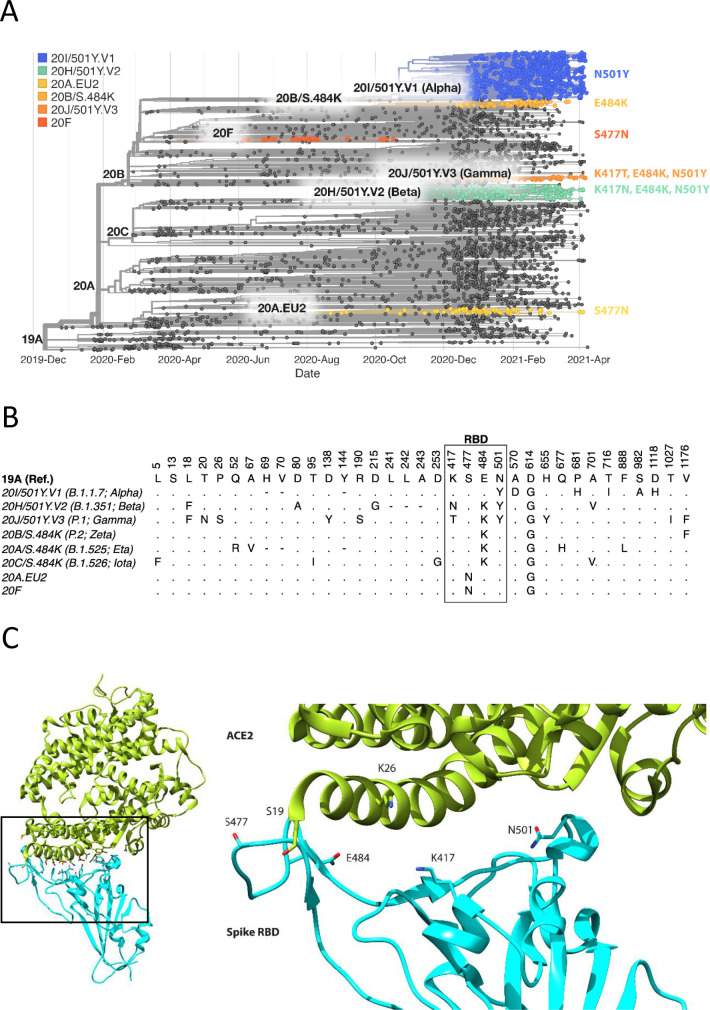
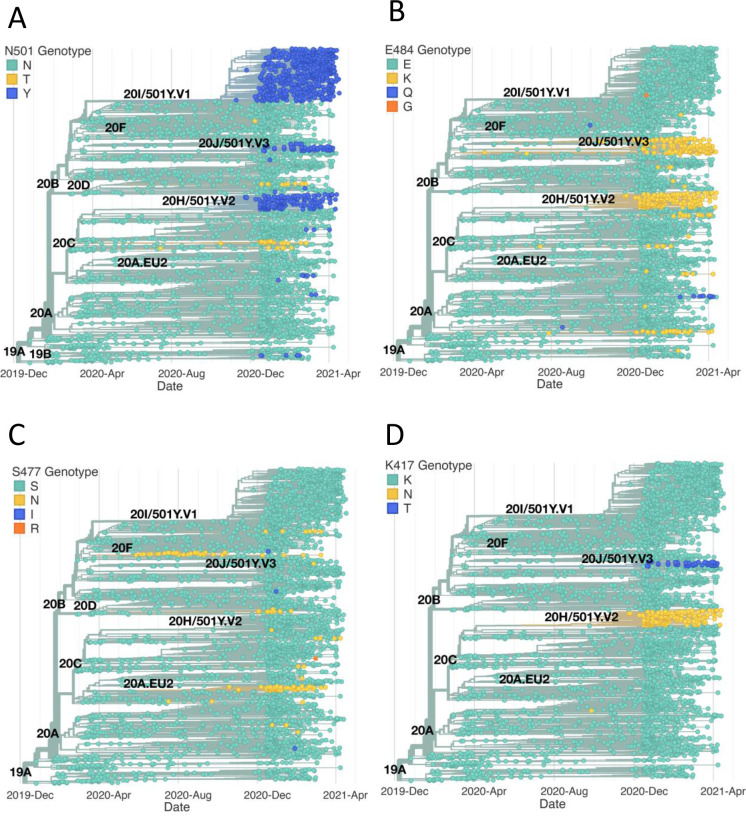
Figure 1. Spike RBD and ACE2 variants analysed in this study.
(A) Phylogenetic tree illustrating the clades containing the RBD mutations investigated in this study. Constructed using TreeTime (Sagulenko et al., 2018) from the Nextstrain Global (Hadfield et al., 2018) sample of SARS-CoV-2 sequences from the GISAID database (Shu and McCauley, 2017) (accessed 15 April 2021, N = 4017). (B) Alignment illustrating the Spike residues that differ between SARS-CoV-2 variants, with the RBD mutants boxed. The variants are labelled with their clade designation from Nextstrain (Hadfield et al., 2018) and/or PANGO lineage (Rambaut et al., 2020), where relevant. The RBD mutations were collated from CoVariants (Hodcroft, 2021) and Nextstrain. (C) The structure of human ACE2 (green) in complex with SARS-CoV-2 Spike RBD (cyan). The area enclosed by the box is shown enlarged on the right, with the residues mutated in this study labelled. Drawn using UCSF Chimera (Pettersen et al., 2004) using coordinates from PDB 6m0j (Lan et al., 2020).