Abstract
Accurate and efficient early diagnosis is crucial for the control of COVID-19 pandemic. However, methods that can balance sensitivity, high throughput, detection speed and automation simultaneously are still scarce. Here, we report an automatic label-free chemiluminescence immunoassay (CLIA) for rapid SARS-CoV-2 nucleocapsid protein (NP) detection with high sensitivity and throughput. N-(4-aminobutyl)-N-ethylisoluminol and Co2+ dual-functionalized chemiluminescent magnetic beads (dfCL-MB) were first applied to the detection of protein by a novel and simple strategy. Sulphydryl polyethylene glycol was coated on the surface of dfCL-MB so as to assemble dfCL-MB and antibody conjugated gold nanoparticles through Au-S bond. Considering the high-risk application scenarios, the immunosensor was integrated with an automatic chemiluminescence analyzer so that the whole testing procedure could be carried out automatically without manual operation. A linear correlation between CL intensities and the logarithm of NP concentration was obtained in the range of 0.1–10,000 pg/mL with a detection limit of 21 fg/mL. The whole process cost 25 min and the sample compartment can bear 24 samples simultaneously. The spiked human serum samples and serum samples from COVID-19 patients were determined with satisfactory recoveries of 91.1–109.4%, suggesting that the proposed label-free CLIA is of great potential for SARS-CoV-2 NP detection in practice.
Keywords: SARS-CoV-2, Nucleocapsid protein, Label-free immunoassay, Chemiluminescence, Magnetic beads, Automation
Graphical Abstract
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes coronavirus disease 2019 (COVID-19) [1] has become a major public health concern all over the world. Until February 2021, more than 2.49 million people have died of COVID-19 and the number is still increasing rapidly [2]. Clinical research indicates that most infected people develop respiratory symptoms in 2–14 days after exposure (namely, incubation period) [3], [4]. To make matters worse, asymptomatic carriers and patients in the incubation period still can transmit the virus effectively [5], [6]. The high infectivity and the indefinite incubation period of SARS-CoV-2 make the pandemic situation even serious. Therefore, accurate and efficient early diagnosis is crucial for patient management and outbreak control.
Pathogenic nucleic acid testing based on quantitative real time polymerase chain reaction (qRT-PCR) and serological antibody testing are the main laboratory methods for clinical diagnosis of COVID-19 currently [7], [8]. However, qRT-PCR requires skilled technicians, expensive reagents and complicated time-consuming amplification steps. A considerable amount of nucleic acid loss is caused in the complex extraction steps, which may lead to a false negative result. Moreover, the high cost of mass population screening by qRT-PCR has resulted in great financial pressure even for well-funded health care systems. Likewise, antibody testing also has some shortcomings. The most important one is that the positive rate of the serum virus-specific IgM and IgG antibody is low when the sampling time is within one week of symptom onset [3], [9], not to mention that of asymptomatic carriers. This means antibody testing is unsuitable for early screening and can only be used as a complement to pathogenic testing [9]. The structural proteins of SARS-CoV-2, such as spike protein (SP) and nucleocapsid protein (NP), as another component of the pathogen, has been declared feasible for the diagnosis of COVID-19 by World Health Organization (WHO) [10]. Both NP and SP have high antigenicities [11], [12]. However, it has been reported that N gene is more conserved and stable than S gene and has fewer mutations over time [11], [13], [14], [15]. Therefore, NP is potentially a better target for a stable and effective SARS-CoV-2 antigen detection method.
Several works for SARS-CoV-2 NP detection have been published. For example, lateral flow immunoassay (LFA) for SARS-CoV-2 NP [16], [17], [18] has gained much attention due to its low cost and portability. Hess et.al. [19] developed a parallel reaction monitoring mass spectrometry assay for detection of SARS-CoV-2 SP and NP, which provided a detection limit of ∼200 aM. Zhang’s group [20] reported an aptamer-assisted proximity ligation assay for COVID-19 diagnosis with a NP detection limit of 37.5 pg/mL. These methods have solved different problems that we are facing in COVID-19 diagnosis, respectively. However, there are still some limitations. LFA is semiquantitative method and the sensitivity is sacrificed, which is an essential trait to patients in the early stage because of the low viral load [5]. The other methods have good performance in sensitivity, but they need hours to complete the whole test and complex manual operation are applied in various degrees, which makes these methods both labor-intensive and time-consuming. Methods that can balance sensitivity, high throughput, detection speed and automation simultaneously for the detection of SARS-CoV-2 NP are still scarce.
In recent years, great attention has been paid to chemiluminescence immunoassays (CLIAs) for disease biomarkers in practical in vitro diagnosis (IVD) due to inherent advantages such as high sensitivity, wide linear range, short detection time and simple devices [21], [22], [23], [24], [25]. While most of current CLIAs used in IVD are mainly labeling-based sandwich method [26], [27]. Furthermore, CL substrate is usually added in liquid phase, which is another error-causing factor. Label-free CLIA [28], [29], [30] based on CL intensity change initiated by the specific interaction between recognition element modified on CL functionalized nanomaterials and target have drawn increasing research interests. They are time-saving, low-cost, and simple, avoiding tedious labeling procedures. N-(4-aminobutyl)-N-ethylisoluminol (ABEI) and cobalt ion dual-functionalized chemiluminescent magnetic beads (dfCL-MB) as powerful interfaces are very attractive for the construction of label-free CLIA due to their rapid and simple separation from matrix, high feasibility of automation and low energy requirement. However, the application of dfCL-MB is restricted to the detection of small molecule [31], [32]. Label-free CLIA for protein biomarkers based on dfCL-MB has rarely been reported.
