Summary
Three limonoid natural products with selective anti-proliferative activity against BRAF(V600E) and NRAS(Q61K)-mutation dependent melanoma cell lines were identified. Differential transcriptome analysis revealed dependency of compound activity on expression of the mitochondrial cytochrome P450 oxidase CYP27A1, a transcriptional target of MITF. We determined that CYP27A1 activity is necessary for the generation of a reactive metabolite that proceeds to inhibit cellular proliferation. A genome-wide siRNA screen in combination with chemical proteomics experiments revealed gene–drug functional epistasis, suggesting that these compounds target mitochondrial biogenesis and inhibit tumor bioenergetics through a covalent mechanism. Our work suggests a strategy for melanoma specific targeting by exploiting the expression of MITF target gene CYP27A1 and inhibiting mitochondrial oxidative phosphorylation in BRAF mutant melanomas.
Introduction
Natural products are a rich source of bioactive compounds for drug discovery and are particularly instrumental as anticancer and antimicrobial therapeutics (Harvey, 2008; Dias, et al., 2012; Harvey, et al., 2015; Shen, 2015). A recent analysis has found that approximately 50% of approved anticancer drugs from 1940 to 2014 originated from natural products or directly derived therefrom (Newman & Cragg, 2016). Key examples are the plant-derived natural products vincristine and paclitaxel, which have been hallmark cancer therapeutics for almost 30 years (da Richa, et al., 2001). As part of an effort to identify new oncology targets and compound leads we profiled a portion of the Novartis natural product collection against a panel of 39 cell lines (lung, ovarian, colon and brain cell lineages) to look for selective cytotoxicity profiles (Shoemaker, 2006), followed by large scale profiling of selected compounds across cancer cell lines from diverse tissue lineages. Herein, we identified several limonoid natural products isolated from the Harrisonia perforata plant from Southeast Asia having a selective cytotoxicity profile. Haperforin G and haperforin C2, previously reported, and a previously uncharacterized limonoid, which we have named harrpernoid D, displayed selective anti-melanoma activity (Khuong-Huu, et al., 2001; Yan, et al., 2011; Zhang, et al., 2020). We identified that the melanoma-specific cytotoxicity is dependent on expression of an MITF (melanogenesis associated transcription factor) target gene, CYP27A1. CYP27A1 converts haperforin G/C2 and harrpernoid D into covalently active metabolite(s) that inhibit mitochondrial biogenesis (mitobiogenesis). This tissue-specific prodrug activation enables targeting the regulation of cancer energetics as a unique opportunity to overcome resistance to BRAFi/MAPKi therapy.
Results
Screening of a chemical library of 15,500 natural products
To identify selective inhibitors of cancer cell proliferation, we screened 15,500 natural products from our internal collection at 1 and 10μM across a panel of 39 cell lines for IC50 determination. The ~1400 active natural products were validated by re-testing across the 39 cell lines to generate dose response curves (SI Figure 1A). From our screen, 113 natural products, demonstrating reasonable potency and selectivity were prioritized for additional cytotoxicity profiling across a larger viability screen containing 498 cell lines from the Cancer Cell Line Encyclopedia (CCLE)( Data S1) (Barretina, et al., 2012). From this intensive phenotypic screening we identified haperforin G (Figure 1A), which exhibits anti-proliferative activity (IC50 of 0.3μM ~ 12μM) against a broad range of melanoma cell lines harboring gain-of-function mutations in BRAF(V600E) and NRAS(Q61K) (Figure 1B; blue circles), and including cell lines with intrinsic resistance to BRAF inhibitors (BRAFi). Haperforin G downregulated MAPK pathway activation and induced proliferative arrest in sensitive cell lines (Figure 1C, 1D, and 1E). It also inhibited proliferation of NRAS(Q61K)-mutation dependent melanoma cell lines which are insensitive to BRAFi (Figure 1F) suggesting that haperforin G is not a BRAF inhibitor; instead exhibiting an uncharacterized mechanism of action.
Figure 1. Identification of limonoid natural products with anti-melanoma activity.
