Abstract
MicroRNAs (miRNAs) are small non-coding RNAs involved in post-transcriptional gene regulation that have a major impact on many diseases and provide an exciting avenue toward antiviral therapeutics. From patient transcriptomic data, we determined that a circulating miRNA, miR-2392, is directly involved with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) machinery during host infection. Specifically, we show that miR-2392 is key in driving downstream suppression of mitochondrial gene expression, increasing inflammation, glycolysis, and hypoxia, as well as promoting many symptoms associated with coronavirus disease 2019 (COVID-19) infection. We demonstrate that miR-2392 is present in the blood and urine of patients positive for COVID-19 but is not present in patients negative for COVID-19. These findings indicate the potential for developing a minimally invasive COVID-19 detection method. Lastly, using in vitro human and in vivo hamster models, we design a miRNA-based antiviral therapeutic that targets miR-2392, significantly reduces SARS-CoV-2 viability in hamsters, and may potentially inhibit a COVID-19 disease state in humans.
Keywords: COVID-19, SARS-CoV-2, microRNA, miRNA, nanoligomers, miR-2392, antiviral therapeutic, biomarker
Graphical abstract
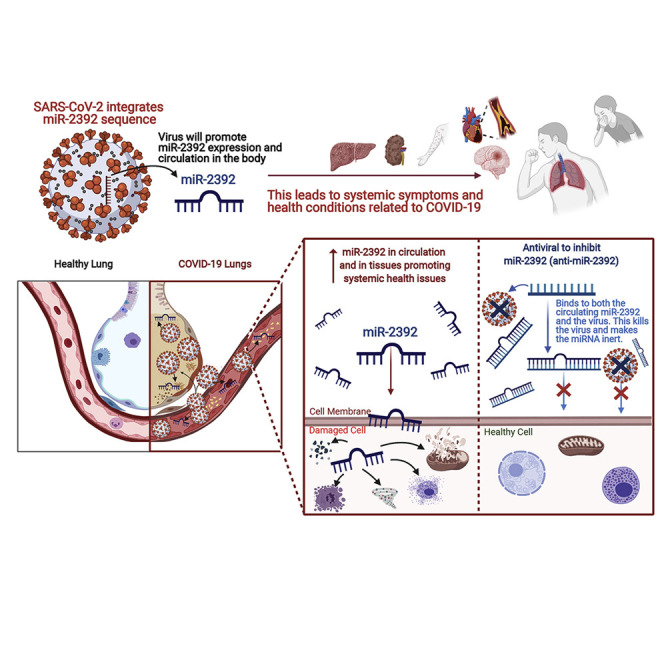
McDonald et al. uncover a role of a circulating microRNA, miR-2392, as a potential biomarker for COVID-19 and begin development of a potential COVID-19 therapeutic and antiviral to inhibit miR-2392.
Introduction
In fall 2019, a zoonotic spillover event led to the first known human infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and subsequent human-to-human transmission triggered a pandemic that led to a worldwide health crisis from the resulting disease, called coronavirus disease 2019 (COVID-19) (Huang et al., 2020; Zhu et al., 2020). COVID-19 causes substantial pulmonary disease but can also cause systemic health risks from extrapulmonary manifestations. Its effects entangle the entire body, including the cardiovascular, gastrointestinal, and hematological systems, and may lead to long-lasting effects after the virus has left the body, known as PASC (post-acute sequela of COVID-19) (Carfì et al., 2020; Jacobs et al., 2020). SARS-CoV-2 is classified as a member of the Coronaviridae family, viruses with a enveloped positive-stranded RNA with the ability to infect cross-species (V’kovski et al., 2021). Although the current vaccines represent a favorable milestone, additional data are required to demonstrate their long-term effectiveness against SARS-CoV-2 and protection against new strains. To prevent an endemic, the global eradication of COVID-19 will require a wide majority of the world’s population to be vaccinated to achieve herd immunity. Unfortunately, a portion of the population will not get vaccinated. Therefore, strategies for therapeutic options against COVID-19 are particularly relevant and important to explore to treat severe illnesses and overcome this global pandemic. Currently, most antivirals are repurposed drugs used for other diseases and have shown limited clinical efficacy, such as remdesivir (Abdelrahman et al., 2021). This brings a needed urgency to develop antivirals specifically designed against SARS-CoV-2.
A potential avenue for an alternative antiviral agent is to target specific microRNAs (miRNAs) associated with SARS-CoV-2 infection and subsequent manifestation of COVID-19. miRNAs are non-coding RNAs involved with the regulation of post-transcriptional gene expression, and they can affect entire pathways related to viruses and diseases (Jiang et al., 2009; Trobaugh and Klimstra, 2017). Each miRNA can target multiple messenger RNAs (mRNAs), and altogether, miRNAs are predicted to regulate more than half of the human transcriptome (Friedman et al., 2009). Recent evidence has shown that different diseases, including COVID-19, lead to distinct complements of miRNAs in the blood (Nersisyan et al., 2020; Saçar Demirci and Adan, 2020; Teodori et al., 2020). These circulating miRNAs are highly stable and have the potential to be used for minimally invasive detection, as biomarkers, and as therapeutic targets (Tribolet et al., 2020). The interactions between miRNAs and viruses have revealed a multifaceted relationship. Specifically, viruses have been shown to avoid the immune response by leveraging cellular miRNAs to complete their replication cycle (Trobaugh and Klimstra, 2017). The following mechanisms are central to the interaction of viruses and miRNAs: (1) miRNA processing can be blocked by viruses interacting with key proteins, such as Dicer and associated proteins; (2) viruses can sequester miRNAs, resulting in dysregulation of specific target mRNAs; (3) viruses can use miRNAs to redirect regulatory pathways of other miRNA targets to provide survival advantages; and (4) viruses can directly encode miRNA precursors that are processed by the canonical miRNA cellular pathway and have well-defined functions to specifically target and regulate the viral replicative cycle (Schult et al., 2018; Trobaugh and Klimstra, 2017).
Here, we report on a miRNA, miR-2392, that may directly regulate and drive a COVID-19 response. This miRNA is predicted from COVID-19 RNA sequencing (RNA-seq) patient data that resulted in multiple miRNAs being suppressed/inhibited (miR-10, miR-10a-5p, miR-1-3p, miR-34a-5, miR-30c-5p, miR-29b-3p, miR-155-5p, and miR-124-3p) and one miRNA being upregulated (miR-2392). With further examination, we hypothesize miR-2392 to be a key miRNA involved with COVID-19 progression. Specifically, miR-2392 drives downstream suppression of mitochondria activity while increasing inflammation, glycolysis, and hypoxia. miR-2392 upregulation is concomitant with symptoms associated with COVID-19 in the host. Patient blood and urine data show that miR-2392 circulates in COVID-19 patients and its concentration increases as a function of viral load. Our results demonstrate that miR-2392 may be used as an effective biomarker of COVID-19. Furthermore, we have started development of a miR-2392 inhibitor and provide evidence that it reduces SARS-CoV-2 viability in targeted viral screens in vitro and reduces the impact of infection in animal models. With further development, this miR-2392 inhibitor may represent an effective antiviral therapeutic and limit a negative host response from COVID-19.
Results
Identification of key miRNAs associated with COVID-19 infection
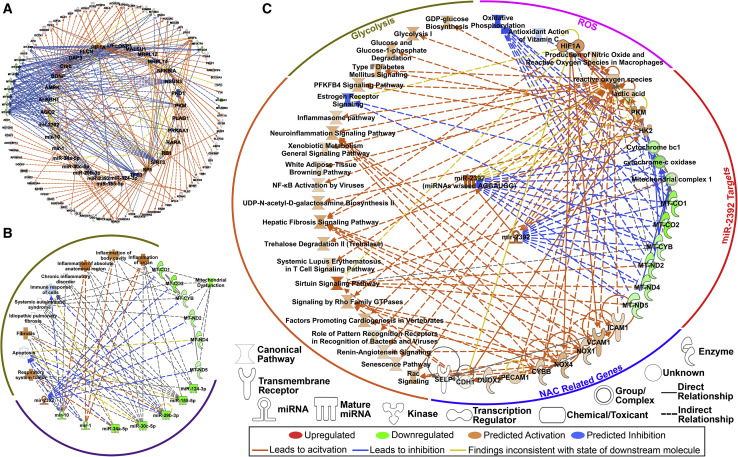
Currently, most publications associated with miRNAs and SARS-CoV-2 are based on in silico predictions. To identify miRNAs that may be involved in driving COVID-19 severity in the host, we examined publicly available bronchial alveolar lavage fluid (BALF) RNA-seq data. Differential gene expression was assessed using a 1.2-fold change for p values less than 0.01, revealing 42 increased and 347 decreased genes compared with controls. Using the upstream regulator analysis from the Ingenuity Pathway Analysis (IPA) knowledge database, miRNAs from differentially expressed genes (false discovery rate [FDR] < 0.05) from patients positive for COVID-19 were inferred. Eight miRNAs were predicted to drive significant changes in patients positive for COVID-19 with the downregulation of seven miRNAs (miR-10, miR-1, miR-34a-5p, miR-30c-5p, miR-29b-3p, miR-124-3p, and miR-155-5p) and upregulation of a single miRNA, miR-2392 (Figure 1 A). Using IPA’s downstream effects analysis to predict biological processes from the combined suppression of the seven miRNAs and upregulation of miR-2392 resulted in increased inflammation, immune suppression, and suppression of mitochondrial activity (Figures 1B and 1C).
Figure 1.
Key miRNA signature as predicted from bronchial alveolar lavage fluid (BALF) RNA-seq data in patients with COVID-19
(A) Predicted upstream regulators determined by IPA consistent with the transcriptional response from differentially expressed genes (FDR < 0.05; outer ring) in patients (n = 13). Eight miRNAs were among the key regulators in response to COVID-19 (inner ring).
(B) Major biological responses from dysregulation of this eight-miRNA signature drive immune- and inflammatory-related pathways, as well as mitochondrial dysfunction determined by IPA.
(C) Pathway regulation by miR-2392 from BALF RNA-seq data determined by IPA.
In support of these findings, previous studies have shown upregulation of miR-10, miR-124, or miR-1 has antiviral roles during infection (Hu et al., 2020; Sardar et al., 2020; Yang et al., 2016) and upregulation of miR-30 and miR-155 causes suppression of other coronaviruses (Dickey et al., 2016; Ma et al., 2018). The one miRNA predicted to be upregulated in COVID-19 patients from the BALF data was miR-2392. Although limited, the existing literature on miR-2392 demonstrates it is related to mitochondrial suppression and increased glycolysis (Fan et al., 2019), as well as circulating factors related to negative health risks (Chen et al., 2013; Fan et al., 2019; Li et al., 2017; Yang et al., 2019).
We performed pathway analysis with miR-2392 targets to determine its potential impact on the host. Upregulation of miR-2392 in the BALF RNA-seq dataset affected many downstream targets and pathways related to negative health outcomes (Figure 1C). This includes mitochondrial suppression and activation of factors related to reactive oxygen species (ROS). Because it is known that miR-2392 directly interacts with mitochondrial DNA (mtDNA) to inhibit the levels of oxidative phosphorylation (OXPHOS) transcripts (Fan et al., 2019), this could be a compensatory response to the inhibition of mitochondrial bioenergetics.
Glycolytic pathways (Figure 1C) are also upregulated in association with increased miR-2392. miR-2392 drives hexokinase 2 (HK2) and pyruvate kinase (PKM), both of which positively regulate glycolysis. HK2 produces a primary regulator of glycolysis, glucose-6-phosphate, and enhances guanosine diphosphate (GDP)-glucose biosynthesis. GDP-glucose is a nucleotide sugar and an essential substrate for all glycosylation reactions (i.e., glycosylation of viral spike proteins). PKM is essential for the production of ATP in glycolysis that catalyzes the transfer of the phosphate group from phosphoenolpyruvate to ADP to make ATP. The mechanism of how miR-2392 is driving these pathways is not clearly understood, but one possibility could be stabilization of glycolytic transcripts.
Overall, the observed upregulation of glycolysis and antiviral effects related to miR-2392 suppression are consistent with the recently documented role of glucose metabolism in the progression of viral infection and poor outcome of COVID-19 (Ardestani and Azizi, 2021). It is also consistent with the reported effects of glycolysis suppression by the glucose analog 2-deoxy-D-glucose (2-DG) that was shown to suppress SARS-CoV-2 replication in vitro (Ardestani and Azizi, 2021). These and miR-2392 data indicate targeting of glucose metabolism might significantly affect SARS-CoV-2 infections.
Targets implicated in antioxidant N-acetyl cysteine (NAC) therapy are to be upregulated (Figure 1C). These include adhesion molecules of activated endothelial cells such as intercellular adhesion molecule 1 (ICAM1), vascular cell adhesion molecule 1 (VCAM1), and E-selectin, which allow attachment of hematopoietic immune and non-immune cells to the endothelial surface and thus contribute to inflammation and activation of the coagulation cascade. Powerful antioxidants such as NAC potentially counteract COVID-19 infections by suppressing viral replication via improving the intracellular thiol redox ratio as a precursor for the major thiol antioxidant glutathione (Ho and Douglas, 1992) and inhibiting the nuclear factor κB (NF-κB) pathway (Poppe et al., 2017). Inhibition of the NF-κB pathway reduces inflammatory damage by altering the glutathione and glutathione disulfide ratio (Aykin-Burns et al., 2005; Jia et al., 2010). These results were in line with the role of miR-2392 in reducing the activities of electron transport chain complexes and enhancing glycolysis. Inflammatory pathways and others that are observed with COVID-19 infection were also seen to be activated downstream of miR-2392.
Conservation of miR-2392 between species and its predicted interactions with the SARS-CoV-2 genome
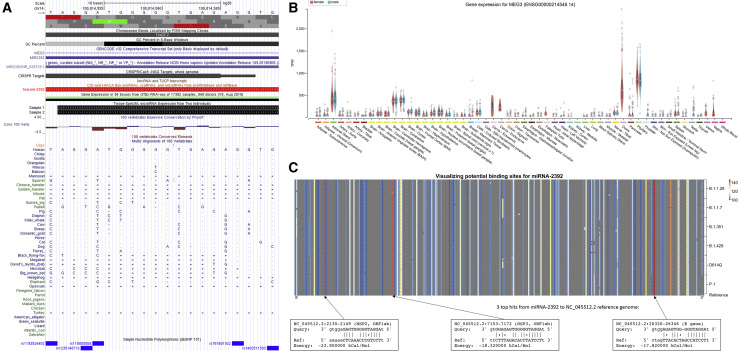
Viral miRNAs can facilitate interspecies viral transmission because of the high conservation of miRNAs among species and the ability of viruses to integrate miRNAs into their own genome (Saçar Demirci and Adan, 2020; Schult et al., 2018). This has been shown to assist viral replication and evasion of the immune system (Islam and Islam, 2021). Thus, we analyzed the conservation of human miR-2392 across species and the integration of miR-2392 into the SARS-CoV-2 genome (Figure 2 ).
