Camillo et al. show that the LPHN2 receptor, upon activation by FLRT2 ligand, inhibits focal adhesion formation and promotes tight junction assembly in endothelial cells. Blood vessels of lphn2a null animals are hyperpermeable, and injected cancer cells extravasate more easily.
Abstract
Dynamic modulation of endothelial cell-to-cell and cell–to–extracellular matrix (ECM) adhesion is essential for blood vessel patterning and functioning. Yet the molecular mechanisms involved in this process have not been completely deciphered. We identify the adhesion G protein–coupled receptor (ADGR) Latrophilin 2 (LPHN2) as a novel determinant of endothelial cell (EC) adhesion and barrier function. In cultured ECs, endogenous LPHN2 localizes at ECM contacts, signals through cAMP/Rap1, and inhibits focal adhesion (FA) formation and nuclear localization of YAP/TAZ transcriptional regulators, while promoting tight junction (TJ) assembly. ECs also express an endogenous LPHN2 ligand, fibronectin leucine-rich transmembrane 2 (FLRT2), that prevents ECM-elicited EC behaviors in an LPHN2-dependent manner. Vascular ECs of lphn2a knock-out zebrafish embryos become abnormally stretched, display a hyperactive YAP/TAZ pathway, and lack proper intercellular TJs. Consistently, blood vessels are hyperpermeable, and intravascularly injected cancer cells extravasate more easily in lphn2a null animals. Thus, LPHN2 ligands, such as FLRT2, may be therapeutically exploited to interfere with cancer metastatic dissemination.
Introduction
The small GTPase Rap1 drives blood vessel formation and function (Chrzanowska-Wodnicka, 2013) by exerting opposite effects on cell adhesion and motility (Coló et al., 2012; Hong et al., 2007; Lagarrigue et al., 2015, 2016; Lyle et al., 2008), likely due to the involvement of different subcellular pools of Rap1 regulators and effectors (Bos and Pannekoek, 2012). Indeed, the Rap1-GTP–interacting adapter molecule (RIAM) can either promote, through talin, the conformational activation of integrin adhesion receptors at the leading edge of migrating cells (Lagarrigue et al., 2016) or support, via the mitogen-activated protein kinase kinase (MEK), the disassembly of centrally located integrin-based focal adhesions (FAs; Coló et al., 2012). In addition, prototypic repulsive guidance cues such as secreted class 3 Semaphorins (SEMA3) control blood vessel patterning (Valdembri et al., 2016; Wälchli et al., 2015) through the cytosolic GTPase-activating protein domain of Plexin receptors (Worzfeld et al., 2014) that negatively regulates Rap1 signaling (Gioelli et al., 2018; Wang et al., 2012). On the contrary, G protein–coupled receptors (GPCRs) promote Rap1 GTP loading, e.g., via the adenylyl cyclase/cAMP/exchange protein directly activated by cAMP (EPAC) guanine nucleotide exchange factor cascade (Bos and Pannekoek, 2012; Christensen et al., 2003). Indeed, cAMP/EPAC/Rap1 is required for the formation of ECM mechanosensing FAs that allow endothelial cells (ECs) to respond to guidance cues (Chrzanowska-Wodnicka, 2017). However, cAMP/EPAC/Rap1 activation by GPCRs can also inhibit EC migration (Avanzato et al., 2016; Hong et al., 2007).
The ability of ECs to modify their behavior in response to ECM patterning and rigidity also relies on the fact that integrin-containing FAs (Karaman and Halder, 2018) signal to suppress the Hippo pathway–dependent inhibitory phosphorylation of the effectors Yes-associated protein (YAP) and WW domain–containing transcription regulator protein 1 (WWTR1) or transcriptional coactivator with PSD-95/discs large/ZO-1 (PDZ)–binding motif (TAZ). In their dephosphorylated active form, YAP and TAZ translocate to the nucleus to control gene transcription (Dupont et al., 2011; Totaro et al., 2018) and angiogenic blood vessel formation and function (Choi et al., 2015; Elaimy and Mercurio, 2018; Kim et al., 2017; Nakajima et al., 2017; Neto et al., 2018; Wang et al., 2016, 2017). While tight junctions (TJs) promote Hippo-dependent phosphorylation, inhibition, and cytosolic sequestration of YAP/TAZ, their dephosphorylation, activation, and nuclear translocation, triggered by FAs, involve integrins and their effectors integrin-linked kinase, Src, and FA kinase (Karaman and Halder, 2018; Moya and Halder, 2019). In addition, myosin-driven contraction of actin stress fibers, which mechanically connect FAs to the nucleus via the linker of nucleoskeleton and cytoskeleton (LINC) complex (Kechagia et al., 2019), open nuclear pores to allow YAP/TAZ nuclear translocation (Elosegui-Artola et al., 2017; Kechagia et al., 2019; Totaro et al., 2018).
To identify novel guidance receptors that may be key in the regulation of blood vessel formation and function, we focused on adhesion GPCRs (ADGRs) that interact with several ECM ligands and integrins (Langenhan et al., 2013), whose adhesive functions are regulated by archetypal guidance cues and their receptors, such as SEMA3 and Plexins (Serini et al., 2003; Worzfeld and Offermanns, 2014). In this context, Latrophilin 2 (LPHN2) emerged as an ideal candidate since, similarly to Plexins, it was originally identified as a neuronal receptor (Südhof, 2001), and its mRNA was reported to be highly expressed in ex vivo isolated blood vascular, but not lymphatic, ECs (Valtcheva et al., 2013). In neurons, the fibronectin (FN) leucine-rich transmembrane (FLRT) proteins act as sheddable chemorepulsive ligands (Yamagishi et al., 2011) that bind and signal via the LPHN receptors (Seiradake et al., 2016). Here, we unveil how in ECs, FLRT2-elicited LPHN2 signals to inhibit ECM adhesion, but to promote TJ assembly, thus limiting YAP/TAZ signaling and vascular permeability.
Results and discussion
To characterize the mechanisms by which LPHN2 receptor controls ECM adhesion, we investigated its subcellular localization and function in cultured human ECs. First, we confirmed that endogenous LPHN2 protein is expressed on the EC surface (Fig. S1 A), and fluorescence confocal microscopy experiments revealed a robust enrichment of endogenous LPHN2 in vinculin containing ECM adhesions of ECs (Fig. 1 A). Due to autocatalytic processing, ADGRs exist as noncovalently associated heterodimers comprising an extracellular and a transmembrane subunit (Langenhan et al., 2013). To verify the relationships of both LPHN2 subunits with ECM adhesions, we transfected ECs with an N-terminally HA-tagged and C-terminally EGFP-tagged mouse Lphn2 construct (HA-Lphn2-EGFP; Fig. S1 B). We found that, similarly to endogenous LPHN2, both the HA-tagged extracellular and the GFP-tagged intracellular moieties of HA-Lphn2-EGFP colocalized with vinculin at cell-to-ECM adhesion sites (Fig. 1 B). Next, we evaluated the outcome of LPHN2 silencing (Fig. S1 C) on ECM-elicited EC motility (Gioelli et al., 2018). Impedance-based time-lapse migration assays revealed that LPHN2 silenced (siLPHN2) ECs migrate much faster toward type I collagen (Coll I; Fig. 1 C) or FN (Fig. S1 D) than control silenced (siCTL) ECs. Thus, in cultured ECs, LPHN2 mediates inhibitory signals that are likely initiated by autocrine loops of endogenous LPHN2 ligands.
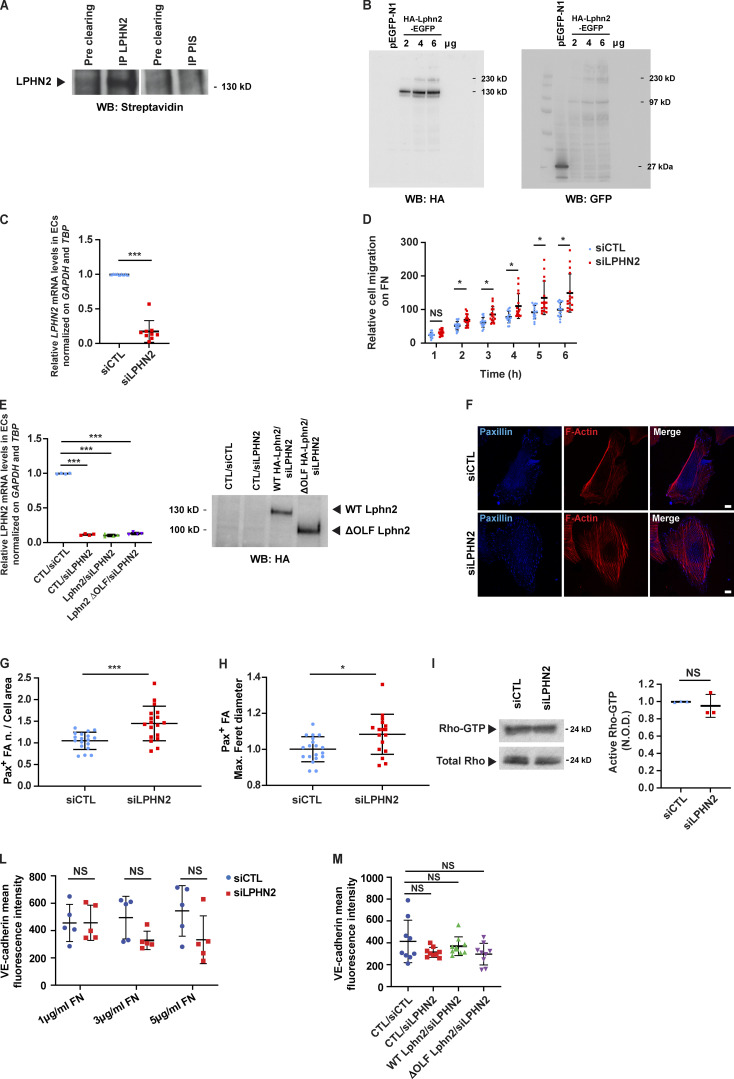
Figure S1.
Endogenous LPHN2 expression, silencing, and rescue impact on endothelial cell adhesion, migration, and signaling. (A) Upon surface biotinylation, LPHN2 was immunoprecipitated (IP) with a rabbit anti-LPHN2 pAb from EC lysates and revealed in Western blot (WB) by means of HRP streptavidin. Pre-immune serum (PIS) was employed for control purposes. The cleaved extracellular portion of LPHN2 appears as an ∼130 kD protein band. (B) HEK 293T cells were transfected with an empty vector or increasing amounts (2, 4, and 6 µg) of HA-Lphn2-EGFP construct. Cell lysates were then analyzed by Western blot with either anti-HA (left) or anti-GFP (right) Ab. The 230-kD, 130-kD, and 97-kD protein bands correspond to uncleaved full-length, cleaved N-terminal extracellular-only portion, and cleaved C-terminal portion of the transfected HA-Lphn2-EGFP protein, respectively. In the anti-GFP Western blot, the 27-kD band corresponds to the GFP whose cDNA was present in the control empty vector. (C) Real-time quantitative PCR analysis of LPHN2 mRNA in siCTL or siLPHN2 human ECs relative to the housekeeping genes GAPDH and TBP and normalized on siCTL levels. Results are themean ± SD of nine independent assays. Statistical analysis: two-tailed heteroscedastic Student’s t test; ***, P ≤ 0.001. (D) Real-time analysis of control (siCTL) or LPHN2 (siLPHN2) silenced EC migration toward FN was assessed with an xCELLigence RTCA DP system. Results are the mean ± SD of four independent assays. Statistical analysis: two-way ANOVA and Bonferroni’s post hoc analysis; NS, P > 0.05; *, P ≤ 0.05. (E) Real-time quantitative PCR analysis of LPHN2 mRNA in siCTL or siLPHN2 human ECs rescued or not (CTL) with mouse WT or ΔOLF Lphn2 relative to the housekeeping genes GAPDH and TBP and normalized on siCTL levels. Results are the mean ± SD of four independent assays. Statistical analysis: one-way ANOVA and Bonferroni’s post hoc analysis; ***, P ≤ 0.001. Right: Transduced and silenced ECs were also lysed and WT HA-Lphn2-pCCL and ΔOLF HA-Lphn2-pCCL protein expression levels analyzed by Western blot with anti-HA Ab. (F–H) Confocal microscopy analysis (F) of endogenous paxillin (Pax; blue), and phalloidin-labeled F-actin (red) reveals how, compared with siCTL ECs, LPHN2 silencing increases the number, normalized on cell area (G) and size (expressed by maximum Feret diameter, H) of paxillin-containing FAs. Scale bars, 10 µm. Results are the mean ± SD of two independent experiments for a total of 18 (siCTL) and 18 (siLPHN2) ECs. Statistical analysis: two-tailed heteroscedastic Student’s t test; *, P ≤ 0.05; ***, P < 0.001. (I) LPHN2 silencing in human ECs does not affect basal GTP loading of RhoA small GTPase. Total Rho was used to calculate the normalized OD (N.O.D.) levels of active Rho-GTP. Results are the mean ± SD of three independent assays. Statistical analysis: two-tailed heteroscedastic Student’s t test; NS, P > 0.05. (L) Mean fluorescence intensity of VE-cadherin (VE-cad+) intercellular staining (in green in Fig. 5 A) in siCTL ECs seeded on 10 kPa substrates coated with increasing amounts of FN (1, 3, and 5 µg/ml). The VE-cadherin intercellular recruitment was not affected by FN density. Results are the mean ± SD of two independent experiments. Statistical analysis: two-way ANOVA with Bonferroni’s post hoc analysis; NS, P > 0.05. (M) Mean fluorescence intensity of VE-cad+ intercellular staining (in green in Fig. 5 B) in lentivirally delivered WT or ΔOLF Lphn2 in ECs seeded on 10 kPa substrates coated with FN (5 µg/ml). The VE-cadherin intercellular recruitment was not affected by LPHN2 silencing. Results are the mean ± SD of two independent experiments. Statistical analysis: two-way ANOVA with Bonferroni’s post hoc analysis; NS, P > 0.05. Max., maximum; n., number.
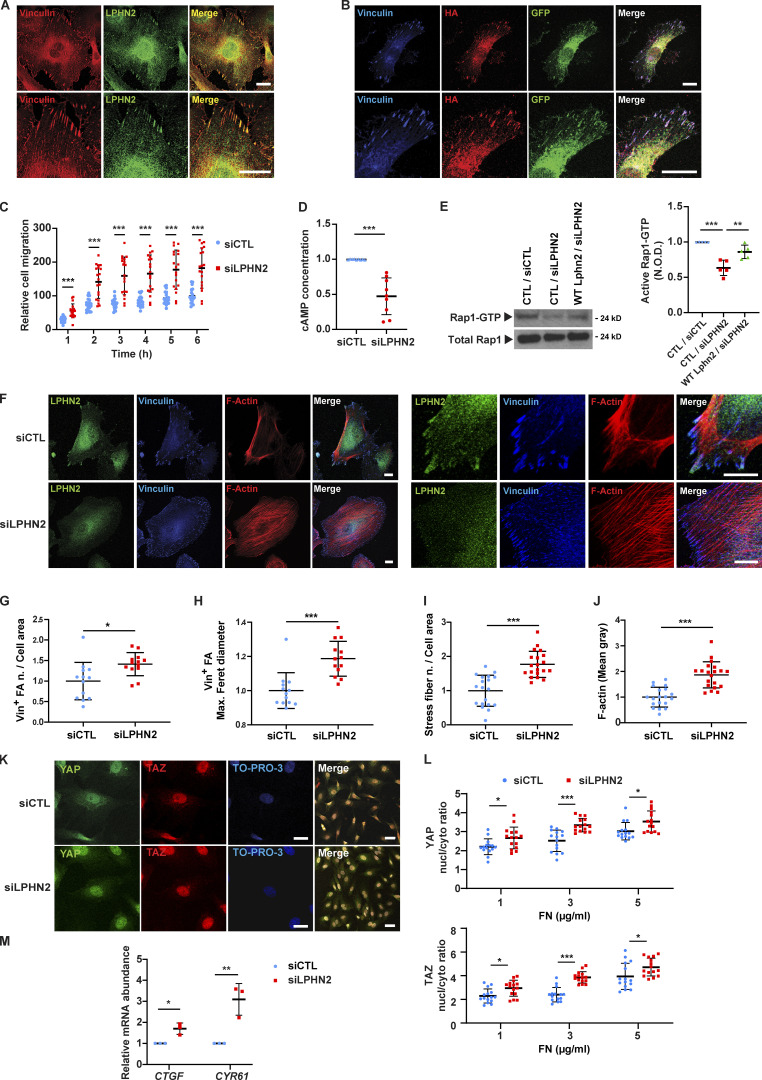
Figure 1.
