Abstract
Tat interactive protein, 60 kDa (TIP60) is an important partner of ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1), ensuring various cellular processes through its acetyltransferase activity. TIP60 is believed to play a tumor suppressive role, partly explained by its downregulated expression in a number of cancers. The aim of the present study was to investigate the role and mechanisms of action of TIP60 in the regulation of UHRF1 expression. The results revealed that TIP60 overexpression downregulated the UHRF1 and DNA methyltransferase 1 (DNMT1) expression levels. TIP60 interfered with USP7-UHRF1 association and induced the degradation of UHRF1 in an auto-ubiquitination-dependent manner. Moreover, TIP60 activated the p73-mediated apoptotic pathway. Taken together, the data of the present study suggest that the tumor suppressor role of TIP60 is mediated by its regulation to UHRF1.
Keywords: cancer, epigenetics, UHRF1, TIP60, USP7, ubiquitination, fluorescence lifetime imaging microscopy, protein-protein interaction, apoptosis
Introduction
The multidomain protein, ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1, also known as ICBP90 in humans) (1,2), is an important epigenetic integrator responsible for the faithful transmission of DNA methylation patterns from parent strands to daughter strands during DNA replication (1,3-8). UHRF1 performs this role by recognizing the CpG motifs in hemi-methylated DNA through its SRA domain (SET and RING-associated domain) and by recruiting DNA methyltransferase 1 (DNMT1) (6-10). TTD and PHD domains help UHRF1 to read the histone marks (11-13). The RING domain of UHRF1 has intrinsic ubiquitin E3 ligase activity by which UHRF1 can ubiquitinate itself (auto-ubiquitination) (14,15) or other proteins including histones (16,17). The ubiquitination of H3K23 and H3K18 by UHRF1 is important for the creation of binding sites for DNMT1 (7,18-21). The N-terminal UBL domain of UHRF1 binds directly to DNMT1 and increases its enzymatic activity towards chromatin by controlling H3 ubiquitination (20-22). UHRF1 is also involved in the DNA damage response (4,23,24) and the regulation of the stability and function of several other proteins, such as p53, promyelocytic leukemia protein (PML) and DNMT1 through the collaboration with other epigenetic partners such as ubiquitin-specific-processing protease 7 (USP7), histone deacetylase 1 (HDAC1) and Tat interactive protein, 60 kDa (TIP60) (4,17,25,26).
The authors of the present study first reported the interaction of UHRF1 with TIP60 in a previous study (27). Indeed, UHRF1 and TIP60 were found to be in the same macromolecular complex and interact with each other (17,25,27). TIP60 was originally recognized as an interacting partner of HIV-1 Tat protein (28). TIP60 (also known as KAT5) belongs to the MYST family (MOZ, YBF2/SAS3, SAS2 and TIP60) having an evolutionary conserved domain which harbors histone acetyltransferase (HAT) activity (29-32). At its N-terminus, TIP60 has a chromodomain (CRD), while its C-terminus contains the conserved enzymatic MYST domain (33). TIP60 reads histone marks (H3K4me2/H3K9me3) through its CRD domain (34) and translates it through MYST domain (35). Inside the MYST domain, there is the catalytic HAT domain which binds to acetyl coenzyme A and catalyzes acetylation of both histone and non-histone proteins (36,37). This acetyltransferase activity is stimulated by a zinc finger which helps TIP60 to interact with the targeted substrates (38-40). Through its enzymatic activity, TIP60 is a central player in many key cellular processes like chromatin remodeling, DNA damage response, transcription regulation, genomic integrity, cell cycle and apoptosis (25,36,39,41-43). For instance, it interacts with and regulates the transcription of nuclear hormone receptors, p53, c-MYC and NF-κB (39,42,44). Of note, it also regulates p53 activity in an acetylation-dependent (K120 of p53) and -independent manner (25). The acetylation of p53 activates p21 and the PUMA pathway, which leads to cell growth arrest and apoptosis, and thus, ensures tumor suppression (25).
The downregulation of TIP60 inhibits both p21 activation and growth arrest (45). During the M-phase, TIP60 is essential for chromosomal segregation (46) and cell cycle progression (47-49). Cells lacking TIP60 acetyltransferase activity lose their ability to repair DNA and ultimately, cell cycle control (41). The heterozygous deletion of the TIP60 gene (HTATIP) has a lethal effect on embryos (50). In a number of cancer types, TIP60 levels are low as compared to normal cells, supporting its tumor suppressive role (25,41,45,51-56). In accordance with this role, high levels of UHRF1 have been shown to interfere with the TIP60-p53 interplay and prevent p53 activation, which leads to tumorigenesis and/or tumor progression (25). Therefore, targeting UHRF1 in cancer cells would permit the 'rescue' of p53 levels and would enhance the coordinated dialogue between p53 and TIP60. In a previous study, the authors demonstrated that UHRF1 interacts with the MYST domain of TIP60 (57). Moreover, UHRF1, through its E3 ligase activity, ubiquitinates DNMT1 and thus affects its expression levels (17,58,59). Although it has already been shown that TIP60 overexpression downregulates UHRF1 levels in HeLa cells (57), the mechanisms responsible for the TIP60-mediated downregulation of UHRF1 in cancer cells remain elusive. The present study demonstrates that TIP60 interferes with the UHRF1-USP7 association. Following dissociation from USP7, UHRF1 is auto-ubiquitinated by its RING domain. The resulting downregulation of UHRF1 activates p73-mediated apoptosis. Taken together, these observations provide new insight into the tumor suppressive role of TIP60 by controlling the UHRF1 levels.
Materials and methods
Materials
MG-132 (C26H41N3O5) was purchased from Selleckchem.com -Bioactive Compounds Expert (cat. no. S2619). MG-132 was dissolved in pure DMSO (Sigma-Aldrich; Merck KGaA) and stored at −80°C. Propidium iodide (PI; cat. no. 130-093-233) was purchased from Miltenyi Biotec GmbH, while Annexin V-iFluor™ 350 conjugate (cat. no. 20090) was purchased from AAT Bioquest. The TIP60 inhibitor, 5-(1,2-thiazol-5-yldisulfanyl)-1,2-thiazole (NU9056), was purchased from Tocris Bioscience (cat. no. 1450644-28-6).
Cells and cell culture
HeLa cells (ATCC, CCL-2, Amp, cervical adenocarcinoma; human) were grown in Dulbecco's modified Eagle's medium (DMEM 1X + GlutaMAX™, Pyruvate, Gibco; Thermo Fischer Scientific, Inc.) which was supplemented with 10% fetal bovine serum (FBS, cat. no. S1810-500; Dominique Dutscher), in addition to mixture of penicillin (100 U/ml) and streptomycin (100 U/ml) (cat. no. 17-602E; Lonza Group, Ltd.), at 37°C with 5% CO2 in a humidified environment. HeLa cell lines stably expressing either GFP-UHRF1 WT or GFP-UHRF1 C724A-H741A protein, were constructed using the pOZ-N plasmid (Addgene) in which the GFP-UHRF1 WT or GFP-UHRF1 C724A-H741A mutant cDNAs had been subcloned as previously described (15). Mycoplasma testing has been performed for the cell lines. Plasmids (TIP60 wild-type and its mutants: ΔHAT and ΔMYST; supplied by EpiGex) were transfected (at a concentration of 1 µg/2 ml of media, for a time period of 24 h) into HeLa cells with either jetPEI™ or jetPRIME (2 µl) (cat. no. 101-10N and cat. no. 114-15; PolyPlus-transfection SA) according to the manufacturer's protocol.
Plasmid constructs
TIP60 wild-type and mutants (ΔHAT and ΔMYST, respectively) were cloned into a pEGFP-N1 plasmid (supplied by EpiGex) to express eGFP-labeled TIP60 proteins in HeLa cells. RFP-Ubiquitin was purchased from Addgene (cat. no. 11935).
Antibodies
Mouse monoclonal anti-UHRF1 (1:2,000) antibody was engineered as previously described (1). Other antibodies used included rabbit polyclonal anti-HAUSP/USP7 (1:5,000; cat. no. ab4080, Abcam), mouse monoclonal anti-DNMT1 (1:5,000; cat. no. PTG-MAB0079, ProteoGenix), mouse monoclonal anti-ubiquitin (1:500; cat. no. 05-944, Sigma-Aldrich; Merck KGaA), mouse monoclonal eGFP (1:1,000; cat. no. 66,002-1-Ig, Proteintech Group, Inc.; and cat. no. A-11120, Thermo Fisher Scientific, Inc.), mouse monoclonal anti-GAPDH (1:5,000; cat. no. MAB374, Merck KGaA), mouse monoclonal anti-GFP (1:1,000; cat. no. 66002-1-Ig, Proteintech Group, Inc.), mouse monoclonal anti-p73 (1:500; cat. no. 558785, BD Biosciences), rabbit polyclonal anti-caspase-3 (1:1,000; cat. no. 9661, Cell Signaling Technology, Inc.), mouse monoclonal anti-BCL2 (1:1,000; cat. no. 05-826, Merck KGaA), mouse monoclonal anti-poly(ADP-ribose) polymerase (PARP; 1:1,000; cat. no. 51-6639GR, BD Biosciences) and rabbit polyclonal anti-BAX (1:1,000; cat. no. AB2930, Merck KGaA).
Western blot analysis
For western blot analysis, cells were collected at 24 h following transfection by trypsinization. For ubiquitination experiments, cells were treated with MG-132 (10 µM) 8 h prior to cell harvesting. Following centrifugation (500 × g for 4 min at 4°C), the medium was discarded, and the cell pellet was washed with PBS. Cells were lysed with ice cold lysis buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 150 mM NaCl and 1% NP40 supplemented with protease inhibitors (cat. no. 1183617000;1 cOmplete mini EDTA-free protease inhibitor cocktail tablets, Roche Diagnostics GmbH). Following denaturation at 95°C for 7 min in 4X Laemmli sample buffer freshly supplemented with β-mercaptoethanol (cat. no. 1610747 Bio-Rad Laboratories, Inc.), 40 µg of the protein from cell lysates were loaded on 7.5 and 10% SDS-PAGE gels which after separation were transferred to previously activated polyvinylidene difluoride (PVDF) membrane (GE Healthcare Life Sciences, Cytiva). Membranes were blocked by 3% blotting-grade blocker (Bio-Rad Laboratories, Inc.) in Tris-Buffered Saline, with Tween®-20, pH 8.0 (TBS-T) (Sigma-Aldrich; Merck KGaA). Proteins were identified by anti-UHRF1 (1:2,000 dilution in blocking buffer), anti-ubiquitin (1:500 dilution in blocking buffer), anti-DNMT1 (1:5,000 dilution in blocking buffer), anti-USP7 (1:5,000 dilution in blocking buffer), anti-eGFP (1:1,000 dilution in blocking buffer) and anti-GAPDH (1:5,000 dilution in blocking buffer) primary antibodies, with overnight incubation at 4°C. Membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies, anti-mouse (1:10,000 dilution in blocking buffer; cat. no. W402B; Promega Corporation) or anti-rabbit (1:10,000 dilution in blocking buffer; cat. no. W401B; Promega Corporation), for 1 h at room temperature, to label the primary antibodies. Signals were detected on an Image Quant LAS 4000 apparatus (GE Healthcare Life Sciences, Cytiva) with chemiluminescent ECL system (Clarity™ ECL western blotting substrate; cat. no. 170-5060, Bio-Rad Laboratories, Inc.). Image Studio Lite (Li-Core Biosciences, Inc.) was used to analyze the images.
