Abstract
Introduction:
The mechanistic basis for the development of esophageal squamous cell carcinoma (ESCC) remains poorly understood. The goal of the present study was thus to characterize mRNA and long noncoding RNA (lncRNA) expression profiles associated with ESCC in order to identify key hub genes associated with the pathogenesis of this cancer.
Materials and Methods:
The GSE26866 and GSE45670 datasets from the Gene Expression Omnibus (GEO) database were used to conduct a weighted gene co-expression network analysis (WGCNA), after which Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted. Cytoscape was additionally used to construct lncRNA-mRNA networks, after which hub genes were identified and validated through the assessment of TCGA datasets and clinical samples.
Results:
Two gene modules were found to be closely linked to ESCC tumorigenesis. These genes were enriched in cell cycle, MAPK signaling, JAK-STAT signaling, pyrimidine metabolism, arachidonic acid metabolism, and P53 signaling pathway activity, all of which are directly linked with the development of cancer. In total, we identified and validated 9 hub genes associated with ESCC (DDX18, DNMT1, NCAPG, WDHD1, PRR11, VOPP1, ZKSCAN5, LC35C2, and PHACTR2).
Conclusion:
In summary, we identified key gene modules and hub genes associated with ESCC development, and we constructed a lncRNA-mRNA network pertaining to this cancer type. These results provide a foundation for future research regarding the mechanistic basis of ESCC.
Keywords: esophageal squamous cell carcinoma, Co-expression, WGCNA, lncRNA-mRNA network
Introduction
Esophageal squamous cell carcinoma (ESCC) is a highly aggressive form of cancer with a poor prognosis. While many advances in the treatment of ESCC have been made in recent years, patient prognosis remains poor. Understanding the mechanistic basis for this disease has the potential to significantly improve patient outcomes.1 As the mechanistic basis for ESCC pathogenesis remains incompletely understood, however, developing novel treatments for this disease remains challenging. To improve treatment efficacy in ESCC patients, it is thus essential that the molecular mechanisms governing ESCC development be clarified.
Many bioinformatics techniques have been developed to effectively analyze high-throughput data. Weighted gene co-expression network analysis (WGCNA) approaches are a particularly powerful means of exploring system-level functionality corresponding to a given transcriptome.2 In prior studies, WGCNA strategies have been employed to identify highly co-expressed genes within a given biological context in order to identify key hub genes associated with a particular condition of interest.3-5
Herein, we therefore conducted a WGCNA-based analysis in order to identify key modules and hub genes associated with ESCC pathogenesis. We additionally analyzed the pathways enriched for genes in 2 key ESCC-related modules, validated these hub genes, and constructed a lncRNA-mRNA network in an effort to gain functional insights into the molecular basis for this cancer type. Together, our study has the potential to serve as a foundation for future research regarding the molecular mechanisms governing ESCC tumorigenesis while highlighting potential diagnostic or therapeutic targets associated with this disease.
Patients and Methods
Gene Expression Data and Preprocessing
The GSE26866 and GSE45670 microarray datasets were downloaded from the NCBI Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). The GSE26866 dataset included 9 ESSC and 19 normal samples, while the GSE45670 dataset contained 28 ESCC and 10 normal samples. The [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array platform was used for both datasets. The affy and annotate R packages were used for data processing and analysis.
Differentially Expressed lncRNA and mRNA Identification
The edgeR package was employed to detect lncRNAs and mRNAs that were differentially expressed when comparing tumor and control tissue samples,6 using |log2(fold change [FC]) |>1 and a false discovery rate (FDR) < 0.01 as cutoff criteria.
WGCNA Construction
Those lncRNAs and mRNAs that were differentially expressed between tumor and control tissue types in the GSE26866 and GSE45670 datasets were utilized for WGCNA. The R WGCNA package was used for this analysis, with the pickSoftThreshold function being utilized to calculate the power parameter. Co-expressed gene modules were identified via the dynamic tree cut method, where a hierarchical clustering approach was used to produce a dendrogram of genes based upon the corresponding dissimilarity of TOM. Genes with comparable expression profiles were grouped into modules.
