Abstract
Advanced imaging techniques provide a powerful tool to assess the intra- and intertumoral heterogeneity of gliomas. Advances in the molecular understanding of glioma subgroups may allow improved diagnostic assessment combining imaging and molecular tumor features, with enhanced prognostic utility and implications for patient treatment. In this article, a comprehensive overview of the physiologic basis for conventional and advanced imaging techniques is presented, and clinical applications before and after treatment are discussed. An introduction to the principles of radiomics and the advanced integration of imaging, clinical outcomes, and genomic data highlights the future potential for this field of research to better stratify and select patients for standard as well as investigational therapies.
Keywords: Tumoral heterogeneity, advanced imaging, gliomas, high-grade gliomas, glioblastomas, MRI, Radiomics, Radiogenomics
Introduction:
Rapid molecular advancements have resulted in changes in the understanding of the pathogenesis and classification of gliomas and in particular, glioblastoma. These tumors were amongst the first to be included in the Cancer Genome Atlas[1, 2]. Revision of the 4th edition of the WHO classification of Central Nervous Tumors in 2016 elaborated a structured reporting scheme of malignant gliomas. Furthermore, in anticipation of a new classification due in 2021, the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy (cIMPACT-NOW) provided further periodic updates in 2018–2020 which simplified and clarified the classification of malignant tumors[3]. These advances indicate the strides which have been made in molecular diagnostics over the last decade.
This review provides an overview of the technical basis for conventional and advanced MRI sequences in assessing the phenotypic heterogeneity associated with varying histologic and molecular glioma subgroups, which thus far has primarily been implemented for diagnostic purposes before treatment, or for response assessment after therapy. The use of these images in the field of radiomics, combined with clinical outcome data and genomics to predict patient outcome, demonstrate how computational approaches incorporating varying imaging phenotypes may permit a more refined diagnostic and prognostic assessment of gliomas.
Pathologic Basis of Imaging Features on Conventional MRI sequences:
Magnetic resonance imaging (MRI) is the standard imaging procedure for diagnostic assessment of gliomas. Conventional MR sequences are based on the relaxation of magnetic spins of hydrogen [4]atoms (protons) perturbed in a uniform magnetic field by external radiofrequency pulse. By employing serial alterations in the application of the radiofrequency pulses and subtle gradient shifts in the magnetic fields, images of tissues can be obtained with excellent delineation of tissues with differing water content. Conventional sequences are foundationally termed as T1 weighted and T2 weighted sequences, each having distinct appearances. The disruption of the blood brain barrier (BBB) with development of “leaky”, damaged vessels secondary to neoangiogenesis related to Vascular Endothelial Growth Factor (VEGF) forms the pathological basis for contrast enhancement of tumors on the T1 contrast enhanced images. Contrast enhanced T1 weighted sequences using gadolinium based intravenous contrast agent add further clarity to lesions and demarcate their margins. T2 weighted Fluid Attenuated Inversion Recovery Sequence (FLAIR) is employed to assess the water content of tissues with reference to the primary lesion and the secondary changes of perilesional edema. The relative intensity of the normal grey matter and white matter are used as references to the pathological alterations seen in the presence of tumors.
Assessment of intertumoral phenotypic imaging heterogeneity between glioma subgroups defined by (a) combined loss of short arm of chromosome 1 and long arm of chromosome 19 or 1p19q codeletions (b) presence or absence of isocitrate dehydrogenase (IDH) mutation and (c) the methylation of methylguanine methyltransferase promoter (MGMT)[5] using conventional MRI sequences has yielded variable findings with only modest correlation with the presence of these molecular markers [6]. In studies correlating MR morphology and tumor location, the presence of 1p19q codeletions were associated with a predilection for the frontal lobe[4], whereas those tumors with the intact chromosome configuration tended to be located in the temporal, insular or temporo-insular regions [7]. In studies correlating tumor margins with the presence or absence of codeletions, an indistinct margin was seen in almost all tumors with codeletions; however, a majority of tumors without codeletions also showed indistinct margins [8]. A weak association between 1p/19q co-deletion and specific MRI morphology has been highlighted in the literature [6, 9, 10].
Correlations between IDH mutation status with geographic localization of tumors have shown characteristic lobar confinement of lesions to a single lobe with IDH mutated tumors. Localization to the frontal lobe with rostral extension along the lateral ventricles has been documented in glioblastomas. This has given rise to the hypothesis that IDH mutated astrocytomas (formerly known as glioblastomas) arise from precursor cells in the subventricular zone [11]. Additionally, preferential location of low-grade IDH mutated tumors in the frontal lobe with extension along the rostral extent of the lateral ventricle and involvement specifically of the left hippocampus has been reported [11, 12]. Several studies have reported the correlation between IDH mutation and patterns of enhancement as well as the morphological patterns of tumor margins in high-grade gliomas and compared them to lower grade tumors. One of the hallmark features of IDH mutated glioblastomas is the relative lack of enhancement. This feature, together with the presence of enhancing satellite lesions, large area of cystic necrosis, and localization to the frontal lobe yielded a sensitivity of 73% and a specificity of 99% with an overall accuracy of more than 99 % in a study by Carrillo and colleagues comprising of 14 IDH mutated tumors and 188 IDH wild type lesions [11]. A similar finding was noted by Qi et al. [13] but in lower grade II and III astrocytomas. The presence of MGMT promoter methylation has not strongly correlated with geographic tumor localization or specific imaging features, although Eoli et al. reported a preponderance of unmethylated tumorsin the temporal lobe in their series, whereas methylated tumors were predominantly located in the parietal and occipital lobes. Ellingson et al. reported methylated lesions to be localized to the left hemisphere with a preponderance of lesions in the temporal lobe [14, 15].
In summary, conventional T1 and T2 weighted MRI images are fairly limited in identifying specific features distinguishing molecular glioma subgroups. Moreover, meaningful assessment of the intratumoral heterogeneity inherent to malignant gliomas and in particular, glioblastoma, is limited with conventional imaging approaches. In the following sections, the physiologic basis and limitations of advanced imaging are reviewed, for diagnostic utility in the pre-treatment setting, for non-invasive response assessment after definitive therapy, and also in combination with imaging features and genomic information for prognostic and potentially predictive assessments. The goals of advanced imaging in the improved diagnosis and treatment of malignant gliomas have been summed up into three major areas by Hu et al. [16]. These are (a) increasing specificity in targeting high and low-grade regions in predominantly non-enhancing malignant gliomas, (b) identifying more specific imaging techniques to differentiate non-enhancing tumor infiltration from non-specific peritumoral edema which appears similarly hyperintense on T2/FLAIR images, and (c) to differentiate treatment related sequelae (post-radiation necrosis, pseudoprogression) from tumor progression and to improve response assessment to treatment.
Advanced Imaging Techniques.
MRI
Diffusion Weighted Imaging:
Diffusion weighted imaging (DWI) measures random motion of water molecules in a voxel of tissue sampled by timed dephasing and rephasing radio frequency pulses in three orthogonal planes in a sequence with rapidly changing gradient fields. Loss of signal relates to bulk motion of water whereas retention of signal indicates impeded motion of water, and attenuation of the T2* signal is based on the ease by which water molecules can diffuse through the tissue. Applications of diffusion weighted imaging are primarily focused on assessment of cellularity of tumors; however, given the physiological basis of restriction of water molecules, applications can also relate to infarcts, hemorrhage, necrosis and post treatment sequelae, all of which can confound the diffusion signal [17]. Diffusion features may be used to assess high cellularity in gliomas [18], to differentiate high-grade and low-grade gliomas [19] and to assess the microenvironment of gliomas[20].(Figure 1 and Figure 2).
Figure 1.
