Abstract
Circular RNAs (circRNAs), a special type of RNAs without 5’- and 3’-ends, are widely present in eukaryotes and known to function as noncoding RNAs to regulate gene expression, including as miRNA sponges. Recent studies showed that many exonic circRNAs, generated by back-splicing of pre-mRNAs, can be translated in a cap-independent fashion through IRESs or m6A RNA methylation. However, the scope of the translatable circRNAs and the biological function of their translation products are still unclear in different cells and tissues. Ribosome footprinting and proteomic analysis were usually used to globally identify translatable circRNAs. However, both methods have low sensitivity due to the low efficiency in the discovery of circRNA specific reads or peptides (i.e., the back-splicing junctions are difficult to recover by the short reads of ribosome footprinting and the limitation of proteomic analysis). Here, we described an alternative method to identify translatable circRNAs using polysome profiling and circRNA-seq. Generally, polysome-associated RNAs were separated with sucrose gradients. Then polysome-bound circRNAs were enriched by an RNase R treatment and identified through paired-end deep sequencing. Thus, this method is more sensitive than ribosome footprint and proteomic analyses for the identification of translatable circRNAs.
Keywords: Polysome profiling, circRNA translation, RNA-seq, back-splicing junction, RNase R
Background
Circular RNAs (circRNAs) are a special type of RNA without 5’- and 3’-ends, which form a covalent close loop. They were first discovered in viroids four decades ago ( Sanger et al., 1976 ). Recently, many studies showed that circRNAs were widely present in eukaryotes and expressed in a tissue-specific manner (Rybak- Wolf et al., 2015 ; Chen, 2020). Several types of circRNAs are found in cells, including exonic circRNA, EIciRNA, and ciRNA ( Kristensen et al., 2019 ); however, the majority of them are exonic circRNAs that are generated through back-splicing of pre-mRNAs ( Salzman et al., 2012 and 2013; Memczak et al., 2013 ; Ashwal- Fluss et al., 2014 ; Zhang, X. O. et al., 2014). Thus, most exonic circRNAs sequences are the same as those of linear mRNAs, and the back-splicing junctions are unique sequences for circRNA detection.
CircRNAs regulate biological processes through their activities as miRNA sponges, protein scaffolds, and transcription regulators ( Kristensen et al., 2019 ). However, the functions of most circRNAs are still unclear. Recently, several studies showed that many circRNAs can be translated through cap-independent translation from IRESs or m6A RNA modifications ( Legnini et al., 2017 ; Pamudurti et al., 2017 ; Yang et al., 2017 ). In addition, some circRNA encode peptides reported to influence cancer proliferation or lifespan in flies ( Weigelt et al., 2020 ; Yang et al., 2018 ; Zhang, M. et al., 2018), indicating that the translation products from circRNAs play important biological roles. However, the roles of translatable circRNAs are still unknown. Therefore, transcriptome-wide identification of translatable circRNAs is an important way to study their biological function.
Two different types of omics approaches are usually used to comprehensively measure the translation of mRNAs at RNA (translatome) and protein (proteome) levels. Ribosome footprinting is an emerging technique to globally measure mRNA translation via high-throughput sequencing of ribosome-protected mRNA fragments; however, it’s difficult to capture the translating circRNAs. Since the ribosome-protected mRNA fragments are short (around 28-35nt), the back-splicing junctions are easy to be missed by this method. Proteomic analysis is a technique to directly determine the translation products of mRNAs through identification of enzymatically digested peptides. Similar to ribosome footprinting, the peptides coded by back-splicing junctions are difficult to identify through tandem mass spectrometry.
We developed a new method to identify translatable circRNAs using polysome profiling and circRNA-seq. The polysome-associated RNAs can be separated using sucrose gradients and then isolated from different fractions. The polysome-bound circRNAs are enriched by an RNase R treatment and used to generate the circRNA-seq libraries. The resulting libraries are pair-end sequenced, and the circRNAs are identified by the back-splicing junction reads. Therefore, our method is easier and more sensitive than ribosome footprint and proteomic analyses for identifying translatable circRNAs.