(a) Chemical structure of the limonoid natural products haperforin G, haperforin C2 and harrpernoid D. (b) Selective cell line profile showing enrichment for BRAF(V600E)/NRAS(Q61K) mutant melanoma cell lines (blue circles) in haperforin G-sensitive cell line group. Crossing point indicates compound concentration where 50% growth inhibition by control (MG132, proteasome inhibitor) is achieved. (c) Immunoblot analysis of phospho-MAPK pathway in cells treated with compound (0.25μM) for 72 hours. Tubulin was used for loading control. (d) Percent viability relative to vehicle (DMSO) control for the indicated melanoma cell lines after treatment for 96 hours with the indicated doses of compound. Error bars indicate mean ± s.d. of 3 replicates. (e) Cell proliferation assay by EdU staining (left, top) and cell cycle analysis by PI staining (left, bottom) in MELHO treated with compound (0.25μM) for 72 hours. Cell cycle analysis from treatment of haperforin G compared with DMSO control (right). (f) Comparison of cell line profiles (skin-lineage) between BRAFi and haperforin G. Indicated cell lines have mutation in NRAS (Q61K/Q61L).
Hit expansion and structural elucidation of harrpernoid D
Our limited supply of haperforin G drove us to attempt the identification of additional small molecules structurally related to haperforin G. This expanded set was generated using 5 different in silico expansion methods; identification of bioisosteres followed by 2D similarity search (Morgan/RDKit), Extended Connectivity Fingerprints (ECFP6) with Tanimoto and Tversky similarity, MDL Public Keys similarity, and Bayesian Affinity Fingerprints (BAFPs) (Bender, et al., 2006). Our expansion to ~1300 structurally similar compounds was tested in a synthetic lethal screen comprised of two cell lines that were sensitive (MELHO & MM603) and three cell lines that were insensitive (MM466, A375, MM127) to haperforin G treatment (SI Figure 1B). In our expansion, we identified two additional compounds with similar cell proliferation selectivity profiles; haperforin C2 (Khuong-Huu, et al., 2001) and a compound we have named harrpernoid D by reason of its structural similarity to harrpernoid B and C (Figure 1A and SI Figure 1C) (Yan, et al., 2011).
Previous reports have described the isolation and structural elucidation of highly rearranged limonoids present in the leaves of Harrisonia perforata (Blanco) Merr. (Simaroubaceae) collected from Southeast Asia (Yan, et al., 2011; Khuong-Huu, et al., 2001; Khuong-Huu, et al., 2000). Harrpernoid D, a compound isolated from Harrisonia perforata, has a molecular formula of C25H28O7 as deduced from HRMS(ESI) (m/z 441.1908 [M + H]+; found 441.1909). The NMR spectra of harrpernoid D showed the presence of a γ-lactone, furan moiety, and a ring skeleton with structural similarity to previously identified harrpernoid B and C (Yan, et al., 2011) except for the presence of a double bond on ring B2, not on ring C (SI Figure 1C). The absolute stereochemistry and structural configuration was confirmed by X-ray crystallographic analysis. All further studies herein were performed with harrpernoid D because of our limited supply of haperforin G and C2.
Global analysis of expression-sensitivity relationships reveals correlation of haperforin G sensitivity to high MITF expression and CYP27A1 activity
To understand the observed selective cytotoxicity of haperforin G we compared transcriptome profiles of the identified sensitive cell lines and insensitive cell lines from all tissue lineages. Microarray and RNA-sequencing (RNA-seq) analyses revealed distinct gene signatures between sensitive and insensitive cell lines (Figure 2A and SI Figure 1D). MITF (Melanogenesis associated transcription factor), a skin lineage survival oncogene amplified in malignant melanoma (Garraway, et al., 2005), was highly expressed in a group of compound sensitive cell lines from all tissue lineages. Next, we compared transcriptome profiles of compound sensitive cell lines and insensitive cell lines with melanoma lineage. MITF was observed to be highly expressed in the compound sensitive cell lines (Figure 2B). Furthermore, downstream target genes of MITF (CYP27A1, TYR, MLANA, SLC45A2, TNFRSF14, LGALS3, PIR, ST6GAL6, ASCL1, and ATP6V1B2) were also highly expressed in compound sensitive melanoma cell lines (Du, et al., 2003; Hou, et al., 2000; Hoek, et al., 2008). Among those MITF target genes that showed differential expression in haperforin G sensitive and insensitive cell lines, CYP27A1 expression was the most significantly correlating feature conferring compound sensitivity (Figure 2A). Gene expression analysis from the CCLE on melanoma cell lines indicated positive correlation (R2 = 0.529) between the mRNA expression levels of MITF and CYP27A1 (Figure 2C).
