Endogenous Alu cDNA triggers inflammasome-mediated cell death in age-related macular degeneration.
Abstract
Long interspersed nuclear element-1 (L1)–mediated reverse transcription (RT) of Alu RNA into cytoplasmic Alu complementary DNA (cDNA) has been implicated in retinal pigmented epithelium (RPE) degeneration. The mechanism of Alu cDNA–induced cytotoxicity and its relevance to human disease are unknown. Here we report that Alu cDNA is highly enriched in the RPE of human eyes with geographic atrophy, an untreatable form of age-related macular degeneration. We demonstrate that the DNA sensor cGAS engages Alu cDNA to induce cytosolic mitochondrial DNA escape, which amplifies cGAS activation, triggering RPE degeneration via the inflammasome. The L1-extinct rice rat was resistant to Alu RNA–induced Alu cDNA synthesis and RPE degeneration, which were enabled upon L1-RT overexpression. Nucleoside RT inhibitors (NRTIs), which inhibit both L1-RT and inflammasome activity, and NRTI derivatives (Kamuvudines) that inhibit inflammasome, but not RT, both block Alu cDNA toxicity, identifying inflammasome activation as the terminal effector of RPE degeneration.
INTRODUCTION
Age-related macular degeneration (AMD) is a vision-threatening disease estimated to affect 200 million people worldwide (1). Geographic atrophy, an advanced form of AMD characterized by progressive retinal pigment epithelium (RPE) cell death (2), has no approved treatments. The accumulation of Alu retrotransposon RNAs in the RPE is implicated in the pathogenesis of geographic atrophy (3, 4). These noncoding RNAs act as an endogenous trigger for NLRP3 inflammasome activation, leading to production of the proinflammatory, cytotoxic cytokines (5).
The replication cycle of Alu involves its reverse transcription (RT) and genomic insertion, a process termed target-primed RT, which involves long interspersed nuclear element-1 (L1) RT and L1 endonuclease (EN) activities (6–8). These events enabling retrotransposition of Alu are thought to be coupled in the nucleus (9, 10). However, we recently demonstrated that Alu RNA can also be reverse-transcribed into Alu complementary DNA (cDNA) via L1-RT activity in the cytoplasm, independent of retrotransposition, and that cytoplasmic Alu cDNA promotes RPE degeneration in mice (11). However, whether the product of this newly discovered shunt pathway in the Alu replication cycle is detectable and enriched in human geographic atrophy is unknown.
Previously, we also determined that Alu RNA–induced RPE degeneration requires both canonical (caspase-1–mediated) and noncanonical (caspase-4–mediated) inflammasome activation (5, 12). The latter is triggered by Alu RNA–induced cytosolic escape of mitochondrial DNA (mtDNA) and its recognition by the DNA sensor cyclic guanosine 3′,5′-monophosphate–adenosine 3′,5′-monophosphate (cGAMP) synthase (cGAS), whose abundance is increased in human geographic atrophy eyes and whose activation promotes RPE degeneration (12). However, it is also unknown how the newly described cytosolic Alu cDNA collaborates with mtDNA for cGAS and subsequent inflammasome activation.
Here, we report an increased abundance of endogenous L1 mRNA and Alu cDNA in the RPE of human geographic atrophy eyes. We also demonstrate that Alu cDNA engages cGAS via a guanosine-rich immunostimulatory motif (ISM) to induce cytosolic mtDNA release, which induces RPE degeneration via subsequent amplification of cGAS activation. Last, we demonstrate the linearity of the Alu RNA–Alu cDNA–mtDNA-cGAS inflammasome activation cascade in triggering RPE degeneration by demonstrating that U.S. Food and Drug Administration (FDA)–approved nucleoside RT inhibitors (NRTIs), which inhibit both L1-RT and inflammasome activation, and Kamuvudines, alkylated NRTI derivatives that inhibit inflammasome activation but not L1-RT, both block Alu cDNA–induced RPE degeneration.
RESULTS
Endogenous Alu cDNA in human geographic atrophy
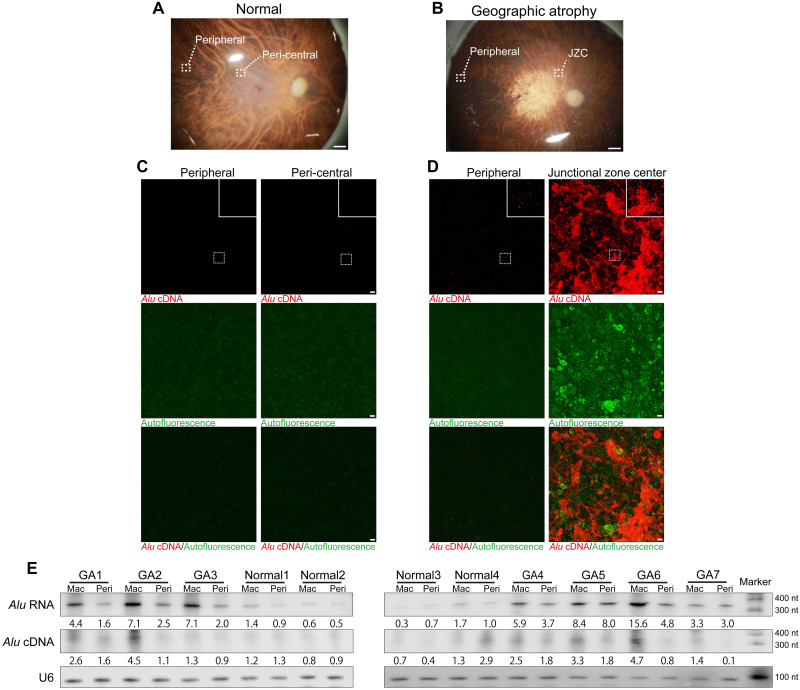
Previously, we demonstrated that Alu RNA–induced RPE degeneration and inflammation are mediated via cytoplasmic L1–reverse-transcribed Alu cDNA independently of retrotransposition in human RPE cells and mice (11). In geographic atrophy, an untreatable, advanced vision-threatening form of AMD (2), Alu RNA expressed from endogenous Alu retrotransposons by RNA polymerase III accumulates in the RPE (3, 4). We performed in situ hybridization of Alu cDNA in ribonuclease A (RNase A)–treated RPE whole mounts of human donor eyes with geographic atrophy. Alu cDNA was enriched at the center of the junctional zone and its border with the atrophic area (Fig. 1, A to D, and fig. S1, A to E), less abundant in peripheral disease-free areas of affected eyes (Fig. 1D), and only faintly detected in normal control eyes (Fig. 1C). Geographic atrophy is topographically heterogeneous within the retina: A junctional zone is interposed between a central area of atrophy and a peripheral area of surviving RPE cells. This metastable region consists of stressed and degenerating RPE cells (13), displays impaired visual function (14), and undergoes atrophy over time as the disease expands centrifugally (15). Thus, Alu cDNA is spatially enriched in the most dynamic zone of disease and nearly undetectable in disease-free regions, suggestive of its contribution to geographic atrophy progression. S1 nuclease abolished the Alu cDNA signal, confirming its single strandedness (fig. S1F).
Fig. 1. Endogenous Alu cDNA in RPE of human geographic atrophy.
(A and B) Photographs of normal human donor eye retina illustrating peripheral and peri-central areas (A) and geographic atrophy (GA) retina illustrating peripheral and junctional zone center (JZC) areas (B). Scale bars, 1 mm. (C and D) In situ hybridization of RPE whole mounts with Alu cDNA–specific probes in peripheral and peri-central areas of normal eyes (C) and in peripheral and JZC areas of GA eyes (D). Insets show higher magnification. Red, Alu cDNA; green, autofluorescence. Scale bars, 10 μm. (E) Northern blotting of Alu RNA and equator blotting of Alu cDNA in macular (Mac) and peripheral (Peri) RPE of human GA (n = 7) and normal (n = 4) eyes. Densitometry of the bands corresponding to Alu RNA and Alu cDNA normalized to loading control (U6) with the mean densitometry ratio of macular RPE of normal eyes set to 1.0.
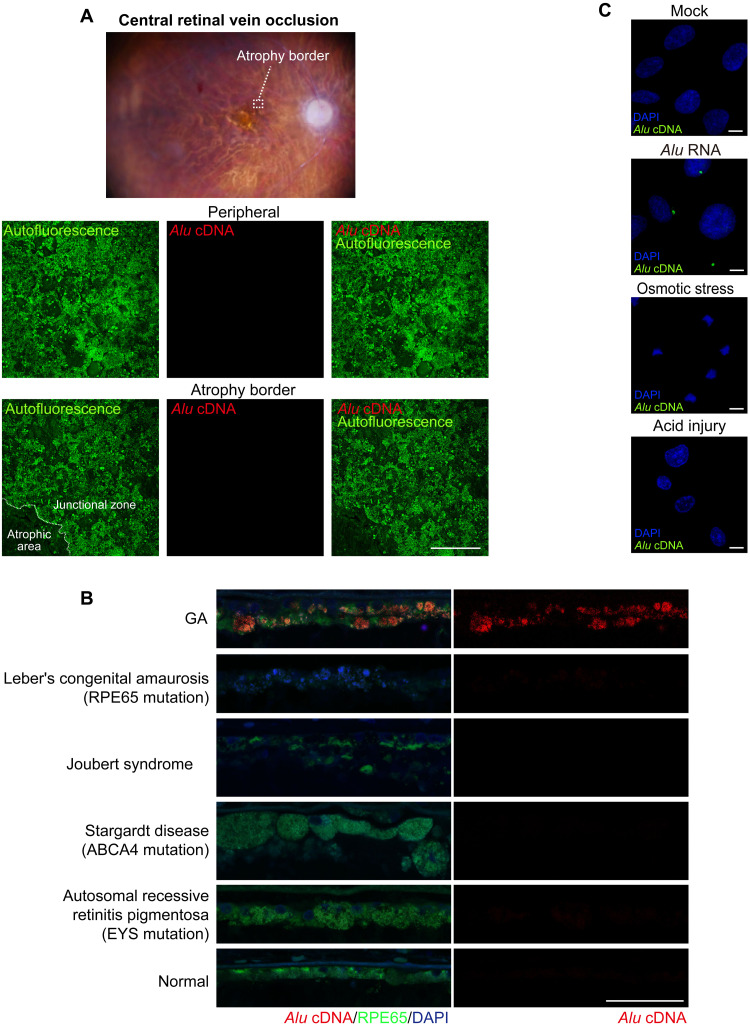
We next used “equator blotting,” a new variation of nucleic acid blotting we recently described (11). This technique combines Southern and Northern blotting functionally to detect extrachromosomal DNAs and their size (11). Equator blotting detected increased abundance of Alu cDNA in the macular RPE of geographic atrophy eyes, and Northern blotting detected increased abundance of Alu RNA in the same region of disease (Fig. 1E and fig. S1G). As the Alu cDNA signal was approximately 300 nts long, it was compatible with bona fide nongenomic Alu cDNA molecules. We did not detect Alu cDNA in numerous other human retinal diseases (Fig. 2, A and B), suggesting a disease specificity to Alu cDNA accumulation in geographic atrophy. Furthermore, primary human RPE cells did not synthesize Alu cDNA when exposed to osmotic stress or acid injury (Fig. 2C), indicating that Alu cDNA generation is not a generic response of dying cells.
Fig. 2. Absence of endogenous Alu cDNA in human retinal diseases other than GA.
(A) Ex vivo fundus photograph of a human eye with RPE atrophy that developed subsequent to treatment of central retinal vein occlusion with anti-angiogenic drugs. In situ hybridization of RPE whole mounts showing no detectable endogenous Alu cDNA in peripheral RPE or at the border of the atrophic RPE. Scale bars, 200 μm. Red, Alu cDNA; green, autofluorescence of RPE cells. (B) Abundant endogenous Alu cDNA detected in the RPE of a human GA eye but not in the RPE of human eyes with Leber congenital amaurosis, Joubert syndrome, Stargardt macular dystrophy, or autosomal recessive retinitis pigmentosa. Red, Alu cDNA; green, RPE65; blue, 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 50 μm. (C) In situ hybridization of Alu cDNA formation in primary human RPE cells subjected to Alu RNA transfection, osmotic stress (distilled H2O), or acid injury (hydrochloric acid; HCl, pH 4.0 medium). Alu cDNA, green; DAPI, blue. Scale bars, 10 μm.