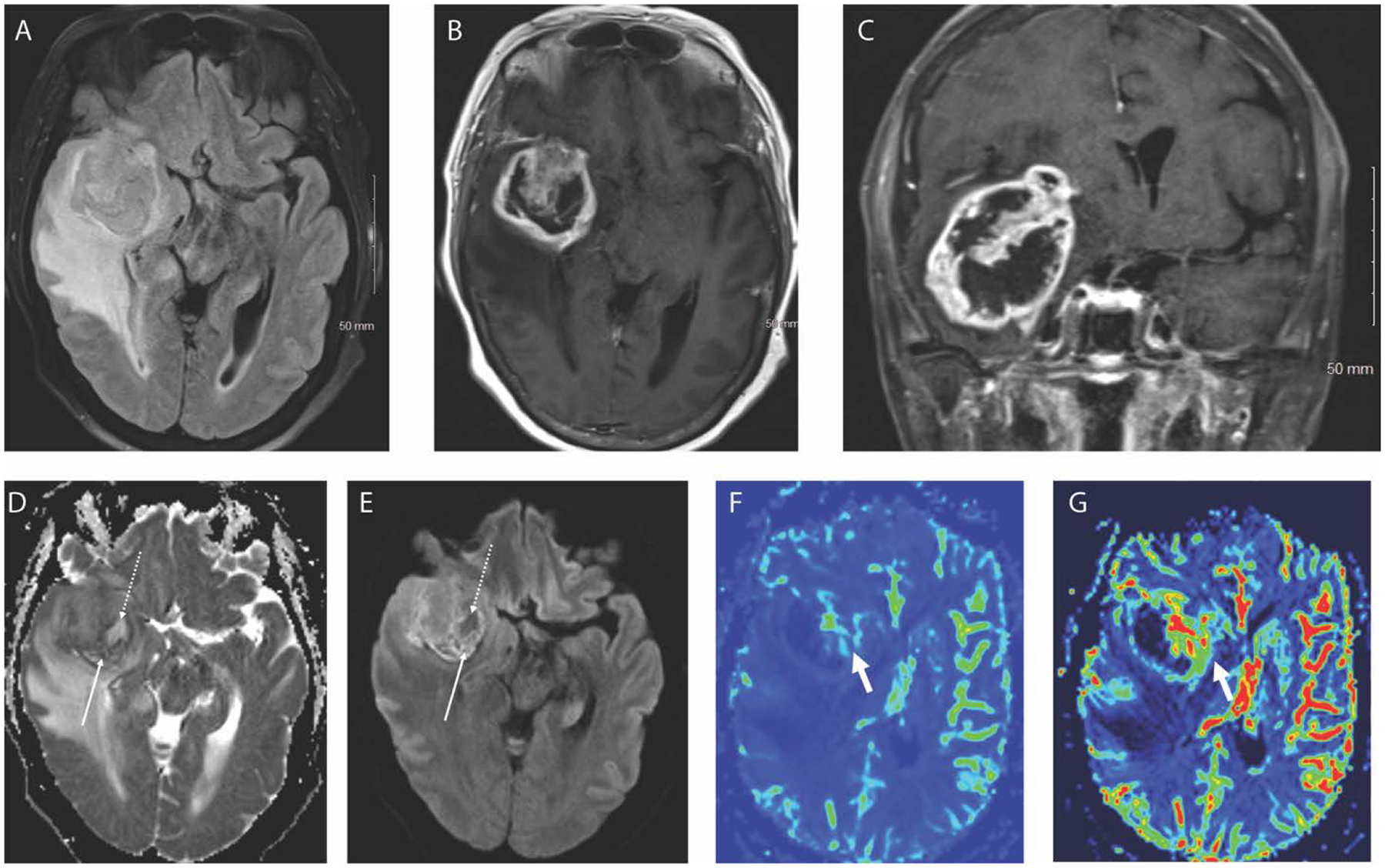
Axial FLAIR (A), axial T1 weighted post contrast (B) and coronal T1 weighted post contrast (C) images in a 66-year-old female patient demonstrate a heterogeneously enhancing right temporal lobe mass with internal necrosis (non-enhancing portions) and surrounding edema. Areas of hypercellularity (solid arrow) and hypocellularity/necrosis (dotted arrow) are well depicted on the DWI trace image (D) and ADC map (E). Corresponding DSC perfusion maps show elevated CBF (F) and CBV (G) along the anteromedial aspects of the mass compared to the lateral aspect. The pathological diagnosis was grade IV glioblastoma.
Figure 2.
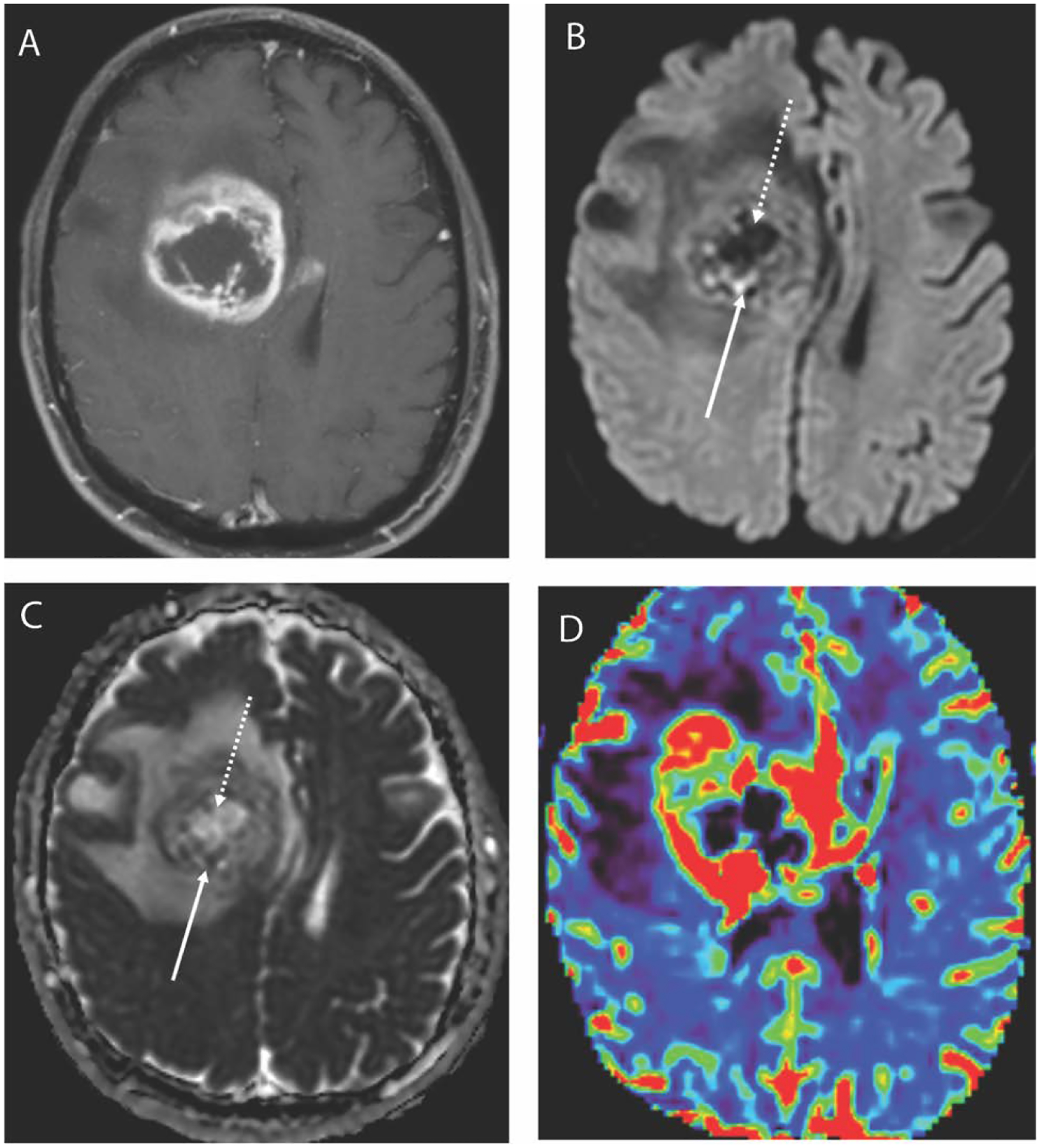
Axial T1 weighted post contrast image (A) in a 67-year-old male with a glioblastoma demonstrates a large centrally necrotic mass in the right frontal lobe. DWI trace image (B) shows peripheral foci of restricted diffusion posteriorly suggestive of hypercellularity and the corresponding ADC map (C) confirms the presence of low ADC in these areas. Similarly, the central portion demonstrates facilitated diffusion with low signal on DWI trace image and elevated ADC.
