Abstract
Acute myeloid leukemia (AML) with rearrangement of the lysine methyltransferase 2a gene (KMT2Ar) has adverse outcomes. However, reports on the prognostic impact of various translocations causing KMT2Ar are conflicting. Less is known about associated mutations and their prognostic impact. In a retrospective analysis, we identified 172 adult patients with KMT2Ar AML and compared them to 522 age-matched patients with diploid AML. KMT2Ar AML had fewer mutations, most commonly affecting RAS and FLT3 without significant impact on prognosis, except for patients with ≥2 mutations with lower overall survival (OS). KMT2Ar AML had worse outcomes compared with diploid AML when newly diagnosed and at relapse, especially following second salvage (median OS of 2.4 vs 4.8 months, P < 0.0001). Therapy-related KMT2Ar AML (t-AML) had worse outcomes compared with de novo KMT2Ar AML (median OS of 0.7 years vs 1.4 years, P < 0.0001). Allogeneic hematopoietic stem cell transplant (allo-HSCT) in first remission was associated with improved OS (5-year, 52 vs 14% for no allo-HSCT, P < 0.0001). In a multivariate analysis, translocation subtypes causing KMT2Ar did not predict survival, unlike age and allo-HSCT. In conclusion, KMT2Ar was associated with adverse outcomes regardless of translocation subtype. Therefore, AML risk stratification guidelines should include all KMT2Ar as adverse.
Subject terms: Leukaemia, Leukaemia
Introduction
Chromosomal translocations involving 11q23 where the lysine methyltransferase 2a gene (KMT2A) is located cause acute leukemias with high rates of resistance and relapse following standard treatments [1]. Despite an increased understanding of the leukemogenic mechanisms caused by KMT2A (also known as MLL) rearrangements (KMT2Ar), less is known about determinants of response and resistance to current treatments. KMT2Ar leukemias affect the myeloid lineage, lymphoid lineage, or both. They are associated with an adverse prognosis when occurring in infants, children, or adults with leukemia [2–4]. The KMT2A-MLLT3 fusion caused by t(9;11)(p21.3;q23.3) is the most common KMT2Ar in adults with AML, but more than 80 different fusion partners have been described [5].
Among chemotherapies associated with therapy-related AML (t-AML), topoisomerase II inhibitors are strongly associated with KMT2Ar leukemias, with a short latency to clinical manifestation [6]. In other genotypes of AML, previous exposure to chemotherapy leads to worse clinical outcomes, however, there have been conflicting reports on the prognostic impact of this factor in KMT2Ar AML [7–9]. The European LeukemiaNet (ELN) classifies t(9;11) as an intermediate-risk abnormality whereas other KMT2Ar AML were classified as adverse [10]. Nonetheless, results across studies on this difference in prognostic impact are not consistent, with some indicating similar adverse outcomes with all KMT2Ar AML [8, 9, 11, 12]. Genomic characterization of AML has refined prognostic models, although less is known about the mutational landscape of KMT2Ar AML and their prognostic impact [13]. Finally, much less is known about the clinical outcomes associated with KMT2Ar in the relapsed or refractory (R/R) setting.
We conducted a retrospective analysis of patients with KMT2Ar AML treated at our institution to characterize their genomic and phenotypic characteristics further and determine the association of these variables with prognosis and response to various lines of therapy.
Methods
Patient selection
We screened adult patients with AML treated at The University of Texas MD Anderson Cancer Center between January 1990 and December 2019. We identified 9465 patients with newly diagnosed AML, of whom 172 (2%) had KMT2Ar. We excluded patients with 11q23 translocations in whom KMT2Ar was not detected by fluorescence in situ hybridization (FISH) (Supplemental Fig. 1). Given that KMT2Ar occur in younger patients with AML, a cohort of patients with diploid karyotype were age-matched at a 3 to 1 ratio using propensity score and were used as a comparator for all subsequent analyses throughout this manuscript [14]. Among patients with KMT2Ar, 117 (68%) had mutational analysis done with targeted next-generation sequencing (NGS) panels. These panels included genes frequently involved in hematologic malignancies (panels of either 28, 53, or 81 genes depending on the time period; Supplemental Table 1) [15]. Fifty patients with KMT2Ar AML presenting to our institution at the time of relapse were included in the analysis of response and outcomes with subsequent lines of therapy (Supplemental Tables 2, 3). Treatment consisted of either high- or low-intensity regimens based on age and comorbidities. High-intensity regimens included combinations of cytarabine and idarubicin with or without a second nucleoside analog (i.e., cladribine, fludarabine, or clofarabine). Low-intensity regimens included either hypomethylating agents (i.e., azacitidine or decitabine) or low-dose cytarabine, with the addition of venetoclax more recently (starting in 2018) (Supplemental Table 4). Targeted therapies were added when available and indicated (Supplemental Table 4). Measurable residual disease (MRD) assessment was performed on bone marrow samples using multicolor flow cytometry as previously described [16]. This study was performed in accordance with the Declaration of Helsinki and was approved by the institutional review board.
Statistical methods
Patient characteristics were summarized using medians and ranges for continuous variables and frequencies or percentages for categorical variables. Continuous variables were compared using the Wilcoxon rank-sum test for pairwise comparisons and the Kruskal–Wallis for multiple comparisons. Categorical variables were compared using Fisher’s exact test. Responses were defined according to the International Working Group recommendations [17]. Overall survival (OS) was calculated from the time of diagnosis in the newly diagnosed population or treatment start date in patients with relapsed disease, to the time of death or last follow-up. Cumulative incidence of relapse (CIR) was calculated from the time of complete response (CR) or CR with incomplete count recovery (CRi) until relapse, censored for death in morphological remission or if the patient was alive at last follow-up. To minimize potential lead-time bias, landmark analyses were used while assessing the impact of allogeneic hematopoietic stem cell transplant (allo-HSCT), where only patients in first remission lasting beyond the median time to transplant were included [18]. The Kaplan–Meier method was used to estimate the probability of OS or CIR and were compared by the log-rank test. Univariate and multivariate Cox proportional hazards models were used to assess the association between patient characteristics and outcomes. Allo-HSCT was included as a time-dependent variable, and variables with P ≤ 0.05 in the univariate analysis were included in the initial multivariate analysis. Backward model selection was used to eliminate variables until all remaining were statistically significant with P < 0.05. Analyses were performed using GraphPad Prism (San Diego, CA, USA) and SAS version 9.4 (Cary, NC, USA).
Results
Patient characteristics and KMT2A rearrangements
The median age at diagnosis of patients with KMT2Ar AML was 52 years (range, 17–85 years), with a relatively higher proportion of females (61 vs 49% in diploid AML, P = 0.01) (Table 1). As previously described, KMT2Ar AML commonly manifested as monocytic in 67% of patients compared with 33% in the diploid karyotype group (P < 0.0001), was associated with markers of proliferation such as a higher percentage of bone marrow blasts at diagnosis and a lower platelet count. In this cohort, 69 patients (40%) had t-AML and KMT2Ar.
