Abstract
Research focusing on the gut-brain axis is growing, but the interplay of ethanol (alcohol molecule), the gut microbiome, the brain and behavior is poorly understood. In the current study, we remodeled the gut microbiota by providing adult male C57BL/6J mice with a non-absorbable antibiotic cocktail (ABX) in the drinking water and tested ethanol consumption behavior in a binge-like “Drinking in the Dark” model. Notably, 2 weeks of ABX pre-treatment significantly increased ethanol consumption during the 6 weeks of ethanol exposure in the DID paradigm. ABX treatment also appeared to prevent anxiety-like behavior during ethanol withdrawal period. ABX-treated mice expressed reduced bacterial diversity and modified microbiota compositions within cecal samples. There were drastically reduced levels of commensal Firmicutes and increases in the Bacteroidetes and Verrucomicrobia populations. Importantly, the relative abundance of Firmicutes inversely correlated to ethanol intake levels regardless of antibiotic treatment, whereas Bacteroidetes and Verrucomicrobia populations positively correlated to ethanol intake levels. This is the first report demonstrating that ABX-induced disruption of the gut commensal microbiota leads to increased ethanol consumption in mice. This work reveals an important relationship between the gut microbiota and ethanol consumption behavior and supports the use of microbial-targeted approaches to study gut-brain interactions during alcohol use disorder.
Keywords: Alcohol use disorder, antibiotics, ethanol intake, ethanol withdrawal-related anxiety behavior, gut-brain axis, microbiome diversity
Introduction
Alcohol use disorder (AUD) is a common and complex disease that is a major contributor to economic and health problems across the world. The cost of excessive alcohol use nears $250 billion, with the majority (~77%) being due to binge drinking (Sacks et al., 2015). The World Health Organization’s Global Status on alcohol and health reported that 5.9% of all global deaths were attributable to alcohol exposure (WHO, 2014). There are also more than 17 million people in the US suffering from AUD, yet only 4% of individuals diagnosed are currently being prescribed any pharmacotherapy (Franck and Jayaram-Lindstrom, 2013). Moreover, among these treated individuals, there is up to a 70% relapse rate within the first year of treatment (Moos and Moos, 2006), partially due to insufficient pharmacotherapies for AUD. These findings illustrate the need for studies identifying mechanisms linked to early signs of excessive use and enhanced susceptibility to alcohol abuse. This information will enable the development of more effective options for preventative and intervention-based AUD treatment strategies.
Building evidence links the commensal microbiota ecosystem to healthy functioning of several host activities, including immune system development and function, mood regulation, and behavior (Foster et al., 2017; Thion et al., 2018). Consequently, disruption to the dynamic balance of the microbiome (dysbiosis) has been related to numerous neuropathologies including Parkinson’s disease and Alzheimer’s disease (Cox and Weiner, 2018; Minter et al., 2016). Individuals who excessively drink alcohol have also been shown to express altered bacterial compositions compared to moderate and non-drinkers (Couch et al., 2015; Dubinkina et al., 2017; Mutlu et al., 2012). Although these studies have shown connections between AUD and the microbiome, there is insufficient information describing how changes in the microbiome may influence alcohol abuse behavior. Chronic exposure to alcohol has been related to microbial dysbiosis, intestinal permeability (“leaky gut”) and changes in immune responses (Engen et al., 2015; Leclercq et al., 2017a). Notably, release of gut bacterial fragments such as endotoxins into circulation and subsequent increases in inflammatory cytokines have been observed in those who consume alcohol excessively and within animal models of ethanol exposure (Mutlu et al., 2012; Qin et al., 2008). Systemic inflammation can lead to depression-like symptoms and sickness behavior, major factors that contribute to self-medication and increased voluntary ethanol consumption and seeking behavior in rodent models (Asatryan et al., 2015; Blednov et al., 2011). Changes in metabolites linked to gut microbiota may also have an influence on ethanol consumption behavior. As such, the plasma levels of short-chain fatty acids (SCFAs), i.e. propionate, acetate and butyrate, were shown to be associated with AUD (Bjorkhaug et al., 2019; Mostafa et al., 2017). Collectively, these findings highly suggest the gut microbiome plays an important role in behaviors and pathologies related to AUD.
Investigations are beginning to utilize microbiome manipulation to modify neuropathology and behavior. For example, studies using sterile-bred, germ-free animals observed reduced anxiety-like symptoms typical for major depressive disorder (Neufeld et al., 2011). Additional studies report that germ-free mice exhibit altered behaviors in models of Parkinson’s disease and Alzheimer’s disease (Cox and Weiner, 2018; Minter et al., 2016; Sampson et al., 2016b). Unfortunately, germ-free animal models are often complicated due to aberrant development of immune systems and other host functions. They insufficiently represent the conditions of bacteria-host interactions. Alternatively, antibiotic-based strategies to reduce and modify the gut microbiota in adult animal subjects have provided insights into gut-brain interactions without compromising normal host development. Antibiotic treatment has been shown to ameliorate neuroinflammation and motor deficits in Parkinson’s disease and anxiety behavior in rodent models of avoidance (Foster et al., 2017; Sampson et al., 2016b). Furthermore, the anthelmintic ivermectin and antibiotics, such as ceftriaxone and tigecycline, have been observed to reduce ethanol intake in animal models (Asatryan et al., 2014; Bergeson et al., 2016; Sari et al., 2011). Most of these studies did not explore connections to changes in the gut microbiome and its possible causative role in mediating observed behaviors. However, a few studies have been investigating the causal link between changes in microbial populations and neuropathology. For example, Sampson et al. established the ability of gut microbiome fecal matter transplantation to enhance motor deficits in a mouse model of Parkinson’s disease (Sampson et al., 2016a), pointing to the importance of considering the gut microbiome as a causal player in contributing to pathological host behavior.
In this study, we set forth a hypothesis that antibiotic-induced alterations in the gut microbiome could affect host ethanol consumption behavior. To test this hypothesis, we remodeled the gut microbiome using ad libitum administration of non-absorbable antibiotics in male C57BL/6J mice and measured drinking behavior in a “Drinking in the Dark” (DID) binge-like model of voluntary ethanol intake. In addition, we identified correlations between changes in specific microbial populations and drinking levels.
Results
ABX treatment did not affect liquid/food intake or body weights but affected cecal and adipose tissue
We treated adult male mice with an antibiotic cocktail (ABX), which has previously been shown to reduce gut microbiome diversity with minimal systemic effects to host tissue (Bercik et al., 2011). ABX was administered ad libitum in the drinking water 2 wks prior to the start of DID exposure (Fig. 1). Additionally, ABX treatment was continued throughout the 6 wk duration of DID to maintain microbiota dysbiosis (Fig. 1). Liquid intake, either water or ABX, did not significantly differ between any of the groups (Fig. 2A; two-way RM ANOVA: F(3,28)=1.1, p=0.35). Food consumption remained similar (Fig. 2B; two-way RM ANOVA: F(3,28)=1.2, p=0.35), and although all mice steadily gained weight throughout the study, average weight did not differ between groups at each week (Fig. 2C; two-way RM ANOVA: F(3,28)=2.5, p=0.08).
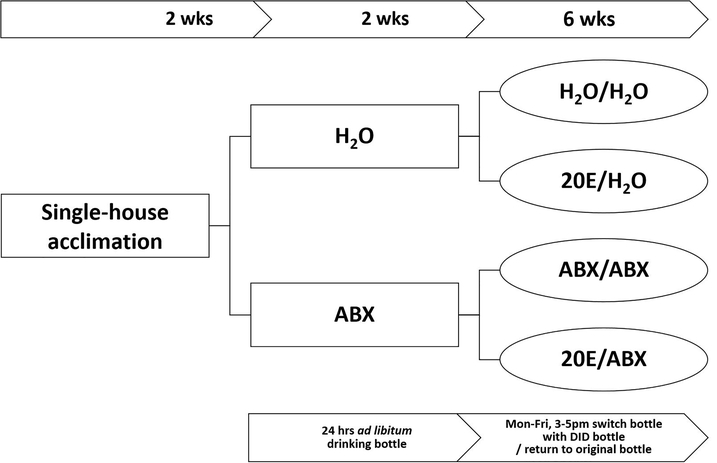
Figure 1. Schematic representation of the animal treatment procedures.