Materials and Reagents
10 cm and 15 cm dish (Thermo ScientificTM, NuncTM EasYDish, catalog numbers: 150464, 150468)
50 ml and 15 ml centrifuge tube (Thermo ScientificTM, Thermo ScientificTM NuncTM, catalog numbers: 339650, 339652)
1.5 ml and 2 ml Eppendorf tube (RNase free) (CORNING, Axygen, catalog numbers: MCT-150-C, MCT-200-C)
10 ml and 25 ml serological pipette (Thermo ScientificTM, NuncTM, catalog numbers: 170357N, 170356N)
10 ml syringe (Sigma-Aldrich, catalog number: Z248029)
Pipetting needle (BioComp, belong to Gradient Master in Equipment)
25 cm cell scraper (JET BIOFIL, catalog number: CSC011025)
Polypropylene centrifuge tube (Beckman coulter, catalog number: 331372)
Vacuum Filtration Systems (Corning, catalog number: 430758)
Cell line: HEK293T
Nuclease free water (Sigma-Aldrich, catalog number: W4502)
TRIzolTM reagent (Thermo Fisher, catalog number: 15596018)
Chloroform (Sinoreagent, catalog number: 10006818)
Isopropanol (Sinoreagent, catalog number: 80109218)
Ethanol (Sigma-Aldrich, catalog number: E7023)
Dulbecco’s modified eagle medium (DMEM) (meilunbio, catalog number: MA0212)
Phosphate buffered saline (PBS) (Hyclone, catalog number: SH30256.01)
Ribonuclease inhibitor (Promega, catalog number: N2518)
RQ1 RNase-free DNase (Promega, catalog number: M6101)
RNase R (Lucigen, catalog number: RNR07250)
cOmpleteTM EDTA-free Protease Inhibitor Cocktail (Roche, catalog number: 11873580001)
Cycloheximide (CHX) (Sigma, catalog number: C7698)
Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43815)
TritonTM X-100 (Sigma-Aldrich, catalog number: T8787)
Bromophenol blue (Sigma-Aldrich, catalog number: B0126)
KAPA Stranded RNA-Seq Kit with RiboErase (HMR) (KAPA, catalog number: KK8483)
RNA clean and concentrator kit (ZYMO RESEARCH, catalog number: R1019)
10× Polysome buffer (see Recipes)
Lysis buffer (see Recipes)
Wash buffer (see Recipes)
70% sucrose solution (see Recipes)
50% sucrose grading solution (see Recipes)
10% sucrose grading solution (see Recipes)
Equipment
Ultracentrifuge (with rotor SW41) (Beckman, model: Optima XPN-100-IVD, catalog number: A99846)
Photo-spectrometer (Thermo Fisher, NanoDrop, catalog number: ND-1000)
Density gradient fractionation system (Complete system includes tube piercing system; syringe pump; UA-6 Detector with 254 and 280 nm filters, R1 Fraction Collector, Density Gradient Flow Cell, cables, and tubing) (BRANDEL, catalog number: BR-188)
Gradient Master (BioComp, catalog number: Biocomp 108)
Software
Peak chart (Data capture software for Density gradient fractionation system, www.brandel.com)