Clinically Significant Module Identification
Correlations between clinical features and module eigengenes (MES) were determined in order to identify clinically relevant modules by utilizing the MES function in the WGCNA package. Gene significance (GS) and module significance (MS) were respectively defined as the correlation between genes and clinical features and the average GS for all genes in a given module. Following calculations of the association between MS and GS, the module exhibiting the top-ranked MS was identified as the module most closely linked to ESCC-related clinical features.
Functional Enrichment Analyses
Gene Ontology (GO) enrichment analyses were conducted using the DAVID platform (DAVID; https://david.ncifcrf.gov/summary.jsp).7,8 Gene lists corresponding to the identified key modules were uploaded and biological processes (BPs), cellular components (CCs), molecular functions (MFs), and KEGG pathways for which these genes were enriched were identified,9 with P < 0.05 as the significance threshold.
Construction of a lncRNA-mRNA Co-expression Network
Those mRNAs and lncRNAs that exhibited the highest degree of interconnectivity within a given module were considered to be hub genes, and were screened based on degree values in 2 key ESCC-related modules identified in the present study using Cytoscape (https://cytoscape.org).10 The top 100 hub genes were identified based upon maximal clique centrality (MCC), with the top 200 paired lncRNA-mRNA and mRNA-mRNA interactions being incorporated into Cytoscape to construct a regulatory network.
TCGA-Mediated Validation of Hub lncRNAs and mRNAs in Key ESCC-Related Modules
The top 50 hub lncRNAs and mRNAs identified in the above module network analyses were selected as candidates for subsequent validation. The R Survival package was used to conduct Kaplan-Meier analyses in order to assess the prognostic relevance of these hub genes using TCGA patient survival data, with log-rank p < 0.05 being indicative of significance. The top 4 genes per module yielding significant results in these survival analyses were then ranked according to node degree and were considered to be hub genes associated with the development of ESCC.
qRT-PCR
To further validate our findings, 10 ESCC patient tumor and matched paracancerous tissue samples were obtained from patients undergoing tumor resection at the Affiliated Hospital of Xi’an Jiaotong University. The institutional review board of the Affiliated Hospital of Xi’an Jiaotong University approved this study, and all patients provided informed consent to participate. RNA was isolated from these samples and used for qRT-PCR analyses as in prior studies.11 Briefly, an RNAfast2000 Total RNA Extraction Kit (Fastgene, Shanghai, China) was used to extract RNA from patient samples, after which a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, MA, USA) was used based on provided directions to prepare cDNA. All qRT-PCR reactions were then conducted using SYBR green using an appropriate instrument (Bio-Rad, CA, USA), with changes in gene expression being calculated via the comparative CT (2-CT) approach. Primers used in the present study are shown in Table 1.
Table 1.
Primers for Validated Genes.
| Gene name | Prime sequence (5′-3′) | |
|---|---|---|
| Forward | Reverse | |
| DNMT1 | CCATCAGGCATTCTACCA | CGTTCTCCTTGTCTTCTCT |
| MPRIP | CTCTCCACACACGAGCTGAC | TCTTCTGGTGCGTTTCTTCC |
| NCAPG | GGCTGCTGTCGATTAAGGAG | TTATCATCCATCGTGCGGTA |
| PHACTR2 | AACCTCGGCAGTATTAGAAAGGA | CCTCTCCGATGGGTATCATGTG |
| PRR11 | AAAGATGGACCCATGCAGATAAC | TGCTTTCGGCGATGGTATAAG |
| VOPP1 | TCGAGGAGCCAGCCTTCAA | TCCTGGGTCGGTGTAATAGGG |
| WDHD1 | GCTCGGTCACCCGGTTTTAT | GGGGCATCATGTCCTCGAAA |
| ZKSCAN5 | TGACCGAATCCCGAGAAGTTA | GCGGGTTGTACTCCTGCAT |
| DDX18 | ATGTCACACCTGCCGATGAAA | CCCTGAAACTTTAGGTTCCGC |
| SLC35C2 | CTCGTTCATCGGTGGCATTC | CCCAGGAACATGAGTGGCTG |
Results
Differentially Expressed lncRNA and mRNA Identification
The overall design of the present study is shown in Figure 1. First, we downloaded the raw GSE26866 and GSE45670 data from the GEO database, after which data pre-processing and normalization were conducted using R. When we compared ESCC tumor and control tissue samples, we identified 1333 and 6344 differentially expressed lncRNAs and mRNAs, respectively, in the GSE26866 dataset, of which 905 lncRNAs and 3863 mRNAs were upregulated in tumor samples and 428 lncRNAs and 2481 mRNAs were downregulated in tumor samples. In addition, 982 lncRNAs and 5091 mRNAs were found to be differentially regulated in the GSE45670 dataset, of which 472 lncRNAs and 2605 mRNAs were upregulated in ESCC samples and 510 lncRNAs and 2489 mRNAs were downregulated in tumor samples. These 2 datasets were then merged, leading to the identification of 211 lncRNAs and 1535 mRNAs that were differentially expressed in both datasets when comparing ESCC tumor tissues to normal esophageal squamous epithelium samples (Figure 2).