In this work, we developed an automatic label-free CLIA with high sensitivity for the rapid detection of SARS-CoV-2 NP. dfCL-MB were taken as the carrier of CL reagent ABEI and catalyst Co2+ as well as the interface of the label-free CLIA. As the carboxyl group of MB was occupied by ABEI and Co2+, it was hard to immobilize antibody directly. Therefore, citrate reduced gold nanoparticles were used to conjugate NP antibody (AuNP-Ab) due to its excellent protein affinity and stability. Moreover, polyethylene glycol with a sulfhydryl group at the end (SH-PEG) was chosen as the bridging molecule between AuNP-Ab and dfCL-MB because PEG as a polymer could be modified on dfCL-MB through non-covalent interactions and AuNP-Ab could be well conjugated via Au-S bond. Finally, the label-free immunosensor was obtained when the non-specific sites were blocked by BSA. It was found that NP had a negative impact on the CL intensity of the as-assembled immunosensor. Under these circumstances, a label-free CLIA was established. Considering the high-risk application scenarios, the label-free CLIA was integrated with an automatic CL analyzer so that the whole testing procedure could be carried out automatically without manual operation. The assembly process was characterized by CL, XPS, TEM, STEM, etc. After the CL measurement conditions were optimized, analytical performance and selectivity of the label-free CLIA were evaluated. Ultimately, the spiked human serum samples with NP and serum samples of COVID-19 patients were employed to investigate the applicability of the immunosensor in practical COVID-19 diagnosis.
2. Experimental section
2.1. Chemicals and reagents
Trisodium citrate (Na3Cit), sodium chloride (NaCl), sodium hydroxide (NaOH), disodium hydrogen phosphate dodecahydrate (Na2HPO4.12H2O), sodium dihydrogen phosphate dehydrate (NaH2PO4.2H2O), hydrogen peroxide (H2O2, 30.0%), cobalt chloride hexahydrate (CoCl2.6H2O) and chloroauric acid tetrahydrate (HAuCl4.4H2O) were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Bovine serum albumin (BSA) was obtained from Solarbio (Beijing, China). 4-(2-hydroxyethyl)−1-piperazineethanesulfonic acid (HEPES) and 10 mM PBS were purchased from Sangon Biotechnology Co., Ltd. (Shanghai, China). Carboxyl MBs were purchased from Biomag Biotechnology Co., Ltd. (Wuxi, China). Thiol and carboxyl modified polyethylene glycol (HS-PEG-COOH, Mw = 2000) was purchased from Ziqi Biotechnology Co., Ltd. (Shanghai, China). 1-(3-(dimethylamino)propyl)−3-ethylcarbodiimidehydrochloride (EDC) and N-Hydroxysuccinimide (NHS) were purchased from Aladdin Reagent (Shanghai, China). 4 mM N-(aminobutyl)-N-(ethylisoluminol) (ABEI, TCI, Japan) was dissolved in pH = 12.0 NaOH solution to prepare ABEI stock solution and stored at 4 °C. A stock solution of 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) was obtained by dissolving MES (Sigma, St. Louis, MO) in ultrapure water and adjusting the pH to 6.0 with 0.5 M NaOH solution. Rabbit-derived nucleocapsid protein (NP), NP antibody of SARS-CoV-2 and inactivated COVID-19 patient serum samples were provided by Jin’s Group (University of Science and Technology of China). All chemicals above were analytical-grade pure. A milli-Q system (Milli-pore, France) was employed to produce ultrapure water which was used throughout the experiment.
2.2. Apparatus
TEM images were recorded on an electronic microscope (JEM-2010, Japan). A high resolution electronic microscope (Talos F200X, USA) were used to collect HAADF images and EDS mapping images. XPS (Al Kα radiation as the X-ray source) was performed on an electronic spectrometer (ESCALAB 250Xi, USA). Inductively coupled plasma mass spectrometry (ICP-MS) measurements were carried out on a mass spectrometry (PlasmaQuad3, USA). UV–vis absorption spectra were conducted on an UV–vis spectrophotometer (Agilent 8453, USA). Magnetization curves were performed on a magnetic measurement system (MPMS3, USA). The zeta potential was detected using a zeta potential analyzer (90Plus PALS, USA). The centrifugation process was carried out on a high-speed refrigerated centrifuge (TGL-16 M, China). The pH of solutions used in this work was controlled by a pH meter (FE-28 standard, China). The citrate reduced gold nanoparticles (cit-AuNPs) were synthesized on a constant temperature heating magnetic stirrer (DF-101S, China). All shaking processes were performed on a DNA rotary mixer (DH-II, China).
2.3. Assembly of label-free immunosensor
First of all, ABEI and Co2+ dual-functionalized CL magnetic beads (dfCL-MB) were prepared according to a previous work of our group [32] with some modifications as shown in S1. Secondly, 0.2 mL of 8 mM HS-PEG-COOH (MW = 2000) was added to 1 mL of dfCL-MB under consecutive shaking for 4 h at room temperature. Afterwards, the obtained PEG/dfCL-MB was washed twice with ultrapure water and re-dispersed in 1 mL of ultrapure water. Thirdly, AuNP-Ab was prepared as shown in S2. Subsequently, 1 mL of AuNP-Ab and 1 mL of PEG/dfCL-MB were mixed and kept shaking for 4 h at room temperature. Then, 0.5 mL of 5% (w/w) BSA solution was added to block non-specific binding sites, followed by shaking for 30 min at room temperature. Finally, the obtained BSA/AuNP-Ab/PEG/dfCL-MB was washed twice with 10 mM PBS and re-dispersed in 2 mL of 1% (w/w) BSA solution. The obtained BSA/AuNP-Ab/PEG/dfCL-MB was stored at 4 °C for further experiment.
2.4. CL measurement and NP detection
CL kinetic curves of the stepwise assembled MBs were obtained by Centro LB 960 microplate luminometer (Berthold, Germany). Typically, 50 μL of MB solution was added into a microwell of 96-well plates, and then 50 μL of 2 mM H2O2 and 100 μL of 0.1 M NaOH were injected into the same well in sequence by the injection pump of Centro LB 960. CL signals varying with time were recorded as the CL kinetic curve.