LPHN2 signals via cAMP/Rap1 and negatively regulates FA turnover, stress fiber formation, and ECM-elicited mechanosensing. (A) Confocal microscopy analysis of ECs indicates how endogenous LPHN2, as detected by an anti-LPHN2 Ab (green), colocalizes with vinculin (Vin; red) in ECM adhesions of ECs as shown in merge (right). Bottom: Magnifications of the corresponding top row panels. Scale bars, 20 µm. (B) Fluorescence confocal microcopy reveals that in ECs transfected with HA-Lphn2-EGFP, both the extracellular, as detected by an anti-HA Ab (red), and the EGFP-fused intracellular (green) moieties colocalize with vinculin (blue) at ECM adhesions, as shown in merge (right). Bottom: Magnifications of the corresponding upper row panels. Scale bars, 20 µm. (C) Real-time analysis of cell migration in siCTL or siLPHN2 ECs toward Coll I, assessed with an xCELLigence RTCA DP system. Results are the mean ± SD of five independent assays. Statistical analysis: two-way ANOVA and Bonferroni’s post hoc analysis; ***, P ≤ 0.001. (D) LPHN2 silencing in ECs decreases basal amount of cAMP. Results are the mean ± SD of nine independent assays. Statistical analysis: two-tailed heteroscedastic Student’s t test; ***, P ≤ 0.001. (E) Rap1-GTP was pulled down on a GST fusion protein carrying the Rap1-binding domain of human Ral guanine nucleotide dissociation stimulator. LPHN2 silencing in human ECs decreases basal GTP loading of Rap1 small GTPase, and pCCL lentivirus-mediated overexpression of silencing-resistant mouse Lphn2 rescues the decrease. Total Rap1 was employed to calculate the normalized OD (N.O.D.) levels of active Rap1-GTP. Results are the mean ± SD of five independent experiments (a representative one is shown). Statistical analysis: one-way ANOVA and Bonferroni’s post hoc analysis; ***, P ≤ 0.001 for CTL/siCTL versus CTL/siLPHN2, and **, P ≤ 0.01 for CTL/siLPHN2 versus Lphn2/siLPHN2. (F–J) Confocal microscopy analysis (F) of endogenous LPHN2 (green), vinculin (blue), and phalloidin-labeled F-actin (red; each high magnification images are on the right) reveals how, compared with siCTL ECs, LPHN2 silencing increases the number, normalized on cell area (G) and size (expressed by maximum Feret diameter, H) of vinculin-containing FAs and the number, normalized on cell area (I) and amount (evaluated as mean gray intensity, J) of F-actin stress fibers in siLPHN2 ECs. Scale bars, 20 µm. Results concerning vinculin-containing FAs and F-actin stress fibers are the mean ± SD of two independent experiments for a total of 13 siCTL and 13 siLPHN2 ECs and two independent experiments for a total of 19 siCTL and 20 siLPHN2 ECs, respectively. Statistical analysis: two-tailed heteroscedastic Student’s t test; *, P ≤ 0.05; ***, P ≤ 0.001. (K and L) Confocal microscopy analysis (K) of endogenous YAP (green) and TAZ (red) reveals how, compared with siCTL ECs, LPHN2 silencing increases the nuclear (nucl)/cytoplasmic (cyto) ratio of both YAP (L, top) and TAZ (L, bottom). Nuclei were stained with TO-PRO3 (blue). Scale bar, 20 µm. Representative images of ECs plated on 10 kPa stiffness on FN (5 µg/ml) are shown. Results are the mean ± SD of two independent experiments for a total of 11 ECs for each condition (10 kPa and increasing FN 1, 3, and 5 µg/ml concentration). Statistical analysis: two-way ANOVA and Bonferroni’s post hoc analysis; *, P ≤ 0.05; ***, P ≤ 0.001. (M) Real-time quantitative PCR analysis of CTGF and CYR61 mRNA in siCTL or siLPHN2 human ECs relative to the housekeeping genes, GAPDH and TBP, and normalized on siCTL levels. Data of one of two independent assays are shown. Results are the mean ± SD of three technical replicates. Statistical analysis: two-tailed heteroscedastic Student’s t test; *, P ≤ 0.05; **, P ≤ 0.01. Max., maximum; n., number.
Several GPCRs activate the cAMP/Rap1 pathway that regulates cell adhesion (Gloerich and Bos, 2011). Thus, we assessed whether in ECs, LPHN2 may impact on basal cAMP production and Rap1 GTP loading by pulling it down with a GST fusion protein carrying the Rap1-binding domain of human Ral guanine nucleotide dissociation stimulator (Franke et al., 1997). Compared with siCTL ECs, siLPHN2 ECs displayed a decrease in the basal amount of both cAMP (Fig. 1 D) and active Rap1 (Fig. 1 E) by 52.6 ± 8.7% and 36.35 ± 4.8% (mean ± SD), respectively. Moreover, the transduction of a silencing-resistant mouse Lphn2 construct (Fig. S1 E) effectively rescued active Rap1 levels in siLPHN2 ECs (Fig. 1 E). These findings were consistent with the notion that cAMP/EPAC/Rap1 activation can inhibit migration by modulating cell adhesion dynamics (Lyle et al., 2008). Therefore, we next evaluated the impact of LPHN2 silencing on FAs and the associated F-actin stress fibers. Confocal microscopy analysis of both vinculin (Fig. 1 F) and paxillin (Fig. S1 F) containing FAs revealed that in ECs, the lack of LPHN2 results in a significant increase of both FA corrected number (Figs. 1 G and S1 G) and size (Figs. 1 H and S1 H). Moreover, the corrected number (Fig. 1 I) and mean gray fluorescence (Fig. 1 J) of F-actin stress fibers were also clearly increased in siLPHN2 compared with siCTL ECs. The small GTPase RhoA controls stress fiber formation and FA turnover (Lawson and Burridge, 2014), and other ADGRs, such as GPR56/ADGRG1, were previously reported to activate RhoA (Paavola and Hall, 2012). Yet we did not detect any reduction in Rho activation after LPHN2 silencing in ECs (Fig. S1 I). Hence, LPHN2 activates cAMP/Rap1 signaling and negatively regulates FA and stress fiber formation as well as EC migration toward the ECM in a RhoA-independent manner.
In cultured ECs, LPHN2 silencing results in a significantly increased number of FAs coupled to F-actin filaments (Fig. 1, F, G, I, and J; and Fig. S1, F and G), with both structures part of the mechanically sensitive system that promotes YAP/TAZ translocation into the cell nucleus (Chang et al., 2018; Elosegui-Artola et al., 2017; Karaman and Halder, 2018; Kechagia et al., 2019; Moya and Halder, 2019; Totaro et al., 2018). Of note, endothelial YAP/TAZ signaling controls angiogenesis, vascular barrier maturation, and blood vessel maintenance (Kim et al., 2017; Nakajima et al., 2017; Wang et al., 2017). To investigate whether LPHN2 may impact on YAP/TAZ subcellular localization and signaling, we employed hydrogels with a soft 10 kPa elastic module, previously found to be optimally stiff for cultured EC monolayers (Birukova et al., 2013; Galie et al., 2015; Janmey et al., 2020). In agreement with the notion that ECM coating density is a key determinant of force transmission at integrin-based adhesion sites (Elosegui-Artola et al., 2017; Stanton et al., 2019; Lee et al., 2019), we found that coating 10 kPa hydrogels with increasing amounts of FN resulted in a significantly higher dose-dependent increase of YAP/TAZ nuclear translocation (Fig. 1, K and L) and transcription of YAP/TAZ target genes CTGF and CYR61 (Fig. 1 M) in siLPHN2 compared with siCTL ECs. Hence, in ECs, LPHN2 inhibits the formation of FAs and stress fibers along with the ensuing nuclear translocation and transcriptional activity of YAP/TAZ.
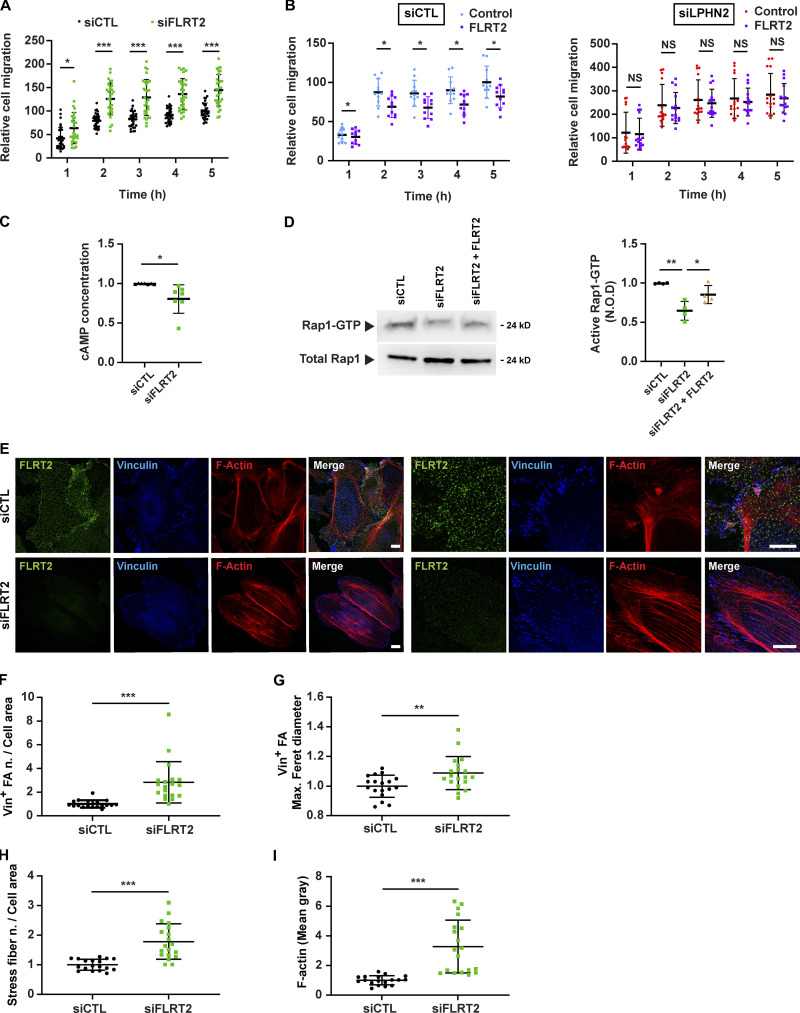
Next, we reasoned that the increase of ECM-driven motility (Fig. 1 C and Fig. S1 D), FAs (Fig. 1, F–H; and Fig. S1, F–H), stress fibers (Fig. 1, F, I, and J), and YAP/TAZ nuclear translocation (Fig. 1, K and L) and signaling (Fig. 1 M) caused by LPHN2 silencing in ECs may be due to a loss of cell responsivity to endogenously produced LPHN2 ligands, such as FLRT1-3 transmembrane proteins (Seiradake et al., 2016), whose ectodomains can be shed (Jackson et al., 2016) and act in vivo as repulsive factors steering both axons (Yamagishi et al., 2011) and blood vessels (Seiradake et al., 2014). In real-time quantitative RT-PCR (qRT-PCR) analyses, we observed that cultured ECs actively transcribe FLRT2, but neither FLRT1 nor FLRT3 genes (Fig. S2 A). In addition, we found that the silencing of LPHN2 in ECs causes a twofold up-regulation of FLRT2 mRNA (Fig. S2 B), hinting that FLRT2 and LPHN2 gene transcription are reciprocally regulated. To explore the potential role of FLRT2 binding in the inhibitory activity of LPHN2 on the formation of FAs and stress fibers, YAP/TAZ accumulation in the nucleus, and the migration rate of ECs, we created a mouse Lphn2 construct devoid of the olfactomedin domain (ΔOLF Lphn2; Fig. S2 C), which mediates the binding of FLRT ligands to LPHN receptors (Seiradake et al., 2016). Fittingly, ligand-receptor in situ binding assay on COS-7 cells demonstrated that FLRT2 interacts with high affinity with WT but not ΔOLF Lphn2 (Fig. S2 C). Next, we compared the abilities of WT and ΔOLF Lphn2 constructs to rescue the aberrant phenotypes of FA corrected number (vinculin+ in Fig. 2, A and B; and paxillin+ in Fig. S2, D and E) and size (vinculin+ in Fig. 2, A and C; and paxillin+ in Fig. S2, D and F), and stress fibers corrected number (Fig. 2, A and D), mean gray fluorescence (Fig. 2, A and E), and mean cross-sectional area (Fig. 2 F), YAP/TAZ nuclear translocation (Fig. 2, G and H), and Coll I-elicited migration (Fig. 2 I) caused by LPHN2 silencing in ECs. Remarkably WT but not ΔOLF Lphn2 rescued these phenotypic abnormalities in siLPHN2 ECs (Fig. 2; and Fig. S2, D–F). We concluded that, in ECs, the role played by LPHN2 on the inhibition of FAs, F-actin stress fibers, and ECM-elicited cell motility relies on its OLF domain binding to endogenous ligand(s), such as FLRT2. Consistently, we found that ECM-elicited EC directional migration is respectively increased upon silencing of endogenous FLRT2 (siFLRT2; mRNA in Fig. S2 G and protein in Fig. S2 H; Fig. 3 A) and decreased by stimulation with exogenous recombinant FLRT2 protein (Fig. 3 B, left), while no response was seen when LPHN2 was silenced (Fig. 3 B, right). Similar to what observed upon LPHN2 silencing (Fig. 1, D and E), siFLRT2 ECs displayed, compared with siCTL cells, a decrease of the basal amount of both cAMP (Fig. 3 C) and active Rap1 (Fig. 3 D) by 12.9 ± 2.9% and 35.1 ± 6%, respectively. In addition, stimulation with exogenous FLRT2 rescued active Rap1 GTP-loading in siFLRT2 ECs (Fig. 3 D). Furthermore, confocal microscopy revealed that, compared with siCTL, siFLRT2 ECs display a significantly higher corrected number (Fig. 3, E and F) and size (Fig. 3, E and G) of vinculin+ FAs and F-actin stress fiber corrected number (Fig. 3, E and H) and mean gray fluorescence (Fig. 3, E and I). Hence, FLRT2 is one of the endogenous ligands sustaining autocrine/paracrine LPHN2-mediated chemorepulsive signals that negatively regulate the formation of FAs and associated F-actin stress fibers in ECs. Yet uncleaved FLRT2 may also activate LPHN2 localized outside ECM adhesions, e.g., in areas of EC-to-EC contact.
