Abstract
Breast implant associated anaplastic large cell lymphoma (BIA-ALCL) is a distinct type of ALCL, and a new provisional entity by the 2016 revision of the World Health Organization (WHO) classification of tumors of hematopoietic and lymphoid tissues. In contrast to systemic and primary cutaneous ALCLs, BIA-ALCLs have been genetically characterized by the absence of fusions and frequent activation of the JAK-STAT3 pathway through mutations in JAK1 and STAT3. In this study, we report the results of the genetic profiling of 9 BIA-ALCL casessupportingthe role of the JAK-STAT pathway activation in this entity, including the identification of an activatingSTAT3-JAK2 fusion similar to those recently reported in T-cell lymphoproliferative disorders of the gastrointestinal tract. To our knowledge, this is the first fusion reported in BIA-ALCL, providing further insight into the overall genetic landscape of this rare entity as well as uncovering potential options for targeted therapy in cases with advanced disease.
Introduction
Breast implant associated anaplastic large cell lymphoma (BIA-ALCL) has been recently incorporated as a provisional category of ALCL in the 2016 revision of the World Health Organization (WHO) classification of tumors of hematopoietic and lymphoid tissues.1This is a rare and distinct entity which arises in the capsule surrounding textured saline or silicone breast implants at a median intervalof 9 years and typically presents as a unilateral peri-implant effusion with no grossly identifiable masslesion. The morphologic and immunophenotypic features are indistinguishablefrom those of systemic or cutaneous ALK negative ALCL, with the presence of large cells with pleomorphic and anaplastic morphology andcharacteristic strong / uniform expression of CD30. The prognosisis generally excellent,provided that the disease is confined to the effusion and capsule,as it can be successfullytreated with complete capsulectomy alone.2–5The presence of an associated mass, invasion through the capsule and lymph node involvementare seen in approximately 20% of patients and are characteristics associated with worseoutcome.6
While BIA-ALCL appears to have distinct biologic features that sets itapart from other ALCL subtypes, the genomic landscape and genetic events specifically responsible for its development remain poorly understood, in part due to the rarity and likely underdiagnosis of the disease.Activating mutations in STAT3 and JAK1 have been recently described as common events,7–10 suggesting that JAK-STAT3 signaling pathway constitutive activation may be at least partially involved in its oncogenesis.11,12To date, however, structural gene rearrangements, including those commonly associated with other ALCLs, such as translocations involving ALK, DUSP22 or TP63, have never been reported in BIA-ALCL.12–15 In this report, we describethe genomic profile of 9 patients with BIA-ALCL using MSK-IMPACT heme, a 400 gene hybrid capture next generation sequencing assay and the identification of an activatingSTAT3-JAK2 fusion not previously described in this entity.
Materials and Methods
Nine cases with BIA-ALCL diagnosed at our institution were studied. Both DNA and RNA were extracted from sections of formalin-fixed paraffin embedded (FFPE)tumor tissue blocks. To address adequacy issues, between10–40 sections (5 μm thick) were used for extraction. Samples were sequenced using the MSK IMPACT HEME assay,a hybridization capture-based next generation sequencing panel capturing all coding regions of 400 genes16and the MSK-PanHeme Fusion assay, an RNA-based genefusion detection assay which utilizes the ArcherDx Anchored Multiplex polymerase chain reaction (PCR) (AMP™) technology and targets199 genes. Matched normal control DNA from all 9 patients(4 FFPE non-neoplastic tissue, 3 nail clippings, 2 blood samples) was used for paired tumor: normal analysis to ensure only somatic variant calling. Copy number alterations were assessed using FACETS.17To determineT-cell receptor (TCR) betaandTCR gammagene rearrangement status,conserved regions within the variable (Vb), diversity (Db), and the joining (Jb) regions of TCR beta and gamma were amplified by PCR in the presence of fluorescently-labeled primers (InVivoScribe BIOMED-2), and the amplified products were detected by capillary electrophoresis on an ABI 3730 DNA analyzer (Thermo FisherScientific). Fluorescent in-situ hybridization (FISH) was further performed on correspondingFFPE tissue sections using a JAK2specific break-apart probe set (Cytocell/Oxford Gene Technology, Tarrytown, NY) following manufacturer’s protocol. Immunohistochemical (IHC) staining for pSTAT3 was performed using M9C6antibody (Cell Signaling Technology, Danvers, MA) at 1:250 dilution on the Leica Bond III staining platform (Leica Biosystems Inc, Buffalo Grove, IL).
