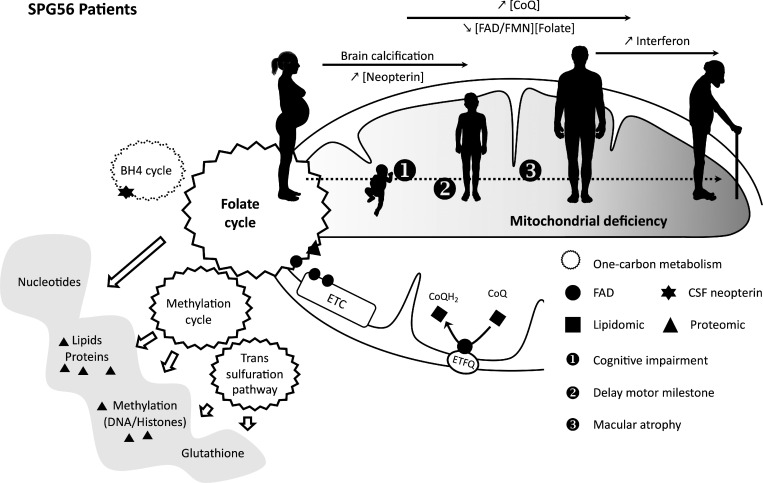
This study characterizes a Cyp2u1−/− mouse model and connects neurodevelopmental deficits to mitochondrial dysfunctions and folate, which could be prevented by folate supplementation in a mouse model. It also validates biochemical and imaging biomarkers in SPG56 patients, which are critical to allow drug development.
Abstract
Hereditary spastic paraplegias are heterogeneous neurodegenerative disorders. Understanding of their pathogenic mechanisms remains sparse, and therapeutic options are lacking. We characterized a mouse model lacking the Cyp2u1 gene, loss of which is known to be involved in a complex form of these diseases in humans. We showed that this model partially recapitulated the clinical and biochemical phenotypes of patients. Using electron microscopy, lipidomic, and proteomic studies, we identified vitamin B2 as a substrate of the CYP2U1 enzyme, as well as coenzyme Q, neopterin, and IFN-α levels as putative biomarkers in mice and fluids obtained from the largest series of CYP2U1-mutated patients reported so far. We also confirmed brain calcifications as a potential biomarker in patients. Our results suggest that CYP2U1 deficiency disrupts mitochondrial function and impacts proper neurodevelopment, which could be prevented by folate supplementation in our mouse model, followed by a neurodegenerative process altering multiple neuronal and extraneuronal tissues.
Graphical Abstract
Introduction
Hereditary spastic paraplegias (HSPs) are genetically and clinically heterogeneous neurodegenerative disorders (Boutry et al., 2019). Clinically, they manifest as a progressive spasticity in the lower limbs, the disorder’s hallmark, potentially in association with additional neurologic or extraneurologic features (Fink, 2013). Recent advances have strongly enhanced the understanding of HSP genetics, but our understanding of the pathogenic mechanisms remains sparse (Parodi et al., 2018).
In the present study, we investigated the pathophysiology of a severe autosomal recessive form of HSP caused by CYP2U1 pathogenic variants, namely spastic paraplegia 56 (SPG56; Tesson et al., 2012). The CYP2U1 gene encodes a member of the cytochrome P450 family, enzymes known to play key roles in tissue-specific conversion of natural substrates into locally active hormones, vitamins, and signaling molecules. So far, loss of function (LoF) mutations in CYP2U1 have been detected in 32 affected individuals (Table S1 and Table S2) who developed early-onset spastic paraplegia with a wide spectrum of clinical signs, including upper limb involvement, dystonia, cognitive impairment, and heterogeneous neuroimaging features of brain calcification and hypomyelination. Mutations in this gene have recently been implicated in a neurocutaneous syndrome (Legrand et al., 2021), and it has also been suggested that they could affect the retina (Leonardi et al., 2016). To better understand the pathophysiology of the disease and dissect its underlying mechanisms, we combined lipidomics and proteomics using biological material from known and novel SPG56 patients and from a genetically modified mouse model mimicking the disease.
Results and discussion
Cyp2u1−/− mouse model recapitulates the cognitive and retinal impairments observed in patients
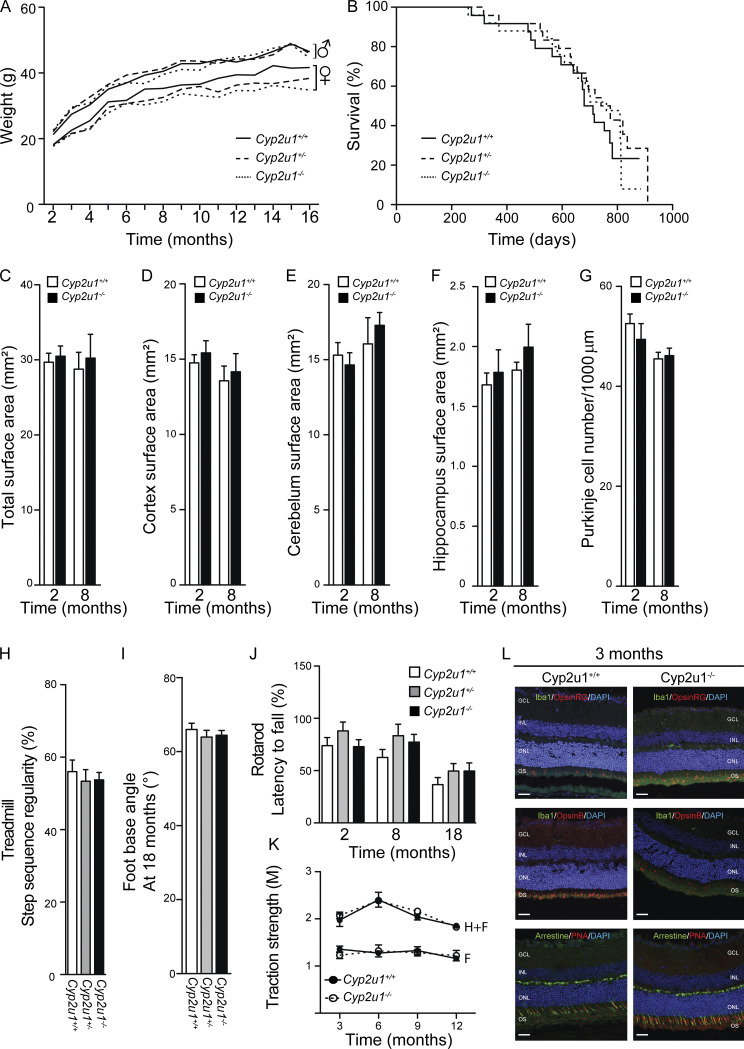
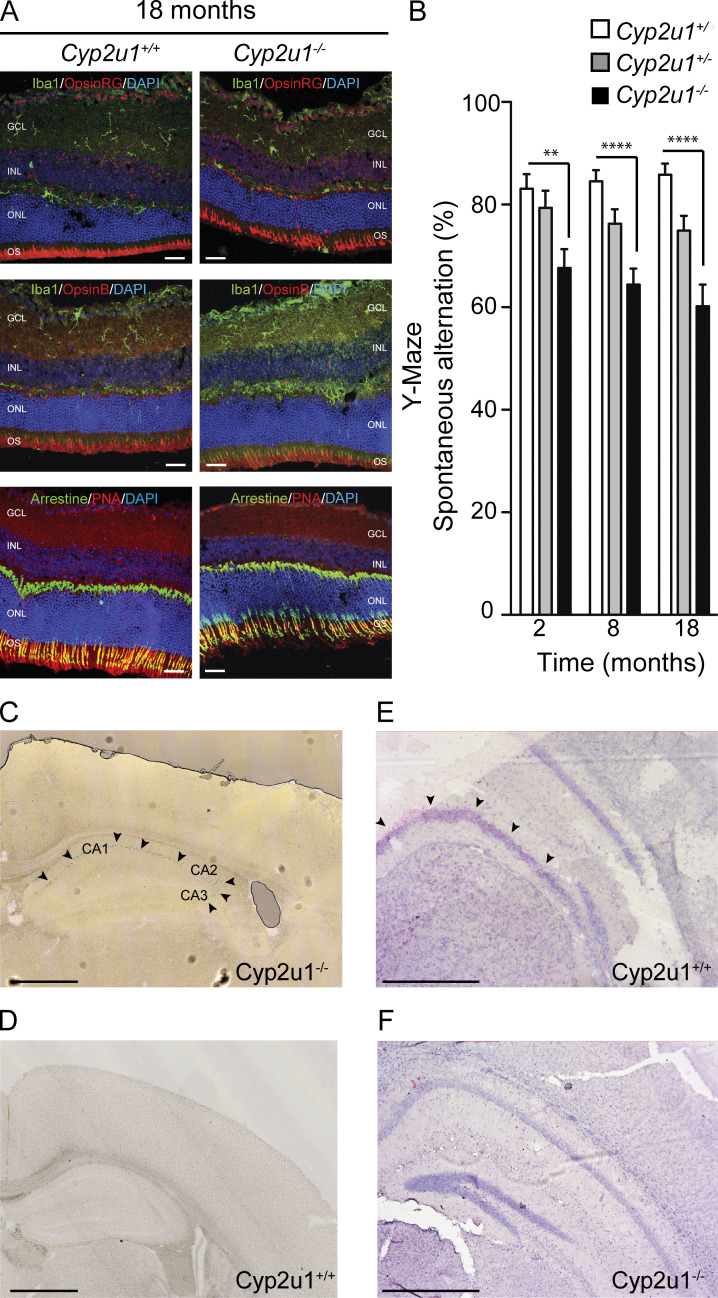
To delve into the cellular mechanisms underlying SPG56-HSP pathophysiology, we characterized the Cyp2u1−/− mouse from the Knockout Mouse Project (Lloyd, 2011) carrying a ubiquitous constitutive deletion in exon 2, resulting in the complete loss of the encoded protein (Tuck et al., 2015). There was no embryonic lethality based on perfect Mendelian ratios of the homozygous, wild-type, and heterozygous animals (n = 769; not depicted). The overall phenotype of Cyp2u1−/− mice was evaluated compared with wild-type and heterozygous littermates. No changes were detected in body weight (Fig. S1 A) or lifespan (Fig. S1 B). At the histological level, no obvious atrophy was detected in the tested brain regions (Fig. S1, C–G). Moreover, as in other HSP mouse models (Fink, 2013), Cyp2u1−/− mice did not display any significant locomotor impairment in four behavioral tests (Fig. S1, H–K). They exhibited, however, a late-onset (18 mo) ophthalmologic phenotype characterized by a cone dystrophy, as highlighted through opsin labeling, as well as an accumulation of microglia, as evidenced by IBA1 labeling at 18 mo (Fig. 1 A), but not in younger mice (Fig. S1 L). Such histologic features were reminiscent of those reported in animal models of macular degeneration (Pennesi et al., 2012). Finally, the Y-Maze test, used to assess short-term memory in mice, showed a significant cognitive deficit in Cyp2u1−/− mice from 2 mo of age onwards that slightly worsened over time (Fig. 1 B).