The CL signals for the immunoassays were measured with Kaeser 1000 automatic CL immunoassay analyzer. Kaeser 1000 is composed of a reaction cup compartment, a sample compartment, a reagent compartment, a built-in bottle compartment, and a waste compartment. The synthetic BSA/AuNP-Ab/PEG/dfCL-MB and different concentrations of NP were put in the reagent compartment and sample compartment, respectively. Substrate A (1 mM H2O2, dispersed in 5 mM PBS), substrate B (pH = 12.0 NaOH) and washing buffer (10 mM PBS) were placed in the built-in bottle compartment. In a typical assay, Kaeser 1000 automatically took 50 μL of NP into the reaction cup, and then added 50 μL of BSA/AuNP-Ab/PEG/dfCL-MB. Next, the mixtures were incubated at 37 °C. The incubation time was set at 20 min for 100-fold diluted serum samples and 30 min for 20-fold diluted serum samples. Subsequently, the obtained immunocomplex was washed once with 10 mM PBS, and 100 μL of substrate A was added after removing the supernatant, followed by placing the immune-complex in the cassette. Finally, the light emission was collected by PMT after injecting 100 μL of substrate B, and presented in integrated CL intensity (CL intensity was the sum of the intensity of 10 points near the peak value with a time interval of 0.1 s). The CL intensity was used for quantitative analysis of NP. The used reaction cups entered to the waste compartment ultimately.
3. Results and discussion
3.1. Assembly strategy of immunosensor
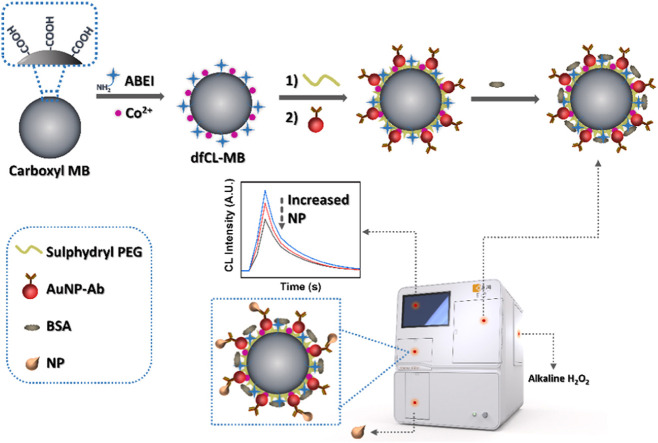
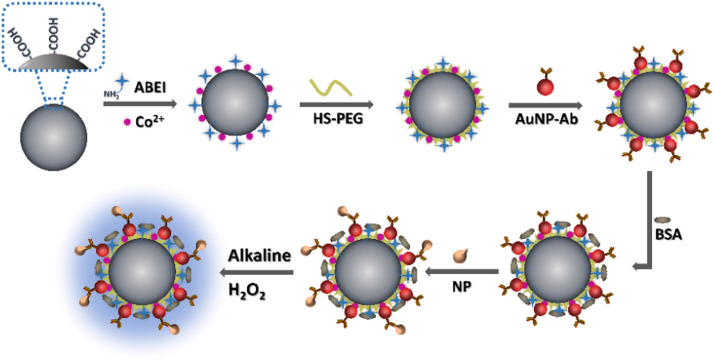
The assembly strategy of the label-free CL immunosensor is shown in Scheme 1. First of all, ABEI and Co2+ dual-functionalized CL magnetic beads (dfCL-MB) were prepared according to our previous work with some modifications [32]. Then, PEG with a sulphydryl group were coated on the surface of dfCL-MB to form PEG/dfCL-MB. Citrate reduced gold nanoparticles (cit-AuNPs) were taken as the carrier of NP antibody through Au-S bond due to its excellent stability and biocompatibility. Subsequently, NP antibody coated gold nanoparticles (AuNP-Ab) were assembled with PEG/dfCL-MB via Au-S bond to form AuNP-Ab/PEG/dfCL-MB. After the non-specific binding sites were blocked by BSA, BSA/AuNP-Ab/PEG/dfCL-MB were obtained as the immunosensor of the label-free CLIA. Finally, in the presence of NP, BSA/AuNP-Ab/PEG/dfCL-MB could capture NP by specific immunoreaction, thus NP/BSA/AuNP-Ab/PEG/dfCL-MB immunocomplex was formed. The immunocomplex could produce a CL emission varying with the concentration of NP when it was mixed with alkaline H2O2.
Scheme 1.
Schematic illustration of assembly strategy of the label-free immunosensor.
3.2. Stepwise characterization of immunosensor
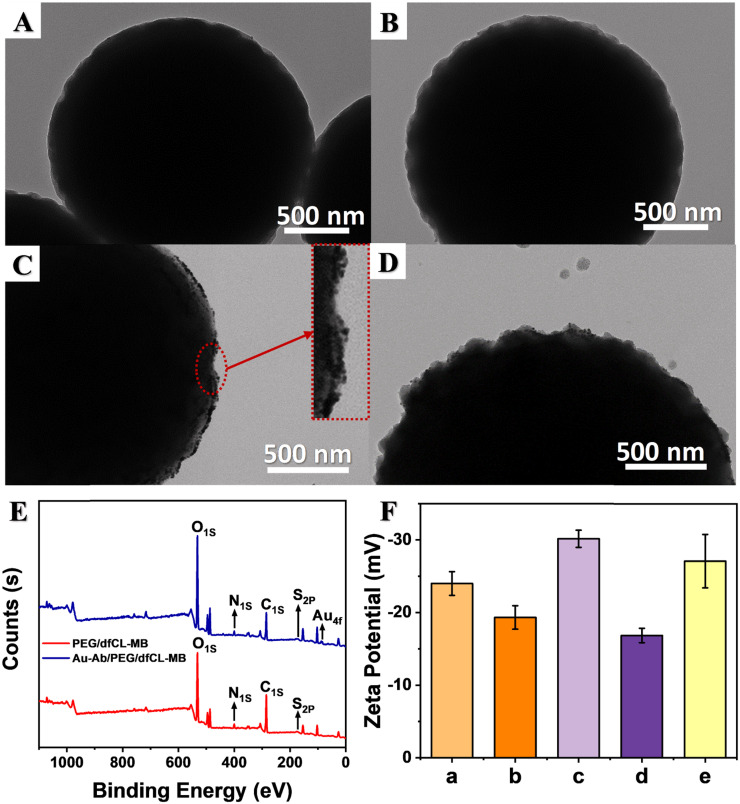
To monitor the assembly process, a series of characterizations were conducted. Fig. 1A-D display typical TEM images of MB surface during the assembly process. The commercial MBs were highly uniform microspheres with an average diameter of 2.17 ± 0.05 µm (Fig. S1). After functionalized with ABEI and Co2+, there was no obvious change in morphology (Fig. 1A). The surface of PEG/dfCL-MB (Fig. 1B) showed no obvious difference compared with that of dfCL-MB (Fig. 1A) due to the low contrast of PEG under TEM. The successful coating of PEG was reflected in XPS results. As shown in Fig. S4, the percentage of sulfur element in both MB and dfCL-MB was 0.12%, but that of PEG/dfCL-MB increased to 0.17%. The interaction between dfCL-MB and PEG was non-covalent bond, including hydrogen bond and Van der Waals' force. There were abundant hydroxyl