L1-RT activity is required for Alu RNA–induced toxicity
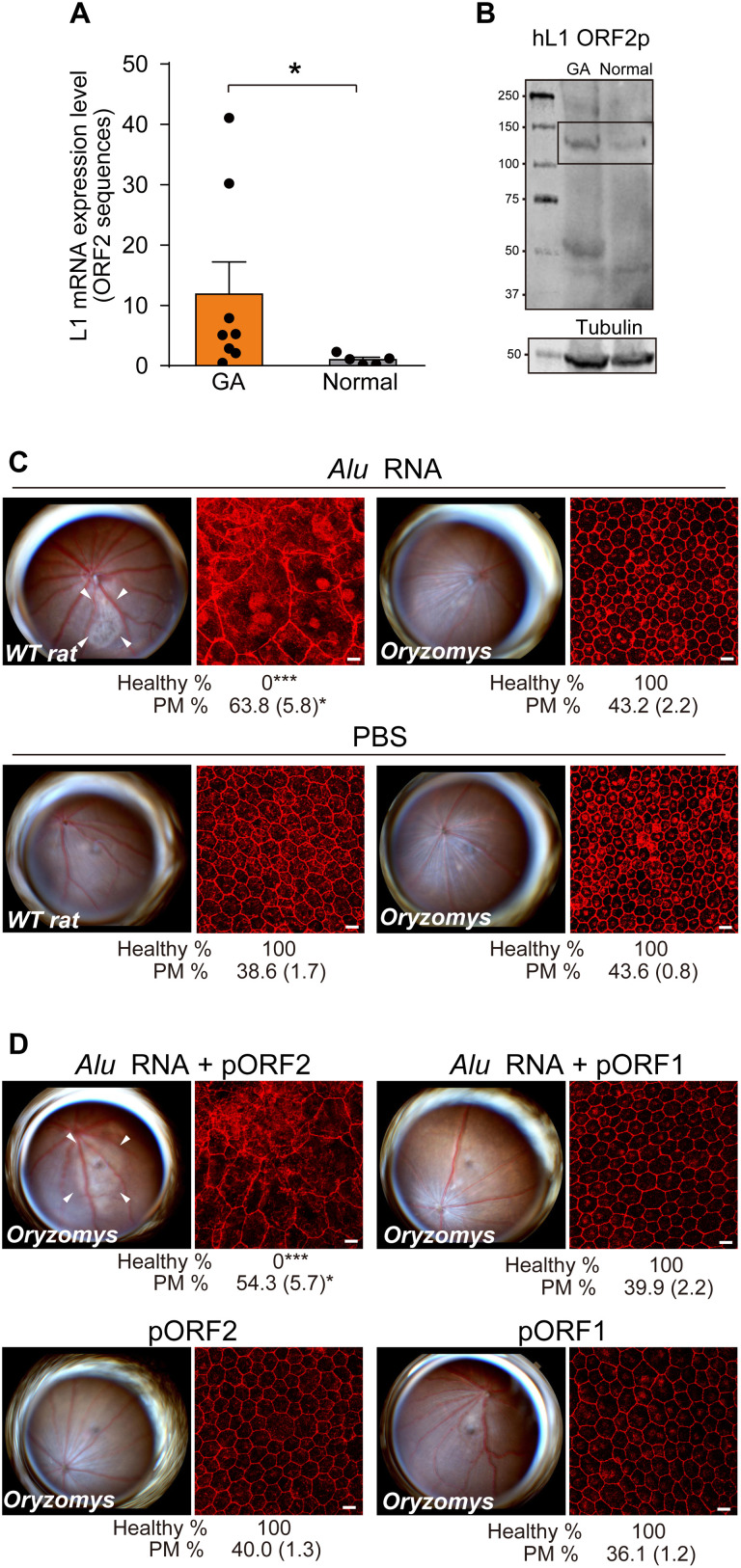
L1-encoded mRNA and ORF2 protein were enriched in the macular RPE of human geographic atrophy eyes (Fig. 3, A and B, and fig. S2, A to E). Therefore, to determine whether L1 activity is required for Alu toxicity, we investigated Alu RNA toxicity in a heterologous system using the rice rat (Oryzomys palustris), an “L1 extinct species” (16–19) whose genome lacks retrotransposition-competent L1 loci due to insertions, deletions, and stop codons within formerly intact L1 sequences. Endogenously expressed rat L1 mRNAs harboring L1 ORF1 or L1 ORF2 sequences were not detected in O. palustris RPE cells (fig. S3A). Alu RNA induced RPE degeneration in L1Rn-competent Brown-Norway rats [Rattus norvegicus also designated as wild-type (WT) rat] but not in L1-incompetent O. palustris (Fig. 3C). In vivo–enforced expression (fig. S3B) of plasmid-encoded rat L1 ORF2p (which encodes L1’s RT and EN activities), but not of rat L1 ORF1p, restored Alu RNA toxicity in O. palustris (Fig. 3D). These data suggest L1Rn-encoded ORF2p activity is required for Alu RNA toxicity.
Fig. 3. Endogenous L1 is required for Alu cDNA synthesis.
(A) Endogenous L1 mRNA abundance, monitored by real-time polymerase chain reaction (PCR) in the macular RPE of human GA eyes (n = 8) compared with normal human eyes (n = 5). *P < 0.05 by Mann-Whitney U test. Error bars show SEM. (B) Immunoblotting analysis of cell extracts from macular RPE from individual human donor eyes. Lysates were used to detect endogenous L1 ORF2p. (C) Fundus photographs and RPE morphology stained for zonula occludens-1 (ZO-1; red) in R. norvegicus (WT rat) or O. palustris (Oryzomys) following Alu RNA administration. n = 5 to 9. (D) Fundus photographs and RPE morphology stained for ZO-1 in O. palustris after Alu RNA administration following enforced expression of rat L1 ORF2p or rat L1 ORF1p. n = 5 to 9. Binary and morphometric quantification of RPE degeneration are shown (*P < 0.05; ***P < 0.001). PM, polymegethism [mean (SEM)]. Arrowheads in fundus images denote the boundaries of RPE hypopigmentation.
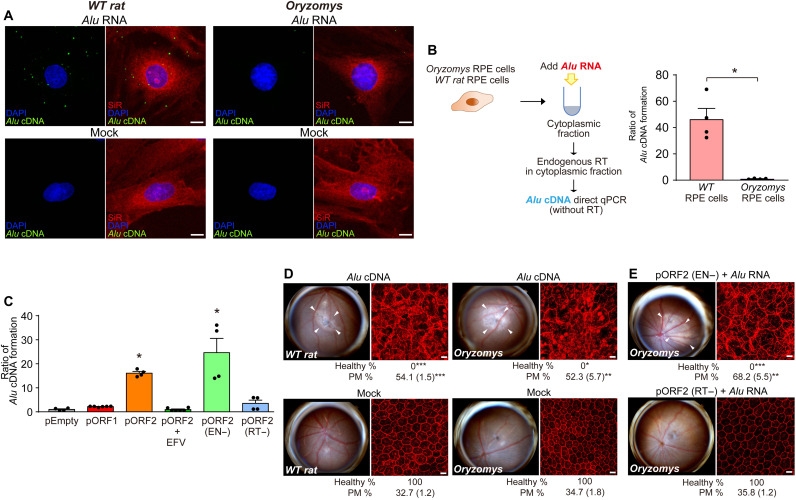
As expected, Alu cDNA formation after Alu RNA transfection was not detectable in L1-extinct O. palustris RPE cells compared with R. norvegicus RPE cells (Fig. 4, A and B). Enforced expression of rat L1 ORF2p, but not of rat L1 ORF1p, increased Alu cDNA formation in O. palustris RPE cells; this was inhibited by high doses of the non–nucleoside reverse transcriptase inhibitor (NonNRTI) efavirenz (EFV) (Fig. 4C), which inhibits L1-RT at high doses (20). Unlike Alu RNA, which did not induce RPE degeneration in O. palustris (Fig. 3C), subretinal administration of Alu cDNA, which bypasses L1 deficiency, induced RPE degeneration in O. palustris just as in R. norvegicus (Fig. 4D). Together, these data are consistent with the concept that L1-mediated formation of Alu cDNA is essential for Alu RNA toxicity.
Fig. 4. Cytoplasmic Alu cDNA is synthesized via retrotransposition-independent RT.
(A) Alu cDNA formation monitored by in situ hybridization of RPE cells of R. norvegicus (WT rat) and O. palustris (Oryzomys) transfected with Alu RNA. Green, Alu cDNA; red, SiR-actin; blue, DAPI. Scale bars, 10 μm. Representative of n = 6. (B) Quantitative PCR (qPCR) of Alu cDNA generated by endogenous RT activity in cytoplasmic fractions of RPE cells of WT rat and O. palustris incubated with Alu RNA. *P < 0.05, Mann-Whitney U test. (C) Quantification of Alu cDNA products in cytoplasmic fractions of O. palustris RPE cells by Alu-specific PCR as described above. Cells were transfected with a control plasmid (pEmpty) or plasmids to enforce expression of rat L1 ORF1p, ORF2p (in the presence or absence of EFV), EN-deficient (EN−) ORF2p, or RT-deficient (RT−) ORF2p. *P < 0.05, Mann-Whitney U test. The error bars in (B) and (C) represent the means ± SEM. (D and E) Fundus photographs (left) and corresponding representative RPE sheet micrographs stained for ZO-1 (red) (right) of WT rat and O. palustris. Scale bars, 10 μm. Arrowheads in fundus images denote the boundaries of RPE hypopigmentation. Binary and morphometric quantification of RPE degeneration are shown. *P < 0.05; **P < 0.01; ***P < 0.001, Fisher’s exact test for binary; two-tailed t test for morphometry. PM, polymegethism [mean (SEM)]. n = 6 to 12. (D) RPE morphology in WT rats and Oryzomys after administration of Alu cDNA or mock injection. (E) RPE morphology in Oryzomys after Alu RNA administration following enforced expression of EN- or RT-deficient L1 ORF2p.
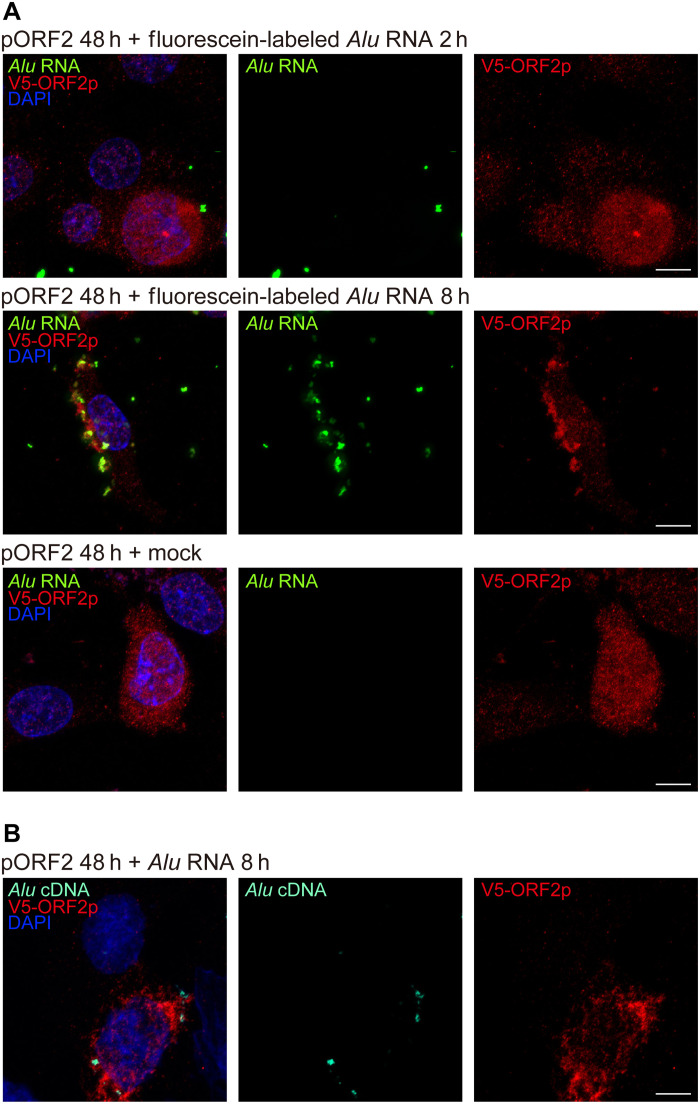
Next, we enforced expression of an EN-deficient (EN−) rat L1 ORF2p mutant and a RT-deficient (RT−) rat L1 ORF2p mutant, both of which were incapable of supporting Alu retrotransposition in a trans-mobilization assay (fig. S4). Alu RNA transfection into cultured O. palustris RPE cells induced Alu cDNA formation in the presence of L1 ORF2p (EN−) but not in the presence of L1 ORF2p (RT−) (Fig. 4C). Similarly, in vivo–enforced expression of the (EN−) L1 ORF2p mutant restored the ability of Alu RNA to induce RPE degeneration in O. palustris (Fig. 4E). In contrast, Alu RNA did not induce RPE degeneration in O. palustris following in vivo–enforced expression of the (RT−) L1 ORF2p mutant (Fig. 4E). These data identify L1 ORF2p’s RT activity as crucial and its EN activity as dispensable for cytoplasmic Alu cDNA synthesis and Alu RNA–induced retinal toxicity. To monitor Alu cDNA formation in the cytoplasm, we probed the association between L1 ORF2p, Alu RNA, and Alu cDNA in O. palustris RPE cells. We transfected O. palustris RPE cells with rat pORF2 and Alu RNA, and we observed colocalization of both Alu RNA and Alu cDNA with ORF2p (Fig. 5, A and B). These data support a model in which Alu RNA associates with L1 ORF2p in the cytoplasm and is reverse-transcribed into Alu cDNA, which is consistent with our previous report that Alu cDNA can be synthesized in the cytoplasm (11).
Fig. 5. Colocalization of L1 ORF2p, Alu RNA, and Alu cDNA.
(A) Fluorescence imaging of O. palustris RPE cells coexpressing ORF2p-V5 (detected by anti-V5 antibody, red) and fluorescein-labeled Alu RNA (transfected 48 hours after V5-ORF2 transfection, green). Images were acquired 2 and 8 hours after Alu RNA transfection. Note the diffuse localization of ORF2p-V5 and punctate foci of fluorescein-Alu RNA after 2 hours and cytoplasmic colocalization of multiple punctate foci 8 hours after Alu RNA transfection. ORF2p-V5 localization remained diffuse in the absence of transfected fluorescein-Alu RNA (mock, bottom). (B) In situ hybridization of O. palustris RPE cells shows coexpression of V5-ORF2 (red) and Alu cDNA (teal), detected by an Alu cDNA specific probe (48 hours after V5-ORF2 transfection and 8 hours after Alu RNA transfection). Note the cytoplasmic colocalization of ORF2p-V5 (red) and Alu cDNA at 8 hours after Alu RNA transfection. DAPI, blue. Scale bars, 10 μm.