Results
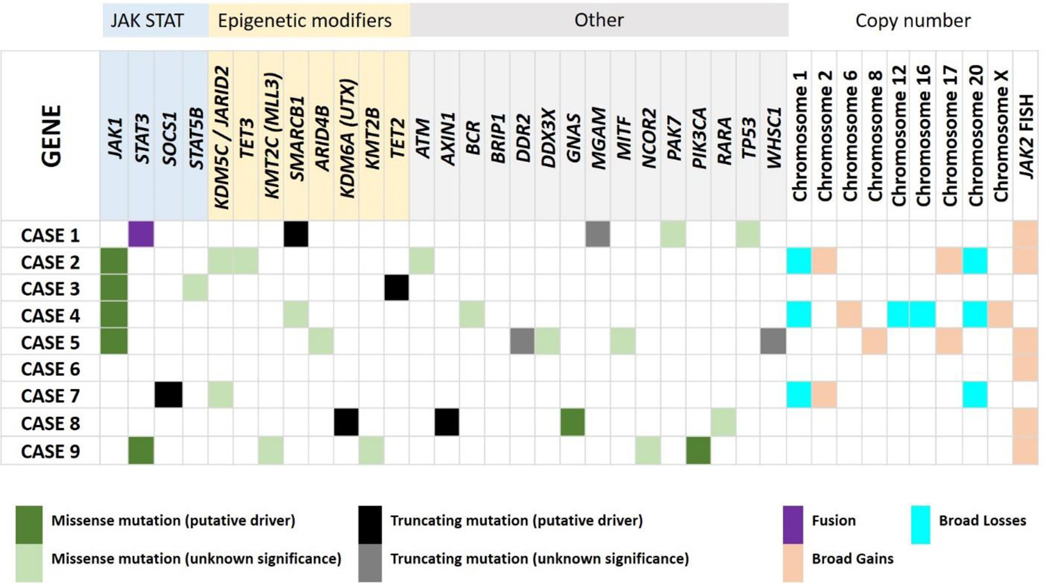
The clinical features of the 9 patients examined in this cohort are provided in Table 1.DNA sequencing was successfully performed on all 9 cases with mean sequencing depthof 461X(range, 184X-678X).Thirty-foursomatic mutations were detected across26genesin 8 of 9 patients (89%), at a median variant allele frequency (VAF) of 10% (range, 2.3–76.9%). The most commonly mutated gene was JAK1, with point mutations involving codon G1097 (D,V or S) identified in 44% (4/9) of the cases. Overall, missense mutations were the most common type of alteration (26/34, 76%) followed by splice site(3/34, 9%), nonsense (3/34, 9%), and frameshift (2/34, 6%) mutations. In all, 78% (7/9) of the cases had a somatic alteration affecting genes in the JAK/STAT pathway including JAK1, JAK2, STAT3, STAT5B and SOCS1and 56% (5/9) had mutations in epigenetic modifiers (Table 2, Figure 1). An in-frame fusion involving exon 24 of STAT3 and exons 19–25 of JAK2 (Figure 2) was detected in 1 case. Analysis of copy-number aberrations (CNAs) showed several losses and gains as summarized in Table 2 and Figure 1.
Table 1.
Clinicopathologic features of 9 patients with breast implant associated anaplastic large cell lymphoma.
| Patient | Age | Reason for Implant | Type of Implant | Number of Years after Implant | Presentation | Stage | Treatment | pSTAT IHC |
|---|---|---|---|---|---|---|---|---|
| 1 | 68 | IDC | Textured | 8 | Left breast swelling | T2 | Bilateral complete capsulectomies | Positive |
| 2 | 77 | IDC | Textured | 12 | Right breast mass | T4, stage IIA | Excision and complete capsulectomy | Positive |
| 3 | 61 | IDC and ILC | Textured | 8 | Right breast swelling, effusion | T1* | Bilateral complete capsulectomies | N/A |
| 4 |
58 | DCIS | Textured | 6 | Firmness above her left breast implant and change in shape of implant | T4N2M0 | Capsulectomy and BV-CHP x 6 cycles + consolidative radiotherapy (3060 cGY in 17 fractions) | Positive |
| 5 | 65 | IDC | Textured | 10 | Right breast effusion | T1* | Complete capsulectomy | Positive |
| 6 | 35 | Cosmetic | Textured | 8 | Right breast swelling, effusion | T1 | Bilateral complete capsulectomies | N/A |
| 7 | 51 | IDC | Textured | 6 | Left breast swelling, effusion | T1, stage IA | Bilateral complete capsulectomies | Positive |
| 8 | 66 | IDC | Textured | 11 | Cutaneous lesion | T4 | XRT (15 fractions, total 3000cGY)-, 6 cycles BV-CHOP, SCT | Negative |
| 9 | 50 | ILC | Textured | 6 | Left breast swelling, fevers, night sweats | T3, stage 1C | Bilateral complete capsulectomies | Positive |
IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; DCIS, ductal carcinoma in situ; SCT, stem cell transplant; IHC, Immunohistochemistry;
lymphoma confined to fluid only
Table 2.