groups and a carboxyl group in PEG, which would form hydrogen bonds with carboxyl groups and amide bonds of dfCL-MB. Moreover, Van der Waals' force as a universal intermolecular force also existed between PEG and ABEI/MB. After AuNP-Ab reacted with PEG/dfCL-MB, number of nanoparticles appeared on MB (Fig. 1C). Meanwhile, the new peak at around 85 eV in survey XPS (Fig. 1E) of AuNP-Ab/PEG/dfCL-MB belonged to Au4f, revealing the successful conjugation of AuNP-Ab. Moreover, the presence of gold element in STEM EDS mapping of AuNP-Ab/PEG/dfCL-MB (Fig. S3) was also a conclusive evidence of the successful conjugation of AuNP-Ab. The TEM image of BSA/AuNP-Ab/PEG/dfCL-MB (Fig. 1D) showed distinct proteins coated on the surface after BSA was introduced to AuNP-Ab/PEG/dfCL-MB, indicating the non-specific binding sites were blocked by BSA.
Fig. 1.
TEM images of dfCL-MB (A), PEG/dfCL-MB (B), AuNP-Ab/PEG/dfCL-MB (C), BSA/AuNP-Ab/PEG/dfCL-MB (D). (E) Survey XPS of AuNP-Ab/PEG/dfCL-MB (blue) and PEG/dfCL-MB (red). (F) Zeta potential of dfCL-MB (a), PEG/dfCL-MB (b), AuNP-Ab/PEG/dfCL-MB (c), BSA/AuNP-Ab/PEG/dfCL-MB (d) and NP/BSA/AuNP-Ab/PEG/dfCL-MB (E).
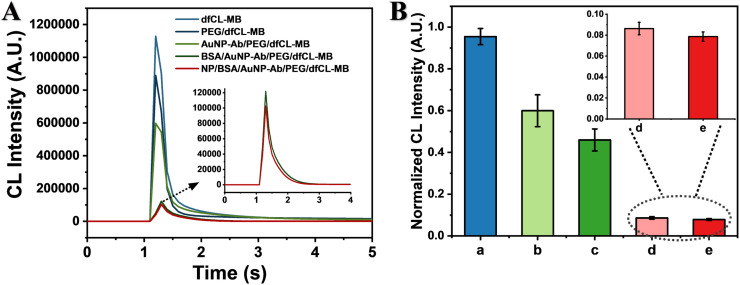
CL kinetic curves obtained by Centro LB 960 luminometer and normalized CL intensities obtained by Kaeser 1000 of MBs in every assembly step are shown in Fig. 2. All these MBs displayed a flash type emission (Fig. 2 A). However, the kinetic curves changed in various degrees during the assembly process. The peak of dfCL-MB and PEG/dfCL-MB appeared within 0.1 s after the injection of H2O2. The CL intensity decreased by over 1/5 after dfCL-MB was assembled with SH-PEG owing to the existence of lots of sulphydryl groups in PEG/dfCL-MB, which competed with ABEI for O2 •− [33]. AuNP-Ab had negative impact on the CL intensity of AuNP-Ab/PEG/dfCL-MB. To investigate the reason of this negative impact, cit-AuNPs and NP antibody were added in the liquid phase of PEG/dfCL-MB solution, respectively. The results (Fig. S7) showed that both cit-AuNPs and NP antibody could decrease the CL intensity of PEG/dfCL-MB. Two reasons were possibly responsible for the CL intensity decrease of AuNP-Ab/PEG/dfCL-MB. First, the maximum emission wavelength of ABEI is 450 nm where cit-AuNPs has a strong absorption, thus CL resonance energy transfer (CRET) may occur between PEG/dfCL-MB and cit-AuNPs, resulting in a decrease in CL intensity [34]. Furthermore, sulphydryl group as a common group in antibodies may also exist in NP antibody, leading to a decrease in CL intensity. Moreover, the peak drop of AuNP-Ab/PEG/dfCL-MB showed a delay of 0.1 s, which was due to a slower diffusion rate of H2O2 when AuNP-Ab was assembled onto PEG/dfCL-MB. After the non-specific binding sites were blocked by BSA, the CL intensity of BSA/AuNP-Ab/PEG/dfCL-MB was only about 1/6 of that of AuNP-Ab/PEG/dfCL-MB and the peak appeared at 0.2 s after the injection of H2O2. Two reasons may lead to these: (1) After AuNP-Ab/PEG/dfCL-MB was blocked by BSA, the diffusion rate of H2O2 decreases, leading to a decrease in CL intensity and the delayed peak; (2) BSA contains 17 disulfide bonds and 1 sulphydryl group, which may compete with ABEI for O2 .- to quench CL. Similarly, the CL intensity of NP/BSA/AuNP-Ab/PEG/dfCL-MB further decreased due to 8 methionine residues in NP and a slower diffusion rate of H2O2. As shown in Fig. 2B, the CL intensities obtained by Kaeser 1000 showed the consistent regulation with that obtained by Centro LB 960, indicating the high feasibility of integrating the as-assembled immunosensor with Kaeser 1000.
Fig. 2.