Immunoprecipitation (IP)
For IP, cells were collected and lysed by freeze shock. Mild sonication was performed in ice-cold PBS freshly supplemented with protease inhibitors cocktail tablet. Input controls were established by taking 40 µg of protein from each lysate. A total of 1,000 to 1,500 µg of protein lysate were incubated with anti-UHRF1 antibody at 4°C for 3 h or with anti-USP7 antibody at 4°C overnight. After washing and equilibration, 60 µl of Dynabeads® protein A (cat. no. 1002D; Thermo Fischer Scientific, Inc.) were added to the lysate-antibody mixture and incubated for 1 h at 4°C. The beads were then collected and washed 3-5 times with ice-cold PBS freshly supplemented with protease inhibitors tablet. Finally, beads were resuspended in Laemmli sample buffer. Proteins were denatured by heating at 95°C for 7 min and examined by western blot analysis.
UHRF1 auto-ubiquitination assay
HeLa cells stably expressing GFP-UHRF1 WT and GFP-UHRF1 C724A-H741A mutant proteins were transfected with either TIP60 WT (1 µg/2 ml of media) or TIP60ΔMYST (1 µg/2 ml of media) mutant using jetPRIME reagent (2 µl), for a time duration of 24 h. Samples were treated with 10 µM MG-132, 8 h before harvesting the cells. IP (as described above) was performed with anti-GFP antibody to immunoprecipitate the GFP-tagged UHRF1 protein. Samples were resolved by western blot analysis.
Confocal microscopy
To examine the effect of TIP60 overexpression on the UHRF1 and DNMT1 levels, HeLa cells were seeded on a cover glass and transfected with eGFP (1 µg/2 ml of media) or TIP60-eGFP (1 µg/2 ml of media) or TIP60ΔMYST-eGFP (1 µg/2 ml of media) plasmids using jetPEI™ reagent (2 µl) as described in the manufacturer's protocol. At 24 h post-transfection, cells were fixed with 4% paraformaldehyde for 10 min and then permeabilized with 0.2% Triton X-100 for 20 min at room temperature. Subsequently, blocking was performed with 1% BSA for 1 h, prior to incubation with a primary antibody against either UHRF1 or DNMT1 for 3 h at 4°C. After washing three times with PBS, cells were incubated with secondary antibody labeled with Alexa Fluor 568 (goat anti-mouse, cat. no. A11031; Invitrogen; Thermo Fischer Scientific, Inc.) for 60 min at room temperature. The cells were then washed three times and labeled with DAPI (Invitrogen Hoechst stain, cat. no. 33258; Thermo Fischer Scientific, Inc.). Finally, cells were imaged with a confocal Leica TCS SPE microscope equipped with a 20X air (0.7 NA) immersion lens objective. For DAPI, Alexa Fluor 568 and eGFP, excitation was performed with a 405 nm laser (25 mW), 561 nm laser (10 mW) and 488 nm laser (25 mW), respectively. The detection range for the three dyes was 430-480, 570-630 and 500-523 nm, respectively.
To examine the effect of TIP60 overexpression on the co-localization of UHRF1 and ubiquitin, HeLa cells were co-transfected with either eGFP (1 µg/2 ml of media) and RFP-Ubiquitin (1 µg/2 ml of media) or TIP60-eGFP (1 µg/2 ml of media) and RFP-Ubiquitin (1 µg/2 ml of media) using jetPEI™ reagent (2 µl), for a time duration of 24 h. One group of samples was treated with MG-132 (10 µM) 8 h before cell fixation, to block the proteasomal degradation of UHRF1. Cells were labeled with anti-UHRF1 as primary antibody for 3 h at 4°C and Alexa Fluor 647-labeled goat anti-mouse, cat. no. A-21237 (Thermo Fischer Scientific, Inc.) as secondary antibody, for 1 h at room temperature. DAPI staining was done to visualize the nucleus. All samples were imaged with a confocal Leica TCS SPE equipped with an oil immersion objective (HXC PL APO 63X/1.40 OIL CS). For DAPI, RFP, Alexa Fluor 647 and eGFP, excitation was performed with a 405 nm laser (25 mW), 561 nm laser (10 mW), 635 nm laser (18 mW) and 405 nm laser (25 mW), respectively. The detection range for the four dyes was 430-480, 570-630, 640-702 and 500-523 nm, respectively.
For the UHRF1-USP7 association analysis, HeLa cells were transfected with either TIP60-eGFP (1 µg) or TIP60ΔMYST-eGFP (1 µg) using jetPEI™ reagent (2 µl), for a time duration of 24 h. One group of samples was treated with MG-132 (10 µM) 8 h before cell fixation, to block the proteasomal degradation of UHRF1 and USP7. Cells were labeled with anti-UHRF1 (mouse) and anti-USP7 (rabbit) antibodies overnight at 4°C. The cells were then incubated with secondary antibody labeled with Alexa Fluor 568 (goat anti-rabbit, cat. no. A11011; Invitrogen; Thermo Fischer Scientific, Inc.) for USP7 and Alexa Fluor 647 (goat anti-mouse) for UHRF1. DAPI staining (100 µg/ml in PBS for 20 min at room temperature) was performed to stain the nuclei. All samples were imaged with a confocal Leica TCS SPE equipped with an oil immersion objective (HXC PL APO 63X/1.40 OIL CS). For DAPI, Alexa Fluor 568, Alexa Fluor 647 and eGFP, excitation was performed with a 405 nm laser (25 mW), 561 nm laser (10 mW), 635 nm laser (18 mW) and 405 nm laser (25 mW), respectively. The detection range for the four dyes was 430-480, 570-625, 644-707 and 500-531 nm, respectively. All the images were processed using ImageJ software (1.52p; Wayne Rasband, NIH; http://imagej.nih.gov/ij).
Fluorescence lifetime imaging microscopy (FLIM)
HeLa cells stably expressing GFP-UHRF1 WT or GFP-UHRF1 C724A-H741A protein, were seeded (105 cells per dish) in a µ-dish (Ibidi) with 35-mm wells. Cells were transfected with 1 µg RFP-Ubiquitin plasmid using jetPEI™ reagent. Cells were fixed with 4% paraformaldehyde. Following fixation, cells were imaged with a homemade two-photon excitation scanning microscope based on an Olympus IX70 inverted microscope with a 60X 1.2 NA water immersion objective operating in the descanned fluorescence collection mode as previously described (60,61). Two-photon excitation at 930 nm was provided by an Insight DeepSee laser (Spectra Physics, Inc.). Fluorescence photons were collected using a short-pass filter with a cut-off wavelength of 680 nm (cat. no. F75-680; Analysentechnik) and a band-pass filter of 520±17 nm (cat. no. F37-520; Analysentechnik). The fluorescence was directed to a fibre-coupled APD (cat. no. SPCM-AQR-14-FC; Perkin Elmer Inc., USA), which was connected to a time-correlated single photon counting module (cat. no. SPC830: Becker & Hickl). FLIM data were analyzed using SPCImage v 7.3 (Becker & Hickel) and the Förster resonance energy transfer (FRET) efficiency was calculated according to E=1-(τDA/τD), where τDA is the lifetime of the donor (GFP) in the presence of acceptor (RFP) and τD is the lifetime of GFP in the absence of acceptor.
Apoptosis analysis
Flow cytometry was used to analyze TIP60-induced apoptosis. HeLa cells were seeded in six-well plates. Cells were transfected with TIP60-eGFP by using jetPEI™ reagent. TIP60 transfected cells were compared to control cells or cells treated with jetPEI™ only. Cells were collected after mild trypsinization and incubated with PI and Annexin V-iFluor™350 conjugate. The samples were then analyzed with a Guava easyCyte™ flow cytometer (Merck KGaA). InCyte Software for Guava (Merck KGaA) was used to analyze the results.
Retrospective TIP60 expression analysis
To evaluate the differential expression of TIP60 in normal and cancer cervical tissues retrospectively, the expression profile of TIP60 was retrieved from GDS3233 (62) at NCBI Gene Expression Omnibus data base (https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS3233). The expression analysis was done by using Affymetrix U133A oligonucleotide microarray (Santa Clara, CA) which contains 14500 probes for analysis. This dataset included expression profile of TIP60 in 24 normal cervical tissues and 28 cervical cancer tissues which were compared by using an unpaired Student's t-test.
RNAseq expression analysis
Raw counts RNAseq expression data of cancer samples were downloaded from the TCGA website (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), normalized using DESeq2 (63) and used to plot the distribution of expression of three genes (UHRF1, TIP60 and USP7) in each cancer type, and represented as box plots in Figs. S4, S5 and S9. When available, the mean expression level of the corresponding non-tumor tissue was calculated (and shown as a red circle in the corresponding figures). Co-expression of TIP60 and UHRF1 was determined by linear regression analysis. Simple linear regression was used between y-axis and x-axis. The delivered score is the R2, also known as the linear association, characterizing the percentage of explained variance. Here caution is advised, R2 score is indicating the behavior/tendency towards the association of two genes.
Survival probability analysis
To investigate the association between the expression of either the TIP60/KAT5, UHRF1 or USP7 gene and the probability of survival of TCGA cancer patients for whom survival data were available (meta data available from the TCGA site, as well as from our custom website http://epimed.univ-grenoble-alpes.fr/database/series), a two-step bioinformatics analysis was performed: i) Seeking for a significant association between the expression value and survival probability, using the Cox model; and ii) when the P-value of the Cox model was significant (P<0.05), samples were grouped by quintiles of expression (from the lowest expression <20th percentile to the highest >80th percentile) and survival probabilities were compared between the groups with a log-rank test. Survival plots are only shown for the cancer types in which an association between the expression of each gene and survival was found.
Statistical analysis
The results were statistically analyzed using GraphPad Prism (version 9; GraphPad Software, Inc.) software. Comparisons among multiple groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. In addition, comparisons between two groups were analyzed using an unpaired Student's t-test. All data are presented as the mean ± standard error of the mean (SEM) of at least three independent experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
TIP60 overexpression induces the ubiquitination of UHRF1
The authors have previously demonstrated that TIP60 overexpression downregulates UHRF1 and DNMT1 expression (57). The present study, using confocal microscopy experiments, confirmed that a significant (P<0.0001) decrease in UHRF1 and DNMT1 fluorescence intensity was detected in the TIP60-eGFP WT-transfected cells, while TIP60ΔMYST-eGFP transfection only marginally affected the UHRF1 and DNMT1 fluorescence intensity (Fig. S1).
