Abstract
Gut homeostasis is maintained by dynamic host-microbiota interactions. Recently, microRNAs (miRNAs) have emerged as key molecular regulators in mediating such interactions. Here, we discuss the role of a host miRNA-microbiome axis in gut homeostasis and colorectal cancer, and the involvement of diet and microbial metabolites in miRNA-mediated intestinal health.
Keywords: miRNA, microbiome, colorectal cancer, diet
Interactions between the host and gut microbiota are crucial for shaping gut homeostasis and immune response. Dysbiosis of gut microbiota has been linked to various gastrointestinal (GI) diseases, including colorectal cancer (CRC). It is clear that host-gut microbiota communication requires a complicated and elaborate regulatory network. Recently, microRNAs (miRNAs) have emerged as important mediators of this communication. MiRNAs are a type of short non-coding RNA (~of 21–25 nucleotides) that play important roles in gene regulation at both post-transcriptional and translational levels. In the past several years, many groups have extensively examined the functions of miRNA in immune cells and intestinal epithelial cells within the GI tract (reviewed by Runtsch, M.C et al. [1]). Moreover, the importance of miRNA in host-microbiota interactions and their impact on intestinal homeostasis and gut-associated cancers have also begun to be appreciated (Fig. 1).
Figure 1. Reciprocal regulation of miRNA and microbiota in CRC.
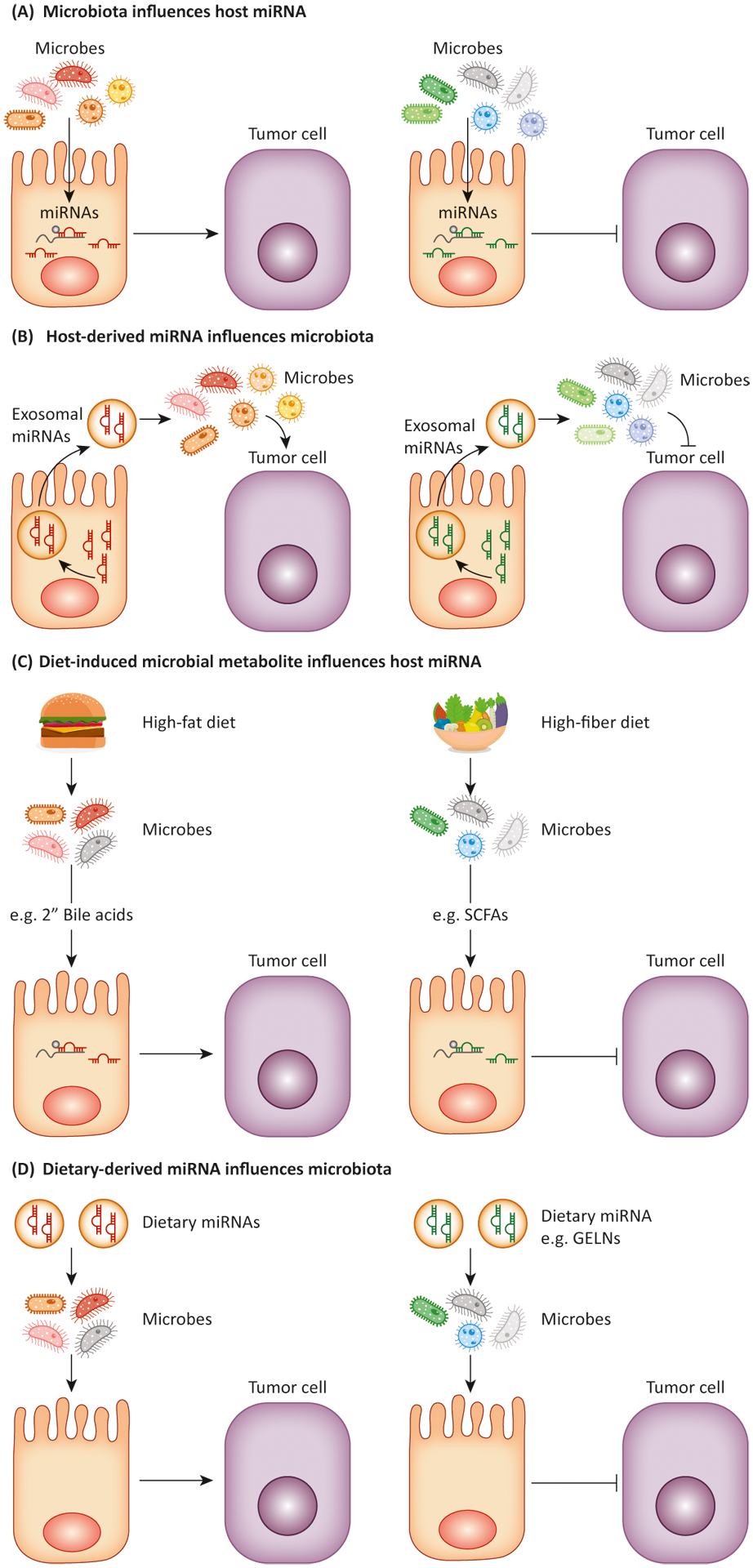
(A) Microbes can influence host miRNA expression and affect tumor cells. (B) Host cells release miRNAs via extracellular vesicles to control intestinal homeostasis and gut microbiota. (C) Diet-induced microbial metabolites, such as short-chain fatty acids (SCFAs) or secondary bile acids, can affect miRNA expression in host cells and to promote or inhibit tumor in CRC. (D) Dietary miRNAs, such as miR- 7267-3p from ginger exosome-like nanoparticles (GELN), have impacts on modulation of gut microbiota and intestinal homeostasis.