(A) CL kinetic curves obtained by Centro LB 960. (B) Normalized CL intensity obtained by Kaeser 1000 of dfCL-MB (a), PEG/dfCL-MB (b), AuNP-Ab/PEG/dfCL-MB (c), BSA/AuNP-Ab/PEG/dfCL-MB (d) and NP/BSA/AuNP-Ab/PEG/dfCL-MB (e, CNP = 10−10 g/mL).
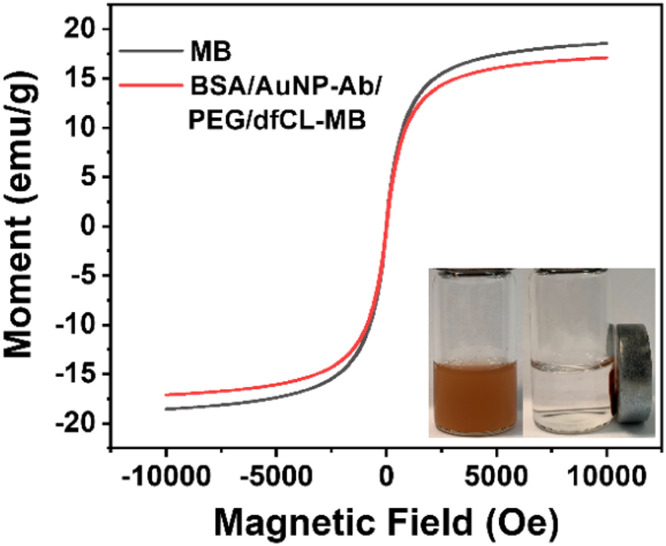
To investigate the impact of the assembly process on the magnetic property of MB, magnetization curves were obtained. As shown in Fig. 3, the commercial carboxyl MB displayed no hysteresis, which indicated that MB possessed good superparamagnetism. The saturation magnetization of MB and BSA/AuNP-Ab/PEG/dfCL-MB were 18.5 emu/g and 17.1 emu/g, respectively. Compared with MB, the saturation magnetization of BSA/AuNP-Ab/PEG/dfCL-MB decreased by 1.4 emu/g, which was ascribed to the introduction of nonmagnetic reagents and materials including ABEI, Co2+, SH-PEG, AuNP-Ab and BSA. The insert picture of Fig. 3 showed that BSA/AuNP-Ab/PEG/dfCL-MB was of good dispersivity without an external magnet and it could be separated from matrix rapidly (within 10 s) under an external magnet. Therefore, the as-assembled immunosensor displayed good magnetic separation property though some nonmagnetic reagents were introduced to MB.
Fig. 3.
Magnetization curves of MB and BSA/AuNP-Ab/PEG/dfCL-MB, insert: photograph of BSA/AuNP-Ab/PEG/dfCL-MB with (right) and without (left) an external magnet.
3.3. Analytical performance of label-free CLIA
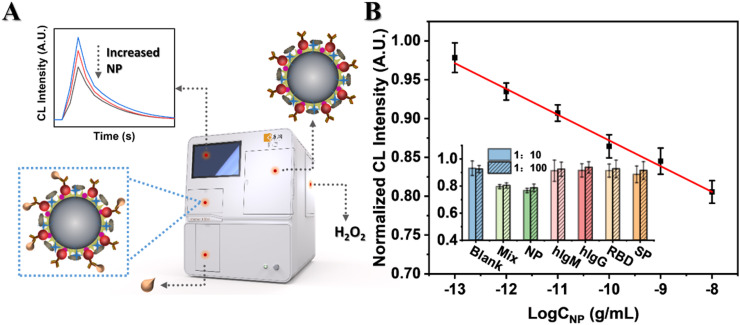
On account of the good magnetic property, an automatic CL analyzer with a magnetic separation system (Kaeser 1000) was employed to carry out the whole testing process. First of all, to acquire the best analytical performance, the concentration of PEG and MB, MB size, CL reaction conditions and incubation time were optimized by Kaeser 1000. Optimal conditions were 8 mM for PEG concentration, 1 mg/mL for MB concentration, 2.8 µm for MB size, 20 min for incubation time, pH = 12.0 and 1 mM H2O2 for the CL reaction, respectively (Fig. S8–10). Subsequently, we studied the analytical performance of the proposed label-free CLIA under these optimized conditions. The CL intensity was negatively correlated with the logarithm of NP concentration. A good linear relationship was observed in the NP concentration range of 0.1–10,000 pg/mL ( Fig. 4B). The linear regressive equation with a correlation coefficient of 0.992 was (I: Normalized CL intensity; C: concentration of NP). Furthermore, , namely the average normalized CL intensity of blank minus three times the standard deviation, was defined as the detection limit and the corresponding concentration of NP was calculated to be 21 fg/mL. When the concentration of NP was 100 pg/mL, the relative standard derivation (RSD) of CL intensity was 2.20% within a day (n = 8) and 5.70% within 15 days (n = 8), respectively, revealing the good reproducibility of the proposed label-free CLIA (Fig. S11).
Fig. 4.
(A) Schematic illustration of the detection process. (B) Linear relationship between normalized CL intensity and logarithm of NP concentration. Insert: A comparison of CL responses of NP (100 pg/mL) with those of different interferents. 1:10, 1000 pg/mL of hIgM, hIgG, RBD and SP, respectively; 1:100, 10,000 pg/mL of hIgM, hIgG, RBD and SP, respectively.
Afterwards, the selectivity of the CLIA was studied. hIgG, hIgM and other proteins including RBD and SP in SARS-CoV-2 were selected as the interferents instead of NP. As shown in the insert picture of Fig. 4B, only CL signals of NP and mixture containing NP decreased obviously and the others were similar to that of blank when the interferents were 10 and 100 times as much as NP. Therefore, the immunosensor could recognize NP specifically and remain unresponsive to other proteins that might coexist in samples even when the concentration of interferents were 100-fold higher than that of NP. In other words, our label-free CLIA showed excellent selectivity to SARS-CoV-2 NP.