Single-housed mice had access to water (H20) or were treated ad libitum in drinking water with an antibiotic cocktail (ABX) for 2 wks prior to DID ethanol exposure and throughout the 6-wk duration of DID. This generated the following treatment groups: Water groups – H2O; H2O-20E; ABX groups – ABX; ABX-20E.
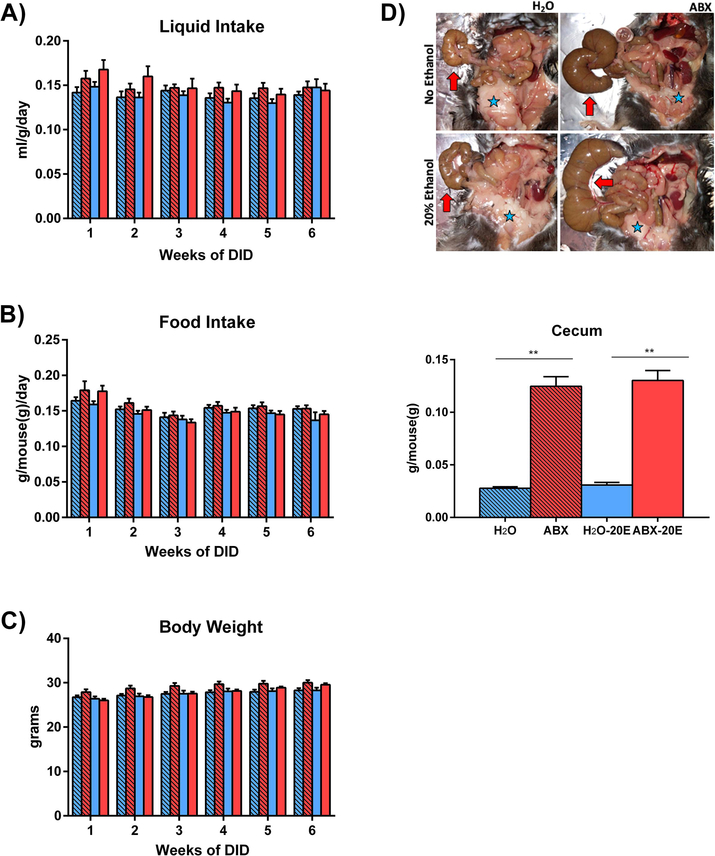
Figure 2. ABX treatment did not alter liquid/food intake and body weight but affected cecum and adipose tissues.
A) Liquid (H2O or ABX) and B) food intake, as well as C) body weight did not significantly differ between any of the four groups at any point during the study: H2O, ABX, H2O-20E, ABX-20E. D) (Top panel) Post-mortem necropsy images demonstrating antibiotic-induced cecum enlargement (arrows) and reduction in white adipose tissue (stars). (Bottom panel) Bar graph demonstrating analyzed cecum weights. ABX-treated mice exhibited significantly increased cecum weights compared to untreated controls. Data are presented as Mean ± SEM, n=8/group; **p<0.01.
Mice with prolonged 8 wks ABX treatment, with and without 20E exposure, were observed to have engorged ceca and less prominent adipose tissue compared to untreated mice (Fig. 2D, Top). ABX-treated mice had significantly heavier ceca compared to controls (Fig. 2D, Bottom; Kruskal-Wallis test: **p<0.01)
ABX treatment increased ethanol consumption in a binge-like ethanol intake model
There was a significant combined effect of ABX treatment and 20E consumption for the total 6 wk duration of 20E consumption (Fig. 3A; two-way RM ANOVA, F(1,14)=59.81, p<0.01; ABX-20E compared to H2O-20E). After 2 wks of ABX treatment alone, ABX-20E mice immediately and consistently consumed significantly higher amounts of 20E, range of 6.1±0.5 – 8.21±1.2 g/kg/2hrs, compared to untreated mice that consumed 4.7±0.5 – 7.24±1.3 g/kg/2hrs (Fig. 3, FDR-Benjamini and Hochberg, *p<0.05, **p<0.01; n=8/group). We also performed daily (Mon-Fri) statistical analysis of ethanol drinking throughout 6 wks of DID. Two-way RM ANOVA tests showed no effect of “Day of Week” or combined “Day of Week” and “Treatment (ABX or untreated); p=0.22 and p=0.93, respectively. Main effect of treatment was found at p<0.01. Post hoc Benjamini-Hochberg tests did not find significant day-to-day (Mon-Friday) differences in 20E intake levels within treatment groups, either within week of DID or between weeks of DID.
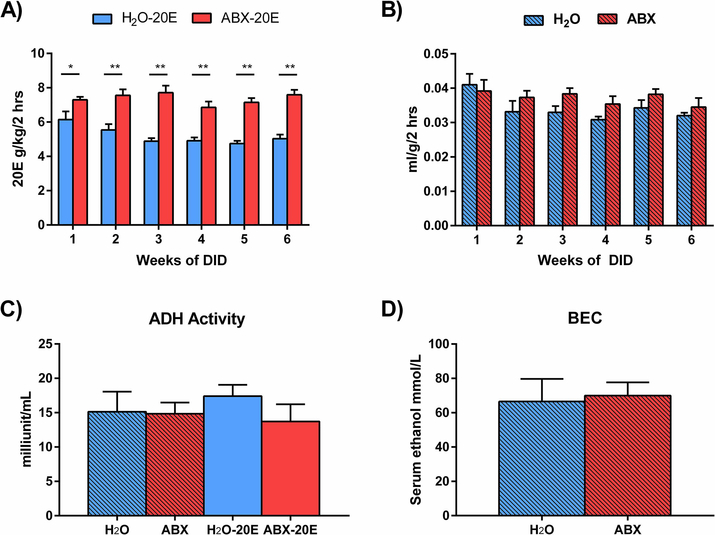
Figure 3. ABX-treated mice exhibited higher levels of ethanol consumption.
A-C) For these experiments, single-housed mice were treated 24 hrs ad libitum with antibiotics (ABX) or H2O for 2 wks prior to and throughout the 6 wks of DID except during the 2 hrs of ethanol exposure. Daily 20E consumption was measured and compared between groups. A) ABX-20E mice consumed significantly more ethanol than H2O-20E mice consistently throughout the 6 wks of the study. B) H2O and ABX groups that were not exposed to 20E did not significantly differ in the amount of H2O or ABX consumed during the 2 hrs of DID, and C) liver alcohol dehydrogenase (ADH) activity did not different between any of the groups at the conclusion of this experiment. D) In a separate cohort, 2 wks of ABX treatment did not significantly affect serum blood ethanol concentration (BEC) 45 mins following IP injection of ethanol (3.5 g/kg per mouse weight). Data are presented as Mean ± SEM, n=4–8/group; *p<0.05, **p<0.01.
We addressed the possibility that the dark-phase time frame chosen (3 pm - 5 pm) may be linked to general increases in liquid intake of ABX mice. Liquid intake, either H2O or ABX, did not significantly differ between non-20E exposed mice (H2O, ABX) during the 2 hrs of DID (Fig. 3B; two-way RM ANOVA, F(1,14)=3, p=0.11; ABX compared to 20E, n=8/group). This suggests the increase in 20E consumption was due to enhanced ethanol seeking behavior rather than a general increase in liquid intake during the 2 hr DID exposure period of the nocturnal mouse model.
There was no apparent effect of ABX on alcohol dehydrogenase (ADH) activity in the liver and no significant differences in ADH activity between treatment groups after DID exposure (Fig. 3C; One-way ANOVA, p=0.65; n=4/group). Additionally, 2 wks of ABX treatment did not appear to have an effect on serum blood ethanol concentrations (BECs) after intraperitoneal injections with a single dose of ethanol at 3.5 g/kg per mouse weight. Following 45 min after ethanol injections, BECs were not significantly different between ABX-treated mice and untreated controls (Fig. 3D; Mann-Whitney test, p=0.64; n=5–7/group).
ABX treatment prevented anxiety-like behavior during ethanol withdrawal
We conducted the elevated plus maze (EPM) behavioral test in a separate experimental cohort in order to test whether the higher ethanol consumption levels with ABX treatment were related to increased anxiety-like phenotypes. At baseline and following 2 wks of ABX treatment or H2O control, there were no significant differences in the duration of time spent in the open arms between H2O and ABX (Fig. 4A; two-way RM ANOVA, Baseline p=0.03 and q=0.06, Week 2 p=0.74 and q=0.74; n=11–13/group). Similarly, there were also no significant differences in the duration of time spent in the closed arms between H2O and ABX at baseline and following 2 wks of treatment (Fig. 4B; two-way RM ANOVA, Baseline p=0.27 and p=0.55, Week 2 p=0.70 and q=0.70; n=11–13/group). Of note, we observed significant reduction in the duration spent in both open and closed warms when comparing baseline and 2 wks of ABX treatment or H2O control within groups (Fig. 4A,B; two-way RM ANOVA, **p<0.01 and **q<0.01 for each treatment group between baseline and wk 2 for open and closed arm durations). These changes are likely increased habituation effects due to mice becoming familiar with the behavioral test apparatus (Schrader et al., 2018).