As shown in Fig. 1, HeLa cells were co-transfected with TIP60-eGFP + RFP-ubiquitin. Untreated HeLa cells and eGFP + RFP-ubiquitin-co-transfected cells served as the controls. Endogenous UHRF1 levels were detected using a specific primary antibody against UHRF1 and Alexa 647-labeled secondary antibody. TIP60, UHRF1 and ubiquitin were well co-localized in the nucleus (Fig. 1). A clearly visible decrease in UHRF1 expression was observed in the TIP60 + ubiquitin-co-transfected cells as compared with the adjacent untransfected cells in the same sample or control samples (Fig. 1A). The quantification of the mean fluorescence intensity of Alexa 647 revealed a significant (P<0.0001) decrease in the UHRF1 fluorescence intensity (57%) in the TIP60 + ubiquitin-co-transfected cells (Fig. 1C). The decrease in fluorescence intensity was comparable with both the control or eGFP + ubiquitin-transfected cells (6%) (Fig. 1C). UHRF1 expression was restored in the TIP60 + ubiquitin-co-transfected cells treated with the proteasomal inhibitor, MG-132 (Fig. 1B). The mean fluorescence intensity of UHRF1 was partially recovered (Fig. 1D).
Figure 1.
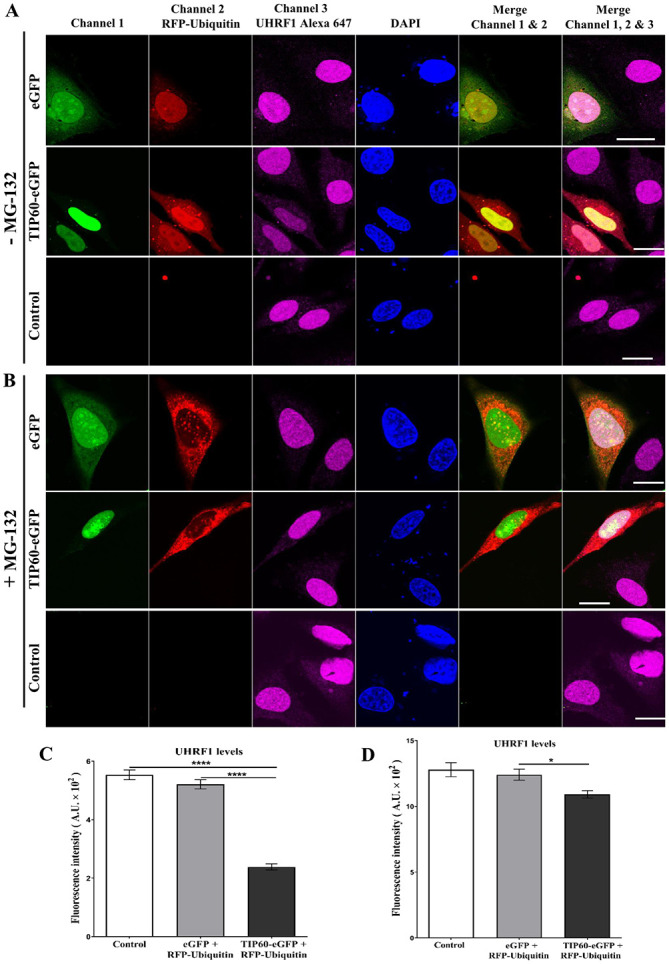
TIP60 and ubiquitin co-transfection induces the downregulation of UHRF1. Cells were co-transfected with either TIP60-eGFP (green) and RFP-Ubiquitin (red) or eGFP and RFP-Ubiquitin. Immunostaining of UHRF1 in HeLa cells without (A) or with treatment by MG-132 (B). Cells were fixed following transfection and labeled with anti-UHRF1 antibody. Endogenous UHRF1 protein was labeled with Alexa 647-labeled secondary antibody before visualization with confocal microscopy. Scale bar, 10 µm. (C and D) Represent mean fluorescence intensities levels of UHRF1 in the different samples. Values are the mean ± SEM for three independent experiments; *P<0.05; ****P<0.0001 (vs. control group), determined by one-way ANOVA with Tukey's post hoc test. UHRF1, ubiquitin-like, containing PHD and RING finger domains 1; TIP60, Tat interactive protein, 60 kDa.
Furthermore, western blot analysis was performed to support the findings of the confocal microscopy experiments (Fig. S2). HeLa cells were transfected with either TIP60 WT or TIP60ΔMYST mutant. One group of samples was treated with MG-132. In the TIP60 WT-transfected sample (Fig. S2A, -MG-132 group, lane 2), the UHRF1 level significantly decreased as compared with either the control or TIP60ΔMYST mutant samples (lane 1 and 3, respectively). Incubation with MG-132 stabilized the UHRF1 levels in the TIP60 WT overexpressing sample (Fig. S2A, + MG-132 group, lane 2). The improvement in the expression levels of UHRF1 was comparable to that of the control and TIP60ΔMYST mutant-transfected samples (Fig. S2B). In the TIP60 WT-transfected sample, a prominent smear and ubiquitinated protein bands were observed (indicated with arrows) over UHRF1 following treatment with MG-132 that the mutant failed to reproduce (Fig. S2A, + MG-132 lanes 2 and 3). Of note, this observation was observed even with a lower expression of TIP60 WT compared to the mutant TIP60ΔMYST (Fig. S2A, + MG-132 lane 2).
Subsequently, the effects of TIP60 overexpression on UHRF1 ubiquitination as a function of time (Fig. 2) were examined. HeLa cells were transfected with either TIP60 WT or TIP60ΔMYST mutant. Cells were collected at different time intervals following transfection. IP was performed with an anti-UHRF1 antibody. A prominent ubiquitination smear was observed with the ubiquitinated UHRF1 bands at 3 and 6 h post-TIP60 WT transfection, suggesting that TIP60-mediated UHRF1 ubiquitination was an early event (Fig. 2B, IP lanes 2 and 3). However, this effect was still observed at 12 and 24 h post-TIP60 WT transfection. In the case of TIP60ΔMYST, no ubiquitination of UHRF1 was observed up to 24 h post-transfection. These results demonstrated that TIP60 overexpression induced ubiquitination that did not lead to the degradation of UHRF1, due to proteasome inhibition by MG-132.
Figure 2.
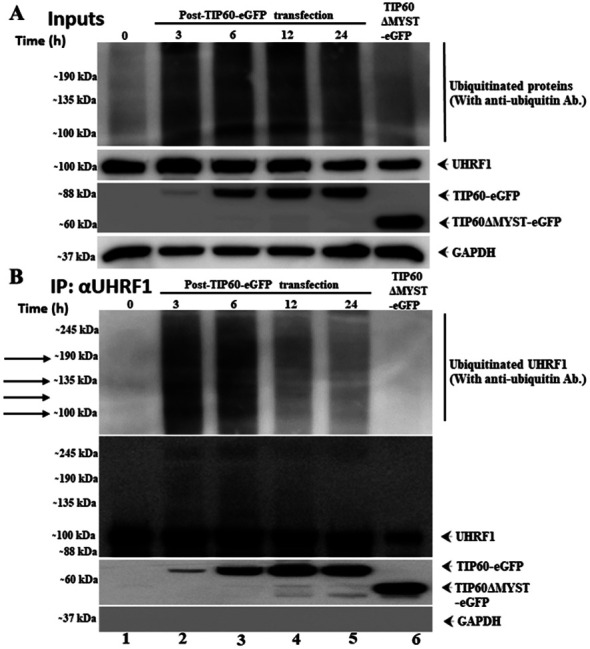
TIP60 induces UHRF1 ubiquitination in HeLa cells. (A and B) Cells were transfected with either TIP60-eGFP WT or TIP60ΔMYST-eGFP mutant. All samples were treated with 10 µM of MG-132 for 8 h before harvesting the cells (In case of 3 and 6 h sample, MG-132 was added 5 and 2 h before transfection, respectively). Cells were collected 3, 6, 12 and 24 h post TIP60 transfection and 24 h in case of TIP60ΔMYST mutant. (A) Represents inputs, while (B) represents immunoprecipitated samples. (B) Immunoprecipitation was performed with anti-UHRF1 antibody. (A) Inputs and (B) immunoprecipitation samples were analyzed by SDS-PAGE and then immunoblotted with anti-UHRF1 and anti-Ubiquitin antibodies. (B) Arrows indicate ubiquitinated UHRF1 bands. Inputs and immunoprecipitation gels were processed in parallel under similar conditions. UHRF1, ubiquitin-like, containing PHD and RING finger domains 1; TIP60, Tat interactive protein, 60 kDa.
TIP60 overexpression induces the auto-ubiquitination of UHRF1
The RING finger domain of UHRF1 has E3 ligase activity through which it can either ubiquitinate itself (auto-ubiquitination) (14,15) or other proteins (16,17). Therefore, the present study investigated whether the downregulation of UHRF1 levels is the consequence of UHRF1 auto-ubiquitination activity or whether other E3 ligases are responsible for this. This experiment was performed using HeLa cells stably expressing either UHRF1 WT protein or UHRF1 C724A-H741A mutant protein having impaired RING finger domain activity. Cells were transfected with either TIP60 WT or TIP60ΔMYST mutant and treated with MG-132. The poly-ubiquitination of UHRF1 WT was observed when TIP60 WT was overexpressed, as compared with either the controls or ΔMYST mutant samples (Fig. 3, IP lane 2). Notably, in the case of UHRF1 C724A-H741A, no ubiquitination smear and bands above the UHRF1 band were observed (Fig. 3, IP lanes 4, 5 and 6). This indicated that following TIP60 overexpression, UHRF1 was auto-ubiquitinated via its RING finger. By contrast, UHRF1 bearing the RING finger domain mutation failed to be auto-ubiquitinated following TIP60 overexpression.
Figure 3.
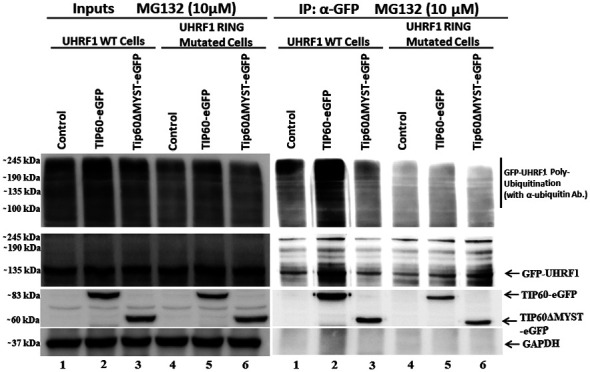
TIP60 induces auto-ubiquitination of UHRF1 in HeLa cells. Cells stably expressing either UHRF1 WT or UHRF1 C724A-H741A mutant were transfected with either TIP60-eGFP WT or TIP60ΔMYST-eGFP mutant. All samples were treated with 10 µM of MG-132, 8 h before harvesting the cells. Whole cell lysates and immunoprecipitated samples were analyzed by SDS-PAGE and then immunoblotted with anti-GFP and anti-Ubiquitin antibodies. Inputs and IP gels were processed in parallel under similar conditions. UHRF1, ubiquitin-like, containing PHD and RING finger domains 1; TIP60, Tat interactive protein, 60 kDa.
UHRF1 interacts with ubiquitin
The interaction between UHRF1 and ubiquitin inside the cell was further confirmed using FRET experiments. FRET between GFP- and RFP-labeled proteins only occurs when they are <8 nm apart, a distance relative to intermolecular protein-protein interactions (60). FRET efficiency is deduced from the decrease in GFP fluorescence lifetime measured by Fluorescence Lifetime Imaging Microscopy (FLIM) (64). FLIM technique allows to derive and color code the fluorescence lifetime (τ) of GFP at each pixel of the image (Fig. 4). In comparison to the fluorescence intensity, τ does not depend on the fluorophore concentration or instrumentation. HeLa cells expressing either GFP-UHRF1 WT or GFP-UHRF1 C724A-H741A mutant were used for the experiments.