Table 1.
Baseline characteristics of newly diagnosed adults with AML.
| Characteristic | KMT2Ar | Age-matched diploid | P |
|---|---|---|---|
| Patients, no. | 172 | 522 | |
| Median age, years (range) | 52 (17–85) | 52 (10–86) | |
| Female, no. (%) | 104 (61) | 256 (49) | 0.01 |
| Monocytic phenotype, no. (%) | 116 (67) | 170 (33) | <0.0001 |
| WBC, median x 109/L (range) | 9.1 (1–270) | 10.9 (0–390) | 0.3 |
| Platelets, median x 109/L (range) | 49 (3–279) | 55 (1–635) | 0.009 |
| BM blast %, median (range) | 76 (20–98) | 52 (10–99) | <0.0001 |
| t-AML, no. (%) | 69 (40) | 35 (7) | <0.0001 |
| High-intensity treatment, no. (%) | 145 (84) | 432 (83) | 0.6 |
| Low-intensity treatment, no. (%) | 27 (16) | 90 (17) | |
| Allo-HSCT, no. (%) | 46 (27) | 118 (23) | 0.3 |
High-intensity treatment includes the combination of cytarabine and idarubicin or the addition of a nucleoside analog to the combination. Low-intensity treatment includes treatment with hypomethylating agents, low-dose cytarabine, or targeted therapies.
WBC white blood cell, BM bone marrow, LDH lactate dehydrogenase, Allo-HSCT allogeneic hematopoietic stem cell transplant, t-AML therapy-related AML.
P: Kruskal–Wallis or Fisher exact test.
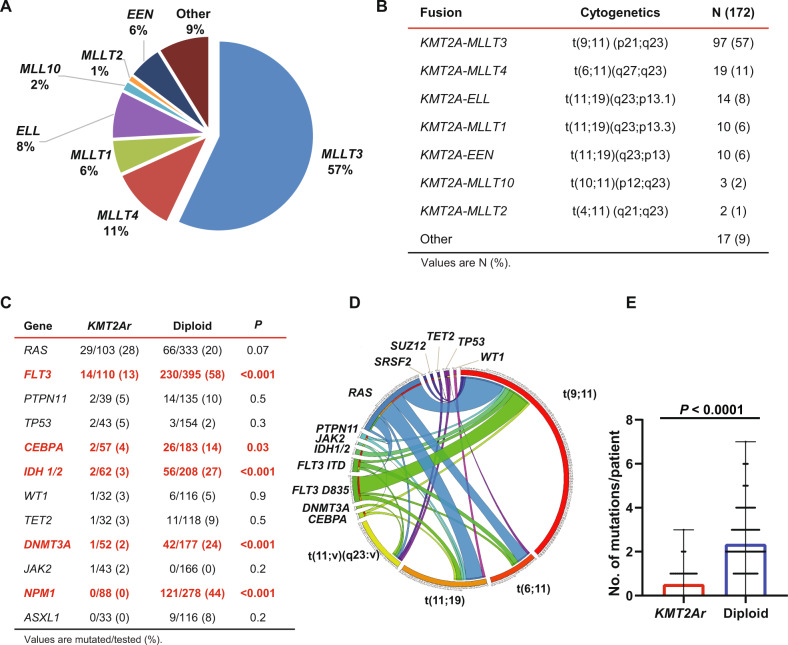
The most common translocations were t(9;11)(p21;q23)/KMT2A-MLLT3 detected in 97 patients (57%), followed by t(6;11)(q27;q23)/KMT2A-MLLT4 detected in 19 patients (11%), t(11;19)(q23;p13.1)/KMT2A-ELL in 14 patients (8%), t(11;19)(q23;p13.3)/KMT2A-MLLT1 in 10 patients (6%), t(11;19)(p23;q13)/KMT2A-EEN in 10 patients (6%), t(10;11)(q12;q23)/KMT2A-MLLT10 in 3 patients (2%), and t(4;11)(q21;q23)/KMT2A-MLLT2 in 2 patients (1%) (Fig. 1A, B). Seventeen patients (9%) had less common translocations occurring in ≤2 patients (Supplemental Table 5). Baseline characteristics by 11q23 translocations were mostly similar (Supplemental Table 6). Notably, t(9;11) presented more commonly with monocytic features compared to t(11:19) (75 vs 56%, P = 0.048) and t(11;v)(q23:v) (75 vs 45%, P = 0.009). No other significant differences in the characteristics were found, including rates of allo-HSCT.
Fig. 1. Fusion partner genes and mutational profile of adults with newly diagnosed KMT2Ar AML.
A Distribution of fusion partner genes. B Cytogenetics and distribution of 11q23 translocations. C Genes most commonly mutated in KMT2Ar AML compared to an age-matched cohort of AML with a diploid karyotype. D Circos plot depicting patterns of co-occurrence between mutations and various translocations leading to KMT2Ar. E Number of mutations per patient comparing KMT2Ar AML to an age-matched cohort of AML with a diploid karyotype.
Mutational profile and immunophenotype
Among patients with newly diagnosed AML, 117 patients (68%) had targeted NGS assessment of their diagnostic bone marrow specimens (Supplemental Fig. 2). Most patients examined had no additional mutations detected (65 of 117 patients or 56%). Overall, the analysis yielded 63 mutations in 52 patients with ≥1 mutation (52 of 117 patients or 44%). The median number of mutations per patient in the KMT2Ar cohort was 0.5 mutation (range, 0–3), which was significantly lower compared to diploid AML (median of two mutations/patient, range 0–7, P < 0.0001) (Fig. 1E). The most common mutations in KMT2Ar AML involved RAS in 29 of 103 patients (28%), followed by FLT3 in 14 of 110 patients (13%). These mutations were predominately D835 FLT3 kinase domain mutations (nine patients, 8%), and less commonly FLT3 internal tandem duplications (five patients, 5%). Mutations in FLT3 and RAS co-occurred in 5 of 110 patients (5%) (Fig. 1D and Supplemental Fig. 2). In addition, KMT2Ar patients had mutations in PTPN11 (5%), TP53 (5%), CEBPA (4%), and IDH1 (3%). Therefore, mutations associated with KMT2Ar most commonly involved the RAS pathway (RAS and PTPN11, with one patient having co-occurrence of these mutations).
