Abstract
Human transcription factor IIIC (hTFIIIC) is a multisubunit complex that directly recognizes promoter elements and recruits TFIIIB and RNA polymerase III. Here we describe the cDNA cloning and characterization of the 90-kDa subunit (hTFIIIC90) that is present within a DNA-binding subcomplex (TFIIIC2) of TFIIIC. hTFIIIC90 has no specific homology to any of the known yeast TFIIIC subunits. Immunodepletion and immunoprecipitation studies indicate that hTFIIIC90 is a bona fide subunit of TFIIIC2 and absolutely required for RNA polymerase III transcription. hTFIIIC90 shows interactions with the hTFIIIC220, hTFIIIC110, and hTFIIIC63 subunits of TFIIIC, the hTFIIIB90 subunit of TFIIIB, and the human RPC39 (hRPC39) and hRPC62 subunits of an initiation-specific subcomplex of RNA polymerase III. These interactions may facilitate both TFIIIB and RNA polymerase III recruitment to the preinitiation complex by TFIIIC. We show that hTFIIIC90 has an intrinsic histone acetyltransferase activity with a substrate specificity for histone H3.
RNA polymerase III transcribes genes encoding small structural RNAs that include 5S RNA, tRNA, adenovirus-associated (VA) RNA, and the U6 and 7SK RNAs. Together with RNA polymerase III, transcription factor IIIC (TFIIIC) and TFIIIB suffice for transcription of tRNA, VA RNA, and yeast U6 RNA genes, whereas expression of the 5S gene is additionally dependent on TFIIIA (reviewed in references 14, 19, 49, and 50). Mammalian U6 and 7SK genes require PTF (SNAPc/PBP), TFIIIC1, and an alternative form of TFIIIB to direct transcription by RNA polymerase III (reviewed in reference 38).
In the simplest cases, preinitiation complex assembly on class III genes involves direct promoter recognition by TFIIIC (A and B boxes in tRNA, VA RNA, and yeast U6 RNA genes) and TFIIIB and RNA polymerase III recruitment through interactions with TFIIIC (reviewed in references 18, 46, and 47). Consistent with conservation of the assembly pathway from yeast to human, there is a corresponding conservation in structure and function of RNA polymerase III, TFIIIB, and a subset of TFIIIC subunits (reviewed in references 18, 46, and 47).
TFIIIC has been most extensively characterized in yeast, where it is composed of six polypeptides (138, 131, 95, 91, 60, and 55 kDa) (reviewed in references 1 and 28) and binds to both A and B boxes, whereas human TFIIIC (hTFIIIC) contains at least nine subunits and can be resolved into a five-subunit (220-, 110-, 102-, 90-, and 63-kDa) complex (TFIIIC2) that binds weakly to the B box and a less well-characterized complex (TFIIIC1) that stabilizes the binding of TFIIIC2 to the A and B boxes (21, 44, 47, 53, 54). Whereas the two largest hTFIIIC subunits are not conserved (24, 27, 34), the 102-kDa subunit (hTFIIIC102) and the hTFIIIC63 subunit are conserved in structure and in their interactions with TFIIIB and RNA polymerase III subunits (18). Thus, interactions of hTFIIIC102 (homologue of yeast TFIIIC131 [yTFIIIC131]) with hTFIIIB90 (homologue of yTFIIIB70) and of hTFIIIB90 with human RPC39 (hRPC39) (homologue of yeast RPC34 [yRPC34] and a subunit of an initiation-specific RNA polymerase III subcomplex) parallel the interactions of yTFIIIC131 with yTFIIIB70 and of yTFIIIB70 with yRPC34 (10, 18, 20, 45, 48), respectively. Moreover, although not yet observed for their yeast counterparts, recently described interactions of hTFIIIC63 (homologue of yTFIIIC95) with hTFIIIC102, hTFIIIB90, and hRPC62 (homologue of yRPC82) further strengthen the notion that TFIIIC acts as an assembly factor for the recruitment of TFIIIB and RNA polymerase III to the promoter (18).
Both yTFIIIC and hTFIIIC can relieve chromatin-mediated repression of preassembled class III gene transcription in vitro (9, 22). A study of the yeast U6 gene showed that TFIIIC is essential for in vitro transcription only in the context of a chromatin template but did not establish the mechanism for TFIIIC-mediated chromatin transcription (9). A more recent study of a tRNA gene showed that highly purified hTFIIIC, at concentrations above those necessary for optimal transcription of naked DNA templates, efficiently relieves nucleosome-mediated repression on cognate chromatin templates (22). It was further shown that TFIIIC alone can bind to the A and B boxes of a tRNA gene within the chromatin template and that TFIIIC has an intrinsic histone acetyltransferase (HAT) activity. An in-gel HAT assay suggested that three components (220, 110, and 90 kDa) of the hTFIIIC complex had intrinsic HAT activities, and this was confirmed, with a recombinant protein, for the 110-kDa subunit. The ability of p-hydroxymercuribenzoic acid to inhibit both TFIIIC HAT activity and TFIIIC-dependent chromatin transcription, but not TFIIIC-dependent DNA transcription, suggested a possible link between TFIIIC HAT activity and chromatin function (22).
To further understand the structure and function of hTFIIIC, we cloned the cognate cDNA for the 90-kDa subunit (hTFIIIC90) of TFIIIC2 and studied interactions of the recombinant subunit with other TFIIIC2 subunits and with TFIIIB and RNA polymerase III subunits. We showed that hTFIIIC90 is indeed a HAT that specifically acetylates histone H3.
MATERIALS AND METHODS
Purification and cloning of hTFIIIC90.