Figure 2.
Cross-species and viral integration of miR-2392
(A) Conservation of miR-2392 across species determined by the UCSC Genome Browser. Boxes (▪) represent aligning and conserved sequence regions. The double horizontal line (=) shows both the genome and the query have an unalignable sequence between regions of the aligned sequence, a double-sided insertion. Single lines (-) indicate gaps.
(B) Expression of MEG3, the miR-2392 host gene, in different tissues from healthy patients.
(C) Potential binding sites of miR-2392 visualized across the SARS-CoV-2 genomes (NC045512.2 from GISAID). Average miRanda scores are for hits within the 100 bp window. The top three hits are shown.
We determined the conservation of miR-2392 across different species using the UCSC (University of California Santa Cruz) Genome Browser (Kent et al., 2002). The mature 20 base-pair miR-2392 is derived from an 84 base-pair region of the 3′ UTR in a long non-coding RNA gene, maternally expressed 3 (MEG3), and is located in an imprinted region, DLK1-DIO3, that contains three clusters with 51 other miRNAs (Figures 2A and 2B). A basewise evolutionary comparison showed miR-2392 is highly conserved among non-human primates such as dogs, cats, and ferrets, species known to be infected with SARS-CoV-2, whereas mice and rats, species not affected by COVID-19 (Johansen et al., 2020), have poor conservation with miR-2392.
The impact of miR-2392’s host gene, MEG3, within normal tissues was determined by GTEx data (GTEx Consortium, 2020). For most healthy tissues, MEG3 either was not detected or was expressed at low levels (Figure 2B). This can imply that miR-2392 does not seem to significantly affect normal tissues.
We used miRanda (Enright et al., 2003) to identify potential miR-2392 binding sites with respect to the SARS-CoV-2 reference genome (Wuhan-Hu-1; NC045512.2) and lineages of concern. The miR-2392 seeding region is heavily integrated and conserved in different viral strains (Figure 2C). The three best scores are located in the NSP2, NSP3, and E genes. Notably, these regions were conserved among 6 concerning lineages represented by 14 recent genomes available from the Global Initiative on Sharing All Influenza Data (GISAID) (Shu and McCauley, 2017).
miR-2392 targets mitochondrial and inflammatory pathways associated with SARS-CoV-2
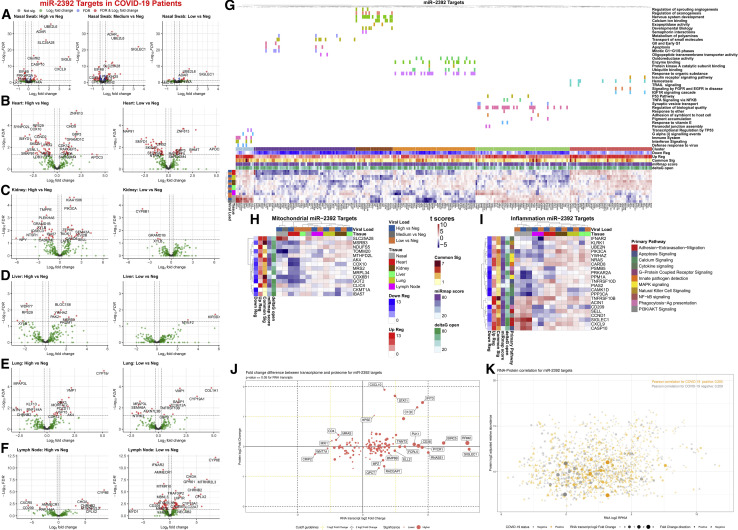
To determine a more comprehensive impact of miR-2392-affected pathways in COVID-19 patients, gene targets were predicted by seed-region base-pairing in the miRmap database (Vejnar and Zdobnov, 2012). This list was refined by overlap in several miRNA databases, including miRmap, miRwalk (Dweep and Gretz, 2015), miRDB (Chen and Wang, 2020), miRnet (Chang et al., 2020), and ClueGo (Bindea et al., 2009). We added RNA-seq analysis of 39 autopsy tissue samples from the heart, lung, kidney, liver, and lymph node of COVID-19-positive patients with high or low viral loads (Park et al., 2021). miR-2392 gene targets (375 genes) were visualized using volcano plots (Figures 3 A–3F).
Figure 3.
Gene targets of miR-2392 in COVID-19 patients, as well as mitochondrial and inflammatory genes
(A–F) Volcano plots for differential gene expression from (A) nasopharyngeal swab and autopsy COVID-19 patient tissues from the (B) heart, (C) kidney, (D) liver, (E) lung, and (F) lymph node separated by viral load.
(G) Heatmaps displaying t score statistics when comparing viral load versus negative patient samples for nasopharyngeal swab and autopsy tissues. Main gene clusters were determined by k-means clustering and annotated using ShinyGO (Ge et al., 2020) above the heatmap.
(H and I) miR-2392 gene targets for (H) mitochondrial or (I) inflammatory genes are displayed. Genes shown have at least one comparison with a FDR < 0.05 when comparing COVID-19 patients (high, medium, or low viral loads) to non-infected patients (none). MiRmap-only heatmaps for miR-2392 mitochondrial and inflammatory gene targets are available in Figures S1 and S2, respectively.
(J) Scatterplot of log2-fold changes in RNA and protein for miR-2392 targets for genes differentially expressed at the RNA level. Student’s t test, RNA p ≤ 0.05, no limitation on the protein p value.
(K) Scatterplot of log2-transformed medians in RNA and protein. COVID-19-positive samples (orange) or COVID-19-negative samples (gray).
Student’s t test is used in fold change calculations. Shape size and opacity represent RNA fold change values (circle, positive; triangle, negative). Pearson correlations are displayed in the top corner.
To ascertain the systemic impact on miR-2392 gene targets in COVID-19, we performed pathway analysis of nasopharyngeal swab samples taken from living donors with and without COVID-19, using viral load as the independent variable (high, medium, low, or other virus). The differentially expressed genes (FDR < 0.05) in at least one comparison of COVID-19-positive patients or other detected viruses were found to separate into six distinct hierarchical clusters annotated using ShinyGO (Ge et al., 2020) (Figure 3G). Most upregulated miR-2392 targets participate in immune and inflammatory pathways. Downregulated targets are involved in mitochondrial function, oxidative stress, the cell cycle, developmental biology, and ubiquitin binding, all pathways recently associated with SARS-CoV-2 infection (Hemmat et al., 2021). These data demonstrate that miR-2392 may target several pathways related to perpetuating SARS-CoV-2 infection. For all tissues excluding the lymph nodes, higher viral loads are associated with greater differential expression of miR-2392-regulated gene targets.
Because miR-2392 was shown to directly target the transcription of mtDNA genes (Fan et al., 2019), we evaluated mitochondrial miR-2392 targets in our datasets using MitoCarta (Rath et al., 2021) (Figure 3H). This revealed 14 genes harboring miR-2392 seed sequences that were significantly dysregulated in the nasal and heart samples. In nasal samples, SLC25A28, a mitoferrin that mediates mitochondrial iron transport, was strongly upregulated, along with IBA57, which is involved in iron sulfur assembly. The mitochondrial outer membrane protein import complex subunit TOMM20, cytochrome c oxidase (complex IV) subunit COX6B1, and mitochondrial transcription factor COT-2 (NR2F2) were strongly downregulated. In the heart, the folate enzyme MTHFD2L (methylenetetrahydrofolate dehydrogenase) was upregulated, whereas all other nuclear-coded mitochondrial genes were downregulated. Downregulated heart mitochondrial genes included NDUFS5 (complex I subunit), COX6B1 and COX10 (complex IV structural and assembly subunits), CKMT1A (mitochondrial creatine kinase), MRPL34 (mitochondrial ribosome small subunit), COT-2 (NR2F2), AK4 and MSRB3 (adenylate kinase 4 and methionine-R-sulfoxide reductase, respectively, which mitigate oxidative stress), MRS2 (magnesium transporter), and CLIC4 (chloride channel). The kidney showed mild upregulation of complex I and single methyl group metabolism but downregulation of complex IV (COX10), regulatory factor (COT-2), and iron sulfur center protein (IBA57). Hence, SARS-CoV-2 seems to downregulate nuclear mitochondrial gene transcription in the more oxidative organs, heart and kidney, as well as in nasal tissues.
Because inflammation is a key component of COVID-19, we overlaid known inflammatory genes determined from Loza et al. (2007) with miR-2392 targets (Figure 3I). From analysis at the mRNA level, most complement pathway genes are upregulated in the tissue samples analyzed. These changes could be compensatory, because proteins encoded by the genes could be downregulated as a function of traditional miRNA effects. The responses reflect the importance of degrees of inflammation for mediation of disease severity in COVID-19 patients and a key modulatory role of miR-2392 in this context.
Proteomic and transcriptomic analysis in blood from COVID-19 patients using COVIDome (Sullivan et al., 2021) revealed interesting patterns between RNA and protein levels for miR-2392 targets from miRmap, ClueGO, miRwalk, miRnet, and miRDB (Figures 3J and 3K). Several miR-2392 targets in tissue show a significant transcriptional increase in COVID-19-positive samples, with small to no changes on the proteomics level: PLK1, CD38, PYCR1, RNASE1, BIRC5, RRM2, and SIGLEC1 (Figure 3J). Interestingly, all these genes were also positively regulated for most tissues when considering only miR-2392 gene targets with miRmap (Figures S1 and S2). The miR-2392 targets CXCL10, STAT1, IFIT3, and C1QC were positively regulated at both the protein and the gene levels for the blood and other tissues. The correlation between RNA and protein expression was close for miR-2392 targets and was slightly stronger in COVID-19-negative samples (correlation score [cor] = 0.209, p = 4e−10) versus COVID-19-positive samples (cor = 0.205, p = 8e−10) (Figure 3K). Further investigation is needed to understand whether increased levels of miR-2392 could potentially bind genes’ mRNAs at a higher rate, and therefore prevent translation to protein, or whether other mechanisms prevent mRNA translation to protein.
Overexpression of miR-2392 simulates a phenotype similar to COVID-19 infection
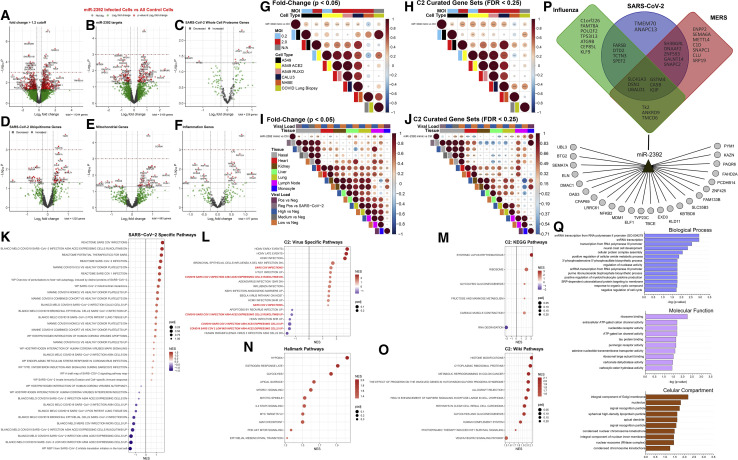
To determine whether miR-2392 upregulation would elicit effects similar to a COVID-19 infection, cells were treated with a miR-2392 mimic. Using RNA-seq, 649 genes had a fold change greater than ±1.2 and a p value less than 0.05 (Figure 4 A), and many genes were miR-2392-predicted targets (Figure 4B). These genes were then compared with whole-cell proteome data from a human-derived cell culture model of a SARS-CoV-2 infection profile (Stukalov et al., 2021), which found 10 overlapping gene/proteins that were significantly altered: KIF22, FKBP14, RAD51, AFAP1, ZCCHC17, ZWINT, MAGED1, CENPF, TMEM70, and NFKB2 (Figure 4C). Because viral infection may alter post-translational modification, including ubiquitination, we analyzed the ubiquitinome of a SARS-CoV-2 human-derived cell culture model and observed several altered proteins in normalized ubiquitin abundance that were also dysregulated genes by miR-2392 overexpression. We also found miR-2392 overexpression affected genes involved with mitochondria and inflammation (Figures 4D–4F).
Figure 4.
Increased miR-2392 expression in vitro mimics a COVID-19 phenotype
(A–F) RNA-seq volcano plots in cells overexpressing miR-2392.
(G–J) Correlation plot between miR-2392 overexpression and related SARS-CoV-2 datasets. Circle size is proportional to the correlation coefficient. Statistical significance was determined using a two-tailed Student’s t test; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
(K–O) Dot plots for statistically significant gene sets determined by fast preranked gene set enrichment analysis (fGSEA). NES, nominal enrichment score.
(P and Q) Predicted miR-2392 targets by the MIRDIP algorithm that are downregulated in the overexpression experiments. The putative miR-2392 mRNA targets belonging to the consensus transcriptomic networks observed in SARS-CoV-2, Middle East respiratory syndrome (MERS), and influenza infections of different human cells are represented in a Venn diagram in the upper part of (P).
To determine whether there was a direct correlation between miR-2392 overexpression and SARS-CoV-2 infection, we compared gene expression changes or overlap in statically significant canonical gene sets with our fast preranked gene set enrichment analysis (fGSEA) analysis. Published data from Blanco-Melo et al. (2020) showed a statistically significant and positive correlation of the miR-2392 treatment to A549 and Calu-3 cells infected with SARS-CoV-2 (Figures 4G and 4H), as well as in lung biopsies post-mortem from two patients positive for COVID-19 (Figure 4H). Using nasal swab samples, there was a significant and positive correlation between patients with medium and low viral loads compared with non-infected patients (Figures 4I and 4J). Additional miR-2392 correlation to SARS-CoV-2 infections used RNA-seq data from multiple tissues (heart, kidney, liver, lymph node, and lung) obtained during autopsies of COVID-19 patients with high or low viral loads (Figures 4I and 4J). There was a positive correlation to lung and lymph node tissues with miR-2392 expression. Interestingly, there was a significant and positive correlation to liver tissue when comparing gene fold change values (Figure 4I), but not when comparing C2 curated biological gene sets (Figure 4J). In contrast, a negative correlation to heart tissue was observed.