Alterations detected by 400 gene hybrid capture next generation sequencing panel in 9 patients with breast implant associated anaplastic large cell lymphoma.
| Patient | Gene Altered | Reference Sequence | Protein Change | cDNA Change | Location | Type of Alteration | VAF (%) | JAK2 FISH Results | FACETS Results |
|---|---|---|---|---|---|---|---|---|---|
| 1 |
TP53
MGAM PAK7 SMARCB1 |
NM_000546 NM_004668 NM_177990 NM_003073 |
p.A307P p.X1449_splice p.P358R p.Q368* |
c.919G>C c.4345+1G>A c.1073C>G c.1102C>T |
exon 8 exon 36 exon 5 exon 8 |
Missense Splice Missense Nonsense |
6.9 9.9 3.9 8.4 |
JAK2 translocation positive; 70%, large cells, complex patterns | No calls |
| STAT3-JAK2 |
NM_139276 NM_004972 |
t(17;9)(q21.1) | Fusion | ||||||
| 2 |
ATM
JAK1 JARID2 TET3 TET3 |
NM_000051 NM_002227 NM_004973 NM_144993 NM_144993 |
p.M900I p.G1097S p.V1019L p.V595L p.V787I |
c.2700G>C c.3289G>A c.3055G>C c.1783G>T c.2359G>A |
exon 18 exon 24 exon 14 exon 1 exon 4 |
Missense Missense Missense Missense Missense |
5.1 27 8.6 15.8 17.1 |
Gain of extra copies; 3–5 signals, 50–60% | No calls |
| 3 |
TET2
JAK1 STAT5B |
NM_001127208 NM_002227 NM_012448 |
p.Q960* p.G1097D p.I174S |
c.2878C>T c.3290G>A c.521T>G |
exon 3 exon 24 exon 5 |
Nonsense Missense Missense |
2.4 10.1 5.6 |
Normal | No calls |
| 4 |
BCR
JAK1 SMARCB1 |
NM_004327 NM_002227 NM_003073 |
p.R162C p.G1097D p.D101N |
c.484C>T c.3290G>A c.301G>A |
exon 1 exon 24 exon 3 |
Missense Missense Missense |
34.4 41.4 25.5 |
Normal |
Gain:1p32–36, 2p (including ALK, REL), 2q11–35, 6p, 17q11–21, Xq25, Xq26.1, Xq26.2 (including STAG2, BCORL1, and PHF6) CNLOH: SP140 Loss: on 1p (JAK1, BCL10), 12q13 (including ARID2, HDAC7 and KMT2D/MLL2), 16q23 (including PLCG2, IRF8 and FANCA), 20p and 20q12–13, Xq12 (AR) |
| 5 |
ARID4B
BRIP1 DDR2 DDX3X JAK1 MITF WHSC1 |
NM_016374 NM_032043 NM_006182 NM_001356 NM_002227 NM_198159 NM_001042424 |
p.P134A p.G763A p.X189_splice p.G533E p.G1097V p.E303V p.X961_splice |
c.400C>G c.2288G>C c.566–2A>G c.1598G>A c.3290G>T c.908A>T c.2882–28_2895del |
exon 7 exon 16 exon 7 exon 14 exon 24 exon 7 exon 16 |
Missense Missense Splice Missense Missense Missense Splice |
11.6 33 9.7 13.4 36 7 11.3 |
Gain of extra copies; 3–4 signals, 30–40% |
Gain: 8q, 17q |
| 6 | None | N/A | N/A | N/A | N/A | N/A | N/A | Gain of extra copies; 3–5 signals, 60–80% |
No calls |
| 7 |
JARID2
SOCS1 |
NM_004973 NM_003745 |
p.A700V p.S32Ffs*44 |
c.2099C>T c.95_142delins TCCTTCCTCCTCCTCGCCCG |
exon 8 exon 2 |
Missense Frameshift, deletion |
37.9 76.9 |
Gain of extra copies; 3 signals, 10–20% |
Gain: 2p, 2q11–35, 17q11–21, ASXL1 (20q11.21) CNLOH: HDAC4, chr22q Loss: 1p22.3-p12, 20p, 20q12–13 |
| 8 |
AXIN1
GNAS KDM6A RARA |
NM_003502 NM_000516 NM_021140 NM_000964 |
p.Q476* p.R201C p.V1113Sfs*8 p.R272W |
c.1426C>T c.601C>T c.3334_3335dupGT c.814C>T |
exon 6 exon 8 exon 23 exon 7 |
Nonsense Missense Frameshift, insertion Missense |
2.3 2.4 14.2 41.1 |
Gain of extra copies; 3–4 signals, 40–50% |
No calls |
| 9 |
PIK3CA
STAT3 KMT2B KMT2C KMT2C NCOR2 |
NM_006218 NM_139276 NM_014727 NM_170606 NM_170606 NM_006312 |
p.H1047R p.S614R S1739F L596V S593L S1518L |
c.3140A>G c.1842C>G c.5216C>T c.1786C>G c.1778C>T c.4553C>T |
exon 21 exon 20 exon 25 exon 13 exon 13 exon 34 |
Missense Missense Missense Missense Missense Missense |
3.2 7.7 14.3 5.9 6.1 5.2 |
Gain of extra copies; 3–4 signals, 50–60% |
No calls |
VAF, variant allele frequency; Chr, chromosome; CNLOH, copy neutral loss of heterozygosity; FISH, fluorescence in situ hybridization.