Figure 1.
Flowchart of the study. Abbreviations: DE mRNA, differentially expressed mRNA; DE IncRNA, differentially expressed IncRNA.
Figure 2.
Co-expressed DE mRNAs and DE IncRNAs were identified in datasets GSE26866 and GSE45670. Abbreviations: DE mRNA, differentially expressed mRNA; DE IncRNA, differentially expressed IncRNA.
Weighted Co-Expression Construction and Key Module Identification
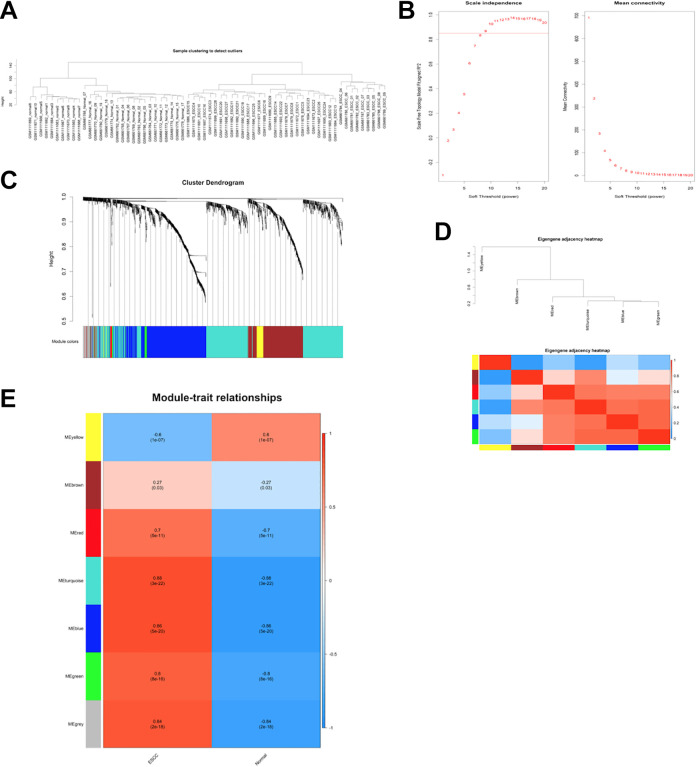
An initial soft-threshold approach was used to implement our WGCNA approach. We first assessed the reliability of the network but detected no outlier samples in need of deletion (Figure 3A). After testing thresholding powers from 1-20, a power value of 9 was selected as at this value the connectivity between genes in the network was consistent with a scale-free network distribution (Figure 3B), leading to the generation of 7 modules (Figure 3C). These modules were designated using colors, with 518, 348, 29, 50, 13, 732, and 54 genes in the blue, brown, green, yellow, red, turquoise, and gray modules, respectively. Genes that were not successfully incorporated into any other modules were incorporated into the gray module, and were omitted from downstream analyses.
Figure 3.