TFIIIC2 was purified as described previously (21) and component subunits were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Microsequence analysis was performed on the 90-kDa polypeptide (hTFIIIC90) as described previously (24). A cDNA clone encoding hTFIIIC90 was obtained by screening human cDNA libraries (Namalwa, BJAB, and HeLa cell libraries) with four degenerate oligonucleotide probes corresponding to the internal peptide sequences of hTFIIIC90. DNA sequences were obtained by the dideoxynucleotide chain termination method (United States Biochemicals).
FLAG epitope-tagged recombinant hTFIIIC90 was expressed and purified by infection of Sf21 cells with recombinant baculovirus followed by affinity chromatography of a whole-cell extract on M2 agarose as described elsewhere (22). Briefly, a whole-cell extract was mixed with previously equilibrated M2 agarose beads in BC400 buffer (20 mM Tris-HCl [pH 7.9], 400 mM KCl, 20% glycerol, 0.01 mM dithiothreitol [DTT]). After being washed with the same buffer, the bound protein was eluted with 0.2 mg of FLAG peptide per ml in BC100 buffer.
Antigen and antiserum preparation.
To produce 10-histidine-tagged recombinant proteins for use as antigens, cDNAs encoding two fragments (residues 146 to 434 and residues 497 to 796) of hTFIIIC90 were subcloned into pET-19b vectors. The resulting plasmids were introduced into Escherichia coli BL21(DE3)pLysS (37). After induction with isopropyl-β-d-thiogalactopyranoside, recombinant proteins were purified by Ni+2-nitrilotriacetic acid-agarose affinity chromatography followed by SDS-PAGE. To prepare antisera, New Zealand White rabbits were injected and boosted subsequently every 3 weeks with excised gel slices emulsified with Freund’s adjuvant. Blood was collected 10 to 15 days after each boost. The antigens were cross-linked to CNBr-activated Sepharose 4B (Pharmacia) according to the manufacturer’s instructions and then used to affinity purify corresponding antisera as described previously (16).
Immunodepletion and immunoprecipitation of HeLa nuclear extracts with anti-hTFIIIC90 antibodies.
Antigen-purified anti-hTFIIIC90 antibodies were first bound to protein A agarose (Oncogene Sciences) and then covalently cross-linked to the beads with dimethyl pimelimidate as described previously (16). As a control, the individual preimmune sera were processed the same way as immune sera. Antibody-coupled beads were incubated for 2 to 3 h at 4°C with HeLa nuclear extract in buffer BC (20 mM Tris-HCl [pH 8], 20% glycerol, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM DTT) containing 500 mM KCl (BC500) and 0.1% NP-40. After centrifugation, the supernatant was dialyzed against BC100 and aliquots were frozen. The beads were washed extensively with the same buffer. The bound proteins were eluted with glycine (pH 2.5) and then neutralized with Tris-HCl (pH 8).
In vitro transcription assays.
Transcription reactions for human fMet-tRNAf, 5S RNA, and adenovirus VAI genes were performed in a 25-μl reaction mixture with 250 ng of each supercoiled DNA template, 60 mM KCl, 6 mM MgCl2, 2 mM DTT, 8% glycerol, 10 mM HEPES (pH 7.9), a 0.6 mM concentration each of ATP, CTP, and UTP, 0.0125 mM GTP, and 2.5 μCi of [α-32P]GTP. Reactions were allowed to proceed for 1 h at 30°C before termination by addition of 50 μl of stop solution (100 mM Na acetate, 2 mM EDTA, 1% SDS, and 1 mg of yeast tRNA per ml). The mixtures were extracted with phenol-chloroform, precipitated with ethanol, and resolved on 8% polyacrylamide-7 M urea gels. Transcription reactions for RNA polymerase II templates (pG5HMC2AT and pMLΔ53) and activators (Gal4-AH) were performed as described previously (13).
Protein-protein interaction assays.
Recombinant proteins (0.5 to 1 μg) were bound to glutathione Sepharose or M2 agarose beads and incubated with 1 to 10 μl of reticulocyte lysate containing the other S35-labelled recombinant protein in BC400–0.5% NP-40. The beads were washed extensively with BC400–0.5% NP-40. Retained proteins were eluted by boiling the beads in SDS gel sample buffer and analyzed by autoradiographic analysis.
Preparation of hTFIIIC, TFIIIB, and a recombinant p300 HAT domain.
hTFIIIC and TFIIIB were prepared as described elsewhere (22). A FLAG epitope-tagged p300 HAT domain (amino acid positions 1195 to 1810) was expressed in E. coli and purified by M2 agarose affinity chromatography as described previously (22).
Preparation of substrates for HAT assays.
Human core histones were prepared from HeLa nuclear pellets as described previously (22). Native HeLa nucleosomes were prepared as described elsewhere (12). H3 N-terminal peptides were a gift from Y. Nakatani.
HAT assays.
HAT assays were performed as described elsewhere (6, 30). For solution assays (filter binding and autoradiography), purified HeLa core histones (1.5 μg) were incubated for 30 min at 30°C with the indicated amount of hTFIIIC90 or control proteins in 30-μl reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM EDTA, 10 mM butyric acid, and 25 nCi of [3H]acetyl coenzyme A (acetyl-CoA). Acetylation of synthetic H3 N-terminal peptides (300 ng) by hTFIIIC90 (100 ng) was monitored by the filter binding assay.
Nucleotide sequence accession number.
The GenBank accession number for the hTFIIIC90 sequence is AF142328.
RESULTS
Cloning and sequence analysis of hTFIIIC90.
