stronger in cis than in trans, the hot spot-binding proteins may diffuse along the DNA molecule or be located near the chromosome in a region in which diffusion is restricted. Alternatively, it is possible that a change in chromatin structure associated with activation of one hot spot is propagated for long distances along the chromosome, inhibiting activation of other hot spots. Long-range changes in chromatin structure affecting gene expression have been seen in a variety of higher eukaryotes (51).
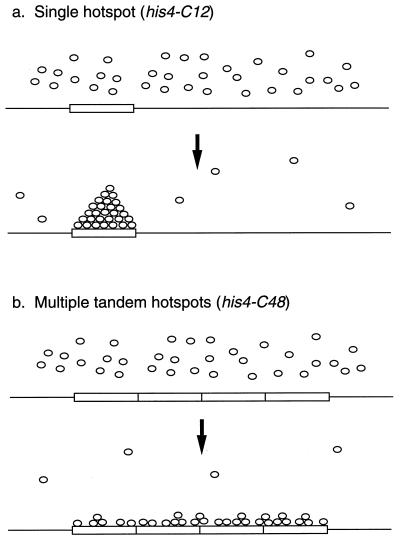
We favor a model in which adjacent hot spots compete for one or more recombination-initiating proteins. We suggest that a critical density of these proteins must assemble noncooperatively within a restricted region of DNA to catalyze a recombination event. Open chromatin regions may be preferred sites for this assembly. By this model (diagrammed in Fig. 7), the (CCGNN)12 insertion functions as one of these sites of assembly. Since the (CCGNN)48 tract is composed of four tandem copies of the (CCGNN)12 insertion, we suggest that the (CCGNN)48 insertion behaves as four adjacent and competing hot spots. Although the putative recombination-initiating proteins bind to the (CCGNN)48 insertion, they do not reach the critical density required for recombination. By this model, one would expect that the DNase I-sensitive region would be greater in strains with the (CCGNN)48 tract than the region in strains with the (CCGNN)12 tract. As discussed previously, this expectation is met (Fig. 2a), although the expanded region is not four times larger in the strain with the (CCGNN)48 insertion.
FIG. 7.
Model to explain hot spot activity of his4-C12 and cold spot activity of his4-C48. Tracts of (CCGNN)12 are indicated by rectangles, and small circles represent a DNA-binding protein involved in the initiation of recombination. (a) Single hot spot (his4-C12). A complex of the DNA-binding proteins forms on the CCGNN repeats required for the initiation of recombination at that site. Complex formation results in local depletion of free DNA-binding proteins, preventing complex formation at adjacent hot spots. (b) Multiple hot spots (his4-C48). Although the DNA-binding proteins attach to the CCGNN repeats, the critical density of these proteins is not achieved and recombination is not initiated. As in panel a, the concentration of free DNA-binding proteins is reduced, suppressing nearby recombination initiation events.
Several additional points concerning this model should be discussed. First, since the suppressive effects of the (CCGNN)48 tract are local, we postulate that the diffusion of the putative recombination-initiating protein is limited. Second, in Fig. 7, we show a single type of protein assembling at the hot spot. It is likely that multiple different proteins are involved in initiating meiotic recombination events (reviewed in reference 32), although a single protein may be rate limiting in the assembly of the recombination complex. Third, we suggested above that the (CCGNN)12 insertions might represent β hot spots (hot spots reflecting the structural properties of the DNA sequence), whereas his4-203 is likely to be an α hot spot (a hot spot requiring the binding of the Rap1p transcription factor [10]). The suppression of his4-203 activity by his4-C48 suggests that these two types of hot spots may compete for proteins required to initiate recombination. Fourth, we cannot exclude models in which the (CCGNN)12 insertion alters the chromatin in order to favor recombination, but the (CCGNN)48 insertion alters chromatin in such a way that recombination is suppressed. One argument against this possibility is the evidence that the suppressive interactions between hot spots do not involve any obvious change in chromatin structure (52).