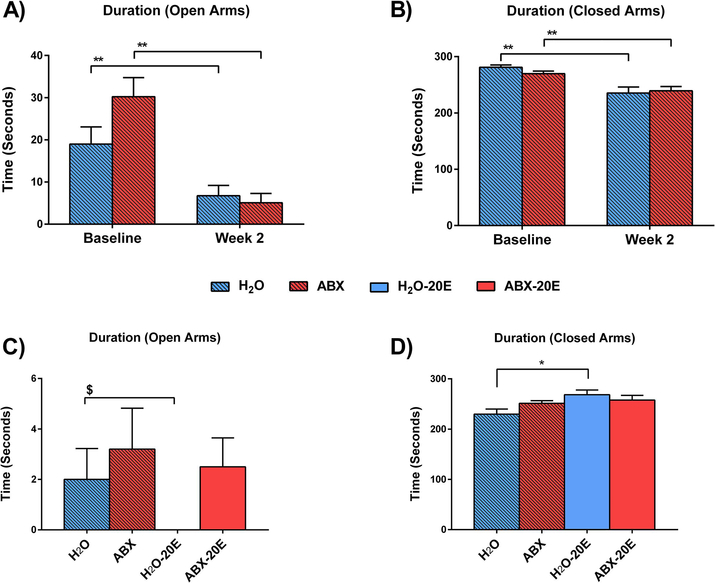
Figure 4. Elevated plus maze (EPM) behavioral test revealed reduced anxiety-related behavior in ABX-20E mice.
(A,B) EPM baseline compared to after 2 wks ABX, (C,D) EPM after 2 weeks DID, with or without ABX treatment. The duration of time spent in the A) open arms and B) closed arms at baseline compared to after 2 wks of ABX treatment. The duration spent within C) open arms and D) closed arms after 2 weeks of DID. Data are presented as Mean ± SEM, two-way RM ANOVA (A,B; n=4–7/group), one-way ANOVA (C,D; n=11–13/group; *p<0.05, **p<0.01) and unpaired t-test (C; $p=0.04).
To further asses anxiety-like behavior due to ethanol withdrawal, we tested mice in EPM after 2 wks of 20E exposure during DID (with and without ABX treatment). The behavioral testing in ethanol-exposed groups (H2O-20E, ABX-20E) occurred during the ethanol withdrawal period, i.e. 16–20 hrs after the last acquisition of 20E. Non-DID groups (H2O, ABX) were tested in parallel to the 20E groups. Mice in the ABX and ABX-20E groups spent similar time in the open as well as closed arms compared to the mice in the H2O group (Fig. 4C,D). H2O-20E mice were observed not to spend any time within the open arms compared to H2O controls and ABX-treated groups. The difference between H2O-20E and H2O mice was statistically significant using the unpaired t-test (Fig. 4C, $p=0.04 for H2O-20E compared to H2O) but did not reach significance according to one-way ANOVA (Fig. 4C). Consequently, mice in the H2O-20E group spent significantly more time in the closed arms compared to H2O mice (Fig. 4D; One-way ANOVA, *p=0.019; n=4–7/group).
Cecal microbiome is significantly altered in ABX-treated mice
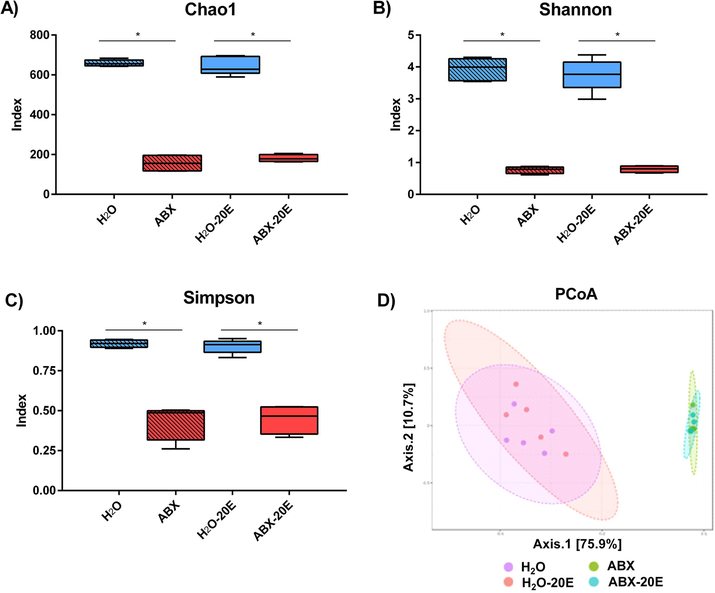
We analyzed the microbiome isolated from cecal samples at the end of DID exposure. As expected, ABX significantly altered the microbiome composition, whereas changes due to ethanol alone were much less pronounced. According to several α-diversity measures (Chao1, Shannon, Simpson indices), ABX treatment drove the observed reductions in diversity. Chao1 index (more emphasis on low abundance species) was significantly altered in ABX-treated groups (ABX and ABX-20E compared to H2O and H2O-20E treated groups, Fig. 5A; Kruskal-Wallis test, *p<0.05, n=4–5/group). Similarly, α-diversity indicating taxa richness and evenness were significantly different between the ABX exposed groups and controls according to Shannon and Simpson indices (Fig. 5B,C; Kruskal-Wallis tests, *p<0.05 for both measures, n=4–5/group). We further compared microbiome β-diversity, the similarities and dissimilarities between different treatment groups, visualized by Principal Coordinates Analysis (PCoA – Fig. 5D). ABX and ABX-20E mice groups clustered together and were significantly dissimilar from both H2O and H2O-20E groups which also clustered together (Fig. 5D; PERMANOVA, F(15.704), p<0.001, r2=0.771, n=4–5/group).
Figure 5. ABX treatment significantly reduced gut microbiome diversity.
Microbiome αdiversity compared from cecal samples of untreated and ABX-treated mice (with and without prolonged ethanol exposure) using A) Chao1, B) Shannon, and C) Simpson Indices. D) β-diversity is measured by Principal Coordinates Analysis (PCoA) of cecal samples at the OTU (observational taxonomic unit: unique species) level and identifies treatment groups which cluster together according to community compositions based on treatment group. Data are presented as Median and Min/Max, Kruskal-Wallis test, n=4–5/group; *p<0.05.
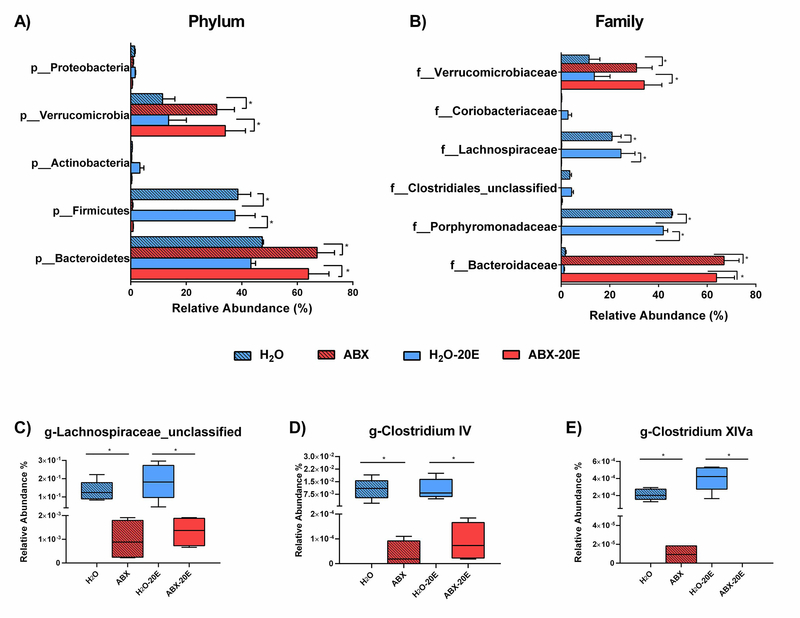
As in the β-diversity clustering of H2O groups (H2O and H2O-20E) and ABX groups (ABX and ABX-20E), similar pairings were observed when investigating specific cecal microbiota taxa differences and relative abundances (%) (Fig. 6). As presented in Fig. 6A, the main phyla constituting the majority for microbiome in H2O and H2O-20E mice were Bacteroidetes (47.4%, 43.3), Firmicutes (38.6%, 37.6%) and Verrucomicrobia (11.5%, 13.62%). ABX and ABX-20E mice also had phyla predominated by Bacteroidetes (67.2%, 64.0% ) and Verrucomicrobia (31.0%, 34.1%) though the relative abundance of these phyla were significantly higher than in H2O groups (Fig. 6A; two-way ANOVA, *p<0.05; n=4–5/group). Additionally, compared to the H2O groups there was a significant reduction of Firmicutes phyla in ABX groups (<1% for both ABX-treated groups) (Fig. 6A). Other phyla such as Proteobacteria and Actinobacteria were present in lower levels in all treatment groups (Fig. 6A).
