Abstract
The present study aimed to explore the role of the long noncoding RNA cytoskeleton regulator (CYTOR) in non-small cell lung cancer (NSCLC) radiosensitivity by manipulating the microRNA (miR)-206/prothymosin α (PTMA) axis. First, 58 pairs of NSCLC and paracancerous tissues, normal human lung epithelial cells and NSCLC cells were collected to analyze CYTOR expression and the relation- ship between CYTOR and NSCLC prognosis. Subsequently, CYTOR expression in radioresistant cells was assessed. Radioresistant cells with low CYTOR expression and parental cells with high CYTOR expression were established. Functional assays were then performed to assess changes in cell radiosensitivity after irradiation treatment. Subsequently, the downstream mechanism of CYTOR was explored. The binding interactions between CYTOR and miR-206 and between miR-206 and PTMA were predicted and certified. Xenograft transplantation was applied to confirm the role of CYTOR in the radiosensitivity of NSCLC. CYTOR was overexpressed in NSCLC and was associated with poor prognosis. CYTOR was further upregulated in NSCLC cells with radioresistance. CYTOR knockdown enhanced the radiosensitivity of NSCLC cells, while overexpression of CYTOR led to the opposite result. Mechanistically, CYTOR specifically bound to miR-206 and silencing CYTOR promoted miR-206 to enhance the radiosensitivity of NSCLC cells. PTMA is a target of miR-206 and silencing CYTOR inhibited PTMA expression via miR-206, thus promoting radiosensitivity of NSCLC cells. CYTOR knockdown also enhanced NSCLC cell radiosensitivity in vivo. CYTOR was highly expressed in NSCLC, while silencing CYTOR potentiated NSCLC cell radiosensitivity by upregulating miR-206 and suppressing PTMA. The present study preliminarily revealed the role of CYTOR in radiotherapy sensitivity of NSCLC and provided a novel potential target for the clinical treatment of NSCLC.
Keywords: non-small cell lung cancer, radiotherapy, long noncoding RNA cytoskeleton regulator, miRNA-206, PTMA, competing endogenous RNA, subcellular localization
Introduction
Lung cancer (LC) is the most fatal type of malignancy world-wide and non-small cell LC (NSCLC) accounts for 85% of patients with LC (1). NSCLC may be a predominant cause of the increasing number of deaths resulting from tumors in all countries (2,3). Older individuals (age, >65 years) constitute the most susceptible group to NSCLC, with a rapidly increasing risk of morbidity and death (4). The known causes of NSCLC include excessive smoking, air contamination and exposure to radon (5). Supportive care, immunologic and biological therapies, chemotherapy and radiotherapy are all promising choices for NSCLC treatment (6). However, resistance derived from a long-term use of chemotherapy markedly hinders its efficiency (7). In addition, the clinical repercussions and survival rate of individuals with NSCLC have merely improved (8). To date, biomarkers have been utilized in the diagnosis, prognosis and attenuation of NSCLC (9). With this background and the requirement to identify reliable biomarkers of NSCLC and possible interventions to induce cell radiosensitivity in NSCLC, the present study was performed to determine the underlying mechanisms of NSCLC.
Long noncoding RNAs (lncRNAs) and microRNAs (miRNAs/miRs) are being widely studied to identify medical solutions targeting radioresistance involved in radiotherapies for NSCLC (10). lncRNAs are the gold standard for cancer diagnosis and prevention due to their wide range of carcinogenic or antitumoral effects on the occurrence and progression of tumors (11). Initially, the lncRNA cytoskeleton regulator (CYTOR) was documented to be sufficiently expressed in NSCLC and associated with cell biological activities and frustrating survival rates (12). In addition, CYTOR overexpression is associated with unsatisfactory clinical results and leads to aggravated cancer cellular mobility, aggressiveness and the epithelial-mesenchymal transition (EMT) of neoplasms, including breast cancer, gastric cancer and colon cancer (13,14). Of note, CYTOR is also responsible for the escalated relapse and resistance of various cancer types, including oral squamous cell carcinoma and breast cancer (15,16). Since CYTOR affects most malignancies by participating in competing endogenous RNA (ceRNA) interactions (17-19), the present study sought to investigate miRs that hold promise in alleviating NSCLC. Similarly, alterations in certain miRNAs significantly influence cellular growth, the EMT, metabolic pathways, apoptosis and radioresistance in NSCLC (20). According to certain previous studies, miR-206 is repeatedly acknowledged as a major cytokine in the ceRNA network involved in cancer progression (21,22). miR-206 is expressed at low levels in NSCLC, resulting in the active aggressiveness, invasiveness and dissemination of cells (23). However, Samaeekia et al (24) revealed that miR-206 suppresses breast tumor stemness and metastasis by inhibiting both self-renewal and invasion. Furthermore, miR-206 was demonstrated to reduce the resistance of LC toward effective drugs (25). When miRNAs are sponged in NSCLC during the treatment process, certain downstream mRNAs may also be regulated (26). Prothymosin α (PTMA) is activated in several cancer types (such as esophageal cancer and ovarian cancer), suggesting that it may represent a target (27,28). Based on these findings, the present study aimed to elucidate the effect of CYTOR on NSCLC radiosensitivity by manipulating the miR-206/PTMA axis.
Materials and methods
Expression analysis of CYTOR
The online tool Starbase (http://starbase.sysu.edu.cn/) (29) was employed for expression analysis of CYTOR on LUSC (n=501) and LUAD (n=526) data from TCGA.
Collection of clinical tissues
Between February 2013 and June 2015, 58 pairs of NSCLC and paracancerous tissue specimens (≥5 cm away from tumor tissue) were excised from patients with NSCLC during surgery at Yantaishan Hospital (Yantai, China); clinically diagnosed and histopathologically confirmed. None of these patients were treated by radiotherapy or chemotherapy prior to surgery. After the operation, each patient underwent radiotherapy and follow-up for at least 60 months and their survival was recorded. If the tumor recurred or the patient died, the follow-up ended; otherwise, the data were recorded up to the last follow-up. The patients who relapsed or died within 60 months were classified as the radiotherapy-resistant group (n=28), and the remaining patients were classified as the radiotherapy-sensitive group (n=30). The present study was approved by the Ethics Committee of Yantaishan Hospital (Yantai, China; accession no. for approval, YSLZ2021037). All study procedures conformed to the Declaration of Helsinki. Written informed consent was obtained from each patient. Supplementary Table SI provides the general information of the clinically included patients.
Cell culture and transfection
A normal human lung epithelial cell line (BEAS-2B) and NSCLC cell lines (H1650, H460 and A549; all from the China Infrastructure of Cell Line Resource) were cultured in RPMI-1640 basic medium containing 10% fetal bovine serum, 100 mg/ml streptomycin and 100 U/ml penicillin (all from Gibco; Thermo Fisher Scientific, Inc.) in an incubator with 5% CO2 at 37°C.
Short hairpin RNA targeting CYTOR (sh-CYTOR), miR-206 mimics, miR-206 inhibitor, mimics-negative control (NC), inhibitor-NC, overexpression vector for PTMA (pcDNA3.1-PTMA) and NC (empty pcDNA3.1 vector; Shanghai GenePharma Co., Ltd.) were transfected with Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol to generate stably transfected cells (30). The plasmids and sequence information are provided in Table SII.
Establishment of radioresistant cells
Radioresistant cells were established as previously described (31). H460 and A549 cells were cultured to 90% confluence and then irradiated at a dose of 0-8 Gy. After the X-ray irradiation, the medium was replaced with fresh medium and the cells were allowed to grow in a 37°C incubator. For the generation of radioresistant cells, the radiation dose applied to the cells (90% confluence) was gradually increased at a rate of 2.0 Gy/fraction until the final dose of 64 Gy was reached. The cells were subcultured 5 times at the same dose and the radiation dose was then increased. The produced radioresistant cell lines were named H460R and A549R and their viability after radiation was tested by the Cell Counting Kit-8 (CCK-8) method.
Reverse transcription-quantitative PCR (RT-qPCR)
According to a previous protocol (30), the total RNA was isolated from the tissues or cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.). Following the instructions of the PrimeScript RT kit (Takara Bio Inc.), the extracted RNA was reverse-transcribed into cDNA, which was subsequently amplified by real-time qPCR (Perfect Real Time; Takara Bio Inc.) to quantify the levels of CYTOR, miR-206 and PTMA. The PCR amplification procedure was set as two steps: The first step was pre-denaturation at 95°C for 10 min; the second step was PCR for 40 cycles with denaturation at 95°C for 10 sec and annealing/extension at 60°C for 30 sec, and each sample was set up in three wells. The expression of CYTOR and PTMA was standardized to that of 18S RNA and GAPDH, respectively, and that of miR-206 was standardized to that of U6. The relative expression of each cytokine was attained using the 2−ΔΔCq method (32). The primer sequences are listed in Table I.
Table I.