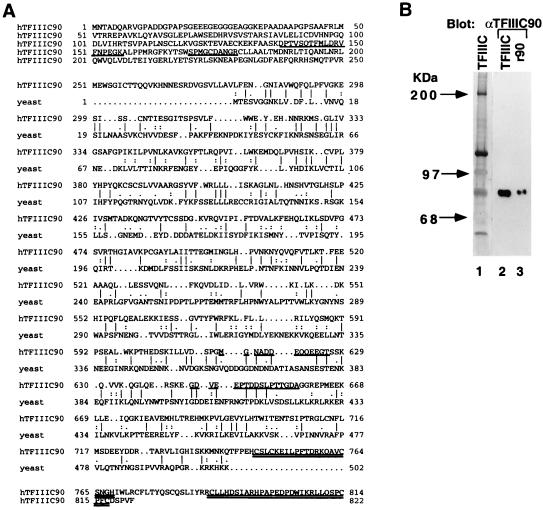
TFIIIC2 was purified as previously described (21), and the derived 90-kDa protein was subjected to microsequencing. The protein sequence information for four internal peptides was used to clone the corresponding cDNA. The deduced open reading frame of hTFIIIC90 cDNA encodes a polypeptide of 822 amino acids with a calculated molecular mass of 92 kDa (Fig. 1A). hTFIIIC90 contains two potential zinc finger motifs at the C terminus (Fig. 1A). A search of the National Center for Biotechnology Information nonredundant databases and the Sanger Centre Caenorhabditis elegans genome databases with the BLAST network service and the Genetics Computer Group-GAP program servicer was performed to determine sequence similarities between hTFIIIC90 and other proteins. hTFIIIC90 shows no significant sequence relationships to the 138-, 131-, 95-, 91-, and 55-kDa subunits of yTFIIIC. However, the C-terminal two-thirds of hTFIIIC90 shows 27% identity and 42% similarity to a hypothetical yeast protein (GenBank accession no. Z73205) (Fig. 1A) with a calculated molecular mass of 58 kDa. hTFIIIC90 also shows similar sequence relationships (overall, 26 to 28% identity and 36 to 38% similarity) to two hypothetical C. elegans proteins (Sanger Centre database accession no. CE12284 and CE02287).
FIG. 1.
(A) Predicted amino acid sequence of hTFIIIC90 and sequence alignments with a hypothetical yeast protein. The peptide sequences obtained by microsequence analyses are underlined. Two potential zinc finger motifs are doubly underlined. (B) Identification of the cDNA-encoded protein as the 90-kDa hTFIIIC2 subunits. The polypeptide components of a highly purified TFIIIC2 preparation were analyzed in a silver-stained gel (lane 1). Anti-hTFIIIC90 antibodies were used to detect hTFIIIC90 in a highly purified TFIIIC2 preparation (lane 2) and in a purified baculovirus-expressed FLAG-hTFIIIC90 (lane 3).
To verify that the cloned cDNA indeed encodes a bona fide subunit of hTFIIIC, antibodies were raised against purified bacterially expressed recombinant proteins corresponding to the C-terminal two-thirds of the hTFIIIC90 cDNA open reading frame. Anti-hTFIIIC90 antibodies reacted strongly and specifically with a protein band of 90 kDa in a highly purified TFIIIC2 fraction (Fig. 1B, lane 2) in an immunoblot assay. To determine whether the cloned cDNA contained the complete coding sequence, the corresponding open reading frame was expressed from a baculovirus vector in Sf21 cells as a recombinant FLAG-tagged protein (FLAG-hTFIIIC90). In an immunoblot analysis, the purified recombinant FLAG-tagged hTFIIIC90 showed a slightly slower mobility in SDS-PAGE than did native hTFIIIC90 (Fig. 1B, lane 3 versus lane 2), indicating that the cDNA clone encodes full-length hTFIIIC90. In further support of this notion, a stop codon was detected upstream of the first methionine codon and a polyadenylation signal followed by a poly(A) sequence was detected downstream of a termination codon in the cDNA.
The cDNA-encoded 90-kDa protein is a bona fide subunit of the TFIIIC2 complex and involved in RNA polymerase III-mediated transcription.
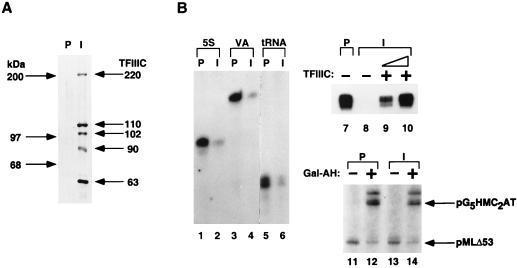
It has been established that hTFIIIC is required for the formation of stable preinitiation complexes on 5S, tRNA, and VA RNA genes (25). To determine whether the hTFIIIC90 cDNA-encoded protein is a component of the TFIIIC complex, anti-hTFIIIC90 antisera, as well as control preimmune sera, were used to immunoprecipitate TFIIIC from HeLa nuclear extracts under stringent conditions (0.5 M KCl–0.1% NP-40). As revealed by immunoblot analysis with antibodies to corresponding recombinant proteins (Fig. 2A), the immune sera precipitate but not the preimmune sera precipitate contained the 220-, 110-, 102-, 90-, and 63-kDa polypeptides that had been shown to copurify and correlate with TFIIIC2 activity (21, 53). These results indicate that hTFIIIC90 is an integral tightly associated component of the TFIIIC2 complex. To determine the requirement for hTFIIIC90 in the transcription of 5S RNA, tRNA, and VA RNA genes, nuclear extract was depleted with anti-hTFIIIC90 antibodies and tested in a transcription assay. The high levels of transcription from 5S RNA, tRNA, and VA RNA genes were dramatically reduced after treatment with anti-TFIIIC90 sera and partial removal (data not shown) of hTFIIIC90 but were unaffected by treatment with preimmune sera (Fig. 2B, lanes 1 to 6). The specificity of TFIIIC90 (and associated components) for RNA polymerase III is indicated by the lack of an effect of immunodepletion on high levels of basal and activated (Gal4-AH) transcription by RNA polymerase II on corresponding (pMLΔ53 and pG5HMC2AT) templates (Fig. 2B, lanes 11 and 13 versus lanes 12 and 14). Significantly, addition to a completely immunodepleted extract (data not shown) of an immunopurified TFIIIC complex containing TFIIIC1 and TFIIIC2 (47), which are both required to restore transcription to extracts treated with anti-hTFIIIC110 sera (34), restored transcription from the VAI template to levels similar to those observed with extracts treated with preimmune sera (Fig. 2B, lanes 7 to 10). Thus, hTFIIIC90 and associated polypeptides within TFIIIC are necessary for RNA polymerase III-mediated transcription.