Figure 6. ABX treatment had a greater effect on the gut microbiome of male C57BL/6J mice than ethanol (20E) exposure.
A) Phylum-level (“p_”) and B) Family-level (“f_”) relative abundance (%) of gut microbiota taxa recovered in cecal samples of untreated mice and Abxtreated mice (8 wks) with and without prolonged 20E exposure (6 wks). C-E) At the genus-level, butyrate-producing bacteria (Lachnospiraceae_unclassified, Clostridium Cluster IV, and Clostridium Cluster XIVa) were significantly reduced in ABX-treated mice compared to untreated controls. Data are presented as Median and Min/Max, Kruskal-Wallis test, n=4–5/group; *p<0.05.
There were also family-level differences between H2O and ABX groups (Fig. 6B). Families within H2O groups consisted of dominating families, Porphyromonadaceae (45.4%, 42.0%) and Lachnospiraceae (20.9%, 24.6%), Verrucomicrobiaceae (11.5%, 13.6%) and minor constituents (Clostridiales-unclassified, Coriobacteriaceae). Whereas, microbiota in ABX groups largely were comprised of two families, Bacteroidaceae (66.9%, 63.8%) and Verrucomicrobiaceae (31.0%, 34.1%). Lower abundances of the other minor families were found in ABX groups. Compared to H2O groups, profound reductions were observed in the abundances of Porphyromonadaceae and Lachnospiraceae and increases in Bacteroidaceae and Verrucomicrobiaceae families (Fig. 6B; two-way ANOVA, *p<0.05; n=4–5/group). Finally, when comparing relative abundances of butyrate-producing bacteria, ABX and ABX-20E treated groups had significantly reduced genus numbers of Lachnospiraceae_unclassified, Clostridium Cluster IV, and Clostridium Cluster XIVa populations (Fig. 6C–E; Kruskal-Wallis tests, *p<0.05; n=4–5/group). Table 1 further describes significant differences between treatment groups when considering absolute number of bacteria for each taxa group in 25 ng of DNA sample (Table 1; two-way ANOVA multiple comparisons, FDR Benjamini and Hochberg corrections, *p<0.05 and q<0.05; n=4–5/group).
Table 1.
Phyla and families significantly different in H2O, H2O-20E, ABX, ABX-20E adult male C57BL/6J mice (FDR, *p>0.05 and q<0.05)
| Phylum | Family | p value | q value | Bacterial counts per 25 ng (DNA) |
|||
|---|---|---|---|---|---|---|---|
| H2O | H2O-20E | ABX | ABX-20E | ||||
| Bacteroidetes* | 0.0033 | 0.0126 | 25815.8 | 23545.8 | 36649 | 34903.5 | |
| Bacteroidaceae* | 0.0047 | 0.0160 | 927 | 624.4 | 36536.75 | 34798 | |
| Porphyromonadaceae* | 0.0038 | 0.0160 | 24717.4 | 22817.6 | 107.75 | 98 | |
| Firmicutes* | 0.0053 | 0.0126 | 21089.2 | 20570.4 | 343 | 465.75 | |
| Clostridiales_unclassified* | 0.0044 | 0.0160 | 1969.8 | 2275.4 | 14.5 | 157.75 | |
| Lachnospiraceae* | 0.0054 | 0.0176 | 11376.8 | 13498.8 | 96.75 | 95.75 | |
| Ruminococcaceae* | 0.0030 | 0.0160 | 6809.4 | 3306.6 | 42.5 | 23.25 | |
| Actinobacteria* | 0.0048 | 0.0126 | 299.4 | 1798.4 | 167 | 207.75 | |
| Coriobacteriaceae* | 0.0019 | 0.0160 | 123.2 | 1526.4 | 1.25 | 3.75 | |
| Verrucomicrobia | 0.0531 | 0.0579 | 6223.2 | 7436 | 16851.5 | 18553 | |
| Verrucomicrobiaceae | 0.0531 | 0.0748 | 6223.2 | 7436 | 16851.5 | 18553 | |
| Proteobacteria* | 0.0367 | 0.0490 | 811.2 | 871.4 | 468.25 | 343 | |
Correlations between total ethanol consumption and relative abundances of cecal microbiome phyla
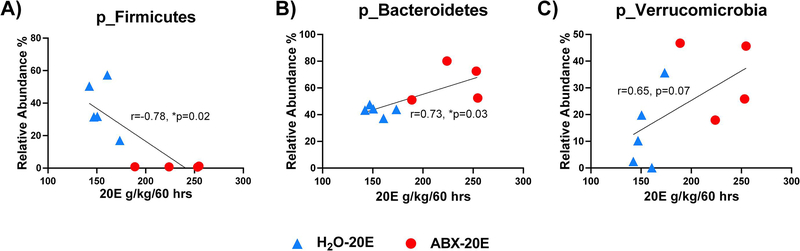
The total 20E intake data (g/kg/60 hrs) was analyzed per mouse over the 6 wks of DID exposure from H2O-20E and ABX-20E groups and correlated to the relative abundance (%) of Firmicutes, Bacteroidetes, and Verrucomicrobia phyla (Fig. 7A–C; Spearman r correlation, *p<0.05, **p<0.01; relative abundance (%) of bacterial population compared to total ethanol consumption 20E g/kg/60 hrs; n=4–5/group). Total 20E intake was found to be significantly inversely correlated with Firmicutes relative abundance (%) levels (Fig. 7A; Spearman r correlation, *p=0.02). Total 20E intake was also found to be significantly positively correlated with Bacteroidetes levels, and, though not reaching significance, showed a positive trend towards enhancement of Verrucomicrobia levels (Fig. 7B,C; Spearman r correlation, *p=0.03 and p=0.07, respectively).
Figure 7. Total ethanol intake correlated with differences in phyla microbiome populations.
Accumulated 20E consumption was calculated per mouse (H2O-20E and ABX-20E data shown together) across the 6 wks (60 hrs) of DID exposure. ABX-20E mice (red dots) clustered similarly and separately from H2O-20E mice (blue triangles). Total 20E intake significantly correlated with relative abundances (%) of phyla A) Firmicutes reductions and B) Bacteroidetes enhancement. C) Though not significant, total 20E intake trended towards an increase in Verrucomicrobia. Spearman r correlation, n=4–5/group; *p<0.05, **p<0.01.
Discussion
The present investigation tested the hypothesis that remodeling the commensal microbiota using antibiotics will significantly influence ethanol consumption levels. In agreement, we found that non-absorbable, oral antibiotic treatment significantly increased voluntary ethanol consumption levels in a binge-like drinking model (DID) in adult male C57BL/6J mice. The increase in ethanol intake was not likely due to differences in ethanol metabolism or absorption. ADH activity measured in liver samples at the end of the ABX and DID exposure was similar between ABX-treated mice and water controls. In addition, BECs measured after a single high dose ethanol challenge did not differ between ABX and water treated mice.