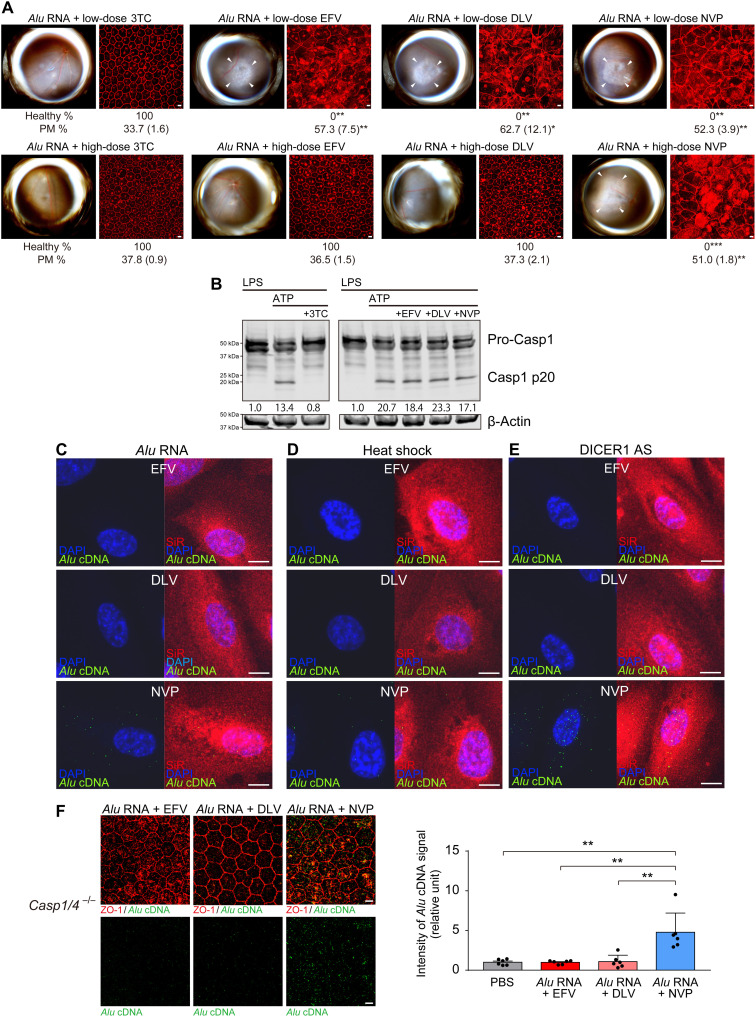
The NonNRTIs EFV and delavirdine (DLV) inhibit L1-RT at high doses but not at low doses (20). At low doses equimolar to the effective concentration of the NRTI lamivudine (3TC) (21), intravitreous administration of EFV or DLV did not block Alu RNA–induced RPE degeneration in WT mice (Fig. 6A). However, EFV and DLV blocked Alu RNA toxicity at high doses (Fig. 6A). At these high doses, EFV and DLV did not inhibit NLRP3 inflammasome activation (as monitored by caspase-1 activation) by lipopolysaccharide (LPS) and adenosine triphosphate (ATP) stimulation (Fig. 6B). Therefore, the cytoprotective effect of high-dose EFV and DLV is attributed to L1-RT inhibition and not to inflammasome inhibition. In contrast, the NonNRTI nevirapine (NVP), which does not inhibit L1-RT (20) or NLRP3 inflammasome activation (Fig. 6B), did not prevent Alu RNA toxicity at low or high doses (Fig. 6A). DLV also blocked Alu RNA toxicity in O. palustris reconstituted with rat L1 ORF2p (fig. S5). The results of these three complementary L1-RT inhibitory strategies further suggest that endogenous L1-RT activity mediates Alu RNA toxicity. High doses of EFV and DLV (20) also prevented Alu cDNA synthesis by endogenous L1-RT in primary human RPE cells after Alu RNA transfection, heat shock, or DICER1 knockdown (Fig. 6, C to E). We injected Alu RNA in Casp1−/− Casp4−/− mice, which are resistant to Alu RNA toxicity (5, 12), to avoid distortions arising from degenerating cells that would impair visualization of Alu cDNA signals in WT mice, and we found that high doses of EFV and DLV also prevented Alu cDNA formation in these mice (Fig. 6F). In contrast, NVP was unable to prevent Alu cDNA formation in any of these settings (Fig. 6, C to F). These findings suggest overactive conversion of Alu RNA to Alu cDNA by endogenous L1-ORF2p contributes to RPE cell death.
Fig. 6. Effects of NonNRTIs on Alu cDNA synthesis.
(A) RPE degeneration (fundus photos, left; ZO-1 flat mounts, right) in WT mice after treatment with low doses (50 pmol) of 3TC, EFV, DLV, or NVP (top row) and with high doses (500 pmol) of 3TC, EFV, DLV, or NVP (bottom row). ZO-1 (red). Scale bars, 10 μm. n = 6. Binary and morphometric quantification of RPE degeneration are shown. (*P < 0.05; **P < 0.01; ***P < 0.001, Fisher’s exact test for binary; two-tailed t test for morphometry). PM, polymegethism [mean (SEM)]. Arrowheads in fundus images denote the boundaries of RPE hypopigmentation. (B) Detection of caspase-1 activation by immunoblotting of primary mouse bone marrow–derived macrophages (BMDMs) treated with LPS and ATP, in the presence of 3TC, EFV, DLV, or NVP. Densitometry of the bands corresponding to active caspase-1 (Casp1 p20) normalized to loading control (β-actin). (C to E) In situ hybridization of Alu cDNA (green) in primary human RPE cells after treatment with in vitro transcribed Alu RNA, DICER1 antisense oligonucleotides (DICER1 AS), or heat shock after treatment with high doses of EFV, DLV, and NVP. SiR (F-actin, red), DAPI (blue). Scale bars, 10 μm. (F) In situ hybridization to detect Alu cDNA (green) of RPE whole mounts from Casp1−/− Casp4−/− mice after subretinal administration of in vitro–transcribed Alu RNA after treatment with EFV, DLV, or NVP. ZO-1 (red). Scale bars, 10 μm. Bar graph (right) of signal intensity. **P < 0.01 by Mann-Whitney U test. Error bars show SEM. n = 6.
Alu cDNA toxicity is mediated by cGAS
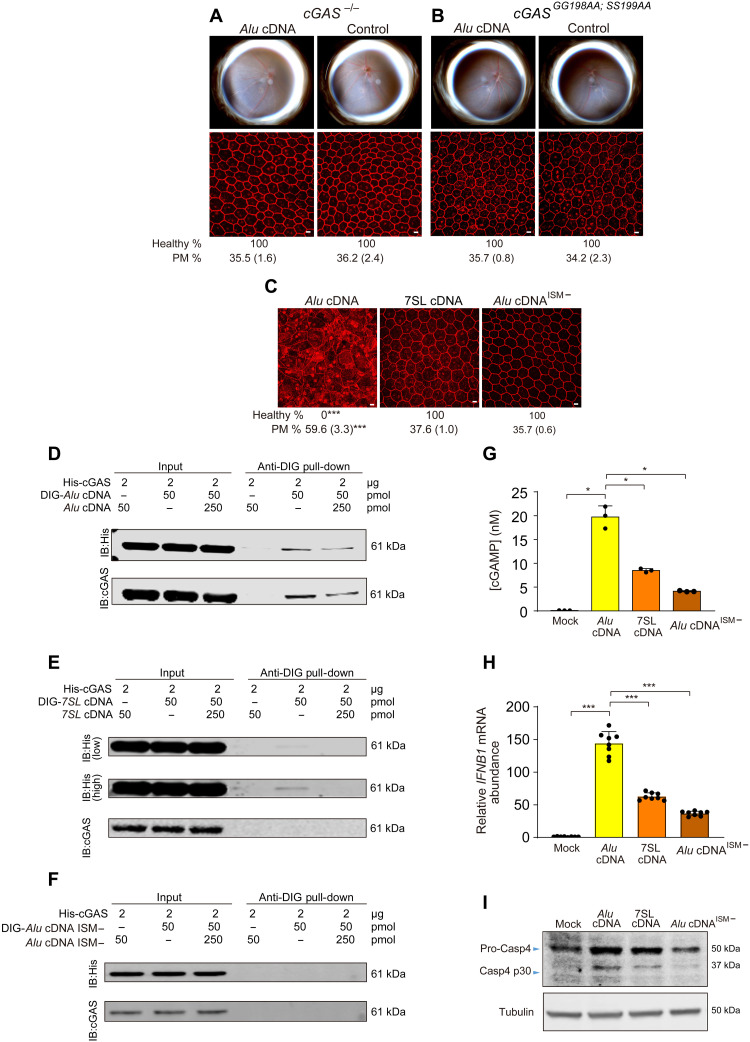
We next investigated whether various DNA sensors mediated Alu cDNA toxicity. Alu cDNA induced RPE degeneration in mice lacking the gene encoding absent in melanoma 2 (AIM2) and in mice lacking the genes encoding multiple Toll-like receptors (fig. S6A). In contrast, mice lacking cGAS were protected (Fig. 7A). To determine whether the catalytic activity of cGAS was essential for Alu cDNA toxicity, we generated a mouse strain containing two homozygous point mutations in cGAS that eliminated its ability to catalyze cGAMP synthesis in human embryonic kidney (HEK) 293T cells (22). In these cGASGG198AA; SS199AA mice, Alu cDNA did not induce RPE degeneration (Fig. 7B).
Fig. 7. Alu cDNA induces RPE degeneration via cGAS.
(A to C) Fundus photographs (top) and corresponding representative RPE sheet micrographs (bottom) of mice. Scale bars, 10 μm. Binary and morphometric quantification of RPE degeneration are shown. ***P < 0.001, Fisher’s exact test for binary; two-tailed t test for morphometry. PM, polymegethism [mean (SEM)]. RPE morphology after Alu cDNA or control administration in cGAS−/− mice (n = 6 to 8) (A) and catalytically null cGASGG198AA; SS199AA mice (n = 6) (B). (C) RPE morphology in WT mice after administration of Alu cDNA, 7SL cDNA, or a mutant form of Alu cDNA lacking a guanosine-rich ISM (Alu cDNAISM−). n = 6 to 8. (D to F) Immunoblotting of recombinant HIS-tagged cGAS using anti-HIS and anti-cGAS antibodies before (input) and after pull-down with anti–digoxigenin (DIG) antibody in the presence or absence of DIG-labeled or unlabeled competitor Alu cDNA (D), DIG-labeled or unlabeled competitor 7SL cDNA (E), or DIG-labeled or unlabeled competitor Alu cDNA lacking an ISM (Alu cDNAISM−) (F). Low- and high-exposure blots are presented in (E). (G) Enzyme-linked immunosorbent assay quantification of cGAMP production by recombinant cGAS in the presence of Alu cDNA, 7SL cDNA, or Alu cDNAISM−. (H) qRT-PCR quantification of IFNB1 mRNA in human RPE cells transfected with Alu cDNA, 7SL cDNA, or Alu cDNAISM−. *P < 0.05, ***P < 0.001, Mann-Whitney U test. The error bars in (G) and (H) represent the means ± SEM. (I) Immunoblots for pro-Casp4 and Casp4 p30 of human RPE cells transfected with Alu cDNA, 7SL cDNA, or Alu cDNAISM−.
cGAS-STING–driven type I interferon (IFN) signaling licenses the caspase-4–dependent noncanonical inflammasome in a gasdermin D–mediated fashion in the context of Alu RNA–induced RPE degeneration (12). Similar to Alu RNA, Alu cDNA did not induce RPE degeneration in mice lacking STING (encoded by Tmem173), the type I IFN-α/β receptor (encoded by Ifnar1), the transcription factor IRF3 that induces IFN-β production, caspase-4, or gasdermin D (fig. S6B).
A synthetically generated cDNA of 7SL RNA, the evolutionary precursor of Alu sequences (23), did not induce RPE degeneration in WT mice (Fig. 7C and fig. S7A) except at a dose 250 times greater than the least toxic dose of Alu cDNA (fig. S7B), possibly due to nonspecific DNA sensing. Using the mfold secondary structure prediction algorithm (24), we observed that Alu cDNA contained a guanosine-rich ISM that has been found to enhance cGAS-mediated IFN-β induction (25), whereas 7SL cDNA did not (fig. S8, A and B). To investigate the importance of this ISM, we generated Alu cDNAISM−, a mutant form of Alu cDNA lacking this motif (fig. S8C), and 7SL cDNAISM+, a mutant form of 7SL cDNA containing this motif (fig. S8D). In WT mice, Alu cDNAISM− did not induce RPE degeneration (Fig. 7C and fig. S8E), whereas 7SL cDNAISM+ did induce RPE degeneration (fig. S8F), suggesting that this ISM motif is indeed critical for Alu cDNA toxicity.
We monitored binding of Alu cDNA, 7SL cDNA, and Alu cDNAISM− to cGAS by incubating digoxigenin (DIG)–labeled cDNAs with recombinant His-tagged cGAS. Labeled Alu cDNA readily bound cGAS and was displaced by unlabeled Alu cDNA (Fig. 7D); in contrast, 7SL cDNA and Alu cDNAISM− did not interact with cGAS as efficiently (Fig. 7, E and F). We also examined cGAS-cDNA interaction in cell culture using cGAS−/− immortalized mouse embryonic fibroblasts (MEFs) reconstituted with hemagglutinin (HA)–tagged cGAS, which express HA-cGAS from a genomic transgene at levels similar to endogenous expression (26). Immunoprecipitation followed by real-time polymerase chain reaction (PCR) revealed enrichment of Alu cDNA in the HA-cGAS immunoprecipitate relative to 7SL cDNA or Alu cDNAISM− (fig. S8, G and H). Consistent with differential cGAS binding, Alu cDNA stimulated greater cGAMP synthesis when incubated with recombinant cGAS compared to 7SL cDNA or Alu cDNAISM− (Fig. 7G) and more robustly induced IFN-β (Fig. 7H) and caspase-4 activation (Fig. 7I) in human RPE cells, which we demonstrated was driven by cGAS-induced IFN signaling (12).