Figure S1.
Characterization of Cyp2u1−/− mice. (A) Mean body mass of female and male Cyp2u1+/+ (black line), Cyp2u1+/− (dashed line), and Cyp2u1−/− (dotted line) at the indicated times (n = 26 animals per genotype). (B) Kaplan–Meier survival curves for overall mortality of Cyp2u1+/+ (black line, n = 26), Cyp2u1+/− (dashed line, n = 30), and Cyp2u1−/− (dotted line, n = 34) mice. (C) Quantification of the total surface areas of brain coronal slices (sections cut between Bregma 0.90 and 0.60 mm) of Cyp2u1+/+ (white bars) and Cyp2u1−/− (black bars) mice at 2 and 8 mo of age (n ≥ 5 slices/animal and n ≥ 5 animals/genotype/age). (D) Quantification of the surface areas of the cortex (sections cut between Bregma 0.90 and 0.60 mm) of Cyp2u1+/+ (white bars) and Cyp2u1−/− (black bars) mice at 2 and 8 mo of age (n ≥ 5 slices/animal and n ≥ 5 animals/genotype/age). (E) Quantification of the cerebellum surface areas of brain coronal slices (sections cut between Bregma −6.00 and −6.30 mm) of Cyp2u1+/+ (white bars) and Cyp2u1−/− (black bars) mice at 2 and 8 mo of age (n ≥ 5 slices/animal and n ≥ 5 animals/genotype/age). (F) Quantification of the hippocampal surface-areas of brain coronal slices (sections cut between Bregma −1.70 and −2.00 mm) of Cyp2u1+/+ (white bars) and Cyp2u1−/− (black bars) mice at 2 and 8 mo of age (n ≥ 5 slices/animal and n ≥ 5 animals/genotype/age). (G) Cerebellar sections immunostained for GFAP (astrocyte marker), Calbindin (Purkinje cell marker), and Hoechst-33258 (nucleus marker) allowed the quantification of Purkinje cells in Cyp2u1−/− mice (black bars) compared with control (white bars). (H) Step sequence regularity values recorded during a forced walk on a treadmill at moderate speed (10 cm/s) of 18-mo-old Cyp2u1+/+ (white bars, n = 7), Cyp2u1+/− (gray bars, n = 6), and Cyp2u1−/− (black bars, n = 6) mice. Values shown are mean ± SEM. (I) Measurement of the foot/base angle for 18-mo-old Cyp2u1+/+ (white bars, n = 8), Cyp2u1+/− (gray bars, n = 7), and Cyp2u1−/− (black bars, n = 7) mice. Values shown are mean ± SEM. (J) Rotarod duration of Cyp2u1+/+ (white bars), Cyp2u1+/− (gray bars), and Cyp2u1−/− (black bars) mice at 2 mo (n = 16/genotype), 8 mo (n = 24/genotype), and 18 mo (n = 18/genotype). Values shown are mean ± SEM. (K) Grip test of Cyp2u1+/+ (black circle) and Cyp2u1−/− (white circle) mice to test the muscular strength in both forelimbs (F) and hindlimbs (H) from 3–12-mo-old compared with control mice (n > 4 for 2/6/9 mo/genotype and n = 2 for 12-mo/genotype). (L) Immunofluorescence analysis on vertical sections of 3-mo-old wild-type and Cyp2u1−/− mice retinas stained for Iba1/Arrestin (green) and OpsinRG/OpsinB/PNA (red) with DAPI counterstaining (blue). Exemplary images are shown from at least three experimental repetitions. With outer nuclear layer (ONL), inner nuclear layer (INL), ganglion cell layer (GCL), and outer segments (OS). Scale bar: 100 µm.
Figure 1.
Characterization of Cyp2u1−/− mice. (A) Immunofluorescence analysis on sections of vertical retinas of 18-mo-old mice stained for Iba1/Arrestin (green) and OpsinRG/OpsinB/PNA (red) with DAPI counterstaining (blue); n = 2. With outer nuclear layer (ONL), inner nuclear layer (INL), ganglion cell layer (GCL), and outer segments (OS). Scale bar: 100 µm. (B) Spontaneous alternation behavior in the Y-Maze task at various times, showing impaired spontaneous alternation worsening with time in Cyp2u1−/− mice. Data pooled from at least two independent experiments with n = 16/genotype. Data are mean ± SEM. Unpaired two-tailed Student t test. **, P < 0.01; ****, P < 0.0001. (C–F) Expression of the Cyp2u1 gene in 2-mo-old mouse hippocampus through histochemical analysis of β-gal expression under Cyp2u1 promoter by X-gal staining, pinpointed by arrows in Cyp2u1−/− mice (C). In D, note the absence of staining in Cyp2u1+/+ mice. RNAscope (pink labeling) targeting Cyp2u1 mRNA transcripts in wild-type (E) or knockout (F) animals. Note the absence of staining in Cyp2u1−/− mice. Exemplary images are shown from at least two independent experiments. Scale bar: 1 mm.
CYP2U1 expression is detectable only in the hippocampus of young mice
We used histochemical staining for LacZ enzymatic activity to analyze CYP2U1 expression, taking advantage of the reporter β-galactosidase (lacZ) gene introduced in our Cyp2u1−/− mouse model downstream of the endogenous promoter of Cyp2u17,8. The protein expression was low and detectable only in the hippocampus cornu ammonis pyramidal cells (CA1–3) at 2 mo of age (Fig. 1 C and D). We then used RNAscope technology to increase the sensitivity and specificity of detection. The restricted hippocampal detection of Cyp2u1 mRNA was confirmed in wild-type mice (Fig. 1 E) and absent in Cyp2u1−/− 2-mo-old mice, as expected (Fig. 1 F). Our results are not in agreement with a more widespread expression of CYP2U1 according to previous reports, probably resulting from different methodologies used. The requirement for CYP2U1 in specific tissues and developmental stages should be compensated by other factors including additional Cytochromes P450 (CYP; Balasubramaniam et al., 2019). The hippocampal expression is also in accordance with the observed cognitive impairment in our mouse model and is in line with previous studies linking hippocampus and cognitive tasks (Albani et al., 2014).
Increased coenzyme Q (CoQ) content and deregulated expression of MTHFD1L in Cyp2u1−/− mouse hippocampus
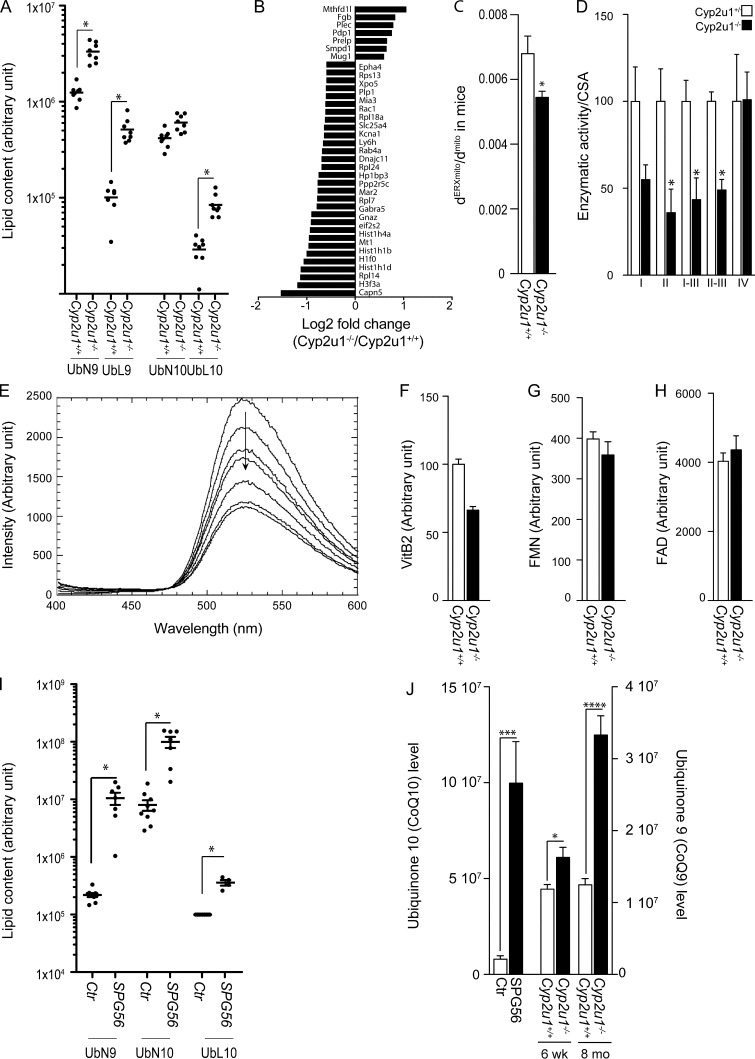
CYP2U1 hydroxylates lipids, particularly arachidonic acid (Chuang et al., 2004). To investigate this aspect, we performed a lipidomic analysis of 2-mo-old mouse hippocampi. We obtained a very peculiar profile exhibiting a significant increase of different CoQ species (Fig. 2 A), whereas the arachidonic acid level remained unchanged. These results were replicated in cerebellar extracts (data available on https://doi.org/10.5281/zenodo.5499883). CoQ is a crucial lipid that serves as an antioxidant and plays a role as an electron carrier in the mitochondrial respiratory chain. This increase was not associated with an increase of its biosynthesis, as we did not detect any significant changes in enzymes involved in the pathway (Fig. S2 B), at either the RNA (Fig. S2 C) or protein level (Fig. S2 D). A decrease of CoQ degradation is unlikely, as the majority of nonmetabolized CoQ is usually excreted in urine and feces, so an increased CoQ biosynthetic rate could be the explanation. While a decrease in CoQ levels has already been associated with several neurodegenerative disorders (Acosta et al., 2016; Salviati et al., 2017), our study is the first to report increased CoQ levels in a neurologic disease.