TopHat 2/ TopHat-Fusion (https://ccb.jhu.edu/software/tophat/index.shtml, https://ccb.jhu.edu/software/tophat/fusion_index.shtml)
CIRCexplorer2 (https://circexplorer2.readthedocs.io/en/latest/)
Procedure
-
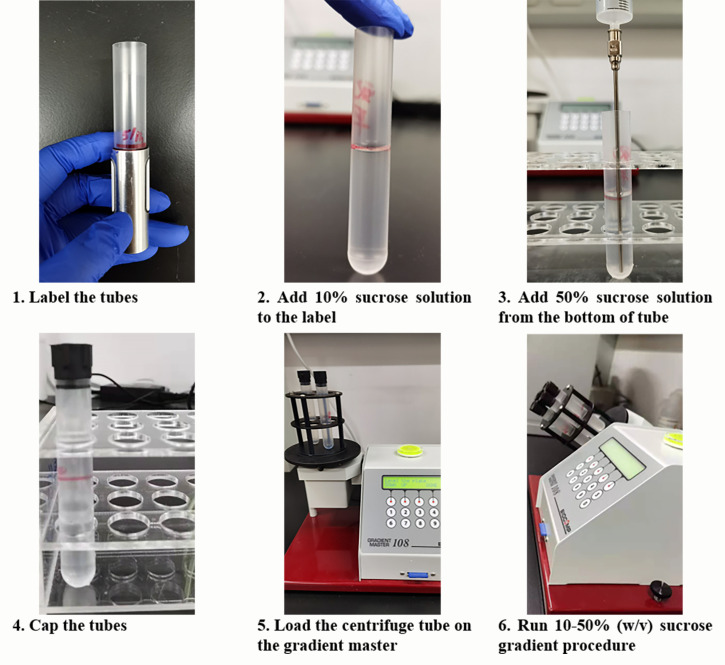
Preparation of sucrose gradient (Figure 1)
Prepare 500 ml of 10× polysome buffer in nuclease free water and 500 ml of 70% (w/v) sucrose solution in 1× polysome buffer (see Recipes). Filter solutions with Vacuum Filtration Systems.
Prepare 10% (w/v) and 50% (w/v) sucrose solution with 70% (w/v) sucrose solution (see Recipes).
-
Add the 10% and 50% sucrose solution, in that order, into a Beckman polypropylene centrifuge tube at room temperature.
Note: Add a half tube of 10% sucrose solution first and then the 50% sucrose solution to the bottom of the tube under the 10% solution using a needle.
Load the centrifuge tube on the gradient master to make a 10-50% (w/v) sucrose gradient. Sucrose gradients can be used immediately or stored long-term at -80°C (no more than 2 weeks) (see Note 1).
-
Sample preparation
Note: Cool all the solutions and buffers before use. Keep the samples on ice all the time if possible.
Seed the cells in 10 cm or 15 cm dishes one day before the experiment (see Note 2 and Table 1).
Add 100 μg/ml cycloheximide into the culture medium and incubate cells at 37°C for 10 min.
Discard the medium and wash the cells with 10 ml ice-cold PBS containing 100 µg/ml cycloheximide twice.
Add 10 ml ice-cold PBS to cells and harvest the cells with cell scraper.
Transfer the cell suspension into 15 ml centrifuge tube and centrifuge at 1,000 × g for 3 min at 4°C.
Remove the supernatant and add 250 μl lysis buffer to 15 ml centrifuge tube.
Lyse the cells by gently pipetting and transfer the lysate into a 1.5 ml Eppendorf tube.
Keep cell lysate on ice for 10 min and centrifuge the lysate at 14,000 × g for 10 min at 4°C to remove cell debris.
Transfer the supernatant into a new 1.5 ml Eppendorf tube.
Dilute 1 μl samples with 999 μl water, and measure the approximate RNA concentration using a NanoDrop photo-spectrometer (see Note 3).
Keep the samples on ice for immediate use or freeze them in liquid nitrogen immediately and store at -80°C until use (no more than one month).
-
Polysome profiling
Remove the same volume of solution as that of the loading sample from the top of the 10-50% sucrose gradient and then load the samples onto the sucrose gradient. The optimal loading volume is 200-500 μl (see Note 3). Keep 10% of lysates as an input.
Centrifuge at 35,000 × g for 2.5h at 4°C using the SW41 rotor in a Beckman Ultracentrifuge.