A comparison of the proposed label-free CLIA with other previously reported methods for SARS-CoV-2 NP detection is displayed in Table 1. We can see that both linear range and detection limit of the proposed label-free CLIA are superior to other immunoassays. Moreover, our label-free immunosensor is fabricated with only one antibody in the whole process, which make it simple, time-saving and low-cost. Besides, the proposed CLIA can be carried out by Kaeser 1000 analyzer automatically without manual operation, which will greatly reduce the infection risk of health care workers from virus-contained samples in practical applications. Additionally, the proposed label-free CLIA also possesses a competitive detection speed compared with other methods.
Table 1.
A comparison of the proposed label-free CLIA with other reported methods for SARS-CoV-2 NP detection.
| Analytical Method for SARS-CoV-2 NP | Labeled or Label-free | Linear Range | LOD | Manual Operation | Test Time |
|---|---|---|---|---|---|
| LFA[16] | Labeled | Semiquantitative | 2 ng | + | 20 min |
| Half-Strip LFA[17] | Labeled | Semiquantitative | 0.65 ng mL−1 | + | 20 min |
| Immunochromatography[18] | Labeled | Semiquantitative | 100 ng mL−1 | + | >15 min |
| Aptamer-assisted Proximity Ligation Assay[20] | Labeled | 50–5000 pg/mL | 37.5 pg/mL | + | 2 h |
| Mass Spectrometry[19] | Label-free | 3 aM to 12.5 fM※ | 200 aM | + | >4 h |
| Electrochemical Immunosensor[35] | Label-free | 1 pg/mL to 1000 ng mL−1 | 0.8 pg/mL | + | >20 min |
| This work | Label-free | 0.1 pg/mL to 10,000 pg/mL | 21 fg mL−1 | – | 25 min |
“+”: need manual operation; “−”: automatic method without manual operation.
“*”: the limit of quantitation was 390 aM.
Finally, the validity of the proposed label-free CLIA in complex matrix was investigated. Earlier studies showed that NP was detectable in serum samples of patients and serum NP was a sensitive and specific early diagnostic marker for COVID-19 [36], [37], [38]. Compared with swab and saliva samples, serum samples tend to have less variations and be better at eliminating sampling errors. Therefore, serum was chosen as the sample matrix. Serum samples were diluted 100 times and 20 times with 10 mM PBS without any other pretreatment. Four concentrations (1, 5, 10, 50 pg/mL) of NP were added to the diluted healthy human serum samples, respectively. As shown in Table 2, the detected concentrations of NP were approximately consistent with the corresponding added concentration in both 100-fold and 20-fold diluted serum samples and the recovery were between 97.4% and 109.4%, demonstrating that the complex serum matrix did not have obvious influence on NP detection. Furthermore, serum samples from COVID-19 patient were also determined to estimate the practicability in real COVID-19 samples. As shown in Table 2, the detected concentrations of NP in diluted patient serum were in agreement with previous reports [36], [39], [40]. Accordingly, the proposed label-free CLIA is of great application potential for the detection of SARS-CoV-2 NP in practical blood samples.
Table 2.
Determination of NP in diluted human serum samples.
| Serum Sample | Initial (pg/mL) | Added (pg/mL) | Found (pg/mL) | Recovery (%) | |
|---|---|---|---|---|---|
| Healthy | 1* | – | 1 | 1.04 ± 0.03 | 103.9 |
| 2* | – | 5 | 4.87 ± 0.22 | 97.4 | |
| 3* | – | 10 | 10.81 ± 0.69 | 108.1 | |
| 4* | – | 50 | 49.25 ± 1.72 | 98.4 | |
| 5# | – | 1 | 1.06 ± 0.06 | 106.3 | |
| 6# | – | 5 | 5.11 ± 0.18 | 102.3 | |
| 7# | – | 10 | 10.94 ± 0.55 | 109.4 | |
| 8# | – | 50 | 52.35 ± 1.26 | 104.7 | |
| COVID-19 Patient | 1* | 5.15 ± 0.34 | 10 | 14.47 ± 0.34 | 95.5 |
| 2* | 1.85 ± 0.12 | 10 | 12.37 ± 0.44 | 104.5 | |
| 3* | 27.59 ± 1.42 | 10 | 40.07 ± 2.53 | 106.6 | |
| 4* | 5.28 ± 0.33 | 10 | 16.42 ± 0.46 | 107.5 | |
| 5# | 37.05 ± 0.96 | 10 | 42.85 ± 1.34 | 91.1 | |
| 6# | 129.8 ± 4.04 | 10 | 146.5 ± 3.72 | 104.8 | |
“-”: undetectable
“*”: 100-fold diluted; “#”: 20-fold diluted
4. Conclusion
In this work, we have successfully developed an automatic label-free CL immunoassay for SARS-CoV-2 nucleocapsid protein detection for the first time. The immunosensor was assembled by dfCL-MB and NP antibody coated AuNPs by virtue of PEG with a sulphydryl group via Au-S bond. It was found that NP could decrease the CL signal of the as-assembled immunosensor. A linear correlation between the CL intensities and the logarithm of NP concentration was obtained in the range of 0.1–10,000 pg/mL. An automatic CL analyzer Kaeser 1000 was employed to carry out the detection procedure, which was a labor-saving and relatively safe way to these potential users, mainly frontline healthcare workers. The proposed label-free CLIA were highly sensitive with a LOD of 21 fg/mL and excellent capacity of resisting disturbance. The spiked human serum with NP and COVID-19 patient serum samples were determined with satisfactory recoveries of 91.1–109.4%. Moreover, the sample compartment of Kaeser 1000 can bear 24 samples simultaneously, which means that the results of 24 samples can be read out simultaneously within 25 min in practical applications, indicating the relatively high throughput of this method. Although this method was not as portable as semiquantitative LFA, it also has its own vital applications since quantitative analysis of SARS-CoV-2 is of great significance for the research of pathology and prognosis, especially with high sensitivity and wide linear range.