FIG. 2.
Immunoprecipitation of the TFIIIC2 complex and immunodepletion of TFIIIC transcription activity by anti-hTFIIIC90 antibodies. (A) Immunoprecipitates from HeLa nuclear extracts treated with preimmune (P) and immune (I) anti-hTFIIIC90 antibodies in BC500–0.1% NP-40 were subjected to Western blot analysis. The polypeptides were detected by a mixture of antibodies against hTFIIIC220, -110, -102, -90, and -63. (B) Nuclear extracts treated with preimmune (P) and immune (I) anti-hTFIIIC90 antibodies were used for in vitro transcription assays with the 5S, VAI, and tRNA templates (lanes 1 to 6) and with the pG5HMC2AT and pMLΔ53 templates (lanes 11 to 14). The immunodepleted extract employed for lanes 1 to 6 retained a small amount (<10%) of hTFIIIC90 (data not shown). The immunodepleted extract employed for lanes 7 to 10 had no detectable hTFIIIC90 (data not shown), and immunopurified TFIIIC (the same preparation analyzed for Fig. 1B) was added in lanes 9 (1 μl) and 10 (2 μl).
hTFIIIC90 interacts with hTFIIIC220, hTFIIIC110, and hTFIIIC63, as well as hTFIIIB90, hRPC62, and hRPC39.
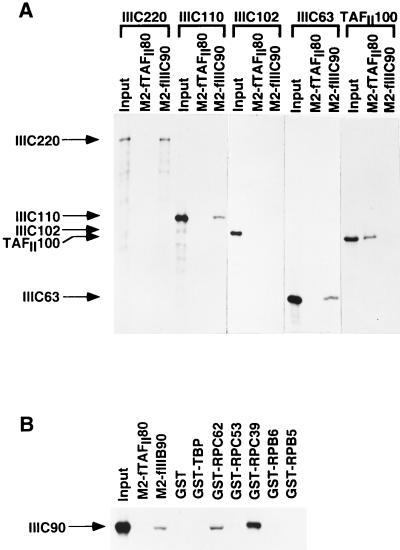
To further understand the architecture of the TFIIIC2 complex, we determined the ability of purified baculovirus-expressed FLAG-hTFIIIC90, immobilized on M2 agarose, to bind to previously cloned TFIIIC2 subunits (hTFIIIC220, hTFIIIC110, hTFIIIC102, and hTFIIIC63) that had been translated in vitro. As shown in Fig. 3A, FLAG-hTFIIIC90 interacted with hTFIIIC220, hTFIIIC110, and hTFIIIC63 but not with hTFIIIC102 or with the human TAFII100 (hTAFII100) subunit of TFIID. As a further indication of the specificity of these interactions, hTFIIIC220, hTFIIIC110, and hTFIIIC63 failed to interact with the FLAG-tagged hTAFII80 subunit of TFIID, whereas the hTAFII100 subunit of TFIID did show the expected interaction with hTAFII80 (Fig. 2A).
FIG. 3.
Interactions of hTFIIIC90 with subunits of TFIIIC2 and TFIIIB and an initiation-specific RNA polymerase III subcomplex. Input samples contained 10% of the amounts used for the interactions. (A) Reticulocyte lysates containing expressed 35S-hTFIIIC220, -hTFIIIC110, -hTFIIIC102, -hTFIIIC63, or -hTAFII100 were incubated with M2 agarose-immobilized FLAG-hTAFII80 or M2 agarose-immobilized FLAG-hTFIIIC90, and samples were washed, eluted, and analyzed as described in Materials and Methods. (B) Reticulocyte lysates containing expressed 35S-hTFIIIC90 were incubated with M2 agarose-immobilized hTAFII80 or M2 agarose-immobilized FLAG-hTFIIIB90 and with glutathione Sepharose-immobilized GST, GST-TBP, GST-RPC62, GST-RPC53, GST-RPC39, GST-RPB6, or GST-RPB5, and samples were washed, eluted, and analyzed as described in Materials and Methods. The amounts of the immobilized tagged recombinant proteins were normalized by SDS-PAGE with Coomassie blue staining. Equivalent amounts of the various FLAG-tagged proteins or the GST fusion proteins were bound to M2 agarose or glutathione Sepharose, respectively.
In vertebrates, preinitiation complex assembly on tRNA genes involves promoter recognition by TFIIIC followed by recruitment of TFIIIB and RNA polymerase III through protein-protein interactions (4, 25). Previously, we had shown interactions of hTFIIIC102 with hTFIIIC63, hTFIIIB90, and TBP, as well as interactions of hTFIIIC63 with hTFIIIB90, TBP, and hRPC62 (18). In order to further explore the interactions of TFIIIC with TFIIIB and RNA polymerase III, we investigated the ability of in vitro-translated hTFIIIC90 to interact with either purified baculovirus-expressed or purified bacterially expressed TFIIIB subunits (hTFIIIB90 and TBP), RNA polymerase III-specific subunits (hRPC62, hRPC53, and hRPC39), and common (I, II, and III) RNA polymerase subunits (human RPB5 [hRPB5] and hRPB6). As shown in Fig. 3B, hTFIIIC90 was bound to M2 agarose-immobilized FLAG-hTFIIIB90 but not to M2 agarose-immobilized FLAG- hTAFII80. Similarly, hTFIIIC90 was bound to glutathione Sepharose-immobilized glutathione S-transferase (GST)- hRPC62 or GST-hRPC39 but not to glutathione Sepharose-immobilized GST alone, GST-TBP, GST-hRPC53, GST-hRPB6, or GST-hRPB5.
hTFIIIC90 is a HAT.