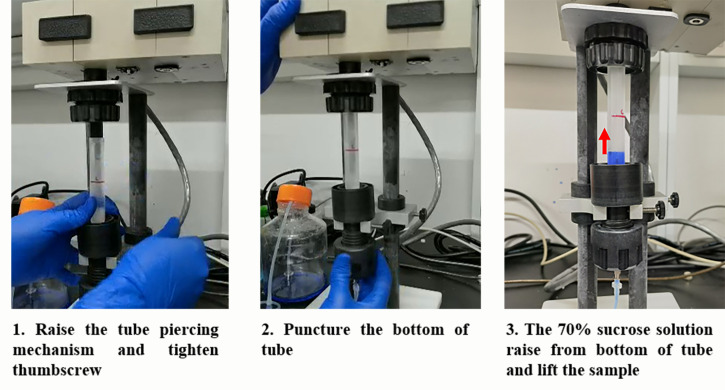
Set up the density gradient fractionation system while the samples are centrifuging. Fill the syringe with chasing solution [70% (w/v) sucrose with bromophenol blue], flush the line, and calibrate the ultraviolet spectrophotometer following the equipment instructions.
Gently remove tubes from the rotor and put them into the density gradient fractionation system. Alternatively, keep them at 4°C until use (no more than 4 h).
Load the sample under the UA-6 Detector and puncture the bottom of the tube, communicating the syringe pump and detector system (see Figure 2).
-
Number the 2 ml Eppendorf tubes and place them onto the R1 Fraction Collector.
Note: About 750 μl of sample will be collected in each tube.
Set syringe pump at 1.5 ml/min flow rate and switch on remote model. Set the fraction collector by time.
Turn on density gradient fractionation system using Peak chart software. Record the data with Peak chart (see Figure 3) and collect the fractions with the fraction collector.
Store the samples at -80°C until use (no more than one month).
-
RNA-seq library preparation
Add 750 μl of TRIzol into each tube, and extract the RNAs according to the manufacturer’s instruction (use chloroform, isopropanol, and 70% ethanol to separate RNAs from samples and elute in about 30 μl water) or store at -80°C.
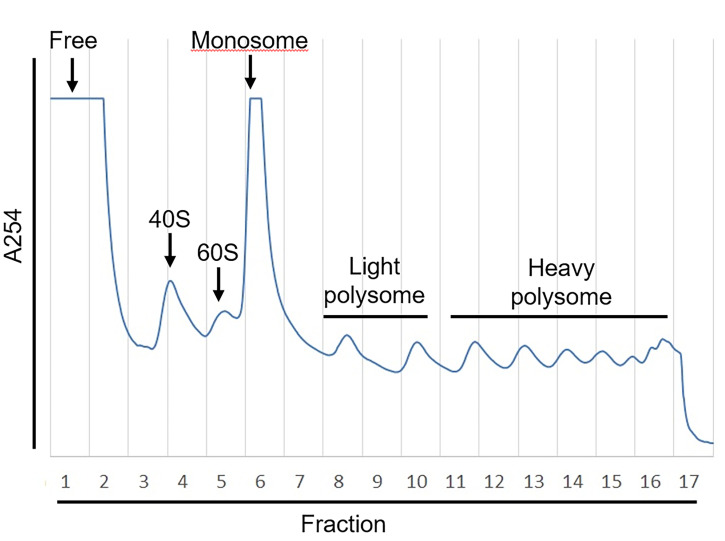
Pool RNA samples in free, monosome, light polysome or heavy polysome fractions with equal volume, respectively (e.g., as shown in figure 3, fraction 1 was termed “free fraction,” fraction 6-7 was termed “monosome fraction,” fractions 8-10 were pooled with equal volumes and termed “light polysome fraction,” and fractions 11-16 were pooled with equal volume and termed “heavy polysome fraction”).
Measure the RNA concentration of each RNA mixture with NanoDrop.
-
Treat the RNAs with DNase I (RNase free) at 37°C for 15 min.
2 μg of RNA x μl
DNase I 1μl
10× DNase I buffer 5 μl
Water 44-x μl
Clean up the RNAs using the ZYMO RNA clean and concentrator kit according to the manufacturer’s instructions.
-
Treat the DNase I treated RNAs with RNase R at 37°C for 30 min (see Note 4).
1 μg of RNA x μl
RNase R 1 μl
10× RNase R buffer 5 μl
Water 44-x μl
Clean up the RNAs using the ZYMO RNA clean and concentrator kit according to the manufacturer’s instruction (eluted RNAs in 20 μl water).