It is important to emphasize that the increase in ethanol intake in ABX treated mice was evident immediately from the first week of ethanol exposure and was maintained throughout the 6 wks of DID procedure. Absence of time-dependent effects of DID suggests the differences between ABX-treated and H2O control mice, with respect to ethanol intake, were not due to plasticity changes that may have been caused by longer ethanol exposure. This effect is likely related to the changed gut microbiome at the start of ethanol exposure due to the prior 2 wks of ABX treatment. We found large reductions in microbiome diversity and shifts in specific microbial populations in ABX-treated mice. Similar changes in gut microbiome diversity and compositions were found in other antibiotic cocktail-based studies (Bercik et al., 2011). In fact, our microbiome analysis findings revealed that ABX treatment, not ethanol exposure, was the main factor affecting changes in microbial populations. Moreover, changes in specific microbial phyla significantly correlated with total ethanol intake. These findings indicate that changes in microbiome diversity and composition may mediate the observed elevated baseline ethanol consumption.
We show that ethanol consumption remained consistent throughout 6 wks of ethanol exposure. There are other studies which have found gradual increases in ethanol intake over time (Hwa et al., 2011). This could be due to variations of the DID procedure that includes differences in ethanol concentration, the length of time into the dark cycle when ethanol was offered and mouse age (Rhodes et al., 2005). The DID procedure adopted in this study was similar to the one previously used by our group, which included access to 20E for 2 hrs in the dark cycle for 5 consecutive days a week. Using this approach, the previous work from our group reported a stable drinking pattern in adult C57BL/6J mice which is consistent with our findings and others (Crabbe et al., 2012; Huynh et al., 2017). In addition, we believe that animal age is another factor in the observed stable drinking pattern. In fact, it has been demonstrated that 8 wks old adult C57BL/6J mice consume stable 20E amounts in the DID model compared to 4 wks old adolescent mice of the same strain (Becker and Lopez, 2004). At the start of our DID study, C57BL/6J mice were 8–10 wks old, which may explain the low fluctuations in ethanol consumption in both ABX-treated and water control mice.
Currently, mechanisms underlying gut-brain associations are not fully established, especially regarding alcohol consumption behavior and AUD. People with alcohol dependence demonstrate shifts in microbiota compositions compared to non-dependent individuals (Leclercq et al., 2017b). Furthermore, anxiety and depression are known risk factors underlying AUD and have been linked to changes in the gut microbiome. Leclercq et al. observed that alcohol-dependent patients with altered microbiome compositions also had increased intestinal permeability, which was linked to higher scores of craving, anxiety, and depression (Leclercq et al., 2014). Another study demonstrated significant correlations between the gut microbiome and behavioral and neurophysiological traits defining AUD, such as impulsivity measures and augmentations in striatal dopamine receptor expressions (Jadhav et al., 2018). Ketamine treatment in rats has also been shown to have microbiome-associated effects on depression and inflammation (Getachew et al., 2019), both of which are implicated in alcohol abuse. The anti-depressive action of ketamine may be related to its ability to enhance Lactobacillus populations known to be significantly reduced in animal models of depression (Zheng et al., 2016).
We investigated the possibility that ABX-induced changes to the gut microbiome could enhance anxiety in mice leading to increased ethanol intake. In a separate cohort, we assessed anxiety-related behavior in an established EPM test. ABX alone did not affect anxiety-related behavior after 2 weeks of treatment. Moreover, ABX treatment appeared to reduce anxiety-like behaviors during ethanol withdrawal after DID exposure. These findings support the increase we observed in ethanol consumption with ABX treatment is not caused by ethanol withdrawal-related anxiety. Other studies support our findings suggesting that antibiotic treatment has been observed to reduce ethanol-related anxiety. For example, treatment with the antibiotic tigecycline reduced the severity of handling-induced convulsions during periods of acute ethanol withdrawal (Martinez et al., 2016). In addition, fecal matter transplantation from ethanol-exposed donor mice transferred behavioral signs of ethanol withdrawal-induced anxiety in alcohol-naïve mice (Xiao et al., 2018). Collectively, these studies and our initial data of modified withdrawal-induced anxiety-related behaviors support the notion that the gut-brain axis is implicated in alcohol consumption and related withdrawal behaviors in ABX-treated mice.
More extensive studies using ABX treatment approach are needed to elucidate the relationship between changes in microbiota and ethanol-induced anxiety and stress-related behaviors. However, our current studies are beginning to shed light into the gut-brain relationship regarding the observed increases in ethanol intake behavior. Although it has been difficult to pinpoint what distinguishes a healthy microbiome from a dysbiotic composition, a reduction in diversity within the population has been regarded as an indication of an unhealthy microbiome. Within the current study, we found shifts in commensal microbiota populations that clustered according to ABX-treated groups (ABX and ABX-20E) compared to non-ABX controls (H2O and H2O-20E). Both ABX-treated groups expressed lower population richness and decreased diversity compared to controls indicated by multiple measures, such as Chao1, Shannon index, and Simpson index, suggesting ABX-induced dysbiosis may play an important role in determining levels of ethanol intake.
Our current studies also identified shifts in specific bacterial populations within the ceca of higher ethanol drinking, ABX-treated mice, compared to water-control counterparts. ABX-treated mice had drastically reduced Firmicutes populations and enriched Bacteroidetes and Verrucomicrobia phyla. Firmicutes and Bacteroidetes represent the large majority of the microbiota, with both phyla representing up to 90% of bacteria containing both beneficial members and pathogenic species (Rinninella et al., 2019). Verrucomicrobia represent important mucin-degrading bacteria within the gastrointestinal tract and may account for the decreased adipose tissue observed during necropsy (Belzer and de Vos, 2012). Changes in these microbial populations correlated with total ethanol consumption levels, with negative correlations for the Firmicutes populations and positive correlations for Bacteroidetes and Verrucomicrobia phyla. Changes in these microbial populations with ABX treatment in our study parallel microbiome changes observed in mice after ethanol exposure (Engen et al., 2015). It is interesting to speculate the possibility that ABX treatment caused a microbiome phenotype that resembles the microbiome of ethanol-exposed mice, which could have primed the immediate high ethanol intake found in ABX treated mice.
Despite the several changes found in different microbial taxa, the most dramatic changes were found within the Firmicutes population with ABX treatment. There was a near absence for Firmicutes bacteria including family and genus members (Clostridiales_unclassified, Lachnospiraceae, Ruminococcaceae, Clostridium cluster IV and Clostridium cluster XIVa). Firmicutes phylum contains known beneficial commensal bacteria dominant in healthy populations (Lopetuso et al., 2013). For example, patients with irritable bowel syndrome have been shown to have significant reductions in the abundance of Lactobacillus and other Firmicutes members (Carroll et al., 2010). Reductions in Firmicutes have been shown in pathologies which appear to be unrelated to the gastrointestinal tract, including Alzheimer’s disease (Vogt et al., 2017) and autism spectrum disorder (Kang et al., 2017). These studies support the importance of Firmicutes populations and their involvement within the gut-brain axis in determining potential relationships between microbiota and host health and behavior.
In order to study the role of the microbiome in neuropathologies and behavior, antibiotic treatment strategies have been gaining popularity as seen within models of Parkinson’s disease (Sampson et al., 2016b) and rodent models of avoidance and anxiety-like behaviors (Foster et al., 2017). In another example, Kiraly et al. used an antibiotic approach to manipulate gut microbiota in a study of cocaine seeking behavior (Kiraly et al., 2016). These models have the advantage of observing animals with normally developed immune systems in contrast to germ-free animals that have major developmental issues (Kennedy et al., 2018). However, similar to germ-free animals, antibiotic-treated mice demonstrate some physiological aberrations, such as cecal engorgement (due to water retention) and reduced adipose tissue deposits (Reikvam et al., 2011). Consistent with these findings, the ABX-treated mice in our study demonstrated similar features of larger and heavier ceca and reduced adipose. Even though it was not within the scope of the current study to determine the consequences of these physiological changes and how they may relate to ethanol intake, we found no significant differences in food consumption between treatment groups. This suggests the differences in adipose tissue did not affect normal nutrition; however, it may be interesting to further look into the possibility that lower adipose mass could affect higher-caloric or sucrose-seeking feeding behaviors related to ethanol intake.