MR perfusion:
MR perfusion techniques employ exogenous tracers (gadolinium based contrast agents) and endogenous tracers (magnetically labelled spins using protons in flowing arterial blood) to assess the vascular bed of the brain in general and tumors in particular[21]. Perfusion scans obtained following administration of gadolinium contrast can be performed by two methods. Dynamic Susceptibility Contrast (DSC) perfusion relies on the transient decrease in the T2*effects due to the passage of contrast in the tissue bed of interest. This results in a drop of signal intensity which, as a function of time, recovers as the paramagnetic effects of the contrast wear off. This is used to calculate differential blood flow in the vascular bed, which in turn is used to estimate the relative cerebral blood flow (rCBF), cerebral blood volume(rCBV), mean transit time and fractional tumor burden (FTB) [22]. Dynamic contrast enhanced (DCE) perfusion scans utilize T1 weighted imaging to track a bolus of contrast through the vascular bed. The technique measures shortening of the T1 signal as the contrast passes through the vascular bed, termed relaxivity. The pharmacokinetic (extended Tofts) model envisages movement of the contrast between two compartments (intravascular and extravascular extracellular) to assess the permeability of the vessels. The quantitative measurement of the contrast results in the generation of maps for several parameters, of which k-trans is the most used parameter to assess the leakiness of the vessels. DCE is best correlated with contrast enhanced T1 images because it is based on disruption of the blood brain barrier and hence the extravasation of contrast whereas the DSC can be used for assessment of both enhancing and non-enhancing components of the tumor given the passage of contrast which causes an inherent loss of signal. Both techniques have been used to differentiate high and low-grade tumors[23, 24], to assess treatment effects [25], to evaluate for tumor progression after radiotherapy[26], and to characterize pseudoprogression[27] and pseudoresponse[28] (Figure 3 and Figure 4).
Figure 3.
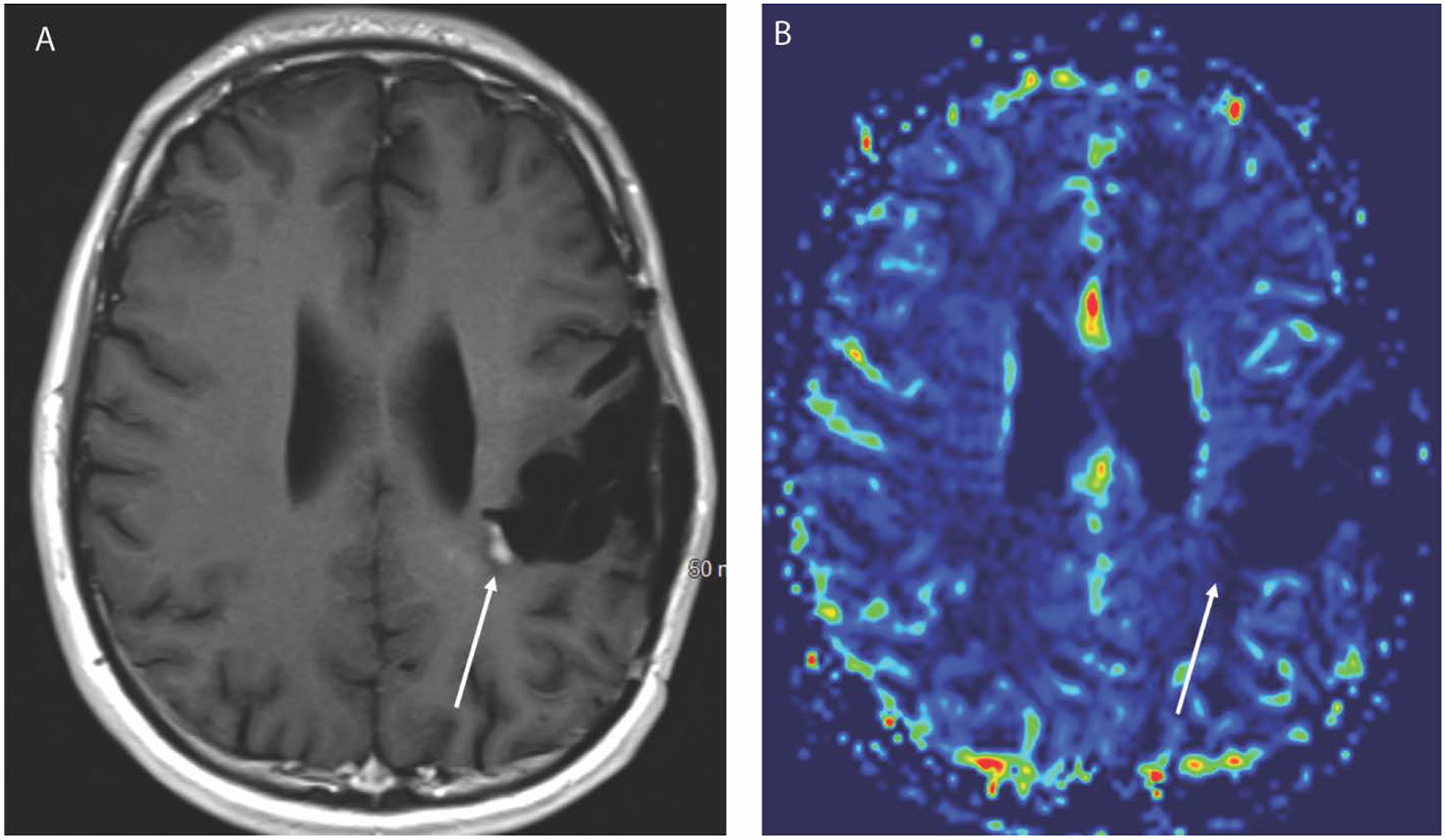
Axial T1 weighted post contrast image (A) in a 27-year-old male with history of IDH mutated anaplastic astrocytoma after treatment with radiation and temozolomide 6 months earlier demonstrates a new focus of enhancement along the medial margin of the post-surgical cavity. The corresponding CBV map (B) from DSC perfusion, however, does not show elevated blood volume, and this was therefore considered to be pseudoprogression. Follow up imaging confirmed resolution of the enhancement.
Figure 4.