The mutational landscape of KMT2Ar AML significantly differed from that of diploid AML (Fig. 1C). Mutations usually seen in clonal hematopoiesis were rare in KMT2Ar AML [19–21]. A DNMT3A mutation was detected in 1 of 52 patients (2 vs 24% in diploid karyotype, P < 0.001), a TET2 mutation in 1 of 32 patients (3 vs 9% in diploid karyotype, P = 0.5), and no ASXL1 mutations were detected (vs 8% in diploid karyotype, P = 0.2). Furthermore, there were no NPM1 mutations among 88 patients with KMT2Ar AML, and mutations in FLT3, IDH, and CEBPA were significantly less common when compared to diploid AML (Fig. 1C and Supplemental Fig. 2). There was no difference in the distribution of these mutations among different 11q23 translocations (Fig. 1D and Supplemental Table 7).
We analyzed by flow cytometry the immunophenotype of patients with KMT2Ar AML compared with diploid AML. We found that KMT2Ar AML was characterized by lower expression of CD7 (median: 4 vs 14%, P < 0.001), CD13 (median: 58 vs 72%, P < 0.001), and CD34 (median: 16 vs 34%, P < 0.001), and higher expression of CD33 (median: 94 vs 85%, P < 0.001) (Supplemental Fig. 3).
Morphologic and cytogenetic responses
Patients with newly diagnosed KMT2Ar AML had lower response rates to therapy compared with diploid AML with a CR/CRi rate of 72 vs 81% respectively (P = 0.01) (Table 2). However, the rate of MRD negative remission in evaluable patients who achieved a morphologic response was similar between the two groups (68 vs 68%, P = 0.9). Notably, patients with KMT2Ar had higher rates of early mortality compared to diploid AML with a 60-day mortality rate of 15 vs 7% (P = 0.004). There was no statistically significant difference in response rates when comparing various 11q23 translocations, although t(6;11) tended to have a lower CR rate compared to t(9;11) (58 vs 65%, P = 0.9) (Supplemental Table 8). As expected, the probability of achieving a response decreased with each line of therapy following relapse. The CR/CRi rates following first, second, or ≥3rd line therapies were 72, 43, and 9% respectively (Supplemental Table 9). However, KMT2Ar was associated with lower response rates at relapse, especially following ≥3rd line of therapy, where the CR/CRi rate was 9 vs 31% for diploid AML (P < 0.001) (Supplemental Table 9).
Table 2.
Response and early mortality rates of newly diagnosed adults with AML.
| Best Response | KMT2Ar | Age-matched diploid | P |
|---|---|---|---|
| N | 172 | 522 | |
| CR | 113 (66) | 403 (77) | 0.01 |
| CRi | 10 (6) | 18 (4) | |
| CR + CRi | 123 (72) | 421 (81) | |
| No response | 49 (28) | 101 (19) | |
| MRD negative by MFC (%) | 17/25 (68) | 90/132 (68) | 0.9 |
| Early mortality rates | |||
| 30-day mortality | 17 (10) | 20 (4) | 0.005 |
| 60-day mortality | 26 (15) | 38 (7) | 0.004 |
Values are n (%).
CR complete remission, CRi complete remission with incomplete hematologic recovery, MRD measurable residual disease assessed by multiparameter flow cytometry (MFC) following induction in evaluable patients.
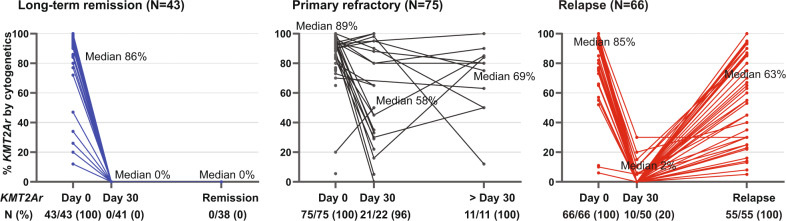
To assess the validity of KMT2Ar as a marker of disease evolution, we tracked the cytogenetic burden following treatment in the frontline and the R/R cohorts (Fig. 2). All patients who achieved a CR/CRi and a sustained remission following treatment had no detectable KMT2Ar by conventional cytogenetics or FISH on monitoring (0 out of 38 patients). Importantly, none of the patients with long-term remission had KMT2Ar detected on assessments done following induction treatment around Day 30, therefore highlighting the importance of achieving early cytogenetic remission in this setting. Conversely, patients who achieved morphologic remission (CR/CRi) and subsequently relapsed had a higher proportion of KMT2Ar detected at Day 30 following induction (10 of 50 patients or 20%). At the time of relapse, all patients with a cytogenetic analysis done had persistence of the KMT2Ar (55 of 55 patients or 100%). This suggests that KMT2Ar are founding events in this leukemia and that relapse following current treatments is not driven by the acquisition of novel drivers in the absence of KMT2Ar.
Fig. 2. Dynamic changes of the cytogenetic burden in adults with KMT2Ar AML following treatment.
Numbers depict the estimated % of KMT2Ar measured by fluorescence in situ hybridization (FISH) or conventional cytogenetics when FISH was not performed. Numbers below the X-axis indicate the proportion (%) of patients with KMT2Ar among those with available cytogenetic data. This analysis included unique patients from the newly diagnosed cohort in addition to patients who presented to our institution with relapsed or refractory disease. The long-term remission graph depicts those who achieved and maintained a morphologic remission whereas the relapse graph depicts those with initial morphologic remission following induction treatment and subsequent relapse.
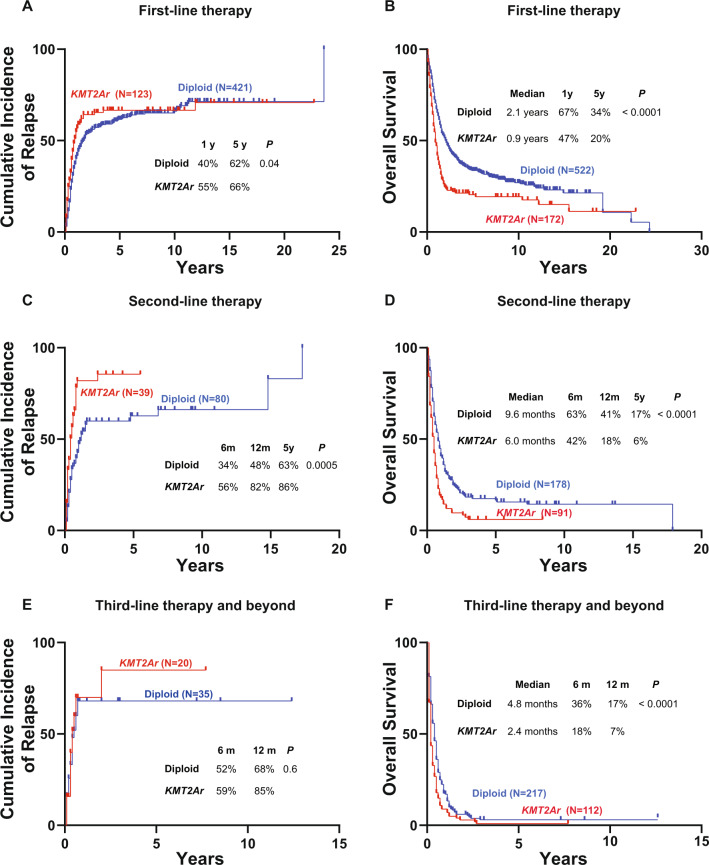
Relapse and overall survival
The median follow-up for this cohort was 8.1 years. Patients with newly diagnosed KMT2Ar AML had a higher risk of relapse compared to diploid AML with a CIR at 5 years of 66 vs 62% (P = 0.04) (Fig. 3A). These patients also had a worse OS rate: median OS of 0.9 years vs 2.1 years for diploid patients and a 5-year OS of 20 vs 34% (P < 0.0001). Following the first relapse, patients with KMT2Ar AML had a higher risk of subsequent relapse and worse OS compared to diploid AML with each line of therapy (Fig. 3C, D, E, F). The median OS of patients with KMT2Ar AML following 1st, 2nd, and ≥3rd lines therapies was 10.8 months, 6 months, and 2.4 months, respectively compared with 2.1 years, 9.6 months, and 4.8 months for diploid AML (P < 0.0001) (Fig. 3B, D, F).