Figure 1.
Alterations detected in 9 patients with breast implant associated anaplastic large cell lymphoma. Detailed list of alteration in each case included in Table 2.
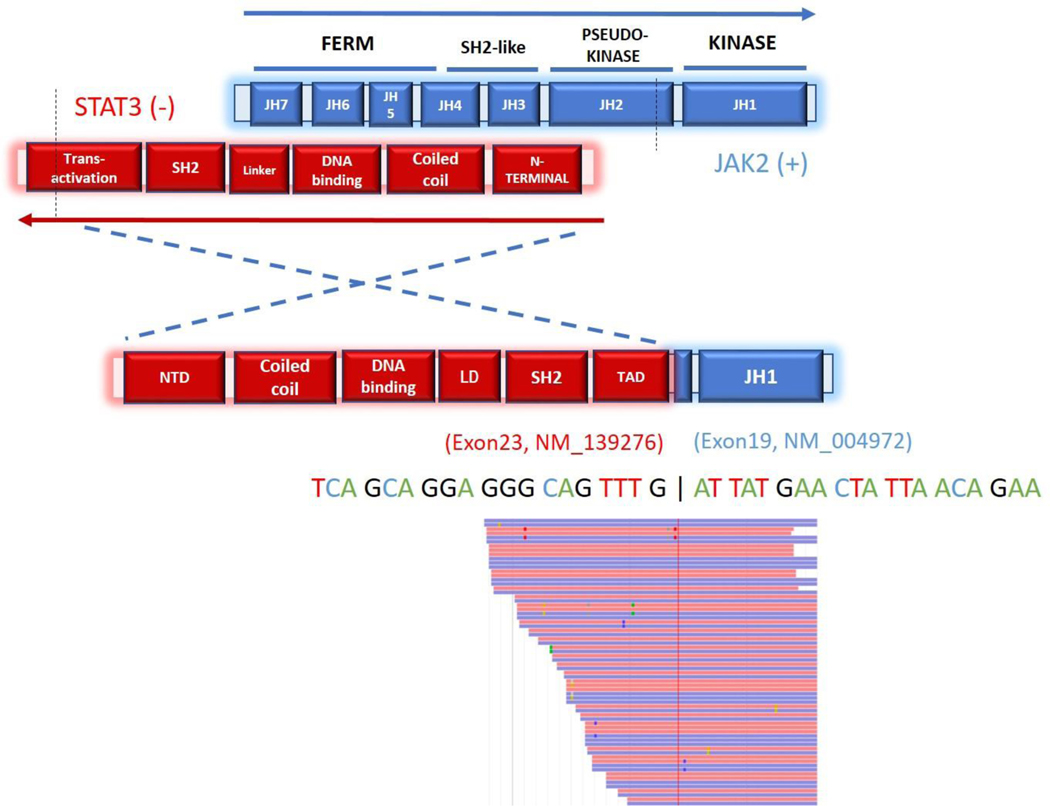
Figure 2.
Schematic illustration of the protein structure and transcript sequence of the STAT3-JAK2 in-frame fusion product. Exons 1–23 of STAT3 are fused to exons 19–25 of JAK2 which include the kinase domain. Red and blue rectangles represent bi-directional RNA sequencing reads supporting the fusion breakpoint.