Primer sequences used for PCR.
| Gene | Primer sequence |
|---|---|
| 18S RNA | F: 5′-CGTTCTTAGTTGGTGGAGCG-3′ |
| R: 5′-CCGGACATCTAAGGGCATCA-3′ | |
| U6 | F: 5′-CGCTTCGGCAGCACATATAC-3′ |
| R: 5′-AATATGGAACGCTTCACGA-3′ | |
| GAPDH | F: 5′-GGGAGCCAAAAGGGTCAT-3′ |
| R: 5′-GAGTCCTTCCACGATACCAA-3′ | |
| miR-206 | F: 5′-GCTTCCCGAGGCCACATGCT-3′ |
| R: 5′-CACTTGCCGAAACCACACAC-3′ | |
| CYTOR | F: 5′-GCGGTGCCTGAGCCCGTGCC-3′ |
| R: 5′-GGGCGGTTGGAACCAGGCC-3′ | |
| PTMA | F: 5′-ATGTCAGACGCAGCCGTAG-3′ |
| R: 5′-CTAGTCATCCTCGTCGGTC-3′ |
F, forward; R, reverse; miR, microRNA; CYTOR, long non-coding RNA cytoskeleton regulator; PTMA, prothymosin α.
CCK-8 assay
Based on the instructions of the CCK-8 kit (cat. no. HY-K0301; MedChemExpress), differentially treated cells (2×104 cells/well) were seeded into 96-well plates and irradiated at 0, 2, 4, 6 or 8 Gy, followed by 24 h of culture at 37°C. Subsequently, the supernatant was discarded and 20 μl CCK-8 solution in 180 μl fresh medium was added to each well, followed by culture at 37°C for 1 h. The optical density of each well at 450 nm was attained using a spectrophotometer.
Colony formation assay
The assay was performed as previously described (31). The 5,000 cells in each group were cultured for 14 days after X-ray irradiation at 4 Gy. The colonies were subjected to 15 min of fixation with 4% paraformaldehyde and 10 min of staining with 1% crystal violet at room temperature. Colonies with >50 cells were scored and counted under the microscope (Olympus Corporation), followed by counting the number of colonies in five randomly selected areas. Each procedure was performed 3 times per group.
Comet assay
A comet assay was applied to differently treated cells according to a previous study (33). Specifically, the cell suspension (1×106 cells/ml) was mixed with low melting point agarose and dripped onto precoated slides (1% normal melting point agarose) incubated with lysis solution (pH 10.0) and electrophoresis buffer (pH >13.0) for 40 min and electrophoresed for 40 min (25 V, 280 mA) (33). Subsequently, the slides were placed in a neutralization solution (pH 7.5) for 10 min, covered with sufficient staining solution (20 μg/ml ethidium bromide) for 10 min and immersed twice in PBS (10 min each time) and distilled water. Subsequently, comets under a 510-560 nm excitation filter and a 590 nm blocking filter were observed and analyzed under a fluorescence microscope. All phases of the comet assay were performed under at 4°C in the dark and all solutions were freshly prepared and cooled for use.
γ-H2AX analysis
Cells with different treatments were cultured in 8-well slides for 24 h, fixed/permeabilized in cold (4°C) methanol for 5 min and blocked with 1% BSA (Thermo Fisher Scientific, Inc.) in PBS containing Tween-20 for 30 min at 37°C. Subsequently, the cells were incubated with γ-H2AX antibody (1:250 dilution; cat. no. ab81299; Abcam) overnight at 4°C, followed by immunoglobulin G (1:250 dilution; cat. no. ab205781; Abcam) for 1 h. The cells were stained with DAPI prior to mounting with coverslips. Finally, images were captured and analyzed under a fluorescence microscope.
Bioinformatics
The downstream target miRNAs of lncRNA CYTOR and the downstream target genes of miR-206 were predicted through the online websites Starbase (http://starbase.sysu.edu.cn/) (29), DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted) (34), miRcode (http://www.mircode.org/?gene=HOTAIR&mirfam=&class=&cons=&trregion=) (35), Targetscan(http://www.targetscan.org/vert_71/?tdsourcetag=s_pcqq_aiomsg) (36) and miRTarBase (http://mirtarbase.cuhk.edu.cn/php/index.php) (37).
RNA fluorescence in situ hybridization (FISH)
In order to detect the localization of CYTOR in cells, RNA FISH was performed using a green fluorescent-labeled CYTOR probe (Guangzhou RiboBio Co., Ltd.). The cells were fixed with 4% paraformaldehyde for 15 min, permeated with 0.5% Triton X-100 on ice for 10 min and then treated with prehybridization buffer at 37°C for 30 min. Next, the cells were hybridized with the fluorescent probe. After 12 h at 37°C, they were stained with DAPI. Finally, FISH results were obtained by confocal microscopy (Sp8; Leica Microsystems).
Dual-luciferase reporter gene assay
CYTOR or PTMA fragments containing the miR-206 binding site were cloned into the pmirGLO dual oligosaccharase vector (Promega Corporation) to construct the pmirGLO-CYTOR-wild-type (WT), pmirGLO-PTMA-WT, pmirGLO-CYTOR-mutant type (MUT) and pmirGLO-PTMA-MUT reporter vectors. After the cells (6×104 cells/well) were seeded into 24-well plates, pmirGLO-CYTOR-WT, pmirGLO-CYTOR-MUT, pmirGLO-PTMA-WT and pmirGLO-PTMA-MUT were cotransfected with miR-206 mimics or mimics-NC for 24 h. Subsequently, luciferase activity was assessed using dual-luciferase assay kits (Promega Corporation). The relative firefly luciferase activity was determined by normalisation to Renilla luciferase activity.
RNA pull-down assay
To clarify the binding relationship between CYTOR and miR-206 (38), biotinylated CYTOR and CYTOR-NC probes (Thermo Fisher Scientific, Inc.) were dissolved in washing/binding buffer according to the manufacturer's protocol and then cultivated with streptavidin-conjugated magnetic beads (Thermo Fisher Scientific, Inc.) for 2 h, followed by cultivation for 2 h with cell lysates supplemented to the buffer to remove RNA complexes conjugated to magnetic beads. Subsequently, miR-206 expression was determined by RT-qPCR.
Xenograft tumors in nude mice
After a week of adaptive feeding (20-22°C, 50-60% humidity, 12-h light/dark cycle; ad libitum access to food and water), 3×106 H460R + sh-NC or H460R + sh-CYTOR cells were subcutaneously inoculated into the right flank of BALB/c nude mice (n=24; age, 5 weeks; body weight, 18-20 g; Beijing Vital River Laboratory Animal Technology Co., Ltd.), with 12 mice per group. The tumor dimensions were measured with calipers every 3 days and the volume was calculated as follows: Tumor volume = (length × width2)/2. When the tumor volume reached 250-300 mm3, pentobarbital (50 mg/kg) was administered via intraperitoneal injection and the tumors of the mice were exposed to a single dose of 20 Gy ionizing radiation. For the tumor irradiation, the anesthetized mice were fixed on a plate with the right hind leg carrying the tumor exposed to the radiation field, while the other parts were protected by a lead plate. After the single irradiation, the tumor volume was examined every 3 days. The health and behavior of the mice were monitored every 2 days. The humane endpoints were as follows: Weight loss of >15% or the nude mice suffering from the tumor load or a tumor length of >1.50 cm and short diameter of >1.15 cm. Upon reaching the humane endpoints, the mice were euthanized by an intraperitoneal injection of an overdose of pentobarbital (>200 mg/kg). Death was confirmed by observation of pupil dilation as well as ceasing of breath (absence of chest fluctuation) and the heartbeat. Subsequently, tumors were removed and weighed, and the tumors from 6 mice per group were washed, and paraffin-embedded sections were prepared for the immunohistochemical analysis, while the remaining tumors were ground into homogenate for the RT-qPCR analysis. The animal experiments were performed in accordance with the requirements of the guidelines for the use of experimental animals (39), with the approval of the Institutional Animal Care and Use Committee at Yantaishan Hospital (Yantai, China). The accession number for this approval was YSLZ2021026. Significant efforts were made to minimize both the number of animals used and their respective suffering.
Immunohistochemistry
The tumor sections were deparaffinized, hydrated, incubated with anti-ki67 (1:200 dilution; cat. no. ab16667; Abcam) at 4°C and reacted with IgG (1:2,000 dilution; cat. no. ab205718; Abcam) at 37°C for 2 h. After washing with PBS, the sections were developed with diaminobenzidine, sealed and then observed and analyzed under a microscope.
Statistical analysis
SPSS 21.0 software (IBM Corporation) was used for data analysis. Values are expressed as the mean ± standard deviation. The normality of distribution of all data was inspected using the Kolmogorov-Smirnov test. The t-test was applied for comparisons between two groups, while one-way or two-way ANOVA was used to compare different groups, with Tukey's multiple-comparisons test applied for pairwise comparisons after the ANOVA. The P-value was attained using a two-tailed test and P<0.05 was considered to indicate a statistically significant difference.