Microbial Impact on Host miRNA in CRC
A plethora of studies suggest a link between intestinal dysbiosis and colorectal cancer (CRC). Some have reported distinct microbiome “fingerprints” associated with different subtypes of CRC [2]. Others showed the specific enrichment of plausibly oncogenic microbes in CRC cases compared to controls. To this end, one microbe that has been heavily linked with CRC is Fusobacterium nucleatum. Recent literature has also implicated F. nucleatum’s effects on the host miRNome as one potential major contributor. In one study, F. nucleatum was shown to increase CRC cell proliferation and tumor growth in mice by up-regulating miR-21 in tumor cells by activating theTLR4-MyD88 signaling cascade [3]. Mechanistically, upon induction, miR-21 targeted and reduced protein levels of RASA1 and PDCD4, both of which are tumor suppressor genes. Consistently, patients with both high amount of tissue F. nucleatum DNA and miR-21 demonstrated a higher risk for poor outcomes. Moreover, F. nucleatum has also been reported to promote chemoresistance to CRC by modulating autophagy in a miRNA-dependent manner [4]. Interestingly, in addition to inducing the expression of miRNA such as the aforementioned miR-21, F. nucleatum infection also led to down-regulation of two miRNAs, miR-18a* and miR-4802, through triggering TLR4-MyD88 activation. As a consequence, two key components of the autophagy pathway, ULK1 and ATG7, which are targeted by miR-18a* and miR-4802 respectively, were up-regulated in CRC cells infected with F. nucleatum, thus preventing them from undergoing chemotherapy-induced apoptosis.
Like F. nucleatum, certain strains of Escherichia coli have also been shown to play a positive role in carcinogenesis of CRC through a miRNA-dependent fashion [5]. While E. coli is a normal component of the gut microbiota, the majority of E. coli isolated from CRC was found to produce colibactin, a genotoxic compound. Colibactin produced by these E. coli strains induced c-Myc expression likely through increasing DNA damage. In turn, high levels of c-Myc expression resulted in the up-regulation of miR-20a-5p which then targeted SENP1, a key negative regulator of p53 Small Ubiquitin-like Modification (SUMOylation). As a result, p53 SUMOylation, a known driver of senescence, drove a senescence-associated secretory phenotype (SASP) and the subsequent release of associated carcinogenic growth factors that promoted colon tumor growth [5].
Host miRNAs Influence Gut Microbiota and Intestinal Homeostasis
The studies discussed above have demonstrated a clear impact of gut microbiome on miRNAs and their target gene expression in CRC. Conversely, host-derived miRNAs, largely transferred via extracellular vesicles, can also influence gut microbiota. For example, it has been shown that CRC cells harboring mutant p53 selectively shed exosomes enriched with miR-1246, a potential biomarker for CRC [6]. Uptake of miR-1246-enriched exosomes by neighboring macrophages triggers their reprogramming into an anti-inflammatory state, which supports tumor survival. Furthermore, fecal miRNAs, especially those derived from intestinal epithelial cells (IECs), can also regulate host control of gut microbiota. MiR-515-5p and miR-1226-5p which are abundant in host fecal samples have been shown to promote the growth of F. nucleatum and E. coli by directly entering gut bacteria and regulating their gene expression [7]. Moreover, mice devoid of miRNAs specifically in IECs exhibited exacerbated colitis which could be rescued by WT fecal miRNA transplantation. As such, these findings revealed an essential role of host-derived miRNAs for the maintenance of normal gut microbiota and intestinal homeostasis.
While the precise role of fecal miRNAs in modulating gut microbes during CRC tumorigenesis remains poorly characterized, clinical studies have identified many CRC-specific fecal miRNAs as potential screening biomarkers [8]. Moreover, considering the aforementioned tumor-promoting role of F. nucleatum and E.coli in CRC, the fact that miR-515-5p and miR-1226-5p can influence the growth of these microbes further suggests that host miRNAs, especially those of which are differentially expressed in CRC, may also impact tumor growth through altering bacterial activities and the resultant intestinal microenvironment. Nevertheless, more extensive studies need to be done to establish the causal relationships between CRC-specific miRNAs and microbiome compositions, and to carefully characterize the mechanism by which individual miRNAs affect different gut microbes during cancer development.