Fig. 3. Cumulative incidence of relapse and overall survival for patients with KMT2Ar AML by a line of therapy compared with an age-matched cohort of AML with diploid karyotype.
A Cumulative incidence of relapse following first-line therapy. B Overall survival following first-line therapy. C Cumulative incidence of relapse following second-line therapy. D Overall survival following second-line therapy. E Cumulative incidence of relapse following third-line therapy. F Overall survival following third-line therapy. Treatment start date for second-line treatment and beyond was used for the calculation of time-to-event.
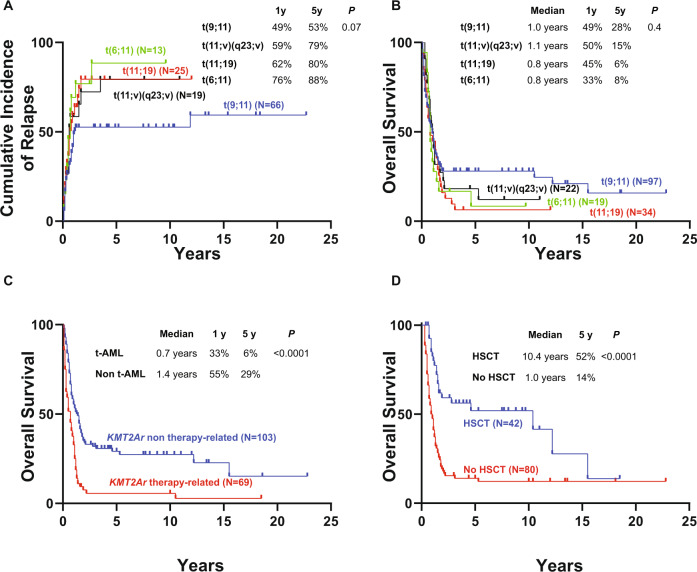
When newly diagnosed, including all patients with KMT2Ar AML regardless of age, t(9;11) was associated with slightly better outcomes compared to other KMT2Ar though these differences were not statistically significant (Fig. 4A, B). The median OS of patients with t(9;11) was 1 year with 5-year CIR and OS rates of 53 and 28%, respectively. t(6;11) was associated with the highest risk of relapse with a 5-year CIR of 88% and a median OS of 0.8 years and a 5-year OS of 8%. Similarly, patients with t(11;19) had a higher risk of relapse and worse OS with a median OS of 0.8 years and 5-year CIR and OS rates of 80 and 6%, respectively. Outcomes were better in younger patients (age <60 years) with newly diagnosed KMT2Ar AML, however, there was no statistically significant difference in OS comparing t(9;11) to other KMT2Ar (Supplemental Fig. 4C, D). However, patients with therapy-related KMT2Ar AML had a significantly higher chance of relapse and a lower likelihood of long-term survival compared to de novo KMT2Ar AML with a median OS of 0.7 years (vs 1.4 years, P < 0.0001) and 5-year CIR and OS rates of 80 (vs 65%, P = 0.009) and 6% (vs 29%, P < 0.0001), respectively (Fig. 4C and Supplemental Fig. 5). The difference in outcomes for the therapy-related disease was also noted in t(9;11) with a median OS of 0.5 years (vs 1.9 years, P < 0.0001) and 5-year CIR and OS rates of 62% (vs 48%, P = 0.1) and 9% (vs 48%, P < 0.0001) respectively (Supplemental Fig. 4A, B). There was no difference in outcomes comparing patients with KMT2A translocations and additional cytogenetic chromosomal abnormalities to those with KMT2A translocations only (Supplemental Figs. 6A, B and 7).
Fig. 4. Risk of relapse and overall survival in newly diagnosed KMT2Ar AML.
A Cumulative incidence of relapse by subtype of KMT2Ar. B Overall survival by subtype of KMT2Ar. C Overall survival of newly diagnosed KMT2Ar AML by therapy-related status. D Landmark analysis comparing overall survival of patients with newly diagnosed KMT2Ar AML who underwent an allogeneic hematopoietic stem cell transplant following the first remission to those who did not undergo transplant.
Prognostic impact of additional mutations in KMT2Ar AML
Given that mutational analysis in AML has improved risk stratification by adding to the established value of cytogenetic abnormalities, we analyzed the impact of additional mutations on outcomes of KMT2Ar AML [13, 22]. Mutations in either RAS or FLT3 did not affect OS in evaluable patients (Supplemental Fig. 8C, D). Compared to patients with KMT2Ar AML without detectable mutations, those who harbored any additional mutation also had a trend for worse OS though not statistically significant (5-year OS 14 vs 29% respectively, P = 0.1) (Supplemental Fig. 8A). However, a minority of patients who harbored ≥2 mutations (8 of 117 patients or 7%) had worse OS with a median of 0.5 years vs 1.1 years in those with fewer mutations (P = 0.01) (Supplemental Fig. 8B). These patients had co-occurrence of either RAS, FLT3, or PTPN11 mutations (six of eight patients), TET2 and TP53 mutations in one patient, and JAK2 and CEBPA in the remaining patient.
HSCT in KMT2Ar AML
In this cohort, 46 patients (27%) with KMT2Ar AML received an allo-HSCT. Among them, 42 (24%) were transplanted in the first remission as consolidation. We performed a landmark analysis to compare outcomes of patients with KMT2Ar AML who had allo-HSCT in the first remission compared to those who did not. Allo-HSCT was associated with significantly improved OS with a median of 10.4 years (vs 1 year for evaluable non-transplanted patients) and 5-year OS of 52% (vs 14% for evaluable non-transplanted patients, P < 0.0001) (Fig. 4D). The median time to relapse for the remaining four patients who received an allo-HSCT as salvage therapy was 7.8 months with an OS of 0.7 years.