Results
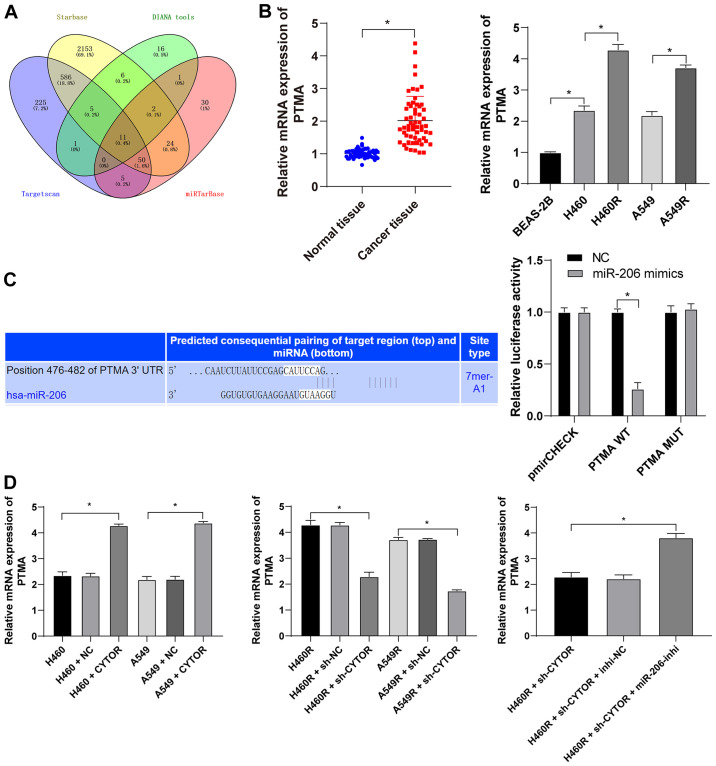
CYTOR is overexpressed in NSCLC and is responsible for the poor prognosis of patients with NSCLC
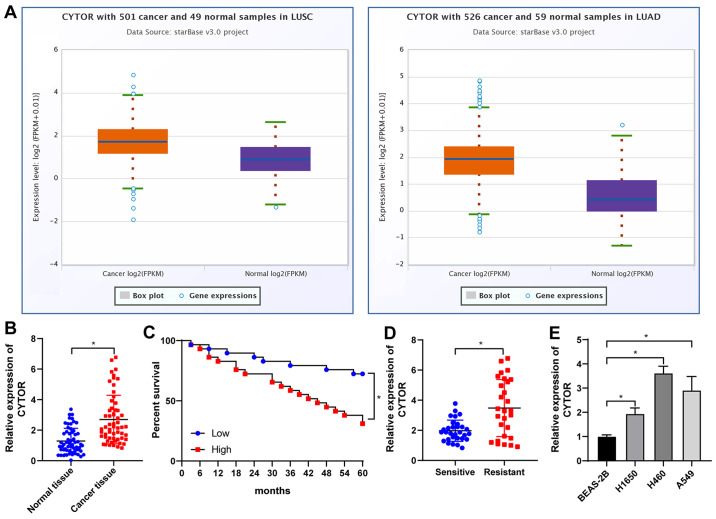
Analysis with the starBase database (http://starbase.sysu.edu.cn/) revealed that CYTOR was overexpressed in lung adenocarcinoma and lung squamous cell carcinoma (Fig. 1A), suggesting that CYTOR may be a possible target for NSCLC treatment. Analysis of the 58 pairs of NSCLC and paracancerous tissues (average age, 51.83±7.22 years; males/females, 29/29). The basic information of the cohort is provided in Table SII. It was revealed that CYTOR was upregulated in NSCLC tissues (P<0.05; Fig. 1B) and after radiotherapy, the patients with high CYTOR expression had a worse prognosis than those with low CYTOR expression (P<0.05; Fig. 1C). The patients were then divided into a sensitive group (n=30) and a resistant group (n=28) according to their sensitivity to radiotherapy and the expression of CYTOR in the two groups was analyzed. The results indicated that the expression level of CYTOR in the resistant group was significantly higher than that in the sensitive group (P<0.05; Fig. 1D). Next, the expression of CYTOR in different cell lines was detected and the results indicated that CYTOR expression in the NSCLC cell lines was higher than that in the non-cancerous cell line (P<0.05; Fig. 1E).
Figure 1.
CYTOR is overexpressed in NSCLC and high expression of CYTOR is associated with poor prognosis of patients with NSCLC. In total, 58 pairs of NSCLC and paracancerous tissues were harvested. (A) CYTOR expression analyzed by the starBase database (http://starbase.sysu.edu.cn/). (B) CYTOR expression in NSCLC tissues tested by RT-qPCR. (C) Survival curve analysis within a 60-month follow-up of 58 patients with patients divided into the CYTOR high- and low-expression group according to the median CYTOR expression level. (D) RT-qPCR was used to detect the expression of CYTOR according to radiosensitivity with patients stratified into sensitive (n=30) and resistant (n=28) groups. (E) CYTOR expression in normal and NSCLC cells. Values are expressed as the mean ± standard deviation. *P<0.05. NSCLC, non-small cell lung cancer; CYTOR, long noncoding RNA cytoskeleton regulator; RT-qPCR, reverse transcription-quantitative PCR; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Silencing of CYTOR enhances radiosensitivity of NSCLC cells
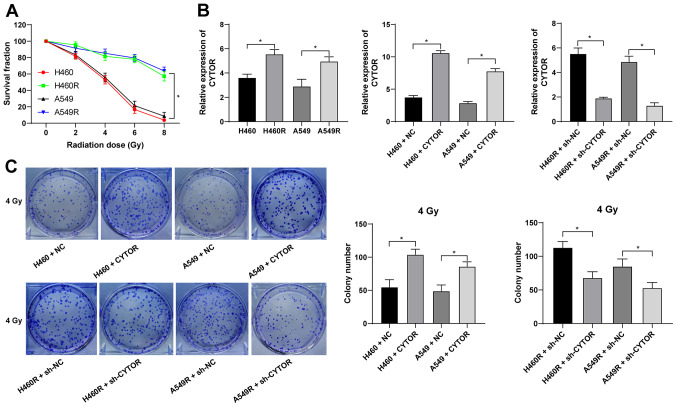
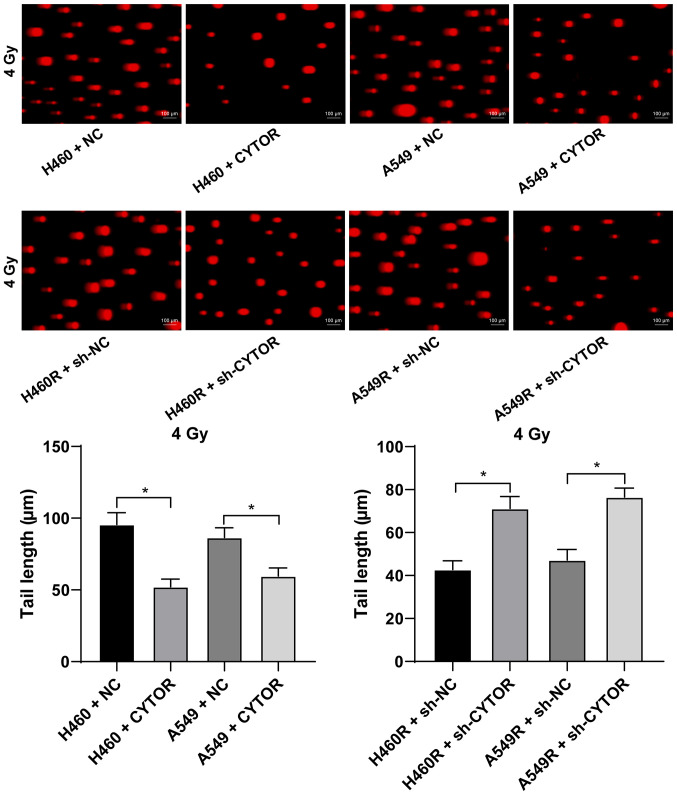
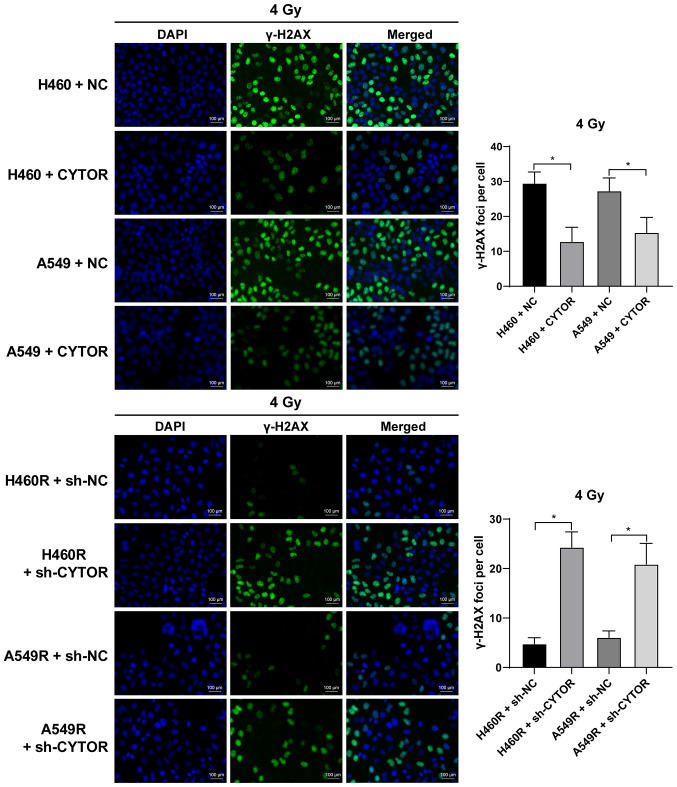
Radiotherapy is used as a prevalent therapy for NSCLC. To investigate the role of CYTOR in the radiosensitivity of NSCLC, radioresistant NSCLC cell lines (H460R and A549R) were established. Compared with the parental cell lines, the radioresistant cell lines had a better survival rate under different doses of irradiation (P<0.05; Fig. 2A). The dose of 4 Gy, under which the survival rate of the parental cells was ~50%, was selected for the subsequent experiments. According to the RT-qPCR results, CYTOR was highly expressed in the radioresistant cell lines (P<0.05; Fig. 2B). CYTOR overexpression vector was transfected into the parental cell lines (H460 and A549) and sh-CYTOR was transfected into the radioresistant cell lines; subsequently, the overexpression and knockdown efficiency was verified (P<0.05; Fig. 2B). Furthermore, it was revealed that overexpression of CYTOR in the parental cell lines impeded cell radiosensitivity (stronger colony formation ability, shorter comet assay tailing and lower γ-H2AX fluorescence intensity), while knockdown of CYTOR in the radioresistant cell lines enhanced cell radiosensitivity (weakened colony formation ability, longer comet assay tailing and increased γ-H2AX fluorescence intensity) (P<0.05; Figs. 2C, 3 and 4). These results suggest that CYTOR influences the radiosensitivity of NSCLC cells.