Figure 4.
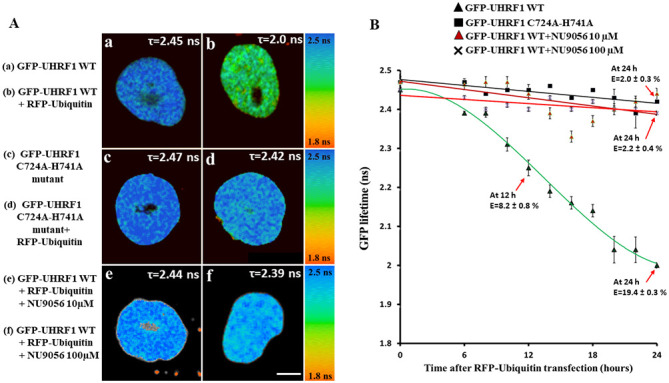
Interaction of UHRF1 and ubiquitin, as determined by FRET-FLIM. (A) Representative 30x30 µm FLIM images of HeLa cells stably expressing (a) GFP-UHRF1 WT and (b) co-transfected with RFP-ubiquitin, HeLa cells (c) stably expressing GFP-UHRF1 C724A-H741A and (d) co-transfected with RFP-ubiquitin. The lifetime values are shown by a color code ranging from red (1.8 ns) to blue (2.5 ns). Scale bar, 10 µm. In comparison to cells (a) expressing only GFP-UHRF1 WT, a marked decrease in the GFP lifetime and thus, a strong FRET efficiency was observed when HeLa cells were (b) transfected with RFP-ubiquitin. By contrast, no substantial difference in lifetime or FRET efficiency was observed when HeLa cells expressing GFP-UHRF1 C724A-H741A were transfected with RFP-ubiquitin (compare panels d and c). No difference in lifetime or FRET efficiency was observed in HeLa cells expressing GFP-UHRF1 WT in the presence of the TIP60 inhibitor NU9056 (e) 10 µM or (f) 100 µM. FLIM data indicate that UHRF1 interacts with ubiquitin while this interaction is impaired in case of UHRF1 having a RING finger domain mutation. (B) Change in GFP lifetime as a function of time. Values are the mean ± SEM from three independent experiments. The fluorescence lifetimes of GFP-UHRF1 WT (without and with TIP60 inhibitor NU9056 10 µM, 100 µM) or GFP-UHRF1 C724A-H741A were measured at different times following transfection with RFP-Ubiquitin. FRET efficiency was calculated according to E=1-(τDA/τD), where τDA is the lifetime of the donor (GFP) in the presence of acceptor (RFP) and τD is the lifetime of GFP in the absence of acceptor. UHRF1, ubiquitin-like, containing PHD and RING finger domains 1; TIP60, Tat interactive protein, 60 kDa.
These cells were co-transfected with RFP-labeled ubiquitin and fixed at different time points, between 6 and 24 h. The fluorescence lifetime of GFP-UHRF1 WT used as a control was 2.45±0.01 ns (n=36 cells) (Fig. 4A-a). The lifetime of GFP-UHRF1 was observed to decrease as a function of time when the GFP-UHRF1 WT cells were transfected with RFP-ubiquitin (Fig. 4B). A substantial decrease (8.2% FRET) in lifetime was observed after 12 h of RFP-ubiquitin transfection (2.25±0.02 ns, n=26 cells) and a further decrease in lifetime was observed after 24 h (2.00±0.01 ns, n=20 cells) (Fig. 4A-bs). The corresponding FRET efficiency (E) was 8.2±0.8 and 19.4±0.3% after 12 and 24 h of RFP-ubiquitin transfection, respectively. Subsequently, the interaction between RFP-labeled ubiquitin and GFP-labeled UHRF1 having a RING Finger domain mutation, as a function of time was examined. The lifetime of GFP-UHRF1 C724A-H741A taken as a control was 2.47±0.01 ns (n=28 cells) (Fig. 4A-c). Notably, no considerable decrease was observed in the lifetime of GFP-UHRF1 C724A-H741A following RFP-ubiquitin transfection. Following 24 h of RFP-ubiquitin transfection, the lifetime was 2.42±0.01 ns (n=18 cells) (Fig. 4A-d), which corresponded to a FRET efficiency of 2.0±0.3%, below the commonly accepted 5% threshold value for protein-protein interaction (65). Thus, these data suggest that mutation in the RING finger domain of UHRF1 can impair its interaction with the ubiquitin.
Furthermore, the present study examined the effects of the inhibition of TIP60 acetylation activity on the interaction between UHRF1 and ubiquitin, by using the specific TIP60 inhibitor, NU9056. First, the effect of NU9056 at various concentrations between 1 and 100 µM after 24 h treatment was examined, and the UHRF1-ubiquitin interaction was analyzed through FLIM, using HeLa cells expressing GFP-UHRF1 WT protein. The interaction between UHRF1 and ubiquitin could be still detected in the presence of 1, 3 and 5 µM of NU9056 (FRET efficiency was 12, 10 and 8.8%, respectively), but was impaired at 10, 30 and 100 µM (FRET efficiency 5.6, 4.8 and 3.6% respectively) (Fig. S3). Further experiments were carried out to examine the effects of NU9056 on the UHRF1-ubiquitin interaction in a time-dependent manner at 10 and 100 µM. Under these conditions, no considerable decrease in the lifetime of GFP-UHRF1 was observed at any time intervals in the presence of TIP60 inhibitor, as shown in Fig. 4A-e and f. Overall, the FLIM data suggest that TIP60 favors UHRF1-Ubiquitin interaction, while the inhibition of acetyltransferase activity of TIP60 results in the impairment of this interaction.
TIP60 overexpression interferes with the USP7-UHRF1 association
In order to decipher the origin of the activation of UHRF1 auto-ubiquitination, an alteration of the protective role of USP7 was hypothesized. Indeed, USP7 interacts with UHRF1 and protects it from ubiquitin-mediated proteasomal degradation (58,66). To assess this hypothesis, HeLa cells were transfected with either TIP60-eGFP or TIP60ΔMYST-eGFP mutant. Anti-UHRF1 antibody was used to immunoprecipitate the endogenous UHRF1 and its associated partner, USP7. The association between USP7 and UHRF1 was observed in the untreated sample (control) as USP7 was co-immunoprecipitated with UHRF1 (Fig. 5D). In the TIP60 overexpressed sample, USP7 was barely detected following co-precipitation with UHRF1 (Fig. 5D, lane 2). By contrast, with TIP60ΔMYST-eGFP mutant this association was not affected (Fig. 5D, lane 3). In a reciprocal experiment, anti-USP7 antibody was used to immunoprecipitate endogenous USP7. It was observed that following TIP60 WT overexpression, reduced levels of endogenous UHRF1 were co-precipitated as compared with the control and TIP60ΔMYST-eGFP mutant sample (Fig. 5E, compare lane 2 to lanes 1 and 3). Taken together, these results suggest that TIP60 regulates the interaction of UHRF1 with USP7, which conditions the auto-ubiquitination activity of UHRF1.
Figure 5.
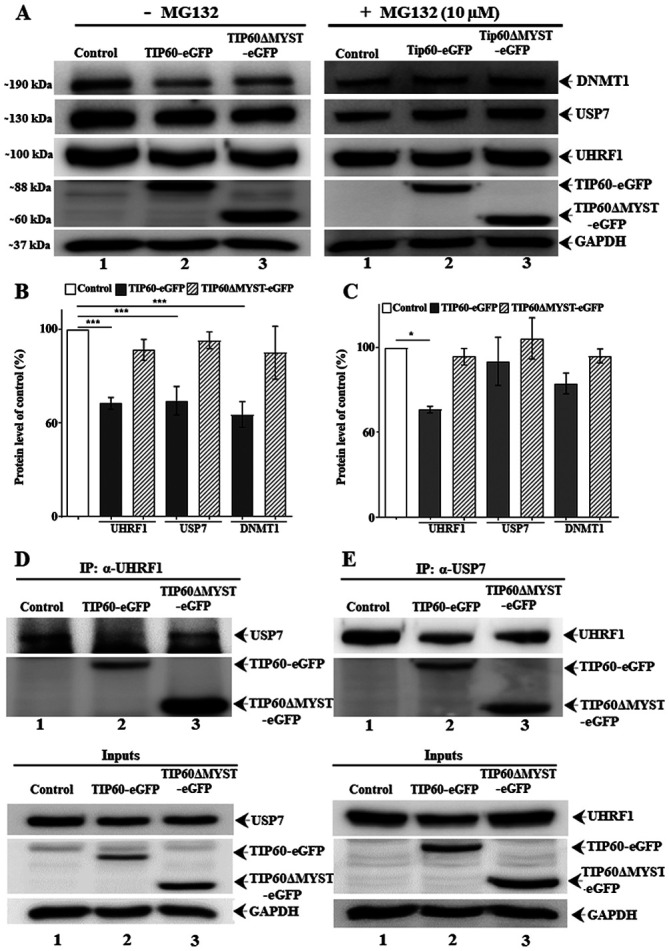
TIP60 interferes with UHRF1-USP7 association and their expression levels. HeLa cells were transfected with either TIP60-eGFP WT or TIP60ΔMYST-eGFP mutant. Western blot and immunoprecipitated samples were resolved by SDS-PAGE and immunoblotted with anti-UHRF1, anti-USP7 and anti-DNMT1 antibodies. (A) Effect of TIP60 on UHRF1, USP7 and DNMT1 levels with or without MG-132 treatment, respectively. (B and C) Quantification of the effect of TIP60 on UHRF1, USP7 and DNMT1 levels with or without MG-132 treatment, respectively. Values are the mean ± SEM from at least three independent experiments which were analyzed statistically by one-way ANOVA with Tukey's post hoc test (*P<0.05; ***P<0.001 vs. control group). (D) Anti-UHRF1 antibody was used to co-immunoprecipitate UHRF1 and its partner USP7. (E) In a reciprocal experiment, anti-USP7 antibody was used to co-immunoprecipitate USP7 and UHRF1. Inputs and IP gels were processed in parallel under similar conditions. UHRF1, ubiquitin-like, containing PHD and RING finger domains 1; TIP60, Tat interactive protein, 60 kDa; USP7, ubiquitin-specific-processing protease 7; DNMT1, DNA methyltransferase 1.