Construct the RNA-seq libraries with the KAPA stranded RNA-seq kit with RiboErase according to the manufacturer’s instruction.
-
Sequence the libraries using a 150 bp paired-end approach with a 100 million read depth per sample.
Note: We recommend Illumina HiSeq® 3000/HiSeq 4000 Sequencing Systems for RNA-seq.
-
Identification of translatable circRNAs
Perform quality control on raw sequencing reads and trim the adaptor sequences. Discard the low-quality reads. Trim the adaptor sequences from the reads and map them to the genome.
Align the reads to the reference genome with TopHat2, and map the unmapped reads with TopHat-Fusion.
Annotate and assembly the circRNA transcripts using the CIRCexplorer2 pipeline (see details in GitHub: https://circexplorer2.readthedocs.io/en/latest/) (Zhang, X. O. et al., 2016).
Figure 1. Protocol for Sucrose Gradient Preparation.
Table 1. Recommend schedule of this protocol.
| Prepare 10× polysome buffer and 70% sucrose solution for long use. | ||
| Day 1 | 1 | Seed cells. |
| Day 2 | 1 | Prepare lysis buffer and wash buffer. |
| 2 | Harvest samples following the steps of Sample preparation in order. | |
| 3 | Prepare sucrose gradient according to step A of the Procedure. | |
| 4 | Run polysome profiling following step C of the Procedure. | |
| 5 | Store the fractionated samples at -80°C. | |
| Day 3 | 1 | Thaw the samples on ice. |
| 2 | Extract RNA and prepare RNA-seq library following Procedure D. | |
| Sequencing | ||
| 7-10 days | Run process for circRNAs identification | |
Figure 2. Protocol for Density Gradient Fractionation System Set-off.
Figure 3. Example of Polysome Profiling.
The absorbance of OD254 (A254) that represents the abundance of RNA fragments was measured by UA-6 detector. The sucrose gradient was fractionated into 17 fractions with Density gradient fractionation system. The value of A254 was recorded by Peak Chart and plotted as shown.
Notes
If there is not Gradient Master for the gradient forming, we suggest an alternative method to make gradient through freezing and thawing. To make a 10-50% (w/v) sucrose gradient, prepare 50% (w/v), 40% (w/v), 30% (w/v), 20% (w/v), and 10% (w/v) sucrose solutions. Put 2 ml of 50% sucrose into a Beckman centrifuge tube and freeze on dry ice-ethanol, then put the 40% solution (2 ml) and also freeze, and repeat with the other solutions. The tubes can be frozen and stored at -80°C. One day before use, thaw the tubes in the refrigerator (4°C) overnight, and they are ready to use.
To keep the cells with active translation, it is better to harvest them at around 70-80% confluency. For example, for HEK293T, cells can be seeded in 10 cm (2.2 × 106 cells) or 15 cm (5.0 × 106 cells) dishes one day before the experiment and harvested at about 7.0 × 106 cells/10 cm dish or 16.0 × 106 cells/ 15cm dish. Since different types of cells have diverse translation activities at different cell confluency, we recommend testing the harvest conditions and loading amount before your experiment.
The optimal RNA amount in loading samples is 80-100 μg (sensitivity of 0.5). In cases where the RNA amount is lower than 80 μg, the sensitivity of the UV-Detector can be set to 0.2. We recommend using 200-600 μl of loading sample to get ideal and reproducible results.
RNase R can digest mostlinear RNAs; however, some RNAs with highly structured 3' ends are resistant to RNase R treatment. In that case, a modified RNase R treatment with an additional polyadenylation step and replacement of K+ with Li+ in the reaction buffer can be used to deplete the linear RNAs (please see Xiao and Wilusz, 2019). In addition, the activity of RNase R varies between different batches; thus, we recommend checking the amount of enzyme and the reaction time before your experiments (Zhang, Y. et al., 2016).