CRediT authorship contribution statement
Aihua Lyu: Methodology, Data curation, Investigation, Writing – original draft. Tengchuan Jin: Resources, Funding acquisition, Writing – review & editing. Shanshan Wang: Data curation, Investigation. Xiaoxue Huang:Resources. Weihong Zeng: Resources. Rui Yang: Conceptualization, Supervision, Project administration, Writing – review & editing. Hua Cui: Conceptualization, Funding acquisition, Supervision, Project administration, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The support of this research by a COVID-19 special task grant supported by Chinese Academy of Sciences Clinical Research Hospital (Hefei, China) with Grant Nos. YD2060002008 and YD2070002017. The support of this research by the National Key Research and Development Program of China (Grant No. 2016YFA0201300) are gratefully acknowledged.
Biographies
Aihua Lyu received her B.S. degree in Chemistry from Sichuan University in 2017, and now is a Ph.D. student in University of Science and Technology of China, Hefei, China. Her research interest is chemiluminescence biosensors for in vitro diagnosis.
Tengchuan Jin is a professor at University of Science and Technology of China (USTC). He received his Ph.D. from Illinois Institute of Technology in 2008. Prof. Jin joined USTC in 2015. Research in his laboratory include structural immunology, host-microbe interactions, antibody engineering.
Shanshan Wang received her M.S. degree in Chemistry from Liaoning University in 2019, and now is a Ph.D. student in University of Science and Technology of China, Hefei, China. Her research interest is chemiluminescence and electrochemiluminescence biosensors for in vitro diagnosis.
Xiaoxue Huang is currently a Ph.D. student at University of Science and Technology of China (USTC). She received her B.S. from USTC in 2017. Her research interests include development of novel heavy-chain antibodies for diagnosis and disease therapy.
Weihong Zeng is currently a postdoctoral fellow at University of Science and Technology of China (USTC). She received her Ph.D. degree from Jinan University in 2019. Her research interests include host immune responses toward SARS-CoV-2 and structural characterization of antibody: antigen complex.
Rui Yang received her Ph.D. in Analytical Chemistry from the University of Science and Technology of China in 2020, and now is a school-appointed associate professor at Anhui Medical University, Hefei, China. Her research interests focus on the synthesis of novel chemiluminescent materials and their applications in vitro and in vivo diagnosis.
Hua Cui is a professor at the University of Science and Technology of China (USTC), Hefei, China. She received her Ph.D. in Analytical Chemistry in 1990 from USTC. Her research interests include novel chemiluminescence and electrochemiluminescence functionalized nanomaterials and their applications in in vitro diagnosis.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2021.130739.
Appendix A. Supplementary material
Supplementary material.
.
References
- 1.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Coronavirus (COVID-19) Dashboard. 〈https://covid19.who.int/〉. (Accessed 2021–2-26).
- 3.Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J., Su K., Zhang F., Gong J., Wu B., Liu X.M., Li J.J., Qiu J.F., Chen J., Huang A.L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 4.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J.-L., Liang Z., Peng Y., Wei L., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N., China Med Treatment Expert G. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wolfel R., Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caruana G., Croxatto A., Coste A.T., Opota O., Lamoth F., Jaton K., Greub G. Diagnostic strategies for SARS-CoV-2 infection and interpretation of microbiological results. Clin. Microbiol Infect. 2020;26(9):1178–1182. doi: 10.1016/j.cmi.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 9.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., Wang D.Q., Hu Y., Ren J.H., Tang N., Xu Y.Y., Yu L.H., Mo Z., Gong F., Zhang X.L., Tian W.G., Hu L., Zhang X.X., Xiang J.L., Du H.X., Liu H.W., Lang C.H., Luo X.H., Wu S.B., Cui X.P., Zhou Z., Zhu M.M., Wang J., Xue C.J., Li X.F., Wang L., Li Z.J., Wang K., Niu C.C., Yang Q.J., Tang X.J., Zhang Y., Liu X.M., Li J.J., Zhang D.C., Zhang F., Liu P., Yuan J., Li Q., Hu J.L., Chen J., Huang A.L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 10.WHO, Antigen-detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays. (2020). 〈https://apps.who.int/iris/bitstream/handle/10665/33425〉.
- 11.Dutta N.K., Mazumdar K., Gordy J.T. The nucleocapsid protein of SARS-CoV-2: a target for vaccine development. J. Virol. 2020;94(13) doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C., Yang Y., Xie J., Ding C., Ma X., Weng J., Gao Y., He H., Jin T. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys. Res Commun. 2020;527(3):618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294e1289. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 15.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H.Y., Lee J.H., Kim M.J., Park S.C., Choi M., Lee W., Ku K.B., Kim B.T., Changkyun Park E., Kim H.G., Kim S.I. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens. Bioelectron. 2021;175 doi: 10.1016/j.bios.2020.112868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S., Weigl B.H., Nichols K.P. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal. Chem. 2020;92(16):11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C., Zhou L., Du K., Zhang Y., Wang J., Chen L., Lyu Y., Li J., Liu H., Huo J., Li F., Wang J., Sang P., Lin S., Xiao Y., Zhang K., He K. Foundation and clinical evaluation of a new method for detecting SARS-CoV-2 antigen by fluorescent microsphere immunochromatography. Front Cell Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.553837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cazares L.H., Chaerkady R., Samuel Weng S.H., Boo C.C., Cimbro R., Hsu H.E., Rajan S., Dall’Acqua W., Clarke L., Ren K., McTamney P., Kallewaard-LeLay N., Ghaedi M., Ikeda Y., Hess S. Development of a parallel reaction monitoring mass spectrometry assay for the detection of SARS-CoV-2 spike glycoprotein and nucleoprotein. Anal. Chem. 2020;92(20):13813–13821. doi: 10.1021/acs.analchem.0c02288. [DOI] [PubMed] [Google Scholar]