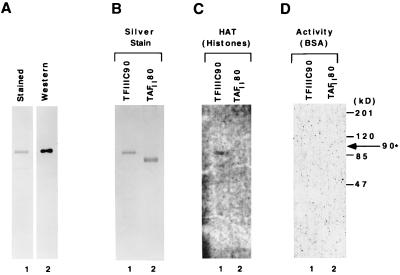
Our recent studies showed that hTFIIIC contains HAT activity and further suggested, on the basis of an in-gel assay, the presence of three independent HATs corresponding to the 220-, 110-, and 90-kDa subunits of TFIIIC2 (22). An analysis of recombinant TFIIIC110 confirmed that this subunit is indeed a HAT. To further document an intrinsic HAT activity in hTFIIIC90, full-length FLAG-hTFIIIC90 was expressed via a baculovirus vector in Sf21 cells and purified (M2 agarose affinity chromatography) to apparent homogeneity as assayed by SDS-PAGE followed by silver staining and immunoblotting (Fig. 4A). Recombinant FLAG-tagged hTFIIIC90 and hTAFII80 (as a negative control) were also subjected to SDS-PAGE in parallel gels containing either no carrier proteins (Fig. 4B), histones (Fig. 4C), or bovine serum albumin (Fig. 4D). Subsequent silver staining (Fig. 4B) and in-gel HAT assays with [3H]acetyl-CoA revealed a radiolabelled 90-kDa band in the presence of histones (Fig. 4C) but not in the presence of bovine serum albumin (Fig. 4D) and only with hTFIIIC90 and not with TAFII80. These results show that hTFIIIC90 contains an intrinsic HAT activity but no apparent autoacetylation activity. However, comparison of the hTFIIIC90 sequence with known HAT domains failed to reveal any significant sequence similarity, consistent with a similar situation for TAFII250 (30).
FIG. 4.
Recombinant hTFIIIC90 possess HAT activity. (A) FLAG epitope-tagged hTFIIIC90 was expressed in Sf21 cells and purified by M2 agarose affinity chromatography. The protein was resolved by SDS–8% PAGE and either subjected to staining (lane 1) or Western blotting (lane 2). (B to D) Baculovirus-expressed hTFIIIC90 and hTAFII80 samples were electrophoresed in an SDS–8% polyacrylamide gel containing either no protein (B), histones (C), or bovine serum albumin (D) and subjected either to silver staining (B) or to an in-gel HAT activity assay (C and D).
Substrate specificity of hTFIIIC90 HAT activity.
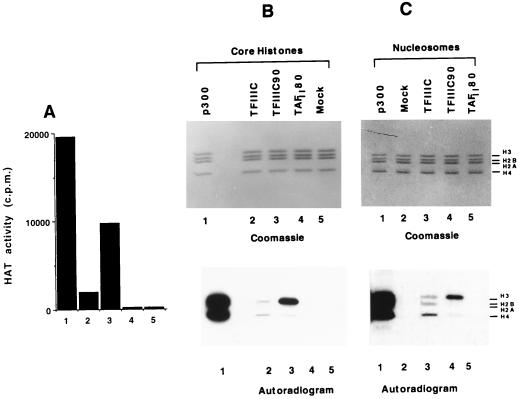
The HAT activity of hTFIIIC90 was further characterized by solution assays that measure acetylation of highly purified HeLa core histones. Purified recombinant FLAG-hTFIIIC90 (200 ng) was compared with a purified recombinant FLAG-p300 fragment (HAT domain) (25 ng) and immunoaffinity-purified hTFIIIC (200 ng) as positive controls and with purified recombinant FLAG-hTAFII80 and a mock-purified hTFIIIC90 fraction (from cells not expressing FLAG-hTFIIIC90) as negative controls. Consistent with previous results (22, 31), p300 showed a high level of activity (Fig. 5A, lane 1), while hTFIIIC showed a modest but significant level of activity (lane 2). hTFIIIC90 showed a high level of HAT activity (Fig. 5A, lane 3) that was 50-fold above the background level observed with the mock-purified hTFIIIC90 fraction (lane 5) and approximately half the level observed with the p300 fragment. FLAG-TAFII80, purified by the same procedure employed for FLAG-hTFIIIC90, did not show HAT activity and served as an additional negative control (Fig. 5A, lane 4).
FIG. 5.
Substrate specificity of hTFIIIC90 HAT activity. (A and B) Baculovirus-expressed hTFIIIC90 shows a strong HAT activity with highly purified HeLa core histones. The HAT activity assay was performed with 1.5 μg of HeLa core histones, [3H]acetyl-CoA, and 25 ng of the p300 HAT domain (lane 1), 200 ng of TFIIIC (lane 2), 200 ng of hTFIIIC90 (lane 3), 200 ng of hTAFII80 (lane 4), or mock immunoprecipitate (lane 5) and subjected to filter binding assay (A) or SDS-PAGE to assess the substrate specificity. The gels were stained with Coomassie brilliant blue (top) and then analyzed by fluorography (bottom) (B and C). (C) hTFIIIC90 acetylates HeLa nucleosomal histones. The HAT assay was performed as described above. Native HeLa nucleosomes were incubated with 25 ng of recombinant p300 HAT domain (lane 1), mock immunoprecipitate (for hTFIIIC90) (lane 2), 0.5 μg of TFIIIC (lane 3), 200 ng of purified recombinant hTFIIIC90 (lane 4), and 200 ng of purified TAFII80 (lane 5).