Figure 2.
Molecular and biochemical characterization of SPG56 patients and Cyp2u1−/− mice. (A) Representation of the relative amounts of various classes of CoQ (ubiquinon 9 and 10 [UbN9, UbN10], ubiquinol 9 and 10 [UbL9, UbL10]) in the hippocampus of 2-mo-old Cyp2u1+/+ and Cyp2u1−/− mice (n = 8/genotype). Arbitrary units. Nonparametric Kruskal–Wallis test without correction for multiple testing from log10-transformed data. *, P values below a threshold set at 0.05. (B) Proteomic analysis in wild-type and knockout mice. Representation of the log2 fold-change to show the functional enrichments of proteins up- and down-regulated in Cyp2u1−/− vs. wild-type hippocampus of 2-mo-old mice (n = 6/genotype). (C) Quantitative analysis of the mitochondrial surface closely apposed to ER in control (n = 2; 170 contacts, white bar) and Cyp2u1−/− (n = 2; 410 contacts, black bar) mice. Unpaired two-tailed Student’s t test. *, P < 0.05. (D) Enzymatic activity of electron transport chain complexes normalized to citrate synthase activity (CSA) in brain mitochondria of 12-mo-old mice (n = 4/genotype). Unpaired two-tailed Student’s t test. *, P < 0.05 between wild-type (white bars) and knockout (black bars) animals. (E) Typical experiment showing quenching of fluorescence of 10 µM riboflavin by increasing concentrations of CYP2U1 in vitro (0–8 µM). (F–H) Metabolomic analysis to measure the relative amounts of vitamin B2 (VitB2; F), FMN (G), and FAD (H) from hippocampus of Cyp2u1+/+ (white bars) and Cyp2u1−/− (black bars) 2-mo-old mice (n = 4/genotype). (I) Representation of the relative amounts of various classes of CoQ (ubiquinon 9 and 10 [UbN9, UbN10], ubiquinol 10 [UbL10]) in plasma samples of 7 SPG56 patients and 9 age/gender-matched controls. For UbL10, the values for the controls were below the detection threshold, and an arbitrary value corresponding to the detection threshold was given to the control for the representation. Arbitrary units. P values using the nonparametric Kruskal–Wallis test without correction for multiple testing from log10-transformed data. *, P values below a threshold set at 0.05. (J) Representation of the main oxidized CoQ form; CoQ10 in 7 SPG56 patients (black bars) and 9 age/gender matched controls (white bars); CoQ9 in hippocampus of Cyp2u1+/+ (white bars) and Cyp2u1−/− (black bars) mice at 6 wk and 8 mo of age (n = 8). *, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Figure S2.
Molecular and biochemical characterization of SPG56 patients and Cyp2u1−/− mice. (A) Quantitative analysis of the mitochondrial surface closely apposed to ER in control and SPG56 patients’ fibroblasts (n = 33/genotype). *, P ≤ 0.05. (B) Schematic representation of CoQ biosynthesis in mammalian cells. (C) Transcript levels of genes encoding enzymes of the CoQ synthesis pathway in Cyp2u1+/+ (white bars) and Cyp2u1−/− (black bars) with n = 4/genotype and normalization with β-2-microglobulin. (D) Levels of the biosynthetic proteins COQ5, COQ6, and COQ9 in hippocampus homogenates of 2- and 8-mo-old mice (n = 3/genotype). Tubulin was used as a loading control. Exemplary images are shown from at least two independent experiments. MW, molecular weight. (E) Steady-state levels of different OXPHOS system subunits determined by Western blot analysis of brain mitochondria from control and Cyp2u1−/− mice at age 12 mo (n = 2/genotype). Exemplary images are shown from at least two independent experiments. (F and G) BN-PAGE using digitonin, followed by in-gel activity assay (F) and immunoblotting (G) used to define brain mitochondrial complexes and SCs in wild-type and Cyp2u1−/− mice (n = 3/genotype). (F) In-gel activity (IGA) of CI (left) and CIV (right). (G) Western blot analysis of levels of NDUFA9 (CI), Fp subunit (CII), subunit core 1 (CIII), subunit I (CIV), and the α-subunit of ATP synthase (complex V). (H) Effects of increased concentrations of recombinant CYP2U1 on riboflavin fluorescence. Experiments were conducted in 50 mM phosphate buffer containing 10 µM of riboflavin, alone (black circles) or in the presence of 1 µM (red squares), 2 µM (empty squares), 4 µM (violet squares), and 8 µM (pink circles) of purified CYP2U1. λex = 385 nm; λem = 520 nm. Excitation and emission slit widths were 15 and 20 nm, respectively. (I) Replot of the percentage of riboflavin fluorescence in the presence of increasing concentrations of CYP2U1. Data are representative of a typical experiment. (J) Representative pose showing the positioning of riboflavin in close proximity with the heme in a 3D model. Distances between the two methyl groups of riboflavin and the iron are 8.38 and 6.03 Å. CDOCKER interaction energy = −54.36 kcal/mol. (K) Representative pose showing the positioning of riboflavin in close proximity with the heme. Distance between the NH group of riboflavin and the iron is 6.35 Å. CDOCKER interaction energy = −52.07 kcal/mol).
To link the pathogenic mechanisms to the increase of CoQ levels, we performed an unbiased proteomic analysis comparing 2-mo-old Cyp2u1−/− to control mouse hippocampi. This analysis showed that 37 proteins were significantly altered. Among them, the expression of MTHFD1L (mitochondrial methylenetetrahydrofolate dehydrogenase 1-like), an important player in the mitochondrial folate cycle, was significantly deregulated in the hippocampus of Cyp2u1−/− mice (Fig. 2 B). Abnormal folate metabolism is associated with neurodevelopmental and neurodegenerative phenotypes, especially psychomotor delay, HSP, epilepsy, and psychiatric symptoms (Zheng and Cantley, 2019). Folate is crucial for many cellular pathways, including mitochondrial respiration and epigenetic regulation (Zheng and Cantley, 2019). Its involvement in all these reactions could explain the dysregulation of seven histones and six cytosolic ribosomal subunits, also observed to be deregulated in our proteomic analysis (Fig. 2 B). Our results point toward a so-far underestimated role of folate in SPG56 pathophysiology.
CYP2U1 LoF impacts mitochondrial functions
At a cellular and molecular level, relatively little is known about CYP2U1. The protein was shown to be localized to both the ER and mitochondria (Tesson et al., 2012). We used electron microscopy (EM) to investigate these organelles (Fowler et al., 2019; Scorrano et al., 2019). A decrease of the ER–mitochondria contacts was observed, both in the brain of Cyp2u1−/− mice (Fig. 2 C) and in patients’ fibroblasts (Fig. S2 A). We then measured the mitochondrial respiratory chain complex activity, known to be essential in the nervous system (Letts and Sazanov, 2017). Even if any of the oxidative phosphorylation (OXPHOS) components can be implicated, complex I (CI) deficiency is the most common cause of mitochondrial disease. Unexpectedly, mitochondria from Cyp2u1−/− mouse brain presented severe deficiencies in the respective capacity to oxidize nicotinamide adenine dinucleotide (NADH) and succinate (Fig. 2 D), although no changes in the steady-state levels of the CI subunit were observed (Fig. S2 E). This mitochondrial deficit associated with the increase in CoQ led us to analyze the organization of supercomplexes (SCs). Indeed, mitochondrial respiratory chain complexes are dynamic entities located in the inner membrane that aggregate in different combinations to form SCs (Letts and Sazanov, 2017). The clinical relevance of SC biology is now evident, since defective assembly of SCs has been correlated with severe cellular dysfunction (McKenzie et al., 2006). We then used blue native PAGE (BN-PAGE), preserving interactions, to explore if rearrangements of the SCs occurred in mitochondria depleted of CYP2U1. No changes were observed in brain extracts from our model when compared with controls (Fig. S2, F and G).
CYP2U1 hydrolyzes vitamin B2
CYP450 has crucial roles in lipid and vitamin metabolism (Balasubramaniam et al., 2019). According to our previous results, we hypothesized that CYP2U1 could be involved in vitamin, and particularly vitamin B2 (riboflavin), metabolism. Yet vitamin B2 is a precursor of flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), key players in critical processes, including the mitochondrial electron transport chain, with FMN acting as a CI cofactor and FAD being involved in complex II (CII) activity (Mosegaard et al., 2020). They are also involved in the metabolism of other essential cofactors and hormones, such as CoQ10 and folate (Balasubramaniam et al., 2019). We then hypothesized that CYP2U1 can hydroxylate vitamin B2. We first performed in vitro binding experiments based on riboflavin fluorescence quenching after binding of riboflavin in close proximity to the heme (Shumyantseva et al., 2004), which led to an apparent dissociation constant of 9.0 ± 0.2 µM, a value close to that observed in the case of CYP2B4 (Fig. 2 E and Fig. S2, H and I; Shumyantseva et al., 2004). We then assessed CYP2U1 capacity to interact with vitamin B2, using a 3D homology model (Dhers et al., 2017), since no crystal structure is yet available for CYP2U1. Docking experiments showed that two families of poses could lead to an oxidation of vitamin B2. In a series of poses, the two methyl groups of riboflavin were found at 8.38 and 6.03 Å of the iron of the heme, whereas in another series of poses, the NH group was found at 6.3 Å of the heme (Fig. S2, J and K). These binding and modeling experiments suggested that vitamin B2 could be oxidized by CYP2U1 at one of these sites. In support of the predicted CYP2U1–vitamin B2 interaction, a decreased tendency of vitamin B2 and FMN levels, but not of FAD ones, was observed in the hippocampi of Cyp2u1−/− mice, although not significant (Fig. 2, F–H).
Extension of the pathologic process to SPG56 patients
To date, only 32 SPG56 patients have been described (Table S2). In this study, we collected a cohort of 23 patients, adding 19 new cases to the literature (Table S1 and Table S2). Most cases presented with an early onset (<3 yr old) and delayed motor milestones. Despite predominant lower limb manifestations, 13 of 23 patients also presented with upper limb involvement. Other neurologic features, such as cognitive impairment (17 of 23), were observed in combination with abnormalities detected through neuroimaging. Interestingly, all adult patients for whom ophthalmologic examination was possible developed macular atrophy (Table S1), generalizing previous occasional observations (Leonardi et al., 2016).