Statistically significant pathways that were enriched due to miR-2392 treatment were examined using fGSEA (Figures 4K–4O). miR-2392 treatment induced a pathway response that was significantly related to SARS-CoV-2 pathways. One obvious relationship shows that Reactome SARS-CoV-2 pathways were significantly activated for the miR-2392-treated cells compared with the controls (Figures 4K and 4L). Significant Hallmark pathways (Figure 4N) show upregulation of hypoxia (Herrmann et al., 2020), glycolysis (Ardestani and Azizi, 2021), and cell-cycle pathways (Su et al., 2020) that have been reported to be associated with COVID-19. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Figure 4M) indicates miR-2392 overexpression highly upregulated systemic lupus erythematosus, which has been reported to occur in COVID-19 patients, who have shown similar pathologies because of the increase of inflammation (Zamani et al., 2021).
Lastly, we determined downregulated targets in the cell lines after miR-2392 overexpression. A regulatory network was built by including the predicted miR-2392 targets in the microRNA Data Integration Portal (MIRDIP) that were also downregulated in the overexpression cell model, as well as from the recently described consensus transcriptional regulatory networks in coronavirus-infected cells (Ochsner et al., 2020) (Figure 4P). The gene enrichment analysis of these putative miR-2392 targets showed the presence of Gene Ontology (GO) terms related to RNA metabolism, transcription, ribosome activity, and the Golgi complex (Figure 4Q).
Circulating miR-2392 and the suppression of other miRNAs in COVID-19-infected patients
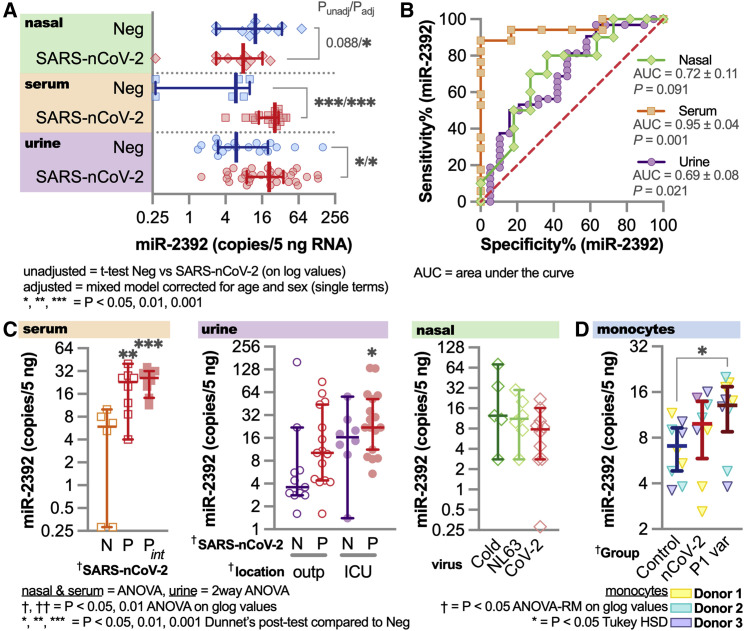
To demonstrate the presence of circulating miR-2392 in COVID-19-infected patients, we quantified miR-2392 by droplet digital PCR (ddPCR) in serum, urine, and nasopharyngeal swab patient samples (Figure 5 ). Serum was obtained from patients positive for COVID-19 with or without intubation or patients negative for COVID-19. Urine samples were from inpatients or outpatients positive for COVID-19, inpatients negative for COVID-19, or COVID-19-negative healthy donors. Nasal swabs were from patients positive for COVID-19, common cold coronavirus (229E, HKU1, and OC43), or respiratory illness/coronavirus (NL63).
Figure 5.
Circulating miR-2392 comparing patients positive and negative for COVID-19
Droplet digital PCR (ddPCR) for miR-2392 on serum, urine, and nasopharyngeal swab samples (including seasonal coronavirus) from COVID-19-positive or COVID-19-negative patients.
(A) miR-2392 levels from COVID-19-positive (SARS-CoV-2) or COVID-19-negative (neg) patients. Unadjusted t tests for each tissue or adjusted statistics with a mixed model corrected for age and sex are shown.
(B) ROC curve for miR-2392 in each tissue.
(C) Tissue comparisons for multiple categories. N, COVID-19 negative; p, COVID-19 positive; Pint, intubated patients; outp, outpatient; ICU, intensive care unit/inpatient; Cold, common cold coronaviruses; NL63, NL63 coronavirus; CoV-2, SARS-CoV-2. For all plots, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Sample numbers: all serum patient groups (n = 10), CoV-2 (n = 10), Cold (n = 6), NL63 (n = 6), inpatient COVID-19-positive (n = 10) or COVID-19-negative (n = 10) urine samples, and outpatient COVID-19-positive (n = 15) or COVID-19-negative (n = 11) urine samples.
(D) miR-2392 in monocytes from healthy donors (n = 3 biological replicates and n = 3 technical replicates for each donor) infected with the SARS-CoV-2 reference strain and P1 variant. Comparisons with three other miRNAs were made: miR-1-3p (Figure S3), miR-155-5p (Figures S4A–S4C), and miR-124-3p (Figure S4D).
We observed a statistically significant increase of miR-2392 in patients positive for COVID-19 from both the serum and the urine samples (Figure 5A). In addition, receiver operating characteristic (ROC) curve analysis revealed that miR-2392 is significantly associated with SARS-CoV-2 infection in patients (Figure 5B) in all tissues. miR-2392 levels were higher for conditions associated with infection of more severely affected patients (i.e., intubated or intensive care unit [ICU]) (Figure 5C). Low levels of miR-2392 appeared in the nasopharyngeal location, with no significant differences occurring between seasonal coronavirus samples. To study more specific tissue origins for miR-2392, we infected monocytes obtained from healthy donors with the SARS-CoV-2 reference strain (i.e., the strain from Wuhan) and P1 variant (Figure 5D). We saw an increase of miR-2392 with the reference SARS-CoV-2-infected cells and a significant increase in the P1 variant-infected cells. Previously, it was shown that these monocytes infected with SARS-CoV-2 have increased pro-inflammatory cytokine and glycolysis expression, with the effects inhibited by glycolysis inhibitors (Codo et al., 2020). Because we hypothesize that miR-2392 is a primary initiator for systemic impact of the infection, this might indicate that miR-2392 does not strongly appear until the virus has established its presence in the body.
We also quantified miR-1-3p (Figure S3), miR-155-5p (Figure S4), and miR-124-3p (Figure S5), which were predicted to be inhibited by COVID-19 infection (Figure 1A). miR-1-3p and miR-155-5p were significantly suppressed in the serum, with no differences in the urine or nasopharyngeal samples (Figures S3 and S4A–S4C). miR-1-3p is known to be beneficial for cardiovascular functions, with its inhibition leading to heart failure and disease (Condorelli et al., 2010). When quantifying miR-1-3p in the SARS-CoV-2-infected monocytes, we observe a significant decrease with the reference strain yet no difference with the P1 variant (Figure S3D). For miR-124-3p, we observed very low amounts (on average, <2 copies/5 ng of RNA) for all conditions, which indicates that miR-124-3p is not circulating for any patients for any conditions observed (Figure S4D). miR-124-3p provides an ideal miRNA-negative control candidate for SARS-CoV-2.
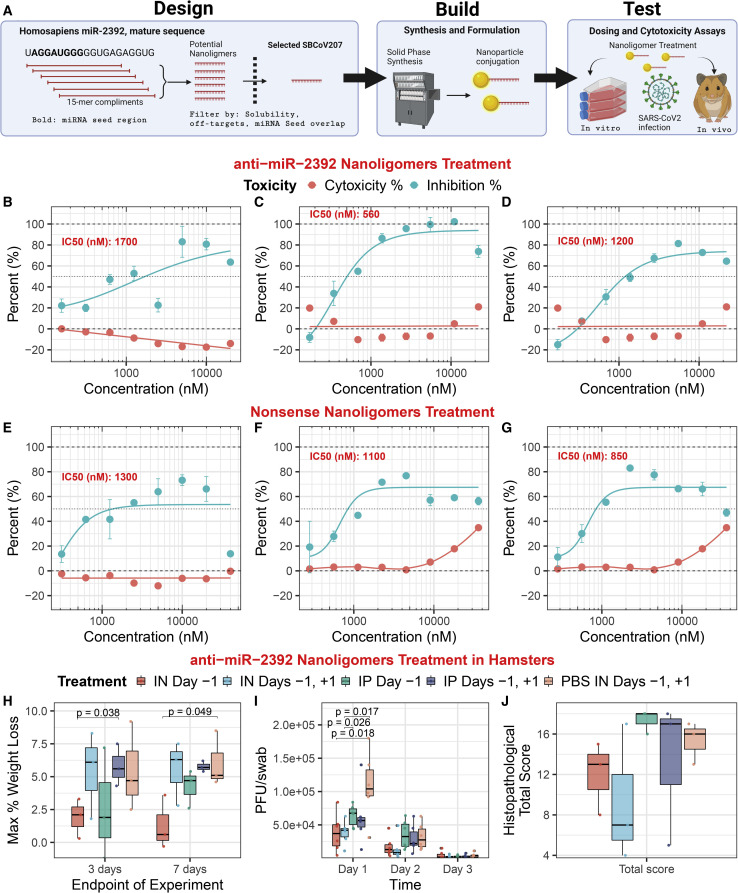
Inhibiting miR-2392: An antiviral COVID-19 therapeutic
The link between miR-2392 and COVID-19 infection prompted the development of an effective antiviral approach for COVID-19 by inhibiting miR-2392. We used the Nanoligomer platform to develop an effective antisense-based therapeutic against human miR-2392 (Eller et al., 2021), termed SBCov207 (Figure 6 A). The anti-miR-2392 nanoligomer was evaluated for efficacy and toxicity against a SARS-CoV-2 infection of the human lung cell line A549 (Figures 6B–6D). Treatment of uninfected A549 cells showed no cytotoxicity up to 20 μM. The control nonsense nanoligomer (SBCoV208) showed no toxicity even up to 40 μM. Treatment of A549 cells infected with SARS-CoV-2 showed drastic improvement in cell viability, with an average of 85% viral inhibition at 10 μM (half maximal inhibitory concentration (IC50) = 1.15 ± 0.33 μM). In contrast, the control nanoligomer showed significantly lower viral suppression (Figures 6E–6G). This human cell line model reaffirms that the anti-miR-2392 (SBCov207) is effective in inhibiting SARS-CoV-2 yet does not exhibit toxicity at the concentrations tested.
Figure 6.
Anti-miR-2392 therapeutic mitigation of SARS-CoV-2 infection with in vitro and in vivo models
(A) Platform schematic for the miR-2392 inhibitor.
(B–G) Anti-miR-2392 nanoligomer inhibitor or (E–G) nonsense control applied to A549 cells infected with SARS-CoV-2 and tested for viral viability and cytotoxicity as biological replicates (n = 3). Error bars represent the standard error for technical replicates (n = 3).
(H–J) Toxicity and efficacy of the anti-miR-2392 nanoligomer inhibitor in an in vivo infection hamster model. Treatments groups: SBCov207 by IP injection or IN instillation 24 h before viral inoculation (IP or IN day −1), 24 h prior and post-viral challenge (IP or IN day −1 and +1, respectively), and 100 μL of PBS 24 h prior and post-viral challenge by IN instillation (PBS IN day −1 and +1, respectively). (H) Pooled weights for each treatment group (n = 6, days 1–3, and n = 3, days 4–7) as the maximum percentage of weight loss. (I) SARS-CoV-2 assayed by plaque assay on Vero E6 cells from oropharyngeal swabs (n = 6). (J) Histopathological total score for lung tissues at day 3.
IN, intranasal; IP, intraperitoneal. Error bars = standard error.
The anti-miR-2392 nanoligomer was then evaluated in a Syrian hamster infection model (Figures 6H–6J). Six hamsters were treated with nanoligomers for 72 h without infection, and there was no observed change in animal behavior, indicating a lack of obvious toxicity. Next, 30 male hamsters were divided into 5 treatment groups. The infected hamsters were given 105 plaque-forming units (PFUs) of the WA01/2020 strain of SARS-CoV-2. The anti-miR-2392 nanoligomer treatment was given by intraperitoneal (IP) injection or intranasal (IN) instillation once at 24 h before or twice at 24 h before and after viral inoculation. Half of the hamsters in each group (n = 3) were euthanized and necropsied on days 3 and 7 post-infection, respectively.
Loss of body weight over the course of the experiment was <10% in all groups and was significantly different for the IN treatment one day before viral inoculation (compared with the control), but not in other groups (Figure 6H). Virus titers from oropharyngeal swabs for IN treatment were significantly lower (p = 0.018) than those from hamsters receiving nanoligomer IP or PBS on day 1 post-challenge, but there were no differences among groups on days 2 and 3 post-challenge (Figure 6I). Although not statistically different from the control, the data indicate a downward trend with nanoligomer treatment (Figure 6J), and the total histopathological score for the IN treatment was lower than that for the controls.
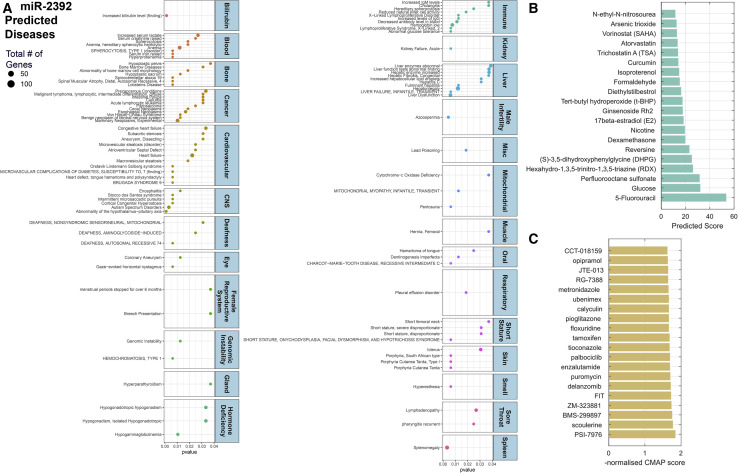
The impact of miR-2392 on diseases, relationship to COVID-19 symptoms, and predicted FDA-approved drugs to target miR-2392
To predict whether miR-2392 has a direct relationship to COVID-19 symptoms, we determined the pathway and disease relevance using miRnet. Among the diseases predicted to be associated with miR-2392 were a surprising number of clinical observations present in individuals with COVID-19 infection (Figure 7 A). These include heart or cardiovascular disease and failure, both known to heavily contribute to morbidity and mortality in patients with COVID-19 (Nishiga et al., 2020); hyperesthesia (Krajewski et al., 2021); and less common COVID-19 symptoms, such as lymphadenopathy and pharyngitis related to sore throat (Edmonds et al., 2021), liver dysfunction (Portincasa et al., 2020), splenomegaly (Malik et al., 2020), CNS failure (Mahajan and Mason, 2021; Rodriguez et al., 2020), and kidney failure (Hultström et al., 2021).
Figure 7.