Figure 2.
Silencing of CYTOR enhances the radiosensitivity of non-small cell lung cancer cells. Radioresistant cell lines were established by gradually increasing the doses of irradiation. (A) Cell survival rate under different doses of irradiation as assessed through the Cell Counting Kit-8 method. (B) CYTOR expression in cells as detected by reverse transcription-quantitative PCR, with CYTOR transfected into the parental cells and sh-CYTOR transfected into the radioresistant cells to determine the overexpression or knockdown efficiency. (C) Colony formation ability of cells under 4 Gy as measured by a colony formation assay. Values are expressed as the mean ± standard deviation. *P<0.05. NC, negative control; CYTOR, long noncoding RNA cytoskeleton regulator; sh-CYTOR, short hairpin RNA targeting CYTOR; A549R, A549 with radioresistance.
Figure 3.
Effect of CYTOR on radiosensitivity of non-small cell lung cancer cells detected by the comet assay. DNA damage to cells under 4 Gy tested via a comet assay (longer tailing indicates more severe damage; scale bar, 100 μm). Values are expressed as the mean ± standard deviation.*P<0.05. Cell experiments were repeated three times. NC, negative control; CYTOR, long noncoding RNA cytoskeleton regulator; sh-CYTOR, short hairpin RNA targeting CYTOR; A549R, A549 with radioresistance.
Figure 4.
Effect of CYTOR on radiosensitivity of non-small cell lung cancer cells determined by detection of γ-H2AX. Cell radiation damage as detected by a γ-H2AX fluorescence assay (stronger fluorescence indicates more severe radiation damage; scale bar, 100 μm). Values are expressed as the mean ± standard deviation. *P<0.05. Cell experiments were repeated three times. CYTOR, long noncoding RNA cytoskeleton regulator; NC, negative control; sh-CYTOR, short hairpin RNA targeting CYTOR; A549R, A549 with radioresistance.
CYTOR targets miR-206 expression in NSCLC cells
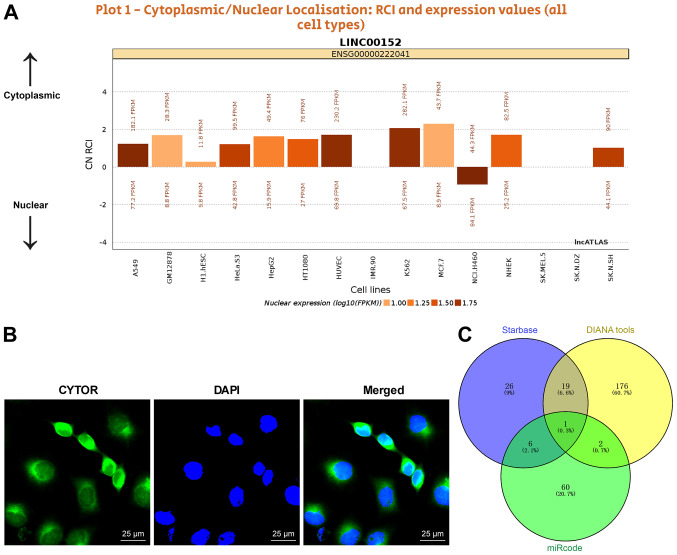
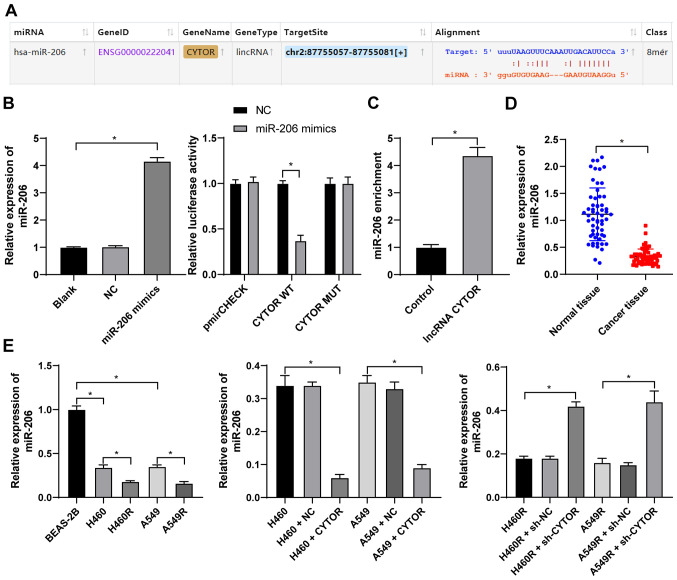
To clarify the specific mechanism of CYTOR to regulate the radiosensitivity of NSCLC cells, the subcellular localization of CYTOR in the cytoplasm was initially predicted by a database (http://lncatlas.crg.eu/?tdsourcetag=s_pcqq_aiomsg) and verified by FISH (Fig. 5A and B), indicating that CYTOR may affect the radiosensitivity of NSCLC cells via the ceRNA mechanism. Thus, multiple databases were employed to search for the downstream targets of CYTOR, and miR-206, the intersecting target, was identified (Fig. 5C). It has been reported that miR-206 improves the radiosensitivity of nasopharyngeal carcinoma (40). Therefore, it may be speculated that CYTOR affects NSCLC cell radiosensitivity by targeting miR-206. The StarBase database (http://starbase.sysu.edu.cn/) predicted the binding site between CYTOR and miR-206 (Fig. 6A), and subsequently, the binding association between CYTOR and miR-206 was confirmed by a dual-luciferase reporter gene assay and an RNA pull-down assay (Fig. 6B and C). Next, compared with the normal adjacent tissues and normal cell line, respectively, miR-206 expression was determined to be decreased in both NSCLC tissues and cells, and it was even further decreased in the radioresistant cells (P<0.05; Fig. 6D and E). Furthermore, miR-206 expression was reduced when CYTOR was overexpressed, while it was promoted when CYTOR was inhibited (P<0.05; Fig. 6E). In summary, the results suggested that CYTOR directly targets miR-206 in NSCLC cells.
Figure 5.
CYTOR targets miR-206 expression in non-small cell lung cancer cells. (A) Subcellular localization of CYTOR predicted by a database (http://lncatlas.crg.eu/?tdsourcetag=s_pcqq_aiomsg). (B) CYTOR was located in the cytoplasm as observed by fluorescence in situ hybridization (scale bar, 25 μm). Cell experiments were performed as three repeats. (C) miRs downstream of CYTOR according to various databases compared in a Venn diagram, with blue representing Starbase (http://starbase.sysu.edu.cn/), yellow representing DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted) and green representing miRcode (http://www.mircode.org/?gene=HOTAIR&mirfam=&class=&cons=&trregion=). CYTOR, long noncoding RNA cytoskeleton regulator; miR, microRNA; RCI, relative concentration index; C, cytoplasmic; N, nuclear.
Figure 6.
CYTOR targets miR-206. (A) The binding site between CYTOR and miR-206 predicted via starBase (http://starbase.sysu.edu.cn/). (B) The transfection of miR-206 mimics was confirmed by RT-qPCR and the binding association between CYTOR and miR-206 was verified by a dual-luciferase reporter assay. (C) The binding relationship between CYTOR and miR-206 in H460 cells was confirmed by an RNA pull-down assay. (D and E) miR-206 expression in (D) patients with NSCLC and (E) NSCLC cell lines as detected by RT-qPCR. Values are expressed as the mean ± standard deviation. Cell experiments were performed as three repeats. *P<0.05. RT-qPCR, reverse transcription-quantitative PCR; NSCLC, non-small cell lung cancer; CYTOR, long noncoding RNA cytoskeleton regulator; NC, negative control; CYTOR, long noncoding RNA cytoskeleton regulator; sh-CYTOR, short hairpin RNA targeting CYTOR; A549R, A549 with radioresistance; miR/miRNA, microRNA; lincRNA, long intergenic non-coding RNA; WT, wild-type; MUT, mutant; hsa, Homo sapiens.