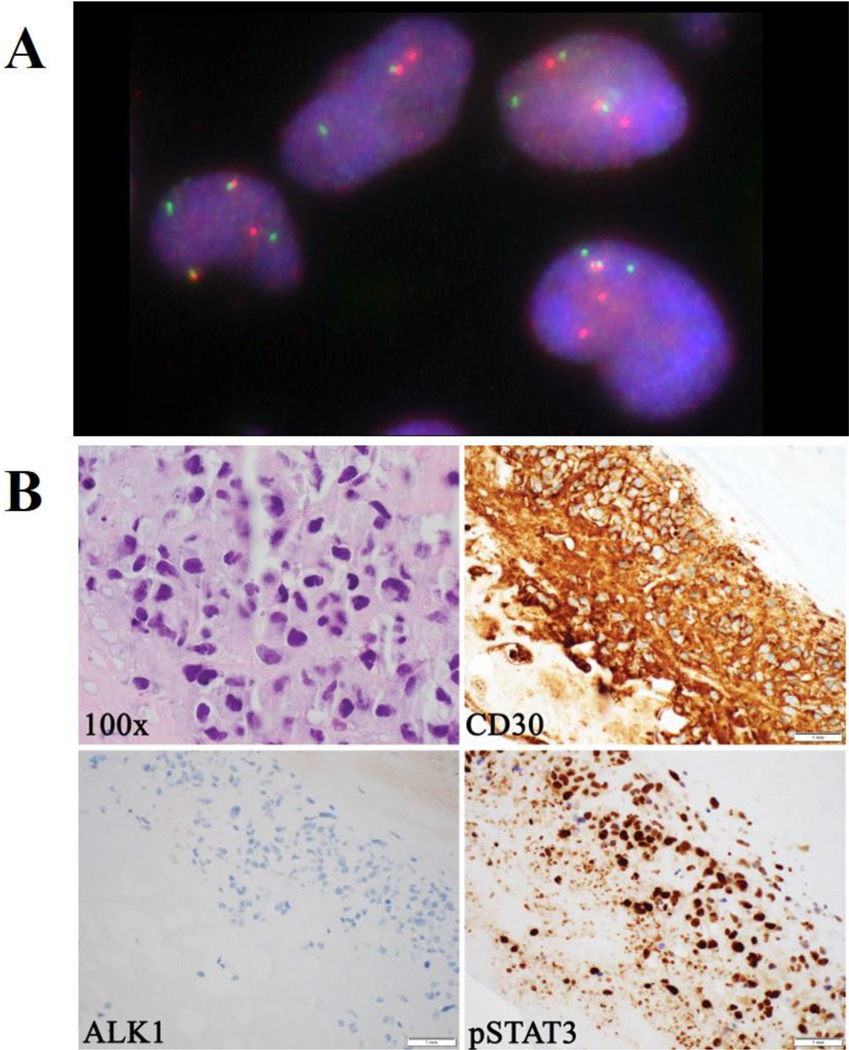
Targeted RNA sequencing (RNASeq)confirmed the fusion transcript and precise breakpoints of theSTAT3-JAK2 rearrangement. No other fusions transcripts were detected by RNAseq on 3 additional cases while the remaining 5 cases could not be successfully sequenced due to low quantity of RNA recovered from the FFPE tissue samples.FISH studies using the JAK2 break-apart probe confirmed the presence of aJAK2rearrangement with a split signal pattern (Figure 3A). No additional JAK2 rearrangements were detected by FISH, however, 6of 9 (67%) cases showed extra signals consistent with copy number gains.
Figure 3.
A, Fluorescence in situ hybridization performed on FFPE using a JAK2 specific break-apart probe set showing a split signal pattern, indicative of a JAK2 rearrangement. B, Large anaplastic cells with irregular and multilobated nuclei with occasional “hallmark” cells (H&E, 1000x, oil immersion); the anaplastic cells strongly and uniformly express CD30 (400x); the cells lack expression of ALK1 (400x); the anaplastic cells show strong expression of pSTAT3 (400x).
TCR gene rearrangement analysis by fragment analysis was successfully performed on all 9 patients and showed clonal rearrangement in 8 of the 9 (89%) cases. One case showed a prominent peak within a polyclonal background.
Immunohistochemical staining for pSTAT3 was performed in 7 of the 9 cases (including the patient with STAT3-JAK2 fusion). Of the patients tested, 6/7(86%) showed strong and diffuse expression of pSTAT3.
Case description
The patient harboring the STAT3-JAK2 fusion is a 69-year-old woman with history of invasive ductal carcinoma diagnosed in 2003 and treated with bilateral mastectomy and chemotherapy. Initial reconstructive surgery with placement of breast silicone implants was performed in 2004,followed by implant replacement in 2010. In 2018,she developed left breast swelling and a pericapsular effusion. Cytologic assessment of aspirated fluidrevealed atypical cells suspicious for lymphoma. Concurrent flow cytometric analysis confirmed the presence of abnormal cells with aberrant immunophenotype (18.9% of the total white cells),includingbright expression of CD30, CD45, and CD56, as well as increased forward and side scatter suggestive of larger size but without expression of CD2, CD3, CD4, CD5, CD7, CD8, CD10, CD19, CD20, CD22, CD23, CD38, CD200, CD279, or FMC7. Subsequent complete capsulectomy confirmed involvement by BIA-ALCL (Figure 3B).4,5 There was no mass formation, extracapsular extension or lymphadenopathy. Immunohistochemicalstains (Figure 3B) showed strong positivity for CD30andlack of ALK1 or pan-cytokeratin expression;pSTAT3 showed strong expression in the neoplastic cells.The patient is currently doing well on expectant monitoring with no adjuvant therapy given.
Discussion
In the present study, we report the sequencing results of 9 BIA-ALCL cases including, to our knowledge, the first fusion identified in this entity. To date, theoverall understanding of the genetic landscape of BIA-ALCL remains limited due to the relatively small number of cases studied. Based on review of literature, only 57 total cases have been previously studied using NGS, of which 24underwent whole exome sequencing7–10,18,19 while others were studied using narrow targeted panels.