Predicted impact of miR-2392 on human disease and top 20 drug compounds predicted to affect miR-2392 expression through a machine learning approach
(A) Dot plot of manually curated diseases associated with miR-2392 predicted from miR-2392 gene targets by miRnet. Values are plotted by p value, and the dot size represents the number of downstream gene targets associated with disease. miR-2392 expression related to patient survival in a pan-cancer analysis is highlighted in Figure S5.
(B) Matrix completion model scores to predict small molecules that affect miRNA expression. Higher scores indicate more predicted associations.
(C) Normalized CMAP scores using transcripts induced by miR-2392 overrepresented genes. Higher negative scores reflect a greater reversal of the miR-2392 transcriptomic signature. Further details on model statistics and performance are found in Figure S6.
miR-2392 was also predicted to affect other diseases that have been linked to COVID-19 infection in some patients. For example, COVID-19 patients have experienced azoospermia, which is linked to male infertility (Younis et al., 2020), an altered menstrual cycle (Li et al., 2021), dental damage (Sirin and Ozcelik, 2021), and deafness or hearing loss (Koumpa et al., 2020). Using the Kaplan-Meier Plotter (Győrffy, 2021) to associate miR-2392 expression with pan-cancer patient survival (Figure S5), high expression of miR-2392 was generally related to poor prognosis with most cancer types (p < 0.05). Intriguingly, one miR-2392-predicted consequence was decreased antibody levels in the blood; this might account for the reported loss of the antibodies overtime (Gudbjartsson et al., 2020; Self et al., 2020).
Finally, using computational models, we predicted small molecules, including FDA-approved drugs, that could inhibit miR-2392. A state-of-the-art machine learning algorithm for predicting missing drug targets (Galeano et al., 2021) was applied to an association dataset between 213 small molecules and 1,519 miRNAs from the SM2miR database (Liu et al., 2013) (see statistics in Figures S6A and S6B), integrating chemical and sequence similarity, respectively. The average area under the ROC curve was of 0.877 when predicting missing small molecule-miRNA associations (Figures S6C–S6F). The top 20 predicted small molecules (Figure 7B) include dexamethasone (Ledford, 2020) and atorvastatin, which have shown protective roles in COVID-19 patients (Rossi et al., 2020). Next, following the ideas presented in Sirota et al. (2011), we performed an analysis on the genomic signature of miR-2392 (i.e., significant up- and downregulated genes) and predicted small molecules that can reverse it. We screened the genomic signature of miR-2392 against 30,000 small molecules contained in the connectivity map (CMAP) (Lamb et al., 2006). The top 20 predicted small molecules include the androgen receptor antagonist enzalutamide and the insulin sensitizer pioglitazone (Carboni et al., 2020), both of which are in clinical trials for COVID-19 (Figure 7C) (ClinicalTrials.gov: NCT04475601 and NCT04604223), as well as evidence for the leukotriene inhibitor ubenimex (Asai et al., 2020) and bacterial DNA inhibitor metronidazole (Gharebaghi et al., 2020).
Discussion
Although the potential eradication of the novel coronavirus through worldwide vaccination is under way, new potent strains of SARS-CoV-2 are constantly evolving, and there remains a major need to develop effective interventional strategies to minimize the damage caused by these infections. Host-mediated lung inflammation is a driver of mortality in critically ill COVID-19 patients. Thus, it is logical to focus on therapeutics that may have immunomodulating properties or disrupt viral replication. Our research uncovers an eight-miRNA signature in patients with COVID-19 viral loads compared with those without disease as predicted from RNA-seq data. The expression of seven miRNAs was decreased (miR-10, miR-1, miR-34a-5p, miR-30c-5p, miR-29b-3p, miR-124-3p, and miR-155-5p), whereas a single miRNA, miR-2392, was significantly increased (Figure 1). This key miRNA signature was involved in major cellular and molecular mechanisms that drive the viral-host response.
Several studies have measured differential expression of miRNAs in COVID-19 patients and proposed their use as biomarkers or therapeutics. Lung biopsies from 9 COVID-19 patients showed miR-26a, miR-29b, and miR-34a correlated to endothelial dysfunction and inflammatory biomarkers (Centa et al., 2020). Sequencing in the blood from patients with moderate or severe COVID-19 identified miR-146a, miR-21, miR-142, and miR-15b as potential biomarkers, as well as contributors to disease pathogenesis (Tang et al., 2020). Although these studies are limited to a specific tissue, our data that correlate miRNA signatures from multiple tissues (Figure 3) suggest miR-2392 is a unique target that is ubiquitously involved in COVID-19 symptoms. To start addressing the multi-tissue impact of miR-2392, we also show miR-2392 is overexpressed in healthy monocyte cells infected with both reference and P1 SARS-CoV-2 variants (Figure 5D). This may indicate isolated cells will overexpress miR-2392 when infected with SARS-CoV-2, and further work is being done to investigate multi-tissue behavior of miR-2392.
Most miR-2392 publications are focused on cancer tissues, where it may drive cellular invasion and metastasis. miR-2392 was one of 6 miRNAs altered in the serum and tissue of cervical cancer patients used to predict metastasis (Chen et al., 2013). Higher levels of miR-2392 in gastric cancer were linked to lower clinical staging and increased patient survival (Li et al., 2017) by inhibiting invasion and metastasis via the loss of Snail1, Slug, and Twist1 expression. Similarly, lower miR-2392 and miR-1587 levels were found in human keloid tissues that led to a loss of inhibition of ZEB2 and promoted cellular proliferation and invasion (Hou et al., 2017). Recently, a role for miR-2392 in tongue squamous cell carcinoma showed partial inhibition of transcription through direct miRNA-mtDNA base-pairing, resulting in reprogramming of tumor cell metabolism and chemoresistance (Fan et al., 2019). These miR-2392 reports establish the significant impact that this miRNA may have on cellular activity. Particularly relevant here is the more than 2-fold increase of miR-2392 in extracellular vesicles of hepatitis B-infected human hepatocytes (Enomoto et al., 2017). Although miR-2392 has a reported impact on tumor cell biology, our study expands the valuable therapeutic potential of targeting miR-2392 to decrease SARS-CoV-2 viral infections (Figure 6). These results warrant further exploration of the mechanistic underpinnings for the role of miR-2392 in driving viral infection.
One therapeutic insight deduced from miR-2392 interactions is the importance of the mitochondrial OXPHOS and glycolytic pathways in COVID-19, which are dramatically highlighted in BALF samples (Figure 1C). In a study of tongue squamous cell carcinoma (Fan et al., 2019), miR-2392 enters the mitochondrion, where it binds Ago2 and nucleotides 4,379 to 4,401 in the mtDNA heavy (H) strand within the MT-TQ (tRNA glutamine) gene (m.4329-4400). MT-TQ is part of a large polycistronic transcript encompassing 12 of the mtDNA H strand polypeptide genes punctuated by tRNAs. Cleavage of the tRNAs releases the mRNAs. Upstream of MT-TQ are the 12S and 16S rRNAs and the complex I gene MT-ND1 gene. Downstream of MT-TQ are MT-ND2, MT-CO1, MT-CO2, MT-ATP6/8, MT-ND3, MT-ND4L, MT-ND4, MT-ND5, and MT-CYB (Lott et al., 2013; Wallace, 2018). The downregulated mtDNA genes are the complex IV (cytochrome c oxidase) genes MT-CO1 and MT-CO2, the complex III (the bc1 complex) gene (MT-CYB), and the complex I genes (MT-ND2, MT-ND4, and MT-ND5) (Figure 1C, right side arc). Because the miR-2392 inhibition of mtDNA OXPHOS genes (Figure 1C) is also reflected in the downregulation of the nuclear DNA-coded mitochondrial transcripts of the complex I and IV genes and the iron-sulfur and heme iron complexes in the nasal, heart, and kidney autopsy samples (Figure 3D), mitochondrial inhibition by miR-2392 appears to be the only physiological function that is common across all tissues in infected individuals, suggesting mitochondrial modulation is a central feature of SARS-CoV-2 pathophysiology.
The inhibition of mitochondrial genes by miR-2392 would impair OXPHOS, which would have the most adverse effects on the high mitochondrial energetic tissues (brain, heart, and kidney), the tissues central to the most severe COVID-19 cases. Inhibition of mitochondrial OXPHOS genes would increase mitochondrial reactive oxygen species (mROS) production and induce glycolysis to compensate for the energy deficit (see the top of Figure 1C). Mitochondrial function is regulated by the sirtuins (Carrico et al., 2018), mitochondrial decline is associated with senescence, and mROS oxidation of mtDNA is linked to activation of the inflammasome and thus NF-κB (West and Shadel, 2017; Zhong et al., 2018), all of which are modulated around miR-2392 (Figure 1C). Thus, SARS-CoV-2 induction of miR-2392 (Figure 5) and its associated inhibition of mtDNA and nuclear DNA OXPHOS genes (Figures 3 and S1) could explain many metabolic disturbances of COVID-19. Conversely, antagonism of miR-2392 function should ameliorate the inhibition of OXPHOS and may explain the therapeutic benefit of the anti-miR-2392 nanoligomer (Figure 6).
Exploiting miRNAs from serum as a biomarker was pioneered in patients to examine diffuse large B cell lymphoma (Lawrie et al., 2008). Using miRNAs as diagnostic biomarkers has several advantages. Circulating miRNAs are readily obtained through a minimally invasive blood draw and are remarkably resistant to degradation in the plasma and serum (Mitchell et al., 2008). They may provide a means to detect asymptomatic individuals (Hou et al., 2017). However, potential confounding diseases that may influence the expression of multiple miRNAs, which requires further evaluation of the targets found in this study (Figure 5).
Advances in RNA chemistry and delivery systems enabled the first miRNA-based agents to enter into clinical trials several years ago (Rupaimoole and Slack, 2017). miR-122 was found to increase the stability and replication of the hepatitis C virus (HCV) via binding that prevented degradation by the Xrn1 exoribonuclease (Thibault et al., 2015). Following a phase I trial that inhibited miR-122 with no adverse reactions, a phase IIa trial showed a dose-dependent decrease in HCV load and one reported grade 3 thrombocytopenia; in addition, only a small set of patients experienced viral rebound that might be linked to mutations of the HCV viral RNA (Janssen et al., 2013; Ottosen et al., 2015). A separate trial targeting miR-122 reduced viral loads in all treated patients within 4 weeks, with a sustained response in 3 patients after 76 weeks (van der Ree et al., 2017); however, trials were suspended due to two cases of severe jaundice. These clinical trials have demonstrated the promising potential of using anti-miRNAs to significantly reduce viral infection with limited adverse effects, and the similarities with miR-2392 with SARS-CoV-2 warrant further investigations to push to clinical trials.
Presently, there remains no specific treatment option for patients presenting with severe COVID-19. Although vaccines provide a promising avenue toward curbing the infection rate and minimizing symptoms, there remains an urgency to successfully develop and implement therapeutic agents to reduce severe consequences and subsequent patient mortality from COVID-19. Testing of antibody-based or drug-targeted therapies is under way, and miRNAs represent a category of therapeutic agents that have previously shown endogenous activity to alter viral infection.