miR-206 knockdown neutralizes the inhibitory effect of CYTOR on radiosensitivity of NSCLC cells
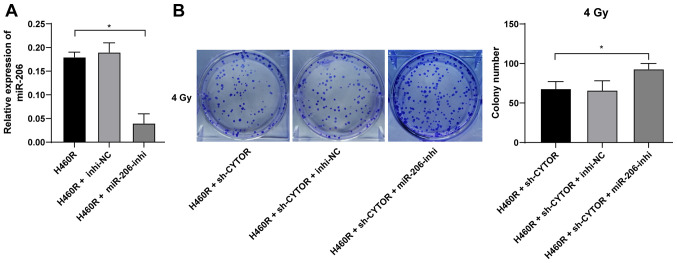
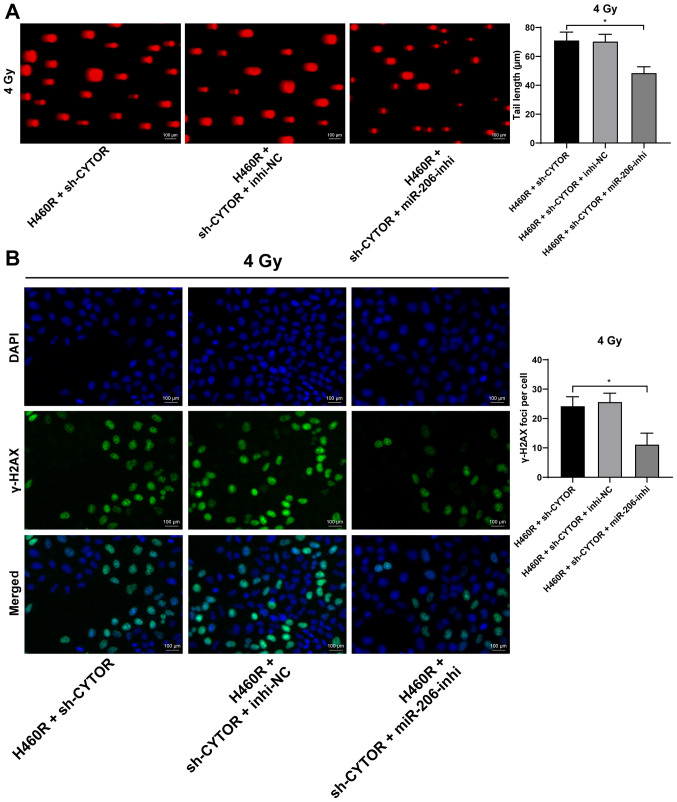
To further validate the role of miR-206 in the CYTOR-regulated radiosensitivity of NSCLC cells, a miR-206 inhibitor was utilized to degrade miR-206 expression in H460R cells (P<0.01; Fig. 7A); subsequently, miR-206 inhibitor was combined with sh-CYTOR for joint application. The results suggested that the radiosensitivity of the H460R cells induced by sh-CYTOR was moderately inhibited (P<0.01; Figs. 7B and 8A and B), indicating that CYTOR suppressed the radiosensitivity of NSCLC cells by targeting miR-206.
Figure 7.
miR-206 regulates the clonal ability of non-small cell lung cancer cells. miR-206 inhibitor was transfected into H460R cells, with inhibitor-NC as a control. (A) Knockdown efficiency was examined by reverse transcription-quantitative PCR. miR-206 inhibitor was combined with sh-CYTOR for the joint experiments. (B) Colony formation ability of cells under 4 Gy as measured by a colony formation assay. Values are expressed as the mean ± standard deviation. Cell experiments were performed as three repeats. *P<0.05. NSCLC, non-small cell lung cancer; CYTOR, long noncoding RNA cytoskeleton regulator; sh-CYTOR, short hairpin RNA targeting CYTOR; NC, negative control; inhi, inhibitor; miR, microRNA; H460R, H460 cells with radioresistance.
Figure 8.
miR-206 regulates radiation damage of non-small cell lung cancer cells after radiotherapy. miR-206 inhibitor and sh-CYTOR were transfected into H460R cells for the joint experiments. (A) DNA damage to cells under 4 Gy as tested via a comet assay (longer tailing indicates more severe damage; scale bar, 100 μm). (B) Cell radiation damage detected by a γ-H2AX fluorescence assay (stronger fluorescence indicates more severe radiation damage; scale bar, 100 μm). Values are expressed as the mean ± standard deviation. Cell experiments were performed as three repeats. *P<0.05. CYTOR, long noncoding RNA cytoskeleton regulator; sh-CYTOR, short hairpin RNA targeting CYTOR; NC, negative control; inhi, inhibitor; miR, microRNA; H460R, H460 cells with radioresistance.
miR-206 targets PTMA expression
Subsequently, the intersection among the target genes downstream of miR-206 predicted from multiple databases was attained (Fig. 9A) and PTMA was identified. PTMA was reported to be highly expressed in a radiation-resistant colorectal cancer cell line (41). PTMA expression in the NSCLC tissues and cell lines was measured and it was indicated that the level of PTMA mRNA was elevated in both the NSCLC tissues and radioresistant cell lines (P<0.05; Fig. 9B). The target binding relationship between miR-206 and PTMA was verified by a dual-luciferase reporter gene assay (P<0.05; Fig. 9C). In addition, increased intracellular CYTOR expression led to upregulation of PTMA mRNA and knockdown of CYTOR expression led to the opposite result; of note, simultaneous CYTOR knockdown and inhibition of miR-206 promoted PTMA mRNA expression (P<0.05; Fig. 9D), indicating that miR-206 targeted PTMA in NSCLC cells and that CYTOR competitively bound to miR-206 to upregulate PTMA expression.
Figure 9.
miR-206 targets PTMA expression. (A) Target genes downstream of miR-206 were analyzed using various databases and their overlapping results were presented in a Venn diagram, with yellow representing starBase (http://starbase.sysu.edu.cn/), green representing DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted), blue representing TargetScan (http://www.targetscan.org/vert_71/?tdsourcetag=s_pcqq_aiomsg) and red representing miRTarBase (http://mirtarbase.cuhk.edu.cn/php/index.php). (B) Level of PTMA mRNA in 58 pairs of non-small cell lung cancer tissues and cell lines as detected by RT-qPCR. (C) The binding site between miR-206 and PTMA was predicted via starBase (http://starbase.sysu.edu.cn/) and their binding interaction was verified by a dual-luciferase reporter assay. (D) The level of PTMA mRNA in differently treated cells as verified by RT-qPCR. Values are expressed as the mean ± standard deviation. Cell experiments were performed as three repeats. *P<0.05. RT-qPCR, reverse transcription-quantitative PCR; miR, microRNA; PTMA, prothymosin α; MUT, mutant; WT, wild-type; hsa, Homo sapiens; CYTOR, long noncoding RNA cytoskeleton regulator; sh-CYTOR, short hairpin RNA targeting CYTOR; NC, negative control; inhi, inhibitor; miR, microRNA; H460R, H460 cells with radioresistance.
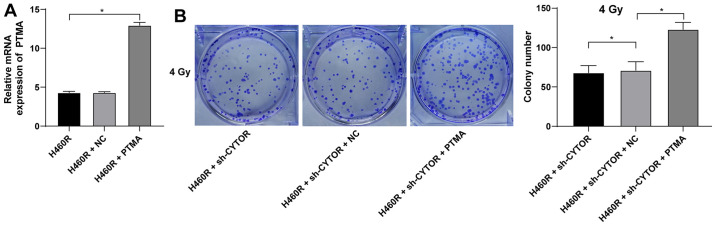
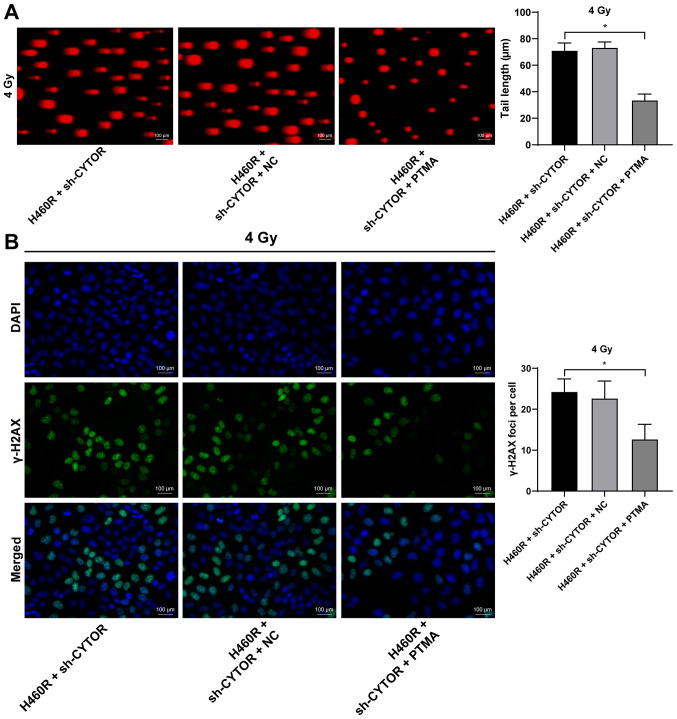
PTMA overexpression debilitates the inhibitory effect of CYTOR depletion on NSCLC cell radiosensitivity
To investigate the role of PTMA in the CYTOR-regulated radiosensitivity of NSCLC cells, pcDNA3.1-PTMA was constructed and transfected into H460R cells to upregulate PTMA expression (P<0.05; Fig. 10A). It was indicated that overexpression of PTMA partially reversed the reduced radioresistance of the sh-CYTOR-treated H460R cells as demonstrated by the enhanced clonogenic ability of the cells (Fig. 10B), shorter comet assay tailing (Fig. 11A) and diminished γ-H2AX immunofluorescence (Fig. 11B). These results suggested that overexpression of PTMA retarded the inhibitory role of sh-CYTOR on NSCLC cell radiosensitivity.