Effects of Diet on miRNAs and Microbiome in Colorectal Carcinogenesis
The western diet, a blanket term used for high-fat, low-fiber diets common in industrialized countries, have been widely recognized as an important risk factor for CRC. However, while the molecular underpinnings responsible for the links between the western diets and colorectal carcinogenesis have been explored for decades, only recently has the role of miRNAs in such processes been appreciated. To this end, high-fat diets stimulate the release of hepatic primary bile acids. Upon entering the colon, bile acids are metabolized by the gut microbiota into genotoxic secondary bile acids, such as deoxycholic acid (DCA) [9]. While DCA can exert its pro-tumor function in many ways, recently, DCA was shown to promote CRC through down-regulation of miR-199a-5p expression [10]. Enforced miR-199a-5p expression in CRC cells could lead to the suppression of tumor cell growth and the restoration of the tumor cell drug sensitivity likely through inhibiting the expression of CAC1, a direct miR-199a-5p target and a driver of the cell cycle that is normally found to be highly expressed in CRC.
On the other hand, fiber can be metabolized by the gut microbiota into short chain fatty acids (SCFAs) such as acetate, propionate, and butyrate. While the anti-tumor role ofbutyrate as an inducer of p21, a potent cyclin-dependent kinase (CDK) inhibitor, has been previously attributed to its repression of histone deacetylases (HDACs) [11],it was reported that butyrate can also promote p21 expression through down-regulating the expression of miR-106b, a miRNA which targets p21[12]. Further investigation of miRNAs differentially expressed in butyrate-treated colon cancer cell line and in CRC tissues has further revealed that the expression of miR-92a, a known oncomiR, is inhibited by butyrate in a c-Myc-dependent manner, allowing p57, another CDK inhibitor that is a target of miR-92a, to exert its control overcell cycle progression in CRC [13].
Finally, in addition to impacting the expression of host-derived miRNAs, many food-derived exogenous miRNAs have been identified. This suggests diets themselves are a source of miRNAs that could regulate gut homeostasis. For example, it has been recently demonstrated that exosome-like nanoparticles (ELNs) containing miRNAs from edible plants can be taken up by the gut microbiota and regulate microbiome composition as well as host-gut barrier function [14]. Specifically, ginger ELN (GELN)-derived miR-7267-3p was shown to affect the expression of Lactobacillaceae rhamnosus (LGG) monooxygenase ycnE, leading to increased production of indole-3-carboxaldehyde (I3A), a ligand for aryl hydrocarbon receptor (AhR). As such, GELN-derived miRNAs induced the production of IL-22 that could ameliorate mouse colitis through improving barrier function. Whereas the impact of dietary miRNAs on CRC tumorigenesis remains to be investigated, these findings provide strong evidence to link food-derived miRNAs to microbiota in regulating host intestinal homeostasis.
Concluding Remarks
To date, accumulating evidence has pointed to miRNA as an important link in host-microbiota interactions to regulate gut health and CRC tumorigenesis [15]. Nevertheless, the causal relationships remain to be established, and whether the mechanisms identified in the previous investigation could have a significant impact on human CRC development require further examination. Moving forward, interdisciplinary research efforts combining the diverse fields of microbiology, immunology, and cancer biology, together with the application of advanced techniques such as next-generation sequencing, imaging and CRISPR/Cas-mediated genome editing are needed to bridge the gaps in current knowledge related to miRNA-mediated host-microbiome communications, and to provide crucial insights for developing novel therapeutic strategies to alleviate human intestinal malignancies.
Acknowledgments:
This work was supported by NIH grant AI108651 (L.-F.L).
References
- 1.Runtsch MC et al. (2014) MicroRNAs and the regulation of intestinal homeostasis. Front Genet 5, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burns MB et al. (2018) Colorectal cancer mutational profiles correlate with defined microbial communities in the tumor microenvironment. PLoS Genet 14 (6), e1007376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y et al. (2017) Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 152 (4), 851–866 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu T et al. (2017) Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 170 (3), 548–563 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cougnoux A et al. (2014) Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 63 (12), 1932–42. [DOI] [PubMed] [Google Scholar]
- 6.Cooks T et al. (2018) Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun 9 (1), 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S et al. (2016) The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe 19 (1), 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu CW et al. (2012) Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut 61 (5), 739–45. [DOI] [PubMed] [Google Scholar]
- 9.Ajouz H et al. (2014) Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol 12, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong Y et al. (2012) The deoxycholic acid targets miRNA-dependent CAC1 gene expression in multidrug resistance of human colorectal cancer. Int J Biochem Cell Biol 44 (12), 2321–32. [DOI] [PubMed] [Google Scholar]
- 11.Archer SY et al. (1998) p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc Natl Acad Sci U S A 95 (12), 6791–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu S et al. (2011) The microbe-derived short chain fatty acid butyrate targets miRNA-dependent p21 gene expression in human colon cancer. PLoS One 6 (1), e16221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu S et al. (2015) Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Mol Cancer 14, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teng Y et al. (2018) Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe 24 (5), 637–652 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan C et al. (2018) Interaction between Host MicroRNAs and the Gut Microbiota in Colorectal Cancer. mSystems 3 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]