Although recombination hot spots have been extensively characterized in S. cerevisiae (24), cold spots have not been extensively studied. A region near the centromere of yeast chromosome III suppresses meiotic recombination (22, 23), although the magnitude of this effect is small (41). In S. pombe, meiotic recombination is suppressed in the region between the mat2 and mat3 genes (6). This suppression of recombination is associated with repressed transcription, and mutations that derepress transcription also increase the level of crossing over (7, 43). Thus, the lack of recombination may reflect formation of a “silencing” type of chromatin structure (reviewed in reference 18). Since the recombination-suppressing effects of the (CCGNN)48 insertion are associated with the activation of transcription, there appear to be at least two different mechanisms for generating a cold spot.
In summary, when the wild-type upstream regulatory sequences of HIS4 are replaced with (CCGNN)48, (CCGNN)12, or (CCGAT)12 sequences, transcription of HIS4 is activated. (CCGNN)12 and (CCGAT)12 insertions also activate meiotic recombination, whereas the (CCGNN)48 sequence strongly represses local recombination. We suggest that the suppression of recombination by the (CCGNN)48 insertion is caused by competitive interactions between adjacent (CCGNN)12 hot spots.
ACKNOWLEDGMENTS
The research was supported by National Institutes of Health grants GM24110 (T.D.P.) and GM31819 (J.D.G.). D.T.K. is a Special Fellow of the Leukemia Society of America.
We thank J. Gerton, H. Moore, G. Fink and K. Struhl for useful discussions.
REFERENCES
- 1.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 2.Arndt K T, Styles C, Fink G R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987;237:874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- 3.Detloff P, Sieber J, Petes T D. Repair of specific base pair mismatches formed during meiotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:737–745. doi: 10.1128/mcb.11.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Detloff P, White M A, Petes T D. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics. 1992;132:113–123. doi: 10.1093/genetics/132.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devlin C, Tice-Baldwin K, Shore D, Arndt K T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991;11:3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egel R. Two tightly-linked silent cassettes in the mating-type region of Schizosaccharomyces pombe. Curr Genet. 1984;8:199–203. doi: 10.1007/BF00417816. [DOI] [PubMed] [Google Scholar]
- 7.Ekwall K, Ruusala T. Mutations in rik1, clr2, clr3, and clr4 asymmetrically derepress the silent mating-type loci in fission yeast. Genetics. 1994;136:53–64. doi: 10.1093/genetics/136.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan Q-Q, Xu F, Petes T D. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol Cell Biol. 1995;15:1679–1688. doi: 10.1128/mcb.15.3.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Q-Q, Petes T D. Relationship between nuclease-hypersensitive sites and meiotic recombination hot spot activity at the HIS4 locus of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2037–2043. doi: 10.1128/mcb.16.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Q-Q, Xu F, White M A, Petes T D. Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae. Genetics. 1997;145:661–670. doi: 10.1093/genetics/145.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goyon C, Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol. 1993;13:373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffith J. Chromatin structure: deduced from a mini-chromosome. Science. 1975;187:1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- 13.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 14.Hartzog G A, Winston F. Nucleosomes and transcription: recent lessons from genetics. Curr Opin Genet Dev. 1997;7:192–198. doi: 10.1016/s0959-437x(97)80128-1. [DOI] [PubMed] [Google Scholar]
- 15.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayasena S D, Behe M J. Competitive nucleosome reconstitution of polydeoxynucleotides containing oligoguanosine tracts. J Mol Biol. 1989;208:297–306. doi: 10.1016/0022-2836(89)90390-2. [DOI] [PubMed] [Google Scholar]
- 17.Kirkpatrick D T, Fan Q-Q, Petes T D. Maximal stimulation of meiotic recombination by a yeast transcription factor requires the transcription activation domain and a DNA binding domain. Genetics. 1999;152:101–115. doi: 10.1093/genetics/152.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klar A J S. Molecular genetics of fission yeast cell type: mating type and mating-type interconversion. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 745–777. [Google Scholar]