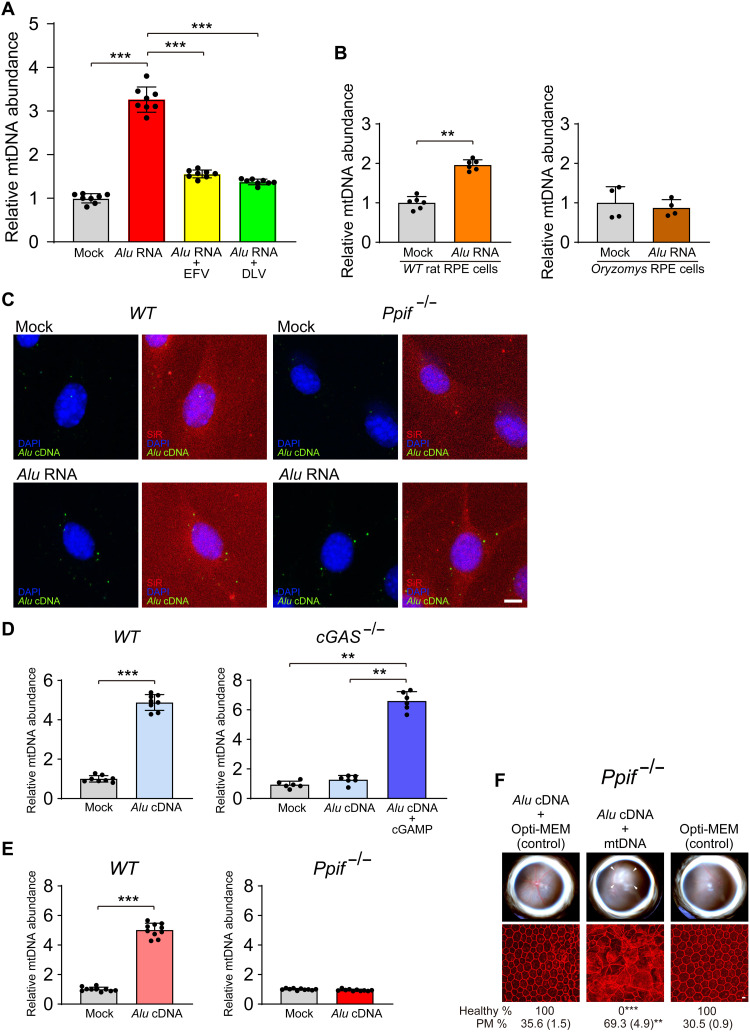
Alu RNA–induced RPE degeneration requires mitochondrial peptidyl-prolyl cis–transisomerase F (PPIF)–mediated cytosolic release of mtDNA and its activation of cGAS (12). We sought to understand the interrelationship of Alu cDNA and mtDNA in this system. Alu RNA–induced mtDNA release was blocked when Alu cDNA formation was disabled by RT inhibitors (EFV/DLV) or L1 inactivity (O. palustris cells) (Fig. 8, A and B). Conversely, disabling mtDNA release by Ppif ablation did not block Alu cDNA formation (Fig. 8C). Thus, Alu cDNA synthesis is upstream of and required for mtDNA release. Alu cDNA–induced mtDNA release in WT MEFs was inhibited in cGAS−/− MEFs but restored after cGAMP transfection, suggesting that cGAS-catalyzed cGAMP promotes Alu cDNA–induced mtDNA release (Fig. 8D). Alu cDNA–induced mtDNA release was also inhibited in Ppif−/− mouse RPE cells (Fig. 8E). Also, Alu cDNA–induced RPE degeneration was inhibited in Ppif−/− mice but was restored by coadministering mtDNA (Fig. 8F). Collectively, these data support a model in which Alu cDNA uses cGAS to induce mtDNA release, which subsequently amplifies cGAS activation to induce RPE toxicity (fig. S9).
Fig. 8. Alu cDNA-induced mtDNA release is cGAS-dependent.
(A and B) mtDNA was quantified in cytoplasmic DNA free of intact mitochondria and normalized to mock-treated cells as described previously (12) in (A) mouse RPE cells after transfection with Alu RNA following treatment with RT inhibitors (EFV or DLV) or (B) O. palustris cells lacking functional genomic L1 loci. This cytoplasmic DNA was subjected to 12S and 16S mtDNA qPCR. n = 4 to 8. **P < 0.01; ***P < 0.001 by Mann-Whitney U test. Error bars show SEM. (C) In situ hybridization of Alu cDNA (green) in primary mouse RPE cells (WT and Ppif−/−) transfected with Alu RNA. DAPI (blue), SiR (F-actin, red). Scale bar, 10 μm. (D) Quantification of mtDNA release induced by Alu cDNA in WT and cGAS−/− MEFs. Transfection of cGAMP into cGAS−/− MEFs restored Alu cDNA–induced mtDNA release (right, dark blue bar); n = 8. **P < 0.01; ***P < 0.001 by Mann-Whitney U test. (E) mtDNA release in WT and Ppif−/− mouse RPE cells. n = 9 to 10. ***P < 0.001 by Mann-Whitney U test. (F) RPE degeneration after Alu cDNA administration in Ppif−/− mice with or without coadministration of mtDNA. Fundus photos, top; flat mounts stained for ZO-1 (red), bottom. White arrowheads indicate degenerated area in the fundus image. Scale bar, 10 μm. Binary and morphometric quantification of RPE degeneration are shown (**P < 0.01; ***P < 0.001, Fisher’s exact test for binary; two-tailed t test for morphometry). PM, polymegethism [mean (SEM)]. Arrowheads in fundus images denote the boundaries of RPE hypopigmentation.
NRTIs block toxicity by inhibiting RT and inflammasome
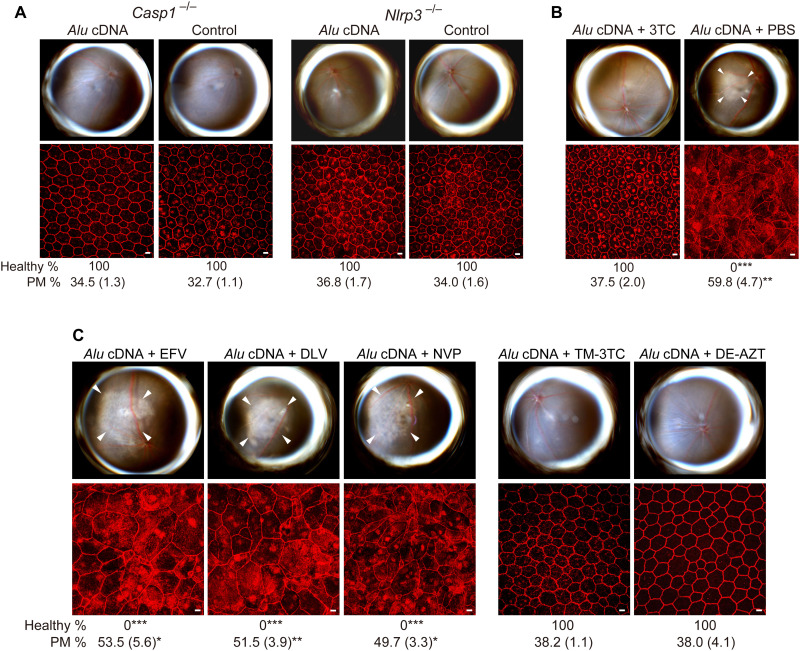
Downstream of cGAS-mediated IFN signaling, Alu RNA induced caspase-1 cleavage via the NLRP3 inflammasome (5, 12). We investigated whether this terminal inflammasome pathway was also involved in Alu cDNA toxicity. Alu cDNA, like Alu RNA, activated caspase-1 in primary human RPE cells (fig. S10A). Alu cDNA transfection led to greater caspase-1 activation compared to an equal quantity of Alu RNA, which was reflected in vivo: On a molar basis, Alu cDNA was at least 100 times more potent in inducing RPE degeneration (fig. S7B) than the doses of Alu RNA required to induce toxicity [figure S3 in (11)]. This inflammasome pathway was critical as Alu cDNA did not induce RPE degeneration in Casp1−/− or Nlrp3−/− mice (Fig. 9A), consistent with the impaired caspase-1 activation induced by Alu cDNA in Nlrp3−/− bone marrow–derived macrophages (BMDMs; fig. S10B).
Fig. 9. Alu cDNA induces RPE degeneration via inflammasome activation.
(A to C) Fundus photos (top) and ZO-1–stained (red) RPE flat mounts (bottom) of mice. White arrowheads indicate degenerated area in the fundus images. Scale bars, 10 μm. Binary and morphometric quantification of RPE degeneration are shown (*P < 0.05, **P < 0.01, ***P < 0.001, Fisher’s exact test for binary; two-tailed t test for morphometry). PM, polymegethism [mean (SEM)]. Arrowheads in fundus images denote the boundaries of RPE hypopigmentation. (A) Casp1−/− (Casp1−/− Casp4129mt/129mt Casp4Tg mice) or Nlrp3−/− mice injected with Alu cDNA or control. n = 6. (B) Effect of 500 pmol of 3TC on Alu cDNA–induced RPE degeneration in WT mice. n = 6. (C) Effect of high doses (500 pmol) of EFV, DLV, or NVP and of trimethyl-3TC (TM-3TC) or diethyl-AZT (DE-AZT) on RPE degeneration in WT mice. n = 4 to 6.
Consistent with its intrinsic anti-inflammatory function (21), 3TC protected against Alu cDNA–induced RPE death (Fig. 9B and fig. S11). Therefore, 3TC appeared to have dual mechanisms of protection against Alu RNA toxicity: (i) blocking conversion of Alu RNA to Alu cDNA and (ii) inhibiting Alu cDNA–mediated NLRP3 inflammasome signaling. Doses of NonNRTIs capable of inhibiting L1-RT (Fig. 6, C to E), however, did not block Alu cDNA toxicity (Fig. 9C). Thus, in contrast to 3TC, the protective effect of high dose NonNRTIs against Alu RNA toxicity is due to their inhibition of RT, whereas their inability to protect against Alu cDNA toxicity is due to their lack of NLRP3 inflammasome antagonism. In contrast, the Kamuvudines TM-3TC and DE-AZT, alkylated NRTI derivatives that inhibit inflammasome activation but not L1-RT (27), blocked Alu cDNA–induced caspase-1 activation in RPE cells (fig. S10C) and RPE degeneration in WT mice (Fig. 9C and fig. S11). Collectively, these data indicate that Alu cDNA is interposed between Alu RNA and inflammasome activation and that inflammasome inhibition is sufficient to block RPE degeneration induced by Alu RNA or Alu cDNA.
DISCUSSION
Our studies reveal the existence of Alu cDNA accumulation in patient samples from a human disease. These findings expand upon our recent description of cytoplasmic synthesis of Alu cDNA reverse-transcribed from Alu RNA via L1-RT activity in experimental systems. Our new data demonstrate the relevance of Alu cDNA to the pathogenesis of geographic atrophy in particular and, possibly, to other human diseases in which Alu RNA dysregulation has been reported (28–30).
We previously also demonstrated that mtDNA release induced by Alu RNA accumulation promotes cGAS activation and RPE degeneration (12). Building on these findings, our new data support a model wherein Alu RNA is first reverse-transcribed into Alu cDNA in the cytoplasm, which then uses cGAS to induce mtDNA release in a PPIF-dependent manner, boosting cGAS activation to induce RPE toxicity. The threshold of cGAS activation required to induce RPE degeneration is crossed only after the first signal—Alu cDNA—induces a second signal—cytosolic escape of mtDNA. These observations suggest that RPE cells have some resistance to endogenous DNA accumulation. The higher cytotoxic potency of Alu cDNA compared to Alu RNA might reflect stoichiometric inefficiencies in L1-mediated RT of Alu RNA. In addition, it is conceivable that some fraction of the pool of Alu RNA does not encounter L1.
We also identify an ISM within Alu cDNA that is responsible for its activation of cGAS, which we demonstrate is an essential mediator of Alu cDNA–induced cytotoxicity. Structural features such as this motif and its lack of fully complementary double-stranded DNA as well as subcellular localization characteristics might explain why Alu cDNA engages cGAS rather than AIM2 or Toll-like receptor 9. Deciphering how Alu cDNA is trafficked or chaperoned to cGAS versus other DNA sensors could provide additional mechanistic insights and therapeutic targets.
Geographic atrophy spreads centrifugally outward from the lesion’s margins (31). Our findings proffer Alu cDNA, which is enriched in the junctional zone interposed between atrophic and healthy regions of the retina, as a pathogenic candidate for the centrifugal expansion of RPE death in geographic atrophy, whose etiology has defied explanation.
Repurposing NRTIs for inflammasome-driven diseases such as AMD has been supported by extensive preclinical data and large observational studies in humans (11, 21, 29). In the present study, we expand our understanding of the mechanisms by which NRTIs inhibit RPE degeneration in animal models of geographic atrophy. Both NRTIs and certain NonNRTIs have activity against L1-RT. However, NonNRTIs lack potent inflammasome inhibition and require much higher doses to prevent RPE degeneration compared to NRTIs. Moreover, modified NRTIs known as Kamuvudines retain the ability to inhibit inflammasome activation and block RPE degeneration in multiple models of geographic atrophy (32) as well as NRTIs despite their inability to inhibit RT. Collectively, our findings establish a new pathway in geographic atrophy’s pathological mechanism and provide a rationale for prospective clinical testing of NRTIs or the less-toxic Kamuvudines (21) for geographic atrophy.
MATERIALS AND METHODS
Animals
WT C57BL/6J mice, Ppif−/− mice, and Brown Norway BN/RijHsd rats (R. norvegicus) were purchased from The Jackson Laboratory (Bar Harbor, ME) and Envigo (Frederick, MD), respectively. Casp1−/− Casp4129mt/129mt (denoted Casp1/4 dko) and Nlrp3−/− mice were obtained from G. Nuñez (University of Michigan School of Medicine), and cGAS−/− mice were obtained from K.A. Fitzgerald (University of Massachusetts Medical School). Casp1−/− Casp4129mt/129mt Casp4Tg mice expressing functional mouse caspase-4 from a bacterial artificial chromosome transgene (denoted Casp1−/−) were obtained from V.M. Dixit (Genentech). Mice lacking Tlr2, Tlr3, Tlr4, Tlr7, and Tlr9 (denoted Tlr23479−/− mice) were obtained from C.J. Kirschning (University of Duisburg-Essen). To generate cGAS catalytic mutant mice (cGASGG198AA; SS199AA), one-cell–fertilized C57BL/6J embryos were microinjected with a mixture of guide RNA (50 ng/μl; 5′-GGTGTGGAGCAGCTGAACACTGG-3′), ssODN (50 ng/μl; 5′-GAATAAAGTTGTGGAACGCCTGCTGCGCAGAATGCAGAAACGGGAGTCGGAGTTCAAAGGTGTGGAGCAGCTGAACACTgccgccTACTATGAACATGTGAAGGTGAGCGTCAAGACCTGCTGGAGGGGCTCCGGCCCCACTCCTCACTTGCCTCCTCA-3′), and Cas9 protein (40 ng/μl; PNA Bio). Injected embryos were implanted into pseudopregnant recipients, and pups were genotyped at weaning via subcloning of amplified fragments and Sanger sequencing. Founder mice were bred to C57BL/6J mice to generate F1s. Rice rats (O. palustris) have been previously described (16–19). For all procedures, anesthesia was achieved by intraperitoneal injection of ketamine hydrochloride (100 mg/kg; Ft. Dodge Animal Health) and xylazine (10 mg/kg; Phoenix Scientific), and pupils were dilated with topical 1% tropicamide and 2.5% phenylephrine hydrochloride (Alcon Laboratories). Mice and rats were treated in accordance with the guidelines of the University of Virginia and University of Kentucky Institutional Animal Care and Use Committees and the Association for Research in Vision and Ophthalmology. Both male and female mice between 6 and 10 weeks of age were used, and male rats between 2 and 3 months of age were used.