In keeping with prior reports, our study supports that the JAK-STAT pathway is frequently dysregulated in BIA-ALCL. Both activating and putative loss-of function mutations were identified within this pathway in 78% of cases. JAK2 copy number gains were identified by FISH analysis in the remaining caseswhich could potentially elicit or enhance aberrant activation of the JAK-STAT3 pathway.20Immunohistochemistryfor expression of pSTAT3 was also positive in all cases tested except for one.In contrast to prior studies, mutationsin the JH1 kinase domain of JAK1, rather than STAT3, were the most common variants in our cohort (44% of cases). Alterations involved codon G1097 exclusively (G1097D, G1097S and G1097V), all of which are well recognized gain-of-function mutations which trigger aberrant phosphorylation of STAT3 downstream in hematologic malignancies. Other alterationsinvolving thispathwayincluded mutations in STAT3, STAT5 and SOCS1.The STAT3mutation, S614R,has been previously described as the most common activating mutation in BIA-ALCL in all previous studies, while the STAT5B mutation is a variant not previously reported. ASOCS1 truncating mutationwith putative loss-of-function effectwas also detected,predicting deregulation through to loss of inhibition on JAK-STAT signaling. Similar mutations in this gene have been identified in various types of lymphomas, including BIA-ALCL.21–24
Of particular interest, theSTAT-JAK2 fusion detected in this study was recentlyreported as a recurrent genetic alterationinindolent T-cell lymphoproliferative disorder of the gastrointestinal tract (GI TLPDs).25To date, only 4 cases have been reported in the literature, exclusively associated with GI TLPDs.The finding of a similar fusion in a similarly indolent process may support the potential role of these rearrangements in a distinct subset of less aggressive lymphoproliferative disorders. Similar to the previously reported fusions, the STAT3-JAK2 rearrangement in BIA-ALCL retains the entire catalytic domain (JH1) of JAK2and shares the same STAT3 RNA breakpoint. Recent functional studies performed by Hu et al, confirm this fusion to be oncogenic both in vitro and in-vivo using various models.26Coincidentally, to specifically assess function in human T-cell lymphoma model, this groupevaluated the effect of the fusion on the growth of TLBR-3 cells derived from a BIA-ALCL, which are dependent on IL2. Fusion expression rescued the cells from IL2 withdrawal causing marked independent growth in vitro as well as large subcutaneous tumor formation in-vivo on NSG mice. Studies also demonstrated that fusion expression activatesSTAT5 and constitutively enhances transcriptional activity within the JAK-STAT pathway. More importantly, preliminary work by the same group using various JAK inhibitors in-vitro and in an animal model, characterize this fusion as a potential targetable event with differential impact on the activity of the fusion protein.26
JAK2 fusions or rearrangements involving other partners have been identified in a variety of myeloid and precursor lymphoid neoplasms27,28 but are rare in mature T-cell lymphomas.29–32 In a previous study by Ehrentraut et al.,for example, the authors used a FISH break apart probe to assess for JAK2 rearrangements in 200 patients with various T-cell lymphomas, including 53 patients with ALCL and ALCL cell lines, finding no evidence of JAK2 fusions in any of the samples studied.31Although our combined assessment for fusions using DNA and RNA in this study was partially limited by the quality and quantity of RNA that could be effectively isolated from some of the samples, targeted FISH analysis did not identify any other JAK2rearrangement outside of our index case.
In addition to alterations involving the JAK-STAT pathway, somatic events involving epigenetic modifiers were the second most common alterations identified. Mutations or gene losses involving TET2, TET3, ARID4B, KDM5C, KDM6A, KMT2C/D, and SMARCB1 were detected in 56% of cases. Alterations in epigenetic modifiers are common in T-cell lymphomas and have also been recently reported as common events in the largest series of BIA-ALCL studied by NGS.19
Taken together, while our current understanding of the genetics of BIA-ALCL still remains limited, thus far, a unifying mechanism of lymphomagenesis in this rare entity seems to be the activation of the JAK-STAT pathway which also offers possibilities for targeted treatment in patients with advanced disease. A more recent finding is the accumulation of alterations in epigenetic modifiers which are also common in other T-cell lymphomas. These findings also have important diagnostic and therapeutic implications. Diagnostically, the presence of mutations, while not necessarily diagnostic of the entity itself, could greatly facilitate the diagnosis of BIA-ALCL in conjunction with the morphologic and immunophenotypic findings and could also provide a molecular marker for staging and monitoring.