We were able to collect blood from seven patients, which was subjected to lipidomic analysis. As previously observed in Cyp2u1−/− mice, SPG56 patients exhibited a significant increase in blood levels of CoQ10, as well as those of its intermediate CoQ9 (Fig. 2 I), whereas arachidonic acid levels were in the normal range. The changes in CoQ levels were not detected in 300 samples of other neurodegenerative conditions. Because CoQ works as an electron carrier, it can reversibly change to either its oxidized (i.e., ubiquinone or CoQ) or reduced (i.e., ubiquinol or CoQH2) form. In humans, CoQ10 mainly exists in its reduced form in mitochondria and plasma. It requires a low potential for its oxidation, thus assuming a pivotal role as an antioxidant. The CoQH2/CoQ ratio is a key indicator of CoQ redox status. In patients, this ratio was massively altered due to a substantial 12-fold increase of the oxidized form as compared with controls (Fig. 2 J), indicating that the reduction of CoQ might not be properly ensured by the FAD/FMN-dependent CoQ oxidoreductases (electron transfer flavoprotein-ubiquinone oxidoreductase).
To assess riboflavin/FMN and FAD levels in SPG56 patient plasma, we performed a metabolomic analysis (Table S1). The values obtained were all in the lowest part of the normal range, suggesting a possible impact of CYP2U1 LoF on vitamin B2 metabolism.
Results obtained in our murine model pointed toward a key role of the folate pathway in the disease, with deregulation of MTHFD1L in proteomic analysis of Cyp2u1−/− mice. FMN/FAD cofactors have been shown to play an important role in one-carbon metabolism, and FAD acts as a MTHFD1L cofactor (Fig. 3 A). Thus, we investigated peripheral and central hallmarks of folate deficiency in patients. Folate levels were reduced in both SPG56 patients (Table S1) and Cyp2u1−/− mouse serum (Fig. 3 B). In agreement with this finding, we observed a high level of neopterin in the cerebrospinal fluid (CSF) of young SPG56 patients, in the absence of infections, as assessed by normal IFN levels (Table S1). Indeed, multiple studies provide indirect evidence of an interconnection between folate and two main metabolites of the tetrahydrobiopterin (BH4) pathway, biopterin and neopterin (Blau et al., 2003; Ghezzi and Zeviani, 2018). The high CSF levels of neopterin are then likely markers of folate deficiency in our patients. The association between the lack of folates and CYP2U1 LoF was further supported by neuroimaging studies. It has previously been reported that folate deficiency may be associated with white matter lesions, cortico-subcortical atrophy, and basal ganglia calcifications (Nicolai et al., 2016). In our cohort, brain magnetic resonance imaging (MRI) revealed white matter lesions at a deep/periventricular level with a radial distribution in 15 patients, similar to earlier studies (Tesson et al., 2012). Patients also presented with cortical and cerebellar atrophy. In addition, we found that computed tomography (CT) revealed the presence of calcifications in all patients imaged in this study, in basal ganglia, periventricular white matter, and cortico-subcortical areas (Table S1 and Fig. 3 C). Notably, the availability of consecutive neuroimaging studies performed in several patients at different stages of the disease provided evidence that those changes were dynamic and could be followed over time, thus serving as an additional marker of disease evolution (Fig. 3 C). These observations reinforce the role of folate deficiency in SPG56 disease, since brain calcifications are a known feature of cerebral folate deficiency (Nicolai et al., 2016).
Figure 3.
Folate implication in disease. (A) Schematic representation of FAD/FMN implications in mitochondria. A, FAD; M, FMN; MTR, methionine synthase reductase; THF, tetrahydrofolate; VitB2, vitamin B2. (B) Folate measurement in the plasma of mice (data pooled from at least three independent experiments with n = 16/genotype). (C) Head CT and brain MRI scans of five SPG56 patients. CT of patient 4 at 1 yr of age (a) showing subcortical calcifications with nodular and linear shape in frontal, parietal, and occipital lobes (arrows). Brain MRI (b–d) of the same patient at age 15. Pathological hypersignal in magnetization transfer T1 axial sequence (b) in bilateral pallidus (arrows), coupled with T2* hyposignal in the same region (c), suggestive of calcium deposition (arrows). Axial FLAIR sequences evidence confluent hyperintensities in deep periventricular white matter (d) with a radial distribution with respect to the lateral ventricles (arrowheads). Larger and confluent calcifications are evident in the brain CT (e) of his sibling (patient 3) at age 2 yr, especially in subcortical white matter at the level of the frontal and occipital horns (arrows). Head CT of patient 13 (f and g) at age 9 yr showing millimetric subcortical calcifications in left frontal and parietal lobes (arrows). Axial FLAIR sequence from brain MRI of patient 12 (h) at age 9 yr showing a nodular white matter hyperintensity in the right frontal lobe (arrowhead). Head CT of patient 12 at age 26 yr (i and j) evidencing minute cortico-subcortical calcifications in frontal lobes and massive confluent calcifications involving the lenticular nucleus bilaterally (arrows). Brain MRI of patient 2 at different time points (k–o). Bilateral pallidal hyposignal, likely due to calcifications, at age 37 yr (axial T2* sequence; k, arrows), along with deep periventricular white matter hyperintensities (axial FLAIR sequence; l, arrowheads), increasing in size and number over time, as seen at age 44 yr (axial FLAIR sequence; m, arrowheads). Moderate cortical atrophy, most notably at frontal level (axial T1 sequence; n), and significant cerebellar atrophy (sagittal T1 sequence; o) were clearly evident at age 44 yr as well (arrows). (D) Spontaneous alternation behavior in the Y-Maze task by 2-mo-old mice complemented with folate. Data pooled from at least two independent experiments with n = 11/genotype. Folate supplementation improved this behavior in Cyp2u1−/− animals only with n (%) spontaneous alternation behavior in Cyp2u1−/− mice without folate complementation. Values are mean ± SEM.
Folate supplementation prevents cognitive deficit in our mouse model
To further confirm our hypothesis that a dysfunctional folate cycle during neuronal development may be associated with SPG56 onset, we supplemented Cyp2u1−/− mice, as previously done (Bryant et al., 2018; Momb et al., 2013). Pregnant dams were given ad libitum access to water containing sodium formate until the litter was weaned and the animals could access the water on their own. At 2 mo of age, behavioral tests were performed. Supplementation prevented the cognitive deficit of Cyp2u1−/− mice, as shown by Y-Maze test results (Fig. 3 D), with no effect in control mice. These results strengthen our assumption that lack of folate during neuronal development is a key feature in the development of cognitive alterations in SPG56.
Animal modeling of human diseases is a cornerstone for basic scientific studies of disease mechanisms and preclinical studies of potential therapies. This is especially true for HSP, whose remarkable genetic heterogeneity is a major challenge to understand its pathophysiology. CYP2U1 has a restricted spatiotemporal expression level and profile, limited to a few cells in the hippocampus of young mice. At a cellular level, its localization at the mitochondria and ER junctions (Chuang et al., 2004) is in agreement with our EM, lipidomic, and proteomic analyses showing a loss of CYP2U1 impacting mitochondrial activity in both rodents and humans, notably with a decrease in CI/II without impact on SCs, as well as reducing ER–mitochondria contacts called mitochondria-associated membranes. Mitochondria-associated membranes are essential for various processes, such as lipid import or apoptosis, whose dysfunction is closely associated with diseases (Scorrano et al., 2019). More importantly, our results link this mitochondrial impairment to a defect in vitamin B2 homeostasis. The findings led us to hypothesize that the disease has at least two components: a neurodevelopmental component related to folate metabolism and a neurodegenerative component related to mitochondrial alterations with age.
Even if our mouse model could not fully reproduce the whole spectrum of the disease, it recapitulates cognitive deficiency and photoreceptor degeneration, and the most significant impact of CYP2U1 deficit was during neurodevelopment. It was related to folate deficiency, as the early cognitive deficit could be corrected through adequate supplementation in the mouse model. Unlike global folate deficiency associated with neural tube defects or craniofacial malformation, loss of CYP2U1 expression led to cognitive impairment, as suggested by the prevention of cognitive deficit when mice were supplemented with folate during development. While mitochondria supply energy by the electron transport chain and ATP synthase, there is a growing body of evidence demonstrating that they also play a critical role in one-carbon metabolism through folate metabolism. Indeed, the folate cycle is the launch point for several interconnected metabolic pathways such as the methylation cycle and the trans-sulfuration pathway (Desai et al., 2016) required for the biosynthesis of a wide range of metabolites. In our model, while some pathways were altered (neopterin, protein translation, and histone), others were unlikely affected (sulfuration pathway with homocysteine level; Table S1). In SPG56 patients, different factors modulating folate metabolism may explain the clinical variability, even for the same mutation (Table S1), including genetic factors, such as common single-nucleotide polymorphisms in genes related to folate metabolism and/or environmental factors, as folate is the naturally occurring form of vitamin B9 present in a wide variety of foods. In this respect, analyzing environmental factors and genetic variants in a large cohort of patients and their mothers could provide prognostic markers and help in adjusting patient care. Of note, ophthalmologic alterations were also observed in old Cyp2u1−/− mice, while not visible earlier in life (Fig. 1 A and Fig. S1 L), a feature also seen in SPG56 patients, especially macular atrophy in all adult patients for whom information was available (Table S1; Legrand et al., 2021). Even though further investigation on these late-onset ophthalmologic alterations should be considered, this aspect of the phenotype may be due to a neurodegenerative part of this disease.
Indeed, a key point of this report is the CoQ alteration in SPG56 patients, who presented a 12-fold increase of the oxidized form compared with controls. We also observed this increase in Cyp2u1−/− mice, and it worsened over time. CoQ constitutes a key component of the mitochondrial respiratory chain but, in its reduced form, also exerts an essential antioxidant role. Then, accumulation of the oxidized form in the absence of CYP2U1 could enhance oxidative stress, leading to neurodegeneration and macular diseases. More recently, dysfunctional mitochondria were also linked to aberrant innate immune responses through the expression of a subset of IFN-stimulated genes (West et al., 2015). Such an IFN-mediated response is also found in the genetic encephalopathy Aicardi–Goutières syndrome (Crow and Manel, 2015). Remarkably, we found an abnormal IFN signature in two adult SPG56 patients that was absent in young cases (Table S1). Altogether, these results allow us to speculate that accumulation of mitochondrial dysfunction in SPG56 patients could lead to excessive activation and induction of the immune response. This needs to be confirmed in larger series of older patients, however.