Figure 2. Global analysis of expression-sensitivity relationships reveals correlation of compound sensitivity to high MITF expression.

(a) Genes showed differential expression in sensitive and insensitive cell lines identified by transcriptome profiling. Cells from all tissue lineages were included in the analysis. Effect size was determined by calculating mean difference between sensitive and insensitive lines divided by standard deviation. (b) MITFhigh cell lines are more sensitive to haperforin G. Expression levels of MITF and its transcriptional target genes were compared between haperforin G-sensitive (36 cell lines) and -insensitive cell lines (19 cell lines) from skin lineage (cell lines used in Data S4). (c) Correlation between the mRNA expression levels of MITF and CYP27A1 in cell lines from skin lineage. The coefficient of determination (R2) is indicated. (d) Cells were infected with either empty lentiviral vector or lentiviral vector expressing MITF. CYP27A1 mRNA level was measured by qRT-PCR. (e) Cells (MITF high) were transfected with siRNA targeting MITF and CYP27A1 mRNA level was measured by qRT-PCR. (f) Binding of MITF to the endogenous CYP27A1 promoter region in melanoma cell line (UACC257). The data shows relative promoter occupancy. TYR is established MITF target and was used as a positive control. RPL11 gene body was used as a negative control.
To validate the correlation between CYP27A1 expression and MITF expression, we expressed MITF in MITFlow melanoma cell lines. Ectopic expression of MITF increased CYP27A1 transcription (~2.5 fold in A375 and ~5.6 fold in Lox Imvi; Figure 2D) which is consistent with reports by others (Hoek, et al., 2008). Likewise, downregulation of MITF mRNA level by siRNA knockdown experiments led to concomitant downregulation of CYP27A1 mRNA levels (Figure 2E). Chromatin immunoprecipitation (ChIP) with MITF followed by qRT-PCR on the promoter region of CYP27A1 revealed that MITF directly binds the CYP27A1 promoter (Figure 2F). Supporting our findings, others have identified through ChIP-seq (chromatin immunoprecipitation coupled to high throughput sequencing) with MITF the putative binding site of MITF within the promoter region of CYP27A1 (Webster, et al., 2014). Together, these findings suggest that CYP27A1 transcription is regulated by MITF and CYP27A1 is a transcriptional target of MITF.
Anti-melanoma activity of harrpernoid D is dependent on CYP27A1
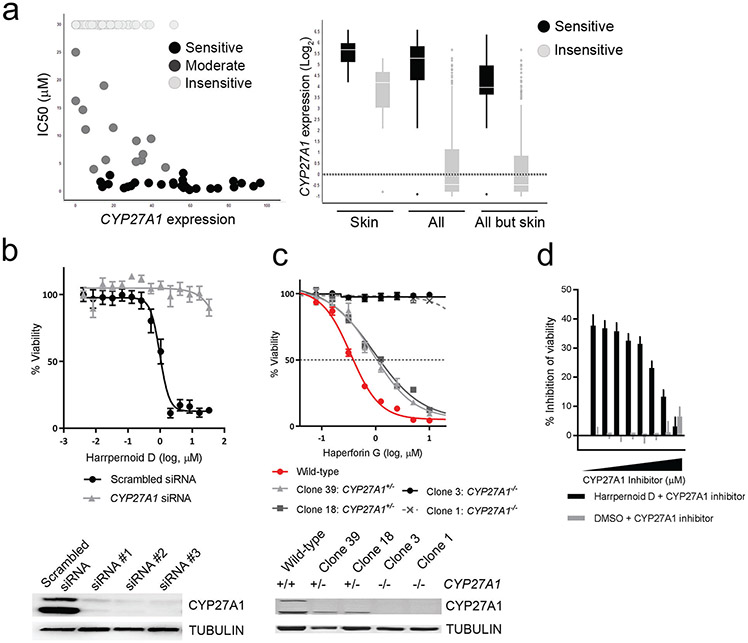
CYP27A1 ranked as a top correlation feature between gene expression and compound sensitivity. CYP27A1 was more highly expressed in the haperforin G sensitive group of cell lines across all tissue lineages and there is a positive correlation between compound response value (lower IC50) and CYP27A1 expression (Figure 3A). To characterize the correlation between CYP27A1 expression and compound sensitivity, we used two genetic approaches. First, we transiently knocked down expression of CYP27A1 using small interfering RNA (siRNA). Transient down-regulation of CYP27A1 expression rescued cells from harrpernoid D mediated inhibition of cell proliferation (Figure 3B). Second, we generated CYP27A1 null cell lines using Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-mediated genome editing. Isolated single clones of biallelic CYP27A1 knock-out cells (CYP27A1−/−) did not show loss of viability compared to wild-type cells (SI Figure 2A). When treated with harrpernoid D, CYP27A1−/− cells were rescued from compound mediated growth inhibition and cells with monoallelic knock-out of CYP27A1 (CYP27A1+/−) showed intermediate phenotype between CYP27A1−/− cells and wild-type cells. This phenotype was confirmed in three different melanoma cell lines (MELHO, UACC257, IGR37) (Figure 3C and SI Figure 2B). These genetic approaches strongly suggest that CYP27A1 is the genetic determinant that confers compound sensitivity. To further confirm that CYP27A1 activity is required for cytotoxic activity, we used combination treatment of harrpernoid D and the CYP27A1 inhibitor, fadrozole (Mast, et al., 2015). Cells co-treated with harrpernoid D and fadrozole were partially rescued from harrpernoid D mediated growth inhibition (Figure 3D), suggesting that CYP27A1 activity is required for harrpernoid D activity.
Figure 3. Global analysis of expression-sensitivity relationships showing harrpernoid D activity is CYP27A1 dependent.
(a) (Left) Correlation of CYP27A1 expression (RNA-seq) and compound sensitivity. Each dot indicates individual cell line from all tissue lineages. (Right) Differential expression of CYP27A1 in compound-sensitive and -insensitive cell lines. Skin: skin lineage, All: all tissue lineage, All but skin: all tissue lineage except for skin lineage. (b) Percent viability (relative to DMSO treated cells) of MELHO treated with indicated concentrations of compound for 96 hours. Cells were transfected with either scrambled siRNA or siRNA against CYP27A1 24 hours prior to compound treatment. (c) Percent viability of wild-type MELHO was compared to monoallelic knock-out clones (CYP27A1+/−) and biallelic knock-out clones (CYP27A1−/−) after compound treatment. (d) Cells were treated with 2.5μM of harrpernoid D (or DMSO) in combination with increasing dose of CYP27A1 inhibitor, fadrozole. Data were shown as percent inhibition of viability.
To determine whether CYP27A1 expression is sufficient for harrpernoid D sensitivity, we overexpressed CYP27A1 in CYP27A1low cell line (MM127) and CYP27A1 null cell line (Cyp27A1−/− MELHO). CYP27A1−/− MELHO complemented with CYP27A1 overexpression restored harrpernoid D sensitivity and MM127 overexpressing CYP27A1 gained sensitivity to harrpernoid D (SI Figure 2C), suggesting CYP27A1 expression is sufficient for harrpernoid D sensitivity in the cell lines tested. A panel of CYP27A1low and CYP27A1mid cell lines (A375, IGR37, Lox Imvi, MEL JUSO, A101d, and WM983b) overexpressed with CYP27A1 also became more sensitive to harrpernoid D activity (SI Figure 2D).