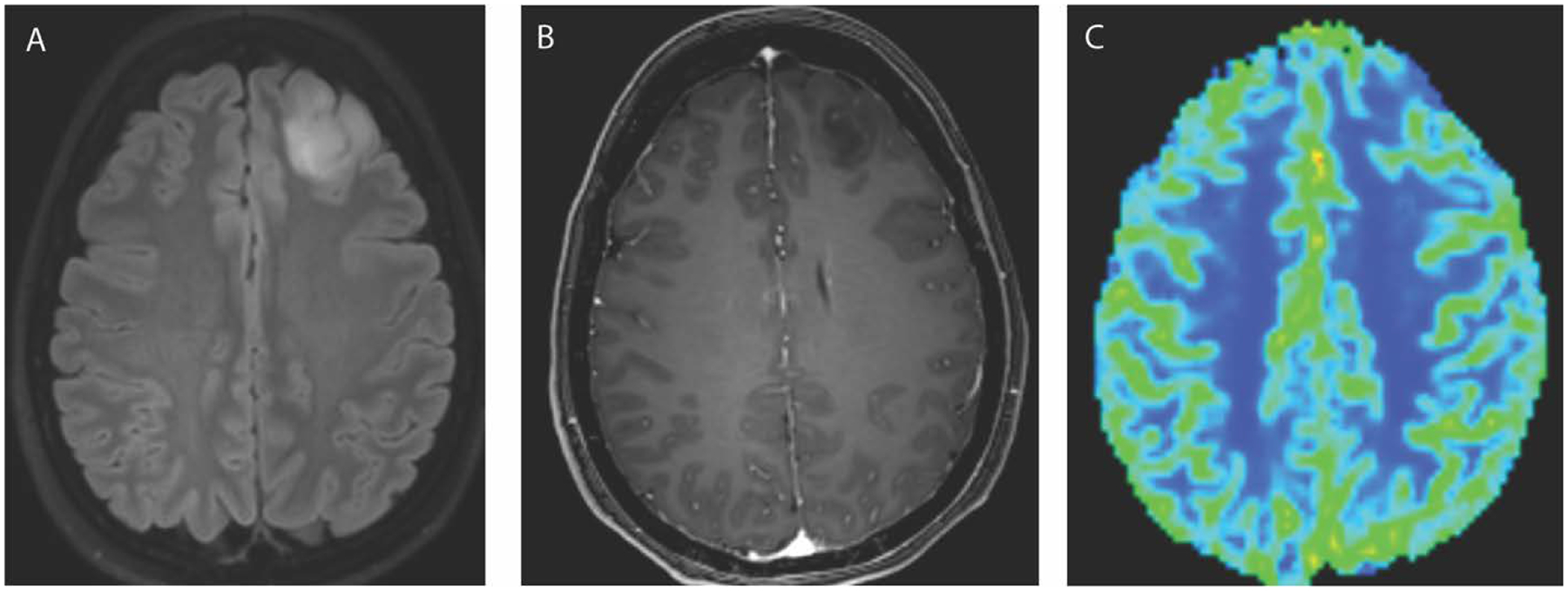
Axial FLAIR (A) and T1 weighted post contrast (B) images in a 33 year old female demonstrate a T2 hyperintense non-enhancing mass in the left frontal lobe. There is normal to low blood volume in this mass on CBV DSC perfusion map. The pathology revealed a grade 2 oligodendroglioma (IDH1 mutated, 1p/19q codeleted).
Diffusion Tensor Imaging:
Diffusion tensor imaging (DTI) is an extension of DWI wherein in the magnitude and directionality of the water molecule are measured along axon bundles. Unlike DWI, measurement of the diffusion signal is made in six or more planes (directions). This allows calculation of the directionality of diffusion surrounding an axonal bundle. The magnitude of diffusion along the primary orientation of the axon bundles is axial diffusivity, whereas radial diffusivity is magnitude perpendicular to the axons. Averaged diffusivity along all measured axonal tracts is mean diffusivity. Three-dimensional color-coded reconstructions can reproduce models of white matter tracts in the brain known as fiber tractography, which is the mainstay in the preoperative assessment of tumors used for intraoperative navigation [29]. DTI assessments of tumor in relation to the adjacent white matter tracts characterizes deviations, displacement or disruption of the tracts[30] Evaluation of perilesional white matter tracts in high-grade gliomas by high-definition fiber tractography has been described [31] with implications for assessment of infiltration or invasion by non-enhanced tumor, or displacements related to the mass and peritumoral edema. Technical advancements have to some extent mitigated the ability to resolve multiple crossing fibers in an imaged voxel and the inability to image fibers which have been infiltrated with tumors or destroyed by the neoplastic processes or obscured due to extensive edema [32, 33]. DTI has been employed to assess the morphological changes in the postoperative period to differentiate pseudoresponse following administration of bevacizumab [34], especially in the absence of enhancement observed during treatment with this agent despite the presence of viable tumor [35].
Magnetic Resonance Spectroscopy:
Magnetic Resonance Spectroscopy (MRS) uses the principles of nuclear magnetic resonance to provide a voxel based metabolic signature of tissues. Clinical application of this advanced technique in the preoperative assessment of tumors has been employed to differentiate tumor from mimics such as primary CNS lymphoma, tumefactive demyelination and metastases. The principal metabolites which have been investigated, validated, and consistently visualized on MRS spectra include choline, creatine, and N-acetyl aspartate[36] (Figure 5). In addition, choline peaks which are increased in neoplastic processes including gliomas are decreased in areas with necrosis and thus play only a moderate role in distinguishing radionecrosis, pseudoprogression and tumor progression [37, 38]. Attempts have been made to utilize MRS in distinguishing low-grade and high-grade gliomas with only modest results[39]. Limitations in the diagnostic accuracy of this techniques stems from the minimal standardization of the technique across vendors and departments. Artifacts from bone and air severely degrade the images. Since the conventional proton H1 spectroscopy technique is voxel dependent, a large voxel size can average out the metabolite signature and smaller lesions are sub-optimally assessed relative to the larger size of the voxel. Incomplete suppression of water and lipid signals can also affect the clarity of the final spectrum. Advances in MRS include whole brain echo-planar spectroscopic imaging (EPSI), which offers a new perspective in non-invasive tissue sampling by identifying areas of greater proliferation linked to greater metabolite concentrations [40]. An ongoing clinical trial is focused on exploring whole brain MRS evaluation for spectroscopic abnormalities beyond the treatment field in resected tumors to identify areas of spectroscopic abnormality [41, 42]
Figure 5A and 5B:
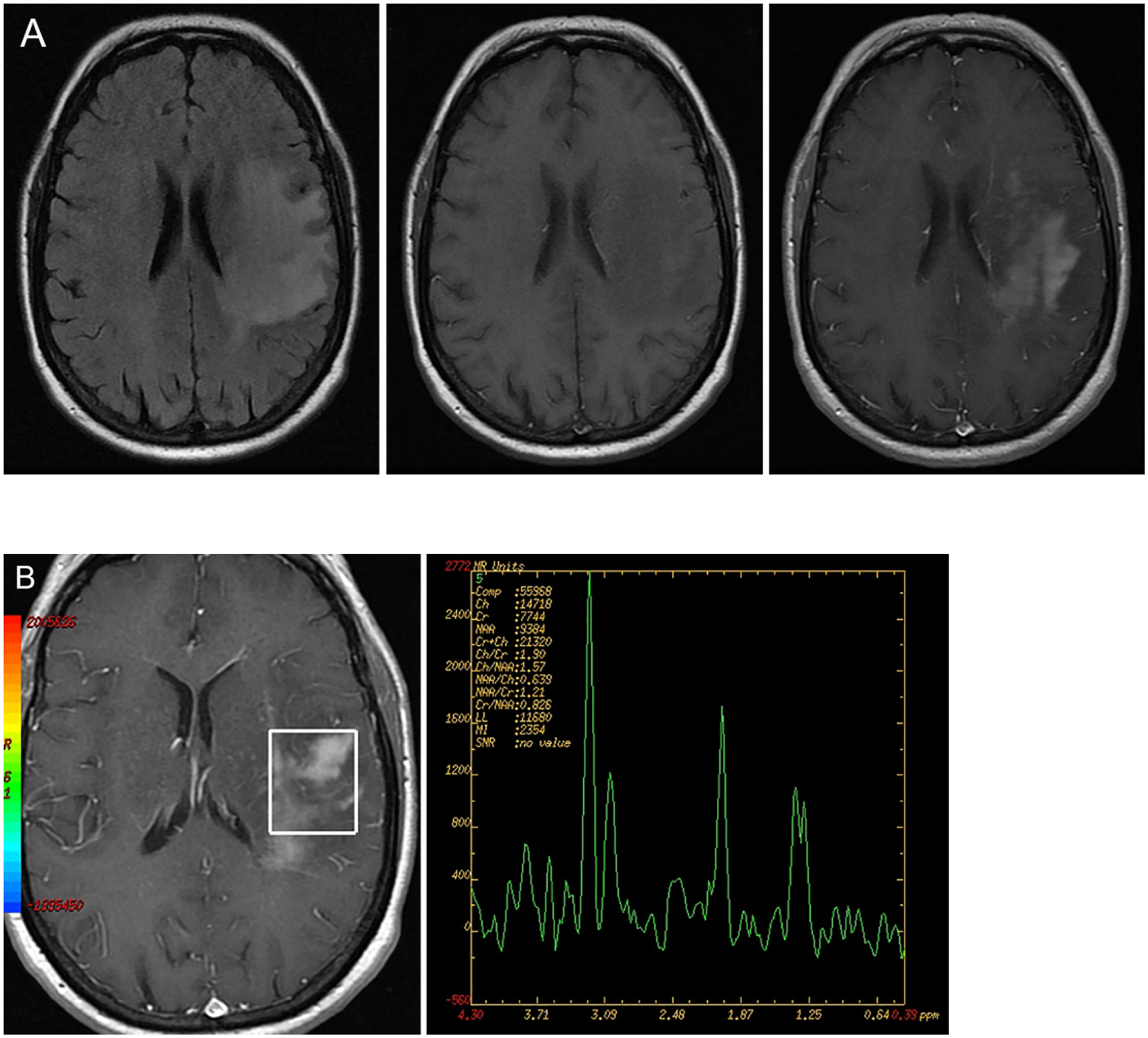
FLAIR hyperintense mass lesion showing predominantly T1 hypointense signal on the pre-contrast T1 weighted image. Heterogenous contrast enhancement present in the post contrast image. Single voxel MRS evaluation shows gross elevation of the choline peak and relatively diminished NAA peak (third from left) consistent with a neoplastic process, in this case of glioblastoma. (Image courtesy Prof. Mohannad Ibrahim, Michigan Medicine)
Chemical Exchange Saturation Transfer:
Chemical Exchange Saturation Transfer (CEST) imaging is a novel technique which depends on the detection of low concentration of molecules with chemically exchangeable protons, which would be otherwise undetectable on conventional MRS studies, saturated with a radiofrequency pulse tagged to water molecules. Measuring the increased concentration of molecules containing amide (−NH), amino (−NH2) or hydroxyl (−OH) radicals in the tagged protons contained in the water molecules, gives a spectrum of the water signal intensities[43]. Several sequences such as 2D Fast spin echo and 2D gradient echo sequences can be employed preferentially on high field strength magnets [44]. The most common CEST technique involves images of the amide proton transfer techniques. The APT technique has been employed for the assessment of heterogeneity within non-enhancing gliomas [45], to differentiate low-grade gliomas from high-grade gliomas, and to assess post treatment changes with residual viable tumor[43].
Positron Emission Tomography:
Positron emission tomography (PET) is an imaging study primarily involving the radiolabeled molecules that are actively metabolized in target organs, which are then imaged using detector-based imaging systems. C-11 methionine has the shortest half-life and is frequently employed for imaging gliomas and CNS metastases. There are more stable F-18 based agents which have a longer half-life and are therefore more amenable for routine clinical imaging. F-18 FDG [2–18 F-fluorodeoxyglucose] is the most widely used for evaluation of neoplasms outside the CNS. F-18 FDG has limited application for CNS imaging given the high baseline glucose metabolism in the brain, which diffusely sequesters this radiotracer with no preferential uptake by a localized neoplastic process within the brain. This aspect is mitigated by tagging F-18 with amino acids rather than a glucose analogue to increase the specificity for CNS neoplasms. Visualization of these F-18 labeled amino acids is linked to plasma membrane transfer via large amino acid transporters (LAT1). The 1–3,4 Dihydroxy-6F18 fluoro-phenylalanine (F18-FDOPA) has been used for detection and evaluation of recurrent gliomas, especially low-grade gliomas [46]. Meta-analyses show only modest pooled sensitivity in tumor evaluation [47]. F-18 FET (O-(2-[F18] fluoroethyl)-L-tyrosine) is a tyrosine analog which has shown promise in localization of tumors for targeted biopsies. F-18 FET has also proven to be cost-effective in biopsy site assessment when coupled with MRI. This tyrosine analog is also considered effective in the assessment of residual tumor following surgery and has been correlated with 5-ALA guided intraoperative excision[48].
Radiomics and Tumor Heterogeneity:
The underlying principle of most imaging studies as has been reviewed here is the information contained in a voxel of digitized data. These units contain information of attenuation of x-rays as in the case of CT studies, radiofrequency signal intensities perturbed, accentuated, or diminished by changing magnetic fields in MRI, or attenuated energies from disintegrating photons in PET images. Each of these alterations are depicted as images in a finite space. The outline of these applications, both basic and advanced, was presented earlier; however, there remains an unknown dimension of the data which is not evident to the human eye. The field of radiomics refers to the computerized extraction of quantifiable data to create an alternate or enhanced interpretation of the images. For tumor heterogeneity, radiomics has an important role to identify features within images to characterize a lesion, and assess its features to predict the response to treatment and prognosis[49]. Images must be converted to mineable data prior to analyses. This stepwise process involves acquisition and reconstruction of the images followed by processing of the images for analyses. Afterwards, images are evaluated for specific regions of interest which are segmented and extracted for quantification. Features of the quantifiable data are then subjected to predictive modeling using machine learning techniques. Some features used for machine learning paradigms include semantics features, morphological radiomics, texture analyses, and functional radiomics.
Semantic features:
Semantic features are the frequently encountered features in radiology reports which can be assessed from imaging data. These include location, volume, enhancement characteristics, presence or absence of necrosis, presence, or absence of impeded diffusion, increased or decreased perfusion, assessment of the peritumoral regions presenting as enhancing and non-enhancing regions. Of these, observations made on conventional MRI sequences are embedded in the Visually AcceSAble Rembrandt Images (VASARI) feature set comprising of 24 common MR observations from the TCGA initiative. These features are linked to the Repository of the Molecular Brain Neoplasia Data Set (REMBRANDT) for correlation between MRI features and tumor genomics [49, 50].
Morphological radiomics:
This aspect of radiomics focuses on tumor surface characteristics and includes measurements of the perimeter, dimensions in the major and minor axes, and surface contours. It also includes how well-defined the margins of the lesion are with respect to the surrounding tissues. These features have been used to distinguish tumor progression from pseudoprogression and differentiation of recurrent tumor and radionecrosis [51, 52].
Texture Analyses:
Since MRI constitutes the most widely used modality in the diagnosis and post-operative assessment of malignant gliomas, the complex interplay between advanced imaging techniques and the genetic make-up of the tumors is best assessed using textural analyses which extract and encode spatially distinct data from imaged voxels, including but not limited to intensities of the voxel but also the surrounding voxels. This analysis brings higher fidelity information beyond what the human eye can recognize [53]. The analyses depend on several mathematical algorithms which can analyze structure (as described above) and statistical variance or a combination of both. Algorithms commonly used for texture analyses include Gabor filter banks, which decompose from original images into sinusoidal waveforms of different frequencies to allow 2-D and 3-D measurements of voxel patterns[54]. The most common example of textural algorithms include the grey level co-occurrence matrix (GLCM) which measures the frequency of pixel-pair distribution in a predefined distance [53]. The algorithm uses second order statistics on the distribution of grey scale intensity levels within a region of interest both in angular and radial dimensions [53, 55]. An algorithm which uses both statistical and structural techniques is local binary patterns (LBP). LBP calculate the pixel value by comparing it with neighboring pixels and assigning them a binary value. These patterns are invaluable in assessing motion degraded images.