Construction of co-expression modules by WGCNA package. (A) The clustering was based on the co-expressed data of GSE26866 and GSE45670. (B) Analysis of the scale-free fit index for various soft-thresholding powers (β) and analysis of the mean connectivity for various soft-thresholding powers. In all, 9 was the most fit power value. (C) The cluster dendrogram of co-expressed DE mRNAs and DE IncRNAs identified in datasets GSE26866 and GSE45670. Each branch in the figure represents 1 gene, and every color below represents 1 co-expression module. (D) Hierarchical clustering of module hub genes that summarize the modules yielded in the clustering analysis and heatmap plot of the adjacencies in the hub gene network. (E) Heatmap of the correlation between module eigengenes and the tissue type. The turquoise module was the most positively correlated with ESCC, and the yellow module was the most negatively correlated with ESCC. Abbreviations: ESCC, Esophageal squamous cell carcinoma; WGCNA, weighted gene co-expression network analysis.
Key Module Identification and Analysis of Relationships between Modules
To explore module co-expression similarity, eigengenes were calculated and clustered based upon correlations. Seven modules were found to be in the same cluster, and an adjacency-based heatmap yielded similar findings (Figure 3D). Following ME calculations, the MEs of the yellow and turquoise modules were found to be more highly correlated with ESCC than were those of the other modules (Figure 3E). Specifically, the yellow module was negatively correlated with ESCC, whereas the turquoise module was positively correlated with ESCC. This suggested that the turquoise module may be associated with ESCC tumorigenesis, whereas the yellow module may be related to the suppression of tumor growth or formation.
Function Enrichment Analyses of Key ESCC-Related Modules
Putative roles of genes within these 2 key modules were next explored through functional enrichment analyses. Genes in the turquoise module were found to be primarily associated with pro-tumorigenic activities including cell division, proliferation, signal transduction, and the suppression of apoptosis (Figure 4A and C). In contrast, genes in the yellow module were closely linked to the induction of apoptosis, which would in turn suppress tumorigenesis.
Figure 4.
GO and KEGG analysis. Top 20 terms from a GO analysis of molecular function, biological process and cellular component in the turquoise module (A) and yellow module (C); DE mRNAs were clustered by KEGG analysis, and the top 20 pathways in the turquoise module (B) and yellow module (D) are shown.
KEGG pathway analyses were additionally conducted to explore the functional pathways in which these key module genes were enriched. Genes in the turquoise module were enriched in pro-tumorigenic pathways such as MAPK signaling, JAK/STAT signaling, and metabolic pathways (Figure 4B and D).12-14 In contrast, genes in the yellow module were enriched in key pro-apoptotic pathways including the P53, arachidonic acid metabolism, and pyrimidine metabolism pathways, all of which can suppress oncogenesis.15,16
Preparation of a lncRNA-mRNA Network
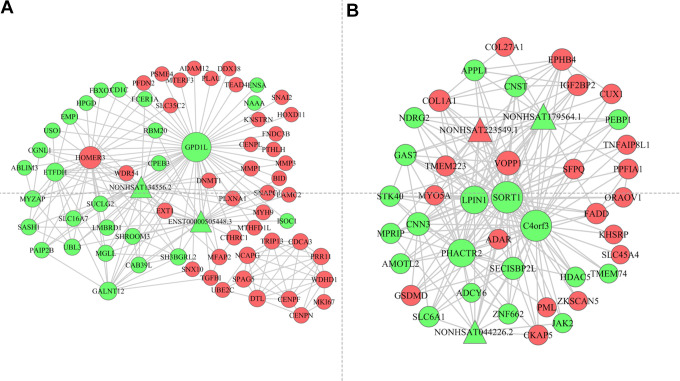
We next leveraged Cytoscape to prepare a lncRNA-mRNA network based on the lncRNAs and mRNAs incorporated into the turquoise and yellow modules. The resultant networks are shown in Figure 5, wherein node sizes correspond to degree values and node color corresponds to expression (green: downregulated; red: upregulated). The turquoise module network was composed of 2 lncRNAs and 98mRNAs, whereas the yellow module network was composed of 3 lncRNAs and 97mRNAs.
Figure 5.