Although the majority of patients diagnosed with BIA-ALCL can be cured with complete capsulectomy alone, those that present with a mass lesion or disease beyond the capsule are considered high risk for recurrence and progression.6Within our small cohort, for instance, patients 4 and 8 presented with advanced disease, requiring systemic therapy and radiotherapy and one ultimately required stem cell transplant as outlined in table 1. Patients with advanced disease often constitute a therapeutic challenge for current treatment modalities, underscoring the need to explore alternate therapeutic options. At present, significant pre-clinical and early clinical data support the potential efficacy of therapies targeting the JAK-STAT pathway and epigenetic regulators, with several clinical trials currently underway,underscoring the importance of comprehensively evaluating patients for potential therapeutic targets.
An important and critical consideration for future studies, is the adequate preservation of material for broad molecular testing. Two recent publications by Jaffe, et al and Lyapichev, et aloutline key strategies in processing of theseroma fluid/effusion surrounding the implant and the handling of capsulectomy specimens following removal ofimplant(s).33,34Of note, based on our own experience, we find that while these guidelines effectively address the basic needs for morphologic and immunophenotypic assessment in clinical practice, comprehensive molecular testing remains a challenge.Due to the nature of this disease,involvement of the capsule is often very focal, making it difficult to obtain sufficient tumor from FFPE capsuletissue. Alternatively, the effusion fluid may contain many more tumor cells than the capsule itself and could provide more material for downstream studies but the recommendations to prioritize the creation of a cell block over fresh material for flow cytometry and other studies, limit the amount and quality of material that can be recovered for molecular assays, particularly for RNA studies. In our small series, despite extensive efforts to maximize tumor and nucleic acid yield by using up to 40 sections of FFPE tissue, most samples could not be tested by RNA seq due to low quantity and quality of RNA. In light of the scant nature of the material, protocols designed for maximal preservation of fresh pericapsular fluid for molecular studies would be highly valuable to obtain high quality sequencing data.The concentration of the cellular content by centrifugation from larger volumesof pericapsular fluid would also greatly facilitate future studies.
Acknowledgments
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Footnotes
Data Availability Statement
Data available on request from the authors.
Conflict of Interest Disclosure: None.
References
- 1.Feldman AL, Harris NL, Stein H, et al. Breast implant-associated anaplastic large cell lymphoma. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J (Eds). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (Revised 4th Edition). IARC: Lyon; 2017. [Google Scholar]
- 2.Miranda RN, Aladily TN, Prince HM, et al. Breast implant-associated anaplastic large-cell lymphoma: long-term follow-up of 60 patients. J Clin Oncol. 2014;32(2):114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemens MW, Medeiros LJ, Butler CE, et al. Complete Surgical Excision Is Essential for the Management of Patients With Breast Implant-Associated Anaplastic Large-Cell Lymphoma. J Clin Oncol. 2016;34(2):160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemens MW, Jacobsen ED, Horwitz SM. 2019 NCCN Consensus Guidelines on the Diagnosis and Treatment of Breast Implant-Associated Anaplastic Large Cell Lymphoma (BIA-ALCL). Aesthet Surg J. 2019;39(Supplement_1):S3–S13. [DOI] [PubMed] [Google Scholar]
- 5.Quesada AE, Medeiros LJ, Clemens MW, Ferrufino-Schmidt MC, Pina-Oviedo S, Miranda RN. Breast implant-associated anaplastic large cell lymphoma: a review. Mod Pathol. 2019;32(2):166–188. [DOI] [PubMed] [Google Scholar]
- 6.Ferrufino-Schmidt MC, Medeiros LJ, Liu H, et al. Clinicopathologic Features and Prognostic Impact of Lymph Node Involvement in Patients With Breast Implant-associated Anaplastic Large Cell Lymphoma. Am J Surg Pathol. 2018;42(3):293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blombery P, Thompson ER, Jones K, et al. Whole exome sequencing reveals activating JAK1 and STAT3 mutations in breast implant-associated anaplastic large cell lymphoma anaplastic large cell lymphoma. Haematologica. 2016;101(9):e387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Napoli A, Jain P, Duranti E, et al. Targeted next generation sequencing of breast implant-associated anaplastic large cell lymphoma reveals mutations in JAK/STAT signalling pathway genes, TP53 and DNMT3A. Br J Haematol. 2018;180(5):741–744. [DOI] [PubMed] [Google Scholar]
- 9.Letourneau A, Maerevoet M, Milowich D, et al. Dual JAK1 and STAT3 mutations in a breast implant-associated anaplastic large cell lymphoma. Virchows Arch. 2018;473(4):505–511. [DOI] [PubMed] [Google Scholar]