Figure 10.
PTMA overexpression enhances the clonal ability of non-small cell lung cancer cells. pcDNA3.1-PTMA was constructed and transfected into H460R cells, with pcDNA3.1 empty vector serving as a control (NC). (A) Transfection efficiency examined by reverse transcription-quantitative PCR. pcDNA3.1-PTMA was combined with sh-CYTOR for the joint experiments. (B) Colony formation ability of cells treated with pcDNA3.1-PTMA at 4 Gy as measured by a colony formation assay. Values are expressed as the mean ± standard deviation. Cell experiments were performed as three repeats. *P<0.05. CYTOR, long noncoding RNA cytoskeleton regulator; NC, negative control; PTMA, prothymosin α; sh-CYTOR, short hairpin RNA targeting CYTOR; H460R, H460 cells with radioresistance.
Figure 11.
Overexpression of PTMA attenuates radiation injury of non-small cell lung cancer cells after radiotherapy. pcDNA3.1-PTMA was combined with sh-CYTOR for the joint experiments. (A) DNA damage to cells treated with pcDNA3.1-PTMA under 4 Gy tested via a comet assay (longer tailing indicates more severe damage; scale bar, 100 μm). (B) Radiation damage in cells treated with pcDNA3.1-PTMA as detected by a γ-H2AX fluorescence assay (stronger fluorescence indicates more severe radiation damage; scale bar, 100 μm). Values are expressed as the mean ± standard deviation. Cell experiments were performed as three repeats. *P<0.05. CYTOR, long noncoding RNA cytoskeleton regulator; sh-CYTOR, short hairpin RNA targeting CYTOR; NC, negative control; PTMA, prothymosin α; H460R, H460 cells with radioresistance.
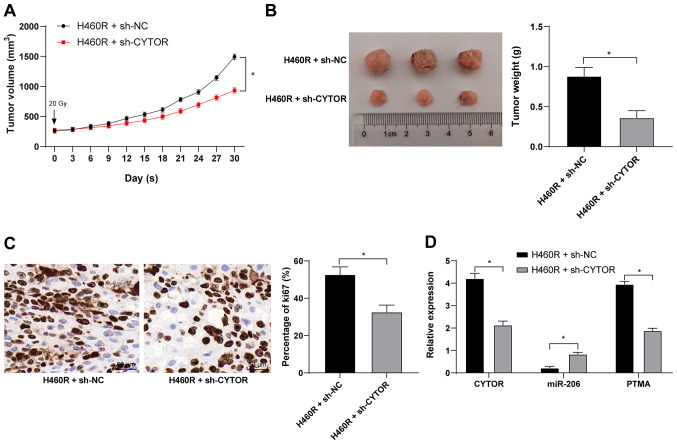
CYTOR knockdown in vivo enhances the radiosensitivity of xenograft tumors in mice
Xenograft tumors were established in mice by subcutaneous injection of differently treated H460R cells. After the tumors were treated by irradiation, compared with the H460R + sh-NC group, the H460R + sh-CYTOR group exhibited a reduced tumor weight and size and ki67-positive rate (P<0.05; Fig. 12A-C), decreased expression of CYTOR and PTMA in the tumors and enhanced miR-206 expression (P<0.05; Fig. 12D). The above results indicated that CYTOR enhances NSCLC radioresistance by competitively binding miR-206 to upregulate PTMA at the posttranscriptional level in vivo.
Figure 12.
CYTOR knockdown in vivo enhances the radiosensitivity of xenograft tumors in mice. H460R + sh-NC and H460R + sh-CYTOR were subcutaneously injected into mice and one dose of ionizing radiation (20 Gy) was administered to the mice once the tumors grew to 250-300 mm3. (A) Tumor volume was measured every three days and the tumors were removed after 30 days. (B) Representative image of the tumor specimens and bar graph displaying tumor weight. (C) Ki67-positive rate as detected by immunohistochemistry (scale bar, 50 μm). (D) Expression of CYTOR, miR-206 and PTMA in tumors as verified by reverse transcription-quantitative PCR. Values are expressed as the mean ± standard deviation. *P<0.05. CYTOR, long noncoding RNA cytoskeleton regulator; sh-CYTOR, short hairpin RNA targeting CYTOR; NC, negative control; PTMA, prothymosin α; miR, microRNA; H460R, H460 cells with radioresistance.
Discussion
Although significant advances have been achieved in the field of medicine targeting LC, the prognostic repercussions and survival rate of NSCLC remain disappointing and mortality is high, partially due to escalated chemoresistance (42). lncRNAs may serve as tumor promotors or inhibitors by modulating the growth and development of a variety of cancer types, such as NSCLC (43). Although the specific regulatory mechanisms of CYTOR in NSCLC have yet to be clarified, studies have indicated that CYTOR is activated in multiple malignancies, including gastric cancer, renal cell carcinoma and hepatocellular carcinoma, and predicts metastatic tumors and unfavorable prognosis (44). Therefore, CYTOR is an oncogenic factor and marker of neoplasms. Hence, the present study attempted to uncover the role of CYTOR in NSCLC, including the molecular signaling involved in the relevant axis.
The most important finding of the present study was that CYTOR was overexpressed in NSCLC and high expression of CYTOR was associated with poor prognosis of patients with NSCLC. Early studies discovered that CYTOR functions as a promoter of various neoplasms, including LC, kidney cancer, gastric cancer, gallbladder cancer and colon cancer, as it expedites cellular metastasis, invasiveness and motility (45). CYTOR acts as a driving factor in a considerable number of malignancies by regulating cell biological behaviors, facilitating lymph node metastasis and incurring relapse, consequently being associated with poor prognosis (46,47). Of note, proactively expressed CYTOR was commonly observed in tumors that received no treatment, with large tumor volume and advanced stage NSCLC (48). However, CYTOR is a well-acknowledged therapeutic target and biomarker in NSCLC and CYTOR knockdown causes cell inactivation and even death (49). Furthermore, silencing of CYTOR enhanced the radiosensitivity of NSCLC cells (50). In a previous study, high CYTOR expression was inextricably linked to limited apoptosis and enhanced drug resistance (13). In addition, CYTOR knockout was able to restore chemosensitivity and enhance the apoptotic rate in epithelial ovarian cancer (51). In summary, silencing CYTOR may represent a strategy for treating NSCLC from the perspective of strengthening cell sensitivity to drugs or radiotherapy.
Of note, CYTOR targeted miR-206 expression in NSCLC cells. CYTOR is involved in ceRNA mechanisms and sponges miR-497 in thyroid tumors, which increases tumor progression and metastasis, while CYTOR depletion, in turn, restrains cancer cell growth, viability and aggressiveness (52). CYTOR functions as an upstream gene to sponge miR-193a/b-3p, augmenting hepatocellular carcinoma (53). In a relevant study, CYTOR was determined to be potent in sponging miR-195 in NSCLC, therefore enhancing malignancy and impaired radiosensitivity of NSCLC (50), suggesting that CYTOR mediates NSCLC development by acting as a sponge in ceRNA interactions. Similarly, miR-206, a crucial cytokine connecting homeobox transcript antisense intergenic RNA and cyclin D1 in the ceRNA network, was reduced in ovarian cancer and catalyzed cell survival, motility and proliferation (54). In addition, when miR-206 was competitively bound by metastasis-associated lung adenocarcinoma transcription 1, cyclin-dependent kinase 9, a direct target of miR-206, was activated to stimulate osteosarcoma augmentation (55), strengthening the feasibility of proceeding with research concerning miR-206 in NSCLC. Subsequently, it was uncovered that miR-206 knockdown neutralized the inhibitory effect of CYTOR on NSCLC cell radiosensitivity. miR-206 contributes to quenching various types of carcinoma, such as thyroid cancer, prostate cancer and ovarian cancer, by restricting cellular pathogenesis, viability and motility (56-58). miR-206 was also beneficial in preventing drug resistance and inducing necrocytosis in papillary thyroid carcinoma (59). In addition, miR-206 inhibited aerobic glycolysis to ameliorate NSCLC (60). Subsequently, the present study indicated that, as a target of miR-206, PTMA overexpression debilitated the inhibitory role of CYTOR depletion in NSCLC cell radiosensitivity, as evidenced by the improved Ki67-positive rate. As a downstream factor in the ceRNA network, the upregulation of PTMA caused the outgrowth of exacerbated colorectal cancer (61). Furthermore, PTMA exhibited a negative association with the radiosensitivity of colorectal cancer (41). In addition, Ki67 is a dependable indicator of cancer cellular duplication and migration (62). Studies have suggested that in diverse cancers, Ki67 activation usually concurs with upregulated CYTOR, low expression of miR-206 or promoted PTMA (63-65), which is consistent with the results of the present study. Based on the above, the CYTOR/miR-206/PTMA axis may be valuable in NSCLC research.
In conclusion, the present study illustrated that silencing CYTOR potentiated NSCLC cell radiosensitivity by upregulating miR-206 and suppressing PTMA. This suggests therapeutic implications for NSCLC alleviation. Experimental revelations and realistic applications in medical practice require extensive validation. It is esteemed that the present results contribute to NSCLC research.