- 20.Liu R., He L., Hu Y.S., Luo Z.F., Zhang J.J. A serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers. Chem. Sci. 2020;11(44):12157–12164. doi: 10.1039/d0sc03920a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giljohann D.A., Mirkin C.A. Drivers of biodiagnostic development. Nature. 2009;462(7272):461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong Y., Tang X., Li J., Lan Q., Min L., Ren C., Hu X., Torrente-Rodriguez R.M., Gao W., Yang Z. A nanozyme tag enabled chemiluminescence imaging immunoassay for multiplexed cytokine monitoring. Chem. Commun. 2018;54(98):13813–13816. doi: 10.1039/c8cc07779g. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Q., Liu Y., Wang L., Adkins G.B., Zhong W. Rapid enrichment and detection of extracellular vesicles enabled by CuS-enclosed microgels. Anal. Chem. 2019;91(24):15951–15958. doi: 10.1021/acs.analchem.9b04485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D., Zhang Y., Xu Y., Shen T., Cheng G., Huang B., Ruan X., Wang C. Comparison of chemiluminescence immunoassay, enzyme-linked immunosorbent assay and passive agglutination for diagnosis of Mycoplasma pneumoniae infection. Ther. Clin. Risk Manag. 2018;14:1091–1097. doi: 10.2147/TCRM.S159227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park I.U., Fakile Y.F., Chow J.M., Gustafson K.J., Jost H., Schapiro J.M., Novak-Weekley S., Tran A., Nomura J.H., Chen V., Beheshti M., Tsai T., Hoover K., Bolan G. Performance of treponemal tests for the diagnosis of syphilis. Clin. Infect. Dis. 2019;68(6):913–918. doi: 10.1093/cid/ciy558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roda A., Mirasoli M., Dolci L.S., Buragina A., Bonvicini F., Simoni P., Guardigli M. Portable device based on chemiluminescence lensless imaging for personalized diagnostics through multiplex bioanalysis. Anal. Chem. 2011;83(8):3178–3185. doi: 10.1021/ac200360k. [DOI] [PubMed] [Google Scholar]
- 27.Ehsania M., Chaichi M.J., Hosseini S.N. Comparison of CuO nanoparticle and CuO/MWCNT nanocomposite for amplification of chemiluminescence immunoassay for detection of the hepatitis B surface antigen in biological samples. Sens. Actuators B-Chem. 2017;247:319–328. [Google Scholar]
- 28.Huang Y., Gao L.F., Cui H. Assembly of multifunctionalized gold nanoparticles with chemiluminescent, catalytic, and immune activity for label-free immunoassays. ACS Appl. Mater. Interfaces. 2018;10(20):17040–17046. doi: 10.1021/acsami.8b02521. [DOI] [PubMed] [Google Scholar]
- 29.Li J., Cao Y., Hinman S.S., McKeating K.S., Guan Y.W., Hu X.Y., Cheng Q., Yang Z.J. Efficient label-free chemiluminescent immunosensor based on dual functional cupric oxide nanorods as peroxidase mimics. Biosens. Bioelectron. 2018;100:304–311. doi: 10.1016/j.bios.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z.J., Cao Y., Li J., Lu M.M., Jiang Z.K., Hu X.Y. Smart CuS nanoparticles as peroxidase mimetics for the design of novel label-free chemiluminescent immunoassay. ACS Appl. Mater. Interfaces. 2016;8(19):12031–12038. doi: 10.1021/acsami.6b02481. [DOI] [PubMed] [Google Scholar]
- 31.Kong W., Li Q., Wang W., Zhao X., Jiang S., Zheng T., Zhang Q., Shen W., Cui H. Rational design of functional materials guided by single particle chemiluminescence imaging. Chem. Sci. 2019;10(21):5444–5451. doi: 10.1039/c9sc00954j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong W., Zhao X., Zhu Q., Gao L., Cui H. Highly chemiluminescent magnetic beads for label-free sensing of 2,4,6-trinitrotoluene. Anal. Chem. 2017;89(13):7145–7151. doi: 10.1021/acs.analchem.7b01111. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z., Cui H., Lai C., Liu L. Gold nanoparticle-catalyzed luminol chemiluminescence and its analytical applications. Anal. Chem. 2005;77(10):3324–3329. doi: 10.1021/ac050036f. [DOI] [PubMed] [Google Scholar]
- 34.Shu J., Han Z., Cui H. Highly chemiluminescent TiO2/tetra(4-carboxyphenyl)porphyrin/N-(4-aminobutyl)-N-ethylisoluminol nanoluminophores for detection of heart disease biomarker copeptin based on chemiluminescence resonance energy transfer. Anal. Bioanal. Chem. 2019;411(18):4175–4183. doi: 10.1007/s00216-019-01821-2. [DOI] [PubMed] [Google Scholar]
- 35.Eissa S., Zourob M. Development of a low-cost cotton-tipped electrochemical immunosensor for the detection of SARS-CoV-2. Anal. Chem. 2021;93(3):1826–1833. doi: 10.1021/acs.analchem.0c04719. [DOI] [PubMed] [Google Scholar]
- 36.Shan D., Johnson J.M., Fernandes S.C., Suib H., Hwang S., Wuelfing D., Mendes M., Holdridge M., Burke E.M., Beauregard K., Zhang Y., Cleary M., Xu S., Yao X., Patel P.P., Plavina T., Wilson D.H., Chang L., Kaiser K.M., Nattermann J., Schmidt S.V., Latz E., Hrusovsky K., Mattoon D., Ball A.J. N-protein presents early in blood, dried blood and saliva during asymptomatic and symptomatic SARS-CoV-2 infection. Nat. Commun. 2021;12(1):1931. doi: 10.1038/s41467-021-22072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T., Wang L., Wang H., Li X., Zhang S., Xu Y., Wei W. Serum SARS-COV-2 nucleocapsid protein: a sensitivity and specificity early diagnostic marker for SARS-COV-2 infection. Front Cell Infect. Microbiol. 2020;10:470. doi: 10.3389/fcimb.2020.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B., Ding J., Li F. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (rnaemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with Coronavirus disease 2019. Clin. Infect. Dis. 2020;71(8):1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torrente-Rodriguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3(6):1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Lillehoj P.B. Microfluidic magneto immunosensor for rapid, high sensitivity measurements of SARS-CoV-2 nucleocapsid protein in serum. ACS Sens. 2021;6(3):1270–1278. doi: 10.1021/acssensors.0c02561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.