In conclusion, SPG56/CYP2U1 is a severe neurological condition for which appropriate clinical diagnosis is both a growing concern and a difficult challenge. In our study, we were able to define more precisely the symptoms found in SPG56 patients, to show the systematic presence of basal ganglia calcifications. We also identified potential biomarkers of the disease, i.e., increased blood CoQ, CSF neopterin, and IFN-α. These possible biomarkers may have a critical role in helping to better understand the affected molecular pathways and improve drug development strategies in SPG56, such as treatment with folic acid, whose efficacy has already been reported in treating movement disorders in adults (Masingue et al., 2019) and is demonstrated in the cognitive aspects of our mouse model. Its effect on motor and retinal aspects of the disease could not be tested in our model, however. Finally, these advances in our understanding of SPG56 could also benefit and provide new insights into other overlapping disorders sharing impaired cellular pathways, such as macular atrophy and interferonopathy.
Materials and methods
Mice
All mice were maintained on a C57BL/6N genetic background. Mice harboring a floxed conditional knockout cassette overlapping exon 2 of Cyp2u1 were obtained from the Knockout Mouse Project (Cyp2u1tm1a(EUCOMM)Wtsi). Floxed mice were mated with mice expressing Cre recombinase under control of the cytomegalovirus promoter (CMV-Cre; genOway) to generate mice lacking exon 2 of Cyp2u1 ubiquitously and where expression of the bacterial β-galactosidase reporter gene (lacZ) is driven by the endogenous promoter of Cyp2u1. Genotyping was performed according to the genotyping protocol. It was performed by PCR with the 5′-GTAGGAGAGGTCTCTACAGAGGTG-3′ and 5′-CCAACAGCTTCCCCACAACGG-3′ primers to amplify a 292-bp amplicon from the KI allele. To detect the wild-type allele, the primers 5′-GGGTAAAGGCAGGTTCTTTGAACCC-3′ and 5′-CCTCTCTACACGCCACTTCTCTAAC-3′ were used. Cre recombinase was detected using 5′-GCATTACCGGTCGATGCAACGAGTGATGAG-3′ and 5′-GAGTGAACGAACCTGGTCGAAATCAGTGCG-3′ to produce a 408-bp amplicon. All mice were given ad libitum access to water and standard mouse chow (LabDiet 5K67).
Study approvals
The care and treatment of animals were in accordance with European Union Directive 2010/63/EU and with national authority (Ministère de l’Agriculture, France) guidelines for the detention, use, and ethical treatment of laboratory animals. All the experiments were approved by the local ethics committee (APAFIS, reference 2018070316423884), and experiments were conducted by authorized personnel in the in-house specific pathogen–free animal facility (agreement reference B75-13-19; CEEA-005).
Human
This study included 23 HSP-affected patients from the SPATAX research network for whom clinical information based on neurologic examinations and genetic counseling was available. All patients included in the present study gave their consent for DNA testing in the research setting, and consent forms (RBM 01-29) were signed by each participant. Patient-derived material (blood and skin biopsies) was obtained through procedures approved by the Paris-Necker ethics committee with the written, informed consent of each family member (approval SST04/11/04052011).
Behavioral evaluation
All tests were performed as previously described (Branchu et al., 2017). In short, all behavioral procedures were performed between 8:00 a.m. and 1:00 p.m. Bias was minimized by randomly testing mice in experimental cohorts, regardless of their genotype.
The foot base angle test was performed as described previously (Irintchev et al., 2005). Mice were accustomed, in three or four trials, to beam walking before operation. In this test, the animal walks unforced from one end of a horizontal beam (length 1,000 mm, width 40 mm) toward its home cage located at the other end of the beam. For all mice, rear views of walking trials were captured at 12 mo. Selected frames in which the animals were seen in defined phases of locomotion were used for measurements performed with ImageJ software. Three to six measurements were performed and averaged per animal.
The treadmill test was performed with an apparatus consisting of a plastic chamber containing a 4 × 16-cm treadmill (CleverSys). The mice were tested individually at a controlled speed of 10 cm/s. After a 1-min habituation phase, the gait of the mice was recorded for 20 s (80 frames/s; BCam). Gait was analyzed using GaitScan software (CleverSys).
The motor deficit was also evaluated with a Rotarod apparatus (accelerating model LE8200; Bioseb). Mice were placed on the accelerating rod at a starting speed of 4 rpm. The final speed of 40 rpm was reached within 5 min. Mice were tested on two consecutive days, in five trials per day, with an interval of 15 min between trials. The duration for which they were able to remain on the rod was recorded.
Muscle strength was recorded with a grip test system (Dual Sensor 1027CD-M, Linton Instrumentation) as previously described (Branchu et al., 2017). The maximum traction force (N) developed by the forelimbs and hindlimbs was recorded.
Cognitive function was monitored using the Y-Maze test. The Y-Maze consists of three transparent arms of equal length separated by the same distance (40 cm long, 20 cm high, 10 cm wide, 120°). Visual cues are placed in the room. One arm of the maze (arm B) was blocked by a removable partition and the mice were placed individually in the starting arm (arm A) of the apparatus facing the center of the maze. The mouse was allowed to walk freely into the maze. When it arrived at the end of the open arm (arm C), the partition was put in place retaining the mouse. After 1 min, the mouse was immediately placed in the starting arm again (arm A), and the partition was removed to offer two choices. Alternation was defined as spontaneous entry into the unvisited arm (arm B). On the contrary, reentry into the already explored arm (arm C) was considered to be an error.
Supplementation with sodium formate
The supplementation began for Cyp2u1+/− matings, where cages were equipped with a water bottle containing 0.55 M sodium formate (71539; Sigma-Aldrich) and lasted until the behavior test, when mice were 2 mo old. The concentration was calculated to deliver 7,500 mg sodium formate/kg/d, based on an average water intake of 5 ml/d for a 25-g C57BL/6 mouse (Bryant et al., 2018; Momb et al., 2013). The supplemented water bottles were changed twice a week.
Histologic and immunohistochemical analyses
Mice were anesthetized by i.p. injection of 1/6 2% xylazine (Rompun), 1/3 ketamine (10 mg/ml; Imalgen 1000) in PBS and were subjected to intracardiac perfusion of 4% paraformaldehyde in PBS.
For immunostaining analysis, the brains were dissected and postfixed by incubation for 24 h in 4% paraformaldehyde and 24 h in 30% sucrose PBS. Slices (20 µm for brain) were cut on a freezing microtome (Microm HM450; Thermo Fisher Scientific).
Evaluation of β-galactosidase enzymatic activity was accomplished by washing the slides with three changes of PBS for 5 min each and then rinsing in distilled water for 5 min. The sections were incubated in X-Gal (0.2%) working solution, pH 7.38, for 18 h (overnight) at 37°C in covered containers. After this incubation, the sections were dehydrated, and coverslips were applied.
For immunohistochemical analyses, after 90-min incubation in blocking solution, sections were incubated with primary antibodies (mouse anti-NeuN; MAB377; Merck Millipore), rabbit anti-glial fibrillary acidic protein (Z0334; Dako), and mouse anti-calbindin (300; Swant) in 2% BSA/0.25% Triton X-100 in PBS overnight at 4°C. After washing, the sections were incubated with the secondary antibodies for 90 min at room temperature and mounted in Fluoromount-G mounting medium (Southern Biotechnology). Staining specificity was determined by incubation in the absence of primary antibodies. Images were obtained with a NanoZoomer 2.0-RS (Hamamatsu) equipped with a 20× objective. Identical brightness, contrast, and color balance adjustments were applied to all groups. The surface areas of the brains or brain regions were determined using NDPview software. Counting of cell populations was performed using ImageJ software.
Single-molecule fluorescence in situ hybridization experiments were performed with RNAscope technology per manufacturer’s protocols for multiplexed fluorescence assays with paraformaldehyde-fixed frozen brain sections (https://acdbio.com). The probes used were “RNAscope probe - Mm-Cyp2u1,” the “RNAscope probe - Mm-Ppib” positive control probe, and the “RNAscope Probe-DapB” negative control probe. The stained slides were visualized under a standard bright-field microscope.
Immunohistochemistry on retinas was conducted as previously described (Trouillet et al., 2018). Briefly, mice were killed by CO2 asphyxiation and enucleated. The globes were fixed in 4% PFA for 30 min, then rinsed in 1× PBS (pH 7.3). Retinal and RPE/choroid tissues were dissected intact from the globe, flat-mounted, and processed for immunohistochemistry using polyclonal goat anti-IBA1 (1:500, ab5076; Merck Millipore), rabbit anti-opsin red/green (1:200, AB5405; Merck Millipore), rabbit anti-opsin blue (1:200, ab5407; Merck Millipore), rabbit anti-cone arrestin (ab15282; Merck Millipore), and donkey anti-rabbit secondary antibody conjugated with Alexa Fluor 594 and donkey anti-goat secondary antibody conjugated with Alexa Fluor 488 (Life Technologies). Flat mounts were stained with the nuclear marker Hoechst (1:1,000) and the lectin from Arachis hypogaea (peanut agglutinin) coupled with a 594-nm fluorophore (1:100, L7381; Sigma-Aldrich). Images were captured with a Leica SP8 X confocal microscope (objective 20× lens).