TIP60 WT overexpression induced the downregulation of UHRF1, USP7 and DNMT1 protein expression in comparison to control or TIP60 ΔMYST mutant (Fig. 5A, left panel). However, treatment with MG-132 (Fig. 5A, right panel) led to a recovery in the expression levels of these proteins. Quantitative analysis of the input fractions revealed a significant (P<0.001) decrease in the UHRF1, USP7 and DNMT1 levels following TIP60 WT overexpression (Fig. 5B). MG-132 treatment fully restored the USP7 and DNMT1 levels, whereas it only partially restored the UHRF1 levels (P<0.05) (Fig. 5C). To examine the expression levels of USP7 and UHRF1 inside the cells following TIP60 overexpression, confocal microscopy experiments were performed. The endogenous levels of UHRF1 and USP7 were examined in the same cells by labeling with respective antibodies. Based on the mean fluorescence intensity of the Alexa 568- and Alexa 647-labeled secondary antibodies, the USP7 and UHRF1 levels were found to be significantly decreased (P<0.0001) following TIP60 overexpression (Fig. 6A and C). As compared with the control samples, decreases in fluorescence of 45 and 60% were observed for USP7 and UHRF1, respectively. By contrast, the overexpression of the TIP60ΔMYST-eGFP mutant only marginally affected the fluorescence intensities of USP7 and UHRF1 (Fig. 6A and C). Thus, these data demonstrate that TIP60 overexpression can downregulate the USP7 and UHRF1 levels simultaneously. Due to its downregulation, USP7 was likely unable to protect the UHRF1 degradation via the proteasomal pathway. As a significant decrease (P<0.0001) was observed in the USP7 levels following TIP60 overexpression, the USP7 levels were further examined following treatment with MG-132. Of note, the expression levels of USP7 were improved in the TIP60 overexpressing samples following treatment with MG-132 (Fig. 6B and D). The expression levels of UHRF1 were also improved, although to a lesser extent as compared with those of USP7, which suggests that once UHRF1 is degraded through the proteasomal degradation pathway, its levels are not restored immediately (Fig. 6B and D).
Figure 6.
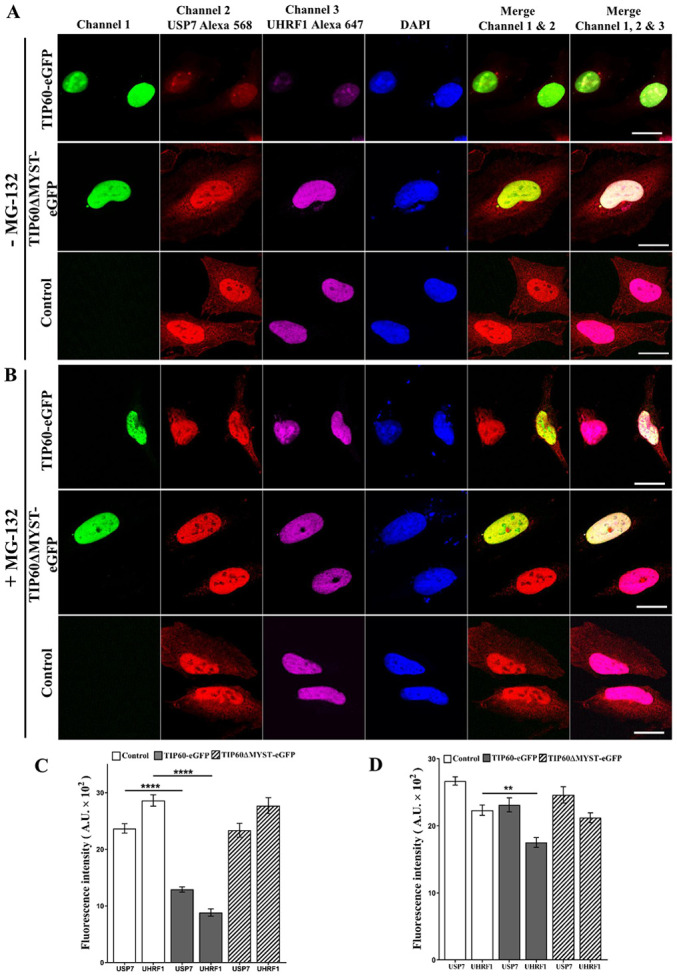
TIP60 downregulates the UHRF1 and USP7 levels in HeLa cells. Cells were either (A) not treated with MG-132 or (B) treated with MG-132 at 10 µM for 8 h. Cells were immunostained with anti-USP7 or anti-UHRF1 antibodies. TIP60-eGFP wild-type (WT) or TIP60ΔMYST-eGFP mutant was transiently overexpressed and their effects were compared with non-transfected control cells. Cells were fixed following transfection and tagged by anti-USP7 and anti-UHRF1 antibodies. Alexa 568 and Alexa 647-labeled secondary antibodies were used as indicated in the figure to visualize the corresponding proteins by confocal microscopy. Scale bar, 10 µm. (C and D) Mean fluorescence intensities representing the levels of USP7 and UHRF1 before and after MG-132 treatment, respectively. Values are the mean ± SEM for three independent experiments; **P<0.01; ****P<0.0001 (vs. control group), determined by one-way ANOVA with Tukey's post hoc test. UHRF1, ubiquitin-like, containing PHD and RING finger domains 1; TIP60, Tat interactive protein, 60 kDa; USP7, ubiquitin-specific-processing protease 7.
TIP60 overexpression induces the activation of p73
Since the experiments indicated that TIP60 regulated UHRF1 expression by governing its auto-ubiquitination, the physiological or physiopathological consequences of this regulation were then investigated. Tumor suppressor protein p73 is important for genomic stability by responding to a number of stress signals and is under the control of UHRF1 (67,68). The p73-mediated apoptosis leads to the activation of the mitochondria-dependent apoptotic pathway through the transactivation of pro-apoptotic proteins (e.g., BAX) and the downregulation of pro-survival proteins (e.g., BCL2) (69,70). Therefore, the present study examined the effect of the TIP60-mediated UHRF1 downregulation on the levels of p73. It was found that the overexpression of TIP60 induced an increase in the expression of p73 (Fig. 7A) and BAX protein (Fig. 7B), whereas it induced a decrease in the expression of the anti-apoptotic BCL2 protein (Fig. 7C). Furthermore, it was observed that TIP60 overexpression induced caspase-3 activation from its precursor pro-caspase-3 (Fig. 7A), which in turn triggered the cleavage of PARP to induce apoptosis.
Figure 7.
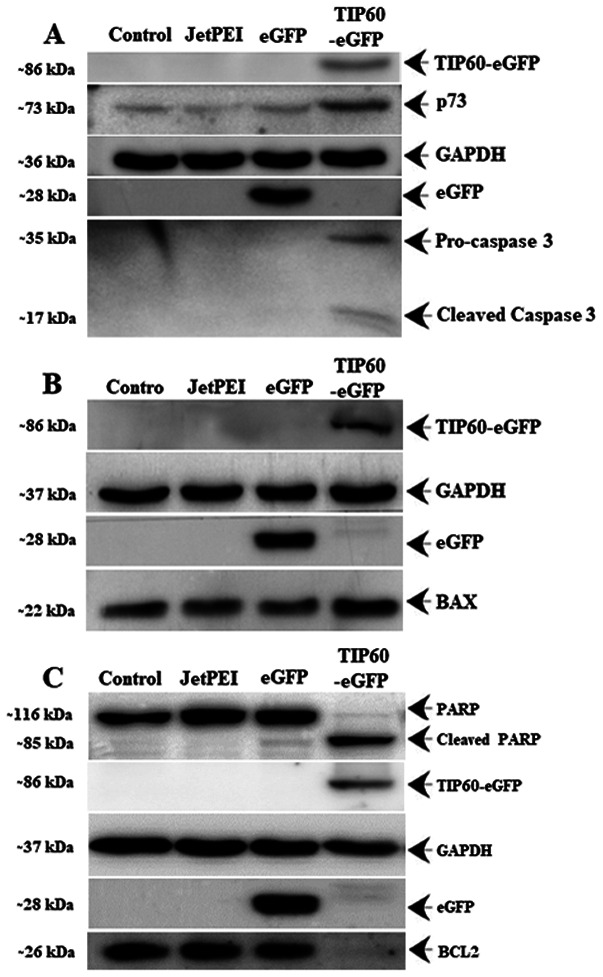
Effect of TIP60 overexpression on p73, and pro- and anti-apoptotic proteins. (A) p73 and caspase-3 (B) BAX, (C) BCL2 and PARP levels were examined by western blot analysis following TIP60 overexpression in HeLa cells. TIP60, Tat interactive protein, 60 kDa.
In order to assess the effect of TIP60 overexpression on downstream signaling pathways of p73, flow cytometric experiments were performed. TIP60-eGFP transfected cells were analyzed by FACS and compared with cells transfected with the vector, jetPEI. PI and Annexin V-iFluor™ 350 staining aided the detection of late and early phases of apoptosis. A significant (P<0.001) decrease (34%) in cell viability was observed in the TIP60-transfected cells as compared with the control cells. Along with the decrease in cell viability following TIP60 overexpression, an increase of 12% (P<0.05) and 16% (P<0.01) in the number of early and late apoptotic cells was also observed, respectively (Fig. 8A-C).
Figure 8.
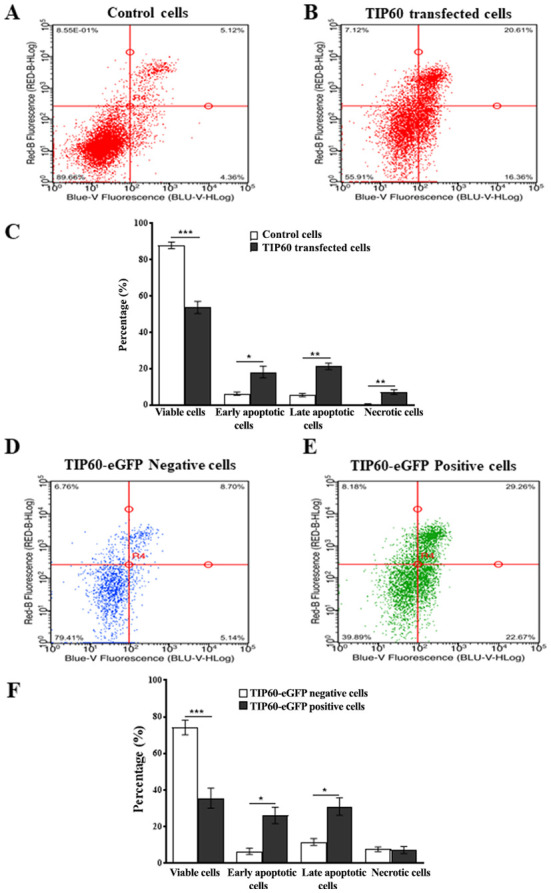
TIP60 overexpression induces the apoptosis of cancer cells. (A) FACS analysis examining Annexin V-iFluor™ 350 and PI labeling in control HeLa cells (treated with jetPEI in an identical manner) and (B) cells transfected with TIP60-eGFP for 24 h. (D and E) FACS analysis examining Annexin V-iFluor™ 350 and PI labeling in TIP60-eGFP-negative cells to TIP60-eGFP-positive cells in TIP60-eGFP transfected samples. (C and F) Graph represents average values from three independent experiments which were statistically analyzed using a Student's t-test (*P<0.05; **P<0.01; ***P<0.001). TIP60, Tat interactive protein, 60 kDa.
To confirm the aforementioned results, the total population of TIP60-eGFP-transfected cells was separated into TIP60-eGFP-positive and TIP60-eGFP-negative cell populations based on eGFP fluorescence. The average transfection efficiency of TIP60-eGFP was 61%, so that significant populations of both types of cells could be obtained. This separation allowed the comparison of apoptosis induction in TIP60-eGFP-expressing cells and non-expressing cells, in the same sample. The viability of the TIP60-eGFP-expressing cells decreased (P<0.001) by 39% as compared with the cells not expressing TIP60-eGFP (Fig. 8D-F). In the TIP60-eGFP-transfected cells, there was also a marked increase in the number of early and late apoptotic cells (Fig. 8E and F). As UHRF1 exhibits anti-apoptotic properties (5,67), targeting UHRF1 expression can thus activate apoptotic pathways in cancer cells. Cumulatively, these data explain the association between the TIP60-mediated downregulation of the epigenetic integrator, UHRF1, and the induction of apoptosis in cancer cells to maintain the cellular and genomic integrity.