The long noncoding RNA-mRNA biomolecular network in the turquoise module (A) and yellow module (B). The nodes represent different molecules and the edges represent connections between the nodes. The size of the node stands for the number of interactions between different molecules. The node color represented the regulation of the genes (Red indicated upregulated genes, and green indicated downregulated genes).
TCGA-Mediated Validation of Hub ESCC-related lncRNAs and mRNAs in Key Modules
The top 50 lncRNAs and mRNAs in these key module networks were next selected for subsequent validation. The TCGA database was used to assess the relationship between these genes and ESCC patient survival, leading to the identification of the top 5 prognosis-related genes in each of these modules, which were designated as hub genes (Figure 6). The hub genes within the turquoise module included DDX18, DNMT1, NCAPG, WDHD1, and PRR11, whereas in the yellow module they included MPRIP, PHACTR2, VOPP1, ZKSCAN5, and SLC35C2.
Figure 6.
Survival analysis of hub genes in the key modules using TCGA database.
Validation of Differential Hub mRNA Expression in ESCC Tumor Samples Via qRT-PCR
To directly validate a subset of these bioinformatics results, we assessed hub mRNA expression levels in a distinct cohort of ESCC and normal tissue samples via qRT-PCR. This analysis revealed that the expression of DDX18, DNMT1, NCAPG, WDHD1, PRR11, VOPP1, ZKSCAN5, and LC35C2 was significantly elevated in ESCC tissues relative to control tissue samples, whereas PHACTR2 expression was reduced in these ESCC tissues (Figure 7). In contrast, no significant differences in MPRIP expression were detected when comparing these 2 sample types (Figure 7).
Figure 7.
Validation of hub genes using clinical samples from ESCC patients.
Discussion
ESCC is a common form of cancer that is highly prevalent in China and other countries. While several therapeutic advances have improved ESCC patient prognosis, this has not corresponded with any significant improvement in patient 5-year survival rate. Prior research has shown that abnormal gene expression is an important regulator of ESCC development,17 emphasizing the value of molecular studies aimed at exploring the etiological basis of this cancer type.
Data mining strategies can be used to explore key biological phenotypes associated with high-dimensional datasets. The advent of microarray and high-throughput sequencing technologies has led to the discovery of myriad genes and expression patterns linked to oncogenesis and tumor progression.5,18,19 Herein, we downloaded 2 extant datasets containing data pertaining to ESCC tumor and non-tumor tissues from the GEO database, after which we analyzed differential patterns of lncRNA and mRNA expression when comparing ESCC and normal tissues.
A WGCNA approach was then leveraged to detect 2 key gene modules associated with ESCC development. WGCNA strategies facilitate systems biology approaches to exploring transcriptomic functionality.20 In a WGCNA analysis, gene networks are constructed based upon correlations among genes across samples, such that modules are formed by grouping genes which are strongly co-expressed. Genes within a given module are more likely to play similar functional or regulatory roles in a given pathological or physiological context, with the most central genes in a given module functioning as hub genes that are likely to be important regulators of overall module functionality. This analytical strategy also assesses the relationship between modules and clinical characteristics, enabling researchers to assess the relationship between patterns of mRNA, miRNA, or lncRNA expression and cancer.21 Herein, we performed WGCNA by assessing co-expressed gene networks in the GSE26866 and GSE45670 datasets, leading to the identification of 7 gene co-expression modules. Of these modules, the yellow and turquoise modules were most closely linked to ESCC, being negatively and positively correlated with ESCC development, respectively. These 2 modules were thus speculated to be key regulators of ESCC, and were therefore analyzed further.
GO and KEGG enrichment analyses were employed to assess the potential functional roles of these gene modules in the context of ESCC. Genes in the turquoise module were primarily associated with the positive regulation of cellular proliferation, signal transduction, division, and the inhibition of apoptosis, all of which are involved in promoting cancer development. These turquoise module genes were also enriched for MAPK signaling, JAK/STAT signaling, and metabolic pathway activity, potentially thereby driving ESCC. Genes in the yellow modules were primarily enriched for the positive regulation of apoptosis, pyrimidine metabolism, arachidonic acid metabolism, cell cycle regulation, and P53 signaling, all of which can constrain tumor development. This suggests that genes in the yellow module may serve antitumor functions.