Limitations of the study
We determined that miR-2392 is a key driver for SARS-CoV-2 infections and could be used as a biomarker of COVID-19 positivity. The specificity of the miRNAs, specifically miR-2392, needs to be expanded by observing expression in multiple tissues from COVID-19-infected patients and animal models. From RNA-seq autopsy data, we determined genes altered by miR-2392 in multiple organs, but further investigation is warranted to quantify the presence of miRNAs in tissues. Although the antiviral studies for miR-2392 are promising, more studies are needed to improve efficacy and determine whether this antiviral will provide an effective therapeutic option for COVID-19 patients. Lastly, the predicted top-ranked drugs to target miR-2392 (Figure 7) need to be validated in a laboratory setting to determine the true effect on SARS-CoV-2 infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SARS-CoV-2 WA-1 | Provided by Dr. Natalie Thornburg at the Centers for Disease Control and Prevention | N/A |
| SARS-CoV-2 B | GenBank: MT126808.1 | N/A |
| SARS-CoV-2 P1 | Provided bu Hematology Amazonas (Fundação Hospitalar de Hematologia e Hemoterapia do Amazonas,Manaus, Brazil | N/A |
| Biological samples | ||
| Human Blood/Serum | Provided by Dr. Mohammad M. Sajadi at the University of Maryland | N/A |
| Human Nasopharyngeal samples for ddPCR | Provided by Dr. Mohammad M. Sajadi at the University of Maryland | N/A |
| Human Urine Samples | Provided by Dr. Matthew C. Wolfgang and the UNC COVID-19 Pathobiology Consortium at the University of North Carolina at Chapel Hill | N/A |
| Human Nasopharyngeal samples for RNA-Seq | Weill Cornell Medicine | N/A |
| Human autopsy samples | Weill Cornell Medicine | N/A |
| Human Peripheral blood mononuclear cells (PBMC) | Provided by Dr. Pedro M. Moraes-Vieira at the Hematology and Hemotherapy Center of the University of Campinas | N/A |
| Critical commercial assays | ||
| miRNeasy serum/plasma kit | QIAGEN | Cat# 217184 |
| miRCURY LNA RT kit | QIAGEN | Cat# 339340 |
| QX200 ddPCR Evagreen Supermix | BioRad | Cat# 1864034 |
| miRNA primers | QIAGEN | Listed under “Oligonucleotides” |
| miRNeasy mini kit | QIAGEN | Cat# 217004 |
| Urine microRNA Purification Kit | Norgen Biotek Corp | Cat# 29000 |
| shMIMIC Human Lentiviral microRNA hsa-miR-2392 hCMV-TurboGFP, 200 μL, 108 TU/mL | Horizon | Cat# VSH6187 |
| SMARTvector Non-targeting hCMV-TurboGFP Control Particles 2, 50 μL, 108 TU/mL | Horizon | Cat# VSC10236 |
| Deposited data | ||
| miR-2392 mimic experiments | SRA (https://www.ncbi.nlm.nih.gov/sra) | BioProject ID: PRJNA76339 |
| Nasopharyngeal and autopsy data | Weill Cornell COVID-19 Genes | https://covidgenes.weill.cornell.edu/ |
| Experimental models: Cell lines | ||
| SH-SY5Y | ATCC | Cat# CRL-2266; RRID:CVCL_YK67 |
| A549-ACE2 | gifted by Dr. Brad Rosenberg | N/A |
| Experimental models: Organisms/strains | ||
| Golden Syrian hamsters | Charles River Laboratories | Crl: LVG(SYR) |
| Oligonucleotides | ||
| hsa-miR-2392 miRCURY LNA miRNA PCR Assay: MIMAT0019043: 5′UAG GAUGGGGGUGAGAGGUG |
QIAGEN | Cat# YP02104616 |
| hsa-miR-155-5p miRCURY LNA miRNA PCR Assay: MIMAT0000646: 5′UUAAU GCUAAUCGUGAUAGGGGU |
QIAGEN | Cat# YP00204308 |
| hsa-miR-124-3p miRCURY LNA miRNA PCR Assay: MIMAT0000422: 5′UAAGGC ACGCGGUGAAUGCC |
QIAGEN | Cat# YP00206026 |
| hsa-miR-1-3p miRCURY LNA miRNA PCR Assay: MIMAT0000416: 5′UGGAAUGUAA AGAAGUAUGUAU |
QIAGEN | Cat# YP00204344 |
| Software and algorithms | ||
| FASTQC version 0.11.8 | N/A | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/; RRID:SCR_014583 |
| Trim Galore! Version 0.50 | N/A | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/; RRID: SCR_011847 |
| STAR version STAR_2.6.1a_08-27 | Dobin et al., 2013 | https://github.com/alexdobin/STAR; RRID: SCR_015899 |
| RSEM version 1.3.1 | Dobin et al., 2013 | https://deweylab.github.io/RSEM/; RRID: SCR_013027 |
| R Version 3.5.1 and 4.0.2 | R Core Team, 2020 | https://www.r-project.org/; RRID: SCR_001905 |
| DESeq2 Version 1.22.2 | Love et al., 2014 | https://github.com/mikelove/DESeq2; RRID: SCR_01568 |
| LIMMA/LIMMA-Voom | Ritchie et al., 2015 | https://www.bioconductor.org/packages/release/bioc/html/limma.html; RRID: SCR_010943 |
| GSEA | Subramanian et al., 2005 | https://www.gsea-msigdb.org/gsea/index.jsp; RRID: SCR_003199 |
| EnhancedVolcano package (Bioconductor) | Blighe et al., 2018 | http://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html; RRID: SCR_018931 |
| Cytoscape | Shannon et al., 2003 | https://cytoscape.org; RRID: SCR_003032 |
| ClueGo/CluePedia | Bindea et al., 2009 | http://www.ici.upmc.fr/cluego/; RRID: SCR_005748 |
| pheatmap R package version 1.0.12 | Kolde, 2015 | https://cran.r-project.org/web/packages/pheatmap/index.html; RRID: SCR_016418 |
| miRNet 2.0 | Chang et al., 2020 | https://www.mirnet.ca/ |
| miRmap | Vejnar and Zdobnov, 2012 | https://mirmap.ezlab.org/; RRID:SCR_016508 |
| miRwalk | Dweep and Gretz, 2015 | http://mirwalk.umm.uni-heidelberg.de/; RRID:SCR_016509 |
| miRDB | Chen and Wang, 2020 | http://mirdb.org/; RRID:SCR_010848 |
| MiRanda | Enright et al., 2003 | http://cbio.mskcc.org/miRNA2003/miranda.html; RRID:SCR_017496 |
| ggplot2 v3.3.1 | Wickham, 2016 | https://ggplot2.tidyverse.org; RRID:SCR_021139 |
| QuantaSoft v1.7.4.0917 | Bio-Rad Laboratories | https://www.bio-rad.com/webroot/web/pdf/lsr/literature/10047467.pdf |
| ShinyGO | Ge et al., 2020 | http://bioinformatics.sdstate.edu/go/; RRID:SCR_019213 |
| fGSEA | Korotkevich et al., 2021 | https://github.com/ctlab/fgsea; RRID:SCR_020938 |
| COVIDome Explorer Researcher Portal | Sullivan et al., 2021 | https://medschool.cuanschutz.edu/covidome |
| ggrepel version 0.8.2 | N/A | https://ggrepel.slowkow.com/; RRID:SCR_017393 |
| Pyrpipe | Singh et al., 2021 | https://github.com/urmi-21/pyrpipe/tree/master/case_studies/Covid_RNA-Seq |
| MetaOmGraph | Singh et al., 2020 | https://github.com/urmi-21/MetaOmGraph/ |
| UCSC Genome Browser | Kent et al., 2002 | https://genome.ucsc.edu/ |
| GTEx | GTEx Consortium, 2020 | https://www.gtexportal.org/home/; RRID:SCR_013042 |
| SM2miR database | Liu et al., 2013 | http://www.jianglab.cn/SM2miR/ |
| RDKit | Open-source cheminformatics | http://www.rdkit.org |
| Ingenuity Pathway Analysis (IPA) | QIAgen | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/; RRID:SCR_008653 |
| Other | ||
| QX200 Automated Droplet Generator | BioRad | Cat# 1864101, RRID:SCR_019714 |
| QX200 Droplet Reader | BioRad | Cat# 1864003; RRID:SCR_019707 |
| NovaSeq 6000 | Illumina | N/A; RRID:SCR_016387 |
| BioAnalyzer | Agilent | Cat# D1000 HS; RRID:SCR_019382 |
| ABI 7500 Real-Time PCR | Applied Biosystems | N/A; RRID:SCR_019334 |
| NanoDrop 2000 Spectrophotometer | ThermoFisher Scientific | N/A; RRID:SCR_020309 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Afshin Beheshti (afshin.beheshti@nasa.gov).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human serum and nasopharyngeal swab sample collection for ddPCR
All plasma and nasal swab samples from those with COVID-19 infection, seasonal coronavirus infection, and controls were collected from inpatients at the University of Maryland Medical Center, in Baltimore, USA, between March and May of 2020. Sample collection obtained through informed consent waiver, which was approved by the University of Maryland, Baltimore IRB.
For serum samples, N = 10 samples from COVID-19 intubated patients, COVID-19 outpatients, and patients negative for COVID-19 were obtained. An equal distribution of N = 5 males and females were used for each group. Also, an equal age distribution of patients from 27 to 85 years old was utilized for each group.
For the nasopharyngeal samples the following patient samples were obtained: N = 10 SARS-CoV-2 positive patients, N = 6 common cold coronavirus samples, and N = 6 Coronavirus NL63 samples. For the common cold coronavirus samples the breakdown was the following for the specific viruses: N = 2 Coronavirus 229E, Coronavirus HKU1, and N = 2 Coronavirus OC43.
Human nasopharyngeal swab sample collection for RNA-seq analysis
Patient specimens were processed as described in Butler et al. (2021). Briefly, nasopharyngeal swabs were collected using the BD Universal Viral Transport Media system (Becton, Dickinson and Company, Franklin Lakes, NJ) from symptomatic patients. Total Nucleic Acid (TNA) was extracted from using automated nucleic acid extraction on the QIAsymphony and the DSP Virus/Pathogen Mini Kit (QIAGEN).
Human autopsy tissue collection for RNA-seq analysis
The full methods of the patient sample collection from the autopsy patients are currently available in the Park et al. (2021). All autopsies are performed with consent of next of kin and permission for retention and research use of tissue. Autopsies were performed in a negative pressure room with protective equipment including N-95 masks; brain and bone were not obtained for safety reasons. All fresh tissues were procured prior to fixation and directly into Trizol for downstream RNA extraction. Tissues were collected from lung, liver, lymph nodes, kidney, and the heart as consent permitted. For GeoMx, RNAscope, trichrome and histology tissue sections were fixed in 10% neutral buffered formalin for 48 hours before processing and sectioning. These cases had a post-mortem interval of less than 48 hours. For bulk RNA-seq tissues, post-mortem intervals ranged from less than 24 hours to 72 hours (with 2 exceptions - one at 4 and one at 7 days - but passing RNA quality metrics) with an average of 2.5 days. All deceased patient remains were refrigerated at 4°C prior to autopsy performance.
Human urine sample collection
Urine was collected from patients and volunteers at the University of North Carolina at Chapel Hill. All patients provided informed consent prior to participation in IRB-approved research protocols (UNC IRB: 20-0822 [RHS] and 20-0792 [NMB]). Mid-stream urine of outpatients and non-critically ill patients was collected by the clean catch method. Urine of intubated critically ill patients was collected from a port on the Foley catheter. Urine was aliquoted into 5 mL aliquots and stored at −80°C. The following patient urine samples were obtained: N = 10 inpatient patients positive for COVID-19, N = 10 inpatient COVID-19 negative patients, N = 15 outpatient patients positive for COVID-19, N = 11 outpatient patients negative for COVID-19 (or healthy donors).
Urine aliquots were thawed, and microRNA was extracted from 1 mL per sample using Norgen Urine microRNA Purification Kit (Cat. 29000). Microalbumin and creatinine levels were assessed using Microalbumin 2-1 Combo strips (CLIAwaived Inc, cat# URS-2M).
Human peripheral blood mononuclear cells collection for ddPCR
Peripheral blood mononuclear cells (PBMC) were obtained from buffy coats (Hematology and Hemotherapy Center of the University of Campinas) from healthy donors provided Study was approved by the Brazilian Committee for Ethics in Human Studies (CAEE: 31622420.0.0000.5404). PBMCs were isolated as described in Codo et al. (2020).
Cell lines used for miR-2392 mimic experiments
Human SH-SY5Y cells were obtained from the ATCC and grown in Minimum Essential Medium (GIBCO) / 10% FBS (Invitrogen) /1% MEM Non Essential Amino Acids (GIBCO) / 1%GlutaMAX -l (GIBCO). Cells were plated in 3.5 cm dishes and incubated with miR-2392 or control lentivirus particles (MOI 1) for 48h. Cells were harvested and lysed in Trizol reagent and RNA was extracted following manufacturers protocol (Invitrogen).
COVID-19 hamster model
Male Syrian hamsters 6-8 weeks old were utilized for efficacy studies with anti-miR-2392 Nanoligomers treatment. Three hamsters were used for each experimental group for a total of 30 hamsters with 10 treatment groups. Hamsters were infected with 105 pfu of the WA01/2020 strain of SARS-CoV-2 passaged twice in Vero E6 cells from the original isolate obtained from BEI Resources. There were 5 major treatment groups (N = 6 per group) with two endpoints at day 3 or 7 post-viral challenge (N = 3 per endpoint). Groups 1 and 3 were given the Nanoligomer treatment by Intraperitoneal (IP) injection while groups 2 and 4 were given by Intranasal (IN) instillation under ketamine-xylazine anesthesia. Groups 1 and 2 were given single Nanoligomer treatment 24 hours before viral challenge. Groups 3 and 4 were given two doses of Nanoligomer at 24 hours before and 24 hours after viral challenge. The control group 5 was treated with PBS 24 hours prior to and 24 hours following viral challenge by IN instillation. A primary focus for the animal tests was to assess treatment efficacy to reduce viral infection in the pulmonary system. Thus, the Nanoligomers were administered directly to lung tissues via the IN route and the primary endpoints were specifically evaluated for oropharyngeal shedding of virus and lung tissue burden for the virus at necropsy. Injecting the Nanoligomer intravenously (IV) may have also provided a suitable route to target the pulmonary system, however there is concern that the blood filtering organs or uptake in the endothelium may have blunted delivery directly to the lungs.
Treatment efficacy was assessed in multiple ways: 1) Change in daily body weight, 2) oropharyngeal shedding of virus on days 1-3 from all groups post-challenge assayed by plaque assay on Vero E6 cells (PFU/swab), 3) tissue burden of the virus at necropsy on day 3 from 2 lung tubes and turbinates assayed by plaque assay (PFU/100mg), and 4) histopathologic scoring on lungs and turbinates from all hamsters; the histopathological score for individual tissues, inflammation score from the interstitial lung inflammation, and total histopathological scores/assessment was made.
The dose of anti-miR-2392 that was used was calculated to raise blood levels to 10 μM if it were given intravenously. The dose was estimated from the average IC50s determined from our in vitro viral screening experiments. The molecular weight of anti-miR-2392 is 15,804. Assuming that hamsters weigh 120 g and have 8% of body weight as blood, blood volume was approximately 0.01 l. The dose per hamster was 1.58 mg in a 100 μL volume from an anti-miR-2392 solution.
In vitro viral screening model
A549-ACE2 cells, gifted by Dr. Brad Rosenberg (MSSM), were maintained in DMEM (Quality Biological, Gaithersburg, MD; #112-014-101) + 10% Fetal Bovine Serum (GIBCO; #26140079) + 1% Penicillin-Streptomycin (Gemini Bio; #400-109). The day prior to treatment, 5,000 A549-ACE2 cells were plated per well in 96-well plates. MiR-2932 was diluted in duplicate in A549-ACE2 media to a starting concentration of 20 μM (Run 1) or 22 μM (Runs 2 and 3), and then an 8-point 1:2 dilution series was prepared. Media was removed from cells and 90 μL of each dilution was transferred to the cells. The plates were incubated for 2 hours at 37°C before being infected with an M.O.I. of 0.1 SARS-CoV-2 WA-1 (provided by Dr. Natalie Thornburg at the Centers for Disease Control and Prevention). Parallel plates were also run and left uninfected to monitor toxicity. Since Runs 2 and 3 were run simultaneously, a single toxicity plate was run for both. All plates were incubated at 37°C for 72 hours before being analyzed via Cell Titer Glo (Promega, Madison, WI; #G7573). Cell viability was compared to uninfected, untreated cells and infected, untreated cells.
Method details
Human peripheral blood mononuclear cell infection with SARS-CoV-2
Monocytes were isolated from PBMCs as described in Toniolo et al. (2015). Cells were infected with SARS-CoV-2 B and P1 lineages as described. B lineage virus was isolated from the second confirmed case in Brazil (GenBank: MT126808.1). P1 was obtained as described by Souza et al. (2021).
miRNA extraction for Droplet Digital PCR (ddPCR)
MiRNA extractions from serum were carried out using the QIAGEN miRNeasy serum/plasma kit (#217184). MiRNA extractions from urine samples were carried out using Norgen urine microRNA Purification Kit (Cat. 29000, Norgen Bioteck Corp. Thorold, ON, Canada). Quantitation of miRNA samples was done using a NanoDrop 2000 Spectrophotometer (ThermoFisher Scientific).
cDNA generation and ddPCR
First, cDNA was synthesized from miRNA samples using the QIAGEN miRCURY LNA RT Kit (Cat. 339340) using a concentration of 5ng/μl for the miRNA per sample. Next, samples were mixed with a 1:20 dilution of the generated cDNA with the BioRad QX200 ddPCR Evagreen Supermix (Cat. 1864034) and the appropriate miRNA primers from miRCURY LNA miRNA PCR Assays (QIAGEN). BioRad QX200 Automated Droplet Generator (Cat. 1864101) was used to create emulsion droplets. With the C1000 Touch Thermal Cycler with 96–Deep Well Reaction Module (Bio-Rad) the following PCR reaction was used for all the primers: 1 cycle 95°C for 5 min, 40 cycles of 95°C for 30 s and 58°C for 1 min (the annealing temperature can change depending on the primer), 1 cycle of 4°C for 5 min, and 1 cycle of 90°C for 5 min. Not all miRNA primers sets for ddPCR will have the same annealing temperature, so optimizing the annealing temperature is required for each primer set. Their respective annealing temperatures are found in Table S1. Finally, the QX200 Droplet Digital PCR System (Bio-Rad) quantified the amount of miRNA for each primer set per sample. QuantaSoft software (Bio-Rad) generated the data for each primer set and sample. The same threshold setting was used for all samples per primer set. The concentration (miRNA copies/μl) value generated by QuantaSoft was converted to miRNA copies/ng of serum. These values were used for all miRNA analysis. For all analysis the miRNA concentrations were log2(x+1) transformed to allow for easy comparison between miRNAs and samples.