There are certain limitations to the present study. First, since the role of CYTOR in the radiosensitivity of NSCLC cells was discussed, patients who did not receive any radiotherapy were not included. This is also a limitation of the present study. This point will also be considered for investigation in the future. Furthermore, due to limitations regarding the sample volume, other possible miRs or mRNAs downstream of CYTOR that may affect NSCLC radiosensitivity were not fully investigated. Future research by our group will aim to further identify the possible downstream genes or axes of PTMA and in-depth experiments targeting other miRs related to CYTOR will be designed. Finally, the present study mainly focused on the effect of CYTOR on the radiosensitivity of NSCLC and its downstream mechanism. Therefore, the present results were all obtained with the background of radiation and the role of CYPOR in common NSCLC cell lines was not explored. Certain studies have reported the effect of CYPOR on NSCLC. One previous study investigated the mechanism of CYTOR in the migration and invasion of NSCLC cells and revealed that CYTOR promoted cell proliferation, migration and invasion ability, and induced radioresistance in NSCLC cells (50). Another study assessed the role of CYTOR in NSCLC, including the cell cycle and apoptosis and found that CYTOR knockdown also promoted cell apoptosis and induced cell cycle arrest in G1 phase (12). Our group aims to further explore the role of the CYPOR/miR-206/PTMA axis in the malignant behavior of NSCLC cells and cancer metastasis in the future.
Supplementary Data
Acknowledgments
Not applicable.
Funding Statement
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GJ and FZ conceived and designed the present study. GJ, HY, ZL and FZ performed the experiments. GJ, HY, ZL and FZ analyzed and interpreted the data. GJ, HY and FZ wrote, reviewed and/or revised the manuscript. GJ and FZ confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Yantaishan Hospital (Yantai, China; accession no. YSLZ2021037). Informed consent was obtained from all subjects. The animal experiments were carried out in accordance with the requirements of the guidelines for the use of experimental animals (38), with the approval of the Institutional Animal Care and Use Committee at Yantaishan Hospital (Yantai, China; accession no. YSLZ2021026). Significant efforts were made to minimize both the number of animals used and their respective suffering.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tandberg DJ, Tong BC, Ackerson BG, Kelsey CR. Surgery versus stereotactic body radiation therapy for stage I non-small cell lung cancer: A comprehensive review. Cancer. 2018;124:667–678. doi: 10.1002/cncr.31196. [DOI] [PubMed] [Google Scholar]
- 3.Rafei H, El-Bahesh E, Finianos A, Nassereddine S, Tabbara I. Immune-based therapies for non-small cell lung cancer. Anticancer Res. 2017;37:377–387. doi: 10.21873/anticanres.11330. [DOI] [PubMed] [Google Scholar]
- 4.Elias R, Morales J, Presley C. Checkpoint inhibitors for non-small cell lung cancer among older adults. Curr Oncol Rep. 2017;19:62. doi: 10.1007/s11912-017-0619-0. [DOI] [PubMed] [Google Scholar]
- 5.Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, Petrella F, Spaggiari L, Rosell R. Non-small-cell lung cancer. Nat Rev Dis Primers. 2015 May 21; doi: 10.1038/nrdp.2015.9. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Skřičková J, Kadlec B, Venclíček O, Merta Z. Lung cancer. Cas Lek Cesk. 2018;157:226–236. [PubMed] [Google Scholar]
- 7.Liang J, Lu T, Chen Z, Zhan C, Wang Q. Mechanisms of resistance to pemetrexed in non-small cell lung cancer. Transl Lung Cancer Res. 2019;8:1107–1118. doi: 10.21037/tlcr.2019.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabchi S, Kassouf E, Rassy EE, Kourie HR, Martin J, Campeau MP, Tehfe M, Blais N. Management of stage III non-small cell lung cancer. Semin Oncol. 2017;44:163–177. doi: 10.1053/j.seminoncol.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Pennell NA, Arcila ME, Gandara DR, West H. Biomarker testing for patients with advanced non-small cell lung cancer: Real-world issues and tough choices. Am Soc Clin Oncol Educ Book. 2019;39:531–542. doi: 10.1200/EDBK_237863. [DOI] [PubMed] [Google Scholar]
- 10.Huang Q. Predictive relevance of ncRNAs in non-small-cell lung cancer patients with radiotherapy: A review of the published data. Biomarkers Med. 2018;12:1149–1159. doi: 10.2217/bmm-2018-0004. [DOI] [PubMed] [Google Scholar]
- 11.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A New Paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H, Li SB. Role of LINC00152 in non-small cell lung cancer. J Zhejiang Univ Sci B. 2020;21:179–191. doi: 10.1631/jzus.B1900312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yue B, Liu C, Sun H, Liu M, Song C, Cui R, Qiu S, Zhong M. A positive feed-forward loop between LncRNA-CYTOR and Wnt/β-catenin signaling promotes metastasis of colon cancer. Mol Ther. 2018;26:1287–1298. doi: 10.1016/j.ymthe.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moradi MT, Hatami R, Rahimi Z. Circulating CYTOR as a potential biomarker in breast cancer. Int J Mol Cell Med. 2020;9:83–90. doi: 10.22088/IJMCM.BUMS.9.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Li M, Yu H, Piao H. lncRNA CYTOR promotes tamoxifen resistance in breast cancer cells via sponging miR 125a 5p. Int J Mol Med. 2020;45:497–509. doi: 10.3892/ijmm.2019.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Yang M, Wang C, Ouyang Y, Chen X, Bai J, Hu Y, Song M, Zhang S, Zhang Q. Forkhead box D1 promotes EMT and chemoresistance by upregulating lncRNA CYTOR in oral squamous cell carcinoma. Cancer Lett. 2021;503:43–53. doi: 10.1016/j.canlet.2020.11.046. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Shan Y, Ge K, Lu J, Kong W, Jia C. LncRNA CYTOR promotes pancreatic cancer cell proliferation and migration by sponging miR-205-5p. Pancreatology. 2020;20:1139–1148. doi: 10.1016/j.pan.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Li M, Wang Q, Xue F, Wu Y. lncRNA-CYTOR works as an Oncogene through the CYTOR/miR-3679-5p/MACC1 axis in colorectal cancer. DNA Cell Biol. 2019;38:572–582. doi: 10.1089/dna.2018.4548. [DOI] [PubMed] [Google Scholar]
- 19.Hu B, Yang XB, Yang X, Sang XT. LncRNA CYTOR affects the proliferation, cell cycle and apoptosis of hepatocellular carcinoma cells by regulating the miR-125b-5p/KIAA1522 axis. Aging (Albany NY) 2020;13:2626–2639. doi: 10.18632/aging.202306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrek H, Yu AM. MicroRNAs in non-small cell lung cancer: Gene regulation, impact on cancer cellular processes, and therapeutic potential. Pharmacol Res Perspect. 2019;7:e00528. doi: 10.1002/prp2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shengnan J, Dafei X, Hua J, Sunfu F, Xiaowei W, Liang X. Long non-coding RNA HOTAIR as a competitive endogenous RNA to sponge miR-206 to promote colorectal cancer progression by activating CCL2. J Cancer. 2020;11:4431–4441. doi: 10.7150/jca.42308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao HL, Liu ZJ, Huang PL, Yue YL, Xi JN. lncRNA-RMRP promotes proliferation, migration and invasion of bladder cancer via miR-206. Eur Rev Med Pharmacol Sci. 2019;23:1012–1021. doi: 10.26355/eurrev_201902_16988. [DOI] [PubMed] [Google Scholar]
- 23.Liao M, Peng L. MiR-206 may suppress non-small lung cancer metastasis by targeting CORO1C. Cell Mol Biol Lett. 2020;25:22. doi: 10.1186/s11658-020-00216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samaeekia R, Adorno-Cruz V, Bockhorn J, Chang YF, Huang S, Prat A, Ha N, Kibria G, Huo D, Zheng H, et al. miR-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway. Clin Cancer Res. 2017;23:1091–1103. doi: 10.1158/1078-0432.CCR-16-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao D, Chen J, Li Y, Tang X, Wang J, Xu W, Song J, Li Y, Tao H, Chen Q. miR-1-3p and miR-206 sensitizes HGF-induced gefitinib-resistant human lung cancer cells through inhibition of c-Met signalling and EMT. J Cell Mol Med. 2018;22:3526–3536. doi: 10.1111/jcmm.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weidle UH, Birzele F, Nopora A. MicroRNAs as potential targets for therapeutic intervention with metastasis of non-small cell lung cancer. Cancer Genomics Proteomics. 2019;16:99–119. doi: 10.21873/cgp.20116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y, Qi X, Yu C, Yu S, Zhang C, Zhang Y, Liu X, Xu Y, Yang C, Jiang W, et al. Identification of prothymosin alpha (PTMA) as a biomarker for esophageal squamous cell carcinoma (ESCC) by label-free quantitative proteomics and Quantitative Dot Blot (QDB) Clin Proteomics. 2019;16:12. doi: 10.1186/s12014-019-9232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandow JJ, Rainczuk A, Infusini G, Makanji M, Bilandzic M, Wilson AL, Fairweather N, Stanton PG, Garama D, Gough D, et al. Discovery and validation of novel protein biomarkers in ovarian cancer patient urine. Proteomics Clin Appl. 2018;12:e1700135. doi: 10.1002/prca.201700135. [DOI] [PubMed] [Google Scholar]