Fundus photography
Fundus imaging of dilated mouse and rat eyes was performed using a TRC-50 IX camera (Topcon) linked to a digital imaging system (Sony).
Human tissue
Following procedures previously described in (12), all studies on human tissue followed the guidelines of the Declaration of Helsinki. The study of de-identified tissue collected from deceased individuals and obtained from various eye banks in the United States was exempted from IRB review by the University of Virginia Institutional Review Board for Health Sciences Research, in accordance with U.S. Health and Human Services human subject regulations. Donor eyes from patients with geographic atrophy or age-matched patients without AMD were obtained from various eye banks. These diagnoses were confirmed through ophthalmic examination of dilated eyes before acquisition of the tissues or eyes or after examination of the eye globes postmortem. Enucleated donor eyes isolated within 6 hours postmortem were immediately preserved in RNAlater (Thermo Fisher Scientific) or formalin. The neural retina and sclera were removed, and tissues comprising both macular RPE and choroidal tissue were snap-frozen in liquid nitrogen. For in situ hybridization, eyes were transferred to 70% (v/v) ethanol after fixation. For eyes with geographic atrophy, the RPE and choroidal tissues were collected and divided into atrophic, junctional, and peripheral areas (13). Frozen section of eyes with Leber congenital amaurosis, Joubert syndrome, Stargardt macular dystrophy, or autosomal recessive retinitis pigmentosa have been previously described (33–36).
Chemicals
The NRTI lamivudine (3TC), as well as the NonNRTIs EFV, DLV, NVP, and ATP, were purchased from Sigma-Aldrich. LPS was purchased from InvivoGen. A trimethyl-modified version of 3TC (TM-3TC) and a diethyl-modified version of AZT (DE-AZT) were synthesized as previously described (27).
In vitro–transcribed Alu RNA
Alu RNA was synthesized from a linearized plasmid containing a consensus AluY sequence with an adjacent 5′ T7 promoter (37), subjected to AmpliScribe T7 Flash Transcription kit (Epicentre) according to the manufacturer’s instructions. Deoxyribonuclease (DNase)–treated RNA was purified using MEGAclear (Ambion), and integrity was confirmed by agarose gel electrophoresis.
Assessment of RPE degeneration
Following procedures previously described in (12), subretinal injections were performed as previously described (3, 5, 12, 21, 37, 38). Seven days after subretinal injection, RPE health was assessed by fundus photography and immunofluorescence staining of zonula occludens-1 (ZO-1) on RPE flat mounts (whole mount of posterior eye cup containing RPE and choroid layers). Mouse RPE/choroid flat mounts were fixed with 2% paraformaldehyde (PFA), stained with rabbit polyclonal antibodies against mouse ZO-1 (1:100; Invitrogen) and visualized with Alexa 594 (Invitrogen). All images were obtained by microscopy (model SP-5, Leica or A1R Nikon confocal microscope system, Nikon). Imaging was performed by an operator blinded to the group assignments.
Quantification of RPE degeneration
Following procedures previously described in (12), quantification of RPE degeneration was performed using two methodologies (binary assignment and cellular morphometry) as described previously (27): Binary assignment (healthy versus unhealthy) (5, 12, 21, 37) was independently performed by two blinded raters (inter-rater agreement = 99.5%; Pearson r2 = 0.98, P < 0.0001; Fleiss κ = 0.99, P < 0.0001). Quantifying cellular morphometry for hexagonally packed cells was performed in semi-automated fashion by three masked graders by adapting our previous analysis of the planar architecture of corneal endothelial cell density (39), which resembles the RPE in its polygonal tessellation. We quantified polymegethism (coefficient of variation of cell size), a prominent geometric feature of RPE cells in geographic atrophy (3, 5, 40–42), using the Konan Cell Check software (ver. 4.0.1), a commercial FDA–cleared software that has been used for registration clinical trials, as previously described (12).
Subretinal and intravitreous injections
Subretinal injections (1 μl for mice, 3 μl for rat) or intravitreous injections (0.5 μl for mice, 2 μl for rats) in mice or rat were performed using a 35-G needle (Ito Co. Fuji, Japan). In vivo transfection of Alu RNA or mutant Alu RNA (3.6 to 300 ng per eye), Alu cDNA (0.0036 to 90 ng per eye), Alu cDNAISM− (3.6 to 90 ng per eye), 7SL cDNA (3.6 to 90 ng per eye), or 7SL cDNAISM+ (3.6 to 90 ng per eye) was performed using 10% NeuroPORTER (Genlantis) as previously described (3, 37). 3TC, TM-3TC, DE-AZT, EFV, DLV, or NVP (50 μM/1 μl or 500 μM/1 μl) was administered by intravitreous injection. Similarly, in O. palustris, rat L1 plasmids expressing ORF1p, ORF2p, reverse transcriptase–deficient ORF2p [pORF2 (RT−)], or endonuclease-deficient ORF2p [pORF2 (EN−)] was delivered via subretinal injection 3 days before administering Alu RNA or vehicle. The choice of eye for experimental versus control injection was chosen randomly. Plasmids encoding rat L1-encoded ORF1 and ORF2 have been described previously (43). The rat L1 ORF2 (EN−) construct contained mutations D207A and H232A, which, based on CLUSTALW alignment, correspond to D205A and H230A (44) in the EN domain of human L1 ORF2p. The rat L1 ORF2 (RT−) construct contained mutation D703A, which, based on CLUSTALW alignment, corresponds to D702A (45) in the RT domain of human L1 ORF2p. Plasmids expressing rat L1 ORF2 (EN−) and L1 ORF2 (RT−) mutant proteins were created by introducing specific point mutations via site-directed mutagenesis (performed by GenScript services) into the rat L1 ORF2p expression plasmid pORF2 [pEF6-ORF2 (43) expressing rat L1 ORF2p-V5]. Generation of both L1 ORF2p mutant expression plasmids included PCR mutagenesis, cloning of the point mutations harboring DNA fragments [Kpn I/Afe I for the ORF2 (EN−) construct and Sna BI/Not I for the ORF2 (RT−) construct], and sequencing to verify the presence of the introduced point mutations in the expression plasmids.
cDNA synthesis
Single-stranded Alu cDNA, Alu cDNAISM−, 7SL cDNA, and 7SL cDNAISM+ cDNA were isolated from biotinylated double-stranded PCR products synthesized from a linearized plasmid containing a consensus Alu Y or 7SL sequence using Dynabeads M-270 Streptavidin (Life Technologies) and then purified using QIAquick PCR purification kit (Qiagen catalog no. 28104) (46). Briefly, PCR products were biotinylated on one strand by synthesis with a biotinylated primer (forward 5′-biotin-GGGCCGGGCGCGGTG-3′ and reverse 5′-GTACCTTTAAAGAGACAGAGTCTCGC-3′ for Alu Y and forward 5′-biotin-CGTGCCTGTAGTCCCAGCTA-3′ and reverse 5′-AGACGGGGTCTCGCTATGTT-3′ for 7SL). Dynabeads M-270 Streptavidin magnetic beads were used to capture the biotin-tagged PCR product. The PCR product was heated at 95°C for 10 min for strand separation, and isolation of the nonbiotinylated strand was performed using a magnetic stand followed by alcohol precipitation according to the manufacturer’s instructions.
Immunoblotting
Cells and tissue were homogenized in radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich) with protease and phosphatase inhibitors (Roche) or lysed directly in Laemmli buffer. Protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Equal quantities of protein (10 to 50 μg) prepared in Laemmli buffer were resolved by SDS–polyacrylamide gel electrophoresis (PAGE) on Novex tris-Glycine Gels (Invitrogen) or Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad) and transferred onto Immobilon-FL polyvinylidene difluoride membranes (0.2 or 0.45 μm) (Millipore). The transferred membranes were blocked in Odyssey Blocking Buffer [phosphate-buffered saline (PBS)] for 1 hour at room temperature and then incubated with primary antibody at 4°C overnight. Immunoreactive bands were visualized using species-specific secondary antibodies conjugated with IRDye. The blot images were captured on Odyssey imaging systems. Rabbit polyclonal anti-human caspase-1 (1:1000; Cell Signaling catalog no. 2225), mouse monoclonal anti-mouse caspase-1 (1:1000; AdipoGen catalog no. AG-20B-0042-C100), anti-human caspase-4 (1:200; Santa Cruz Biotechnology, 1229), rabbit monoclonal anti-human lamin B1 (1:1000; CST catalog no. 12586), rabbit monoclonal anti human L1 ORF2p (MT9 antibody; 1:1000; gift of K.H. Burns, Dana-Farber Cancer Institute) (47), rabbit polyclonal anti-human vinculin (1:2000; Sigma-Aldrich catalog no. V4139), mouse monoclonal anti–β-actin (1:50,000; Sigma-Aldrich catalog no. A2228), goat-polyclonal anti–β-actin (1:50,000; Abcam catalog no. ab8229), chicken polyclonal anti–β-actin (1:50,000; Abcam catalog no. ab13822), chicken polyclonal anti-tubulin (1:50,000; Abcam catalog no. ab89984), or mouse monoclonal anti-tubulin (1:5000; Sigma-Aldrich catalog no. T6199) was used.
Cell lysates isolated from HAP1, HeLa, hFF1 (human foreskin fibroblasts), and HEK-293TLD cells expressing L1-ORF2p-3xFlag (48) were separated on 4 to 12% NuPAGE bis-tris gradient gels, and subsequent immunoblotting was performed by wet blot transfer. After protein transfer, the membranes was blocked for 2 hours at room temperature in a 10% solution of nonfat milk powder in 1× PBS-T [137 mM NaCl, 3 mM KCl, 16.5 mM Na2 HPO4, 1.5 mM KH2PO4, 0.05% Tween 20 (Sigma-Aldrich Chemie GmbH, Germany)], washed in 1× PBS-Tween (PBS-T), and incubated overnight with the monoclonal α-L1 ORF2p antibody MT9 (fig. S2A) (48) or with the monoclonal anti-FLAG M2 antibody (Sigma-Aldrich; product number: F1804) (fig. S2B) in a 1:1000 dilution in 1× PBS-T containing 5% milk powder at 4°C. Subsequently, both membranes were washed thrice in 1× PBS-T. As secondary antibody, we used horseradish peroxidase (HRP)–conjugated anti-rabbit immunoglobulin G (IgG) antibody from donkey (GE Healthcare NA934-1ML) in the case of the MT9 primary antibody and HRP-conjugated anti-mouse IgG antibody from sheep (GE Healthcare NXA931V) in the case of the anti-FLAG primary antibody at a 1:10,000 dilution in 1× PBS-T/1.67% milk powder each and incubated the membrane for 2 hours at 4°C. After stripping the membranes and subsequent blocking for 2 hours at room temperature as described above, the membrane of fig. S2A was probed with anti-FLAG M2 antibody to confirm the presence of the FLAG-tagged L1 ORF2p. To detect L1 ORF1p (fig. S2E), the membrane of fig. S2B was probed with the polyclonal rabbit-anti-L1 ORF1p antibody #984 (49) at a 1:2000 dilution and HRP-conjugated donkey anti-rabbit IgG antibody (Amersham Biosciences) at a 1:30,000 dilution. After stripping and subsequent blocking for 2 hours at room temperature had been repeated, β-actin expression was detected using a monoclonal anti–β-actin antibody (clone AC-74, Sigma-Aldrich Chemie GmbH, Steinheim, Germany) at a dilution of 1:30,000 as primary antibody and an anti-mouse HRP-linked species-specific antibody (from sheep) at a dilution of 1:10,000 as secondary antibody (fig. S2F). Immunocomplexes were visualized using lumino-based ECL immunoblot reagent (Amersham Biosciences Europe GmbH, Freiburg, Germany).
Cell culture and transfection
Primary mouse, human, R. norvegicus, and O. palustris RPE cells were isolated by adapting previously described protocols (11, 12). All cells were maintained at 37°C in a 5% CO2 environment. Mouse RPE cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum (FBS) and penicillin/streptomycin antibiotics at standard concentrations; primary human RPE cells were maintained in DMEM supplemented with 10% FBS and antibiotics. The human RPE cell line ARPE19 was cultured as previously described (12) and maintained in DMEM-F12 containing penicillin/streptomycin, Fungizone, and gentamicin. HEK293T cells were cultured in DMEM with 10% FBS with penicillin/streptomycin (100 U/ml) and 2 mM l-glutamine. Primary WT mouse BMDMs were isolated and cultured in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 30% L929 supernatant containing macrophage-stimulating factor, nonessential amino acids, sodium pyruvate, 10% FBS and antibiotics, and 50 μM β-mercaptoethanol (50) and serum-starved in IMDM with 1% FBS and penicillin/streptomycin (100 U/ml) overnight before LPS stimulation. NRTIs or NonNRTIs were added 30 min before LPS stimulation and again 30 min before ATP activation. LPS (100 ng/ml) was added for 3 to 4 hours before the addition of ATP. Cell lysates were collected 30 min after addition of ATP (5 mM). Transfections were performed according to the manufacturer’s instructions (Lipofectamine 2000, Invitrogen). NRTIs were administered 60 min before transfection and added again upon replacement of media at 8 hours. ARPE-19 cells were pretreated with 3TC or TM-3TC (200 μM) for 1 hour and again after transfection with in vitro–transcribed Alu cDNA. Proteins were extracted 48 hours after transfection. To induce acid injury or osmotic stress, primary human RPE cells were treated with HCl (pH 4.0 media) or H2O for 30 min, 1, and 2 hours at 37°C in 5% CO2, respectively. NIH3T3 Tet-ON cells were cultured in DMEM with 10% tetracycline-free FBS with penicillin/streptomycin (100 U/ml). O. palustris RPE cells were transfected with pL1Md-Gf (p99-GFP-TGF21; full-length mouse L1GF element; gift of J.L. García-Pérez, University of Edinburgh) (51), and mouse L1 ORF2p abundance was measured. O. palustris RPE cells were transfected with prtTA and pLD401, and human L1 ORF2p abundance was measured after 24 hours of doxycycline exposure.