Long non-coding RNAs (lncRNAs) are > 200 nucleotides in length, and function as key epigenetic regulators of cellular physiology.22,23 Prior work has highlighted a role for lncRNAs in the regulation of ESCC development.24 The specific relationship between lncRNAs and mRNAs in the context of ESCC, however, remains to be defined. As such, we explored the potential post-transcriptional crosstalk between lncRNAs and mRNAs in this oncogenic context, generating an integrated lncRNA-mRNA regulatory network based upon the hub genes identified within the yellow and turquoise module hub genes. Only the top 100 mRNAs and lncRNAs from these 2 modules were selected to ensure accuracy, and the network was composed of the top 200 lncRNA-mRNA and mRNA-mRNA pairs. Future work will be essential, however, in order to establish the functional roles of these genes in ESCC.
Given that the turquoise and yellow gene modules were found to be closely associated with the development of ESCC, we next screened for hub genes within these modules that were closely associated with patient survival and that were validated in an independent sample cohort. We first identified DDX18, DNMT1, NCAPG, WDHD1, and PRR11 as hub genes within the turquoise module, and MPRIP, PHACTR2, VOPP1, ZKSCAN5, and SLC35C2 as hub genes within the yellow module. We then assessed the expression of these genes via qRT-PCR in a separate set of ESCC patient clinical samples, confirming that DDX18, DNMT1, NCAPG, WDHD1, PRR11, VOPP1, ZKSCAN5, and SLC35C2 expression levels were increased in ESCC tumor tissue samples whereas PHACTR2 expression was reduced in these samples relative to normal control tissue levels. Several of these hub genes have been previously linked to oncogenesis including VOPP1, WDHD1, PRR11, DNMT1, and NCAPG. DNMT1 is a DNA methyltransferase that preserves DNA methylation patterns during DNA replication, and that is upregulated in a range of cancer types including colorectal cancer (CRC), gastric cancer, breast cancer, and pancreatic cancer.25 DNMT1 is also a key regulator of methylation status in the context of stem cell maintenance and embryogenesis.26 NCAPG is a condensin I complex subunit that regulates chromosome condensation and stabilization in the context of mitosis and meiosis. NCAPG overexpression has been detected in cancers including melanoma, lung cancer, prostate cancer, and HCC, suggesting that this gene plays an important oncogenic role.27-29 WDHD1 is a DNA-binding protein that interacts with DNA via an HMG domain,30 controlling mitosis, DNA damage responses, and DNA repair.31 In prior research, WDHD1 has been closely linked to malignancies including ESCC, lung cancer, and cervical cancer.32 PRR11, which is encoded on chromosome 17q22, is composed of a zinc finger domain, 2 proline-rich regions, and a bivalent nuclear localization signal.33 PRR11 has been highlighted as a potential oncogene associated with HCC, ovarian cancer, breast cancer, pancreatic cancer, and non-small cell lung cancer.34,35 VOPP1 is localized encoded by the 7p11.2 locus, and is frequently amplified along with EGFR.36 The overexpression of VOPP1 has bene detected in glioblastoma and gastric, head and neck, lung, and breast cancers.37 Few prior studies have identified any relationship between DDX18, ZKSCAN5, PHACTER2, SLC35C2, and cancers.
While the results of this study are interesting, it nonetheless has certain limitations. For one, these results were primarily derived from data mining and bioinformatics approaches, and further validation is thus essential. Second, we were only able to utilize a limited number of datasets as most studies of ESCC samples lacked corresponding normal controls, thus constraining our findings. Third, the association between key genes and survival outcomes in this study are not causal relationship. In further studies, whether these relationship are causal relationship still need to be explored. At the same time, the relationship key genes and other prognostic factors such as stage, grade and response to therapy are also need to be further studied. At last, WCGNA approaches are inherently limited by the criteria used for module selection and the thresholds for network culling, potentially impacting the final study results.