Recipes
-
10× polysome buffer
1 M KCl
50 mM MgCl2·6H2O
0.1 M HEPES (pH 7.4)
Nuclease free water
-
Wash buffer
1× PBS
100 μg/ml CHX
-
Lysis buffer
1× polysome buffer
100 units/ml Ribonuclease inhibitor
25 units/ml DNase I
1× Protease Inhibitor cocktail (EDTA-free)
2 mM DTT
0.5% Sodium Deoxycholate
0.5% Triton X-100
100 μg/ml CHX
-
70% (w/v) sucrose solution
70%(w/v) sucrose
1× polysome buffer
filtered with filter system
-
50% (w/v) sucrose grading solution
71.4%(v/v) 70% sucrose solution
1× polysome buffer
100 units/ml Ribonuclease inhibitor
1× Protease Inhibitor cocktail (EDTA-free)
100 μg/ml CHX
-
10% (w/v) sucrose grading solution
1.43% (v/v) 70% sucrose solution
1× polysome buffer
100 units/ml Ribonuclease inhibitor
1× Protease Inhibitor cocktail (EDTA-free)
100 μg/ml CHX
Acknowledgments
We want to thank all the members of Wang lab. This work was supported by the National Key Research and Development Program of China (MOST grant# 2018YFA0107602 to ZW), the National Natural Science Foundation of China (NSFC grant #31730110, #91940303, and #32030064 to ZW, #31870814 to YY), the Strategic Priority Research Program of Chinese Academy of Sciences(grant #XDB38040100 to ZW andYY) and the Science and Technology Commission of Shanghai Municipality (STCSM grant # 19QA1410500 to YY). The present protocol was developed and applied in Yang et al. (2017).
Competing interests
The authors declare no competing financial interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Ashwal-Fluss R., Meyer M., Pamudurti N. R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N. and Kadener S.(2014). circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1): 55-66. [DOI] [PubMed] [Google Scholar]
- 2.Chen L. L.(2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 21(8): 475-490. [DOI] [PubMed] [Google Scholar]
- 3.Kristensen L. S., Andersen M. S., Stagsted L. V. W., Ebbesen K. K., Hansen T. B. and Kjems J.(2019). The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20(11): 675-691. [DOI] [PubMed] [Google Scholar]
- 4.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al.(2017). Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 66(1): 22-37e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S. D., Gregersen L. H., Munschauer M., et al.(2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441): 333-338. [DOI] [PubMed] [Google Scholar]
- 6.Pamudurti N. R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., et al.(2017). Translation of CircRNAs. Mol Cell 66(1): 9-21e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R., et al.(2015). Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 58(5): 870-885. [DOI] [PubMed] [Google Scholar]
- 8.Salzman J., Chen R. E., Olsen M. N., Wang P. L. and Brown P. O.(2013). Cell-type specific features of circular RNA expression. PLoS Genet 9: e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salzman J., Gawad C., Wang P. L., Lacayo N. and Brown P. O.(2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7: e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanger H. L., Klotz G., Riesner D., Gross H. J. and Kleinschmidt A. K.(1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 73(11): 3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weigelt C. M., Sehgal R., Tain L. S., Cheng J., Esser J., Pahl A., Dieterich C., Gronke S., and Partridge L.(2020). An Insulin-Sensitive Circular RNA that Regulates Lifespan in Drosophila . Mol Cell 79(2): 268-279e265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao M. S., and Wilusz J. E.(2019). An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3' ends. Nucleic Acids Res 47(16): 8755-8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y., et al.(2017). Extensive translation of circular RNAs driven by N6-methyladenosine . Cell Res 27(5): 626-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H., et al.(2018). Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 110(3): 304-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H., et al.(2018). A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 9(1): 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X. O., Dong R., Zhang Y., Zhang J. L., Luo Z., Zhang J., Chen L. L., and Yang L.(2016). Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 26(9): 1277-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X. O., Wang H. B., Zhang Y., Lu X., Chen L. L. and Yang L.(2014). Complementary sequence-mediated exon circularization. Cell 159(1): 134-147. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y., Yang L. and Chen L. L.(2016). Characterization of Circular RNAs. Methods Mol Biol 1402: 215-227. [DOI] [PubMed] [Google Scholar]