The function of gut bacteria and their metabolites on host activities may give insight into their links to alcohol abuse and related behaviors. Some members of the microbiota are important for fermenting undigested dietary materials to produce metabolites, which interact with the host to modulate health (Sharon et al., 2014). A major metabolic pathway involved in health modulation is the production of SCFAs, fatty acids with less than six carbon atoms such as butyrate and acetate. SCFAs can affect colonocytes, energy storage, metabolism, gut-barrier function, and the immune system (Chambers et al., 2018). SCFAs were also shown to modulate levels of free dopamine and norepinephrine (Asano et al., 2012) and impact behavior such as stress-responsivity, anxiety, and depression (van de Wouw et al., 2018). Specifically, butyrate is the major energy source for colonocytes and is linked to the maintenance of intestinal health and host metabolism (Chakraborti, 2015). Our findings demonstrate reductions in concentrations of butyrate-producing bacteria, such as Lachnospiraceae_unclassified, Clostridium Cluster IV, and Clostridium Cluster XIVa populations in ABX and ABX-20E treated groups (van de Wouw et al., 2018). Therefore, we are currently investigating potential changes in butyrate levels as a result of the decreases in the levels of Firmicutes populations and the possibility that these changes may contribute to the observed ABX-induced increases in ethanol intake behavior in our voluntary binge-like drinking model.
Collectively, our findings support the hypothesis that there is a link between specific gut microbial populations and voluntary alcohol consumption behavior. Further work is necessary to uncover potential mechanisms between the gut-brain interactions and effects on alcohol abuse and related behaviors such as withdrawal-induced anxiety. This knowledge would allow for the discovery and development of less invasive, microbiome-targeted strategies to address alcohol abuse and mood disorders, such as anxiety and depression, commonly associated with AUD.
Methods and Materials
Animals and antibiotic treatment
Adult male C57BL/6J mice, aged 6–8 weeks (wks), were purchased from Jackson Laboratories (JAX, USA) and housed under a reversed 12 hour (hr) light/dark cycle (lights on 12:00 am-12:00 pm). All animals were single-housed and allowed to acclimate to the facility for at least 2 wks before being randomly assigned to a specific treatment group. In order to determine the effects of continuous disruption to the microbiome on ethanol consumption levels, an antibiotic-cocktail (ABX), comprised of 0.5 mg/ml bacitracin (Sigma, USA), 2.0 mg/ml neomycin (GoldBio, USA), 0.2 mg/ml vancomycin (Thermo Fisher, USA) and 1.2 μg/ml pimaricin (Molekula, USA), was provided to mice ad libitum in the drinking water for the duration of the experiment. We changed the antibiotic mixture every two days to ensure consistent concentrations. Animals were weighed 5 days a week during the light cycle to monitor body weight and normal feeding was maintained. We chose this ABX treatment as it has been shown to significantly alter commensal gut microbiota composition in adult rodents while having low absorbance and minimal direct effects on host tissues when given orally (Bercik et al., 2011; Foster et al., 2017; Kiraly et al., 2016). To maintain microbiome dysbiosis, we continued to administer ABX throughout the 6 wk duration of DID while observing ethanol intake behaviors (Fig. 1). All animals were treated in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals and protocols approved by the USC Institutional Animal Care and Use Committee.
“Drinking in the Dark (DID)” ethanol intake model
The DID model is widely used to assess differences in binge-like drinking behaviors. A modified version of this procedure was utilized in the current study where mice had daily limited access (2 hr) to one bottle containing 20% ethanol (20E) beginning at 3 hrs into the circadian dark phase, which we have previously shown to maintain consistent ethanol intake levels during the 5 consecutive days of 20E exposure (Huynh et al., 2017). Ethanol intake was recorded for the 2 hr drinking period for 5 consecutive days with 2 days off (Saturdays and Sundays) for the duration of 6 wks. Vivarium-provided rodent chow was available at all times, and a single bottle of water (H2O) or ABX was continuously available between ethanol access periods. This paradigm was shown to result in high blood ethanol concentrations (BECs) in C57BL/6J mice in a short period of time (>100 mg/dL; Thiele and Navarro, 2014). As this was our first study, we chose 6 wks of DID exposure to determine the effects of prolonged ethanol exposure.
Alcohol dehydrogenase (ADH) activity
Following ABX treatment and DID exposure, liver tissues were processed and analyzed for ADH activity according to the standard ADH activity kit (catalog no. MAK053; Sigma-Aldrich). Total protein concentrations in the samples for standardization were determined using the Pierce BCA protein assay kit and an albumin standard calibration curve (Thermo Scientific, USA). ADH activity was measured according to the manufacturer’s specifications using 50 ul of liver tissue supernatant standardized to a concentration of 3 mg/ml per mouse, n=4/group.
Blood ethanol concentrations (BECs)
In a separate cohort, control mice (H2O, n=5) and mice treated with ABX for 2 wks (ABX, n=7), were given an intraperitoneal injection of ethanol at a concentration of 3.5 g/kg per mouse weight. At 45 minutes (min) after each injection, mice were euthanatized, blood collected, and serum prepared immediately for BEC analysis. Serum samples were processed on the ANALOX AM1 machine according to manufacturer’s instructions (Analox Instruments Ltd., UK).
Elevated plus maze (EPM)
Anxiety-like behavior was assessed in a separate cohort of mice using EPM at 3 separate phases: at baseline, after 2 wks of ABX treatment (3rd wk), and after 5 wks of ABX treatment with 2 wks of DID exposure (6th wk). Anxiety-related parameters were measured for 4 consecutive days during behavior-testing weeks. The test was conducted by individuals who were blind to the treatments received by each mouse. Mice were handled daily for at least one week for several mins before initial behavioral test started in order to allow acclimation to more frequent handling during the behavioral trial. Mice were transferred in their home cage to the behavioral room and allowed to acclimate for at least 30 mins prior to behavior testing.
The EPM apparatus was shaped like a plus sign and consisted of two opposite open-arms (30 cm × 5 cm) with no walls, two opposite closed-arms (30 cm × 5 cm) surrounded by 20 cm black high walls, and a central area (5 cm × 5 cm). The maze was elevated 50 cm above the ground and illuminated by dim light. The experiment started by placing the mouse in the center of the maze and allowing free exploration for 5 mins. Arm entries were recorded when all four paws were inside any individual arm. The individual trials were recorded for 5 mins using a video camera, and the frequency and time of closed/open arm entries were measured. Time spent in open arms suggest possible reductions in anxiety-like behaviors, because it indicates mice exploratory inclination is stronger than the tendency to avoid vulnerable environments, such as those of the open arms.
Cecal tissue collection and DNA isolation
Cecum samples were immediately excised during necropsies, flash frozen, and stored in −80°C until further processing. Cecal DNA was extracted using the E.Z.N.A. ®Stool DNA Kit (D4015, Omega, Inc., USA) according to the manufacturer’s instructions. Ultrapure nuclease-free water was used as the blank to prevent contamination. The total DNA was eluted in 50 μL of elution buffer and stored at −80°C until sequencing.
16S rRNA gene sequence processing
16S rRNA gene sequencing of ceca samples was performed at LC Sciences (Houston, TX). Modified versions of primers 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) were used to target the variable V3-V4 region of the prokaryotic (bacterial and archaeal) ribosomal small-subunit (16S) rRNA gene. The 5’ ends of the primers were tagged with sequencing universal primers and barcodes specific to each sample.
PCR amplification reaction mixtures (25 μL total) contained 25 ng of template DNA, 12.5 μL PCR premix, 2.5 μL of each primer, and PCR-grade water to adjust the volume. To amplify the prokaryotic 16S fragments, PCR conditions consisted of initial denaturation at 98°C for 30 seconds; 35 cycles of denaturation at 98°C for 10 seconds, annealing at 54°C/52°C for 30 seconds, and extension at 72°C for 45 seconds; with a final extension at 72°C for 10 mins. The PCR products were confirmed with 2% agarose gel electrophoresis. Throughout the DNA amplification process, ultrapure water was used as a negative control to exclude the possibility of false-positive PCR results. The PCR products were purified using AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, USA) and quantified by Qubit (Invitrogen, USA). The amplicon pools were prepared for sequencing, and the size (Agilent 2100 Bioanalyzer; Agilent, USA) and quantity (Library Quantification Kit for Illumina; Kapa Biosciences, Woburn, MA, USA) of the amplicon library were assessed. The PhiX Control library (v3) (Illumina) was combined with the amplicon library (expected 30%) in order to improve quality of the sequencing reads. The libraries were sequenced on Illumina MiSeq platform (300bp, pair-ended).