Publicly available Bronchial Alveolar Lavage Fluid (BALF) COVID-19 RNA-sequencing data
Fastq files were downloaded from SRA (NCBI BioProject PRJNA605907 (Shen et al., 2020) and NCBI BioProject: PRJNA390194 (Ren et al., 2018)). Fastq data files were trimmed using TrimGalore v (0.6.4) with a quality cutoff of 30. Data were then aligned using STAR (v2.7.3) two pass mode to the Human reference genome (GRCh38 v99 downloaded 04-27-2020). Unaligned data were written to a fastq file, and then realigned to the GRCh38 reference genome using Bowtie 2 (v2.3.4.1), and output sam file converted to a bam file using samtools (v1.7). The resultant Bam files were merged, sorted, and read groups added using picard tools (v2.21.3).
Publicly available RNA-seq data: A549, Calu-3, NHBE, and COVID-19 lung biopsy
Raw RNA-seq read counts from the publication by Blanco-Melo et al. (2020) for the A549, Calu-3, and NHBE cell lines as well as post-mortem lung biopsies from two COVID-19 patients were downloaded from the Gene Expression Omnibus (series accession GSE147507).
RNA-seq of Nasopharyngeal Swab COVID-19 patient samples
RNA isolation and library preparation is fully described in Butler et al. (2021). Briefly, library preparation on the all nasopharyngeal swab samples’ total nucleic acid (TNA) were treated with DNase 1 (Zymo Research, Catalog # E1010). Post-DNase digested samples were then put into the NEBNext rRNA depletion v2 (Human/Mouse/Rat), Ultra II Directional RNA (10 ng), and Unique Dual Index Primer Pairs were used following the vendor protocols from New England Biolabs. Kits were supplied from a single manufacturer lot. Completed libraries were quantified by Qubit or equivalent and run on a Bioanalyzer or equivalent for size determination. Libraries were pooled and sent to the WCM Genomics Core or HudsonAlpha for final quantification by Qubit fluorometer (ThermoFisher Scientific), TapeStation 2200 (Agilent), and qRT-PCR using the Kapa Biosystems Illumina library quantification kit.
RNA-seq of COVID-19 autopsy tissue samples
RNA isolation and library preparation is fully described in Park et al. (2021). Briefly, autopsy tissues were collected from lung, liver, lymph nodes, kidney, and the heart and were placed directly into Trizol, homogenized and then snap frozen in liquid nitrogen. At least after 24 hours these tissue samples were then processed via standard protocols to isolate RNA. New York Genome Center RNA sequencing libraries were prepared using the KAPA Hyper Library Preparation Kit + RiboErase, HMR (Roche) in accordance with manufacturer’s recommendations. Briefly, 50-200ng of Total RNA were used for ribosomal depletion and fragmentation. Depleted RNA underwent first and second strand cDNA synthesis followed by adenylation, and ligation of unique dual indexed adapters. Libraries were amplified using 12 cycles of PCR and cleaned-up by magnetic bead purification. Final libraries were quantified using fluorescent-based assays including PicoGreen (Life Technologies) or Qubit Fluorometer (Invitrogen) and Fragment Analyzer (Advanced Analytics) and sequenced on a NovaSeq 6000 sequencer (v1 chemistry) with 2x150bp targeting 60M reads per sample.
miR-2392 mimic experiments in SH-SY5Y cells and RNA-seq
RNA was dissolved in nuclease free water and concentration determined spectrometrically at 260nm using a Biotek plate reader (Biotek). 500ng RNA was used as input for a whole transcriptome library preparation (ThermoFisher Total RNA). Libraries were quantified using a bioanalyzer chip reader (nanoDNA chips: Aglient Technologies) and diluted to 100 pM final concentration. Barcoded libraries were combined and use to seed a OneTouch bead templating reaction (OneTouch2). Cloned libraries were enriched and loaded on 540 Ion Torrent chips. Data were sequenced using the Ion Torrent RNA-seq workflow. Unaligned Bam files were converted to fatsq and aligned to the Grch 38 reference genome using STAR Two pass approach (Dobin et al., 2013) to create gene count tables as described in Overbey et al. (2021). The raw RNA-sequence data was deposited on SRA (https://www.ncbi.nlm.nih.gov/sra) with BioProject ID: PRJNA76339.
Anti-miR-2392 Nanoligomer inhibitor design and construction
The Nanoligomer platform was used to design Nanoligomer inhibitors, which are composed of a nanobiohydrd molecule based on antisense peptide nucleic acid (PNA) moiety conjugated to nanoparticle for improved delivery and membrane transport. The PNA moiety was chosen to be 15 bases long (in order to maximize both solubility and specificity), which yielded six potential target sequences within the 20-nucleotide mature human miR-2392. These potential targets were screened using FAST for solubility, self-complementing sequences, and off-targeting within the human genome (GCF_000001405.26) and SARS-CoV-2 viral genome (NC_045512). The antisense sequence complementing miR-2392 nucleotides 2 to 16 (TCTCACCCCCATCCT) was chosen in order to minimize off-targeting while maximizing coverage of the miR-2392 seed region. The Nanoligomer was synthesized (with an N-terminal histidine tag and a 2-(2-(2-aminoethoxy)ethoxy)acetic acid linker) on an Apex 396 peptide synthesizer (AAPPTec, LLC) with solid-phase Fmoc chemistry. Fmoc-PNA monomers were obtained from PolyOrg Inc., with A, C, and G monomers protected with Bhoc groups. Following synthesis, the peptides were conjugated with nanoparticles and purified via size-exclusion filtration. Conjugation and concentration of the purified solution was monitored through measurement of absorbance at 260 nm (for detection of PNA) and 400 nm (for quantification of nanoparticles).
Quantification and statistical analysis
Analysis of BALF RNA-seq data
Bam files were imported into Partek Genome Studio v7.0, and gene expression values quantified versus the Grch38 reference annotation guide (Ensembl v99). Samples with fewer than 2 million aligned reads were excluded from further analysis. Genes with fewer than 10 reads in 25% of samples were excluded, and differential gene expression determined using ANOVA with infection status as contrast. Differentially expressed gene files were used in GSEA and IPA to determine biological significance and pathways being regulated.
Analysis of Nasopharyngeal Swab RNA-seq data
The nasopharyngeal swab samples were analyzed comparing COVID-19 viral infection to the negative patients and was as previously described in Butler et al. (2021) and the DESeq2 (Love et al., 2014) was utilized to generate the differential expression data. Heatmaps were displayed using pheatmap (Kolde, 2015). Volcano plots were made use R program EnhancedVolcano (Blighe et al., 2018).
Analysis of Autopsy RNA-seq data
The full methods of the analysis from the autopsy patients is currently available in Park et al. (2021). Briefly, RNA-seq data was processed through the nf-core/rnaseq pipeline (Ewels et al., 2020). This workflow involved adaptor trimming using Trim Galore! (https://github.com/FelixKrueger/TrimGalore), read alignment with STAR (Dobin et al., 2013), gene quantification with Salmon (Patro et al., 2017), duplicate read marking with Picard MarkDuplicates (https://github.com/broadinstitute/picard), and transcript quantification with StringTie (Kovaka et al., 2019). Other quality control measures included RSeQC, Qualimap, and dupRadar. Alignment was performed using the GRCh38 build native to nf-core and annotation was performed using Gencode Human Release 33 (GRCH38.p13). FeatureCounts reads were normalized using variance-stabilizing transform (vst) in DESeq2 package in R for visualization purposes in log-scale (Love et al., 2014). Differential expression of genes were calculated by DESeq2. Differential expression comparisons were done as either COVID+ cases versus COVID- controls for each tissue specifically, correcting for sequencing batches with a covariate where applicable, or pairwise comparison of viral levels from the lung as determined by nCounter data. Volcano plots were made use R program EnhancedVolcano (Blighe et al., 2018).
Analysis Combining Autopsy and Nasopharyngeal Swab RNA-seq data
To combine the results from the autopsy and nasopharyngeal swab RNA-seq data, we utilized the t-score values from the DESeq2 analysis. Heatmaps were displayed using pheatmap (Kolde, 2015).
Gene Set Enrichment Analysis (GSEA)
For pathway analysis on the miR-2392 targets (Figure 3) we utilized ShinyGO (Ge et al., 2020) to determine the significantly regulated pathways for each main cluster in the heatmap. The clustering was determined through k-mean statistics.
For pathway analysis on the miR-2392 mimic RNA-seq data, we utilized fast Gene Set Enrichment Analysis (fGSEA) (Korotkevich et al., 2021). Pathway analysis was done comparing miR-2392 mimics to all controls and the ranked list of genes were defined by the t-score statistics. The statistical significance was determined by 1000 permutations of the genesets (Subramanian et al., 2005).
Analysis of proteomic and transcriptomic blood datasets from COVID-19 patients
For the analysis of the miR-2392 targets in the blood tissue, we downloaded whole blood transcriptome data and plasma proteome data from The COVIDome Explorer Researcher Portal (Sullivan et al., 2021). For Transcriptome data we used the following filters: Category “Effect of COVID-19 status,” Platform “Blood,” Statistical test “Student’s t-test,” Adjustment method “none,” Sex “male” and “female,” Age Group “All.” For Proteome data we used the following filters: Category “Effect of COVID-19 status,” Platform “SOMAscan,” Statistical test “Student’s t-test,” Adjustment method “none,” Sex “male” and “female,” Age Group “All.” We created the list of the intersecting genes from both datasets. We analyzed the list using RStudio Desktop 1.3.1093 (R Studio Team, 2020). RStudio: Integrated Development Environment for R. RStudio, PBC, Boston, MA URL https://www.rstudio.com/), and visualized data using ggplot2 version 3.3.2 (Wickham, 2016) and ggrepel version 0.8.2 (https://ggrepel.slowkow.com/).
Analysis of Monocyte RNA-seq data
The monocyte COVID-19 RNA-Seq data, published under the accession GSE159678 (Rother et al., 2020), was downloaded from SRA and gene expression was quantified using Salmon’s selective alignment approach (Srivastava et al., 2020). The RNA-Seq processing pipeline was implemented using pyrpipe (Singh et al., 2021) (https://github.com/urmi-21/pyrpipe/tree/master/case_studies/Covid_RNA-Seq). Exploratory data analysis and differential expression analysis were performed using MetaOmGraph (Singh et al., 2020).
Analysis of A549, Calu-3, NHBE, and COVID lung biopsy data
Each data series was normalized and filtered for low-expressed genes (counts < 1). Cell culture samples treated with SARS-CoV-2 were compared to untreated controls and COVID-19-positive patient samples were compared to healthy lung biopsies. Differentially expressed genes were determined from the R-program Limma-Voom (Ritchie et al., 2015) using a linear model with weighted least-squares for each gene and the false discovery rate adjusted p values were calculated.
Analysis of miR-2392 mimic RNA-seq data
Differential gene expression was determined using LIMMA-voom (Ritchie et al., 2015). Data were filtered to ensure data contained at least 5 million aligned reads, and average gene counts of > 10. Cell treatments we used as contrasts for differentially expressed gene calculations. These results were then uploaded to GSEA for further analysis. (R script in Supplemental information)
Conservation of miR-2392 between species
To determined conservation of miR-2392 among different species we utilized UCSC Genome Browser (Kent et al., 2002). Hsa-miR-2392 was entered as input and target species were chosen to include common models for SARS-CoV-2 in vivo studies (e.g., mice, ferrets, and hamsters) as well as primates and other animals to provide a wide spectrum of species to observe conservation of miR-2392. The USCS Genome Browser provides the host gene for miR-2392 (i.e., MEG3) and redirects to GTEx (GTEx Consortium, 2020) to provide a plot of MEG3 levels based on RNA-seq data on normal tissues.
Mapping miR-2392 sequence to SARS-CoV-2 sequences
To explore potential binding sites for miR-2392 we used miRanda software (Enright et al., 2003) to identify all potential binding sites with respect to the SARS-CoV-2 reference genome (Wuhan-Hu-1; NC045512.2) and representative genomes from lineages of concern. The lineages of concern were selected from Global Initiative on Sharing All Influenza Data (GISAID) with each lineage being represented by 14 recent genomes.
In silico predictions of genes from miRNAs
Through the use of a Cytoscape (Shannon et al., 2003) plug-in called ClueGo/CluePedia (Bindea et al., 2009), we were able to predict genes targeted by the miRNAs determined. This involved entering all miRNAs in ClueGo and allowing the software to determine the top 50 genes that were significantly regulated and connected to the miRNAs. The predictions only reflect the functions that will be regulated by the miRNAs and do not show whether the function will be activated or inhibited. Lastly, miRNet 2.0 was utilized to predict the diseases and pathways that are associated with the miRNAs (Chang et al., 2020). This was plotted as a dot plot utilizing the R-program ggplot2 (v3.2.1) (Wickham, 2016).
ddPCR analysis of miRNA levels in patient samples
First, we normalized the amount of each miRNA measured per body location (nasal, serum, and urine) using the general logarithm transformation. We compared miRNA levels in samples from patients either positive or negative for SARS-nCoV-2 using the Student’s t test (unadjusted) as well as controlling for sex and age using least-squares adjustment. Next, we generated receiver operating characteristic curves from each body location to show the performance of a classification model (SARS-nCoV-2 positive versus negative) at all classification thresholds using the absolute, non-transformed levels (miRNA copies per 5 ng RNA). Finally, we performed a subanalysis on each location to compare the variance of each miRNA in SARS-nCoV-2 negative patients compared to other patient groups. For serum and nasal samples, 1-way ANOVA was used to identify variation associated with the patient classification. For urine samples, 2-way ANOVA was used with location (outpatient versus inpatient) and SARS-nCoV-2 positivity as the main factors. If the ANOVA yielded a p < 0.05, Dunnett’s post-test was used to compare subgroup means to the negative patient sample mean.