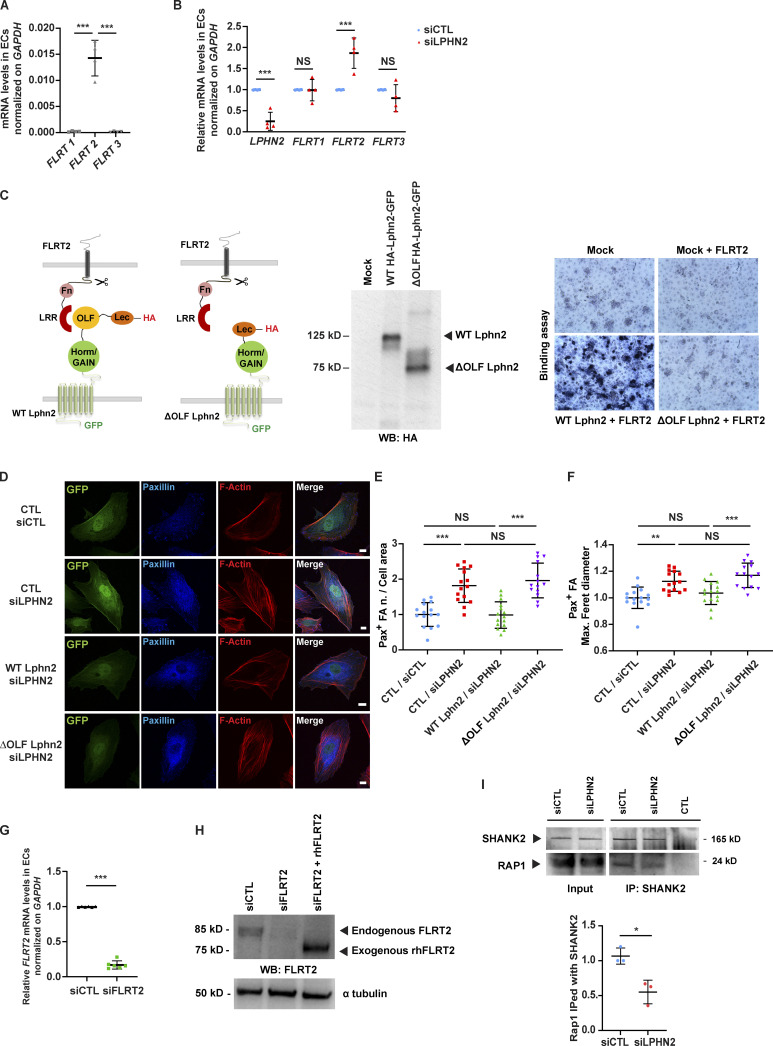
Figure S2.
Endogenous FLRT2 expression, silencing, and LPHN2-mediated signaling impacts on endothelial cell adhesion and migration. (A) Real-time quantitative PCR analysis of FLRT1-3 mRNAs in human ECs relative to the housekeeping gene GAPDH. Results are the mean ± SD of four independent experiments. Statistical analysis: one-way ANOVA and Bonferroni’s post hoc analysis; ***, P ≤ 0.001. (B) Real-time quantitative PCR analysis of LPHN2 and FLRT1-3 mRNAs in siCTL and siLPHN2 human ECs. Results are normalized on siCTL values and are the mean ± SD of four independent experiments. Statistical analysis: one-way ANOVA and Bonferroni’s post hoc analysis; NS, P > 0.05; ***, P ≤ 0.001. (C) COS-7 cells transfected with empty vector or WT HA-Lphn2-EGFP or ΔOLF HA-Lphn2-EGFP constructs and their expression verified by Western blot (WB) analysis using a rat mAb anti-HA (middle). Next, COS-7 cells transfected with empty vector or WT HA-Lphn2-EGFP or ΔOLF HA-Lphn2-EGFP constructs were incubated or not with 6xHis-tagged rhFLRT2. The binding between the Lphn2 constructs and the rhFLRT2 ligand was revealed through sequential incubation with a mouse mAb anti-6xHis, an AP-conjugated goat anti-mouse pAb, and the AP substrate nitro blue tetrazolium–5-bromo-4-chloro-3-indolyl-phosphate (right). Results show how WT but not ΔOLF Lphn2 binds rhFLRT2. (D–F) Confocal microscopy analysis (D) of paxillin (Pax; blue), and phalloidin-labeled F-actin (red). Cells were first transduced with pCCL lentivirus (carrying GFP)-mediated overexpression (green) of silencing-resistant mouse WT or ΔOLF Lphn2 and then oligofected with either siCTL or siLPHN2 siRNAs. Scale bar, 10 µm. Confocal microscopy analysis reveals how lentiviral delivery of WT Lphn2, but not ΔOLF Lphn2 mutant, restores the phenotype of paxillin-containing FAs both considering the number (E) and the size (expressed by maximum Feret diameter, F). Results are the mean ± SD of two independent experiments for a total of 15 ECs for each condition. Statistical analysis: one-way ANOVA and Bonferroni’s post hoc analysis; NS, P > 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (G) Real-time quantitative PCR analysis of FLRT2 mRNA in siCTL or siFLRT2 human ECs relative to the housekeeping gene GAPDH and normalized on siCTL levels. Results are the mean ± SD of six independent assays. Statistical analysis: two-tailed heteroscedastic Student’s t test; ***, P ≤ 0.001. (H) Western blot analysis with an anti-FLRT2 Ab of lysates of siCTL or siFLRT2 ECs or siFLRT2 ECs treated with exogenous rhFLRT2 (800 ng/ml). Endogenous FLRT2 appears as an ∼85 kD protein, while the soluble extracellular portion of exogenous rhFLTR2 appears as an ∼75 kD protein band. (I) LPHN2 silencing in human ECs decreased SHANK2 interaction with Rap1 small GTPase. Western blot analysis of Rap1 coimmunoprecipitated (IPed) with SHANK2 in cultured ECs. Results are the mean ± SD of three independent experiments. Statistical analysis: two-tailed heteroscedastic Student’s t test; *, P ≤ 0.05. Max., maximum; n., number.
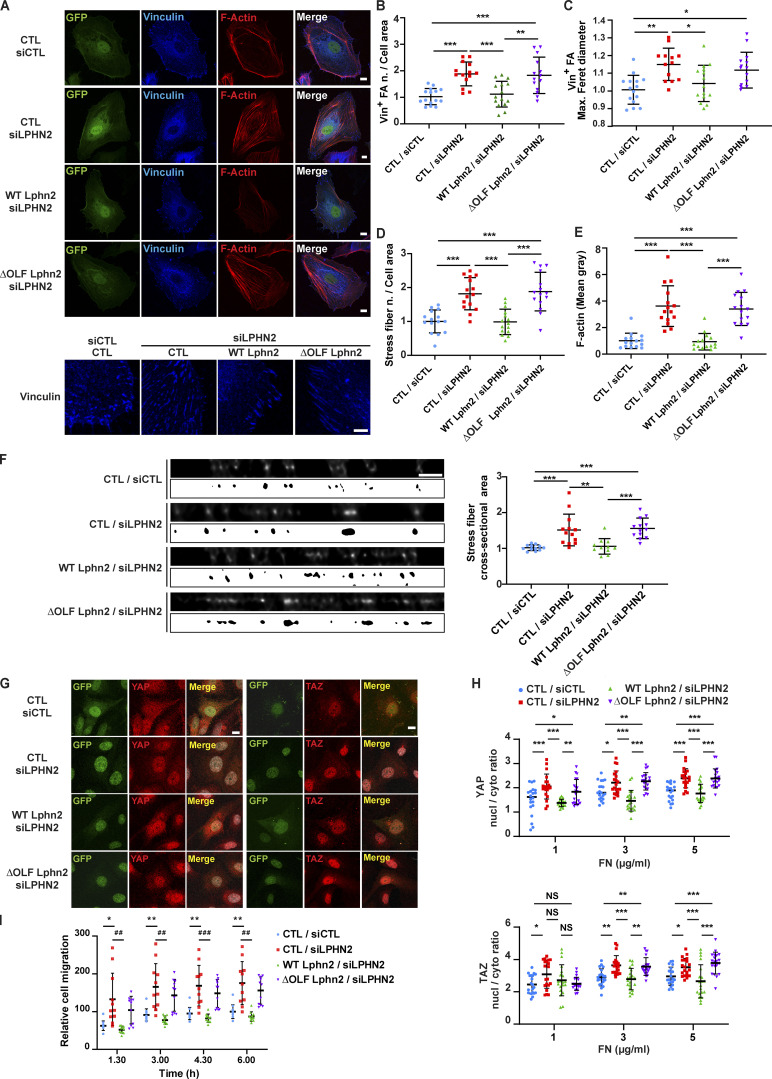
Figure 2.
Negative regulation of FA turnover, stress fiber formation, and ECM-elicited mechanosensing relies on FLRT2-binding OLF domain of LPHN2. (A–E) Confocal microscopy analysis (A) of vinculin (Vin; blue), and phalloidin-labeled F-actin (red; vinculin high-magnification images are on the bottom). Cells were first transduced with pCCL lentivirus (carrying GFP)-mediated overexpression (green) of silencing-resistant mouse WT or ΔOLF Lphn2 and then oligofected with either siCTL or siLPHN2 siRNAs. Scale bars, 10 µm; magnification scale bar, 5 µm. Confocal microscopy analysis reveals how lentiviral delivery of WT LPHN2, but not ΔOLF LPHN2 mutant, restores the phenotype of vinculin-containing FAs number, normalized on cell area (B) and size (expressed by maximum Feret diameter, C). The same rescue effect of WT LphnN2, but not ΔOLF Lphn2 mutant, occurs on stress fiber number, normalized on cell area (D) and amount (evaluated as mean gray intensity, E). Results are the mean ± SD of two independent experiments for a total of 15 ECs for each condition. Statistical analysis: one-way ANOVA and Bonferroni’s post hoc analysis; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (F) XZ-STED confocal microscope cross-sectioning of stress fibers reveals how siLPHN2 ECs have thicker stress fibers in comparison to siCTL ECs, and the transduction of WT, but not ΔOLF Lphn2, rescues this phenotype. The cross-sectional area of stress fibers was measured with a mask, shown (image with white background) at the bottom each image, obtained with ImageJ starting from XZ STED confocal images after deconvolution (image with black background). Scale bar, 1 µm. Results are the mean ± SD of two independent experiments for a total of 11 ECs for each condition. Statistical analysis: one-way ANOVA and Bonferroni’s post hoc analysis; **, P ≤ 0.01; ***, P ≤ 0.001. (G and H) Confocal microscopy analysis (G) of endogenous YAP (in red on the left) and TAZ (in red on the right) reveals how pCCL lentivirus (carrying GFP)-mediated delivery (green) of WT Lphn2, but not ΔOLF Lphn2 mutant, restores the nuclear (nucl)/cytoplasmic (cyto) ratio of both YAP (H, top panel) and TAZ (H, bottom panel). Scale bar, 20 µm. Representative images of ECs plated on FN (5 µg/ml)-coated 10 kPa stiff substrate are shown. Results are the mean ± SD of two independent experiments for a total of 22 ECs for each experimental condition (10 kPa and increasing 1, 3, and 5 µg/ml FN concentration). Statistical analysis: two-way ANOVA and Bonferroni’s post hoc analysis; NS, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (I) Real-time analysis of EC migration toward Coll I (xCELLigence RTCA DP system) reveals how only transduction with WT (green), but not ΔOLF LPHN2 (purple) rescues the higher migration rate of siLPHN2 ECs. Results are the mean ± SD of three independent experiments. Results were analyzed with two-way ANOVA and Bonferroni’s post hoc analysis; *, P ≤ 0.05; ##, P ≤ 0.01; ###, P ≤ 0.001. Max., maximum; n., number.
Figure 3.
FLRT2 negatively regulates EC response to the ECM via LPHN2 and triggers cAMP/Rap1 signaling (A and B) Real-time analysis of EC migration toward Coll I, assessed with an xCELLigence RTCA DP system. (A) Endogenous FLRT2 silencing (siFLRT2) increases migration compared with siCTL ECs. Results are the mean ± SD of seven independent assays. Statistical analysis: two-way ANOVA and Bonferroni’s post hoc analysis; *, P ≤ 0.05; ***, P ≤ 0.001. (B) Exogenous rhFLRT2 (800 ng/ml) inhibits siCTL but not siLPHN2 EC migration. For sake of simplicity, data from the same experiments are plotted in two separate graphs. Results are the mean ± SD of three independent assays. Statistical analysis: two-way ANOVA and Bonferroni’s post hoc analysis; *, P ≤ 0.05 for siCTL ECs (left), whereas all differences in siLPHN2 ECs (right) were not significant; NS, P > 0.05. (C) Endogenous FLRT2 silencing in ECs decreases basal cAMP amount. Results are the mean ± SD of seven independent assays. Statistical analysis: two-tailed heteroscedastic Student’s t test; *, P ≤ 0.05. (D) Endogenous FLRT2 silencing (siFLRT2) in ECs decreases basal Rap1-GTP levels, and treatment of siFLRT2 ECs with exogenous rhFLRT2 (800 ng/ml) rescues the decrease in Rap1-GTP levels. Total Rap1 was employed to calculate the normalized OD (N.O.D.) levels of active Rap1-GTP. Results are the mean ± SD of four independent experiments (a representative one is shown). Statistical analysis: one-way ANOVA and Bonferroni’s post hoc analysis; **, P ≤ 0.01 for siCTL versus siFLRT2 and *, P ≤ 0.05 for siFLRT2 versus siFLRT2 + FLRT2. (E–I) Confocal microscopy analysis (E) of endogenous FLRT2 (green), vinculin (Vin; blue), and phalloidin-labeled F-actin (red; high-magnification images are on the right) reveals how, compared with siCTL ECs, FLRT2 silencing increases the number, normalized on cell area (F) and size (expressed by maximum Feret diameter, G) of vinculin-containing FAs and the number, normalized on cell area (H), and amount (evaluated as mean gray intensity, I) of F-actin stress fibers in siFLRT2 ECs. Scale bar, 10 µm (siCTL), 15 µm (siFLRT2). Results are the mean ± SD of four independent experiments for a total of 18 (siCTL) and 19 (siFLRT2) ECs. Statistical analysis: two-tailed heteroscedastic Student’s t test; **, P ≤ 0.01; ***, P < 0.001. Max., maximum; n., number.