Taxonomic analysis of the gut microbiome metagenomes
Samples were sequenced on the Illumina MiSeq platform according to the manufacturer’s recommendations (Illumina, Inc. San Diego, CA). Paired-end reads were assigned to samples based on their unique barcode and truncated by cutting off the barcode and primer sequence. Paired-end reads were merged using FLASH. Quality filtering on the raw tags were performed under conditions to obtain the high-quality clean tags according to the Fqtrim (Version 0.94). Chimeric sequences were filtered, and sequences with at least 97% similarity were assigned to the same operational taxonomic unit (OTU) by Vsearch (Version 2.3.4). Representative sequences were chosen for each OTU and assigned taxonomic data using the RDP (Ribosomal Database Project) classifier. Approximately 75,000 valid reads per sample were obtained. In order to determine the differences in dominant species of distinct groups, multiple sequence alignments were conducted using the Mafft software (Version 7.310) to study phylogenetic relationships of different OTUs. Rarefaction was standardized based on the sample with the least number of valid reads, determining OTU abundance.
Different indices of diversity were calculated using QIIME software (Version 1.8.0). Alpha diversity analyzes the complexity of species diversity within a sample through Chao1, Shannon, and Simpson measurements. Principal Coordinates Analysis (PCoA) enables beta diversity to be visualized between treatment groups.
Data analyses
For DID, ethanol intake was calculated as g/kg [g of pure ethanol per kg of body weight; 20% ethanol (20E) intake = (volume of 20E consumed in mL x 0.15786 g/mL)/body weight in kg]. The dependent variables included ethanol intake (g/kg), ABX (mL), H2O (mL), food intake (g), and change in mouse weight (g), compared using two-way repeated measures (RM) ANOVA. Two-way RM ANOVA was used to assess the main effect of daily ad libitum antibiotic cocktail administration on 20E consumption between ABX mice and control mice. One-way ANOVA, Kruskal-Wallis test, and non-parametric Mann-Whitney tests were performed to determine differences in ADH and BEC activity, respectively. For anxiety-like behavioral trials, two-way ANOVA and Benjamini-Hochberg tests were used to determine the differences between baseline time point and second week of behavior tests. One-way ANOVA with Dunnett’s multiple comparison and t-tests were used in analyzing the behavioral parameters after DID, significance was set at *p<0.05 (indicating q<0.05 for two-way ANOVA tests), **p<0.01 and $p<0.05 for t-tests.
In order to determine significant differences in microbiome composition between groups, post hoc multiple comparisons False Discovery Rate (FDR q-values) was determined using the Benjamini-Hochberg correction set at 5% (q<0.05). Treatment comparisons on 16S rRNA gene sequencing data were performed using two-way ANOVA and non-parametric, one-way ANOVA, Kruskal-Wallis tests, for individual microbiota phylum and species. Significance was set at p<0.05. OTU-level β-diversity significance was analyzed by permutational multivariate ANOVA (PERMANOVA) of distance between treatment groups measured as Bray Curtis Index. Data are presented as Mean ± Standard Error of Mean (SEM) for each group with post hoc multiple comparisons as *p<0.05 and **p<0.01 after confirming FDR q<0.05. Non-parametric Spearman r correlations, two-tailed with 95% CI, between microbiota phyla populations (p_Firmicutes, p_Bacteroidetes, p_Verrucomicrobia) and total ethanol consumption, 6 wks DID (20E g/kg/60 hrs) were also performed. Graphs and statistical analyses were generated using GraphPad Prism (GraphPad Software Inc., San Diego, CA) and MicrobiomeAnalyst (Dhariwal et al., 2017).
HIGHLIGHTS.
First report on oral antibiotic (ABX) treatment causing increased voluntary ethanol intake
ABX treatment appeared to prevent anxiety-like behavior during ethanol withdrawal
ABX-treated mice expressed reduced bacterial diversity within cecal samples
Changes in Firmicutes, Bacteroidetes and Verrucomicrobia correlated with ethanol intake
Acknowledgements
This work was funded by Rose Hills foundation Innovator Award (USC; to LA), NIAAA R01AA022448 (NIAAA, to DLD), USC School of Pharmacy and USC Good Neighbors. We want to thank our graduate, undergraduate and high school students, Zeyu Zhang, Lei Gao, Kabir Ahluwalia, Vida Andoun, Catalina Vu, Rachel Paik, Carlos Delgado, Eli Bosnoyan, Greg Havton, Elizabeth Bocina, and Sina Kiamehr, for their contributions to data acquisition and continual assistance in animal studies.
Abbreviations
- ABX
antibiotic cocktail
- AUD
alcohol use disorder
- DID
“Drinking in the Dark” ethanol intake model
- 20E
20% ethanol
- BEC
blood ethanol concentration
- ADH
alcohol dehydrogenase
- SCFA
short-chain fatty acid
- GI
gastrointestinal
- PCR
polymerase chain reaction
- OTU
operational taxonomic unit
- RDP
Ribosomal Database Project
- PCoA
Principal Coordinates Analysis
- EPM
elevated plus maze
- LDB
light-dark behavioral box test
- GF
(sterile-bred) germ-free
Footnotes
Conflict of interest
All authors have declared no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asano Y, Hiramoto T, Nishino R, Aiba Y, Kimura T, Yoshihara K, Koga Y, Sudo N, 2012. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am J Physiol Gastrointest Liver Physiol. 303, G1288–G1295. [DOI] [PubMed] [Google Scholar]
- Asatryan L, Yardley MM, Khoja S, Trudell JR, Huynh N, Louie SG, Petasis NA, Alkana RL, Davies DL, 2014. Avermectins differentially affect ethanol intake and receptor function: Implications for developing new therapeutics for alcohol use disorders. Int J Neuropsychopharmacol. 17, 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asatryan L, Khoja S, Rodgers KE, Alkana RL, Tsukamoto H, Davies DL, 2015. Chronic ethanol exposure combined with high fat diet up-regulates P2X7 receptors that parallels neuroinflammation and neuronal loss in C57BL/6J mice. J Neuroimmunol. 285, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HC, Lopez MF, 2004. Increased ethanol drinking after repeated chronic ethanol exposure and withdrawal experience in C57BL/6 mice. Alcohol Clin Exp Res. 28, 18291838. [DOI] [PubMed] [Google Scholar]
- Belzer C, de Vos WM, 2012. Microbes inside--from diversity to function: the case of Akkermansia. ISME J. 6, 1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, MacRi J, McCoy KD, Verdu EF, Collins SM, 2011. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterol. 141, 599–609. [DOI] [PubMed] [Google Scholar]
- Bergeson SE, Nipper MA, Jensen J, Helms ML, Finn DA, 2016. Tigecycline reduces ethanol intake in dependent and nondependent male and female C57BL/6J mice. Alcoholism: Clinic and Experi Res. 40, 2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkhaug ST, Aanes H, Neupane SP, Bramness JG, Malvik S, Henriksen C, Skar V, Medhus AW, Valeur J, 2019. Characterization of gut microbiota composition and functions in patients with chronic alcohol overconsumption. Gut Microbes. 10, 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Benavidez JM, Geil C, Perra S, Morikawa H, Harris RA, 2011. Activation of inflammatory signaling by lipopolysaccharide produces a prolonged increase of voluntary alcohol intake in mice. Brain Beh Immun. 25S92–S105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll IM, Chang YH, Park J, Sartor RB, Ringel Y, 2010. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog. 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborti CK, 2015. New-found link between microbiota and obesity. World J of Gastrointl Pathophys. 6, 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers ES, Preston T, Frost G, Morrison DJ, 2018. Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr Nutri Rep. 7, 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch RD, Dailey A, Zaidi F, Navarro K, Forsyth CB, Mutlu E, Engen PA, Keshavarzian A, 2015. Alcohol induced alterations to the human fecal VOC metabolome. PLOS ONE. 10, e0119362–e0119362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LM, Weiner HL, 2018. Microbiota signaling pathways that influence neurologic disease. Neurotherap. 15, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Harkness JH, Spence SE, Huang LC, Metten P, 2012. Intermittent availability of ethanol does not always lead to elevated drinking in mice. Alcohol Alcohol. 47, 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J, 2017. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 45, W180–W188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubinkina VB, Tyakht AV, Odintsova VY, Yarygin KS, Kovarsky BA, Pavlenko AV, Ischenko DS, Popenko AS, Alexeev DG, Taraskina AY, Nasyrova RF, Krupitsky EM, Shalikiani NV, Bakulin IG, Shcherbakov PL, Skorodumova LO, Larin AK, Kostryukova ES, Abdulkhakov RA, Abdulkhakov SR, Malanin SY, Ismagilova RK, Grigoryeva TV, Ilina EN, Govorun VM, 2017. Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease. Microbiome. 5, 141–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen PA, Green SJ, Voigt RM, Forsyth CB, Keshavarzian A, 2015. The gastrointestinal microbiome: alcohol effects on the composition of intestinal microbiota. Alcohol Res: Curr Rev. 37, 223–236. [PMC free article] [PubMed] [Google Scholar]