- 29.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan F, Zhao W, Xu X, Li C, Li X, Liu S, Shi L, Wu Y. LncRNA DHRS4-AS1 inhibits the stemness of NSCLC cells by sponging miR-224-3p and upregulating TP53 and TET1. Front Cell Dev Biol 2020. 2020;8:585251. doi: 10.3389/fcell.2020.585251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Qu Z, Wang Y, Qin M, Zhang H. miR-101-3p sensitizes non-small cell lung cancer cells to irradiation. Open Med (Wars) 2020;15:413–423. doi: 10.1515/med-2020-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Zamani M, Etebari M, Moradi SH. The increment of genoprotective effect of melatonin due to 'Autooptic' effect versus the genotoxicity of mitoxantrone. J Biomed Phys Eng. 2020;10:771–782. doi: 10.31661/jbpe.v0i0.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P, Floros E, Dalamagas T, et al. DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44:D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeggari A, Marks DS, Larsson E. miRcode: A map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:2015. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang HY, Lin YC, Li J, Huang KY, Shrestha S, Hong HC, Tang Y, Chen YG, Jin CN, Yu Y, et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48:D148–D154. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, Liu Z, Zeng Y, Li C, Zhao J, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer. 2018;17:140. doi: 10.1186/s12943-018-0889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones-Bolin S. Guidelines for the care and use of laboratory animals in biomedical research. Curr Protoc Pharmacol 2012 Appendix. 2012;4:4B. doi: 10.1002/0471141755.pha04bs59. [DOI] [PubMed] [Google Scholar]
- 40.Wang T, Dong XM, Zhang FL, Zhang JR. miR-206 enhances nasopharyngeal carcinoma radiosensitivity by targeting IGF1. Kaohsiung J Med Sci. 2017;33:427–432. doi: 10.1016/j.kjms.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 41.Ojima E, Inoue Y, Miki C, Mori M, Kusunoki M. Effectiveness of gene expression profiling for response prediction of rectal cancer to preoperative radiotherapy. J Gastroenterol. 2007;42:730–736. doi: 10.1007/s00535-007-2089-x. [DOI] [PubMed] [Google Scholar]
- 42.Chiu YH, Hsu SH, Hsu HW, Huang KC, Liu W, Wu CY, Huang WP, Chen JY, Chen BH, Chiu CC. Human non small cell lung cancer cells can be sensitized to camptothecin by modulating autophagy. Int J Oncol. 2018;53:1967–1979. doi: 10.3892/ijo.2018.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osielska MA, Jagodziński PP. Long non-coding RNA as potential biomarkers in non-small-cell lung cancer: What do we know so far? Biomed Pharmacother. 2018;101:322–333. doi: 10.1016/j.biopha.2018.02.099. [DOI] [PubMed] [Google Scholar]
- 44.Liang J, Wei X, Liu Z, Cao D, Tang Y, Zou Z, Zhou C, Lu Y. Long noncoding RNA CYTOR in cancer: A TCGA data review. Clin Chim Acta. 2018;483:227–233. doi: 10.1016/j.cca.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Yu Y, Yang J, Li Q, Xu B, Lian Y, Miao L. LINC00152: A pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017;50:2017. doi: 10.1111/cpr.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quan FY, Jiang J, Zhai YF, Li B, Wu XH, Nie W. The prognostic effect of LINC00152 for cancer: A meta-analysis. Oncotarget. 2017;8:75427–75433. doi: 10.18632/oncotarget.20130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao Y, Tie Y, Du J, He J. LINC00152-promotes the proliferation of gastric cancer cells by regulating B-cell lymphoma-2. J Cell Biochem. 2019;120:3747–3756. doi: 10.1002/jcb.27655. [DOI] [PubMed] [Google Scholar]
- 48.Li N, Feng XB, Tan Q, Luo P, Jing W, Zhu M, Liang C, Tu J, Ning Y. Identification of circulating long noncoding RNA Linc00152 as a novel biomarker for diagnosis and monitoring of non-small-cell lung cancer. Dis Markers. 2017;2017:7439698. doi: 10.1155/2017/7439698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng S, Zhang J, Su W, Bai S, Xiao L, Chen X, Lin J, Reddy RM, Chang AC, Beer DG, Chen G. Overexpression of LINC00152 correlates with poor patient survival and knockdown impairs cell proliferation in lung cancer. Sci Rep. 2017;7:2982. doi: 10.1038/s41598-017-03043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Li W. Long noncoding RNA CYTOR sponges miR-195 to modulate proliferation, migration, invasion and radiosensitivity in nonsmall cell lung cancer cells. Biosci Rep. 2018;38:2018. doi: 10.1042/BSR20181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou H, Li H. Knockdown of long non-coding RNA LINC00152 increases cisplatin sensitivity in ovarian cancer cells. Exp Ther Med. 2019;18:4510–4516. doi: 10.3892/etm.2019.8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Z, Guo X, Zang M, Wang P, Xue S, Chen G. Long non-coding RNA LINC00152 promotes cell growth and invasion of papillary thyroid carcinoma by regulating the miR-497/BDNF axis. J Cell Physiol. 2019;234:1336–1345. doi: 10.1002/jcp.26928. [DOI] [PubMed] [Google Scholar]
- 53.Ma P, Wang H, Sun J, Liu H, Zheng C, Zhou X, Lu Z. LINC00152 promotes cell cycle progression in hepatocellular carcinoma via miR-193a/b-3p/CCND1 axis. Cell Cycle. 2018;17:974–984. doi: 10.1080/15384101.2018.1464834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang L, Guo R, Yuan Z, Shi H, Zhang D. LncRNA HOTAIR regulates CCND1 and CCND2 expression by sponging miR-206 in ovarian cancer. Cell Physiol Biochem. 2018;49:1289–1303. doi: 10.1159/000493408. [DOI] [PubMed] [Google Scholar]
- 55.Ren D, Zheng H, Fei S, Zhao JL. MALAT1 induces osteosarcoma progression by targeting miR-206/CDK9 axis. J Cell Physiol. 2018;234:950–957. doi: 10.1002/jcp.26923. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Xu H, Si L, Li Q, Zhu X, Yu T, Gang X. MiR-206 inhibits proliferation and migration of prostate cancer cells by targeting CXCL11. Prostate. 2018;78:479–490. doi: 10.1002/pros.23468. [DOI] [PubMed] [Google Scholar]
- 57.Wang P, Gu J, Wang K, Shang J, Wang W. miR-206 inhibits thyroid cancer proliferation and invasion by targeting RAP1B. J Cell Biochem. 2019;120:18927–18936. doi: 10.1002/jcb.29213. [DOI] [PubMed] [Google Scholar]
- 58.Dai C, Xie Y, Zhuang X, Yuan Z. MiR-206 inhibits epithelial ovarian cancer cells growth and invasion via blocking c-Met/AKT/mTOR signaling pathway. Biomed Pharmacother. 2018;104:763–770. doi: 10.1016/j.biopha.2018.05.077. [DOI] [PubMed] [Google Scholar]
- 59.Liu F, Yin R, Chen X, Chen W, Qian Y, Zhao Y, Jiang Y, Ma D, Hu T, Yu T, et al. Over-expression of miR-206 decreases the Euthyrox-resistance by targeting MAP4K3 in papillary thyroid carcinoma. Biomed Pharmacother. 2019;114:108605. doi: 10.1016/j.biopha.2019.108605. [DOI] [PubMed] [Google Scholar]
- 60.Jia KG, Feng G, Tong YS, Tao GZ, Xu L. miR-206 regulates non-small-cell lung cancer cell aerobic glycolysis by targeting hexokinase 2. J Biochem. 2020;167:365–370. doi: 10.1093/jb/mvz099. [DOI] [PubMed] [Google Scholar]
- 61.Yang L, Sun H, Liu X, Chen J, Tian Z, Xu J, Xiang B, Qin B. Circular RNA hsa_circ_0004277 contributes to malignant phenotype of colorectal cancer by sponging miR-512-5p to upregulate the expression of PTMA. J Cell Physiol. 2020 Jan 21; doi: 10.1002/jcp.29484. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 62.Yang C, Zhang J, Ding M, Xu K, Li L, Mao L, Zheng J. Ki67 targeted strategies for cancer therapy. Clin Transl Oncol. 2018;20:570–575. doi: 10.1007/s12094-017-1774-3. [DOI] [PubMed] [Google Scholar]
- 63.Wierzbicka-Tutka I, Sokołowski G, Bałdys-Waligórska A, Adamek D, Radwańska E, Gołkowski F. Prothymosin-alpha and Ki-67 expression in pituitary adenomas. Postepy Hig Med Dosw. 2016;70:1117–1123. doi: 10.5604/17322693.1223796. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Li M, Dong C, Ma Y, Xiao L, Zuo S, Gong Y, Ren T, Sun B. Linc00152 knockdown inactivates the Akt/mTOR and Notch1 pathways to exert its anti-hemangioma effect. Life Sci. 2019;223:22–28. doi: 10.1016/j.lfs.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 65.Du L, Huang GH, Mou KJ, Xiang Y, Tang JH, Xu W, Xia SL, Zhao JN, Lv SQ. MiR-206 is down-regulated and suppresses cell proliferation by targeting FOXP1 in brain gliomas. Int J Clin Exp Pathol. 2018;11:3405–3415. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.