Conclusion
In summary, we utilized a WGCNA approach to identify key gene modules and hub genes linked to ESCC development. We also constructed a lncRNA-mRNA regulatory network in an effort to better understand the onset of this cancer type. Together, our findings offer a foundation for future research regarding the mechanistic basis for ESCC tumorigenesis, and the identified hub genes may represent viable diagnostic biomarkers or therapeutic targets that can be leveraged to treat this deadly cancer type.
Footnotes
Author Contributions: All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Statement: This study did not require an ethical board approval because it did not contain human or animal trials.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by Key Research and Development Program of Shaanxi Province (2018SF-236).
ORCID iD: Qifei Wu, MD, PhD  https://orcid.org/0000-0002-7439-2831
https://orcid.org/0000-0002-7439-2831
References
- 1.Huang F-L, Yu S-J.Esophageal cancer: risk factors, genetic association, and treatment. Asian J Surg. 2018;41(3):210–215. [DOI] [PubMed] [Google Scholar]
- 2.Zhou X-G, Huang X-L, Liang S-Y, et al. Identifying miRNA and gene modules of colon cancer associated with pathological stage by weighted gene co-expression network analysis. Onco Targets Ther. Dove Press; 2018;11:2815–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Liu W, Li H, et al. Identification of key genes and pathways associated with cholangiocarcinoma development based on weighted gene correlation network analysis. PeerJ. 2019;7:e7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi X, Li Y, Yan P, Shi Y, Lai J. Weighted gene co-expression network analysis to explore the mechanism of heroin addiction in human nucleus accumbens. J Cell Biochem. 2019;24:135. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Feng H, Li Z, et al. Application of weighted gene co-expression network analysis to identify key modules and hub genes in oral squamous cell carcinoma tumorigenesis. Onco Targets Ther. 2018;11:6001–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 8.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou L, Du Y, Kong L, Zhang X, Chen Q. Identification of molecular target genes and key pathways in hepatocellular carcinoma by bioinformatics analysis. Onco Targets Ther. 2018;11:1861–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Li J, Zhang R, et al. Development of a metabolic pathway-based pseudo-targeted metabolomics method using liquid chromatography coupled with mass spectrometry. Talanta. 2019;192:160–168. [DOI] [PubMed] [Google Scholar]
- 13.Wang M, An S, Wang D, Ji H, Guo X, Wang Z. Activation of PAR4 upregulates p16 through inhibition of DNMT1 and HDAC2 expression via MAPK signals in esophageal squamous cell carcinoma cells. J Immunol Res. 2018;2018:4735752–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian F, Yang X, Liu Y, et al. Constitutive activated STAT3 is an essential regulator and therapeutic target in esophageal squamous cell carcinoma. Oncotarget. 2017;8(51):88719–88729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karagoz K, Lehman HL, Stairs DB, Sinha R, Arga KY. Proteomic and metabolic signatures of esophageal squamous cell carcinoma. Curr Cancer Drug Targets. 2016. [PubMed] [Google Scholar]
- 16.Liu B, Pan C-F, Yao G-L, Wei K, Xia Y, Chen Y-J. The long non-coding RNA AK001796 contributes to tumor growth via regulating expression of p53 in esophageal squamous cell carcinoma. Cancer Cell Int. 2018;18:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy G, McCormack V, Abedi-Ardekani B, et al. International cancer seminars: a focus on esophageal squamous cell carcinoma. Ann Oncol. 2017;28(9):2086–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Zhao X, Wang M, Ji W. Key modules and hub genes identified by coexpression network analysis for revealing novel biomarkers for larynx squamous cell carcinoma. J Cell Biochem. 2019;120(12):19832–19840. [DOI] [PubMed] [Google Scholar]