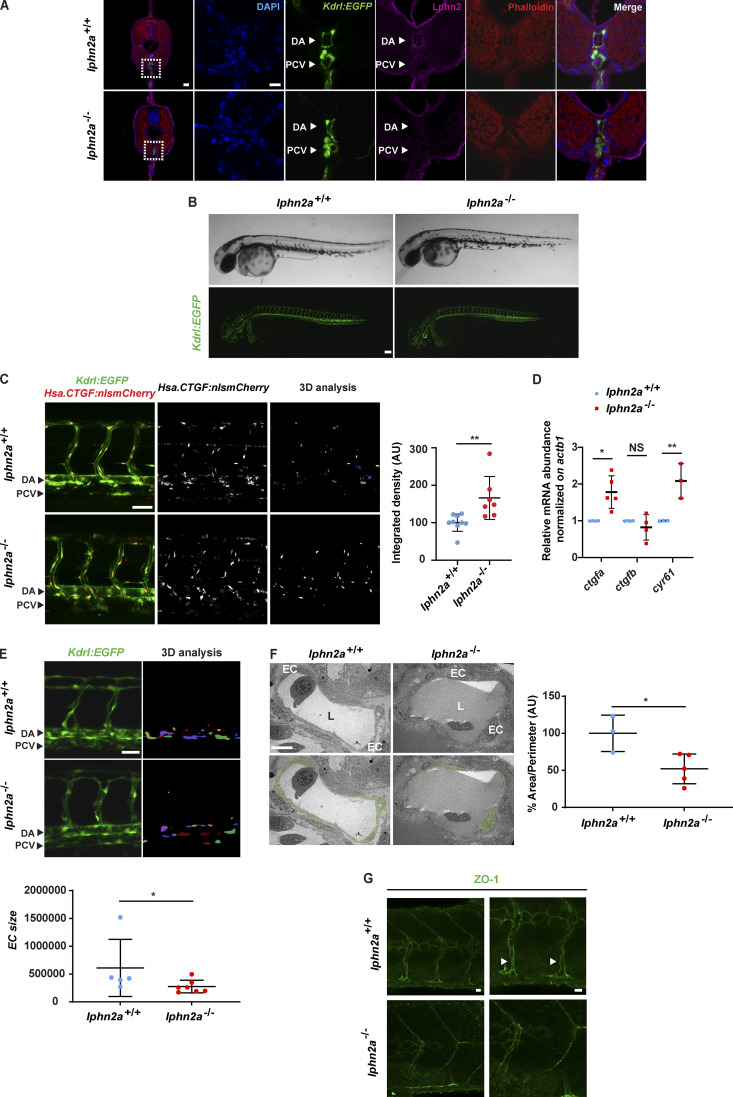
In agreement with what we (Fig. 1 A and Fig. S1 A) and others (Valtcheva et al., 2013) observed in cultured ECs, we found that Lphn2a protein is enriched in vascular ECs of dorsal aorta (DA) and posterior cardinal vein (Fig. 4 A) of developing transgenic Tg(kdrl:EGFP)s843 zebrafish embryos. Hence, to directly investigate the functional role of LPHN2 in the in vivo vasculature, we generated CRISPR/Cas9-mediated lphn2a zebrafish knock-out embryos with Tg(kdrl:EGFP)s843 genetic background (Fig. S3, A–D). The loss of Lphn2a (Fig. 4 A) did not grossly affect vascular patterning or blood circulation (Fig. 4 B). In accord with what was observed in cultured human ECs (Fig. S2, A and B), real-time qRT-PCR in vivo confirmed that flrt2 gene is actively transcribed in ECs sorted from WT zebrafish embryos, and its mRNA levels significantly increase upon lphn2a gene knock-down (Fig. S3 E). In vitro, siLPHN2 ECs display a greatly increased number and size of FAs (Fig. 1, F–H; Fig. 2, A–C; and Fig. S1, F–H) coupled to F-actin filaments (Fig. 1, F, I, and J; and Fig. 2, A, D, and E), YAP/TAZ nuclear translocation (Fig. 1, K and L; and Fig. 2, G and H), and transcription of CTGF and CYR61 target genes (Fig. 1 M). Therefore, we assessed the impact of lphn2a knock-down on the in vivo activation of the Hippo pathway in ECs by exploiting the Tg(Hsa.CTGF:nlsmCherry)ia49/Tg(kdrl:EGFP)s843 double transgenic zebrafish fluorescent reporter line of Yap1/Taz activity (Astone et al., 2018). Of note, lphn2a null embryos displayed a significantly increased Yap1/Taz activation in trunk vascular ECs (Fig. 4 C). Moreover, real-time qRT-PCR revealed a robust up-regulation of ctgfa and cyr61 target gene mRNAs in lphn2a−/− sorted ECs compared with controls (Fig. 4 D). Altogether, these in vivo findings confirm our in vitro model in which FLRT2-activated LPHN2 GPCR inhibits YAP/TAZ signaling.
Figure 4.
LPHN2 controls EC YAP/TAZ mechanosensing and vascular morphogenesis in zebrafish embryo. (A) Confocal fluorescence microscopy analysis of trunk cross-sections at 72 hpf of Tg(Kdrl:EGFP)s843 WT (lphn2a+/+) and CRISPR/Cas9-mediated lphn2a knock-out (lphn2a−/−) zebrafish embryos carrying EC-specific EGFP expression (green) and stained for Lphn2 (purple) and phalloidin (red). Nuclei are stained with DAPI (blue). Lphn2 is enriched in ECs of DA and posterior cardinal vein (PCV). The first panel on the left (scale bar, 30 µm) displays a whole cross-section whose boxed area is magnified and depicted in the five panels located on its right (scale bar, 10 µm). (B) Lateral view in brightfield (top) and fluorescence confocal (bottom) microscopy of WT (lphn2a+/+) and CRISPR/Cas9-mediated lphn2a−/− zebrafish embryos in the Tg(kdrl:EGFP)s843 background. Scale bar, 100 µm. (C) Yap1/Taz reporter activity is prominent in the endothelium trunk vasculature of zebrafish embryos. Left: Representative confocal images of Tg(Hsa.CTGF:nlsmCherry)ia49 /Tg(kdrl:EGFP)s843 double-transgenic lphn2a+/+ and lphn2a−/− siblings at 60 hpf. Yap1/Taz activation signal was automatically segmented on fluorescent confocal mCherry image z-stacks, inside a EGFP fluorescent mask identifying ECs. Scale bar, 50 µm. The EC-restricted Yap1/Taz signal intensity only was represented in 3D analysis and quantified. Right: Relative quantification of integrated density of Yap1/Taz Hsa.CTGF:nlsmCherry reporter activity signal colocalized with kdrl:GFP in lphn2a+/+ (n = 9) and lphn2a−/− (n = 7) zebrafish embryos (56–72 hpf). Results are the mean ± SD. Statistical analysis: Mann–Whitney test; **, P ≤ 0.01. (D) Real-time quantitative PCR analysis of ctgfa, ctgfb, and cyr61 mRNAs of FACS-sorted ECs isolated from lphn2a+/+ or lphn2a−/− zebrafish embryos, at 48 hpf, relative to the housekeeping gene actb1 and normalized on the mRNA levels measured in lphn2a+/+ animals. Results are the mean ± SD of three or more independent assays (n > 80 embryos for condition). Statistical analysis: one-way ANOVA followed by Tukey’s multiple comparison test; NS, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01. (E) Representative confocal images of kdrl:GFP positive cells (left) and represented in the 3D analysis (right) used to quantify EC size (bottom) in lphn2a+/+ (n = 5) and lphn2a−/− (n = 7). Scale bar, 20 µm. Results are the mean ± SD. Statistical analysis: Mann–Whitney test; *, P ≤ 0.05. (F) TEM analysis of endothelium area (left), used to determine the EC area normalized on the diameter of the vessel (right) in lphn2a+/+ (n = 3) and lphn2a−/− (n = 5). Color code identifies the ECs facing the vascular lumen (L). Scale bar, 10 µm. Results are the mean ± SD. Statistical analysis: Mann–Whitney test; *, P ≤ 0.05. (G) Representative confocal images of ZO-1 staining in intersomitic blood vessels (ISVs) of lphn2a+/+ and lphn2a−/− at 48 hpf. Arrowheads point at continuous ZO-1–stained intercellular contacts between ISV ECs of lphn2a+/+ zebrafish embryos. The ZO-1 intercellular staining between ISV ECs is instead reduced, discontinuous, and fragmented in lphn2a−/− zebrafish embryos. Scale bars, 20 µm (left) and 30 µm (right).
Figure S3.
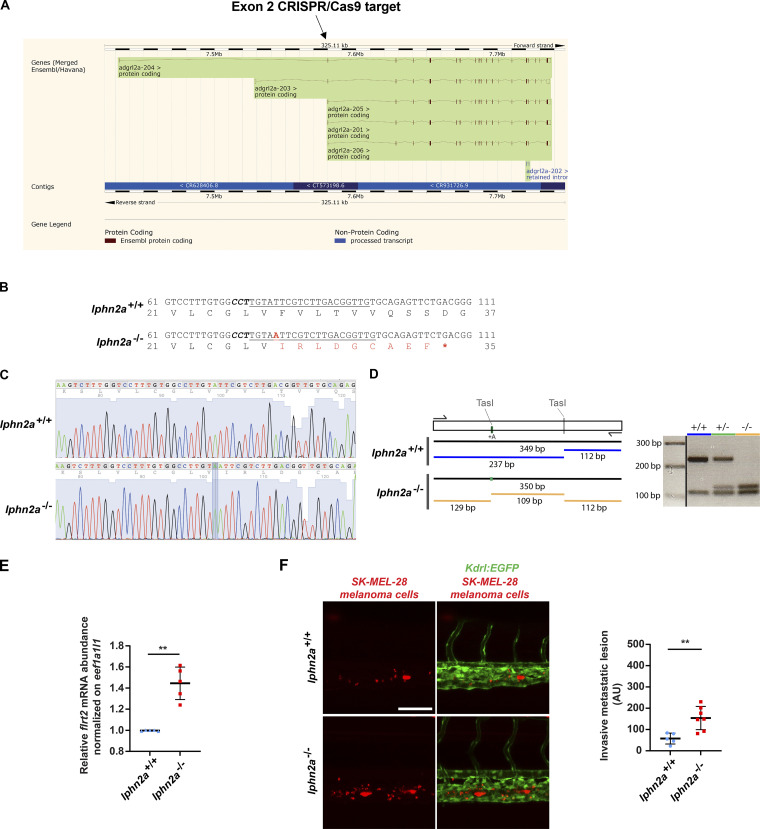
Generation and characterization of lphn2a null zebrafish embryos. (A) lphn2a (aka adgrl2a) zebrafish mRNA splicing variants displaying all exons. The black arrow outlines the lphn2a gene targeted region located in the second exon, which is common to all splicing variants. (B) DNA sequence details of the exon 2 lphn2a gene targeted region in lphn2a+/+ and lphn2a−/− zebrafish embryos. Locus-specific sgRNA is underlined, and the protospacer-adjacent motif sequence is labeled in bold italics. A one-nucleotide insertion (A, red bold underscored), revealed by sequencing, and the consequent nine new amino acids (red) and STOP codon (red asterisk) are shown. (C) Alignment of lphn2a+/+ and lphn2a−/− sequence obtained by Sanger sequencing. (D) Schematic representation of genotyping. The one base insertion generates a second TasI restriction site in the diagnostic PCR, which has been used for genotyping. (E) Real-time qRT-PCR analysis of flrt2 mRNA in lphn2a+/+ or lphn2a−/− zebrafish embryos relative to the housekeeping gene eef1a1l1 and normalized on the mRNA levels measured in lphn2a+/+ animals. Results are the mean ± SD of five independent assays (n > 80 embryos for condition). Statistical analysis: Mann–Whitney test; **, P ≤ 0.01. (F) MemBright-560-labeled melanoma cells were microinjected into the duct of Cuvier of 48 hpf lphn2a+/+ or lphn2a−/− Tg(Kdrl:EGFP) zebrafish embryos. After 36 h, extravasated metastatic melanoma cells were imaged by confocal analysis of the caudal plexus. Human SK-MEL-28 melanoma cell extravasation is enhanced in lphn2a−/− compared with lphn2a+/+ zebrafish embryos. Results are the mean ± SD of two independent assays, in which 12 SK-MEL-28 melanoma cell–injected animals were analyzed. Statistical analysis: Mann–Whitney test; **, P ≤ 0.01.
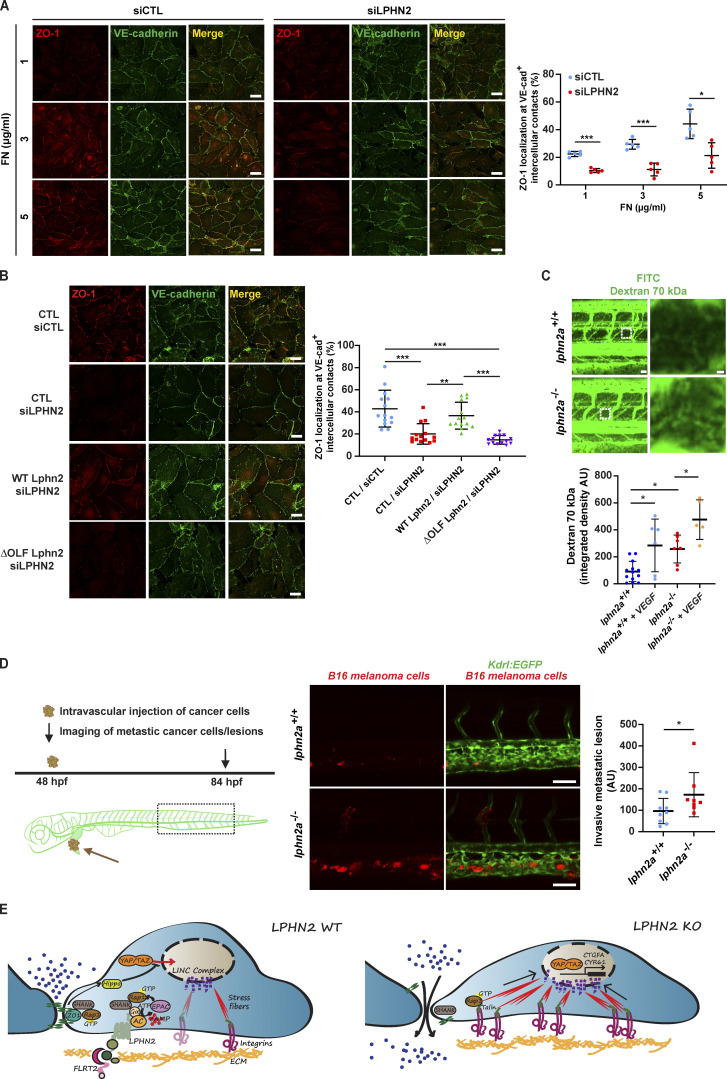
Then, to better define the function of LPHN2 in blood vessels, we thoroughly examined the morphology of vascular ECs of lphn2a knock-out Tg(kdrl:EGFP)s843 embryos. Quantitative confocal microscopy showed that ECs forming the trunk vasculature were enlarged, were thinner, and stretched along the main blood vessel axis in lphn2a−/− compared with WT embryos (Fig. 4 E). Moreover, transmission EM (TEM) on ultra-thin DA cross-sections of 48 h post-fertilization (hpf) lphn2a−/− zebrafish embryos confirmed that, despite a normal lumen, the lining ECs are extremely thin (Fig. 4 F). In addition, confocal microscopy revealed a reduced, discontinuous, and fragmented staining of the TJ marker ZO-1 at the intercellular contacts of ECs of intersomitic blood vessels of lphn2a−/− embryos compared with WT controls (Fig. 4 G). Since, in cultured ECs, LPHN2 stimulates the activation of Rap1 (Fig. 1 E), which promotes TJ formation (Sasaki et al., 2020), LPHN2-elicited Rap1 GTP loading may simultaneously hamper the assembly of FAs and support the developing of TJs. In addition, the recruitment of the mechanosensitive adaptor protein ZO-1 at TJs (Spadaro et al., 2017) was recently described to be under the control of forces regulated by ECM stiffness (Haas et al., 2020). To explore the involvement of LPHN2 in the biochemical and physical crosstalk between FAs and TJs, we plated human ECs on 10 kPa hydrogels coated with increasing FN amounts and assessed, by confocal microscopy, ZO-1 and VE-cadherin (for control purposes) targeting at intercellular contacts. We observed that increasing amounts of FN result in a progressive dose-dependent targeting of ZO-1 at EC-to-EC contacts in control, but not in siLPHN2 ECs (Fig. 5 A). The VE-cadherin intercellular recruitment was instead not affected by either FN density or LPHN2 silencing (Fig. 5 A; and Fig. S1, L and M). Furthermore, the lack of FN density-dependent ZO-1 translocation at cell-to-cell contacts in siLPHN2 ECs was rescued by the introduction of exogenous WT, but not ΔOLF Lphn2 mutant construct that does not bind FLRT2 (Fig. 5 B). In sum, LPHN2 controls EC mechanosensing, TJ assembly, and cell shape in living blood vessels. As substantiated in cultured ECs, LPHN2 favors the mechanochemical crosstalk by which FA-sensed ECM stiffness and density promote the formation of TJs.