CNS disease
KMT2Ar leukemias are frequently associated with central nervous system (CNS) involvement. Among newly diagnosed patients with KMT2Ar AML, 21 patients (12%) had CNS disease, three of whom had it at presentation (2%), whereas the rest developed it later in their treatment (17 patients or 10%). We next assessed the characteristics of these patients and whether there were predictors at baseline of CNS disease. When compared to KMT2Ar AML without CNS involvement (CNS-), we found no difference in variables usually thought to confer an increased risk of CNS involvement such as elevated white blood cell count, a monocytic phenotype (72 vs 67% in CNS- disease, P = 0.8) or presence of extramedullary involvement (35 vs 36% in CNS- disease, P = 0.8) (Supplemental Table 10). However, there was a higher proportion of patients with mutations in FLT3 among those with CNS involvement, though not statistically significant (22 vs 11% in CNS−, P = 0.2). Among KMT2Ar patients with CNS involvement, 13 of 21 patients (62%) successfully cleared their spinal fluid from leukemia cells following intrathecal and systemic therapies. There was no statistically significant difference in OS between the KMT2Ar CNS + and CNS− groups, though CNS + patients had a trend for worse long-term survival (5-year OS 0 vs 23%, P = 0.09) (Supplemental Fig. 9).
Predictors of relapse or death in KMT2Ar AML
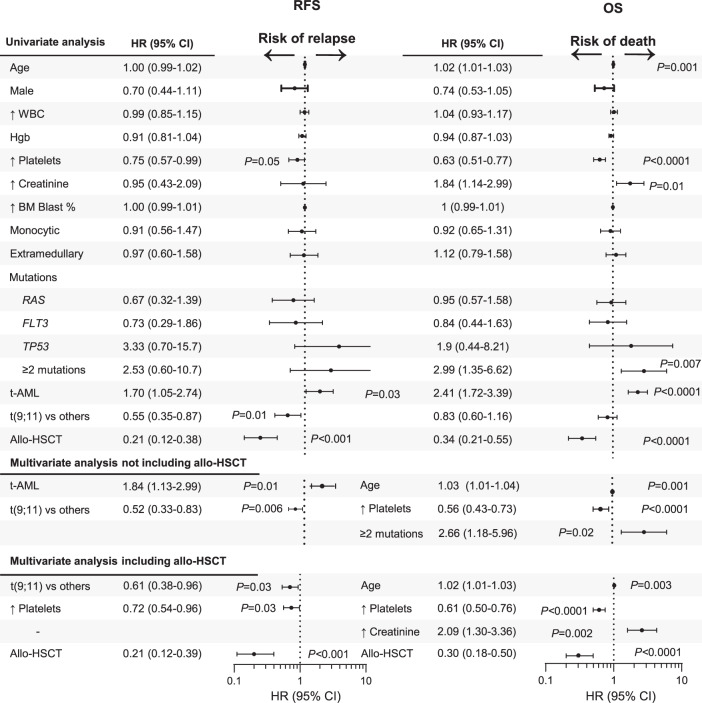
In order to assess the impact of confounding variables on prognosis, we conducted univariate and multivariate analyses predicting risks of relapse or death in patients with KMT2Ar AML (Fig. 5). Allo-HSCT was identified as an independent factor significantly associated with a decreased risk of relapse with a hazard ratio (HR) of 0.21 (95% CI 0.12–0.39, P < 0.001) and decreased risk of death with an HR of 0.30 (95% CI 0.18–0.50, P < 0.0001) (Fig. 5). Despite a decreased risk of relapse, t(9;11) was not associated with improved risk of death compared to other KMT2Ar, even when the transplant was not included in the multivariate analysis, which was done in order to address the potential beneficial effect of allo-HSCT on all confounding adverse features. Therapy-related status in KMT2Ar AML independently increased the risk of relapse when the transplant was not considered in the multivariate analysis with an HR of 1.84 (95% CI 1.13–2.99, P = 0.01), however, this prognostic impact was abrogated by the addition of allo-HSCT to the model. Similarly, having ≥2 mutations in addition to KMT2Ar independently predicted for higher risk of death with an HR of 2.66 (95% CI, 1.18–5.96, P = 0.02), but was not a predictor when allo-HSCT was added to the model. The only other factors identified as independent predictors of the risk of death were age, low platelet count, and an elevated creatinine at diagnosis.
Fig. 5. Univariate and multivariate analyses of factors predicting risks of relapse or death in newly diagnosed KMT2Ar AML.
Variables with P ≤ 0.05 were included in the multivariate analysis. RFS relapse-free survival, OS overall survival, HR hazard ratio, WBC white blood cell count, Hgb hemoglobin, BM bone marrow, t-AML therapy-related AML, allo-HSCT allogeneic hematopoietic stem cell transplant.
Discussion
In this study, we show that the mutational landscape of KMT2Ar AML is unique and characterized by a relative paucity of mutations, where almost all cells at diagnosis have KMT2Ar by cytogenetic analysis, which persists following resistance or relapse. This confirms previous clinical observations and laboratory studies indicating that KMT2Ar is the founding event and a potent driver in this leukemia with a minimal contribution by additional mutations [23–28]. Mutations most commonly involved the RAS or FLT3 genes and did not affect prognosis. These mutations are likely subclonal and occur later in the pathogenesis of the disease but possibly confer some proliferative advantage [29]. The rare occurrence of mutations frequently detected in clonal hematopoiesis indicates that KMT2Ar are not preceded by a precursor state, a common feature of non-fusion-driven AML [20]. However, a minority of patients had two or three mutations in addition to KMT2Ar which was associated with an adverse prognosis. These patients had co-occurrence of mutations in either RAS, FLT3, or PTPN11.
The incidence of KMT2Ar in this cohort of adults with newly diagnosed AML was 2% (172 of 9465 patients), which is lower than the incidence reported in other studies of 3–7% [3, 8, 9]. This is likely because we chose to focus our analysis on confirmed rearrangements when FISH was performed, while other studies have included other 11q23 abnormalities such as deletions. These 11q23 abnormalities without a clear translocation partner identified by conventional cytogenetics could harbor cryptic KMT2Ar translocations in up to 45% of cases in one estimate [30]. We identified 4 of 50 patients (8%) in the relapsed cohort with a cryptic KMT2Ar, while none were detected in the frontline cohort (Supplemental Fig. 10). In addition to routine FISH use at AML diagnosis, incorporation of a novel clinical assay based on RNA-sequencing or whole genome sequencing would improve the detection of cryptic KMT2A translocations [31–33]. Targeted sequencing performed in this cohort did not include identification of KMT2A partial tandem duplications, a well-described prognostic alteration involving this gene [34, 35].