The histone substrate specificity of hTFIIIC90 was assessed by electrophoretic resolution of radiolabelled proteins followed by fluorography. Interestingly, among all of the four free core histones, hTFIIIC90 acetylated predominantly H3 (Fig. 5B, lane 3). hTFIIIC90 also acetylated H3 in native HeLa nucleosomes (Fig. 5C, lane 4). In agreement with the filter binding assay data, no radiolabelled histone bands were detected in assays containing the mock immunoprecipitate or TAFII80 (Fig. 5B, lanes 4 and 5, and C, lanes 2 and 5). As reported previously (31), the p300 fragment acetylates both nucleosomal and free histones (Fig. 5B and C, lanes 1).
Taken together, these results show that hTFIIIC90 is a HAT with a unique specificity for free and nucleosomal H3.
Site specificity of hTFIIIC90 HAT activity.
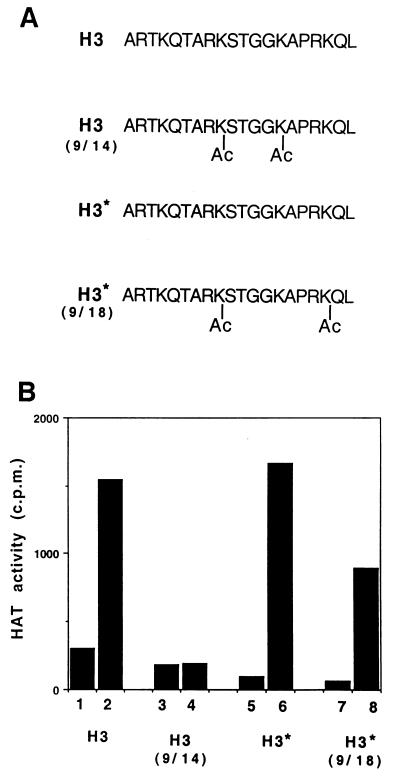
Acetylation of histone H3 has been closely linked to transcription activation. The acetylation of sites in the H3 N-terminal tail is nonrandom and correlates with specific biological processes such as replication and transcription (35, 41). In order to elucidate a possible site specificity for the hTFIIIC90 HAT activity, differentially acetylated H3 N-terminal peptides were employed as substrates for highly purified recombinant hTFIIIC90. The peptide sequences and the positions of acetate groups are shown in Fig. 6A. In agreement with the results described above for intact histones, significant levels of acetylation by hTFIIIC90 were observed for unacetylated peptides (Fig. 6B, lanes 2 and 6). The 9,18-diacetylated H3 peptide was also a good substrate for hTFIIIC90 (Fig. 6B, lane 7 versus lane 8). However, hTFIIIC90 showed no acetylation of the 9,14-diacetylated H3 peptide (Fig. 6, lane 3 versus lane 4). These results suggest that Lys14 is a preferred site for acetylation by hTFIIIC90, although a possible acetylation of Lys9 has not been examined. It is noteworthy that Lys14 is also a preferred acetylation site by other transcriptional coactivators (yeast and human GCN5 and Drosophila TAFII230) with HAT activities (30, 33).
FIG. 6.
Site specificity of hTFIIIC90 histone H3 acetylation. (A) The sequences of the peptides used for panel B are shown. Sites where ɛ-N acetyllysine was incorporated during peptide synthesis in order to mimic sites that are acetylated in vivo are indicated by Ac. Peptides H3 and the diacetylated H3(9/14) are straight chain, and H3* and H3*(9/18) were synthesized by the multiple antigen peptide method. (B) Histone H3 amino-terminal peptides were incubated either with only FLAG-elution buffer (lanes 1, 3, 5, and 7) or with highly purified hTFIIIC90 (lanes 2, 4, 6, and 8). Acetylation of the peptides was assessed by measuring [3H]acetate incorporation by using the filter binding assay.
DISCUSSION
TFIIIC is a multifunctional protein complex required for promoter recognition, recruitment of TFIIIB, and RNA polymerase III and chromatin-mediated transcription. Toward a more detailed analysis of the central role of hTFIIIC in these processes, we report the cloning and characterization of a cDNA encoding the 90-kDa subunit (hTFIIIC90) of the TFIIIC2 subcomplex that directly binds to tRNA and VA RNA gene promoters. This cDNA has been used to further investigate TFIIIC structure and function and documents novel interactions with TFIIIB and RNA polymerase III (18) and a novel histone-specific HAT activity (22).
Cloning and characterization of hTFIIIC90.
Amino acid sequence information from the 90-kDa polypeptide present in purified TFIIIC2 was used to obtain a cDNA encoding the corresponding protein. The encoded protein is a bona fide subunit of TFIIIC2 and involved in RNA polymerase III transcription since anti-hTFIIIC90 antibodies are able to immunoprecipitate all five subunits of the TFIIIC2 subcomplex and to inhibit TFIIIC function in transcription assays. Thus, these data indicate that hTFIIIC90 is an integral part of the TFIIIC2 complex.
The cloned hTFIIIC90 cDNA encodes a protein of 822 amino acids with a calculated molecular mass of 92 kDa. The deduced sequence of hTFIIIC90 contains two potential zinc finger motifs, in the C-terminal region, that might be involved in protein-DNA and/or protein-protein interactions. Database searches revealed no significant sequence similarities between hTFIIIC90 and any of the five cloned subunits (138, 131, 95, 91, and 55 kDa) of yTFIIIC. However, hTFIIIC90 shows significant sequence relationships with two hypothetical C. elegans proteins and with a 58-kDa hypothetical yeast protein. The latter could be the 60-kDa yTFIIIC subunit (3, 5, 32), whose sequence has not yet been reported.
Interactions of hTFIIIC90 with TFIIIC, TFIIIB, and RNA polymerase III.