Lipidomic analyses (Colsch et al., 2015)
8-mo-old Cyp2u1+/+, Cyp2u1+/−, and Cyp2u1−/− mice were killed by CO2 asphyxiation, and the cerebral cortices were immediately dissected and frozen in isopentane. Samples were extracted according to a modified Folch method (Folch et al., 1957). Briefly, 10 mg of hippocampus was added to 190 µl of chloroform/methanol 2:1 (vol/vol). Samples were vortexed for 60 s and then sonicated for 30 s using a sonication probe. Extraction was performed after 2 h at 4°C with mixing. In addition, 40 µl of ultrapure water was added, and samples were vortexed for 60 s before centrifugation at 10,000 rpm for 10 min at 4°C. The upper phase (aqueous phase), containing ganglioside species and several lysophospholipids, was transferred into a glass tube and then dried under a stream of nitrogen. The interphase, which consists of a protein disk, was discarded, and the lower, lipid-rich phase (organic phase) was pooled with the dried upper phase. Samples were then reconstituted with 200 µl of chloroform/methanol 2:1 (vol/vol), vortexed for 30 s, sonicated for 60 s, and diluted 1/100 in MeOH/IPA/H2O 65:35:5 (vol/vol/vol) before injection. All samples were processed and analyzed as described previously (Seyer et al., 2016). After liquid chromatography–high-resolution mass spectrometry analysis, samples were reinjected for higher-energy collisional dissociation tandem mass spectrometry experiments in the negative ion mode, with the instrument set in the targeted mode, using inclusion lists. The isolation width was set to m/z 0.4, the normalized collision energy was 26%, and mass resolution was set to 17,500 FWHM at m/z 200. Higher-energy collisional dissociation mass spectra were inspected manually to confirm the identity of the ganglioside species. The relative amount of each lipid was quantified as the area of its chromatographic peak, and it was normalized to the exact weight of each hippocampus to consider the difference in weight between samples. Blood samples from patients and controls were submitted to the same protocol. No special procedure to avoid the oxidation of the samples were used in the lipidomic analyses.
Proteomic analyses
Hippocampi from 2-mo-old Cyp2u1+/+ and Cyp2u1−/− mice were used for label-free quantification of the proteome. The tissues were disrupted using liquid nitrogen and dissolved in 8 M urea buffer, with the addition of 1× proteinase inhibitor. After acetone precipitation, 50 mg of proteins was used for the digestion. Proteins were reduced by 1-h incubation with 5 mM dithiothreitol at 37°C, and thiols were carboxymethylated with 10 mM iodoacetamide for 30 min. This was followed by digestion with Lys-C at 37°C for 4 h, and samples were then left overnight with trypsin at 37°C. The digestion was stopped with 1% formic acid the next day. StageTip purification of the samples was performed according to the CECAD proteomics facility instructions.
Label-free proteomic profiling was performed in the CECAD proteomics facility according to the provided protocols. The liquid chromatography–tandem mass spectrometry device was made up of an EASY n-LC1000 (Thermo Fisher Scientific) linked via a nano-electrospray ionization source (Thermo Fisher Scientific) to an ion trap–based benchtop Q-Executive Plus (Thermo Fisher Scientific).
Mitochondrial enzyme activity measurements
Mitochondria were isolated from brain of Cyp2u1+/+ and Cyp2u1−/− 1-yr-old mice. For enzyme activity measurements, the mitochondrial proteins obtained (15–50 µg) were diluted in phosphate buffer (50 mM KH2PO4, pH 7.4) followed by spectrophotometric analysis of isolated respiratory chain complex activities at 37°C using a Hitachi UV-3600 spectrophotometer. Citrate synthase activity was measured at 412 nm (ε = 13,600 M−1cm−1) after addition of 0.1 mM acetyl-CoA, 0.5 mM oxaloacetate, and 0.1 mM 5,5′-dithiobis (2-nitrobenzoic acid). NADH dehydrogenase activity was determined at 340 nm (ε = 6,220 M−1cm−1) after addition of 0.25 mM NADH, 0.25 mM decylubiquinone, and 1 mM KCN and monitoring of rotenone sensitivity. Succinate dehydrogenase activity was measured at 600 nm (ε = 21,000 M−1cm−1) after addition of 40 mM succinate, 35 µM dichlorophenol indophenol, and 1 mM KCN. Cytochrome c activity was assessed using a classic N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD)/ascorbate assay. Briefly, homogenized tissue (65 µg/ml) was suspended in mitochondrial respiration buffer (220 mM mannitol, 70 mM sucrose, 10 mM Tris, 2 mM EDTA, pH 7.4). Oxygen consumption was assessed in the presence of TMPD (0.2 mM), ascorbate (1 mM), and antimycin A (0.5 µM). After a few minutes of stationary respiration, KCN (2 mM) was injected into the chamber. Cytochrome c activity corresponds to the KCN-sensitive respiration. Catalase activity was assessed using an Oroboros oxygraph. Catalase activity of homogenized tissues (65 µg/ml) was followed by recording the oxygen production in the presence of 0.01% H2O2. All chemicals were obtained from Sigma-Aldrich.
Quantitative RT-PCR
RNA was isolated with TRIzol Reagent (Invitrogen) and resuspended in nuclease-free water (Ambion), and 2 µg of RNAs were reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR was performed on the obtained cDNAs using the following Taqman probes: b2μglobulin (Mm00437762_m1), Pdss1 (Mm00450958_m1), Pdss2 (Mm01190168_m1), Coq2 (Mm01203260_g1), Coq3 (Mm01220614_m1), Coq5 (Mm00518239_m1), and Coq7 (Mm00501588_m1), all from Life Technologies.
Western blot analyses
Proteins from mitochondrial and whole-cell lysates were separated by SDS-PAGE and blotted onto polyvinylidene fluoride membranes (GE Healthcare). The following primary antibodies were used: rabbit anti-α-actin (1:5,000, ab15246; Abcam), mouse anti-COQ6 (1:100, sc-393932; Santa Cruz Biotechnology), mouse anti-COQ9 (1:1,000, sc-271892; Santa Cruz Biotechnology), mouse anti-COQ5 (1:500, 17453-1-AP; Proteintech), and mouse anti-OXPHOS antibody cocktail (1:1,000, ab110413; Abcam). The following secondary antibodies were used: donkey anti-rabbit IgG (NA9340V; GE Healthcare) and sheep anti-mouse (NXA931; GE Healthcare). Detection was done by incubation with HRP-conjugated secondary antibodies and conversion to chemiluminescence with ECL (GE Healthcare).
Vitamin B2 analysis
Blood was collected in tripotassium EDTA Vacutainer blood collection tubes (BD UK Limited) for FAD, FMN, and riboflavin measurements. Briefly, aliquots of whole blood were directly frozen at less than −18°C before FAD, FMN, and riboflavin analysis. FAD, FMN, and riboflavin were then extracted from whole blood, and proteins were removed by precipitation using Chromsystems reagent kits (Chromsystems Instruments & Chemicals) and a step of centrifugation. Supernatants containing FAD, FMN, and riboflavin were then injected into an isocratic ultra performance liquid chromatography system coupled to fluorimetric detection (Waters). Quantification was performed using Empower3_HF1_Enterprise software (v7.30.00.00; Waters). Mobile phase and chromatographic columns were provided by Chromsystems. Analysis of FAD, FMN, and riboflavin used a specific excitation wavelength of 465 nm and a specific emission wavelength of 525 nm, allowing specific determination of these three components. Due to their native fluorescence, riboflavin and its active metabolites, FMN and FAD, could be detected directly, without derivatization. This method allowed reliable and precise quantification of these components. Intra-assay coefficients of variation were 0.96% at 138.2 nmol/liter and 0.90% at 441.4 nmol/liter for FAD analysis, 7.28% at 9.0 nmol/liter and 5.20% at 20.0 nmol/liter for FMN analysis, and 3.72% at 27.4 nmol/liter and 1.38% at 103.7 nmol/liter for riboflavin analysis. Interassay coefficients of variation were 2.27% at 126.1 nmol/liter and 2.02% at 424.4 nmol/liter for FAD analysis, 9.38% at 7.8 nmol/liter and 6.59% at 14.2 nmol/liter for FMN analysis, and 5.10% at 23.6 nmol/liter and 3.56% at 90.5 nmol/liter for riboflavin analysis.
Folate measurement
Human and mouse serum samples were processed and analyzed using an electrochemiluminescence immunoassay on an automated analyzer (Cobas 8000 e801; Roche Diagnostics).
Homocysteine measurement
Detection and quantification of homocysteine extracted from plasma were accomplished by performing liquid chromatography–tandem mass spectrometry coupled with isotope dilution quantification.
BN-PAGE
BN-PAGE was performed with the NativePAGE Novex Bis-Tris Mini Gel system (Invitrogen) according to the manufacturer’s specifications. CI in-gel activity was measured by incubating BN-PAGE gel in 0.1 mg/ml NADH and 2.5 mg/ml nitrotetrazolium blue in 5 mM Tris (pH 7.4) for 1 h. Complex IV in-gel activity was measured by incubating BN-PAGE gel at 37°C in 0.24 U/ml catalase, 10% cytochrome C, and 0.1% diaminobenzidine tetrahydrochloride in 50 mM Tris (pH 7.4) for 1 h. One set of samples was transferred to polyvinylidene difluoride membrane with the iBlot system (Invitrogen), and immunodetection of mitochondrial protein complexes was performed.
Riboflavin binding to CYP2U1
Recombinant human CYP2U1 was overexpressed in Escherichia coli DH5α and purified as previously described (Dhers et al., 2017). A previously described method to register formation of a complex between CYP2U1 and riboflavin was followed (Shumyantseva et al., 2004). The method is based on fluorescence quenching of the fluorophore after the association with the hemoprotein. Complex formation was measured by monitoring the decrease in the fluorescence intensity of riboflavin (kept at constant concentration) in the presence of increasing concentrations of CYP2U1. Experiments were performed at 30°C in 1-ml quartz cuvettes containing 50 mM phosphate buffer, pH 7.4. The quenching of riboflavin fluorescence emission spectra (λmax = 520 nm) was recorded on an F7000 Hitachi fluorescence spectrometer using the FL Solutions 4.0. Emission spectra were recorded in the range 400–600 nm with excitation at 385 nm.
Docking procedures
A 3D model of CYP2U1 deprived of its predicted membrane-spanning domain (residues 57 to 544) was constructed as previously described (Ducassou et al., 2015). In all modeling experiments, the parameters applied for the heme (defined as FeIII protoporphyrin IX) were obtained from Oda et al. (2005). Molecular docking experiments of riboflavin at the active site were performed as previously described using the CDOCKER protocol from the software package Discovery Studio 2016, with default parameters and a sphere radius of 10 Å (DS2016). Molecular graphics and analyses were performed with DS2016.