The following cell lines were used in immunoblotting analyses. The lymphoblastic tumor cell line HAP1 (52) harbors a PRPF19 transgene expression cassette under transcriptional control of the constitutive RPBSA-EF1a promoter (53). The expressed 55-kDa PRPF19 protein is N-terminally tagged with the 3xFLAG peptide (DYKDHDG-DYKDHDI-DYKDDDD) and served as positive control for the detection of Flag-tagged proteins. HEK-293TLD cells express a full-length L1 element ectopically from the expression plasmid pMT302 (54, 55). The resulting protein machinery includes L1-ORF1p and a Flag-tagged full-length L1-ORF2p (ORF2p-3xFlag) (48, 54). Handling of cryo-milled HEK-293TLD cells ectopically expressing L1 from pMT302 was previously described (54, 56).
Induction of Alu RNA by various stimuli
In vitro–transcribed Alu RNA, DICER1 antisense (AS) oligonucleotide (5′- GCUGACCTTTTTGCTUCUCA-3′), or control scrambled AS (5′- TTGGTACGCATACGTGTTGACTGTGA-3′) (Integrated DNA Technologies) was transfected into human and mouse RPE cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Heat shock was induced by placing cells in a 42°C incubator for 20 min and then allowed to recover at 37°C for 1 hour (57).
In situ hybridization
RPE from mice was collected at 24 hours after subretinal injection. Cells in culture were collected after 6 to 8 hours after Alu RNA transfection. RPE flat mounts or cells were fixed in 4% PFA/PBS for 20 min. For Alu cDNA detection, all samples were treated with RNase A. To confirm whether the target was single-stranded DNA, S1 nuclease (Thermo Fisher Scientific) was treated for 30 min at room temperature. RNA probes, prepared from linearized Alu cDNA templates using a T7 fluorescein RNA labeling kit or T7 DIG RNA labeling kit (Roche), were hybridized overnight at 37°C in a mixture containing 10% dextran sulfate, 2 mM vanadyl-ribonucleoside complex, 0.02% RNase-free bovine serum albumin (BSA), 40 μg of Escherichia coli tRNA, 2× SSC, 50% formamide, and RNA probe. Cells were then subjected twice to stringent washing at 50°C in 50% formamide, 0.1× SSC for 30 min. Following washing, samples were incubated with a HRP-conjugated anti-fluorescent antibody (PerkinElmer catalog no. NEF710001EA) or HRP-conjugated anti-DIG antibody (PerkinElmer catalog no. NEF832001EA) at a 1:200 dilution. Visualization of fluorescein-labeled probe was performed with the TSA plus fluorescence system or the TSA plus Cy5 system (PerkinElmer). The fluorescent or Cy5 signals were detected using a Leica SP-5 or A1R Nikon confocal microscope system. For L1 overexpression, ARPE-19 cells were transfected with the L1 expression vector pES2TE1 (gift of J.V. Moran, University of Michigan Medical School) (58). Then, Alu cDNA synthesis was monitored by in situ hybridization after Alu RNA transfection.
Equator blotting
We term “equator blot” as a combination of classic Southern and Northern blotting procedures. An equator blot is similar to a Southern blot in that it probes for a target DNA sequence, yet unlike a typical Southern blot, it does not involve restriction enzyme digest of the DNA. Instead, the DNA is separated by agarose gel electrophoresis without prior enzyme digestion according to the typical Northern blot procedure. Hence, we refer the procedure of hybridization of undigested DNA as an equator blot. Total nucleic acid or nuclear and cytoplasmic fractions were extracted from cells as described below. Primary human RPE cells were collected after AluY RNA transfection, and the cytoplasmic fraction was treated with RNase A (Invitrogen). To confirm whether the target was single-stranded DNA or double-strand DNA, S1 nuclease (Thermo Fisher Scientific; 30 min at room temperature) or double-strand DNase (Thermo Fisher Scientific; 2 min at 37°C) was added after RNase treatment, according to the manufacturer’s instructions. For human tissue, DNA and RNA were extracted using the DNA and RNA Purification Kit (Epicentre); RNase A was added for DNA isolation. DNA samples were run on 10% tris-borate EDTA (TBE)–urea gels (Bio-Rad) according to the manufacturer’s instructions. Samples were transferred and ultraviolet (UV) cross-linked to a HyBond N+ nylon membrane and blotted for Alu cDNA. Alu cDNA–biotinylated oligonucleotide probe was synthesized by PCR from a linearized plasmid containing a consensus Alu Y element as described above using the following primers for Alu cDNA detection (forward 5′-biotin-GGGCCGGGCGCGGTG-3′ and reverse 5′-GTACCTTTAAAGAGACAGAGTCTCGC-3′) and then purified. Blots were developed with the Pierce chemiluminescent nucleic acid detection kit (Thermo Fisher Scientific). The blot images were captured on Odyssey imaging systems.
Northern blotting
Total nucleic acid or nuclear and cytoplasmic fractions were extracted from cells as described below. For human tissue, DNA and RNA were extracted using the DNA and RNA Purification Kit (Epicentre); DNase I was added for RNA isolation. RNA samples were run on 10% TBE-urea gels (Bio-Rad) according to the manufacturer’s instructions. Samples were transferred and UV cross-linked to a HyBond N+ nylon membrane and blotted for Alu RNA and U6 RNA. U6 biotinylated oligonucleotide probe was synthesized by Integrated DNA Technologies (5′-CACGAATTTGCGTGTCATCCTT-biotin-3′). Alu RNA–biotinylated oligonucleotide probe was synthesized by PCR from a linearized plasmid containing a consensus Alu Y element as above using the following primers for Alu RNA detection (forward 5′-GGGCCGGGCGCGGTG-3′ and reverse 5′-biotin-GTACCTTTAAAGAGACAGAGTCTCGC-3′) and then purified. Blots were developed with the Pierce chemiluminescent nucleic acid detection kit (Thermo Fisher Scientific). The blot images were captured on Odyssey imaging systems.
Nuclear and cytoplasmic fractionation
Briefly, cells were collected and lysed with gentle extraction buffer prepared in 1× PBS containing 1% (v/v) Triton X-100 (Sigma-Aldrich) and 1 mM EDTA for 15 min on ice. Lysates were vortexed and centrifuged at 1000g for 10 min at 4°C. For cytoplasmic fractionation, the supernatant was collected, subjected to repeated centrifugation four times, and then purified using a DNA purification column (Enzymax). Lysis buffer was added to the pellet for reconstitution. The lysate supernatant was vortexed and further centrifuged at 13,000g for 2 min at room temperature. The lysate supernatant was used as the nuclear fraction and purified using a DNA purification column (Enzymax). For cDNA detection, samples were treated with RNase A (Ambion) for 30 min at 37°C.
Alu cDNA detection by real-time PCR
Cells were collected after counting the cell number and the cytoplasmic fraction was treated with RNase A (Ambion). The RNase-treated cytoplasmic fraction was purified with a PCR clean-up kit (QIAquick, Qiagen). Then, samples were directly amplified by real-time quantitative PCR (qPCR) (Applied Biosystems 7900 HT Fast Real-Time PCR system) with Power SYBR green Master Mix. Primers were specific for human Alu cDNA (forward 5′-TTAGCCGGGAGTGGTGTCGG-3′ and reverse 5′-ACCTCCCGGGTTCACGCCATT-3′). The copy number of Alu cDNA was calculated using standard curves that were obtained using serial dilutions of the plasmids containing an AluY sequence. Alu cDNA copy number was normalized to cell number.
Real-time PCR
For human tissue, total RNA was extracted using the MasterPure Complete DNA and RNA Purification Kit (Epicentre) according to the manufacturer’s recommendation. We exhaustively digested RNA samples with RNase-free DNase (Turbo DNase) before cDNA preparation for L1 expression quantification. Effectiveness of the DNase digestion was assessed using controls that omitted the RT enzyme. The RT products (cDNA) were amplified by real-time qPCR (Applied Biosystems 7900 HT Fast Real-Time PCR system) with Power SYBR green Master Mix. Relative gene expression was determined by 2–ΔΔCt method using 18S ribosomal RNA (rRNA) as an internal control. Primers specific for human L1 ORF1 (forward 5′-AGGAACAGCTCCGGTCTACA-3′ and reverse 5′-GATGAACCCGGTACCTCAGA-3′), human L1 ORF2 (forward 5′-ACTGGCCATCAGAGAAATGC-3′ and reverse 5′-CAGCACCTGTTGTTTCCTGA-3′), human 18S rRNA (forward 5′-CGCAGCTAGGAATAATGGAATAGG-3′ and reverse 5′-GCCTCAGTTCCGAAAACCAA-3′), Rat L1 ORF1 (forward 5′-GCCAGAAGATCCTGGACT GAT-3′ reverse 5′-GTAACCTGGGCTGGCATTTG-3′), Rat L1 ORF2 (forward 5′-GCAGATCGATCCATGCTTATCAC-3′ and reverse 5′-GATGTGGAGGTCCTTGATCCA-3′), and human IFNB1 (forward 5′-GCGACACTGTTCGTGTTGTC-3′ and reverse 5′-GCCTCCCATTCAATTGCCAC-3′) were used.
Ex vivo RT activity assay
Ex vivo RT activity in nuclear and cytoplasmic protein fractions was assessed using an Alu RNA–templated reaction. Nuclear and cytoplasmic fractionation was prepared using an NE-PER nuclear cytoplasmic extraction kit (Thermo Fisher Scientific), as per the manufacturer’s instructions. Briefly, in this assay, exogenous Alu RNA and Alu-R primer (5′- ACCTCCCGGGTTCACGCCATT-3′) were incubated with nuclear or cytoplasmic protein lysate containing endogenous L1 ORF2p, which interacts with Alu RNA in trans giving rise to an Alu cDNA. The RT reaction was carried out in a 20-μl reaction mix containing Alu RNA template (10 ng), Alu primer (10 pmol), dNTPs mix, cytoplasmic or nuclear protein, and Quantiscript RT Buffer (Qiagen). The reaction mixture was incubated at 42°C for 30 min. The resulting cDNA was quantified by qPCR using Alu RNA template-specific primers. Heat denaturation of nuclear or cytoplasmic fraction was performed by heat inactivation at 95°C for 10 min.
Alu retrotransposition reporter assay
Retrotransposition reporter assays were carried out as follows. Briefly, 2 × 105 HeLa-HA cells were plated in a six-well tissue culture dish and, 1 day later, were transfected in triplicate using FuGene 6 (Promega) with 1 μg of the WT L1 reporter plasmid pJM101/L1.3Δneo as described previously (59), pORF2, pORF2 (RT−), or pORF2 (EN−), along with 1 μg of Alu retrotransposition indicator construct Alu neo (gift of J.V. Moran, University of Michigan Medical School) and Alu RNA with G25C/G159C mutations. After 72 hours, growth medium was replaced by DMEM containing G418 (600 μg/ml) and penicillin/streptomycin (100 μg/ml; Cellgro). Fourteen days later, the plates were washed with methanol, Giemsa-stained, and photographed. Colonies were counted manually using ImageJ (National Institutes of Health). A total of 1 μg of Alu-neo plasmid with 1-μg empty driver vector was used as a negative control.
Immunofluorescence staining
To test for Alu RNA and L1 ORF2p colocalization, fluorescein-labeled Alu RNA was transfected into O. palustris RPE cells at 48 hours after V5-tagged rat L1 ORF2 plasmid (43) transfection. Cells were fixed in 4% PFA, and L1 ORF2p were detected using Dylight 549–conjugated anti-V5 antibody (1:5000; Rockland, catalog no. 600-442-378). To test for Alu cDNA and L1 ORF2p colocalization, Alu RNA was transfected into O. palustris RPE cells at 48 hours after V5-tagged rat L1 ORF2 plasmid transfection. Alu cDNA formation was monitored by in situ hybridization and L1 ORF2p was detected using anti-V5 antibody. RPE65 and Alu cDNA in human tissue were monitored by in situ hybridization staining of Alu cDNA followed by immunostaining with anti-human RPE65 antibody conjugated with Dylight 488 (1:250; Thermo Fisher Scientific, catalog no. MA5-16042). Slides were mounted in ProLong Gold (Thermo Fisher Scientific), and images were acquired using a A1R Nikon confocal microscope system. For confirmation of in vivo–enforced expression of pORFeus-Hs or rat pORF2, mouse RPE/choroid flat mounts were fixed with 2% and stained with fluorescein isothiocyanate–conjugated anti-FLAG F4049 (10 μg/ml; Sigma-Aldrich) or Dylight 549–conjugated anti-V5 antibody (1:5000; Rockland, catalog no. 600-442-378). To monitor RNA-DNA hybrids, primary human RPE cells were seeded on chamber slides (Thermo Fisher Scientific, catalog no. 154941) and transfected with 12.5 pmol of Alu RNA for 2 hours and then washed with fresh medium to be collected with 2% PFA fixation at the indicated time points. Cells were blocked and incubated with S9.6 antibody (1:200; Kerafast, catalog no. ENH001), followed by incubation with secondary antibody and DAPI (4′,6-diamidino-2-phenylindole) before image acquisition using confocal microscopy (A1R Nikon).