Discussion
UHRF1 and TIP60 are within the same epigenetic complex with other partners, such as DNMT1, USP7, HDAC1, proliferating cell nuclear antigen (PCNA) and euchromatic histone-lysine N methyltransferase 2 (EHMT2, also known as G9a) (17,25,27,66,71). Higher expression levels of UHRF1 have been reported in the majority of cancers (4,72) and are related to suppression of TSGs expression, tumor invasion, poor prognosis and resistance towards chemotherapy (4,73-77). In contrast to UHRF1, TIP60 expression is low in cancer cells. TIP60 is considered to play a tumor suppressor role by maintaining the cellular and genomic stability (24,41,45,51-56). UHRF1 directly interacts with the MYST domain of TIP60 (57) and regulates TIP60 expression and activity (25,27). There is thus a fragile balance between UHRF1 and TIP60 broken in favor of UHRF1 in cancers. Thus, it is considered that the role of TIP60 is to maintain UHRF1 at physiological levels in the UHRF1/DNMT1 macromolecular complex.
The present study performed bioinformatics analysis to investigate expression of TIP60 (Fig. S4) and UHRF1 (Fig. S5) in various types of cancer (Table SI) which revealed that TIP60 expression was mostly downregulated in the majority of cancers, while on the other hand, UHRF1 expression was upregulated in the majority of cancers. Further analysis revealed that a higher TIP60 expression was associated with a better prognosis (Fig. S6) and a higher UHRF1 expression was associated with a poor prognosis (Fig. S7) of cancer patients. The investigation of the co-expression of both genes revealed that they were expressed independently in the majority of cancer types. However, in kidney renal clear cell carcinoma (KIRC) and brain lower grade glioma (LGG) cancers, both genes were found to have the tendency towards an opposite association (Fig. S8). In addition, a higher TIP60 and a lower UHRF1 expression in these two cancer types was associated with a better prognosis. The distribution of USP7 expression in various cancer types did not seem to change in correspondence to the non-tumor samples (Fig. S9) with the exception of glioblastoma multiforme (where USP7 expression was lower) and kidney chromophobe (where USP7 expression was higher). The association between USP7 expression and the probability of patient survival was found only in LGG, where a higher USP7 expression was associated with a better prognosis (Fig. S10). However, a limitation to the present study is that more thorough bioinformatics analyses are required for the validation of the results. Caution is advised, only mRNA levels were monitored, while protein stability may be important at the protein final level.
In a previous study by the authors (57), HeLa cells were used to investigate the interaction between UHRF1 and TIP60. It was reported that UHRF1 interacts with the MYST domain of TIP60, and that TIP60 overexpression leads to the downregulation of UHRF1 and DNMT1 levels (57). The objective of the present study was to investigate the mechanism behind the TIP60-mediated downregulation of UHRF1. Therefore, experiments were performed within the same cell line. Furthermore, the basal level of TIP60 is low in HeLa cells due to the presence of viral oncoproteins (HPV E6 and E7). E6 protein leads to the proteasomal mediated degradation of TIP60 by EDD1 E3 ligase (55,78). Notably, this matches observations of cervical cancer where these viral proteins induce the downregulation of TIP60, leading to apoptosis inhibition (79). Additionally, retrospective data analysis comparing the differential expression of TIP60 gene in a dataset (GDS3233) of normal cervix vs. cervical cancer samples (62) revealed that TIP60 expression was significantly (P<0.01) downregulated in cancerous tissues (Fig. S11). Taken together, these data validate the relevance of HeLa cells for the present study, while it would be of interest to observe if whether the TIP60/UHRF1 pathway is a general mechanism relevant in other cancer cell lines.
In the present study, using western blot analysis and confocal microscopy, it was confirmed that TIP60 downregulates the UHRF1/DNMT1 tandem. Of note, the ΔMYST mutant (lacking acetyltransferase activity) was unable to affect the expression of both proteins, indicating that the acetyltransferase activity of TIP60 is required for downregulating both proteins. It is thus suggested that TIP60 drives the degradation of UHRF1 and consequently DNMT1, considering that this latter has been shown to be under the control of UHRF1 (80). A direct control of TIP60 on DNMT1, in an USP7-dependent way, may also occur (17).
Ubiquitination is a post-translational modification which adds single or multiple ubiquitin molecules to proteins marking them for proteasomal degradation, cellular trafficking, autophagy, DNA repair, receptor internalization or regulation of enzymatic activity (81,82). USP7 is a deubiquitinating enzyme which protects many proteins from ubiquitination including p53, UHRF1, PTEN, MDM2 and Myc. Its expression levels are high in a number of cancers. Dysregulation in ubiquitination/deubiquitination can play a critical role in several diseases, including cancer (81). USP7 interacts with UHRF1 and protects it from degradation (58,83) while during the M phase, UHRF1 is degraded as a result of its dissociation from USP7 (66). Zhang et al (58) reported that TIP60 acetylates UHRF1 at K659, which decreases the interaction of USP7-UHRF1. The data of the present study indicated that the overexpression of TIP60, but not of its ΔMYST mutant, interfered with the association and expression levels of USP7 and UHRF1. The dissociation of USP7 from UHRF1 likely condemns this latter becoming a prey for E3 ligases that have been reported to ubiquitinate UHRF1 (84,85) or belong to the UHRF1 complex (15). The role of USP7 as an oncogene or tumor suppressor gene is still a matter of debate. Indeed, USP7 protects XPC, a crucial damage recognition factor in DNA repair, from proteolysis (86,87). USP7 also interacts with the tumor suppressor gene p53 (88) and regulates its stability (89). However, the overexpression of USP7 and MDM2 leads to the inactivation of p53, resulting in cancer initiation and progression (90). This appears to be a result of a protection of the MDM2 E3 ligase which ubiquitinates p53 by proteosomal degradation. The inhibition of USP7 can reactivate p53 (90). In the majority of cancers, the overexpression of USP7 is observed (91). Consistently, almost all inhibitors of USP7 lead to cancer cell proliferation arrest which favors the idea that USP7 rather plays a role of oncogene. Such a role for USP7 has also been supported by the study of Felle et al (83) on the colon cancer cell line, HCT116, in which it was shown that USP7 favors de novo and maintenance DNA methylation activity of DNMT1. This suggests that DNA methylation patterns, particularly those of tumor suppressor silenced genes, are transmitted throughout mitosis. It is not excluded that this mechanism may also be involved in the onset of tumorigenesis by the de novo hypermethylation of the promoters of tumor suppressor genes. Indeed, the downregulation of UHRF1 via the downregulation of USP7 allows the re-expression of tumor suppressor genes (15). In the present study, the TIP60-mediated interference with USP7-UHRF1 association was observed in HeLa cells. However, it would be of interest to investigate and validate the role of USP7 and its association with UHRF1 in other cancer cell lines.
In the present study, the ubiquitination of UHRF1 was observed following TIP60 overexpression, which is likely a consequence of TIP60-mediated UHRF1-USP7 dissociation, as it has been reported for DNMT1 (17,92). Due to this dissociation, USP7 is no more able to protect UHRF1 from degradation through the proteasomal pathway. This hypothesis was confirmed by use of MG-132 that helped recovering initial UHRF1 levels. The RING domain of UHRF1 has E3 ligase activity through which it can either ubiquitinate itself or other proteins (15,17). The present study demonstrated that TIP60 overexpression controlled the auto-ubiquitination of UHRF1, but not that of UHRF1 C724A-H741A mutant, having impaired RING domain activity. The data further indicated that the interaction between UHRF1 and ubiquitin occurred in a time-dependent manner and that UHRF1 mutant, having impaired E3 ligase activity, was not able to interact with ubiquitin. It was also found that TIP60 favored the UHRF1/ubiquitin interaction, while the inhibition of its acetyltransferase activity impaired this interaction. Subsequently, UHRF1 was degraded via the proteasome, as treatment with MG132 was able to recover the initial UHRF1 levels.
A downregulation of UHRF1 induces a recovery in numerous tumor suppressor genes including RB1, p16INK4A (CDKN2A), CDH13, SOCS3, BRCA1, CDX2, RUNX3, FOXO4, PPGARG, PML and p73 (4,24). The present study focused on p73, as it is known that TIP60 positively regulates apoptosis (41) and that UHRF1 positively regulates p73 (68). It was found that TIP60 overexpression induced an enhanced p73 expression. Therefore, it is suggested that TIP60-mediated apoptosis occurs via the upregulation of p73. However, it cannot be excluded that p73 is involved upstream of TIP60, since it has been previously observed that p73 also negatively regulates UHRF1 (54,93). Furthermore, in agreement with the present study, it has been observed that p73-mediated apoptosis involves a caspase-dependent pathway (93).
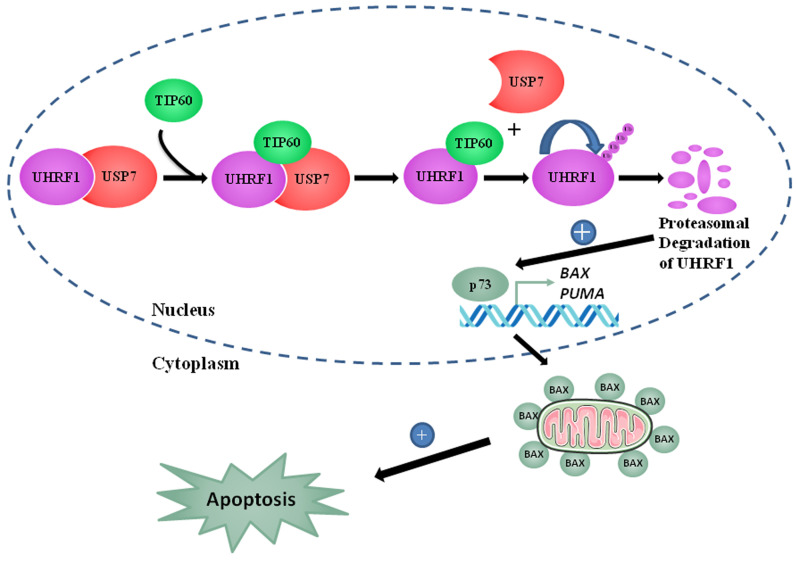
In conclusion, the present study proposes a model (Fig. 9) depicting the tumor suppressor role of TIP60 as a tumor suppressor gene. TIP60 upregulation induced apoptosis by the activation of the p73-mediated downstream signaling pathway. TIP60 overexpression led to a decrease in BCL2 and an increase in BAX expression, which activated caspase-3. Caspase-3 activated the cleavage of PARP and induced apoptosis. Overall, these observations support a tumor suppressor role of TIP60 through the regulation of the auto-ubiquitination activity of UHRF1. This interplay directly governs the expression of TSGs, such as p73, explaining why TIP60 plays a role in apoptosis and cell cycle regulation.