Transmission EM: Imaging
Initial grid screening and generation of ultra-large high-resolution EM maps were acquired using a TECNAI F20 transmission electron microscope with a field emission gun as an electron source, operated at 200 kV, and equipped with a GATAN Ultrascan US4000 charge-coupled device camera. SerialEM software (Mastronarde, 2005; Schorb et al., 2019) was used for multiscale mapping, as follows: initially, a full grid map was acquired at 190× magnification (pixel size = 551.75 nm). Middle-magnification maps at 2,500× (pixel size = 35.98 nm) were acquired in areas with cells. Finally, high-magnification maps (14,500×, pixel size = 6.194 nm) were collected at areas of interest, usually covering areas of the cellular cytoplasm around the nucleus (maps consisted of 50–150 micrographs/pieces). For these high-magnification maps, the stage was allowed to settle for 7 s before the acquisition of a new image, while SerialEM’s autofocus routine was used every 4 micrographs, arranged as 2 × 2 submontages, to maintain a defocus value of approximately −4 µm. Overlapping between neighboring micrographs was set to 12%.
Blending montages to images
Stacks of montages were displayed using the 3dmod interface of IMOD (Kremer et al., 1996; Mastronarde and Held, 2017). The initial piece coordinates for each micrograph were extracted from the header of the mrc stack using the Extract Pieces function of IMOD. Blending was then performed using the Blendmont function of IMOD. In cases of difficult montages, Blendmont was used to determine the XY shifts for each pair of horizontally or vertically adjacent montage pieces through cross-correlation in overlapping zones, before proceeding with the blending of the overlapping edges, using linear and warping transformations. A home-made script was used to permit the batch blending of the maps collected by SerialEM.
Online supplemental material
Fig. S1 provides extra data acquired for the extensive phenotypic characterization of Cyp2u1−/− mice. Fig. S2 shows molecular and biochemical characterization of SPG56 patients and Cyp2u1−/− mice, focused on the mitochondrial impact of CYP2U1 defect deficiency. Table S1 lists clinical characterization of SPG56 patients analyzed in this study. Table S2 summarizes the phenotypes relevant to this study from all the SPG56 patients reported in the literature.
Supplementary Material
lists clinical characterization of SPG56 patients analyzed in this study.
summarizes the phenotypes relevant to this study from all the SPG56 patients reported in the literature.
Acknowledgments
We thank all patients and their families for their unevaluable help in this study, as well as Benoit Colsch for advice on lipidomic analyses and Guillaume Banneau for genetic validation of one patient. We gratefully acknowledge Claire Lovo for her precious advice. We also thank the HISTOMICS, IGENSEQ, ICM.QUANT, and DNA and Cell Bank facilities of the Institut du Cerveau. All animal work was conducted at the PHENO-ICMice facility. The Core is supported by “Investissements d’avenir” (ANR-10-IAIHU-06 and ANR-11-INBS-0011-NeurATRIS) and the “Fondation pour la Recherche Médicale.” Docking studies were performed at the Macromolecular Modeling Platform core facility of BioMedTech Facilities, Institut National de la Santé et de la Recherche Médicale US36, Centre National de la Recherche Scientifique UMS2009, Université de Paris. Lenker and Berninger provided the musical support (C. Pujol).
This study was supported financially by the Association Strümpell-Lorrain–Hereditary Spastic Paraplegia patient association (to C. Pujol and G. Stevanin), the Tom Wahlig–Stiftung Foundation (to K.H. El Hachimi and G. Stevanin), the EMBO short-term fellowship (C. Pujol), French state funds through the Agence Nationale de la Recherche under the framework programme Investissements d’Avenir (ANR-10-INBS-07 PHENOMIN), and the European Union through an internal call under the Seventh Framework Programme (NEUROMICS, to G. Stevanin).
Author contributions: Conceptualization, C. Pujol, S. Steculorum, A. Trifunovic, and G. Stevanin; Investigation, C. Pujol, J-L. Boucher, L. Le Corre; P. Giavalisco, S. Perin, A. Legrand, L. Parodi, P. Thomas, M. Croon, M. Popovic, M. Valet, and A. Mourier; Resources, F. Mochel, D. Saracino, G. Coarelli, N. Villain, S. Elshafie, M. Issa, S. Zuily, M. Renaud, C. Marelli-Tosi, M. Legendre, A. Trimouille, I. Kemlin, S. Mathieu, J.G. Gleeson, F. Lamari, D. Galatolo, R. Alkouri, C. Tse, D. Rodriguez, C. Ewenczyk, F. Fellmann, T. Kuntzer, E. Blond, F. Darios, A. Seyer, A. Gazi, F.M. Santorelli, C. Goizet, M.S Zaki, S. Picaud, A. Mourier, C. Mignot, A. Trifunovic, and A. Durr; Writing – Original Draft, C. Pujol; Writing – Review & Editing, C. Pujol and G. Stevanin; Funding Acquisition, C. Pujol, K.H. El Hachimi, and G. Stevanin.
Data availability
Data from lipidomic and proteomic have been deposited in Zenodo at https://doi.org/10.5281/zenodo.5499883.
References
- Acosta, M.J., Vazquez Fonseca L., Desbats M.A., Cerqua C., Zordan R., Trevisson E., and Salviati L.. 2016. Coenzyme Q biosynthesis in health and disease. Biochim. Biophys. Acta. 1857:1079–1085. 10.1016/j.bbabio.2016.03.036 [DOI] [PubMed] [Google Scholar]
- Albani, S.H., McHail D.G., and Dumas T.C.. 2014. Developmental studies of the hippocampus and hippocampal-dependent behaviors: insights from interdisciplinary studies and tips for new investigators. Neurosci. Biobehav. Rev. 43:183–190. 10.1016/j.neubiorev.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniam, S., Christodoulou J., and Rahman S.. 2019. Disorders of riboflavin metabolism. J. Inherit. Metab. Dis. 42:608–619. 10.1002/jimd.12058 [DOI] [PubMed] [Google Scholar]
- Bibi, F., Efthymiou S., Bourinaris T., Tariq A., Zafar F., Rana N., Salpietro V., Houlden H., Raja G.K., Saeed S., and Minhas N.M.. SYNaPS Study Group . 2020. Rare novel CYP2U1 and ZFYVE26 variants identified in two Pakistani families with spastic paraplegia. J. Neurol. Sci. 411:116669. 10.1016/j.jns.2020.116669 [DOI] [PubMed] [Google Scholar]
- Blau, N., Bonafé L., Krägeloh-Mann I., Thöny B., Kierat L., Häusler M., and Ramaekers V.. 2003. Cerebrospinal fluid pterins and folates in Aicardi-Goutières syndrome: a new phenotype. Neurology. 61:642–647. 10.1212/01.WNL.0000082726.08631.E7 [DOI] [PubMed] [Google Scholar]
- Boutry, M., Morais S., and Stevanin G.. 2019. Update on the Genetics of Spastic Paraplegias. Curr. Neurol. Neurosci. Rep. 19:18. 10.1007/s11910-019-0930-2 [DOI] [PubMed] [Google Scholar]
- Branchu, J., Boutry M., Sourd L., Depp M., Leone C., Corriger A., Vallucci M., Esteves T., Matusiak R., Dumont M., et al. 2017. Loss of spatacsin function alters lysosomal lipid clearance leading to upper and lower motor neuron degeneration. Neurobiol. Dis. 102:21–37. 10.1016/j.nbd.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, J.D., Sweeney S.R., Sentandreu E., Shin M., Ipas H., Xhemalce B., Momb J., Tiziani S., and Appling D.R.. 2018. Deletion of the neural tube defect-associated gene Mthfd1l disrupts one-carbon and central energy metabolism in mouse embryos. J. Biol. Chem. 293:5821–5833. 10.1074/jbc.RA118.002180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, S.S., Helvig C., Taimi M., Ramshaw H.A., Collop A.H., Amad M., White J.A., Petkovich M., Jones G., and Korczak B.. 2004. CYP2U1, a novel human thymus- and brain-specific cytochrome P450, catalyzes ω- and (ω-1)-hydroxylation of fatty acids. J. Biol. Chem. 279:6305–6314. 10.1074/jbc.M311830200 [DOI] [PubMed] [Google Scholar]
- Citterio, A., Arnoldi A., Panzeri E., D’Angelo M.G., Filosto M., Dilena R., Arrigoni F., Castelli M., Maghini C., Germiniasi C., et al. 2014. Mutations in CYP2U1, DDHD2 and GBA2 genes are rare causes of complicated forms of hereditary spastic paraparesis. J. Neurol. 261:373–381. 10.1007/s00415-013-7206-6 [DOI] [PubMed] [Google Scholar]
- Colsch, B., Seyer A., Boudah S., and Junot C.. 2015. Lipidomic analysis of cerebrospinal fluid by mass spectrometry-based methods. J. Inherit. Metab. Dis. 38:53–64. 10.1007/s10545-014-9798-0 [DOI] [PubMed] [Google Scholar]
- Crow, Y.J., and Manel N.. 2015. Aicardi-Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 15:429–440. 10.1038/nri3850 [DOI] [PubMed] [Google Scholar]
- Desai, A., Sequeira J.M., and Quadros E.V.. 2016. The metabolic basis for developmental disorders due to defective folate transport. Biochimie. 126:31–42. 10.1016/j.biochi.2016.02.012 [DOI] [PubMed] [Google Scholar]
- Dhers, L., Pietrancosta N., Ducassou L., Ramassamy B., Dairou J., Jaouen M., André F., Mansuy D., and Boucher J.-L.. 2017. Spectral and 3D model studies of the interaction of orphan human cytochrome P450 2U1 with substrates and ligands. Biochim. Biophys. Acta, Gen. Subj. 1861:3144–3153. 10.1016/j.bbagen.2016.07.018 [DOI] [PubMed] [Google Scholar]
- Ducassou, L., Jonasson G., Dhers L., Pietrancosta N., Ramassamy B., Xu-Li Y., Loriot M.-A., Beaune P., Bertho G., Lombard M., et al. 2015. Expression in yeast, new substrates, and construction of a first 3D model of human orphan cytochrome P450 2U1: Interpretation of substrate hydroxylation regioselectivity from docking studies. Biochim. Biophys. Acta. 1850:1426–1437. 10.1016/j.bbagen.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Durand, C.M., Dhers L., Tesson C., Tessa A., Fouillen L., Jacqueré S., Raymond L., Coupry I., Benard G., Darios F., et al. 2018. CYP2U1 activity is altered by missense mutations in hereditary spastic paraplegia 56. Hum. Mutat. 39:140–151. 10.1002/humu.23359 [DOI] [PubMed] [Google Scholar]