To monitor expression of ORF2 and ORF2 mutant proteins, HeLa-HA cells were plated in an eight-well chamber slide (Thermo Fisher Scientific, catalog no. 154941) and transfected overnight with plasmids encoding green fluorescent protein (GFP), V5-tagged rat L1 ORF2, V5-tagged rat L1 ORF2 (EN−), or V5-tagged L1 ORF2 (RT−) mutant proteins using Lipofectamine 3000 (Thermo Fisher Scientific). Then, the cells were rinsed with prechilled PBS, fixed in 4% PFA, permeabilized with 0.1% Triton X-100, and blocked with 3% normal donkey serum in PBS. The cells were stained with rabbit monoclonal antibody anti-V5 antibody (1:200; D3H8Q, Cell Signaling Technology) and washed twice with PBST and visualized with fluorescent dye–conjugated secondary antibodies.
Pull-down assays
To monitor the association of L1 ORF2p with Alu RNA and Alu cDNA, RNase H–deficient HeLa cells (gift of A.P. Jackson and M.A. Reijns) (60) transfected with V5-tagged rat L1-ORF2p expression plasmid and biotinylated Alu RNA (versus mock) were used. Briefly, biotinylated Alu RNA–transfected cells were cross-linked with 1% formaldehyde for 15 min at room temperature and lysed to collect cytosolic fractions. Streptavidin Dynabeads blocked with 1% BSA (for 18 hours) were incubated with the cytosolic lysates diluted in BC200 buffer [20 mM Hepes (pH 7.9), 0.2 mM EDTA, 0.5 mM dithiothreitol, 20% glycerol, 0.2% NP-40, and 200 mM KCl] for 2 hours at 4°C. The beads were then washed twice with BC200, heated at 95°C for 30 min. The pull-down samples were subsequently analyzed by immunoblotting with anti-V5 antibody.
Using the same experimental system, a reverse pull-down assay was performed to detect the presence of Alu RNA and Alu cDNA in the V5-L1 ORF2p immunoprecipitate. Briefly, cytosolic lysates (1 mg at 500 ng/μl) in BC200 buffer, prepared as above, were precleared by incubating with beads for 6 hours. Precleared cytosolic lysates were subjected to immunoprecipitation using 15 μg of anti-V5 antibody for overnight at 4°C. The immune complexes were captured by incubation with preblocked beads (4 hours at 4°C). The bead-captured immune complexes were washed twice with BC200. Last, the bead-captured immune complexes were resuspended in either 1 ml of TRIzol reagent followed by RNA purification (for detecting Alu RNA) or subjected to Proteinase K treatment and reverse cross-linking (overnight) followed by ethanol precipitation of DNA (for detecting Alu cDNA). The Alu RNA was detected by direct blotting of biotinylated Alu RNA. DNA purified from these assays was analyzed by equator blotting to detect Alu cDNA.
Binding assay for cDNA and cGAS interaction
Recombinant His-tagged cGAS (Cayman Chemical catalog no. 22810) was incubated with Alu cDNA, Alu cDNAISM−, or 7SL cDNA prepared as described above. cDNA-protein pull-down was performed according to the instruction of Pierce Magnetic RNA-protein pull-down kit (catalog no. 20164) with modifications. Briefly, 2 μg of His-tagged cGAS was added to 50 pmol of cDNA, mixed in 100 μl of binding buffer [0.2 M tris-HCl (pH 7.4), 0.5 M NaCl, 20 mM MgCl2, and 1% Tween 20], and incubated on ice for 1 hour. Anti-His magnetic beads (Thermo Fisher Scientific, catalog no. 88845) were added to the preincubated cDNA/protein mixture, and the resulting preparation was gently rotated overnight at 4°C and then washed with washing buffer thrice. Last, magnetic beads were eluted with elution buffer (pH 2.8) at 70°C for 10 min. All eluted supernatants were subjected to qPCR and immunoblot analysis. DNA that associated with the recombinant protein was analyzed by qPCR using Alu-specific or 7SL-specific primers.
Reverse cGAS–Alu cDNA interaction assay
Reverse pull-down assay was carried out using DIG-labeled Alu cDNA, Alu cDNAISM−, or 7SL cDNA. Briefly, 2 μg of recombinant His-tagged cGAS was incubated with 50 pmol of DIG-labeled cDNA within 100 μl of binding buffer [0.2 M tris-HCl (pH 7.4), 0.5 M NaCl, 20 mM MgCl2, and 1% Tween 20] on ice for 1 hour, and then the complex was pulled down using anti- DIG magnetic beads (Thermo Fisher Scientific, catalog no. MA5-14746) by gently rotating overnight at at 4°C. Subsequently, the precipitate was washed with washing buffer thrice. Last, magnetic beads were eluted with elution buffer (pH 2.8) at 70°C for 10 min. The eluted supernatant was separated on a protein gel and submitted to immunoblot analysis.
Immunoprecipitation assays for Alu cDNA and cGAS interaction
Immortalized cGAS−/− MEF cells were reconstituted with HA-tagged mouse cGAS (HA-cGAS) were as previously described (26). The interaction between Alu cDNA and cGAS was monitored by a UV–cross-linked pull-down assay. Briefly, mock-, Alu cDNA–, and 7SL cDNA–transfected HA-cGAS–reconstituted cGAS−/− MEFs and cGAS−/− MEFs were washed twice with prechilled PBS and then UV–cross-linked (Spectrolinker, 200 mJ/cm2). The cells were then lysed by sonication in RIPA buffer with protease inhibitors and centrifuged for 10 min at 18,000g in a 4°C microfuge. The supernatant containing cell lysate was collected, and cGAS was immunoprecipitated using antibodies against HA (Abcam, catalog no. ab9110). cDNA/protein complex in the immunoprecipitate was eluted as described above. The supernatant of elution was analyzed by qPCR using Alu- or 7SL-specific primers.
cGAS activity assay
In vitro reaction for cGAS activity was performed by mixing recombinant human cGAS protein (13 nM) with cDNA (0.2 nM) in buffer containing ATP (50 μM), guanosine triphosphate (50 μM), MgCl2 (0.5 mM), ZnCl2 (0.1 mM), BSA (0.2 mg/ml), and tris-HCl (pH 7.5, 20 mM). After incubation at 37°C for 1 hour, the reaction was terminated by heat inactivation at 95°C for 5 min and followed by centrifugation at 20,000g for 5 min. cGAMP abundance in the supernatant was quantified by cGAMP enzyme-linked immunosorbent assay kit (Arbor Assays).
Statistical analysis
Following procedures previously described in (12), the binary readouts of RPE degeneration (i.e., the presence or absence of RPE degeneration on fundus and ZO-1–stained flat-mount images) were analyzed with Fisher’s exact test. Cell morphometry data were assessed with two-tailed Mann-Whitney U test. All other data were expressed as means ± SEM and were analyzed with Mann-Whitney U test. P < 0.05 were deemed statistically significant. Sample sizes were selected on the basis of power analysis α = 5%; 1 − β = 80%, such that we were able to detect a minimum of 50% change, assuming a sample SD based on Bayesian inference. Outliers were assessed with Grubbs’ test. On the basis of this analysis, no outliers were detected and no data were excluded. Fewer than 5% of subretinal injection recipient tissues were excluded on the basis of predetermined exclusion criteria (including hemorrhage and animal death due to anesthesia complications) relating to the technical challenges of this delicate procedure.
Acknowledgments
We thank G. Pattison, D. R. Robertson, J. Hu, K. A. Fox, and A. Nafziger for technical assistance. Funding: J.A. was supported by the UVA Strategic Investment Fund, U.S. National Institutes of Health (NIH) grants (DP1GM114862, R01EY022238, R01EY024068, R01EY028027, R01EY029799, and R01EY031039), John Templeton Foundation Grant 60763, DuPont Guerry, III, Professorship, and a gift from Mr. and Mrs. Eli W. Tullis; S.F. by JSPS Fund for the Promotion of Joint International Research (Home-Returning Researcher Development Research) and Research Grant of Japan Eye Bank Association; N.K. by NIH grants (K99EY024336, R00EY024336, R01AI48741, and R21EY030651) and Beckman Initiative for Macular Research grant; B.J.F. by NIH grants (T32HL091812 and UL1RR033173); R.Y. by Association for Research in Vision and Ophthalmology (ARVO)/Alcon Early Career Clinician-Scientist Research Award; T.Y. by Fight for Sight postdoctoral award; V.L.B. by NIH grants R01EY027750 and P30EY025585, an unrestricted grant from Research to Prevent Blindness, and the FFB and Cole Eye Institute for the establishment and maintenance of the collection of eyes with retinal degenerations; D.R.H. by NIH R01EY001545 and an unrestricted departmental grant from Research to Prevent Blindness; B.D.G. by NIH grants (R01EY028027, R01EY031039, and R01EY032512), the BrightFocus Foundation, and the Owens Family Foundation; S.F. by Japan Eye Bank Association Research Grant. The funders had no role in study design or conduct, data collection, analysis, or interpretation, manuscript writing, or the decision to submit the manuscript for publication. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Author contributions: J.A. conceived and directed the study. S.F., S.N., A.V., Y.N., S.W., K.A., I.A., F.P., B.J.F., T.Y., S.H., R.Y., P.H., P.Y., R.D.M., M.W., K.L.B., K.M.M., X.H., E.B., M.A., V.L.A., D.B., V.L.B., G.V.T., U.H., Y.O., H.T., T.O., D.B., K.B.K., S.H.F., J.I.A., D.R.H., N.K., S.R.S., G.G.S., B.D.G., and J.A. acquired, analyzed, or interpreted the data. J.A., S.F., S.N., A.V., F.P., G.G.S., and B.D.G. wrote the manuscript with assistance from M.A., N.K., S.R.S., and B.D.G. All authors had the opportunity to read and approve the final version of the manuscript. Competing interests: J.A. is a cofounder of iVeena Holdings, iVeena Delivery Systems, and Inflammasome Therapeutics and has been a consultant for Allergan, Biogen, Boehringer- Ingelheim, Immunovant, Janssen, Olix Pharmaceuticals, Retinal Solutions, and Saksin LifeSciences unrelated to this work. J.A. and B.D.G. are cofounders of DiceRx. J.A., B.J.F., or K.A. are inventors on patents related to this work filed by the University of Kentucky [no. 10294220, filed (26 February 2016), published (15 February 2018); no. 9707235, filed (14 January 2013), published (18 July 2017); no. 9464289, filed (3 July 2014), published (20 November 2014); no. 9453226, filed (17 January 2014), published (27 September 2016); no. 9326983, filed (1 August 2014), published (3 May 2016); no. 8809517, filed (1 June 2011), published (8 December 2011). J.A., N.K., B.D.G., or S.W. are inventors on patent applications related to this work filed by the University of Virginia (no. 20200345756, filed (9 October 2018), published (5 November 2020); no. WO2021097088, filed (12 November 2020), published (20 May 2021); no. WO2021041317, filed (24 August 2020), published (4 March 2021)]. S.R.S. has been a consultant for 4DMT, Allergan, Amgen, Centervue, Heidelberg, Roche/Genentech, Novartis, Optos, Regeneron, and Thrombogenics and has received research funding from Carl Zeiss Meditec, all unrelated to this work. All other authors declare they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Materials can be provided pending scientific review and a completed material transfer agreement. Requests for materials should be submitted to J.A. (ja9qr@virginia.edu).