- 10.Oishi N, Brody GS, Ketterling RP, et al. Genetic subtyping of breast implant-associated anaplastic large cell lymphoma. Blood. 2018;132(5):544–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crescenzo R, Abate F, Lasorsa E, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27(4):516–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oishi N, Miranda RN, Feldman AL. Genetics of Breast Implant-Associated Anaplastic Large Cell Lymphoma (BIA-ALCL). Aesthet Surg J. 2019;39(Supplement_1):S14–S20. [DOI] [PubMed] [Google Scholar]
- 13.Parrilla Castellar ER, Jaffe ES, Said JW, et al. ALK-negative anaplastic large cell lymphoma is a genetically heterogeneous disease with widely disparate clinical outcomes. Blood. 2014;124(9):1473–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldman AL, Dogan A, Smith DI, et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood.117(3):915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasmatzis G, Johnson SH, Knudson RA, et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood.120(11):2280–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ptashkin RN, Benayed R, Ziegler J, et al. MSK-IMPACT Heme: Validation and clinical experience of a comprehensive molecular profiling platform for hematologic malignancies [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2019; 2019 Mar 29-Apr 3; Atlanta, GA. Philadelphia (PA): AACR;. Cancer Res. 2019;79(13 Suppl):Abstract nr 3409. [Google Scholar]
- 17.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44(16):e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laurent C, Delas A, Gaulard P, et al. Breast Implant Associated Anaplastic Large cell Lymphoma: two distinct clinicopathological variants with different outcomes. Ann Oncol. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurent C, Nicolae A, Laurent C, et al. Gene alterations in epigenetic modifiers and JAK-STAT signaling are frequent in breast implant-associated ALCL. Blood. 2020;135(5):360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quintas-Cardama A, Verstovsek S. Molecular pathways: Jak/STAT pathway: mutations, inhibitors, and resistance. Clin Cancer Res. 2013;19(8):1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mottok A, Renne C, Seifert M, et al. Inactivating SOCS1 mutations are caused by aberrant somatic hypermutation and restricted to a subset of B-cell lymphoma entities. Blood. 2009;114(20):4503–4506. [DOI] [PubMed] [Google Scholar]
- 22.Weniger MA, Melzner I, Menz CK, et al. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene. 2006;25(18):2679–2684. [DOI] [PubMed] [Google Scholar]
- 23.Juskevicius D, Lorber T, Gsponer J, et al. Distinct genetic evolution patterns of relapsing diffuse large B-cell lymphoma revealed by genome-wide copy number aberration and targeted sequencing analysis. Leukemia. 2016;30(12):2385–2395. [DOI] [PubMed] [Google Scholar]
- 24.Ehrentraut S, Schneider B, Nagel S, et al. Th17 cytokine differentiation and loss of plasticity after SOCS1 inactivation in a cutaneous T-cell lymphoma. Oncotarget. 2016;7(23):34201–34216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma A, Oishi N, Boddicker RL, et al. Recurrent STAT3-JAK2 fusions in indolent T-cell lymphoproliferative disorder of the gastrointestinal tract. Blood. 2018;131(20):2262–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu G, Phillips JL, Dasari S, et al. Targetability of STAT3-JAK2 fusions: implications for T-cell lymphoproliferative disorders of the gastrointestinal tract. Leukemia. 2020;34(5):1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lacronique V, Boureux A, Valle VD, et al. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278(5341):1309–1312. [DOI] [PubMed] [Google Scholar]
- 28.Smith CA, Fan G. The saga of JAK2 mutations and translocations in hematologic disorders: pathogenesis, diagnostic and therapeutic prospects, and revised World Health Organization diagnostic criteria for myeloproliferative neoplasms. Hum Pathol. 2008;39(6):795–810. [DOI] [PubMed] [Google Scholar]
- 29.Adelaide J, Perot C, Gelsi-Boyer V, et al. A t(8;9) translocation with PCM1-JAK2 fusion in a patient with T-cell lymphoma. Leukemia. 2006;20(3):536–537. [DOI] [PubMed] [Google Scholar]
- 30.Davis TH, Morton CC, Miller-Cassman R, Balk SP, Kadin ME. Hodgkin’s disease, lymphomatoid papulosis, and cutaneous T-cell lymphoma derived from a common T-cell clone. N Engl J Med. 1992;326(17):1115–1122. [DOI] [PubMed] [Google Scholar]
- 31.Ehrentraut S, Nagel S, Scherr ME, et al. t(8;9)(p22;p24)/PCM1-JAK2 activates SOCS2 and SOCS3 via STAT5. PLoS One. 2013;8(1):e53767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panagopoulos I, Gorunova L, Spetalen S, et al. Fusion of the genes ataxin 2 like, ATXN2L, and Janus kinase 2, JAK2, in cutaneous CD4 positive T-cell lymphoma. Oncotarget. 2017;8(61):103775–103784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaffe ES, Ashar BS, Clemens MW, et al. Best Practices Guideline for the Pathologic Diagnosis of Breast Implant-Associated Anaplastic Large-Cell Lymphoma. J Clin Oncol. 2020;38(10):1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyapichev KA, Pina-Oviedo S, Medeiros LJ, et al. A proposal for pathologic processing of breast implant capsules in patients with suspected breast implant anaplastic large cell lymphoma. Mod Pathol. 2020;33(3):367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]