Figure 5.
LPHN2 promotes EC TJ assembly and impairs vascular permeability and cancer cell extravasation. (A) Confocal microscopy analysis of TJs stained with ZO-1 (red) and VE-cadherin (green) reveals how in siCTL ECs seeded on 10 kPa substrates coated with increasing amounts of FN (1, 3, and 5 µg/ml) ZO-1 is progressively accumulating at VE-cadherin+ (VE-cad+) cell-to-cell junctions. Compared with siCTL ECs, LPHN2 silencing impairs ZO-1 but not VE-cadherin accumulation at intercellular contacts on increasing FN amounts. Scale bar, 25 µm. Results concerning the percentage of VE-cad+ intercellular area covered with ZO-1 are the mean ± SD of two independent experiments and a total of five confocal microscopy images for each condition. Statistical analysis: two-way ANOVA with Bonferroni’s post hoc analysis; *, P ≤ 0.05; ***, P ≤ 0.001. (B) Confocal microscopy analysis reveals how, compared with siCTL ECs, LPHN2 silencing impairs ZO-1 accumulation to VE-cad+ intercellular contacts of ECs plated on FN (5 µg/ml)-coated coverslips. Lentiviral delivery of WT Lphn2 restores ZO-1 localization to TJs, while the ΔOLF Lphn2 mutant does not rescue the phenotype. Scale bar, 25 µm. Results concerning the percentage of VE-cad+ intercellular area covered with ZO-1 are the mean ± SD of three independent experiments for a total of 14 confocal microscopy images for each condition. Statistical analysis: one-way ANOVA and Bonferroni’s post hoc analysis; **, P ≤ 0.01; ***, P ≤ 0.001. (C) Top: Representative images of vascular permeability in lphn2a+/+ versus lphn2a−/− zebrafish embryos. 70 kD FITC-dextran was injected with or without 1 ng of VEGF-A (#V7259; Sigma). 70 kD Dextran is in green. Scale bars, 30 µm (left) and 3 µm (right). Bottom: Quantification of relative extravascular fluorescence. For each embryo, the fluorescence intensity of the dextran was measured in two intervascular areas between the intersegmental vessels (dashed box and shown in the zoom images on the right). lphn2a+/+ (n = 13), lphn2a+/+ with VEGF (n = 6), lphn2a−/− (n = 7), lphn2a−/− with VEGF (n = 5) embryos from two independent experiments. Results are the mean ± SD. Statistical analysis: one-way ANOVA followed by Tukey’s multiple comparison test; *, P ≤ 0.05. (D) MemBright-560–labeled mouse B16F10 melanoma cells were microinjected into the duct of Cuvier of 48 hpf lphn2a+/+ or lphn2a−/− Tg(Kdrl:EGFP)s843 zebrafish embryos. After 36 h postinjection, extravasated metastatic melanoma cells were imaged by confocal analysis of the caudal plexus. Mouse B16F10 melanoma cell extravasation is enhanced in lphn2a−/− compared with lphn2a+/+ zebrafish embryos. Results are the mean ± SD of two independent assays, in which 17 animals were analyzed. Scale bars, 50 µm. Statistical analysis: Mann–Whitney test; *, P ≤ 0.05. (E) LPHN2 signaling controls endothelial FAs, TJs, and vascular permeability. Vascular ECs synthesize the FLRT2 ligand of LPHN2 that localizes at integrin-based ECM adhesion sites. FLRT2-activated LPHN2 triggers a canonical heterotrimeric G-protein α subunit (Gα)/adenylate cyclase (AC)/cAMP pathway that in turn, likely via the guanine nucleotide exchange factor EPAC, activates the small GTPase Rap1, which is a well-known regulator of cell-to-ECM adhesions (Coló et al., 2012; Lagarrigue et al., 2016). Furthermore, Rap1 promotes the formation of TJs (Sasaki et al., 2020), which in ECs are crucial for the control of vascular permeability. Hence, LPHN2 activation of Rap1 may act both to inhibit the formation of FAs and to promote the assembly of TJs, which increase EC barrier function. LPHN2 also binds the central PDZ domain of SHANK adaptor protein that in turn, through its N-terminal SPN domain, binds Rap1-GTP, suppressing talin-mediated integrin activation and FA development (Lilja et al., 2017) and promoting the assembly of TJs (Sasaki et al., 2020). Therefore, LPHN2 may favor the turnover of FAs and the formation of TJs by funneling Rap1-GTP toward SHANK. In addition, while TJs inhibit the nuclear translocation of YAP and TAZ through their Hippo pathway–dependent phosphorylation, FAs and the associated F-actin stress fibers exert exactly the opposite effect, promoting YAP/TAZ nuclear localization and transcriptional function (Karaman and Halder, 2018; Moya and Halder, 2019). Thus, the nuclear translocation and functional activation of YAP/TAZ caused by LPHN2 silencing or knock-down likely lie downstream of both the disassembly of TJs and the increased formation of FAs and stress fibers. In addition, the myosin II–mediated contraction of FA-linked stress fibers releases YAP/TAZ from their binding to the inhibitory switch/sucrose non-fermentable complex (not depicted) and transmits force from the ECM to the nucleus, changing nuclear pore conformation, finally promoting the translocation of YAP/TAZ into the nucleus and the transcription of target genes, such as CTGFA and CYR61. The lack of LPHN2 also results in an abnormal ECM-driven intercellular targeting of ZO-1 and assembly of TJs, which increases vascular permeability and favors cancer cell extravasation. KO, knock-out.
The entry and exit of solutes, leukocytes, and cancer cells from the bloodstream are regulated by the endothelial barrier that relies on the controlled remodeling of TJs (Wettschureck et al., 2019; Zihni et al., 2016). Since we found that the lack of LPHN2 results in the abnormal organization of TJs between ECs in vivo (Fig. 4, G and H) and in vitro (Fig. 5, A and B), we sought to investigate the impact of LPHN2 on both basal and VEGF-A–elicited blood vessel permeability. First, we injected or not lphn2a+/+ or lphn2a−/− zebrafish embryos with VEGF-A. Next, we measured vascular leakage by intravascularly inoculating and measuring the amount of extravasated FITC-Dextran 70 kD. Both in basal conditions and upon VEGF-A stimulation, FITC-Dextran 70 kD extravasated significantly more in lphn2a−/− than in lphn2a+/+ zebrafish embryos (Fig. 5 C). Then we measured the extravasation of cancer cells that can be effectively tracked and quantified by using the zebrafish embryo (Follain et al., 2018; Osmani and Goetz, 2019). To this aim, fluorescently labeled mouse (B16F10) or human (SK-MEL-28) melanoma cells were injected in the duct of Cuvier of Tg(kdrl:EGFP)s843 WT or lphn2a−/− zebrafish embryos, and their metastatic extravasation potential was quantified by measuring the number of tumor cells found outside the vessels and in the caudal plexus where circulating tumor cells preferentially arrest and extravasate (Follain et al., 2018; Hyenne et al., 2019). We discovered that, likely due to their abnormal inter-EC TJs, lphn2a null zebrafish embryos are more permissive to extravasation of both mouse B16F10 (Fig. 5 D) and human SK-MEL-28 (Fig. S3 F) melanoma cells compared with WT embryos.
To conclude, we identified the endothelial ADGR LPHN2 as a novel repulsive guidance receptor that controls in vivo blood vessel structure and function. Our data suggest that, upon activation by ligands, such as FLRT2, LPHN2 elicits the synthesis of cAMP and Rap1-GTP pools that, in turn, promote the disassembly of integrin-based FAs and the assembly ZO-1–containing TJs (Fig. 5 E). All LPHNs contain a C-terminal motif that specifically binds the PDZ domain of SH3 and multiple ankyrin repeat domains (SHANK) scaffold proteins (Kreienkamp et al., 2000), which were found to compete with talin for binding to Rap1-GTP and impair Rap1-GTP/talin–driven integrin activation (Lilja et al., 2017). Of note, we observed that LPHN2 silencing significantly reduces the physical interaction between Rap1 small GTPase and the scaffold protein SHANK2 in cultured ECs (Fig. S2 I). Hence, LPHN2 may promote FA turnover by locally funneling Rap1-GTP toward SHANK rather than talin. Interestingly, Rap1-GTP–bound SHANK2 was recently reported to support TJ formation in epithelial cells (Sasaki et al., 2020). Although further work is required, it is tempting to speculate that in ECs, LPHN2 may couple FA disruption and TJ formation by promoting Rap1 GTP loading and its binding to SHANK proteins rather than with talin.
We also uncovered that LPHN2 signals by inhibiting the Hippo effectors YAP and TAZ in vascular ECs, both in vitro (Fig. 1, K and L) and in vivo (Fig. 4, C and D). Although other GPCRs were described to modulate YAP/TAZ activation via Rho GTPase-regulated F-actin dynamics (Totaro et al., 2018; Yu et al., 2015), we did not detect any effect of LPHN2 silencing on Rho activation (Fig. S1 I). Hence, the inhibition of the formation of FAs and F-actin stress fibers along with the stimulation of TJ assembly that we report here appear as the most conceivable mechanisms by which LPHN2 negatively regulates YAP/TAZ signaling in ECs (Fig. 5 E). Indeed, while TJs promote their Hippo-dependent inhibition and cytosolic sequestration, FAs stimulates the activation and nuclear translocation of YAP and TAZ (Karaman and Halder, 2018; Moya and Halder, 2019). Finally, our data show that, likely due to the loosening of endothelial TJs, the lack of Lphn2a favors vascular permeability and the extravasation of tumor melanoma cells (Fig. 5 E). Such findings indicate that LPHN2 ligands, e.g., FLRT2, might be therapeutically exploited to strengthen the vascular barrier and to counteract cancer cell metastatic dissemination.
Materials and methods
DNA constructs
Mouse HA-tagged Lphn2 construct was synthesized by GeneArt Gene Synthesis (Thermo Fisher Scientific) and cloned into the Gateway system pENTR221 vector (Thermo Fisher Scientific). HA-Lphn2 cDNA was then subcloned into the pEGFP-N1 (EGFP) mammalian expression vector by standard PCR protocols. HA-Lphn2 ΔOLF pEGFP-N1 mutant construct lacking the Olfactomedin domain (amino acids 142–398) was obtained by PCR, according to the Taq polymerase manufacturer’s instructions, using the Phusion Site-Directed Mutagenesis Kit (Thermo Fisher Scientific). WT and ΔOLF HA-Lphn2 were also subcloned into the pCCL.sin.cPPT.polyA.CTE.eGFP.minhCMV.hPGK.Wpre (pCCL) lentiviral vector. Lentiviral particles were produced as previously described (Gagliardi et al., 2012). Briefly, transduction of cells was performed with a multiplicity of infection equal to 3 in the presence of 8 µg/ml Polybrene (H-9268; Sigma-Aldrich). Cells were then selected with 2.5 µg/ml puromycin for 2 d, and the surviving cell population was used for the experiments.
Cell culture
Primary human ECs were isolated from the umbilical cords as previously described (Jaffe et al., 1973). Briefly, umbilical vein was cannulated with a blunt 17-gauge needle that was secured by clamping. The umbilical vein was then perfused with 50 ml of PBS to wash out the blood. Next, 10 ml of 0.2% collagenase A (Cat. #11088793001; Roche Diagnostics) diluted in cell culture medium was infused into the umbilical vein and incubated 30 min at room temperature. The collagenase solution containing the ECs was flushed from the cord by perfusion with 40 ml of PBS, collected in a sterile 50-ml centrifuge tube, and centrifuged 5 min at 800 g. Cells were first resuspended in M199 medium completed with cow brain extract, heparin sodium salt from porcine intestinal mucosa (0.025 mg/500 ml), penicillin/streptomycin solution, and 20% FBS (Sigma-Aldrich), and subsequently plated in cell culture dishes that had been previously adsorbed with 1% gelatin from porcine skin (G9136; Sigma-Aldrich). Cells were tested for mycoplasma contamination by means of the Venor GeM Mycoplasma Detection Kit (MP0025-1KT; Sigma-Aldrich) and grown in M199 complete medium. The isolation of primary venous ECs from human umbilical cords was approved by the Office of the General Director and Ethics Committee of the Azienda Sanitaria Ospedaliera Ordine Mauriziano di Torino hospital (protocol approval no. 586, October 22, 2012, and no. 26884, August 28, 2014), and informed consent was obtained from each patient. COS-7, HEK 293T, and B16F10 cells (American Type Culture Collection) were grown in DMEM medium completed with glutamine, penicillin/streptomycin solution, and 10% FBS (Sigma-Aldrich). SK-MEL-28 (American Type Culture Collection) were instead grown in Eagle’s minimum essential medium completed with glutamine, penicillin/streptomycin solution, and 10% FBS (Sigma-Aldrich). Both COS-7 and HEK 293T were transfected by means of Lipofectamine and PLUS reagent (Thermo Fisher Scientific). The calcium phosphate transfection method was instead used to transfect WT or ΔOLF Lphn2-HA-pCCL in HEK 293T cells and to produce the corresponding viral particles subsequently employed to transduce ECs. To measure cell signaling response to matrix stiffness, ECs were plated on FN-coated polyacrylamide gels (6% [∼10 kPa] of gel diluted from 30% Protogel in PBS, 37.5:1 fixed ratio of acrylamide:bis-acrylamide; EC-890, National Diagnostics), previously prepared on glass slide with removable silicon chamber (IBIDI) according to published methods, with modifications (Wang and Pelham, 1998; Zhang et al., 2013).
Antibodies (Abs) and reagents
2Y4824 rabbit polyclonal Ab (pAb) anti-LPHN2 was produced by Eurogentec by immunizing animals with peptide GGKTDIDLAVDENGL (amino acids 259–274; Herberth et al., 2005). Rabbit 2Y4824 anti-LPHN2 was diluted 1:300 for immunofluorescence analysis. 2Y4824 rabbit anti–preimmune serum (Eurogentec) was used for control purposes. Mouse mAb anti-HA tag (clone F-7) and goat polyclonal VE-cadherin (clone C19) Abs, used 1:200 for immunofluorescence analysis, and mAb anti-Shank2 (clone A11) used in immunoprecipitation, were from Santa Cruz Biotechnology. Mouse mAbs anti-vinculin (hVIN-1) and anti–α-tubulin (clone B-5-1-2) and rabbit polyclonal anti-paxillin (HPA051309) were from Sigma-Aldrich and were diluted 1:400, 1:8,000, and 1:200, respectively, for immunofluorescence and Western blot analyses. Rabbit polyclonal anti-ZO-1 (clone D6L1E) used in immunofluorescence 1:100 on cultured ECs was from Cell Signaling Technology. Mouse mAb anti-ZO-1 (clone ZO1-1A12) used in immunofluorescence 1:300 on zebrafish embryos was from Invitrogen. Rat mAb anti-HA High Affinity (clone 3F10) was from Roche, and it was diluted 1:1,000 for Western blot analysis. Mouse mAb anti-6xHis tag (clone HIS.H8) was from OriGene Technologies. Goat pAb anti-FLRT2 (AF2877), from R&D Systems, was diluted 1:100 and 1:1,000 for immunofluorescence and Western blot analysis, respectively.