Functional Radiomics:
As defined by Singh et al, “Functional radiomics markers are a new class of markers which specifically target the issue of interpretability’ by modelling features that directly capture the underlying physiological process such as angiogenesis” [49]. According to these authors, a biomarker should not only be measurable but should reflect underlying anatomy and physiology. An example they suggest, are studies of tumor vascularity and the imaging processes, such as perfusion and vessel architecture imaging. Alic L et al [56] utilized DCE MR perfusion studies to assess the heterogeneity of limb sarcomas. They utilized the k-trans perfusion maps and heuristic model based parametric maps (AUC, slope, and Max. enhancement) to differentiate between tumors to treatment or not. Their analysis extracted texture features data for evaluation the enhancing fraction, coherence, and fractal dimensions for both parametric Ktrans maps and heuristic model maps. They concluded that as a function of the tumor vascularity, both data types obtained from the perfusion studies, irrespective of mathematical modelling, could predict tumor treatment response. Similarly, Stadlbauer, and colleagues [57] assessed heterogeneity of tumor vascularity by demonstrating model-based differentiation of arterial and venous microvasculature in high-grade gliomas, shifts in contrast bolus peak time related to the vascular beds and parameters based on curvature, microvascular radii, and density. In doing so, they were dependent on the intrinsic heterogeneity of the vascular beds of malignant gliomas.
Machine Learning:
The combination of the complex histological architecture of malignant gliomas, advanced imaging techniques, each with unique diagnostic features, and the disparate genomics of tumors, falls in the realm of machine learning. Machine learning combines the aspects of radiomics, clinical data, and genomics, to give a meaningful output which can model treatment effects and outcomes. Machine learning pipelines should have a training and testing data set where models are developed using the former and subsequently evaluated in the latter, following development of logical associations and pattern recognition. The input data can include not only finite parametric outputs from imaging studies, but also genetic profiles including deletions, gene amplification and other nuanced genomic features.
Some of the algorithms most employed common in radiologic machine learning include (a) support vector machines (SVM), (b) decision trees, (c) sparse regression models, (d) convolutional neural networks. SVMs are supervised models used for classification and regression analysis, that maximize space between classes with a high dimensional hyperplane. Decision trees predict a class by recursively partitioning the sample space using one variable at a time in a tree structure. These trees are often used in ensembles to create random forests (RF) by using random subsets of the data to create an array of trees and aggregate their predictions. Statistical regression models, such as linear or logistic regression, often struggle not to overfit high dimensional datasets [58, 59]. Sparse regression models, such as LASSO, ridge regression, or the generalized form of elastic nets, put a penalty on model dimensionality, and can deal better with these datasets that have more signal dimensions than input examples. Convolutional neural networks are machine learning models that use many layers of convolutional filters which are learned through optimizing the model on the training dataset [60]. These complex models have been used for many tumor imaging tasks including genetic prediction, subtype prediction, and tumor segmentation. However, despite their performance it is difficult to understand and interpret how a deep learning model made its prediction (Figure 6).
Figure 6.
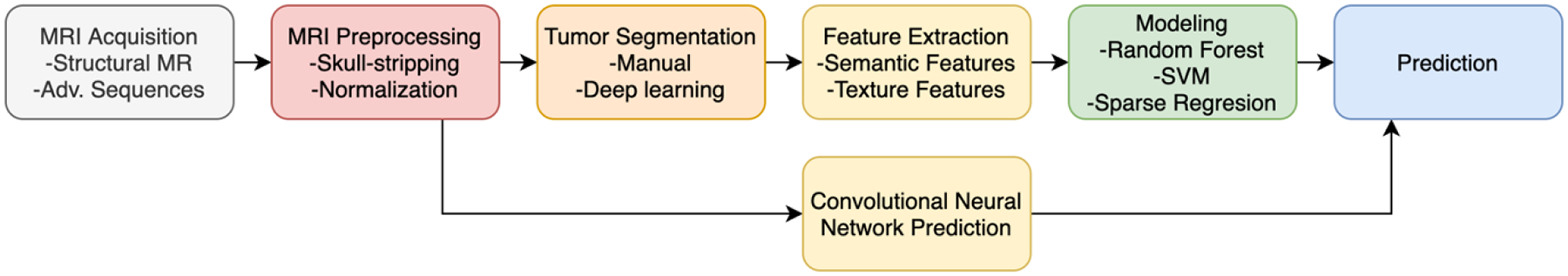
Example schematic of MRI algorithm pipeline. Different methods will extract different features or attempt to predict different tumor characteristics or outcomes. Unless an end-to-end model such as a convolutional neural network is used, tumors are generally segmented to extract features.
Current and future trends in imaging applications for assessing tumor heterogeneity:
Intersections of tumor heterogeneity and advanced imaging techniques have made rapid strides since the last decade. A review of the recent literature shows that the thrust of recent research is toward a personalized and granular approach to malignant gliomas. In his commentary, Timo Auer succinctly summarized the role of MRI as the gold standard for assessment of tumor heterogeneity of glioblastomas. He indicated that novel and advanced MRI techniques will form the ground truth upon which artificial intelligence-based image processing will enable radiomics data to be integrated for noninvasive diagnosis and treatment. He and his colleagues evaluated quantitative volumetric tissue assessment of enhancing tumor volumes to predict overall survival in GBM patients. In this study, they concluded that the cut-off percentage of enhancing tumor volume of >84.78% samples were dichotomized, and overall and progression-free survival was evaluated. Multivariate analyses also included gender, KPS scores and MGMT status, and resection status. Volumes greater than the cut-off value showed significantly shortened survival [61, 62]. Advanced image processing of conventional MR sequences based on quantitative analyses of peritumoral edema on T1 and T2 sequences was described by Blystad et al. [63]. A mechanistic mathematical model based on histopathologic evidence for the presence of EGFR and PDGRA genes from image guided biopsy samples of glioblastoma patients has been proposed by Morris and colleagues [64]. A cohort of 135 de novo GBM patients from the TCGA-GBM archive were studied for quantitative texture and spatial patterns and were correlated with gene expressions by Elsheikh and colleagues [65]. They concluded that various quantitative imaging features extracted from MRI could be effectively correlated with multi-stage genome-wide test at the gene-level using a non-parametric correlation framework.
Fathi Kazerooni and colleagues [66] did a prospective study to characterize active and infiltrative tumorous sub-regions from normal tissue in samples from excised gliomas, by correlating the imaging parameters including diffusion, DTI and DSC perfusion with histopathological evaluation. The authors were able to discriminate between infiltrative edema, active margin of the tumor, and normal brain. They deduced that the maximum accuracy for distinction between infiltrative edema and normal tissue, and active tumor and normal tissue, was found with diffusion-based parameters, whereas perfusion scans were most accurate in distinguishing infiltrative edema and active tumor. A combination of CBV, mean diffusivity, FLAIR and T2- relaxometry showed the highest diagnostic accuracy in distinguishing all regions of the tumor brain interface. Fathi Kazerooni et al. [67]also reviewed the imaging signatures of glioblastoma in a comprehensive review of tumor radiogenomics. They covered exploratory research areas in relation to the existing knowledge base of glioma genomics, including evaluation of the tumor core, peritumoral edema, tumor infiltrated tissues, presence of central necrosis and diffusion abnormalities. They also highlighted studies regarding association of angiogenesis and metabolic signatures on MRS. The focus areas in hypothesis driven studies included association of MRI phenotypes with radiogenomic studies which included studies related to IDH mutation status, MGMT methylation, VEGF mutation, and EGFR amplification. Apart from these correlative studies from the armamentarium of MRI techniques, emerging fields of tissue conductivity measurements made with electrical properties tomography (EPT) have been used to assess tumor progression in glioblastomas[68]. EPT measures the electrical distortions induced in a tissue volume after application of a radiofrequency pulse during the acquisition of a three-dimensional steady state free precession sequence. This technique together with diffusion and perfusion parameters showed promising results in areas of hypo-vascularity, and low electric conductance showed the highest performance for indicating tumor progression.