We show that KMT2Ar AML in adults is characterized by high expression of CD33 (median expression of 94%), the target of gemtuzumab ozogamicin (GO). This is in line with a previous report on the immunophenotype of 17 of 19 KMT2Ar AML expressing CD33 (median expression 77%) [36]. This finding could bolster efforts investigating the addition of GO to the treatment of KMT2Ar AML in adults. A recent study by the Children’s Oncology Group demonstrated improved outcomes with the addition of GO in pediatric KMT2Ar AML [37]. As previously described, we found in this study a high rate of CNS involvement in adults with KMT2Ar AML (10 vs 3% in a general AML population). The NCCN (National Comprehensive Cancer Network) guidelines recommend a screening lumbar puncture (LP) to assess for CNS involvement in asymptomatic patients with monocytic AML, mixed phenotype acute leukemia, extramedullary disease, WBC >40 × 109/L, or presence of FLT3 mutations [38, 39]. Using these criteria, most adult patients with KMT2Ar AML would need a screening LP. We attempted to discriminate further predictors of CNS disease in KMT2Ar AML and found no clear predictors of CNS involvement in this entity.
There is a paucity of data on the outcomes of patients with relapsed KMT2Ar AML. This is the first study to our knowledge to examine outcomes for these patients with each line of therapy. In this study, patients with KMT2Ar AML had significantly worse outcomes following relapse compared to an age-matched population with a diploid karyotype. We found that the median OS for KMT2Ar AML after second-line therapy was 6 months and 2.4 months only after third-line therapy. This highlights the need to improve outcomes for patients with KMT2Ar AML, and the results of this study could be used when comparing available treatments to promising novel therapies in clinical development. Notably, early results from clinical trials investigating menin inhibitors, which are novel targeted agents for KMT2Ar leukemias or leukemias with other susceptible genotypes are highly encouraging [40–42].
A recent study by the Cancer and Leukemia Group B (CALGB) found that outcomes of patients with de novo AML, t(9;11) and age <60 years, excluding those who underwent allo-HSCT in first CR, were comparatively better than those with other 11q23 translocations (108 patients in this analysis) [25]. This was consistent with previous studies by the German Acute Myeloid Leukemia Intergroup where a cohort of 180 patients with 11q23 aberrations and age <60 years, were analyzed and t(9;11) had comparatively better outcomes [9]. We show similar results in our cohort when applying the same analysis criteria by restricting it to age <60 years, and de novo AML (Supplemental Fig. 4E). However, patients with these characteristics form only 26% (44 of 172 patients in our cohort) of adult patients with newly diagnosed KMT2Ar AML. Therefore, these results should not be applied to the general population with KMT2Ar AML. This study shows that the prognosis of KMT2Ar in adults with AML is adverse, regardless of the translocation, when looking at a diverse (all ages, de novo, and t-AML) large cohort of patients. Despite a lower risk of relapse in t(9;11) compared to other KMT2Ar, the difference in OS was not significant albeit a trend and was not an independent predictor of survival in the multivariate analysis. Therefore, risk stratification criteria such as the ones by the NCCN or the ELN, should reflect this data when indicating that among KMT2A rearrangements, t(9;11) is in the intermediate-risk category [38, 43]. The intermediate-risk assignment should apply only to patients with age <60 years, and non-therapy related t(9;11). Or alternatively, revise the criteria to include all KMT2Ar in the adverse risk category which would better reflect the characteristics of patients seen in the clinic. This is critical as we have shown that allo-HSCT in first CR is associated with significantly improved outcomes in all KMT2Ar AML. Certainly, no prognostic model is perfect, however, these risk models are used in clinical practice to determine which patients should undergo a consolidative allo-HSCT. Including all KMT2Ar in the adverse risk group would emphasize the importance of transplant for these patients. Current risk stratification models were developed largely based on cohorts of younger, fit patients with de novo AML, treated with intensive chemotherapy despite the fact that AML is largely a disease of older age (median age at diagnosis of 68). The advent of highly effective venetoclax-based therapies for older patients with AML invites for more inclusive risk stratification models regardless of age [44].
In conclusion, KMT2Ar is associated with adverse outcomes in adults with AML. It has a low mutational burden with a minimal associated prognostic impact. Adverse risk in KMT2Ar AML is worst in older patients and those with t-AML. Consolidation with an allo-HSCT following the first remission is associated with significantly improved outcomes leading to long-term survival. Outcomes of patients with relapsed KMT2Ar AML are dismal, highlighting the need for novel therapeutic strategies.
Supplementary information
Acknowledgements
GCI received funding through the K12 Paul Calabresi Clinical Scholarship Award (NIH/NCI K12 CA088084).
Author contributions
G.C.I., J.Z. and F.R. designed the study and wrote the manuscript. J.Z., G.C.I., K.S. and F.H. analyzed the data. W.Q., D.P. and J.N. performed the statistical analysis. Z.T. assisted with cytogenetic analysis. N.J.S., K.P.P., B.C., N.D., C.D.D., E.J., T.K., G.B., G.G.-M., M.K., M.A., H.M.K. and F.R. provided suggestions and revisions. G.C.I. and F.R. supervised the analysis. All authors read and approved the final manuscript.
Funding
This work was supported by an MD Anderson Cancer Center Support Grant (CA016672 from the National Institutes of Health, National Cancer Institute) and a Specialized Programs of Research Excellence grant.