HRP-conjugated donkey anti-goat (sc-2056), goat anti-rabbit (sc-2054), and goat anti-rat secondary Abs were from Santa Cruz Biotechnology, while HRP-goat anti-mouse (115–035-003) was from Jackson ImmunoResearch Laboratories. Alexa Fluor 555 donkey anti-mouse (A31570), Alexa Fluor 555 donkey anti-rabbit (A31572), Alexa Fluor 555 Phalloidin (A34055), Alexa Fluor 488 donkey anti-rabbit (A21206) and donkey anti-goat (A11055), Alexa Fluor 647 donkey anti-mouse (A31571), and DAPI (D3571) were from Thermo Fisher Scientific. AP-labeled goat anti-mouse IgG (H+L) cross-adsorbed secondary Ab (G-21060) was from Thermo Fisher Scientific.
Recombinant proteins
Human FN was from R&D Systems. Coll I from calf skin was produced by Sigma-Aldrich. Recombinant human carrier-free FLRT2 protein (2877-FL) was from R&D Systems. Streptavidin-conjugated HRP was from GE Healthcare.
Gene silencing in cultured ECs
The day before oligofection, ECs were seeded in 6-well plates at a concentration of 12 × 104 cells per well. Oligofection of siRNA duplexes was performed according to the manufacturer’s protocols. Briefly, human ECs were transfected twice (at 0 and 24 h) with 200 pmol of siGENOME Non-Targeting siRNA #1 (as control) or siGENOME SMART pools siRNA oligonucleotides (GE Healthcare Dharmacon), using Oligofectamine Transfection Reagent (Thermo Fisher Scientific). 24 h after the second oligofection, ECs were lysed or tested in functional assays. Knock-down of human LPHN2 was achieved through the siGENOME SMART pool siLPHN2 M-005651-02 (siLPHN2 #1, 5′-GGAAUGGGCUUGCAAAGUU-3′; siLPHN2 #2, 5′-GGCAAGAACUCAAUUGAUU-3′; siLPHN2 #3, 5′-GAAAGAACGAGGAAUAUUG-3′; siLPHN2 #4, 5′-CGACAAACGUGCCGCAUCA-3′), while, in the case of human FLRT2, the siGENOME SMART pool was siFLRT2 M-009104-00 (siFLRT2 #1, 5′-GCAACCAACUGGACGAAUU-3′; siFLRT2 #2, 5′-CAGAUCGUCUCCUUAAAUA-3′; siFLRT2 #3, 5′-GGAACUUUGUCUACUGUAA-3′; siFLRT2 #4, 5′-UCAAAUAUAUCCCUUCAUC-3′). For silencing and rescue experiments, endogenous LPHN2 was silenced by transfecting the human-specific siGENOME SMART pool siLPHN2 M-005651-02 in ECs previously transduced with pCCL lentiviral vectors carrying or not (CTL) silencing-resistant (see Fig. S1 E) mouse WT or ΔOLF Lphn2 constructs.
Western blot analysis
Cells were lysed in buffer containing 25 mM Tris–HCl, pH 7.4, 100 mM NaCl, 5% glycerol, 0.5 mM EGTA, 2 mM MgCl2, 1 mM PMSF, 1 mM Na3VO4, protease inhibitor cocktail (Sigma-Aldrich), and detergent 1% NP-40. Cellular lysates were incubated for 20 min on ice, and then centrifuged at 15,000 g, 20 min, at 4°C. The total protein amount was determined using the bicinchoninic acid protein assay reagent (Thermo Fisher Scientific). Specific amounts of protein were separated by SDS–PAGE with precast Bolt 4–12% Bis-Tris gel (Thermo Fisher Scientific) or Mini PROTEAN TGX precast 7.5% gel (Bio-Rad). Proteins were then transferred to a nitrocellulose membrane (Bio-Rad), probed with Abs of interest, and detected by enhanced chemiluminescence technique (PerkinElmer).
Immunoprecipitation
To immunoprecipitate and analyze by Western blot endogenous LPHN2, 5 × 106 HUVEC were transferred to ice, washed three times in cold PBS 1X, and surface-labeled at 4°C with 0.2 mg/ml sulfo-NHS-SS-biotin (Thermo Fisher Scientific) in PBS 1X for 30 min. Cells were then washed three times and lysed with a buffer containing 25 mM Tris–HCl, pH 7.4, 100 mM NaCl, 1% Triton X-100, 5% glycerol, 0.5 mM EGTA, 2 mM MgCl2, 1 mM PMSF, 1 mM Na3VO4, and protease inhibitor cocktail (Sigma-Aldrich). Cellular lysates were incubated for 20 min on ice and then centrifuged at 15,000 g, 20 min, at 4°C. The total protein amount was determined using the bicinchoninic acid protein assay reagent (Thermo Fisher Scientific). Equivalent amounts (1 mg) of protein were precipitated for 1 h at 4°C with protein A–Sepharose beads and then centrifuged for 1 min, 15,000 g, at 4°C. Protein A–Sepharose beads were collected to prepare the preclearing samples, and the lysates were then immunoprecipitated for 1 h at 4°C with the rabbit pAb anti-LPHN2 or preimmune serum. Immunoprecipitates were washed four times with lysis buffer and then separated by SDS-PAGE. Proteins were then transferred to a nitrocellulose membrane (Bio-Rad), probed with Abs of interest, and detected by enhanced chemiluminescence technique (PerkinElmer).
Rap1 and Rho-GTP pull-down assay
Rap1-GTP and Rho-GTP were respectively analyzed by means of the active Rap1 (Cat. #16120; Thermo Fisher Scientific) and Rho (Cat. #16116; Thermo Fisher Scientific) pull-down and detection kits. The assays were performed according to the manufacturer’s protocols. Total Rap1 or Rho proteins, detected in the input fractions, were used to calculate the normalized amount of active Rap1-GTP or active Rho-GTP. Bands were quantified with ImageJ software, and normalized OD was calculated relative to control.
Confocal microscopy on cultured ECs
Cells were plated on 0.17-mm glass coverslips (no. 1.5) precoated with FN (5 µg/ml) and allowed to adhere for 3 h. Cells were washed in PBS, fixed in 4% or 2% PFA, permeabilized in 0.1% Triton X-100 or 0.01% Saponin PBS 1X for 2 min or 5 min on ice respectively, incubated with different primary Abs, and revealed by appropriate Alexa Fluor–tagged secondary Abs (Thermo Fisher Scientific). Slides were mounted in Fluoromount-G (Cat. #0100–01; SouthernBiotech). Cells were analyzed at room temperature by using a Leica TCS SP8 AOBS confocal microscope equipped with two hybrid detectors that, by combining classical photomultipliers with highly sensitive avalanche photodiodes, provides higher signal-to-noise ratio, image contrast, and sensitivity. A PL APO 63×/1.4 NA immersion objective was employed. 1,024 × 1,024–pixel images were acquired, and a z-stack was acquired. For nucleus staining in immunofluorescence images taken with the Leica TCS SP8, TO-PRO-3 Fluorescent Nuclear Stain (Thermo Fisher Scientific) was used. The acquisition was performed by adopting laser power, gain, and offset settings that allowed maintaining pixel intensities (grayscale) within the 0–255 range and hence avoiding saturation. Analysis of confocal images was performed using the ImageJ software package. FA and stress fiber number, FA size, and stress fiber intensity were measured, applying for each cell a default threshold. FA and stress fibers number was corrected on cell area. FA size was expressed as maximum Feret diameter. The fluorescence intensity of F-actin has been calculated, adding the mean gray value of all the measured stress fiber within a cell.
To acquire super-resolved images, a Leica TCS SP8 gated–stimulated emission depletion (STED) 3X laser-scanning microscope equipped with a HC PL APO 100×/1.40 objective was employed (Leica Microsystems). Alexa Fluor 555 fluorochrome was excited at the optimal wavelength by means of 80 MHz pulsed white light laser (470–670 nm), allowing time gating of fluorescence lifetimes. For STED, a 660-nm depletion laser was used, and emission was revealed by means of hybrid spectral detectors (HyD SP Leica Microsystems). Pixel size was maintained equal in all images. STED images were deconvolved to reduce noise using the mathematical algorithm Classic Maximum Likelihood Estimation included in Huygens Deconvolution Software. Stress fiber sagittal section and FA area were measured using ImageJ software.
Nuclear/cytoplasmic YAP and TAZ ratios were calculated by measuring the mean fluorescence intensity of each protein coming from the nucleus, divided by its cytoplasmic localization (cell area – nuclear signal).
ZO-1 localization to TJs was evaluated as percentage of VE-cadherin+ intercellular contact areas covered by ZO-1 staining.
Binding assays
Empty vector, WT Lphn2, and ΔOLF Lphn2 expression constructs were transfected in COS-7 cells to be used in the binding assay. In situ binding assays were performed as described previously (Tamagnone et al., 1999). Briefly, Lphn2-expressing COS-7 cells were seeded in wells of 48-well cluster dishes. They were then incubated for 1 h at 37° with or without recombinant 6xHis-tagged human FLRT2, mouse mAb anti-6xHis, and AP-labeled goat anti-mouse. After five washes in DMEM diluted 1:2 in PBS, cells were fixed, heated for 10 min at 65°C to inactivate endogenous phosphatases, and incubated with nitro blue tetrazolium–5-bromo-4-chloro-3-indolyl-phosphate AP substrate (Cat. #S3771; Promega) for in situ cell staining. For a quantitative assessment of ligand binding, receptor-expressing cells were incubated with increasing concentrations of AP-conjugated ligands (with predetermined specific activity/µg); cell-bound AP activity was eventually revealed by incubation with the chromogenic soluble substrate p-nitrophenylphosphate (Cat. #P7998; Sigma-Aldrich) and measured by a multi-well spectrophotometer (absorbance at 405 nm).
EC migration assays
Real-time directional EC migration was monitored with an xCELLigence RTCA DP instrument (ACEA Biosciences/Agilent Technologies) as previously described (Camillo et al., 2017; Gioelli et al., 2018). In detail, the bottom side of the upper chamber (the side facing the lower chamber) of CIM-Plate 16 was coated with 30 µl of 1 µg/ml Coll I or 3 µg/ml FN for 30 min at room temperature. Each lower chamber well was first filled with 160 µl of M199 1% FBS (containing or not 800 ng/ml of rhFLRT2 protein) and then assembled to the upper chamber. Each upper chamber well was then filled with 30 µl of M199 1% FBS. The plate was incubated for 1 h at 37°C. The experiment file was set up using RTCA Software 1.2. ECs were detached and resuspended to a final concentration of 30,000 cells/100 µl. The BLANK step was started to measure the background impedance of cell culture medium, which was then used as reference impedance for calculating CI values. 100 µl of cell suspension (30,000 cells) was then added to each well of the upper chamber. The CIM-Plate 16 was placed in the RTCA DP Instrument equilibrated in a CO2 incubator. EC migration was continuously monitored using the RTCA DP instrument. Mean, SD, and P value were calculated on the CI data exported from the RTCA instrument for the technical replicates of each experimental condition in the time. Migration data are represented as a percentage considering the control samples as 100%.
cAMP assay
The cellular amounts of cAMP were quantified by using the nonacetylated cAMP competitive ELISA Kit (Cat. #EMSCAMPL; Thermo Fisher Scientific) following the manufacturer’s instructions. The concentration of cAMP in samples was expressed as picomoles of cAMP per milligram of total proteins.
mRNA isolation from cultured ECs and real-time RT-PCR analysis
Cells were washed three times with PBS and frozen at −80°C. For RNA isolation and RT, cells were thawed on ice, and total RNA was extracted following the manufacturer’s recommended protocol (ReliaPrep RNA Miniprep Systems; Cat. #Z6011; Promega). The quality and integrity of the total RNA were quantified by the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). cDNAs were generated from 1 µg of total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). For TaqMan real-time RT-PCR assay, mRNA expression of LPHN2, FLRT1-3, and endogenous housekeeping control genes, i.e., GAPDH and TATA-binding protein (TBP), was measured by real-time RT-PCR using TaqMan Gene Expression Assays (Applied Biosystems) run on a C1000 Touch thermal cycler (Bio-Rad). The following assays were used: Hs00202347_m1 (LPHN2), Hs00534771_s1 (FLRT1), Hs00544171_s1 (FLRT2), Hs01922255_s1 (FLRT3), Hs00170014_m1 (CTGF), Hs00155479_m1 (CYR61), Hs99999905_m1 (GAPDH), and Hs00427620_m1 (TBP). For each sample, three technical replicates of each gene were run in a 96-well plate (cDNA concentration, 50 ng/well) according to the manufacturer’s protocol. Between the two measured housekeeping genes, we chose a normalization factor calculation based on the geometric mean of GAPDH and TBP gene transcript for most of the real-time RT-PCR experiments, while GAPDH only was employed in Fig. S2 A (Vandesompele et al., 2002). The experimental threshold was calculated using the algorithm provided by the Bio-Rad CFX Manager 3.1 software (Bio-Rad). Experimental threshold values were converted into relative quantities using the method described by Pfaffl (2001), and the amplification efficiency of each gene was calculated using a dilution curve and the slope calculation method (Pfaffl, 2001).
Zebrafish embryo lines and handling
Zebrafish were handled according to established protocols and maintained under standard laboratory conditions. Experimental procedures related to fish manipulation followed previously reported recommendations (Workman et al., 2010) and conformed to the Italian regulations for protecting animals used in research, including DL 116/92. The ethics committee of the University of Torino approved this study. Larvae were anesthetized and then sacrificed by ice chilling. The following zebrafish lines were used for these studies: WT AB, Tg(kdrl:EGFP)s843, carrying the endothelial-specific expression of the EGFP, and Tg(Hsa.CTGF:nlsmCherry)ia49/Tg(kdrl:EGFP)s843.
Generation of lphn2a null zebrafish embryos
Lphn2a−/− zebrafish mutants were generated by CRISPR/Cas9-mediated genome editing at ZeClinics (Barcelona, Spain). A single guide RNA (sgRNA) was designed using the online tool http://crispor.tefor.net/, based on exon site, high efficacy, and not off-target published algorithms to specifically target an optimal CRISPR sequence on exon 2 of lphn2a gene (ENSDARG00000069356). The lphn2a-targeting sgRNA, with the specific targeting sequence 5′-CAACCGTCAAGACGAATACAAGG-3′, was injected in one-cell stage embryos in a solution containing Nls-CAS9 protein (PNA BIO). The mutagenesis efficacy was evaluated on pools of 30 injected embryos, whose genomic DNA was PCR-amplified and analyzed via T7 endonuclease system. F0 injected embryos were raised to adulthood and screened, by genotyping the F1, for germline transmission of the mutation. Heterozygous mutants harboring the mutation were then incrossed to obtain homozygous mutants (F5 generation). The genomic region surrounding the CRISPR target site was PCR-amplified using the following primers: lphn2a-Fw (5′- TCTCAGAGTGACTTCCCCGGATC-3′), and lphn2a-Rv (5′-GCAGCCATTATTTATCCCAGCTACC-3′). Lphn2a−/− heterozygous and homozygous mutants were identified by analyzing in agarose gel the PCR product digested with TasI restriction enzyme (TSP509I; Thermo Fisher Scientific) and by sequencing.
TEM
Zebrafish embryos were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, overnight at 4°C, following a standard TEM sample preparation protocol (Santoro et al., 2009). Briefly, the samples were postfixed with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer for 2 h at 4°C. After three water washes, samples were dehydrated in a graded ethanol series and embedded in an epoxy resin (Sigma-Aldrich). Ultrathin sections (60–70 nm) were obtained with an Ultrotome V (LKB) ultramicrotome, counterstained with uranyl acetate and lead citrate, and viewed with a Tecnai G2 (FEI) transmission electron microscope operating at 100 kV. Images were captured with a Veleta (Olympus Soft Imaging System) digital camera.