Computational drug repositioning model
Using the SM2miR database (Lui et al., 2007), we assembled an n × m binary matrix (X) containing 3,593 associations between small molecules (n = 213, rows) and miRNAs (m = 1,519, columns). Each matrix entry (X ij) was assigned a value of 1 where a small molecule is known to be associated to miRNA, and was 0 otherwise. The chemical notation as a simplified molecular input line entry system (SMILES) was obtained for each small molecule from PubChem. We then calculated the 2D Tanimoto chemical similarity between pairs of small molecules using the MACCS key binary fingerprints with RDKit (RDKit: Open-source cheminformatics; http://www.rdkit.org). Similarly, for each miRNA, we obtained its sequence from miRbase (Kozomara et al., 2019) and computed sequence similarity between miRNAs as the score of their Needleman-Wunsch alignment. We used the binary matrix, together with the chemical and sequence similarities, as input to our state-of-the-art drug target prediction model to predict missing associations in X (Galeano et al., 2021). The model parameters where: p = 1/2, βChem = 1, and αseq = 0. To assess the prediction performance of the model, we performed ten-fold cross-validation simulations.
Consortia
The UNC COVID-19 Pathobiology Consortium comprises Shannon M. Wallet, Robert Maile, Matthew C. Wolfgang, Robert S. Hagan, Jason R. Mock, Natalie M. Bowman, Jose L. Torres-Castillo, Miriya K. Love, Suzanne L. Meinig, Will Lovell, Colleen Rice, Olivia Mitchem, Dominique Burgess, Jessica Suggs, and Jordan Jacobs.
Acknowledgments
This work was supported by supplemental funds for COVID-19 research from Translational Research Institute of Space Health through NASA Cooperative Agreement NNX16AO69A (T-0404 to A.B. and T-0406 to A.C.) and further funding from KBR, Inc. (to A.B.). This work used resources, services, and support provided via the COVID-19 HPC Consortium (https://covid19-hpc-consortium.org/), specifically by the NASA High-End Computing (HEC) Program through the NASA Advanced Supercomputing (NAS) Division at Ames Research Center (to A.B.); DOD W81XWH-21-1-0128 (to D.C.W.); NIGMS P20 GM1009005 (to N.A.-B.); Individual National Research Service award F32-AI147587 (to J.M.D.); NIH/NHBLI K08 HL143271 and NIH/NHLBI R03 HL155249 (to R.S.H.); NIH/NCI U54 CA260543 (supporting R.S.H., N.M.B., and M.C.W.); and NSF 1956233 (to R.M.). A.P. was supported by Biotechnology and Biological Sciences Research Council (https://bbsrc.ukri.org/) grants BB/K004131/1, BB/F00964X/1, and BB/M025047/1; Consejo Nacional de Ciencia y Tecnología Paraguay (CONACyT) (https://www.conacyt.gov.py/) grants 14-INV-088 and PINV15-315; and National Science Foundation Advances in Bio Informatics (https://www.nsf.gov/) grant 1660648. E.S.W. was supported by NSF IOS 1546858. D.G. and A.P. were supported by CONACYT grant PINV20-337 and the Fundação Getulio Vargas. N.M.B. was supported by the National Center for Advancing Translational Sciences (NCATS) and NIH UL1TR002489, 2KR1272005, and 550KR242003. P.M.M.-V. was supported by FAPESP grants 20/04579-7 and 16/18031-8; Fundo de Apoio ao Ensino, Pesquisa e Extensão (FAEPEX); Unicamp grant 2274/20; and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brazil (CAPES) (finance code 001). Thanks to Dr. Richard Bowen for his assistance with the hamster studies and Dr. Matthew Frieman for his assistance with the in vitro A549 SARS-CoV-2 studies. The graphical abstract was created with BioRender.com and partially adapted from “Bevacizumab: Potential Repurposed Drug Candidate for COVID-19” retrieved from https://app.biorender.com/biorender-templates.
Author contributions
Conceptualization: A.B.; methodology: A.B.; formal analysis: A.B., R.M., C.V., D.T., F.J.E., C. Meydan, C. Mozsary, J.C.S., J.T.M., J.M.D., D.G., U.S., E.S.W., A.S.-B., J.F., V.Z., N.S., and T.J.T.; investigation: A.B., C.V., R.M., C.E.M., A.C., P.N., S.L.M., A.Y., A.N.P., T.R.A., P.M.M.-V., and G.G.D.; sample collection: M.M.S., M.C.W., R.S.H., N.M.B., UNC COVID-19 Pathobiology Consortium, A.D.H., J.C., P.M.M.-V., and G.G.D.; resources: A.B., R.M., C.E.M., A.C., and M.C.W.; writing – original draft: A.B. and J.T.M.; writing – review & editing: A.B., J.T.M., F.J.E., R.J.G., W.P., M.M.S., J.M.D., J.W.G., D.C.W., S.S., S.B.B., V.Z., E.S.W., S.V.C., N.A.-B., A.P., D.G., P.M.C., M.R.E., J.C.S., S.A., A.C., R.M., N.S., T.J.T., B.C., L.N.S., P.G., M.C.W., and P.M.M.-V.; visualization: A.B., J.C.S., F.J.E., D.G., N.S., and V.Z.; supervision: A.B.; funding acquisition: A.B.
Declaration of interests
A.C., P.N., S.V.C., and A.B. have a provisional patent based on the antiviral discovery and design. A.C., P.N., and S.S. are part of the company (Sachi Bioworks Inc.) that has filed a patent on the Nanoligomer technology.
Published: September 30, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109839.
Contributor Information
UNC COVID-19 Pathobiology Consortium:
Shannon M. Wallet, Robert Maile, Matthew C. Wolfgang, Robert S. Hagan, Jason R. Mock, Natalie M. Bowman, Jose L. Torres-Castillo, Miriya K. Love, Suzanne L. Meinig, Will Lovell, Colleen Rice, Olivia Mitchem, Dominique Burgess, Jessica Suggs, and Jordan Jacobs
Supplemental information
Data and code availability
The published article includes all datasets generated and analyzed during this study. Processed bulk RNA-seq data for the human related data from the nasopharyngeal and autopsy data is available online with dbGaP Study Accession number: phs002258.v1.p1 and online here at: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002258.v1.p1 and also https://covidgenes.weill.cornell.edu/. The RNA-sequence data related for the miR-2392 mimic experiments is deposited on SRA (https://www.ncbi.nlm.nih.gov/sra) with BioProject ID: PRJNA76339.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abdelrahman Z., Liu Q., Jiang S., Li M., Sun Q., Zhang Y., Wang X. Evaluation of the Current Therapeutic Approaches for COVID-19: A Systematic Review and a Meta-analysis. Front. Pharmacol. 2021;12:607408. doi: 10.3389/fphar.2021.607408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardestani A., Azizi Z. Targeting glucose metabolism for treatment of COVID-19. Signal Transduct. Target. Ther. 2021;6:112. doi: 10.1038/s41392-021-00532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai A., Konno M., Ozaki M., Otsuka C., Vecchione A., Arai T., Kitagawa T., Ofusa K., Yabumoto M., Hirotsu T., et al. COVID-19 Drug Discovery Using Intensive Approaches. Int. J. Mol. Sci. 2020;21:2839. doi: 10.3390/ijms21082839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aykin-Burns N., Franklin E.A., Ercal N. Effects of N-acetylcysteine on lead-exposed PC-12 cells. Arch. Environ. Contam. Toxicol. 2005;49:119–123. doi: 10.1007/s00244-004-0025-0. [DOI] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blighe K., Rana S., Lewis M. 2018. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling.https://github.com/kevinblighe/EnhancedVolcano [Google Scholar]
- Butler D., Mozsary C., Meydan C., Foox J., Rosiene J., Shaiber A., Danko D., Afshinnekoo E., MacKay M., Sedlazeck F.J., et al. Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Nat. Commun. 2021;12:1660. doi: 10.1038/s41467-021-21361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carboni E., Carta A.R., Carboni E. Can pioglitazone be potentially useful therapeutically in treating patients with COVID-19? Med. Hypotheses. 2020;140:109776. doi: 10.1016/j.mehy.2020.109776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carfì A., Bernabei R., Landi F., Gemelli Against COVID-19 Post-Acute Care Study Group Persistent Symptoms in Patients After Acute COVID-19. JAMA. 2020;324:603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrico C., Meyer J.G., He W., Gibson B.W., Verdin E. The Mitochondrial Acylome Emerges: Proteomics, Regulation by Sirtuins, and Metabolic and Disease Implications. Cell Metab. 2018;27:497–512. doi: 10.1016/j.cmet.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centa A., Fonseca A.S., Ferreira S.G.D.S., Azevedo M.L.V., Vaz de Paula C.B., Nagashima S., Machado-Souza C., Miggiolaro A.F.R.D.S., Baena C.P., de Noronha L., Cavalli L.R. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020;320:L405–L412. doi: 10.1152/ajplung.00457.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Zhou G., Soufan O., Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48(W1):W244–W251. doi: 10.1093/nar/gkaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Yao D., Li Y., Chen H., He C., Ding N., Lu Y., Ou T., Zhao S., Li L., Long F. Serum microRNA expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. Int. J. Mol. Med. 2013;32:557–567. doi: 10.3892/ijmm.2013.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codo A.C., Davanzo G.G., Monteiro L.B., de Souza G.F., Muraro S.P., Virgilio-da-Silva J.V., Prodonoff J.S., Carregari V.C., de Biagi Junior C.A.O., Crunfli F., et al. Elevated Glucose Levels Favor SARS-CoV-2 Infection and Monocyte Response through a HIF-1α/Glycolysis-Dependent Axis. Cell Metab. 2020;32:437–446.e5. doi: 10.1016/j.cmet.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli G., Latronico M.V., Dorn G.W., 2nd microRNAs in heart disease: putative novel therapeutic targets? Eur. Heart J. 2010;31:649–658. doi: 10.1093/eurheartj/ehp573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey L.L., Worne C.L., Glover J.L., Lane T.E., O’Connell R.M. MicroRNA-155 enhances T cell trafficking and antiviral effector function in a model of coronavirus-induced neurologic disease. J. Neuroinflammation. 2016;13:240. doi: 10.1186/s12974-016-0699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H., Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- Edmonds C.E., Zuckerman S.P., Conant E.F. Management of Unilateral Axillary Lymphadenopathy Detected on Breast MRI in the Era of COVID-19 Vaccination. AJR Am. J. Roentgenol. 2021;217:831–834. doi: 10.2214/AJR.21.25604. [DOI] [PubMed] [Google Scholar]
- Eller K.A., Aunins T.R., Courtney C.M., Campos J.K., Otoupal P.B., Erickson K.E., Madinger N.E., Chatterjee A. Facile accelerated specific therapeutic (FAST) platform develops antisense therapies to counter multidrug-resistant bacteria. Commun. Biol. 2021;4:331. doi: 10.1038/s42003-021-01856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto Y., Takagi R., Naito Y., Kiniwa T., Tanaka Y., Hamada-Tsutsumi S., Kawano M., Matsushita S., Ochiya T., Miyajima A. Identification of the novel 3′ UTR sequences of human IL-21 mRNA as potential targets of miRNAs. Sci. Rep. 2017;7:7780. doi: 10.1038/s41598-017-07853-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P.A., Peltzer A., Fillinger S., Patel H., Alneberg J., Wilm A., Garcia M.U., Di Tommaso P., Nahnsen S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020;38:276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- Fan S., Tian T., Chen W., Lv X., Lei X., Zhang H., Sun S., Cai L., Pan G., He L., et al. Mitochondrial miRNA Determines Chemoresistance by Reprogramming Metabolism and Regulating Mitochondrial Transcription. Cancer Res. 2019;79:1069–1084. doi: 10.1158/0008-5472.CAN-18-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeano D., Noto S., Jimenez R., Paccanaro A. Interpretable Drug Target Predictions using Self-Expressiveness. bioRxiv. 2021 doi: 10.1101/2021.03.01.433365. [DOI] [Google Scholar]
- Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharebaghi R., Heidary F., Moradi M., Parvizi M. Metronidazole; a Potential Novel Addition to the COVID-19 Treatment Regimen. Arch. Acad. Emerg. Med. 2020;8:e40. [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., Arnthorsson A.O., Helgason D., Bjarnadottir K., Ingvarsson R.F., et al. Humoral Immune Response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021;19:4101–4109. doi: 10.1016/j.csbj.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmat N., Asadzadeh Z., Ahangar N.K., Alemohammad H., Najafzadeh B., Derakhshani A., Baghbanzadeh A., Baghi H.B., Javadrashid D., Najafi S., et al. The roles of signaling pathways in SARS-CoV-2 infection; lessons learned from SARS-CoV and MERS-CoV. Arch. Virol. 2021;166:675–696. doi: 10.1007/s00705-021-04958-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J., Mori V., Bates J.H.T., Suki B. Modeling lung perfusion abnormalities to explain early COVID-19 hypoxemia. Nat. Commun. 2020;11:4883. doi: 10.1038/s41467-020-18672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho W.Z., Douglas S.D. Glutathione and N-acetylcysteine suppression of human immunodeficiency virus replication in human monocyte/macrophages in vitro. AIDS Res. Hum. Retroviruses. 1992;8:1249–1253. doi: 10.1089/aid.1992.8.1249. [DOI] [PubMed] [Google Scholar]
- Hou X., Liang Y., Chen J., Wei Y., Zeng P., Wang L., Lu C., Diao H. Expression Profiling of Cellular MicroRNA in Asymptomatic HBsAg Carriers and Chronic Hepatitis B Patients. BioMed Res. Int. 2017;2017:6484835. doi: 10.1155/2017/6484835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Li Z., Lan Y., Guan J., Zhao K., Chu D., Fan G., Guo Y., Gao F., He W. MiR-10a-5p-Mediated Syndecan 1 Suppression Restricts Porcine Hemagglutinating Encephalomyelitis Virus Replication. Front. Microbiol. 2020;11:105. doi: 10.3389/fmicb.2020.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultström M., Lipcsey M., Wallin E., Larsson I.M., Larsson A., Frithiof R. Severe acute kidney injury associated with progression of chronic kidney disease after critical COVID-19. Crit. Care. 2021;25:37. doi: 10.1186/s13054-021-03461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.S., Islam A.B.M.M.K. Viral miRNAs confer survival in host cells by targeting apoptosis related host genes. Inform. Med. Unlocked. 2021;22:100501. [Google Scholar]
- Jacobs L.G., Gourna Paleoudis E., Lesky-Di Bari D., Nyirenda T., Friedman T., Gupta A., Rasouli L., Zetkulic M., Balani B., Ogedegbe C., et al. Persistence of symptoms and quality of life at 35 days after hospitalization for COVID-19 infection. PLoS ONE. 2020;15:e0243882. doi: 10.1371/journal.pone.0243882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Jia D., Koonce N.A., Griffin R.J., Jackson C., Corry P.M. Prevention and mitigation of acute death of mice after abdominal irradiation by the antioxidant N-acetyl-cysteine (NAC) Radiat. Res. 2010;173:579–589. doi: 10.1667/RR2030.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., Li M., Wang G., Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen M.D., Irving A., Montagutelli X., Tate M.D., Rudloff I., Nold M.F., Hansbro N.G., Kim R.Y., Donovan C., Liu G., et al. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 2020;13:877–891. doi: 10.1038/s41385-020-00340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. Comprehensive R Archive Network; 2015. pheatmap: Pretty heatmaps. [Google Scholar]