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
REFERENCES AND NOTES
- 1.Wong W. L., Su X., Li X., Cheung C. M. G., Klein R., Cheng C. Y., Wong T. Y., Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2, e106–e116 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Ambati J., Ambati B. K., Yoo S. H., Ianchulev S., Adamis A. P., Age-related macular degeneration: Etiology, pathogenesis, and therapeutic strategies. Surv. Ophthalmol. 48, 257–293 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Kaneko H., Dridi S., Tarallo V., Gelfand B. D., Fowler B. J., Cho W. G., Kleinman M. E., Ponicsan S. L., Hauswirth W. W., Chiodo V. A., Karikó K., Yoo J. W., Lee D. K., Hadziahmetovic M., Song Y., Misra S., Chaudhuri G., Buaas F. W., Braun R. E., Hinton D. R., Zhang Q., Grossniklaus H. E., Provis J. M., Madigan M. C., Milam A. H., Justice N. L., Albuquerque R. J. C., Blandford A. D., Bogdanovich S., Hirano Y., Witta J., Fuchs E., Littman D. R., Ambati B. K., Rudin C. M., Chong M. M. W., Provost P., Kugel J. F., Goodrich J. A., Dunaief J. L., Baffi J. Z., Ambati J., DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471, 325–330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dridi S., Hirano Y., Tarallo V., Kim Y., Fowler B. J., Ambati B. K., Bogdanovich S., Chiodo V. A., Hauswirth W. W., Kugel J. F., Goodrich J. A., Ponicsan S. L., Hinton D. R., Kleinman M. E., Baffi J. Z., Gelfand B. D., Ambati J., ERK1/2 activation is a therapeutic target in age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 109, 13781–13786 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarallo V., Hirano Y., Gelfand B. D., Dridi S., Kerur N., Kim Y., Cho W. G., Kaneko H., Fowler B. J., Bogdanovich S., Albuquerque R. J. C., Hauswirth W. W., Chiodo V. A., Kugel J. F., Goodrich J. A., Ponicsan S. L., Chaudhuri G., Murphy M. P., Dunaief J. L., Ambati B. K., Ogura Y., Yoo J. W., Lee D. K., Provost P., Hinton D. R., Núñez G., Baffi J. Z., Kleinman M. E., Ambati J., DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 149, 847–859 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dewannieux M., Esnault C., Heidmann T., LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Hagan C. R., Sheffield R. F., Rudin C. M., Human Alu element retrotransposition induced by genotoxic stress. Nat. Genet. 35, 219–220 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Cost G. J., Feng Q., Jacquier A., Boeke J. D., Human L1 element target-primed reverse transcription in vitro. EMBO J. 21, 5899–5910 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazazian H. H., Mobile elements: Drivers of genome evolution. Science 303, 1626–1632 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Kazazian H. H. Jr., Moran J. V., Mobile DNA in health and disease. N. Engl. J. Med. 377, 361–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda S., Varshney A., Fowler B. J., Wang S. B., Narendran S., Ambati K., Yasuma T., Magagnoli J., Leung H., Hirahara S., Nagasaka Y., Yasuma R., Apicella I., Pereira F., Makin R. D., Magner E., Liu X., Sun J., Wang M., Baker K., Marion K. M., Huang X., Baghdasaryan E., Ambati M., Ambati V. L., Pandey A., Pandya L., Cummings T., Banerjee D., Huang P., Yerramothu P., Tolstonog G. V., Held U., Erwin J. A., Paquola A. C. M., Herdy J. R., Ogura Y., Terasaki H., Oshika T., Darwish S., Singh R. K., Mozaffari S., Bhattarai D., Kim K. B., Hardin J. W., Bennett C. L., Hinton D. R., Hanson T. E., Röver C., Parang K., Kerur N., Liu J., Werner B. C., Sutton S. S., Sadda S. R., Schumann G. G., Gelfand B. D., Gage F. H., Ambati J., Cytoplasmic synthesis of endogenous Alu complementary DNA via reverse transcription and implications in age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 118, e2022751118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerur N., Fukuda S., Banerjee D., Kim Y., Fu D., Apicella I., Varshney A., Yasuma R., Fowler B. J., Baghdasaryan E., Marion K. M., Huang X., Yasuma T., Hirano Y., Serbulea V., Ambati M., Ambati V. L., Kajiwara Y., Ambati K., Hirahara S., Bastos-Carvalho A., Ogura Y., Terasaki H., Oshika T., Kim K. B., Hinton D. R., Leitinger N., Cambier J. C., Buxbaum J. D., Kenney M. C., Jazwinski S. M., Nagai H., Hara I., West A. P., Fitzgerald K. A., Sadda S. V. R., Gelfand B. D., Ambati J., cGAS drives noncanonical-inflammasome activation in age-related macular degeneration. Nat. Med. 24, 50–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarks J. P., Sarks S. H., Killingsworth M. C., Evolution of geographic atrophy of the retinal pigment epithelium. Eye 2, 552–577 (1988). [DOI] [PubMed] [Google Scholar]
- 14.Hariri A. H., Tepelus T. C., Akil H., Nittala M. G., Sadda S. R., Retinal sensitivity at the junctional zone of eyes with geographic atrophy due to age-related macular degeneration. Am. J. Ophthalmol. 168, 122–128 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Holz F. G., Bellman C., Staudt S., Schütt F., Völcker H. E., Fundus autofluorescence and development of geographic atrophy in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 42, 1051–1056 (2001). [PubMed] [Google Scholar]
- 16.Casavant N. C., Scott L. A., Cantrell M. A., Wiggins L. E., Baker R. J., Wichman H. A., The end of the LINE?: Lack of recent L1 activity in a group of South American rodents. Genetics 154, 1809–1817 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grahn R. A., Rinehart T. A., Cantrell M. A., Wichman H. A., Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet. Genome Res. 110, 407–415 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Rinehart T. A., Grahn R. A., Wichman H. A., SINE extinction preceded LINE extinction in sigmodontine rodents: Implications for retrotranspositional dynamics and mechanisms. Cytogenet. Genome Res. 110, 416–425 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Yang L., Brunsfeld J., Scott L., Wichman H., Reviving the dead: History and reactivation of an extinct L1. PLOS Genet. 10, e1004395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai L., Huang Q., Boeke J. D., Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochem. 12, 18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fowler B. J., Gelfand B. D., Kim Y., Kerur N., Tarallo V., Hirano Y., Amarnath S., Fowler D. H., Radwan M., Young M. T., Pittman K., Kubes P., Agarwal H. K., Parang K., Hinton D. R., Bastos-Carvalho A., Li S., Yasuma T., Mizutani T., Yasuma R., Wright C., Ambati J., Nucleoside reverse transcriptase inhibitors possess intrinsic anti-inflammatory activity. Science 346, 1000–1003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun L., Wu J., Du F., Chen X., Chen Z. J., Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ullu E., Tschudi C., Alu sequences are processed 7SL RNA genes. Nature 312, 171–172 (1984). [DOI] [PubMed] [Google Scholar]
- 24.Zuker M., Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herzner A. M., Hagmann C. A., Goldeck M., Wolter S., Kübler K., Wittmann S., Gramberg T., Andreeva L., Hopfner K. P., Mertens C., Zillinger T., Jin T., Xiao T. S., Bartok E., Coch C., Ackermann D., Hornung V., Ludwig J., Barchet W., Hartmann G., Schlee M., Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat. Immunol. 16, 1025–1033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.West A. P., Khoury-Hanold W., Staron M., Tal M. C., Pineda C. M., Lang S. M., Bestwick M., Duguay B. A., Raimundo N., MacDuff D. A., Kaech S. M., Smiley J. R., Means R. E., Iwasaki A., Shadel G. S., Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.J. Ambati, B. J. Fowler, K. Ambati, Compositions and methods for treating retinal degradation. Patent WO2016138425A1 (2016).
- 28.Hung T., Pratt G. A., Sundararaman B., Towsend M. J., Chaivorapol C., Bhangale T., Graham R. R., Ortmann W., Criswell L. A., Yeo G. W., Behrens T. W., The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science 350, 455–459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambati J., Magagnoli J., Leung H., bin Wang S., Andrews C. A., Fu D., Pandey A., Sahu S., Narendran S., Hirahara S., Fukuda S., Sun J., Pandya L., Ambati M., Pereira F., Varshney A., Cummings T., Hardin J. W., Edun B., Bennett C. L., Ambati K., Fowler B. J., Kerur N., Röver C., Leitinger N., Werner B. C., Stein J. D., Sutton S. S., Gelfand B. D., Repurposing anti-inflammasome NRTIs for improving insulin sensitivity and reducing type 2 diabetes development. Nat. Commun. 11, 4737 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinrich M. J., Purcell C. A., Pruijssers A. J., Zhao Y., Spurlock C. F., Sriram S., Ogden K. M., Dermody T. S., Scholz M. B., Crooke P. S. III, Karijolich J., Aune T. M., Endogenous double-stranded Alu RNA elements stimulate IFN-responses in relapsing remitting multiple sclerosis. J. Autoimmun. 100, 40–51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindner M., Böker A., Mauschitz M. M., Göbel A. P., Fimmers R., Brinkmann C. K., Schmitz-Valckenberg S., Schmid M., Holz F. G., Fleckenstein M., Directional kinetics of geographic atrophy progression in age-related macular degeneration with foveal sparing. Ophthalmology 122, 1356–1365 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Narendran S., Pereira F., Yerramothu P., Apicella I., Wang S. B., Ambati K., Hirahara S., Kim Y., Ambati M., Ambati V. L., Huang P., Varshney A., Nagasaka Y., Fukuda S., Baker K. L., Marion K. M., Deussing J. M., Sadda S. R., Gelfand B. D., Ambati J., Nucleoside reverse transcriptase inhibitors and Kamuvudines inhibit amyloid-β induced retinal pigmented epithelium degeneration. Signal Transduct. Target. Ther. 6, 149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonilha V. L., Rayborn M. E., Bell B. A., Marino M. J., Fishman G. A., Hollyfield J. G., Retinal histopathology in eyes from a patient with Stargardt disease caused by compound heterozygous ABCA4 mutations. Ophthalmic Genet. 37, 150–160 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Bonilha V. L., Rayborn M. E., Bell B. A., Marino M. J., Pauer G. J., Beight C. D., Chiang J., Traboulsi E. I., Hollyfield J. G., Hagstrom S. A., Histopathological comparison of eyes from patients with autosomal recessive retinitis pigmentosa caused by novel EYS mutations. Graefes Arch. Clin. Exp. Ophthalmol. 253, 295–305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonilha V. L., Rayborn M. E., Bell B. A., Marino M. J., Traboulsi E. I., Hagstrom S. A., Hollyfield J. G., Histopathology of the retina from a three year-old suspected to have Joubert syndrome. Austin J. Clin. Ophthalmol. 2, 1057 (2015). [PMC free article] [PubMed] [Google Scholar]
- 36.Bonilha V. L., Rayborn M. E., Li Y., Grossman G. H., Berson E. L., Hollyfield J. G., Histopathology and functional correlations in a patient with a mutation in RPE65, the gene for retinol isomerase. Invest. Ophthalmol. Vis. Sci. 52, 8381–8392 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerur N., Hirano Y., Tarallo V., Fowler B. J., Bastos-Carvalho A., Yasuma T., Yasuma R., Kim Y., Hinton D. R., Kirschning C. J., Gelfand B. D., Ambati J., TLR-independent and P2X7-dependent signaling mediate Alu RNA-induced NLRP3 inflammasome activation in geographic atrophy. Invest. Ophthalmol. Vis. Sci. 54, 7395–7401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y., Tarallo V., Kerur N., Yasuma T., Gelfand B. D., Bastos-Carvalho A., Hirano Y., Yasuma R., Mizutani T., Fowler B. J., Li S., Kaneko H., Bogdanovich S., Ambati B. K., Hinton D. R., Hauswirth W. W., Hakem R., Wright C., Ambati J., DICER1/Alu RNA dysmetabolism induces caspase-8–mediated cell death in age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 111, 16082–16087 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J., Maram J., Tepelus T. C., Sadda S. R., Chopra V., Lee O. L., Comparison of noncontact specular and confocal microscopy for evaluation of corneal endothelium. Eye Contact Lens 44, S144–S150 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Ach T., Tolstik E., Messinger J. D., Zarubina A. V., Heintzmann R., Curcio C. A., Lipofuscin redistribution and loss accompanied by cytoskeletal stress in retinal pigment epithelium of eyes with age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 56, 3242–3252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grossniklaus H. E., Nickerson J. M., Edelhauser H. F., Bergman L. A. M. K., Berglin L., Anatomic alterations in aging and age-related diseases of the eye. Invest. Ophthalmol. Vis. Sci. 54, ORSF23–ORSF27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarau I. S., Berlin A., Curcio C. A., Ach T., The cytoskeleton of the retinal pigment epithelium: From normal aging to age-related macular degeneration. Int. J. Mol. Sci. 20, 3578 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirilyuk A., Tolstonog G. V., Damert A., Held U., Hahn S., Löwer R., Buschmann C., Horn A. V., Traub P., Schumann G. G., Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells. Nucleic Acids Res. 36, 648–665 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Q., Moran J. V., Kazazian H. H. Jr., Boeke J. D., Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Moran J. V., Holmes S. E., Naas T. P., DeBerardinis R. J., Boeke J. D., Kazazian H. H. Jr., High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Wakimoto Y., Jiang J., Wakimoto H., Isolation of single-stranded DNA. Curr. Protoc. Mol. Biol. 107, 1–9 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Ardeljan D., Wang X., Oghbaie M., Taylor M. S., Husband D., Deshpande V., Steranka J. P., Gorbounov M., Yang W. R., Sie B., Larman H. B., Jiang H., Molloy K. R., Altukhov I., Li Z., McKerrow W., Fenyö D., Burns K. H., LaCava J., LINE-1 ORF2p expression is nearly imperceptible in human cancers. Mob. DNA 11, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ardeljan D., Steranka J. P., Liu C., Li Z., Taylor M. S., Payer L. M., Gorbounov M., Sarnecki J. S., Deshpande V., Hruban R. H., Boeke J. D., Fenyö D., Wu P. H., Smogorzewska A., Holland A. J., Burns K. H., Cell fitness screens reveal a conflict between LINE-1 retrotransposition and DNA replication. Nat. Struct. Mol. Biol. 27, 168–178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raiz J., Damert A., Chira S., Held U., Klawitter S., Hamdorf M., Löwer J., Strätling W. H., Löwer R., Schumann G. G., The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40, 1666–1683 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo S.-U., Kuffa P., Kitamoto S., Nagao-Kitamoto H., Rousseau J., Kim Y.-G., Núñez G., Kamada N., Intestinal macrophages arising from CCR2+ monocytes control pathogen infection by activating innate lymphoid cells. Nat. Commun. 6, 8010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Perez J. L., Morell M., Scheys J. O., Kulpa D. A., Morell S., Carter C. C., Hammer G. D., Collins K. L., O’Shea K. S., Menendez P., Moran J. V., Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 466, 769–773 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kotecki M., Reddy P. S., Cochran B. H., Isolation and characterization of a near-haploid human cell line. Exp. Cell Res. 252, 273–280 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Kowarz E., Löscher D., Marschalek R., Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 10, 647–653 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Taylor M. S., LaCava J., Mita P., Molloy K. R., Huang C. R. L., Li D., Adney E. M., Jiang H., Burns K. H., Chait B. T., Rout M. P., Boeke J. D., Dai L., Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 155, 1034–1048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai L., LaCava J., Taylor M. S., Boeke J. D., Expression and detection of LINE-1 ORF-encoded proteins. Mob. Genet. Elements 4, e29319 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor M. S., La Cava J., Dai L., Mita P., Burns K. H., Rout M. P., Boeke J. D., Characterization of L1-ribonucleoprotein particles. Methods Mol. Biol. 1400, 311–338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu W. M., Chu W. M., Choudary P. V., Schmid C. W., Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 23, 1758–1765 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doucet A. J., Hulme A. E., Sahinovic E., Kulpa D. A., Moldovan J. B., Kopera H. C., Athanikar J. N., Hasnaoui M., Bucheton A., Moran J. V., Gilbert N., Characterization of LINE-1 ribonucleoprotein particles. PLOS Genet. 6, e1001150 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei W., Gilbert N., Ooi S. L., Lawler J. F., Ostertag E. M., Kazazian H. H., Boeke J. D., Moran J. V., Human L1 Retrotransposition: Cis preference versus trans complementation. Mol. Cell. Biol. 21, 1429–1439 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mackenzie K. J., Carroll P., Lettice L., Tarnauskaitė Ž., Reddy K., Dix F., Revuelta A., Abbondati E., Rigby R. E., Rabe B., Kilanowski F., Grimes G., Fluteau A., Devenney P. S., Hill R. E., Reijns M. A., Jackson A. P., Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J. 35, 831–844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11