Larvae dissociation and FACS
WT and Tg(kdrl:EGFP)s843 larvae at 2 d after fertilization were dissociated as previously described (Zancan et al., 2015) using 1X PBS, 0.25% trypsin phenol red free, 1 mM EDTA, pH 8.0, and 2.2 mg/ml Collagenase P (Sigma-Aldrich). Digestion was stopped by adding CaCl2 to a final concentration of 1 mM and fetal calf serum to 10%. Dissociated cells were rinsed once in PBS and resuspended in Opti-MEM (Gibco), 1% fetal calf serum, and 1X penicillin-streptomycin solution (Sigma-Aldrich). Cells were filtered through a 40-µm nylon membrane. For sorting, we used a FACS Aria IIIu sorter (BD Biosciences) with the following settings for EGFP: argon-ion Innova Laser (Coherent; 488 nm, 100 mW); 100 µM nozzle; and sorting speed 500 events/s in 0–32-0 sort precision mode. We performed data acquisition and analysis with the BD FACSDiva software (BD Biosciences). GFP+ and GFP− cells were separately collected in resuspension medium, and RNA was extracted using the RNA isolation kit Nucleospin RNA XS (Macherey-Nagel). cDNA was made with an RT High Capacity kit (Applied Biosystems) according to the manufacturer’s protocol. Quantitative real-time RT-PCR on ECs from zebrafish was performed with the CFX384 Touch Real-time PCR Detection System (Bio-Rad) using 5X HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne). The following primer sequences were used: lphn2-F: 5′-AGTATCCCTCATCTGCCTGG-3′, lphn2-R: 5′-AGCTGAACTCCTTCCAGACA-3′; flrt2-F: 5′-CATTGCATGGCTCAGGTCTC-3′, flrt2-R: 5′-ATGAGTTGGCCAGGGATGAA-3′; cyr61-F: 5′-GCGGAGACTCGGAGAAAGAAC-3′, cyr61-R: 5′-CGATGCACTTCTCCATCTGATG-3′; ctgfa-F: 5′-CTCCCCAAGTAACCGTCGTA-3′, ctgfa-R: 5′-CTACAGCACCGTCCAGACAC-3′; ctgfb-F: 5′-CCCACAAGAAGACACCTTCC-3′, and ctgfb-R: 5′-ATTCGCTCCATTCAGTGGTC-3′. Results are expressed as relative mRNA abundance and normalized to actin β1 (actb1) or eukaryotic translation elongation factor 1 α 1, like 1 (eef1a1l1) as endogenous reference genes, which were amplified by using the following primer sequences: actb1-F: 5′-GTATCCACGAGACCACCTTC-3′, actb1-R: 5′-GAGGAGGGCAAAGTGGTAAAC-3′; eef1a1l1-F: 5′-GACAAGAGAACCATCGAG-3′, and eef1a1l1-R: 5′-CCTCAAACTCACCGACAC-3′.
Assessment of vascular permeability in zebrafish embryos
Lphn2a+/− fish were incrossed, and the progeny were incubated in 0.003% 1-phenyl-2-thiourea to inhibit pigment formation. The vascular permeability experiment was performed as previously described (Hoeppner et al., 2012). Microangiography was performed on anesthetized 3 d post-fertilization embryos by injecting in the duct of Cuvier a solution containing FITC-dextran (70 kD; Life Technologies, Inc.) at 1 mg/ml concentration. Human VEGF (Cat. #V7259; Sigma) was injected in the duct of Cuvier. The visualization and real-time imaging were performed after 3 h on a Leica SP8 confocal microscope. The mean of the dextran fluorescence in the intervascular areas was normalized to the mean dextran fluorescence inside the vessels and was quantified after the embryo genotyping.
Intravascular injection of cancer cells in zebrafish embryos
At 48 hpf, embryos were dechorionated, anesthetized with tricaine 0.16 mg/ml, and placed along plastic lanes immersed in 2% methylcellulose/PBS. Sub-confluent cells (B16F10 or SK-MEL-28) in 10-cm culture dishes were rinsed twice with warm serum-free medium and then incubated for 30 min at 37°C with 200 nM MemBright-560 (Lipilight-IDYLLE). To eliminate all possible traces of unbound MemBright-560, cells were rinsed three times with serum-free medium, and then cells were harvested using trypsin-EDTA solution. Stained cells (at 100 ∙ 106 cells per ml) were loaded in a glass capillary needle and microinjected into the duct of Cuvier of the embryos under the stereomicroscope using a WPI PicoPump apparatus. Xenotransplanted embryos were grown at 32°C, monitored daily, and analyzed starting from 1 d after injection up to 3 d.
Image acquisition and analysis on zebrafish embryos
Double-transgenic fluorescence of Tg(kdrl:EGFP)s843 and Tg (Hsa.CTGF:nlsmCherry) in lphn2a−/− mutant background was visualized under a AZ100 stereomicroscope equipped with an AxioCam (Zeiss) dissecting microscope and then with a Leica SP8 confocal microscope at room temperature. An HC PL APO 20×/0.75 IMM CORR CS2 objective was employed. Larvae were anaesthetized and mounted in 1% low-melting-point agarose 1.5% gel. ECs targeted EGFP and nuclear mCherry fluorescence (Yap/Taz) was visualized by using 488-nm and 561-nm lasers. All images were analyzed with the 3D ImageJ Suite of ImageJ/Fiji (Ollion et al., 2013). The 3D analysis of Fig. 4, C and E, was performed using the Gaussian Blur 3D filters followed by the 3D simple segmentation using methods of the 3D ImageJ Suite. Yap/Taz signals were automatically segmented on the mCherry fluorescence image stacks, inside a EGFP fluorescent mask identifying ECs, and total signal intensity was calculated, according to previous work (Facchinello et al., 2016). After confocal acquisition, heterozygous and homozygous siblings were genotyped by PCR on DNA previously extracted from a single larva, as described by Gagnon et al. (2014).
Statistical analysis
For statistical evaluation of in vitro experiments, data distribution was assumed to be normal, but this was not formally tested. Parametric two-tailed heteroscedastic Student’s t test was used to assess the statistical significance when two groups of unpaired normally distributed values were compared; when more than two groups were compared, parametric one-way or two-way ANOVA with Bonferroni’s post hoc analysis was applied. For all quantifications, SD is shown. All data were analyzed with Prism software (GraphPad Software).
For statistical evaluation of in vivo experiments, neither randomization nor blinding was applied for samples or zebrafish embryo analyses. No statistical method or criteria were used to predetermine sample size or to include/exclude samples or animals. The Shapiro–Wilk normality test was used to confirm the normality of the data. The statistical difference of Gaussian datasets was analyzed using the one-way ANOVA with Tukey’s multiple comparison test, in case of unequal variances. For data not following a Gaussian distribution, the Mann–Whitney test was used. Illustrations of statistical analyses of in vivo experiments are presented as the mean ± SD.
For both in vitro and in vivo analysis, statistical differences were considered NS at P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001.
Online supplemental material
Fig. S1 shows endogenous LPHN2 expression, silencing, and rescue impact on endothelial cell adhesion, migration, and signaling. Fig. S2 shows endogenous FLRT2 expression, silencing, and LPHN2-mediated signaling impacts on endothelial cell adhesion and migration. Fig. S3 shows generation and characterization of lphn2a null zebrafish embryos.
Supplementary Material
Acknowledgments
We are grateful to Prof. Alberto Puliafito (University of Torino) for help with data analysis and to Prof. Georg Halder (University of Leuven) for fruitful discussion on YAP/TAZ and cell adhesion. We thank the zebrafish facility at the University of Padova.
The research leading to these results has received funding from Fondazione Associazione Italiana per la Ricerca sul Cancro IG grants #13016, #16702, and #21315 (to G. Serini), #19923 (to L. Tamagnone), #20366 (to D. Valdembri), and #20119 (to M.M. Santoro); Fondazione Associazione Italiana per la Ricerca sul Cancro under 5 per Mille 2018 grant – ID. 21052 program – principal investigator Paolo Comoglio, group leader L. Tamagnone, group leader G. Serini; Fondazione Piemontese per la ricerca sul cancro Onlus Grant “MIUR 2010 Vaschetto - 5 per mille 2010 MIUR” (to G. Serini); Telethon Italy (GGP09175; to G. Serini); Università di Torino, Bando Ricerca Locale 2019 (CUP D84I19002940005; to G. Serini); Associazione “Augusto per la Vita” (to G. Serini); and European Research Council Consolidator project 647057 - rEnDOx (to M.M. Santoro).
The authors declare no competing financial interests.
Author contributions: G. Serini conceived the project; G. Serini, C. Camillo, G. Villari, G. Mana, D. Valdembri, M.M. Santoro, and N. Facchinello designed the experiments; G. Serini, G. Villari, C. Sandri, D. Valdembri, and M.M. Santoro supervised the research; L. Tamagnone and M. Arese provided key reagents, methods, and technologies; C. Camillo, N. Facchinello, G. Villari, G. Mana, C. Sandri, R.E. Oberkersch, D. Tortarolo, F. Clapero, D. Gays, and N. Gioelli performed the experiments; C. Camillo, N. Facchinello, G. Villari, G. Mana, C. Sandri, R.E. Oberkersch, D. Tortarolo, M. Arese, F. Clapero, D. Gays, N. Gioelli, D. Valdembri, M. Arese, L. Tamagnone, M.M. Santoro, and G. Serini analyzed the data; C. Camillo, N. Facchinello, G. Villari, G. Mana, C. Sandri, R.E. Oberkersch, D. Tortarolo, M. Arese, F. Clapero, D. Gays, N. Gioelli, D. Valdembri, M. Arese, L. Tamagnone, M.M. Santoro, and G. Serini interpreted the results; C. Camillo, N. Facchinello, G. Villari, D. Valdembri, M.M. Santoro, and G. Serini wrote the paper; and all authors read and approved the manuscript.
Data Availability
Raw data of all graphs and uncropped scans of Western blots are publicly available on Figshare public repository (https://doi.org/10.6084/m9.figshare.15164292).
References
- Astone, M., Lai J.K.H., Dupont S., Stainier D.Y.R., Argenton F., and Vettori A.. 2018. Zebrafish mutants and TEAD reporters reveal essential functions for Yap and Taz in posterior cardinal vein development. Sci. Rep. 8:10189. 10.1038/s41598-018-27657-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanzato, D., Genova T., Fiorio Pla A., Bernardini M., Bianco S., Bussolati B., Mancardi D., Giraudo E., Maione F., Cassoni P., et al. 2016. Activation of P2X7 and P2Y11 purinergic receptors inhibits migration and normalizes tumor-derived endothelial cells via cAMP signaling. Sci. Rep. 6:32602. 10.1038/srep32602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birukova, A.A., Tian X., Cokic I., Beckham Y., Gardel M.L., and Birukov K.G.. 2013. Endothelial barrier disruption and recovery is controlled by substrate stiffness. Microvasc. Res. 87:50–57. 10.1016/j.mvr.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, J.L., and Pannekoek W.J.. 2012. Semaphorin signaling meets rap. Sci. Signal. 5:pe6. 10.1126/scisignal.2002913 [DOI] [PubMed] [Google Scholar]
- Camillo, C., Gioelli N., Bussolino F., and Serini G.. 2017. An Electrical Impedance-Based Method for Quantitative Real-Time Analysis of Semaphorin-Elicited Endothelial Cell Collapse. Methods Mol. Biol. 1493:195–207. 10.1007/978-1-4939-6448-2_14 [DOI] [PubMed] [Google Scholar]
- Chang, L., Azzolin L., Di Biagio D., Zanconato F., Battilana G., Lucon Xiccato R., Aragona M., Giulitti S., Panciera T., Gandin A., et al. 2018. The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature. 563:265–269. 10.1038/s41586-018-0658-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H.J., Zhang H., Park H., Choi K.S., Lee H.W., Agrawal V., Kim Y.M., and Kwon Y.G.. 2015. Yes-associated protein regulates endothelial cell contact-mediated expression of angiopoietin-2. Nat. Commun. 6:6943. 10.1038/ncomms7943 [DOI] [PubMed] [Google Scholar]
- Christensen, A.E., Selheim F., de Rooij J., Dremier S., Schwede F., Dao K.K., Martinez A., Maenhaut C., Bos J.L., Genieser H.-G., and Døskeland S.O.. 2003. cAMP analog mapping of Epac1 and cAMP kinase. Discriminating analogs demonstrate that Epac and cAMP kinase act synergistically to promote PC-12 cell neurite extension. J. Biol. Chem. 278:35394–35402. 10.1074/jbc.M302179200 [DOI] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka, M. 2013. Distinct functions for Rap1 signaling in vascular morphogenesis and dysfunction. Exp. Cell Res. 319:2350–2359. 10.1016/j.yexcr.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka, M. 2017. Rap1 in endothelial biology. Curr. Opin. Hematol. 24:248–255. 10.1097/MOH.0000000000000332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coló, G.P., Hernández-Varas P., Lock J., Bartolomé R.A., Arellano-Sánchez N., Strömblad S., and Teixidó J.. 2012. Focal adhesion disassembly is regulated by a RIAM to MEK-1 pathway. J. Cell Sci. 125:5338–5352. 10.1242/jcs.105270 [DOI] [PubMed] [Google Scholar]
- Dupont, S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature. 474:179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Elaimy, A.L., and Mercurio A.M.. 2018. Convergence of VEGF and YAP/TAZ signaling: Implications for angiogenesis and cancer biology. Sci. Signal. 11:eaau1165. 10.1126/scisignal.aau1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola, A., Andreu I., Beedle A.E.M., Lezamiz A., Uroz M., Kosmalska A.J., Oria R., Kechagia J.Z., Rico-Lastres P., Le Roux A.L., et al. 2017. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell. 171:1397–1410.e14. 10.1016/j.cell.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Facchinello, N., Schiavone M., Vettori A., Argenton F., and Tiso N.. 2016. Monitoring Wnt Signaling in Zebrafish Using Fluorescent Biosensors. In Wnt Signaling: Methods and Protocols. Barrett Q., and Lum L., editors. NY. 81–94. 10.1007/978-1-4939-6393-5_9 [DOI] [PubMed] [Google Scholar]
- Follain, G., Osmani N., Azevedo A.S., Allio G., Mercier L., Karreman M.A., Solecki G., Garcia Leòn M.J., Lefebvre O., Fekonja N., et al. 2018. Hemodynamic Forces Tune the Arrest, Adhesion, and Extravasation of Circulating Tumor Cells. Dev. Cell. 45:33–52.e12. 10.1016/j.devcel.2018.02.015 [DOI] [PubMed] [Google Scholar]
- Franke, B., Akkerman J.W., and Bos J.L.. 1997. Rapid Ca2+-mediated activation of Rap1 in human platelets. EMBO J. 16:252–259. 10.1093/emboj/16.2.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi, P.A., di Blasio L., Orso F., Seano G., Sessa R., Taverna D., Bussolino F., and Primo L.. 2012. 3-phosphoinositide-dependent kinase 1 controls breast tumor growth in a kinase-dependent but Akt-independent manner. Neoplasia. 14:719–731. 10.1593/neo.12856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon, J.A., Valen E., Thyme S.B., Huang P., Akhmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., and Schier A.F.. 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 9:e98186. 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galie, P.A., van Oosten A., Chen C.S., and Janmey P.A.. 2015. Application of multiple levels of fluid shear stress to endothelial cells plated on polyacrylamide gels. Lab Chip. 15:1205–1212. 10.1039/C4LC01236D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioelli, N., Maione F., Camillo C., Ghitti M., Valdembri D., Morello N., Darche M., Zentilin L., Cagnoni G., Qiu Y., et al. 2018. A rationally designed NRP1-independent superagonist SEMA3A mutant is an effective anticancer agent. Sci. Transl. Med. 10:eaah4807. 10.1126/scitranslmed.aah4807 [DOI] [PubMed] [Google Scholar]