- Foster JA, Rinaman L, Cryan JF, 2017. Stress & the gut-brain axis: Regulation by the microbiome. Neurobiol Stress 7, 124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck J, Jayaram-Lindstrom N, 2013. Pharmacotherapy for alcohol dependence: status of current treatments. Curr Opin Neurobiol. 23, 692–699. [DOI] [PubMed] [Google Scholar]
- Getachew B, Reyes RE, Davies DL, Tizabi Y, 2019. Moxidectin effects on gut microbiota of Wistar-Kyoto rats: relevance to depressive-like behavior. Clin Pharmacol Translation Med. 3, 134–142. [PMC free article] [PubMed] [Google Scholar]
- Huynh N, Arabian N, Naito A, Louie S, Jakowec MW, Asatryan L, Davies DL, 2017. Preclinical development of moxidectin as a novel therapeutic for alcohol use disorder. Neuropharmacol. 113, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwa LS, Chu A, Levinson SA, Kayyali TM, DeBold JF, Miczek KA, 2011. Persistent escalation of alcohol drinking in C57BL/6J mice with intermittent access to 20% ethanol. Alcohol Clin Exp Res. 35, 1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav KS, Peterson VL, Halfon O, Ahern G, Fouhy F, Stanton C, Dinan TG, Cryan JF, Boutrel B, 2018. Gut microbiome correlates with altered striatal dopamine receptor expression in a model of compulsive alcohol seeking. Neuropharmacol. 141, 249–259. [DOI] [PubMed] [Google Scholar]
- Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, Khoruts A, Geis E, Maldonado J, McDonough-Means S, Pollard EL, Roux S, Sadowsky MJ, Lipson KS, Sullivan MB, Caporaso JG, Krajmalnik-Brown R, 2017. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome. 5, 10–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EA, King KY, Baldridge MT, 2018. Mouse microbiota models: Comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol. 9, 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiraly DD, Walker DM, Calipari ES, Labonte B, Issler O, Pena CJ, Ribeiro EA, Russo SJ, Nestler EJ, 2016. Alterations of the host microbiome affect behavioral responses to cocaine. Sci Rep. 6, 35455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Starkel P, Windey K, Tremaroli V, Backhed F, Verbeke K, de Timary P, Delzenne NM, 2014. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci USA. 111, E4485–E4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, De Timary P, Delzenne N, Stärkel P, 2017a. The link between inflammation, bugs, the intestine and the brain in alcohol dependence. Translat Psych. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, de Timary P, Delzenne NM, Stärkel P, 2017b. The link between inflammation, bugs, the intestine and the brain in alcohol dependence. Translat Psych. 7, e1048–e1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A, 2013. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 5, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JM, Groot JA, Curtis DC, Allison CL, Marquardt PC, Holmes AN, Edwards DS, Trotter DRM, Syapin PJ, Finn DA, Bergeson SE, 2016. Effective reduction of acute ethanol withdrawal by the tetracycline derivative, tigecycline, in female and male DBA/2J mice. Alcoholism: Clin and Exper Res. 40, 2499–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minter MR, Zhang C, Leone V, Ringus DL, Zhang X, Oyler-Castrillo P, Musch MW, Liao F, Ward JF, Holtzman DM, Chang EB, Tanzi RE, Sisodia SS, 2016. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer’s disease. Sci Rep. 6, 30028–30028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moos RH, Moos BS, 2006. Rates and predictors of relapse after natural and treated remission from alcohol use disorders. Addiction. 101, 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa H, Amin AM, Teh CH, Murugaiyah VA, Arif NH, Ibrahim B, 2017. Plasma metabolic biomarkers for discriminating individuals with alcohol use disorders from social drinkers and alcohol-naive subjects. J Subst Abuse Treat. 77, 1–5. [DOI] [PubMed] [Google Scholar]
- Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, Kwasny M, Lau CK, Keshavarzian A, 2012. Colonic microbiome is altered in alcoholism. American J of Phys. Gastroint Liver Phys. 302, G966–G978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld KM, Kang N, Bienenstock J, Foster JA, 2011. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 23, 255–264, e119. [DOI] [PubMed] [Google Scholar]
- Qin L, He J, Hanes R, Pluzarev O, Hong J-S, Crews F, 2008. Increased systemic and brain cytokine production and neuroinflammation by endotoxin following ethanol treatment. J of Neuroinflamm. 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, McCoy KD, Macpherson AJ, Meza-Zepeda LA, Johansen FE, 2011. Depletion of murine intestinal microbiota: Effects on gut mucosa and epithelial gene expression. PLoS ONE. 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JS, Best K, Belknap JK, Finn DA, Crabbe JC, 2005. Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiol Behavior. 84, 53–63. [DOI] [PubMed] [Google Scholar]
- Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, Mele MC, 2019. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 7, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, Brewer RD, 2015. 2010 national and state costs of excessive alcohol consumption. Am J Prev Med. 49, e73–e79. [DOI] [PubMed] [Google Scholar]
- Sampson T, Debelius Justine W., Thron T, Janssen S, Shastri Gauri G., Ilhan Z, Challis C, Schretter C, Rocha S, Gradinaru V, Chesselet M-F, Keshavarzian A, Shannon K, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian Sarkis K., 2016a. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s Disease. Cell. 167, 1469–1480.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson Timothy R., Debelius Justine W., Thron T, Janssen S, Shastri Gauri G., Ilhan Zehra E., Challis C, Schretter Catherine E., Rocha S, Gradinaru V, Chesselet M-F, Keshavarzian A, Shannon Kathleen M., Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian Sarkis K., 2016b. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s Disease. Cell. 167, 1469–1480.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sari Y, Sakai M, Weedman JM, Rebec GV, Bell RL, 2011. Ceftriaxone, a beta-lactam antibiotic, reduces ethanol consumption in alcohol-preferring rats. Alcohol Alcoholism. 46, 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader AJ, Taylor RM, Lowery-Gionta EG, Moore NLT, 2018. Repeated elevated plus maze trials as a measure for tracking within-subjects behavioral performance in rats (Rattus norvegicus). PLoS One. 13, e0207804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, Mazmanian SK, 2014. Specialized metabolites from the microbiome in health and disease. Cell Metab. 20, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele TE, Navarro M, 2014. “Drinking in the dark” (DID) procedures: A model of binge-like ethanol drinking in non-dependent mice. Alcohol 48, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thion MS, Low D, Silvin A, Chen J, Grisel P, Schulte-Schrepping J, Blecher R, Ulas T, Squarzoni P, Hoeffel G, Coulpier F, Siopi E, David FS, Scholz C, Shihui F, Lum J, Amoyo AA, Larbi A, Poidinger M, Buttgereit A, Lledo P-M, Greter M, Chan JKY, Amit I, Beyer M, Schultze JL, Schlitzer A, Pettersson S, Ginhoux F, Garel S, 2018. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell. 172, 500–516.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wouw M, Boehme M, Lyte JM, Wiley N, Strain C, O’Sullivan O, Clarke G, Stanton C, Dinan TG, Cryan JF, 2018. Short-chain fatty acids: microbial metabolites that alleviate stress-induced brain-gut axis alterations. J Physiol. 596, 4923–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt NM, Kerby RL, Dill-McFarland KA, Harding SJ, Merluzzi AP, Johnson SC, Carlsson CM, Asthana S, Zetterberg H, Blennow K, Bendlin BB, Rey FE, 2017. Gut microbiome alterations in Alzheimer’s disease. Sci Rep. 7, 13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2014. Global status report on alcohol and health. WHO Press, World Health Organization, Geneva, Switzerland. [Google Scholar]
- Xiao HW, Ge C, Feng GX, Li Y, Luo D, Dong JL, Li H, Wang H, Cui M, Fan SJ, 2018. Gut microbiota modulates alcohol withdrawal-induced anxiety in mice. Toxicol Lett. 287, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, Zeng L, Chen J, Fan S, Du X, Zhang X, Yang D, Yang Y, Meng H, Li W, Melgiri ND, Licinio J, Wei H, Xie P, 2016. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psych. 1–11. [DOI] [PubMed] [Google Scholar]