Competing interests
G.C.I. received research funding from Celgene, Kura Oncology, Syndax, and Novartis, and received consultancy fees from Novartis and Kura Oncology. K.S. received research funding from Novartis and consulting or advisory fees from Otsuka, Novartis, Pfizer, and Takeda. N.J.S. has served as a consultant for Takeda Oncology, AstraZeneca, and Jazz Pharmaceuticals, reports receiving research grants from Takeda Oncology and Astellas Pharma Inc. and has received honoraria from Amgen. C.D.D. received research funding from Abbvie, Agios, Calithera, Cleave, BMS/Celgene, Daiichi-Sankyo, Forma, ImmuneOnc, Loxo, and received consultancy or advisory board fees from AbbVie, Agios, Novartis, Aprea, Celgene/BMS, ImmuneOnc, Notable Laboratories, and Takeda. E.J. received research funding from Abbvie, Adaptive Biotechnologies, Amgen, Bristol Myers Squibb, Pfizer, and Takeda and received advisory board fees from Genetech. N.D. has received research funding from Daiichi-Sankyo, Bristol Myers Squibb, Pfizer, Gilead, Sevier, Genentech, Astellas, Abbvie, Hanmi, Trovagene, FATE therapeutics, Amgen, Novimmune, Glycomimetics, Trillium, and ImmunoGen and has served in a consulting or advisory role for Daiichi-Sankyo, Bristol Myers Squibb, Arog, Pfizer, Novartis, Jazz, Celgene, AbbVie, Astellas, Genentech, Immunogen, Servier, Syndax, Trillium, Gilead, Amgen, Shattuck Labs, and Agios. T.K. has received research funding from Bristol Myers Squibb, Celgene, Sanofi, Amgen, BiolineRx, Incyte, Genentech/AbbVie, Pfizer, Jazz Pharmaceuticals, AstraZeneca, Astellas Pharma, Ascentage Pharma, Genfleet, Cyclacel and received consulting or advisory board fees from Novartis, Jazz Pharmaceuticals, Pfizer, AbbVie/Genentech, and Agios. G.B. has received research funding from Incyte, GlaxoSmithKline, Cyclacel, BiolineRx, MedImmune, Lilly, Oncoceutics, Ryvu Therapeutics, Janssen Scientific Affairs, Bristol Myers Squibb, AbbVie, Novartis, AstraZeneca, Mundipharma Research, PTC Therapeutics, BioTheryX, XBiotech, Arvinas, Astex Pharmaceuticals, TCR2 Therapeutics, Nkarta, Treadwell Therapeutics, Cellestia Biotech, and consulting or advisory board fees from Argenx, PTC Therapeutics, BiolineRx, BioTheryX, Nkarta, Treadwell Therapeutics, Novartis, Catamaran Bio, and Takeda. G.G.-M. received research funding from Celgene, Astex Pharmaceuticals, Amphivena, Helsinn Therapeutics, Novartis, AbbVie, Bristol Myers Squibb, Onconova Therapeutics, H3 Biomedicine, Merck and consulting or advisory board fees from Celgene, Astex Pharmaceuticals, Acceleron Pharma, Helssin, and AbbVie. M.K. has received research funding from AbbVie, Genentech, Roche, Lilly, Cellectis, Calithera Biosciences, Ablynx, Agios, Ascentage Pharma, AstraZeneca, Sanofi and received honoraria or advisory board fees from AbbVie, Genentech, Roche, Amgen, Stemline Therapeutics, KisoJi Biotechnology, Stemline Therapeutics, Forty Seven, and Janssen. M.A. received research funding from Daiichi-Sankyo, and consultancy honoraria from Syndax, Jazz, Celgene, Amgen, AstraZeneca, Dimensions Capital, and equity ownership from Reata, Aptose, Europics, Senti Bio, Chimerix, and Oncolyze. H.M.K. received research funding from Ariad, Astex, Bristol Myers Squibb, Cyclacel, Daiichi-Sankyo, Pfizer, Immunogen, Jazz, Novartis and honoraria from Pfizer, Immunogen, Actinium, and Takeda. F.R. received research funding from Amgen, Bristol Myers Squibb, Sunesis Pharmaceuticals, Pfizer, Xenecor, Macrogenetics, Taiho, Astex, Abbvie and honoraria for consulting or advisory role from Jazz, Amgen, Celgene, Astellas, Syros, Taiho, Novartis, AstraZeneca, Agios, and Bristol Myers Squibb.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ghayas C. Issa, Jabra Zarka.
Contributor Information
Ghayas C. Issa, Email: gcissa@mdanderson.org
Farhad Ravandi, Email: fravandi@mdanderson.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-021-00557-6.
References
- 1.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 2.Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–36. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 3.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 4.Hilden JM, Dinndorf PA, Meerbaum SO, Sather H, Villaluna D, Heerema NA, et al. Analysis of prognostic factors of acute lymphoblastic leukemia in infants: report on CCG 1953 from the Children’s Oncology Group. Blood. 2006;108:441–51. doi: 10.1182/blood-2005-07-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer C, Burmeister T, Gröger D, Tsaur G, Fechina L, Renneville A, et al. The MLL recombinome of acute leukemias in 2017. Leukemia. 2018;32:273–84. doi: 10.1038/leu.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Super H, McCabe N, Thirman M, Larson R, Le Beau M, Pedersen-Bjergaard J, et al. Rearrangements of the MLL gene in therapy-related acute myeloid leukemia in patients previously treated with agents targeting DNA-topoisomerase II. Blood. 1993;82:3705–11. doi: 10.1182/blood.V82.12.3705.3705. [DOI] [PubMed] [Google Scholar]
- 7.Menghrajani K, Zhang Y, Famulare C, Devlin SM, Tallman MS. Acute myeloid leukemia with 11q23 rearrangements: A study of therapy-related disease and therapeutic outcomes. Leuk Res. 2020;98:106453. doi: 10.1016/j.leukres.2020.106453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, Haferlach T. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102:2395–402. doi: 10.1182/blood-2003-02-0434. [DOI] [PubMed] [Google Scholar]
- 9.Krauter J, Wagner K, Schäfer I, Marschalek R, Meyer C, Heil G, et al. Prognostic factors in adult patients up to 60 years old with acute myeloid leukemia and translocations of chromosome band 11q23: individual patient data-based meta-analysis of the German Acute Myeloid Leukemia Intergroup. J Clin Oncol. 2009;27:3000–6. doi: 10.1200/JCO.2008.16.7981. [DOI] [PubMed] [Google Scholar]