- 19.Kon N, Krawchuk M D, Warren B G, Smith G R, Wahls W P. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1997;94:13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornberg R D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 21.Kornberg R D, Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–589. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- 22.Lambie E, Roeder G S. Repression of meiotic crossing-over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics. 1986;114:769–789. doi: 10.1093/genetics/114.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambie E J, Roeder G S. A yeast centromere acts in cis to inhibit meiotic gene conversion of adjacent sequences. Cell. 1988;52:863–873. doi: 10.1016/0092-8674(88)90428-x. [DOI] [PubMed] [Google Scholar]
- 24.Lichten M, Goldman A S H. Meiotic recombination hotspots. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 25.Mavrothalassitis G J, Watson D K, Papas T S. The human ETS-2 gene promoter: molecular dissection and nuclease hypersensitivity. Oncogene. 1990;5:1337–1342. [PubMed] [Google Scholar]
- 26.Nag D K, White M A, Petes T D. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989;340:318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- 27.Nagawa F, Fink G R. The relationship between the “TATA” sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1985;82:8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohta K, Shibata T, Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perkins D. Biochemical mutants in the smut fungus Ustilago maydis. Genetics. 1949;34:607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J R, Jones E W, Pringle J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- 31.Rhodes D. Nucleosome cores reconstituted from poly(dA-dT) and the octamer of histones. Nucleic Acids Res. 1979;6:1805–1816. doi: 10.1093/nar/6.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roeder G S. Meiotic chromosomes: it takes two to tango. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Schultes N P, Szostak J W. A poly(dA-dT) tract is a component of the recombination initiation site at the ARG4 locus in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:322–328. doi: 10.1128/mcb.11.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimada T, Inokuchi K, Nienhuis A W. Chromatin structure of the human dihydrofolate reductase gene promoter. J Biol Chem. 1986;261:1445–1452. [PubMed] [Google Scholar]
- 36.Shrader T E, Crothers D M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solomon F, Connell L, Kirkpatrick D, Praitis V, Weinstein B. Methods for studying the cytoskeleton in yeast. In: Carraway K L, Carraway C A C, editors. The cytoskeleton: a practical approach. New York, N.Y: IRL Press; 1992. pp. 197–221. [Google Scholar]
- 38.Stapleton A, Petes T D. The Tn3 β-lactamase gene acts as a hotspot for meiotic recombination in yeast. Genetics. 1991;127:39–51. doi: 10.1093/genetics/127.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Struhl K. Yeast transcriptional regulatory mechanisms. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- 40.Sun H, Treco D, Schultes N P, Szostak J W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 41.Symington L S, Petes T D. Meiotic recombination within the centromere of a yeast chromosome. Cell. 1988;52:237–240. doi: 10.1016/0092-8674(88)90512-0. [DOI] [PubMed] [Google Scholar]
- 42.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break-repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 43.Thon G, Cohen A, Klar A J S. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics. 1994;138:29–38. doi: 10.1093/genetics/138.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tice-Baldwin K, Fink G R, Arndt K T. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science. 1989;246:931–935. doi: 10.1126/science.2683089. [DOI] [PubMed] [Google Scholar]
- 45.Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y-H, Griffith J D. The [(G/C)3NN]n motif: a common DNA repeat that excludes nucleosomes. Proc Natl Acad Sci USA. 1996;93:8863–8867. doi: 10.1073/pnas.93.17.8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y-H, Gellibolian R, Shimizu M, Wells R D, Griffith J D. Long CCG triplet repeats exclude nucleosomes: a possible mechanism for the nature of fragile sites in chromosomes. J Mol Biol. 1996;263:511–516. doi: 10.1006/jmbi.1996.0593. [DOI] [PubMed] [Google Scholar]
- 48.White M A, Detloff P, Strand M, Petes T D. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr Genet. 1992;21:109–116. doi: 10.1007/BF00318468. [DOI] [PubMed] [Google Scholar]
- 49.White M A, Dominska M, Petes T D. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:6621–6625. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White M A, Wierdl M, Detloff P, Petes T D. DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc Natl Acad Sci USA. 1991;88:9755–9759. doi: 10.1073/pnas.88.21.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 52.Wu T-C, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu L, Kleckner N. Sequence non-specific double-strand breaks and interhomology interactions prior to double-strand break formation at a meiotic recombination hotspot in yeast. EMBO J. 1995;16:5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]