- Korotkevich G., Sukhov V., Budin N., Shpak B., Artyomov M.N., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2021 doi: 10.1101/060012. [DOI] [Google Scholar]
- Koumpa F.S., Forde C.T., Manjaly J.G. Sudden irreversible hearing loss post COVID-19. BMJ Case Rep. 2020;13:e238419. doi: 10.1136/bcr-2020-238419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S., Zimin A.V., Pertea G.M., Razaghi R., Salzberg S.L., Pertea M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019;20:278. doi: 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski P.K., Maj J., Szepietowski J.C. Cutaneous Hyperaesthesia in SARS-CoV-2 Infection: Rare but not Unique Clinical Manifestation. Acta Derm. Venereol. 2021;101:adv00366. doi: 10.2340/00015555-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.P., Subramanian A., Ross K.N., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lawrie C.H., Gal S., Dunlop H.M., Pushkaran B., Liggins A.P., Pulford K., Banham A.H., Pezzella F., Boultwood J., Wainscoat J.S., et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582:469. doi: 10.1038/d41586-020-01824-5. [DOI] [PubMed] [Google Scholar]
- Li J., Li T., Lu Y., Shen G., Guo H., Wu J., Lei C., Du F., Zhou F., Zhao X., et al. MiR-2392 suppresses metastasis and epithelial-mesenchymal transition by targeting MAML3 and WHSC1 in gastric cancer. FASEB J. 2017;31:3774–3786. doi: 10.1096/fj.201601140RR. [DOI] [PubMed] [Google Scholar]
- Li K., Chen G., Hou H., Liao Q., Chen J., Bai H., Lee S., Wang C., Li H., Cheng L., Ai J. Analysis of sex hormones and menstruation in COVID-19 women of child-bearing age. Reprod. Biomed. Online. 2021;42:260–267. doi: 10.1016/j.rbmo.2020.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang S., Meng F., Wang J., Zhang Y., Dai E., Yu X., Li X., Jiang W. SM2miR: a database of the experimentally validated small molecules’ effects on microRNA expression. Bioinformatics. 2013;29:409–411. doi: 10.1093/bioinformatics/bts698. [DOI] [PubMed] [Google Scholar]
- Lott M.T., Leipzig J.N., Derbeneva O., Xie H.M., Chalkia D., Sarmady M., Procaccio V., Wallace D.C. mtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr. Protoc. Bioinformatics. 2013;44:1.23.1–1.23.26. doi: 10.1002/0471250953.bi0123s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza M.J., McCall C.E., Li L., Isaacs W.B., Xu J., Chang B.L. Assembly of inflammation-related genes for pathway-focused genetic analysis. PLoS ONE. 2007;2:e1035. doi: 10.1371/journal.pone.0001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui W.O., Pourmand N., Patterson B.K., Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- Ma Y., Wang C., Xue M., Fu F., Zhang X., Li L., Yin L., Xu W., Feng L., Liu P. The Coronavirus Transmissible Gastroenteritis Virus Evades the Type I Interferon Response through IRE1α-Mediated Manipulation of the MicroRNA miR-30a-5p/SOCS1/3 Axis. J. Virol. 2018;92:e00728-18. doi: 10.1128/JVI.00728-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A., Mason G.F. A sobering addition to the literature on COVID-19 and the brain. J. Clin. Invest. 2021;131:148376. doi: 10.1172/JCI148376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Z.R., Razaq Z., Siff M., Sheikh S. COVID-19 Presenting as Banti’s Syndrome. Cureus. 2020;12:e9096. doi: 10.7759/cureus.9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nersisyan S., Engibaryan N., Gorbonos A., Kirdey K., Makhonin A., Tonevitsky A. Potential role of cellular miRNAs in coronavirus-host interplay. PeerJ. 2020;8:e9994. doi: 10.7717/peerj.9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiga M., Wang D.W., Han Y., Lewis D.B., Wu J.C. COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 2020;17:543–558. doi: 10.1038/s41569-020-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner S.A., Pillich R.T., McKenna N.J. Consensus transcriptional regulatory networks of coronavirus-infected human cells. Sci. Data. 2020;7:314. doi: 10.1038/s41597-020-00628-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottosen S., Parsley T.B., Yang L., Zeh K., van Doorn L.J., van der Veer E., Raney A.K., Hodges M.R., Patick A.K. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob. Agents Chemother. 2015;59:599–608. doi: 10.1128/AAC.04220-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbey E.G., Saravia-Butler A.M., Zhang Z., Rathi K.S., Fogle H., da Silveira W.A., Barker R.J., Bass J.J., Beheshti A., Berrios D.C., et al. NASA GeneLab RNA-seq consensus pipeline: standardized processing of short-read RNA-seq data. iScience. 2021;24:102361. doi: 10.1016/j.isci.2021.102361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Foox J., Hether T., Danko D., Warren S., Kim Y., Reeves J., Butler D.J., Mozsary C., Rosiene J., et al. Systemic Tissue and Cellular Disruption from SARS-CoV-2 Infection revealed in COVID-19 Autopsies and Spatial Omics Tissue Maps. bioRxiv. 2021 doi: 10.1101/2021.03.08.434433. [DOI] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe M., Wittig S., Jurida L., Bartkuhn M., Wilhelm J., Müller H., Beuerlein K., Karl N., Bhuju S., Ziebuhr J., et al. The NF-κB-dependent and -independent transcriptome and chromatin landscapes of human coronavirus 229E-infected cells. PLoS Pathog. 2017;13:e1006286. doi: 10.1371/journal.ppat.1006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portincasa P., Krawczyk M., Machill A., Lammert F., Di Ciaula A. Hepatic consequences of COVID-19 infection. Lapping or biting? Eur. J. Intern. Med. 2020;77:18–24. doi: 10.1016/j.ejim.2020.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T.J., Goodman R.P., Grabarek Z., Haas M.E., Hung W.H.W., et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49(D1):D1541–D1547. doi: 10.1093/nar/gkaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L., Zhang R., Rao J., Xiao Y., Zhang Z., Yang B., Cao D., Zhong H., Ning P., Shang Y., et al. Transcriptionally Active Lung Microbiome and Its Association with Bacterial Biomass and Host Inflammatory Status. mSystems. 2018;3:e00199-18. doi: 10.1128/mSystems.00199-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M., Soler Y., Perry M., Reynolds J.L., El-Hage N. Impact of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in the Nervous System: Implications of COVID-19 in Neurodegeneration. Front. Neurol. 2020;11:583459. doi: 10.3389/fneur.2020.583459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi R., Talarico M., Coppi F., Boriani G. Protective role of statins in COVID 19 patients: importance of pharmacokinetic characteristics rather than intensity of action. Intern. Emerg. Med. 2020;15:1573–1576. doi: 10.1007/s11739-020-02504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother N., Yanginlar C., Lindeboom R.G.H., Bekkering S., van Leent M.M.T., Buijsers B., Jonkman I., de Graaf M., Baltissen M., Lamers L.A., et al. Hydroxychloroquine Inhibits the Trained Innate Immune Response to Interferons. Cell Rep. Med. 2020;1:100146. doi: 10.1016/j.xcrm.2020.100146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- Saçar Demirci M.D., Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8:e9369. doi: 10.7717/peerj.9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar R., Satish D., Gupta D. Identification of Novel SARS-CoV-2 Drug Targets by Host MicroRNAs and Transcription Factors Co-regulatory Interaction Network Analysis. Front. Genet. 2020;11:571274. doi: 10.3389/fgene.2020.571274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schult P., Roth H., Adams R.L., Mas C., Imbert L., Orlik C., Ruggieri A., Pyle A.M., Lohmann V. microRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat. Commun. 2018;9:2613. doi: 10.1038/s41467-018-05053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self W.H., Tenforde M.W., Stubblefield W.B., Feldstein L.R., Steingrub J.S., Shapiro N.I., Ginde A.A., Prekker M.E., Brown S.M., Peltan I.D., et al. CDC COVID-19 Response Team. IVY Network Decline in SARS-CoV-2 Antibodies After Mild Infection Among Frontline Health Care Personnel in a Multistate Hospital Network—12 States, April–August 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1762–1766. doi: 10.15585/mmwr.mm6947a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., Zhou Z., Yang J., Zhong J., Yang D., et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data—from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U., Hur M., Dorman K., Wurtele E.S. MetaOmGraph: a workbench for interactive exploratory data analysis of large expression datasets. Nucleic Acids Res. 2020;48:e23. doi: 10.1093/nar/gkz1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U., Li J., Seetharam A., Wurtele E.S. pyrpipe: a python package for RNA-Seq workflows. bioRxiv. 2021 doi: 10.1093/nargab/lqab049. 2020.2003.2004.925818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirin D.A., Ozcelik F. The relationship between COVID-19 and the dental damage stage determined by radiological examination. Oral Radiol. 2021;37:600–609. doi: 10.1007/s11282-020-00497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirota M., Dudley J.T., Kim J., Chiang A.P., Morgan A.A., Sweet-Cordero A., Sage J., Butte A.J. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci. Transl. Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza W.M., Amorim M.R., Sesti-Costa R., Coimbra L.D., Brunetti N.S., Toledo-Teixeira D.A., de Souza G.F., Muraro S.P., Parise P.L., Barbosa P.P., et al. Neutralisation of SARS-CoV-2 lineage P.1 by antibodies elicited through natural SARS-CoV-2 infection or vaccination with an inactivated SARS-CoV-2 vaccine: an immunological study. Lancet Microbe. 2021 doi: 10.1016/S2666-5247(21)00129-4. Published online July 8, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Malik L., Sarkar H., Zakeri M., Almodaresi F., Soneson C., Love M.I., Kingsford C., Patro R. Alignment and mapping methodology influence transcript abundance estimation. Genome Biol. 2020;21:239. doi: 10.1186/s13059-020-02151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukalov A., Girault V., Grass V., Karayel O., Bergant V., Urban C., Haas D.A., Huang Y., Oubraham L., Wang A., et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246–252. doi: 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]
- Su M., Chen Y., Qi S., Shi D., Feng L., Sun D. A Mini-Review on Cell Cycle Regulation of Coronavirus Infection. Front. Vet. Sci. 2020;7:586826. doi: 10.3389/fvets.2020.586826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K.D., Galbraith M.D., Kinning K.T., Bartsch K.W., Levinsky N.C., Araya P., Smith K.P., Granrath R.E., Shaw J.R., Baxter R.M., et al. The COVIDome Explorer Researcher Portal. Cell Rep. 2021;36:109527. doi: 10.1016/j.celrep.2021.109527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Gao Y., Li Z., Miao Y., Huang Z., Liu X., Xie L., Li H., Wen W., Zheng Y., Su W. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin. Transl. Med. 2020;10:e200. doi: 10.1002/ctm2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodori L., Sestili P., Madiai V., Coppari S., Fraternale D., Rocchi M.B.L., Ramakrishna S., Albertini M.C. MicroRNAs Bioinformatics Analyses Identifying HDAC Pathway as a Putative Target for Existing Anti-COVID-19 Therapeutics. Front. Pharmacol. 2020;11:582003. doi: 10.3389/fphar.2020.582003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault P.A., Huys A., Amador-Cañizares Y., Gailius J.E., Pinel D.E., Wilson J.A. Regulation of Hepatitis C Virus Genome Replication by Xrn1 and MicroRNA-122 Binding to Individual Sites in the 5′ Untranslated Region. J. Virol. 2015;89:6294–6311. doi: 10.1128/JVI.03631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toniolo P.A., Liu S., Yeh J.E., Moraes-Vieira P.M., Walker S.R., Vafaizadeh V., Barbuto J.A., Frank D.A. Inhibiting STAT5 by the BET bromodomain inhibitor JQ1 disrupts human dendritic cell maturation. J. Immunol. 2015;194:3180–3190. doi: 10.4049/jimmunol.1401635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribolet L., Kerr E., Cowled C., Bean A.G.D., Stewart C.R., Dearnley M., Farr R.J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020;11:1197. doi: 10.3389/fmicb.2020.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobaugh D.W., Klimstra W.B. MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Mol. Med. 2017;23:80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ree M.H., de Vree J.M., Stelma F., Willemse S., van der Valk M., Rietdijk S., Molenkamp R., Schinkel J., van Nuenen A.C., Beuers U., et al. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: a phase 1B, double-blind, randomised controlled trial. Lancet. 2017;389:709–717. doi: 10.1016/S0140-6736(16)31715-9. [DOI] [PubMed] [Google Scholar]
- Vejnar C.E., Zdobnov E.M. MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012;40:11673–11683. doi: 10.1093/nar/gks901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D.C. Mitochondrial genetic medicine. Nat. Genet. 2018;50:1642–1649. doi: 10.1038/s41588-018-0264-z. [DOI] [PubMed] [Google Scholar]
- West A.P., Shadel G.S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 2017;17:363–375. doi: 10.1038/nri.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Yang S., Pei Y., Li X., Zhao S., Zhu M., Zhao A. miR-124 attenuates Japanese encephalitis virus replication by targeting DNM2. Virol. J. 2016;13:105. doi: 10.1186/s12985-016-0562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Li C., Li H., sChangyong E. LncRNA CACNA1G-AS1 facilitates hepatocellular carcinoma progression through the miR-2392/C1orf61 pathway. J. Cell. Physiol. 2019;234:18415–18422. doi: 10.1002/jcp.28477. [DOI] [PubMed] [Google Scholar]
- Younis J.S., Abassi Z., Skorecki K. Is there an impact of the COVID-19 pandemic on male fertility? The ACE2 connection. Am. J. Physiol. Endocrinol. Metab. 2020;318:E878–E880. doi: 10.1152/ajpendo.00183.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamani B., Moeini Taba S.M., Shayestehpour M. Systemic lupus erythematosus manifestation following COVID-19: a case report. J. Med. Case Reports. 2021;15:29. doi: 10.1186/s13256-020-02582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Liang S., Sanchez-Lopez E., He F., Shalapour S., Lin X.J., Wong J., Ding S., Seki E., Schnabl B., et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 2018;560:198–203. doi: 10.1038/s41586-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated and analyzed during this study. Processed bulk RNA-seq data for the human related data from the nasopharyngeal and autopsy data is available online with dbGaP Study Accession number: phs002258.v1.p1 and online here at: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002258.v1.p1 and also https://covidgenes.weill.cornell.edu/. The RNA-sequence data related for the miR-2392 mimic experiments is deposited on SRA (https://www.ncbi.nlm.nih.gov/sra) with BioProject ID: PRJNA76339.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.