- 10.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mrózek K, Heinonen K, Lawrence D, Carroll AJ, Koduru PR, Rao KW, et al. Adult patients with de novo acute myeloid leukemia and t(9; 11)(p22; q23) have a superior outcome to patients with other translocations involving band 11q23: a cancer and leukemia group B study. Blood. 1997;90:4532–8. doi: 10.1182/blood.V90.11.4532. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Kantarjian H, Pierce S, Faderl S, O’Brien S, Qiao W, et al. Prognostic significance of 11q23 aberrations in adult acute myeloid leukemia and the role of allogeneic stem cell transplantation. Leukemia. 2013;27:836–42. doi: 10.1038/leu.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Austin PC. Statistical criteria for selecting the optimal number of untreated subjects matched to each treated subject when using many-to-one matching on the propensity score. Am J Epidemiol. 2010;172:1092–7. doi: 10.1093/aje/kwq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luthra R, Patel KP, Reddy NG, Haghshenas V, Routbort MJ, Harmon MA, et al. Next-generation sequencing-based multigene mutational screening for acute myeloid leukemia using MiSeq: applicability for diagnostics and disease monitoring. Haematologica. 2014;99:465–73. doi: 10.3324/haematol.2013.093765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Jorgensen JL, Wang SA. How do we use multicolor flow cytometry to detect minimal residual disease in acute myeloid leukemia? Clin Lab Med. 2017;37:787–802. doi: 10.1016/j.cll.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Cheson BD, Bennett JM, Kopecky KJ, Büchner T, Willman CL, Estey EH, et al. Revised recommendations of the International Working Group for diagnosis, standardization of response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol. 2003;21:4642–9. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 18.Stelljes M, Beelen DW, Braess J, Sauerland MC, Heinecke A, Berning B, et al. Allogeneic transplantation as post-remission therapy for cytogenetically high-risk acute myeloid leukemia: landmark analysis from a single prospective multicenter trial. Haematologica. 2011;96:972–9. doi: 10.3324/haematol.2011.041004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059-74. [DOI] [PMC free article] [PubMed]
- 24.Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:330–7. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bill M, Mrózek K, Kohlschmidt J, Eisfeld AK, Walker CJ, Nicolet D, et al. Mutational landscape and clinical outcome of patients with de novo acute myeloid leukemia and rearrangements involving 11q23/<em>KMT2A < /em. Proc Natl Acad Sci USA. 2020;117:26340–6. doi: 10.1073/pnas.2014732117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milne TA. Mouse models of MLL leukemia: recapitulating the human disease. Blood. 2017;129:2217–23. doi: 10.1182/blood-2016-10-691428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grossmann V, Schnittger S, Poetzinger F, Kohlmann A, Stiel A, Eder C, et al. High incidence of RAS signalling pathway mutations in MLL-rearranged acute myeloid leukemia. Leukemia. 2013;27:1933–6. doi: 10.1038/leu.2013.90. [DOI] [PubMed] [Google Scholar]
- 28.Lavallée VP, Baccelli I, Krosl J, Wilhelm B, Barabé F, Gendron P, et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet. 2015;47:1030–7. doi: 10.1038/ng.3371. [DOI] [PubMed] [Google Scholar]
- 29.Hyrenius-Wittsten A, Pilheden M, Sturesson H, Hansson J, Walsh MP, Song G, et al. De novo activating mutations drive clonal evolution and enhance clonal fitness in KMT2A-rearranged leukemia. Nat. Commun. 2018;9:1770. doi: 10.1038/s41467-018-04180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox MC, Panetta P, Lo-Coco F, Del Poeta G, Venditti A, Maurillo L, et al. Chromosomal aberration of the 11q23 locus in acute leukemia and frequency of MLL gene translocation: results in 378 adult patients. Am J Clin Pathol. 2004;122:298–306. doi: 10.1309/RX27R8GJQM330C22. [DOI] [PubMed] [Google Scholar]
- 31.van der Burg M, Beverloo HB, Langerak AW, Wijsman J, van Drunen E, Slater R, et al. Rapid and sensitive detection of all types of MLL gene translocations with a single FISH probe set. Leukemia. 1999;13:2107–13. doi: 10.1038/sj.leu.2401595. [DOI] [PubMed] [Google Scholar]
- 32.Afrin S, Zhang CRC, Meyer C, Stinson CL, Pham T, Bruxner T, et al. Targeted next-generation sequencing for detecting MLL gene fusions in leukemia. Mol Cancer Res. 2018;16:279–85. doi: 10.1158/1541-7786.MCR-17-0569. [DOI] [PubMed] [Google Scholar]
- 33.Duncavage EJ, Schroeder MC, O’Laughlin M, Wilson R, MacMillan S, Bohannon A, et al. Genome sequencing as an alternative to cytogenetic analysis in myeloid cancers. N Engl J Med. 2021;384:924–35. doi: 10.1056/NEJMoa2024534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Döhner K, Tobis K, Ulrich R, Fröhling S, Benner A, Schlenk RF, et al. Prognostic significance of partial tandem duplications of the MLL gene in adult patients 16 to 60 years old with acute myeloid leukemia and normal cytogenetics: a study of the acute myeloid leukemia study group Ulm. J Clin Oncol. 2002;20:3254–61. doi: 10.1200/JCO.2002.09.088. [DOI] [PubMed] [Google Scholar]
- 35.Libura M, Asnafi V, Tu A, Delabesse E, Tigaud I, Cymbalista F, et al. FLT3 and MLL intragenic abnormalities in AML reflect a common category of genotoxic stress. Blood. 2003;102:2198–204. doi: 10.1182/blood-2003-01-0162. [DOI] [PubMed] [Google Scholar]
- 36.Baer MR, Stewart CC, Lawrence D, Arthur DC, Mrózek K, Strout MP, et al. Acute myeloid leukemia with 11q23 translocations: myelomonocytic immunophenotype by multiparameter flow cytometry. Leukemia. 1998;12:317–25. doi: 10.1038/sj.leu.2400933. [DOI] [PubMed] [Google Scholar]
- 37.Pollard JA, et al. Gemtuzumab ozogamicin improves event-free survival and reduces relapse in pediatric KMT2A-rearranged AML: results from the phase III children’s oncology group trial AAML0531. J. Clin. Oncol. 2014;32:3021–32. doi: 10.1200/JCO.2014.55.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollyea DA, Bixby D, Perl A, Bhatt VR, Altman JK, Appelbaum FR, et al. NCCN guidelines insights: acute myeloid leukemia, version 2.2021. J Natl Compr Cancer Netw. 2021;19:16–27. doi: 10.6004/jnccn.2021.0002. [DOI] [PubMed] [Google Scholar]
- 39.Jabbour E, Guastad Daver N, Short NJ, Huang X, Chen HC, Maiti A, et al. Factors associated with risk of central nervous system relapse in patients with non-core binding factor acute myeloid leukemia. Am J Hematol. 2017;92:924–8. doi: 10.1002/ajh.24799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang ES, Altman JK, Pettit K, De Botton S, Walter RP, Fenaux P, et al. Preliminary data on a phase 1/2A first in human study of the Menin-KMT2A (MLL) inhibitor KO-539 in patients with relapsed or refractory acute myeloid leukemia. Blood. 2020;136:7–8. doi: 10.1182/blood-2020-134942. [DOI] [Google Scholar]
- 41.McGeehan J. A first-in-class Menin-MLL1 antagonist for the treatment of MLL-r and NPM1 mutant leukemias. Proceedings of the 111th Annual Meeting of the American Association for Cancer Research; American Association for Cancer Research, Virtual Meeting, April; 2020. Abstract DDT01-01.
- 42.Issa GC, Ravandi F, DiNardo CD, Jabbour E, Kantarjian HM, Andreeff M. Therapeutic implications of menin inhibition in acute leukemias. Leukemia. 2021;35:2482–95. doi: 10.1038/s41375-021-01309-y. [DOI] [PubMed] [Google Scholar]
- 43.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Short NJ, Tallman MS, Pollyea DA, Ravandi F, Kantarjian H. Optimizing risk stratification in acute myeloid leukemia: dynamic models for a dynamic therapeutic landscape. J. Clin. Oncol. 2021;39:2535–38. doi: 10.1200/JCO.21.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.