Conclusion
Developments in imaging techniques over the past few decades have run in parallel with our increased understanding of the role of molecular analysis in malignant gliomas. While conventional imaging is likely to remain the workhorse for radiographic patient assessment, both in the pre-treatment and post-treatment settings, integration of advanced imaging techniques, supplemented by machine learning data extraction, is pushing the boundaries of non-invasive imaging towards individualized treatment paradigms for patients.
References:
- 1.Levin VA, L. S, Gutin PH, Neoplasms of the central nervous system, in Cancer: Principles and Practice of Oncology., H. S DeVita VT Jr, Rosenberg SA, Editor. 2001, Williams & Wilkins: Philadelphia, Pa. p. 2100–60. [Google Scholar]
- 2.Brennan CW, et al. , The somatic genomic landscape of glioblastoma. Cell, 2013. 155(2): p. 462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez Castro LN and Wesseling P, The cIMPACT-NOW updates and their significance to current neuro-oncology practice. Neurooncol Pract, 2021. 8(1): p. 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fellah S, et al. , Multimodal MR imaging (diffusion, perfusion, and spectroscopy): is it possible to distinguish oligodendroglial tumor grade and 1p/19q codeletion in the pretherapeutic diagnosis? AJNR Am J Neuroradiol, 2013. 34(7): p. 1326–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smits M and van den Bent MJ, Imaging Correlates of Adult Glioma Genotypes. Radiology, 2017. 284(2): p. 316–331. [DOI] [PubMed] [Google Scholar]
- 6.Kanekar S and Zacharia BE, Imaging Findings of New Entities and Patterns in Brain Tumor: Isocitrate Dehydrogenase Mutant, Isocitrate Dehydrogenase Wild-Type, Codeletion, and MGMT Methylation. Radiol Clin North Am, 2021. 59(3): p. 305–322. [DOI] [PubMed] [Google Scholar]
- 7.Kim JW, et al. , Relationship between radiological characteristics and combined 1p and 19q deletion in World Health Organization grade III oligodendroglial tumours. J Neurol Neurosurg Psychiatry, 2011. 82(2): p. 224–7. [DOI] [PubMed] [Google Scholar]
- 8.Jenkinson MD, et al. , Histological growth patterns and genotype in oligodendroglial tumours: correlation with MRI features. Brain, 2006. 129(Pt 7): p. 1884–91. [DOI] [PubMed] [Google Scholar]
- 9.Patel SH, et al. , T2-FLAIR Mismatch, an Imaging Biomarker for IDH and 1p/19q Status in Lower-grade Gliomas: A TCGA/TCIA Project. Clin Cancer Res, 2017. 23(20): p. 6078–6085. [DOI] [PubMed] [Google Scholar]
- 10.Sherman JH, et al. , MR imaging characteristics of oligodendroglial tumors with assessment of 1p/19q deletion status. Acta Neurochir (Wien), 2010. 152(11): p. 1827–34. [DOI] [PubMed] [Google Scholar]
- 11.Carrillo JA, et al. , Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR Am J Neuroradiol, 2012. 33(7): p. 1349–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, et al. , Anatomical localization of isocitrate dehydrogenase 1 mutation: a voxel-based radiographic study of 146 low-grade gliomas. Eur J Neurol, 2015. 22(2): p. 348–54. [DOI] [PubMed] [Google Scholar]
- 13.Qi S, et al. , Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol Lett, 2014. 7(6): p. 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eoli M, et al. , Methylation of O6-methylguanine DNA methyltransferase and loss of heterozygosity on 19q and/or 17p are overlapping features of secondary glioblastomas with prolonged survival. Clin Cancer Res, 2007. 13(9): p. 2606–13. [DOI] [PubMed] [Google Scholar]
- 15.Ellingson BM, et al. , Anatomic localization of O6-methylguanine DNA methyltransferase (MGMT) promoter methylated and unmethylated tumors: a radiographic study in 358 de novo human glioblastomas. Neuroimage, 2012. 59(2): p. 908–16. [DOI] [PubMed] [Google Scholar]
- 16.Hu LS, et al. , Imaging of intratumoral heterogeneity in high-grade glioma. Cancer Lett, 2020. 477: p. 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiroishi MS, Boxerman JL, and Pope WB, Physiologic MRI for assessment of response to therapy and prognosis in glioblastoma. Neuro Oncol, 2016. 18(4): p. 467–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellingson BM, et al. , Validation of functional diffusion maps (fDMs) as a biomarker for human glioma cellularity. J Magn Reson Imaging, 2010. 31(3): p. 538–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, et al. , The utility of diffusion MRI with quantitative ADC measurements for differentiating high-grade from low-grade cerebral gliomas: Evidence from a meta-analysis. J Neurol Sci, 2017. 373: p. 9–15. [DOI] [PubMed] [Google Scholar]
- 20.Nabavizadeh SA, Ware JB, and Wolf RL, Emerging Techniques in Imaging of Glioma Microenvironment. Top Magn Reson Imaging, 2020. 29(2): p. 103–114. [DOI] [PubMed] [Google Scholar]
- 21.Griffith B and Jain R, Perfusion Imaging in Neuro-Oncology: Basic Techniques and Clinical Applications. Magn Reson Imaging Clin N Am, 2016. 24(4): p. 765–779. [DOI] [PubMed] [Google Scholar]
- 22.Iv M, et al. , Perfusion MRI-Based Fractional Tumor Burden Differentiates between Tumor and Treatment Effect in Recurrent Glioblastomas and Informs Clinical Decision-Making. AJNR Am J Neuroradiol, 2019. 40(10): p. 1649–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law M, et al. , Low-grade gliomas: dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging--prediction of patient clinical response. Radiology, 2006. 238(2): p. 658–67. [DOI] [PubMed] [Google Scholar]
- 24.Boxerman JL, Schmainda KM, and Weisskoff RM, Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol, 2006. 27(4): p. 859–67. [PMC free article] [PubMed] [Google Scholar]
- 25.Bag AK, et al. , Survival analysis in patients with newly diagnosed primary glioblastoma multiforme using pre- and post-treatment peritumoral perfusion imaging parameters. J Neurooncol, 2014. 120(2): p. 361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, et al. , Evaluation of perfusion MRI value for tumor progression assessment after glioma radiotherapy: A systematic review and meta-analysis. Medicine (Baltimore), 2020. 99(52): p. e23766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young RJ, et al. , MRI perfusion in determining pseudoprogression in patients with glioblastoma. Clin Imaging, 2013. 37(1): p. 41–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Essock-Burns E, et al. , Assessment of perfusion MRI-derived parameters in evaluating and predicting response to antiangiogenic therapy in patients with newly diagnosed glioblastoma. Neuro Oncol, 2011. 13(1): p. 119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu R and Hoch MJ, Application of Diffusion Weighted Imaging and Diffusion Tensor Imaging in the Pretreatment and Post-treatment of Brain Tumor. Radiol Clin North Am, 2021. 59(3): p. 335–347. [DOI] [PubMed] [Google Scholar]
- 30.Mori S, et al. , Brain white matter anatomy of tumor patients evaluated with diffusion tensor imaging. Ann Neurol, 2002. 51(3): p. 377–80. [DOI] [PubMed] [Google Scholar]
- 31.Abhinav K, et al. , High-definition fiber tractography for the evaluation of perilesional white matter tracts in high-grade glioma surgery. Neuro Oncol, 2015. 17(9): p. 1199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, et al. , Differences between generalized q-sampling imaging and diffusion tensor imaging in the preoperative visualization of the nerve fiber tracts within peritumoral edema in brain. Neurosurgery, 2013. 73(6): p. 1044–53; discussion 1053. [DOI] [PubMed] [Google Scholar]