As an additional step toward an understanding of the architecture of TFIIIC2, it was shown that hTFIIIC90 directly interacts with hTFIIIC220, hTFIIIC110, and hTFIIIC63 but not with hTFIIIC102. A further analysis of a possible role for hTFIIIC90 in TFIIIC-mediated recruitment of TFIIIB and RNA polymerase III demonstrated interactions of hTFIIIC90 with the hTFIIIB90 component of TFIIIB and the hRPC62 and hRPC39 components of the initiation-specific subcomplex of RNA polymerase III. Our previous study (18) showed that (i) hTFIIIC63 interacts with hTFIIIB90 and hRPC62 but not (in isolated form) with native RNA polymerase III, (ii) hTFIIIC102 interacts with hTFIIIB90 and hTFIIIC63, providing an additional link between TFIIIB and TFIIIC, but not with any of the components of the initiation-specific RNA polymerase III subcomplex, and (iii) isolated hTFIIIB90 interacts not only with hTFIIIC102 and hTFIIIC63 but also with hRPC39 (45). Thus, altogether, the past and present observations suggest that hTFIIIC90, like hTFIIIC63, may be localized close to the A box and that hTFIIIC90 cooperates with hTFIIIC63 in facilitating the recruitment of TFIIIB and RNA polymerase III through direct interactions with hTFIIIB90, hRPC62, and hRPC39. These interactions of hTFIIIC90, like certain interactions of hTFIIIC102 and hTFIIIC63, may reflect mechanisms for hTFIIIB and RNA polymerase III recruitment beyond those operative in yeast.
Possible significance of hTFIIIC90 HAT activity.
The presence of hyperacetylated histones is a diagnostic feature of transcriptionally active chromatin (7, 40). Recent discoveries showing that transcriptional regulatory proteins, including GCN5, PCAF, p300, CBP, TAFII250, and the nuclear hormone receptor coactivators ACTR and SRC1, possess intrinsic HAT activities have suggested that targeted histone acetylation facilitates transcription (2, 8, 11, 31, 36, 52). Although an involvement of these proteins in transcription activation has been established, direct roles for the intrinsic HAT activities in transcription are not broadly established. The most notable exception is yeast GCN5, whose modification of promoter proximal histones also appears causal for transcription, and human CBP (15, 23, 29, 42, 43). These results are further supported by in vitro transcription experiments from nucleosomal templates showing acetyl-CoA-dependent activation by the GCN5-containing SAGA complex (42).
Although earlier studies of the incorporation of acetylated histones into 5S gene-containing chromatin templates have reported the facilitation of both TFIIIA binding and TFIIIA-dependent transcription on chromatin templates (17, 26, 39, 41, 51), the effects of HAT-containing factors on the transcription of class III genes were reported only very recently. Thus, we were able to show that hTFIIIC has an intrinsic HAT activity that might play a role in transcription of a tRNA gene within a reconstituted chromatin template. The current demonstration of an intrinsic HAT activity in hTFIIIC90, coupled with a similar analysis of hTFIIIC110 (22), confirms the presence of multiple HATs in the hTFIIIC complex. hTFIIIC can acetylate both free and nucleosomal H3 and H4, as well as nucleosomal H2A, whereas hTFIIIC90 predominantly acetylates both free and nucleosomal H3. The strong H3 specificity of hTFIIIC90 suggests that the HAT-containing polypeptides of hTFIIIC may have different substrate specificities and functions in vivo. It also suggests a functional significance for the presence of multiple HATs in the hTFIIIC complex.
Acetylation of different lysine residues in the H3 tail is nonrandom and closely linked to various biological phenomena (35). hTFIIIC90 preferentially acetylates Lys14 in the H3 N-terminal tail. Lys14 in H3 is also a preferred substrate for acetylation by both human and yeast GCN5 (30). Since a role in transcription for GCN5-mediated histone acetylation is well established (15, 23, 42), preferential acetylation of Lys14 by hTFIIIC90 also suggests a critical role in the transcription of class III genes. To date, no HAT activity has been reported for yTFIIIC, consistent with the absence in yTFIIIC (or the yeast genome database) of counterparts to two of the hTFIIIC subunits with HAT activity (hTFIIIC220 and hTFIIIC110). However, the presence in the yeast genome database of a protein with significant sequence relationships to hTFIIIC90 raises the possibility that this hypothetical protein is a previously unrecognized yTFIIIC component and/or that it has a HAT activity.
ACKNOWLEDGMENTS
Y.-J.H. and T.K.K. contributed equally to this work.
We thank A. J. Berk for an hTFIIIC220 plasmid, Y. Nakatani for the p300 HAT domain clone and H3 N-terminal peptides, M. Guermah for TAFII80 baculoviruses and a TAFII100 plasmid, S. Malik for materials and technical advice on the transcription of RNA polymerase II templates, V. Palhan for a TAFII80 protein, and M. Teichmann for critical comments.
This work was supported by a grant (CA42567) from the National Institutes of Health to R.G.R.