- 19.Zou Y, Jing L. Identification of key modules and prognostic markers in adrenocortical carcinoma by weighted gene co-expression network analysis. Oncol Lett. 2019;18(4):3673–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song Z-Y, Chao F, Zhuo Z, Ma Z, Li W, Chen G. Identification of hub genes in prostate cancer using robust rank aggregation and weighted gene co-expression network analysis. Aging (Albany NY). 2019;11(13):4736–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. Weighted gene coexpression network analysis: state of the art. J Biopharm Stat. 2010;20(2):281–300. [DOI] [PubMed] [Google Scholar]
- 22.Xie Y, Dang W, Zhang S, et al. The role of exosomal noncoding RNAs in cancer. Mol Cancer. 2019;18(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenzi L, Avila Cobos F, Decock A, et al. Long noncoding RNA expression profiling in cancer: Challenges and opportunities. Calin G, editor. Genes Chromosomes Cancer. 2019;58(4):191–199. [DOI] [PubMed] [Google Scholar]
- 24.Song J, Lu Y, Sun W, Han M, Zhang Y, Zhang J. Changing expression profiles of lncRNAs, circRNAs and mRNAs in esophageal squamous carcinoma. Oncol Lett. 2019;18(5):5363–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu R, Gu J, Jiang P, et al. DNMT1-microRNA126 epigenetic circuit contributes to esophageal squamous cell carcinoma growth via ADAM9-EGFR-AKT signaling. Clin Cancer Res. 2015;21(4):854–863. [DOI] [PubMed] [Google Scholar]
- 26.Teng Y, Yu X, Yuan H, Guo L, Jiang W, Lu S-H. DNMT1 ablation suppresses tumorigenesis by inhibiting the self-renewal of esophageal cancer stem cells. Oncotarget. 2018;9(27):18896–18907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao C, Gong J, Jie Y, et al. NCAPG is a promising therapeutic target across different tumor types. Front Pharmacol. 2020;11:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arai T, Okato A, Yamada Y, et al. Regulation of NCAPG by miR-99a-3p (passenger strand) inhibits cancer cell aggressiveness and is involved in CRPC. Cancer Med. 2018;7(5):1988–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu K, Li Y, Yu B, Wang F, Mi T, Zhao Y. Silencing non-SMC chromosome-associated polypeptide G inhibits proliferation and induces apoptosis in hepatocellular carcinoma cells. Can J Physiol Pharmacol. 2018;96(12):1246–1254. [DOI] [PubMed] [Google Scholar]
- 30.Guan C, Li J, Sun D, Liu Y, Liang H. The structure and polymerase-recognition mechanism of the crucial adaptor protein AND-1 in the human replisome. J Biol Chem. 2017;292(23):9627–9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Li Z, Wu R, Han Z, Zhu W. And-1 is required for homologous recombination repair by regulating DNA end resection. Nucleic Acids Res. 2017;45(5):2531–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato N, Koinuma J, Fujita M, et al. Activation of WD repeat and high-mobility group box DNA binding protein 1 in pulmonary and esophageal carcinogenesis. Clin Cancer Res. 2010;16(1):226–239. [DOI] [PubMed] [Google Scholar]
- 33.Qiao W, Wang H, Zhang X, Luo K. Proline-rich protein 11 silencing inhibits hepatocellular carcinoma growth and epithelial-mesenchymal transition through β-catenin signaling. Gene. 2019;81:7–14. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Zhang C, Mai L, Niu Y, Wang Y, Bu Y. PRR11 and SKA2 gene pair is overexpressed and regulated by p53 in breast cancer. BMB Rep. 2019;52(2):157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu J, Hu H, Wang J, Yang Y, Yi P. PRR11 overexpression facilitates ovarian carcinoma cell proliferation, migration, and invasion through activation of the PI3K/AKT/β-Catenin pathway. Cell Physiol Biochem. 2018;49(2):696–705. [DOI] [PubMed] [Google Scholar]
- 36.Baras AS, Solomon A, Davidson R, Moskaluk CA. Loss of VOPP1 overexpression in squamous carcinoma cells induces apoptosis through oxidative cellular injury. Lab Invest. 2011;91(8):1170–1180. [DOI] [PubMed] [Google Scholar]
- 37.Gao C, Pang M, Zhou Z, et al. Epidermal growth factor receptor-coamplified and overexpressed protein (VOPP1) is a putative oncogene in gastric cancer. Clin Exp Med. 2015;15(4):469–475. [DOI] [PubMed] [Google Scholar]