- 33.Kuhnt D, et al. , Fiber tractography based on diffusion tensor imaging compared with high-angular-resolution diffusion imaging with compressed sensing: initial experience. Neurosurgery, 2013. 72Suppl 1: p. 165–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arevalo OD, et al. , Assessment of Glioblastoma Response in the Era of Bevacizumab: Longstanding and Emergent Challenges in the Imaging Evaluation of Pseudoresponse. Front Neurol, 2019. 10: p. 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Artzi M, et al. , Differentiation between vasogenic-edema versus tumor-infiltrative area in patients with glioblastoma during bevacizumab therapy: a longitudinal MRI study. Eur J Radiol, 2014. 83(7): p. 1250–1256. [DOI] [PubMed] [Google Scholar]
- 36.Brandao LA and Castillo M, Adult Brain Tumors: Clinical Applications of Magnetic Resonance Spectroscopy. Magn Reson Imaging Clin N Am, 2016. 24(4): p. 781–809. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, et al. , Role of magnetic resonance spectroscopy for the differentiation of recurrent glioma from radiation necrosis: a systematic review and meta-analysis. Eur J Radiol, 2014. 83(12): p. 2181–2189. [DOI] [PubMed] [Google Scholar]
- 38.Siu A, et al. , Radiation necrosis following treatment of high grade glioma--a review of the literature and current understanding. Acta Neurochir (Wien), 2012. 154(2): p. 191–201; discussion 201. [DOI] [PubMed] [Google Scholar]
- 39.Wang Q, et al. , The diagnostic performance of magnetic resonance spectroscopy in differentiating high-from low-grade gliomas: A systematic review and meta-analysis. Eur Radiol, 2016. 26(8): p. 2670–84. [DOI] [PubMed] [Google Scholar]
- 40.Cordova JS, et al. , Whole-brain spectroscopic MRI biomarkers identify infiltrating margins in glioblastoma patients. Neuro Oncol, 2016. 18(8): p. 1180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gurbani S, et al. , The Brain Imaging Collaboration Suite (BrICS): A Cloud Platform for Integrating Whole-Brain Spectroscopic MRI into the Radiation Therapy Planning Workflow. Tomography, 2019. 5(1): p. 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinberg BD, et al. , Clinical Applications of Magnetic Resonance Spectroscopy in Brain Tumors: From Diagnosis to Treatment. Radiol Clin North Am, 2021. 59(3): p. 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J, et al. , APT-weighted MRI: Techniques, current neuro applications, and challenging issues. J Magn Reson Imaging, 2019. 50(2): p. 347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamimura K, et al. , Amide proton transfer imaging of tumors: theory, clinical applications, pitfalls, and future directions. Jpn J Radiol, 2019. 37(2): p. 109–116. [DOI] [PubMed] [Google Scholar]
- 45.Warnert EAH, et al. , Mapping tumour heterogeneity with pulsed 3D CEST MRI in non-enhancing glioma at 3 T. MAGMA, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen W, et al. , 18F-FDOPA PET imaging of brain tumors: comparison study with 18F-FDG PET and evaluation of diagnostic accuracy. J Nucl Med, 2006. 47(6): p. 904–11. [PubMed] [Google Scholar]
- 47.Xiao J, et al. , Diagnostic and grading accuracy of (18)F-FDOPA PET and PET/CT in patients with gliomas: a systematic review and meta-analysis. BMC Cancer, 2019. 19(1): p. 767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klasner B, et al. , Early [18F]FET-PET in Gliomas after Surgical Resection: Comparison with MRI and Histopathology. PLoS One, 2015. 10(10): p. e0141153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh G, et al. , Radiomics and radiogenomics in gliomas: a contemporary update. Br J Cancer, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diehn M, et al. , Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A, 2008. 105(13): p. 5213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tiwari P, et al. , Computer-Extracted Texture Features to Distinguish Cerebral Radionecrosis from Recurrent Brain Tumors on Multiparametric MRI: A Feasibility Study. AJNR Am J Neuroradiol, 2016. 37(12): p. 2231–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ismail M, et al. , Shape Features of the Lesion Habitat to Differentiate Brain Tumor Progression from Pseudoprogression on Routine Multiparametric MRI: A Multisite Study. AJNR Am J Neuroradiol, 2018. 39(12): p. 2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soni N, Priya S, and Bathla G, Texture Analysis in Cerebral Gliomas: A Review of the Literature. AJNR Am J Neuroradiol, 2019. 40(6): p. 928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jain AK and Farrokhnia F, Unsupervised Texture Segmentation Using Gabor Filters. Pattern Recognition, 1991. 24(12): p. 1167–1186. [Google Scholar]
- 55.Haralick RM, Shanmugam K, and Dinstein I, Textural Features for Image Classification. Ieee Transactions on Systems Man and Cybernetics, 1973. Smc3(6): p. 610–621. [Google Scholar]
- 56.Alic L, et al. , Heterogeneity in DCE-MRI parametric maps: a biomarker for treatment response? Phys Med Biol, 2011. 56(6): p. 1601–16. [DOI] [PubMed] [Google Scholar]
- 57.Stadlbauer A, et al. , Magnetic resonance imaging biomarkers for clinical routine assessment of microvascular architecture in glioma. J Cereb Blood Flow Metab, 2017. 37(2): p. 632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Friedman JH, Fast sparse regression and classification. International Journal of Forecasting, 2012. 38(3): p. 722–738. [Google Scholar]
- 59.Tibshirani R High-dimensional regression. 2014. [cited 2020 6/23/2021]; Available from: https://www.stat.cmu.edu/~ryantibs/advmethods/notes/highdim.pdf.
- 60.Valueva MV, N. NN, Lyakhov PA, Valuev GV, Chervyakov NI,, Application of the residue number system to reduce hardware costs of the convolutional neural network implementation,. Mathematics and Computers in Simulation, 2020. 177: p. 232–243. [Google Scholar]
- 61.Auer TA, Advanced MR techniques in glioblastoma imaging-upcoming challenges and how to face them. Eur Radiol, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Auer TA, et al. , Quantitative volumetric assessment of baseline enhancing tumor volume as an imaging biomarker predicts overall survival in patients with glioblastoma. Acta Radiol, 2020: p. 284185120953796. [DOI] [PubMed] [Google Scholar]
- 63.Blystad I, et al. , Quantitative MRI using relaxometry in malignant gliomas detects contrast enhancement in peritumoral oedema. Sci Rep, 2020. 10(1): p. 17986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morris B, et al. , Identifying the spatial and temporal dynamics of molecularly-distinct glioblastoma sub-populations. Math Biosci Eng, 2020. 17(5): p. 4905–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elsheikh SSM, et al. , Multi-stage Association Analysis of Glioblastoma Gene Expressions with Texture and Spatial Patterns. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Brainles 2018, Pt I, 2019. 11383: p. 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kazerooni AF, et al. , Characterization of Active and Infiltrative Tumorous Subregions From Normal Tissue in Brain Gliomas Using Multiparametric MRI. Journal of Magnetic Resonance Imaging, 2018. 48(4): p. 938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kazerooni AF, et al. , Imaging signatures of glioblastoma molecular characteristics: A radiogenomics review. Journal of Magnetic Resonance Imaging, 2020. 52(1): p. 54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park JE, et al. , Low conductivity on electrical properties tomography demonstrates unique tumor habitats indicating progression in glioblastoma. European Radiology, 2021. [DOI] [PubMed] [Google Scholar]