- Gloerich, M., and Bos J.L.. 2011. Regulating Rap small G-proteins in time and space. Trends Cell Biol. 21:615–623. 10.1016/j.tcb.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Haas, A.J., Zihni C., Ruppel A., Hartmann C., Ebnet K., Tada M., Balda M.S., and Matter K.. 2020. Interplay between Extracellular Matrix Stiffness and JAM-A Regulates Mechanical Load on ZO-1 and Tight Junction Assembly. Cell Rep. 32:107924. 10.1016/j.celrep.2020.107924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberth, G., Stein A., Glienke J., Taudien S., Klaman I., Herr A., Thierauch K.-H., and Sommer A.. 2005. Human Latrophilin-2 is Expressed in the Cytotrophoblast and Syncytiotrophoblast of Placenta and in Endothelial Cells. Am. J. Biochem. Biotechnol. 1:135–144. 10.3844/ajbbsp.2004.135.144 [DOI] [Google Scholar]
- Hoeppner, L.H., Phoenix K.N., Clark K.J., Bhattacharya R., Gong X., Sciuto T.E., Vohra P., Suresh S., Bhattacharya S., Dvorak A.M., et al. 2012. Revealing the role of phospholipase Cβ3 in the regulation of VEGF-induced vascular permeability. Blood. 120:2167–2173. 10.1182/blood-2012-03-417824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, J., Doebele R.C., Lingen M.W., Quilliam L.A., Tang W.J., and Rosner M.R.. 2007. Anthrax edema toxin inhibits endothelial cell chemotaxis via Epac and Rap1. J. Biol. Chem. 282:19781–19787. 10.1074/jbc.M700128200 [DOI] [PubMed] [Google Scholar]
- Hyenne, V., Ghoroghi S., Collot M., Bons J., Follain G., Harlepp S., Mary B., Bauer J., Mercier L., Busnelli I., et al. 2019. Studying the Fate of Tumor Extracellular Vesicles at High Spatiotemporal Resolution Using the Zebrafish Embryo. Dev. Cell. 48:554–572.e7. 10.1016/j.devcel.2019.01.014 [DOI] [PubMed] [Google Scholar]
- Jackson, V.A., Mehmood S., Chavent M., Roversi P., Carrasquero M., Del Toro D., Seyit-Bremer G., Ranaivoson F.M., Comoletti D., Sansom M.S., et al. 2016. Super-complexes of adhesion GPCRs and neural guidance receptors. Nat. Commun. 7:11184. 10.1038/ncomms11184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe, E.A., Nachman R.L., Becker C.G., and Minick C.R.. 1973. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J. Clin. Invest. 52:2745–2756. 10.1172/JCI107470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey, P.A., Fletcher D.A., and Reinhart-King C.A.. 2020. Stiffness Sensing by Cells. Physiol. Rev. 100:695–724. 10.1152/physrev.00013.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman, R., and Halder G.. 2018. Cell Junctions in Hippo Signaling. Cold Spring Harb. Perspect. Biol. 10:a028753. 10.1101/cshperspect.a028753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechagia, J.Z., Ivaska J., and Roca-Cusachs P.. 2019. Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 20:457–473. 10.1038/s41580-019-0134-2 [DOI] [PubMed] [Google Scholar]
- Kim, J., Kim Y.H., Kim J., Park D.Y., Bae H., Lee D.H., Kim K.H., Hong S.P., Jang S.P., Kubota Y., et al. 2017. YAP/TAZ regulates sprouting angiogenesis and vascular barrier maturation. J. Clin. Invest. 127:3441–3461. 10.1172/JCI93825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreienkamp, H.J., Zitzer H., Gundelfinger E.D., Richter D., and Bockers T.M.. 2000. The calcium-independent receptor for alpha-latrotoxin from human and rodent brains interacts with members of the ProSAP/SSTRIP/Shank family of multidomain proteins. J. Biol. Chem. 275:32387–32390. 10.1074/jbc.C000490200 [DOI] [PubMed] [Google Scholar]
- Lagarrigue, F., Vikas Anekal P., Lee H.S., Bachir A.I., Ablack J.N., Horwitz A.F., and Ginsberg M.H.. 2015. A RIAM/lamellipodin-talin-integrin complex forms the tip of sticky fingers that guide cell migration. Nat. Commun. 6:8492. 10.1038/ncomms9492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarrigue, F., Kim C., and Ginsberg M.H.. 2016. The Rap1-RIAM-talin axis of integrin activation and blood cell function. Blood. 128:479–487. 10.1182/blood-2015-12-638700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenhan, T., Aust G., and Hamann J.. 2013. Sticky signaling--adhesion class G protein-coupled receptors take the stage. Sci. Signal. 6:re3. 10.1126/scisignal.2003825 [DOI] [PubMed] [Google Scholar]
- Lawson, C.D., and Burridge K.. 2014. The on-off relationship of Rho and Rac during integrin-mediated adhesion and cell migration. Small GTPases. 5:e27958. 10.4161/sgtp.27958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Stanton A.E., Tong X., and Yang F.. 2019. Hydrogels with enhanced protein conjugation efficiency reveal stiffness-induced YAP localization in stem cells depends on biochemical cues. Biomaterials. 202:26–34. 10.1016/j.biomaterials.2019.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja, J., Zacharchenko T., Georgiadou M., Jacquemet G., De Franceschi N., Peuhu E., Hamidi H., Pouwels J., Martens V., Nia F.H., et al. 2017. SHANK proteins limit integrin activation by directly interacting with Rap1 and R-Ras. Nat. Cell Biol. 19:292–305. 10.1038/ncb3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyle, K.S., Raaijmakers J.H., Bruinsma W., Bos J.L., and de Rooij J.. 2008. cAMP-induced Epac-Rap activation inhibits epithelial cell migration by modulating focal adhesion and leading edge dynamics. Cell. Signal. 20:1104–1116. 10.1016/j.cellsig.2008.01.018 [DOI] [PubMed] [Google Scholar]
- Moya, I.M., and Halder G.. 2019. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 20:211–226. 10.1038/s41580-018-0086-y [DOI] [PubMed] [Google Scholar]
- Nakajima, H., Yamamoto K., Agarwala S., Terai K., Fukui H., Fukuhara S., Ando K., Miyazaki T., Yokota Y., Schmelzer E., et al. 2017. Flow-Dependent Endothelial YAP Regulation Contributes to Vessel Maintenance. Dev. Cell. 40:523–536.e6. 10.1016/j.devcel.2017.02.019 [DOI] [PubMed] [Google Scholar]
- Neto, F., Klaus-Bergmann A., Ong Y.T., Alt S., Vion A.C., Szymborska A., Carvalho J.R., Hollfinger I., Bartels-Klein E., Franco C.A., et al. 2018. YAP and TAZ regulate adherens junction dynamics and endothelial cell distribution during vascular development. eLife. 7:e31037. 10.7554/eLife.31037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollion, J., Cochennec J., Loll F., Escudé C., and Boudier T.. 2013. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics. 29:1840–1841. 10.1093/bioinformatics/btt276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani, N., and Goetz J.G.. 2019. Multiscale Imaging of Metastasis in Zebrafish. Trends Cancer. 5:766–778. 10.1016/j.trecan.2019.10.003 [DOI] [PubMed] [Google Scholar]
- Paavola, K.J., and Hall R.A.. 2012. Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol. Pharmacol. 82:777–783. 10.1124/mol.112.080309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro, M.M., Pesce G., and Stainier D.Y.. 2009. Characterization of vascular mural cells during zebrafish development. Mech. Dev. 126:638–649. 10.1016/j.mod.2009.06.1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, K., Kojitani N., Hirose H., Yoshihama Y., Suzuki H., Shimada M., Takayanagi A., Yamashita A., Nakaya M.A., Hirano H., et al. 2020. Shank2 Binds to aPKC and Controls Tight Junction Formation with Rap1 Signaling during Establishment of Epithelial Cell Polarity. Cell Rep. 31:107407. 10.1016/j.celrep.2020.02.088 [DOI] [PubMed] [Google Scholar]
- Seiradake, E., del Toro D., Nagel D., Cop F., Härtl R., Ruff T., Seyit-Bremer G., Harlos K., Border E.C., Acker-Palmer A., et al. 2014. FLRT structure: balancing repulsion and cell adhesion in cortical and vascular development. Neuron. 84:370–385. 10.1016/j.neuron.2014.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiradake, E., Jones E.Y., and Klein R.. 2016. Structural Perspectives on Axon Guidance. Annu. Rev. Cell Dev. Biol. 32:577–608. 10.1146/annurev-cellbio-111315-125008 [DOI] [PubMed] [Google Scholar]
- Serini, G., Valdembri D., Zanivan S., Morterra G., Burkhardt C., Caccavari F., Zammataro L., Primo L., Tamagnone L., Logan M., et al. 2003. Class 3 semaphorins control vascular morphogenesis by inhibiting integrin function. Nature. 424:391–397. 10.1038/nature01784 [DOI] [PubMed] [Google Scholar]
- Spadaro, D., Le S., Laroche T., Mean I., Jond L., Yan J., and Citi S.. 2017. Tension-Dependent Stretching Activates ZO-1 to Control the Junctional Localization of Its Interactors. Curr. Biol. 27:3783–3795.e8. 10.1016/j.cub.2017.11.014 [DOI] [PubMed] [Google Scholar]
- Stanton, A.E., Tong X., Lee S., and Yang F.. 2019. Biochemical Ligand Density Regulates Yes-Associated Protein Translocation in Stem Cells through Cytoskeletal Tension and Integrins. ACS Appl. Mater. Interfaces. 11:8849–8857. 10.1021/acsami.8b21270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof, T.C. 2001. alpha-Latrotoxin and its receptors: neurexins and CIRL/latrophilins. Annu. Rev. Neurosci. 24:933–962. 10.1146/annurev.neuro.24.1.933 [DOI] [PubMed] [Google Scholar]
- Tamagnone, L., Artigiani S., Chen H., He Z., Ming G.I., Song H., Chedotal A., Winberg M.L., Goodman C.S., Poo M., et al. 1999. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 99:71–80. 10.1016/S0092-8674(00)80063-X [DOI] [PubMed] [Google Scholar]
- Totaro, A., Panciera T., and Piccolo S.. 2018. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 20:888–899. 10.1038/s41556-018-0142-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdembri, D., Regano D., Maione F., Giraudo E., and Serini G.. 2016. Class 3 semaphorins in cardiovascular development. Cell Adhes. Migr. 10:641–651. 10.1080/19336918.2016.1212805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtcheva, N., Primorac A., Jurisic G., Hollmén M., and Detmar M.. 2013. The orphan adhesion G protein-coupled receptor GPR97 regulates migration of lymphatic endothelial cells via the small GTPases RhoA and Cdc42. J. Biol. Chem. 288:35736–35748. 10.1074/jbc.M113.512954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele, J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., and Speleman F.. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wälchli, T., Wacker A., Frei K., Regli L., Schwab M.E., Hoerstrup S.P., Gerhardt H., and Engelhardt B.. 2015. Wiring the Vascular Network with Neural Cues: A CNS Perspective. Neuron. 87:271–296. 10.1016/j.neuron.2015.06.038 [DOI] [PubMed] [Google Scholar]
- Wang, Y.L., and Pelham R.J. Jr. 1998. Preparation of a flexible, porous polyacrylamide substrate for mechanical studies of cultured cells. Methods Enzymol. 298:489–496. 10.1016/S0076-6879(98)98041-7 [DOI] [PubMed] [Google Scholar]
- Wang, Y., He H., Srivastava N., Vikarunnessa S., Chen Y.B., Jiang J., Cowan C.W., and Zhang X.. 2012. Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci. Signal. 5:ra6. 10.1126/scisignal.2002636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Luo J.Y., Li B., Tian X.Y., Chen L.J., Huang Y., Liu J., Deng D., Lau C.W., Wan S., et al. 2016. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature. 540:579–582. 10.1038/nature20602 [DOI] [PubMed] [Google Scholar]
- Wang, X., Freire Valls A., Schermann G., Shen Y., Moya I.M., Castro L., Urban S., Solecki G.M., Winkler F., Riedemann L., et al. 2017. YAP/TAZ Orchestrate VEGF Signaling during Developmental Angiogenesis. Dev. Cell. 42:462–478.e7. 10.1016/j.devcel.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Wettschureck, N., Strilic B., and Offermanns S.. 2019. Passing the Vascular Barrier: Endothelial Signaling Processes Controlling Extravasation. Physiol. Rev. 99:1467–1525. 10.1152/physrev.00037.2018 [DOI] [PubMed] [Google Scholar]
- Workman, P., Aboagye E.O., Balkwill F., Balmain A., Bruder G., Chaplin D.J., Double J.A., Everitt J., Farningham D.A., Glennie M.J., et al. Committee of the National Cancer Research Institute . 2010. Guidelines for the welfare and use of animals in cancer research. Br. J. Cancer. 102:1555–1577. 10.1038/sj.bjc.6605642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worzfeld, T., and Offermanns S.. 2014. Semaphorins and plexins as therapeutic targets. Nat. Rev. Drug Discov. 13:603–621. 10.1038/nrd4337 [DOI] [PubMed] [Google Scholar]
- Worzfeld, T., Swiercz J.M., Sentürk A., Genz B., Korostylev A., Deng S., Xia J., Hoshino M., Epstein J.A., Chan A.M., et al. 2014. Genetic dissection of plexin signaling in vivo. Proc. Natl. Acad. Sci. USA. 111:2194–2199. 10.1073/pnas.1308418111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi, S., Hampel F., Hata K., Del Toro D., Schwark M., Kvachnina E., Bastmeyer M., Yamashita T., Tarabykin V., Klein R., and Egea J.. 2011. FLRT2 and FLRT3 act as repulsive guidance cues for Unc5-positive neurons. EMBO J. 30:2920–2933. 10.1038/emboj.2011.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F.X., Zhao B., and Guan K.L.. 2015. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 163:811–828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancan, I., Bellesso S., Costa R., Salvalaio M., Stroppiano M., Hammond C., Argenton F., Filocamo M., and Moro E.. 2015. Glucocerebrosidase deficiency in zebrafish affects primary bone ossification through increased oxidative stress and reduced Wnt/β-catenin signaling. Hum. Mol. Genet. 24:1280–1294. 10.1093/hmg/ddu538 [DOI] [PubMed] [Google Scholar]
- Zhang, J., Guo W., Rape A., and Wang Y.. 2013. Micropatterning Cell Adhesion on Polyacrylamide Hydrogels. In Cell-Cell Interactions. Baudino T.A., editor. Humana Press, Totowa, NJ. 147–156. 10.1007/978-1-62703-604-7_13 10.1007/978-1-62703-604-7_13 [DOI] [PubMed] [Google Scholar]
- Zihni, C., Mills C., Matter K., and Balda M.S.. 2016. Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 17:564–580. 10.1038/nrm.2016.80 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data of all graphs and uncropped scans of Western blots are publicly available on Figshare public repository (https://doi.org/10.6084/m9.figshare.15164292).