Figure 9.
Schematic model of TIP60-mediated apoptosis in cancer cells. Association of UHRF1 with USP7 protects UHRF1 protein from proteasomal degradation. TIP60 overexpression disrupts the tandem UHRF1-USP7 which leads UHRF1 to commit suicide via auto-ubiquitination. UHRF1, ubiquitin-like, containing PHD and RING finger domains 1; TIP60, Tat interactive protein, 60 kDa; USP7, ubiquitin-specific-processing protease 7.
Supplementary Data
Acknowledgements
The authors would like to thank Mr. Romain Vauchelles (engineer, Plate-forme d'Imagerie Quantitative-PIQ) and Dr Ludovic Richert (University of Strasbourg) for providing assistance with the Confocal and FLIM experiments, respectively. The authors are also thankful to Dr Sophie Rousseaux (MD, PhD, INSERM Research Director), Dr Florent Chuffart (PhD, INSERM Research Engineer) and Dr Ekaterina Flin (University Grenoble Alpes Research Engineer) (EpiMed core facility; http://epimed.univ-grenoble-alpes.fr) for their assistance with the bioinformatics analysis. The authors also wish to thank Dr Nicolas Reynoird (Institute for Advanced Biosciences, University Grenoble Alpes) for the critical reading of the manuscript.
Abbreviations
- UHRF1
ubiquitin-like, containing PHD and RING finger domains 1
- TIP60
Tat interactive protein, 60 kDa
- FLIM
fluorescence lifetime imaging microscopy
- DNMT1
DNA methyltransferase 1
- USP7
ubiquitin-specific-processing protease 7
- HDAC1
histone deacetylase 1
- IP
immunoprecipitation
- WT
wild-type
- ΔMYST
MYST domain mutant
- FRET
Förster Resonance Energy Transfer
- PCNA
proliferating cell nuclear antigen
- EHMT2
euchromatic histone-lysine N methyltransferase 2
Funding Statement
The present study was supported by ANR (SMFLUONA, ANR-17-CE11-0036-01). TA was supported by fellowships from Higher Education Commission (HEC), Pakistan. YM is grateful to the Institut Universitaire de France (IUF) for support and providing additional time to be dedicated to research.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
TA conducted all the experiments with the assistance of WA, AI and LZ under the supervision of MM and CB. MM, CB, AH and YM were involved in the conception and design of the study. CDM assisted with the flow cytometry experiments. TA, CB and MM wrote most of the manuscript and confirm the authenticity of all raw data with the guidance and assistance of AH and YM. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hopfner R, Mousli M, Jeltsch JM, Voulgaris A, Lutz Y, Marin C, Bellocq JP, Oudet P, Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expression. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- 2.Hopfner R, Mousli M, Garnier JM, Redon R, du Manoir S, Chatton B, Ghyselinck N, Oudet P, Bronner C. Genomic structure and chromosomal mapping of the gene coding for ICBP90, a protein involved in the regulation of the topoisomerase IIalpha gene expression. Gene. 2001;266:15–23. doi: 10.1016/S0378-1119(01)00371-7. [DOI] [PubMed] [Google Scholar]
- 3.Krifa M, Alhosin M, Muller CD, Gies JP, Chekir-Ghedira L, Ghedira K, Mély Y, Bronner C, Mousli M. Limoniastrum guyonianum aqueous gall extract induces apoptosis in human cervical cancer cells involving p16 INK4A re-expression related to UHRF1 and DNMT1 down-regulation. J Exp Clin Cancer Res. 2013;32:30. doi: 10.1186/1756-9966-32-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashraf W, Ibrahim A, Alhosin M, Zaayter L, Ouararhni K, Papin C, Ahmad T, Hamiche A, Mély Y, Bronner C, Mousli M. The epigenetic integrator UHRF1: On the road to become a universal biomarker for cancer. Oncotarget. 2017;8:51946–51962. doi: 10.18632/oncotarget.17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: Oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol Ther. 2007;115:419–434. doi: 10.1016/j.pharmthera.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 7.Bronner C, Alhosin M, Hamiche A, Mousli M. Coordinated dialogue between UHRF1 and DNMT1 to ensure faithful inheritance of methylated DNA patterns. Genes (Basel) 2019;10:65. doi: 10.3390/genes10010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 9.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 10.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 11.Nady N, Lemak A, Walker JR, Avvakumov GV, Kareta MS, Achour M, Xue S, Duan S, Allali-Hassani A, Zuo X, et al. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem. 2011;286:24300–24311. doi: 10.1074/jbc.M111.234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu L, Li Z, Wang P, Lin Y, Xu Y. Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell Res. 2011;21:1374–1378. doi: 10.1038/cr.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins Y, Markovtsov V, Lang W, Sharma P, Pearsall D, Warner J, Franci C, Huang B, Huang J, Yam GC, et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol Biol Cell. 2005;16:5621–5629. doi: 10.1091/mbc.e05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahim A, Alhosin M, Papin C, Ouararhni K, Omran Z, Zamzami MA, Al-Malki AL, Choudhry H, Mély Y, Hamiche A, et al. Thymoquinone challenges UHRF1 to commit auto-ubiquitination: A key event for apoptosis induction in cancer cells. Oncotarget. 2018;9:28599–28611. doi: 10.18632/oncotarget.25583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tauber M, Fischle W. Conserved linker regions and their regulation determine multiple chromatin-binding modes of UHRF1. Nucleus. 2015;6:123–132. doi: 10.1080/19491034.2015.1026022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502:249–253. doi: 10.1038/nature12488. [DOI] [PubMed] [Google Scholar]
- 19.Qin W, Wolf P, Liu N, Link S, Smets M, La Mastra F, Forné I, Pichler G, Hörl D, Fellinger K, et al. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 2015;25:911–929. doi: 10.1038/cr.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster BM, Stolz P, Mulholland CB, Montoya A, Kramer H, Bultmann S, Bartke T. Critical role of the UBL domain in stimulating the E3 ubiquitin ligase activity of UHRF1 toward chromatin. Mol Cell. 2018;72:739–752.e9. doi: 10.1016/j.molcel.2018.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishima Y, Brueckner L, Takahashi S, Kawakami T, Otani J, Shinohara A, Takeshita K, Garvilles RG, Watanabe M, Sakai N, et al. Enhanced processivity of Dnmt1 by monoubiquitinated histone H3. Genes Cells. 2020;25:22–32. doi: 10.1111/gtc.12732. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Wang L, Du Y, Xie S, Yang X, Lian F, Zhou Z, Qian C. Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation. Nuclic Acids Res. 2018;46:3218–3231. doi: 10.1093/nar/gky104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alhosin M, Omran Z, Zamzami MA, Al-Malki AL, Choudhry H, Mousli M, Bronner C. Signalling pathways in UHRF1-dependent regulation of tumor suppressor genes in cancer. J Exp Clin Cancer Res. 2016;35:174. doi: 10.1186/s13046-016-0453-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alhosin M, Sharif T, Mousli M, Etienne-Selloum N, Fuhrmann G, Schini-Kerth VB, Bronner C. Down-regulation of UHRF1, associated with re-expression of tumor suppressor genes, is a common feature of natural compounds exhibiting anti-cancer properties. J Exp Clin Cancer Res. 2011;30:41. doi: 10.1186/1756-9966-30-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai C, Shi D, Gu W. Negative regulation of the acetyltransferase TIP60-p53 interplay by UHRF1 (ubiquitin-like with PHD and RING finger domains 1) J Biol Chem. 2013;288:19581–19592. doi: 10.1074/jbc.M113.476606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan D, Factor D, Liu Y, Wang Z, Kao HY. The epigenetic regulator UHRF1 promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene. 2013;32:3819–3828. doi: 10.1038/onc.2012.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Achour M, Fuhrmann G, Alhosin M, Rondé P, Chataigneau T, Mousli M, Schini-Kerth VB, Bronner C. UHRF1 recruits the histone acetyltransferase Tip60 and controls its expression and activity. Biochem Biophys Res Commun. 2009;390:523–528. doi: 10.1016/j.bbrc.2009.09.131. [DOI] [PubMed] [Google Scholar]
- 28.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 30.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee KK, Workman JL. Histone acetyltransferase complexes: One size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 32.Doyon Y, Selleck W, Lane WS, Tan S, Côté J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voss AK, Thomas T. MYST family histone acetyltransferases take center stage in stem cells and development. Bioessays. 2009;31:1050–1061. doi: 10.1002/bies.200900051. [DOI] [PubMed] [Google Scholar]
- 34.Sheikh BN, Akhtar A. The many lives of KATs-detectors, integrators and modulators of the cellular environment. Nat Rev Genet. 2019;20:7–23. doi: 10.1038/s41576-018-0072-4. [DOI] [PubMed] [Google Scholar]
- 35.Kim CH, Kim JW, Jang SM, An JH, Seo SB, Choi KH. The chromodomain-containing histone acetyltransferase TIP60 acts as a code reader, recognizing the epigenetic codes for initiating transcription. Biosci Biotechnol Biochem. 2015;79:532–538. doi: 10.1080/09168451.2014.993914. [DOI] [PubMed] [Google Scholar]
- 36.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: Many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Kimura A, Matsubara K, Horikoshi M. A decade of histone acetylation: Marking eukaryotic chromosomes with specific codes. J Biochem. 2005;138:647–662. doi: 10.1093/jb/mvi184. [DOI] [PubMed] [Google Scholar]
- 38.Kim MY, Ann EJ, Kim JY, Mo JS, Park JH, Kim SY, Seo MS, Park HS. Tip60 histone acetyltransferase acts as a negative regulator of Notch1 signaling by means of acetylation. Mol Cell Biol. 2007;27:6506–6519. doi: 10.1128/MCB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int J Biochem Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Putnik J, Zhang CD, Archangelo LF, Tizazu B, Bartels S, Kickstein M, Greif PA, Bohlander SK. The interaction of ETV6 (TEL) and TIP60 requires a functional histone acetyltransferase domain in TIP60. Biochim Biophys Acta. 2007;1772:1211–1224. doi: 10.1016/j.bbadis.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;104:463–473. doi: 10.1016/S0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 42.Judes G, Rifaï K, Ngollo M, Daures M, Bignon YJ, Penault-Llorca F, Bernard-Gallon D. A bivalent role of TIP60 histone acetyl transferase in human cancer. Epigenomics. 2015;7:1351–1363. doi: 10.2217/epi.15.76. [DOI] [PubMed] [Google Scholar]
- 43.Idrissou M, Rifaï K, Daures M, Penault-Llorca F, Bignon YJ, Bernard-Gallon D. Exciting history of Tip60 and its companions in carcinogenesis across the heterochromatin landscapes. OMICS. 2018;22:626–628. doi: 10.1089/omi.2018.0122. [DOI] [PubMed] [Google Scholar]