- Fink, J.K. 2013. Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 126:307–328. 10.1007/s00401-013-1115-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch, J., Lees M., and Sloane Stanley G.H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497–509. 10.1016/S0021-9258(18)64849-5 [DOI] [PubMed] [Google Scholar]
- Fowler, P.C., Garcia-Pardo M.E., Simpson J.C., and O’Sullivan N.C.. 2019. NeurodegenERation: The Central Role for ER Contacts in Neuronal Function and Axonopathy, Lessons From Hereditary Spastic Paraplegias and Related Diseases. Front. Neurosci. 13:1051. 10.3389/fnins.2019.01051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi, D., and Zeviani M.. 2018. Human diseases associated with defects in assembly of OXPHOS complexes. Essays Biochem. 62:271–286. 10.1042/EBC20170099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irintchev, A., Simova O., Eberhardt K.A., Morellini F., and Schachner M.. 2005. Impacts of lesion severity and tyrosine kinase receptor B deficiency on functional outcome of femoral nerve injury assessed by a novel single-frame motion analysis in mice. Eur. J. Neurosci. 22:802–808. 10.1111/j.1460-9568.2005.04274.x [DOI] [PubMed] [Google Scholar]
- Iodice, A., Panteghini C., Spagnoli C., Salerno G.G., Frattini D., Russo C., Garavaglia B., and Fusco C.. 2017. Long-term follow-up in spastic paraplegia due to SPG56/CYP2U1: age-dependency rather than genetic variability? J. Neurol. 264:586–588. 10.1007/s00415-017-8393-3 [DOI] [PubMed] [Google Scholar]
- Kariminejad, A., Schöls L., Schüle R., Tonekaboni S.H., Abolhassani A., Fadaee M., Rosti R.O., and Gleeson J.G.. 2016. CYP2U1 mutations in two Iranian patients with activity induced dystonia, motor regression and spastic paraplegia. Eur. J. Paediatr. Neurol. 20:782–787. 10.1016/j.ejpn.2016.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kumar, K.R., Wali G.M., Kamate M., Wali G., Minoche A.E., Puttick C., Pinese M., Gayevskiy V., Dinger M.E., Roscioli T., et al. 2016. Defining the genetic basis of early onset hereditary spastic paraplegia using whole genome sequencing. Neurogenetics. 17:265–270. 10.1007/s10048-016-0495-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand, A., Pujol C., Durand C.M., Mesnil A., Rubera I., Duranton C., Zuily S., Sousa A.B., Renaud M., Boucher J.L., et al. 2021. Pseudoxanthoma elasticum overlaps hereditary spastic paraplegia type 56. J. Intern. Med. 289:709–725. 10.1111/joim.13193 [DOI] [PubMed] [Google Scholar]
- Leonardi, L., Ziccardi L., Marcotulli C., Rubegni A., Longobardi A., Serrao M., Storti E., Pierelli F., Tessa A., Parisi V., et al. 2016. Pigmentary degenerative maculopathy as prominent phenotype in an Italian SPG56/CYP2U1 family. J. Neurol. 263:781–783. 10.1007/s00415-016-8066-7 [DOI] [PubMed] [Google Scholar]
- Letts, J.A., and Sazanov L.A.. 2017. Clarifying the supercomplex: the higher-order organization of the mitochondrial electron transport chain. Nat. Struct. Mol. Biol. 24:800–808. 10.1038/nsmb.3460 [DOI] [PubMed] [Google Scholar]
- Lloyd, K.C.K. 2011. A knockout mouse resource for the biomedical research community. Ann. N. Y. Acad. Sci. 1245:24–26. 10.1111/j.1749-6632.2011.06311.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masciullo, M., Tessa A., Perazza S., Santorelli F.M., Perna A., and Silvestri G.. 2016. Hereditary spastic paraplegia: Novel mutations and expansion of the phenotype variability in SPG56. Eur. J. Paediatr. Neurol. 20:444–448. 10.1016/j.ejpn.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Masingue, M., Benoist J.-F., Roze E., Moussa F., Sedel F., Lubetzki C., and Nadjar Y.. 2019. Cerebral folate deficiency in adults: A heterogeneous potentially treatable condition. J. Neurol. Sci. 396:112–118. 10.1016/j.jns.2018.11.014 [DOI] [PubMed] [Google Scholar]
- Mastronarde, D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Mastronarde, D.N., and Held S.R.. 2017. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197:102–113. 10.1016/j.jsb.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie, M., Lazarou M., Thorburn D.R., and Ryan M.T.. 2006. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J. Mol. Biol. 361:462–469. 10.1016/j.jmb.2006.06.057 [DOI] [PubMed] [Google Scholar]
- Minase, G., Miyatake S., Nabatame S., Arai H., Koshimizu E., Mizuguchi T., Nakashima M., Miyake N., Saitsu H., Miyamoto T., et al. 2017. An atypical case of SPG56/CYP2U1-related spastic paraplegia presenting with delayed myelination. J. Hum. Genet. 62:997–1000. 10.1038/jhg.2017.77 [DOI] [PubMed] [Google Scholar]
- Momb, J., Lewandowski J.P., Bryant J.D., Fitch R., Surman D.R., Vokes S.A., and Appling D.R.. 2013. Deletion of Mthfd1l causes embryonic lethality and neural tube and craniofacial defects in mice. Proc. Natl. Acad. Sci. USA. 110:549–554. 10.1073/pnas.1211199110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosegaard, S., Dipace G., Bross P., Carlsen J., Gregersen N., and Olsen R.K.J.. 2020. Riboflavin Deficiency-Implications for General Human Health and Inborn Errors of Metabolism. Int. J. Mol. Sci. 21:3847. 10.3390/ijms21113847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolai, J., van Kempen M.J.A., and Postma A.A.. 2016. Teaching NeuroImages: White matter hypomyelination and progressive calcifications in cerebral folate deficiency. Neurology. 87:e4–e5. 10.1212/WNL.0000000000002805 [DOI] [PubMed] [Google Scholar]
- Oda, A., Yamaotsu N., and Hirono S.. 2005. New AMBER force field parameters of heme iron for cytochrome P450s determined by quantum chemical calculations of simplified models. J. Comput. Chem. 26:818–826. 10.1002/jcc.20221 [DOI] [PubMed] [Google Scholar]
- Parodi, L., Coarelli G., Stevanin G., Brice A., and Durr A.. 2018. Hereditary ataxias and paraparesias: clinical and genetic update. Curr. Opin. Neurol. 31:462–471. 10.1097/WCO.0000000000000585 [DOI] [PubMed] [Google Scholar]
- Pennesi, M.E., Neuringer M., and Courtney R.J.. 2012. Animal models of age related macular degeneration. Mol. Aspects Med. 33:487–509. 10.1016/j.mam.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salviati, L., Trevisson E., Doimo M., and Navas P.. 2017. Primary Coenzyme Q10 Deficiency. In GeneReviews. Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., and Amemiya A., editors. University of Washington, Seattle, WA. pp. 1993–2021. [PubMed] [Google Scholar]
- Schorb, M., Haberbosch I., Hagen W.J.H., Schwab Y., and Mastronarde D.N.. 2019. Software tools for automated transmission electron microscopy. Nat. Methods. 16:471–477. 10.1038/s41592-019-0396-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano, L., De Matteis M.A., Emr S., Giordano F., Hajnóczky G., Kornmann B., Lackner L.L., Levine T.P., Pellegrini L., Reinisch K., et al. 2019. Coming together to define membrane contact sites. Nat. Commun. 10:1287. 10.1038/s41467-019-09253-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyer, A., Boudah S., Broudin S., Junot C., and Colsch B.. 2016. Annotation of the human cerebrospinal fluid lipidome using high resolution mass spectrometry and a dedicated data processing workflow. Metabolomics. 12:91. 10.1007/s11306-016-1023-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumyantseva, V.V., Bulko T.V., Petushkova N.A., Samenkova N.F., Kuznetsova G.P., and Archakov A.I.. 2004. Fluorescent assay for riboflavin binding to cytochrome P450 2B4. J. Inorg. Biochem. 98:365–370. 10.1016/j.jinorgbio.2003.10.024 [DOI] [PubMed] [Google Scholar]
- Tesson, C., Nawara M., Salih M.A.M., Rossignol R., Zaki M.S., Al Balwi M., Schule R., Mignot C., Obre E., Bouhouche A., et al. 2012. Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am. J. Hum. Genet. 91:1051–1064. 10.1016/j.ajhg.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouillet, A., Dubus E., Dégardin J., Estivalet A., Ivkovic I., Godefroy D., García-Ayuso D., Simonutti M., Sahly I., Sahel J.A., et al. 2018. Cone degeneration is triggered by the absence of USH1 proteins but prevented by antioxidant treatments. Sci. Rep. 8:1968. 10.1038/s41598-018-20171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck, E., Estabel J., Oellrich A., Maguire A.K., Adissu H.A., Souter L., Siragher E., Lillistone C., Green A.L., Wardle-Jones H., et al. Sanger Institute Mouse Genetics Project . 2015. A gene expression resource generated by genome-wide lacZ profiling in the mouse. Dis. Model. Mech. 8:1467–1478. 10.1242/dmm.021238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, A.P., Khoury-Hanold W., Staron M., Tal M.C., Pineda C.M., Lang S.M., Bestwick M., Duguay B.A., Raimundo N., MacDuff D.A., et al. 2015. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 520:553–557. 10.1038/nature14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., and Cantley L.C.. 2019. Toward a better understanding of folate metabolism in health and disease. J. Exp. Med. 216:253–266. 10.1084/jem.20181965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulfiqar, S., Tariq M., Ali Z., Fatima A., Klar J., Abdullah U., Ali A., Ramzan S., He S., Zhang J., et al. 2019. Whole exome sequencing identifies novel variant underlying hereditary spastic paraplegia in consanguineous Pakistani families. J. Clin. Neurosci. 67:19–23. 10.1016/j.jocn.2019.06.039 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists clinical characterization of SPG56 patients analyzed in this study.
summarizes the phenotypes relevant to this study from all the SPG56 patients reported in the literature.
Data Availability Statement
Data from lipidomic and proteomic have been deposited in Zenodo at https://doi.org/10.5281/zenodo.5499883.