REFERENCES
- 1.Arrebola R, Manaud N, Rozenfeld S, Marsolier M C, Lefebvre O, Carles C, Thuriaux P, Conesa C, Sentenac A. τ91, an essential subunit of yeast transcription factor IIIC, cooperates with τ138 in DNA binding. Mol Cell Biol. 1998;18:1–9. doi: 10.1128/mcb.18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister A J, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomew B, Kassavetis G A, Braun B R, Geiduschek E P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieker J J, Martin P L, Roeder R G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985;40:119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- 5.Braun B R, Bartholomew B, Kassavetis G A, Geiduschek E P. Topography of transcription factor complexes on the Saccharomyces cerevisiae 5S RNA gene. J Mol Biol. 1992;228:1063–1077. doi: 10.1016/0022-2836(92)90315-b. [DOI] [PubMed] [Google Scholar]
- 6.Brownell J E, Allis C D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 8.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homologue of yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 9.Burnol A F, Margottin F, Huet J, Almouzni G, Prioleau M N, Mechali M, Sentenac A. TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature. 1993;362:475–477. doi: 10.1038/362475a0. [DOI] [PubMed] [Google Scholar]
- 10.Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Lin R J, Schlitz R I, Chakaroborty D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyl transferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 12.Cote J, Utley R T, Workman J L. Basic analysis of transcription factor binding to nucleosomes. Methods Mol Genet. 1995;6:108–152. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 13.Ge H, Roeder R G. Purification, cloning and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 14.Geiduschek E P, Kassavetis G A. RNA polymerase III transcription complexes. In: Yamamoto K, editor. Transcription regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 247–280. [Google Scholar]
- 15.Gregory P D, Schmid A, Zavari M, Lui L, Berger S L, Horz W. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol Cell. 1998;1:495–505. doi: 10.1016/s1097-2765(00)80050-7. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 17.Howe L, Ranalli T A, Allis C D, Ausio J. Transcriptionally active Xenopus laevis somatic 5S ribosomal RNA genes are packaged with hyperacetylated histone H4, whereas transcriptionally silent oocyte genes are not. J Biol Chem. 1998;273:20693–20696. doi: 10.1074/jbc.273.33.20693. [DOI] [PubMed] [Google Scholar]
- 18.Hsieh Y-J, Wang Z, Kovelman R, Roeder R G. Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol Cell Biol. 1999;19:4944–4952. doi: 10.1128/mcb.19.7.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kassavetis G A, Bardeleben C, Bartholomew B, Braun B R, Joazeiro C A P, Pisano M, Geiduschek E P. Transcription by RNA polymerase III. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 107–126. [Google Scholar]
- 20.Khoo B, Brophy B, Jackson S P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 21.Kovelman R, Roeder R G. Purification and characterization of two forms of human transcription factor IIIC. J Biol Chem. 1992;267:24446–24456. [PubMed] [Google Scholar]
- 22.Kundu T K, Wang Z, Roeder R G. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol Cell Biol. 1999;19:1605–1615. doi: 10.1128/mcb.19.2.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo M H, Zhou J, Jambeck P, Churchill M E A, Allis C D. Histone acetyltransferase activity of yeast Gcn5 is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagna G, Kovelman R, Sukegawa J, Roeder R G. Cloning and characterization of an evolutionarily divergent DNA-binding subunit of mammalian TFIIIC. Mol Cell Biol. 1994;14:3053–3064. doi: 10.1128/mcb.14.5.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lassar A B, Martin P L, Roeder R G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983;222:740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- 26.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role of histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–74. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 27.L’Etoile N D, Fahnestock M L, Shen Y, Aebersold R, Berk A. Human transcription factor TFIIIC box B binding subunit. Proc Natl Acad Sci USA. 1994;91:1652–1656. doi: 10.1073/pnas.91.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manaud N, Arrebola R, Buffin-Meyer B, Lefebvre O, Voss H, Riva M, Conesa C, Sentenac A. A chimeric subunit of yeast transcription factor IIIC forms a subcomplex with τ95. Mol Cell Biol. 1998;18:3191–3200. doi: 10.1128/mcb.18.6.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 31.Ogryzko V V, Schiltz R L, Russanvoa V, Howard B H, Naktani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 32.Parsons M C, Weil P A. Purification and characterization of Saccharomyces cerevisiae transcription factor TFIIIC. J Biol Chem. 1990;265:5095–5103. [PubMed] [Google Scholar]
- 33.Schiltz R L, Mizzen C A, Vassilev A, Cook R G, Allis C D, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 34.Sinn E, Wang Z, Kovelman R, Roeder R G. Cloning and characterization of a TFIIIC2 subunit (TFIIICβ) whose presence correlates with activation of RNA polymerase III mediated transcription by adenovirus E1A expression and serum factors. Genes Dev. 1995;9:675–685. doi: 10.1101/gad.9.6.675. [DOI] [PubMed] [Google Scholar]
- 35.Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Conservation of depletion-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, Mckenna N J, Onate S A, Tasi S Y, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 37.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 38.Teichmann M, Dieci G, Huet J, Ruth J, Sentenac A, Seifart K H. Functional interchangeability of TFIIIB components from yeast and human cells in vitro. EMBO J. 1997;16:4708–4716. doi: 10.1093/emboj/16.15.4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tse C, Sera T, Wolffe A P, Hansen J C. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner B M, O’Neill L P. Histone acetylation in chromatin and chromosomes. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 41.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe A P. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 43.Wang L, Lin L, Berger S L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Roeder R G. TFIIIC1 acts through a downstream region to stabilize TFIIIC2 binding to RNA polymerase III promoters. Mol Cell Biol. 1996;16:6841–6850. doi: 10.1128/mcb.16.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Roeder R G. Three human RNA polymerase III specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Luo T, Roeder R G. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Roeder R G. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol Cell. 1998;1:749–751. doi: 10.1016/s1097-2765(00)80074-x. [DOI] [PubMed] [Google Scholar]
- 48.Werner M, Chaussivert N, Willis I M, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- 49.White R J. RNA polymerase III transcription in molecular biology intelligence unit. R. G. Austin, Tex: Landes Co.; 1994. [Google Scholar]
- 50.Willis I M. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 51.Wolffe A P, Wong J, Dmitry P. Activator and repressor: making use of chromatin to regulate transcription. Genes Cells. 1997;2:291–302. doi: 10.1046/j.1365-2443.1997.1260323.x. [DOI] [PubMed] [Google Scholar]
- 52.Yang X, Ogryzko V, Nishikawa J, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 53.Yoshinaga S K, L’Etoile N D, Berk A J. Purification and characterization of transcription factor IIIC2. J Biol Chem. 1989;264:10726–10731. [PubMed] [Google Scholar]
- 54.Yoshinaga S K, Boulanger P A, Berk A J. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci USA. 1987;84:3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]