- 44.Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 46.Mo F, Zhuang X, Liu X, Yao PY, Qin B, Su Z, Zang J, Wang Z, Zhang J, Dou Z, et al. Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat Chem Biol. 2016;12:226–232. doi: 10.1038/nchembio.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeRan M, Pulvino M, Greene E, Su C, Zhao J. Transcriptional activation of histone genes requires NPAT-dependent recruitment of TRRAP-Tip60 complex to histone promoters during the G1/S phase transition. Mol Cell Biol. 2008;28:435–447. doi: 10.1128/MCB.00607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niida H, Katsuno Y, Sengoku M, Shimada M, Yukawa M, Ikura M, Ikura T, Kohno K, Shima H, Suzuki H, et al. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev. 2010;24:333–338. doi: 10.1101/gad.1863810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y, Fisher JB, Koprowski S, McAllister D, Kim MS, Lough J. Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev Dyn. 2009;238:2912–2921. doi: 10.1002/dvdy.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakuraba K, Yasuda T, Sakata M, Kitamura YH, Shirahata A, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G, et al. Down-regulation of Tip60 gene as a potential marker for the malignancy of colorectal cancer. Anticancer Res. 2009;29:3953–3955. [PubMed] [Google Scholar]
- 52.Sakuraba K, Yokomizo K, Shirahata A, Goto T, Saito M, Ishibashi K, Kigawa G, Nemoto H, Hibi K. TIP60 as a potential marker for the malignancy of gastric cancer. Anticancer Res. 2011;31:77–79. [PubMed] [Google Scholar]
- 53.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 54.Kim JH, Kim B, Cai L, Choi HJ, Ohgi KA, Tran C, Chen C, Chung CH, Huber O, Rose DW, et al. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature. 2005;434:921–926. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- 55.Jha S, Vande Pol S, Banerjee NS, Dutta AB, Chow LT, Dutta A. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol Cell. 2010;38:700–711. doi: 10.1016/j.molcel.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown JA, Bourke E, Eriksson LA, Kerin MJ. Targeting cancer using KAT inhibitors to mimic lethal knockouts. Biochem Soc Trans. 2016;44:979–986. doi: 10.1042/BST20160081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashraf W, Bronner C, Zaayter L, Ahmad T, Richert L, Alhosin M, Ibrahim A, Hamiche A, Mely Y, Mousli M. Interaction of the epigenetic integrator UHRF1 with the MYST domain of TIP60 inside the cell. J Exp Clin Cancer Res. 2017;36:188. doi: 10.1186/s13046-017-0659-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang ZM, Rothbart SB, Allison DF, Cai Q, Harrison JS, Li L, Wang Y, Strahl BD, Wang G, Song J. An allosteric interaction links USP7 to deubiquitination and chromatin targeting of UHRF1. Cell Re. 2015;12:1400–1406. doi: 10.1016/j.celrep.2015.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng J, Yang H, Fang J, Ma L, Gong R, Wang P, Li Z, Xu Y. Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation. Nat Commun. 2015;6:7023. doi: 10.1038/ncomms8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clamme JP, Azoulay J, Mély Y. Monitoring of the formation and dissociation of polyethylenimine/DNA complexes by two photon fluorescence correlation spectroscopy. Biophys J. 2003;84:1960–1968. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.El Meshri SE, Dujardin D, Godet J, Richert L, Boudier C, Darlix JL, Didier P, Mély Y, de Rocquigny H. Role of the nucleocapsid domain in HIV-1 Gag oligomerization and trafficking to the plasma membrane: A fluorescence lifetime imaging microscopy investigation. J Mol Biol. 2015;427:1480–1494. doi: 10.1016/j.jmb.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 62.Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, Murty VV. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: Potential role in progression. Genes Chromosomes Cancer. 2008;47:755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Becker W, editor. Advanced time-correlated single photon counting applications. Springer; Heidelberg: 2015. [DOI] [Google Scholar]
- 65.Voss TC, Demarco IA, Day RN. Quantitaive imaging of protein interactions in the cell nucleus. Biotechniques. 2005;38:413–424. doi: 10.2144/05383RV01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma H, Chen H, Guo X, Wang Z, Sowa ME, Zheng L, Hu S, Zeng P, Guo R, Diao J, et al. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci USA. 2012;109:4828–4833. doi: 10.1073/pnas.1116349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alhosin M, Abusnina A, Achour M, Sharif T, Muller C, Peluso J, Chataigneau T, Lugnier C, Schini-Kerth VB, Bronner C, Fuhrmann G. Induction of apoptosis by thymoquinone in lymphoblastic leukemia Jurkat cells is mediated by a p73-dependent pathway which targets the epigenetic integrator UHRF1. Biochem Pharmacol. 2010;79:1251–1260. doi: 10.1016/j.bcp.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 68.Achour M, Mousli M, Alhosin M, Ibrahim A, Peluso J, Muller CD, Schini-Kerth VB, Hamiche A, Dhe-Peganon S, Bronner C. Epigallocatechin-3-gallate up-regulates tumor suppressor gene expression via a reactive oxygen species-dependent down-regulation of UHRF1. Biochem Biophys Res Commun. 2013;430:208–212. doi: 10.1016/j.bbrc.2012.11.087. [DOI] [PubMed] [Google Scholar]
- 69.León-González AJ, Jara-Palacios MJ, Abbas M, Heredia FJ, Schini-Kerth VB. Role of epigenetic regulation on the induction of apoptosis in Jurkat leukemia cells by white grape pomace rich in phenolic compounds. Food Nut. 2017;8:4062–4069. doi: 10.1039/c7fo00263g. [DOI] [PubMed] [Google Scholar]
- 70.Sharif T, Alhosin M, Auger C, Minker C, Kim JH, Etienne-Selloum N, Bories P, Gronemeyer H, Lobstein A, Bronner C, et al. Aronia melanocarpa juice induces a redox-sensitive p73-related caspase-3-dependent apoptosis in human leukemia cells. PLoS One. 2012;7:e32526. doi: 10.1371/journal.pone.0032526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim JK, Estève PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Polepalli S, George SM, Valli Sri Vidya R, Rodrigues GS, Ramachandra L, Chandrashekar R, DN M, Rao PPN, Pestell RG, Rao M. Role of UHRF1 in malignancy and its function as a therapeutic target for molecular docking towards the SRA domain. Int J Biochem Cell Biol. 2019;114:105558. doi: 10.1016/j.biocel.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 73.Boukhari A, Alhosin M, Bronner C, Sagini K, Truchot C, Sick E, Schini-Kerth VB, André P, Mély Y, Mousli M, Gies JP. CD47 activation-induced UHRF1 over-expression is associated with silencing of tumor suppressor gene p16INK4A in glioblastoma cells. Anticancer Res. 2015;35:149–157. [PubMed] [Google Scholar]
- 74.Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD, Bronner C. The retinoblastoma gene and its product are targeted by ICBP90: A key mechanism in the G1/S transition during the cell cycle. Oncogene. 2005;24:7337–7345. doi: 10.1038/sj.onc.1208878. [DOI] [PubMed] [Google Scholar]
- 75.Unoki M, Brunet J, Mousli M. Drug discovery targeting epigenetic codes: The great potential of UHRF1, which links DNA methylation and histone modifications, as a drug target in cancers and toxoplasmosis. Biochem Pharmacol. 2009;78:1279–1288. doi: 10.1016/j.bcp.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 76.Wang F, Yang YZ, Shi CZ, Zhang P, Moyer MP, Zhang HZ, Zou Y, Qin HL. UHRF1 promotes cell growth and metastasis through repression of p16(ink4a) in colorectal cancer. Ann Surg Oncol. 2012;19:2753–2762. doi: 10.1245/s10434-011-2194-1. [DOI] [PubMed] [Google Scholar]
- 77.Xue B, Zhao J, Feng P, Xing J, Wu H, Li Y. Epigenetic mechanism and target therapy of UHRF1 protein complex in malignancies. Onco Targets Ther. 2019;12:549–559. doi: 10.2147/OTT.S192234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subbaiah VK, Zhang Y, Rajagopalan D, Abdullah AN, Yeo-Teh NS, Tomaić V, Banks L, Myers MP, Chow EK, Jha S. E3 ligase EDD1/UBR5 is utilized by the HPV E6 oncogene to destabilize tumor suppressor TIP60. Oncogene. 2016;35:2062–2074. doi: 10.1038/onc.2015.268. [DOI] [PubMed] [Google Scholar]
- 79.Rajagopalan D, Pandey AK, Xiuzhen MC, Lee KK, Hora S, Zhang Y, Chua BH, Kwok HS, Bhatia SS, Deng LW, et al. TIP60 represses telomerase expression by inhibiting Sp1 binding to the TERT promoter. PLoS Pathog. 2017;13:e1006681. doi: 10.1371/journal.ppat.1006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Achour M, Jacq X, Rondé P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, et al. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008;27:2187–2197. doi: 10.1038/sj.onc.1210855. [DOI] [PubMed] [Google Scholar]
- 81.Gao Y, Wang Y, Zhou C, Kong S, Lu J, Wang H, Yang J. Ubiquitin-specific protease 7 (USP7) is essential for endometrial stromal cell decidualization in mice. Dev Growth Differ. 2019;61:176–185. doi: 10.1111/dgd.12594. [DOI] [PubMed] [Google Scholar]
- 82.Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20:1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 83.Felle M, Joppien S, Németh A, Diermeier S, Thalhammer V, Dobner T, Kremmer E, Kappler R, Längst G. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011;39:8355–8365. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mistry H, Gibson L, Yun JW, Sarras H, Tamblyn L, McPherson JP. Interplay between Np95 and Eme1 in the DNA damage response. Biochem Biophys Res Commun. 2008;375:321–325. doi: 10.1016/j.bbrc.2008.07.146. [DOI] [PubMed] [Google Scholar]
- 85.Chen H, Ma H, Inuzuka H, Diao J, Lan F, Shi YG, Wei W, Shi Y. DNA damage regulates UHRF1 stability via the SCF(β-TrCP) E3 ligase. Mol Cell Biol. 2013;33:1139–1148. doi: 10.1128/MCB.01191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.He J, Zhu Q, Wani G, Sharma N, Han C, Qian J, Pentz K, Wang QE, Wani AA. Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis. J Biol Chem. 2014;289:27278–27289. doi: 10.1074/jbc.M114.589812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.He M, Zhu Z, Shah AA, Zou H, Tao J, Chen Q, Wan Y. The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci. 2016;6:62. doi: 10.1186/s13578-016-0127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kwon SK, Saindane M, Baek KH. p53 stability is regulated by diverse deubiquitinating enzymes. Biochim Biophys Acta Rev Cancer. 2017;1868:404–411. doi: 10.1016/j.bbcan.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 89.Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, Frappier L. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- 90.Hou H, Sun D, Zhang X. The role of MDM2 amplification and overexpression in therapeutic resistance of malignant tumors. Cancer Cell Int. 2019;19:216. doi: 10.1186/s12935-019-0937-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nininahazwe L, Liu B, He C, Zhang H, Chen ZS. The emerging nature of ubiquitin-specific protease 7 (USP7): A new target in cancer therapy. Drug Discov Today. 2021;26:490–502. doi: 10.1016/j.drudis.2020.10.028. [DOI] [PubMed] [Google Scholar]
- 92.Bronner C. Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Sci Sign. 2011;4:pe3. doi: 10.1126/scisignal.2001764. [DOI] [PubMed] [Google Scholar]
- 93.Jang SY, Hong D, Jeong SY, Kim JH. Shikonin causes apoptosis by up-regulating p73 and down-regulating ICBP90 in human cancer cells. Biochem Biophys Res Commun. 2015;465:71–76. doi: 10.1016/j.bbrc.2015.07.131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.