CYP27A1 biotransformation of harrpernoid D
The observed dependence of anti-melanoma activity on CYP27A1 suggested that harrpernoid D might be a prodrug converted by the cytochrome P450 oxidase, CYP27A1, into an active metabolite responsible for cytotoxicity. To determine if harrpernoid D was converted into an active metabolite we expressed human CYP27A1 in B. megaterium and incubated with harrpernoid D (Ehrhardt, et al., 2016). Testing cholestenone in the B. megaterium system expressing CYP27A1 gave clear conversion to a number of hydroxylated products (SI Figure 3A). When harrpernoid D was incubated with wild-type B. megaterium no conversion or degradation was observed over a 48h time period (SI Figure 3B). However, when harrpernoid D was incubated with B. megaterium expressing human CYP27A1 there was an observed ~30% loss of the parent compound after 8h and complete disappearance after 24h (SI Figure 3C). Unexpectedly, no identifiable metabolites of harrpernoid D were isolated despite several extraction attempts (SI Figure 3D). We presumed the furan ring of harrpernoid D is activated by CYP27A1 to generate a reactive enedial. Related natural products teucrin A, 8-epidiosbulbin E acetate, and diosbulbin B similarly convert into reactive enedials, which covalently modifies cysteine and lysine residues (Druckovia & Marnett, 2006; Lin, et al., 2016; Lin, et al., 2014; Druckova, et al., 2007; Kornienko & La Clair, 2017).
As a chemically controlled mimetic of P450 oxidation, harrpernoid D was treated with dimethyldioxirane (DMDO) which induced oxidation and ring opening of the furan to a chemically sensitive enedial which was observed by 1H NMR spectroscopy and high-resolution mass spectrometry (Figure 4A and SI Figure 3E). Treatment of this electrophilic species with N-acetyl cysteine methyl ester generated a complex mixture of labile products in analogy to prior observations with teucrin A (Druckovia & Marnett, 2006). Thus, if such a reactive enedial of harrpernoid D is generated biologically by CYP27A1, rapid formation of protein adducts may have obscured our ability to directly detect any metabolites from the B. megaterium biotransformation experiments.
Figure 4. Genome-wide siRNA-compound epistasis screening reveals mitochondrial pathway-specific MoA of harrpernoid D.
(a) Chemical oxidation of harrpernoid D with dimethyldioxirane (DMDO) generates a reactive α,β-unsaturated enedial electrophile. (b) isoTOP-ABPP of harrpernoid D (50 μM) in wild-type, CYP27A1high HCT116 cells. (c & d) Scatter plot of Q3 or Q1 of Robust z-score versus RSA for eight individual siRNAs per gene. Each dot represents an individual target gene in the siRNA library, shaded according to expression level (RNA-seq) in the cell line used (MELHO). Cutoff lines indicate selected thresholds of p-value 10E-5(red) and 10E-4(black). Marked genes are selected hits, which increased resistance to compound (c) or increased sensitivity to compound (d). (e) Sensitivity to antimycin A (ETCi), dabrafenib (BRAFi), haperforin G and harrpernoid D was determined in UACC257 cells cultured in growth media containing glucose or galactose to promote glycolysis or mitochondrial OXPHOS, respectively. Viability (left 4 panels) determined with CellTiter-Glo and membrane integrity was determined using CellTox Green (right 4 panels) 96 hours after addition of compounds.
ABPP to map harrpernoid D targets in melanoma cells
To capture the putative targets that might be covalently modified by the active metabolite of harrpernoid D we applied an isotopic tandem orthogonal proteolysis-enabled activity-based protein profiling (isoTOP-ABPP) approach pioneered by Cravatt and Weerapana (Weerapana, et al., 2010) and subsequently adapted for use in context of a competitive assay (Wang, et al., 2014; Backus, et al., 2016). When used in a competitive manner, covalent small molecules can be competed against binding of a broadly reactive probe to facilitate the discovery of proteins and ligandable sites targeted by the covalently acting compound. In this study we treated either wild-type and CYP27A1−/− HCT116 cells with vehicle or 50μM harrpernoid D followed by competitive labeling of proteomes with cysteine-reactive alkyne-functionalized iodoacetamide probe (IA-alkyne) (SI Figure 4A). Isotopically light or heavy cleavable enrichment handles were appended to probe-labeled proteins for isoTOP-ABPP analysis. Enriched probe-modified tryptic peptides were analyzed by liquid chromatography–mass spectrometry (LC–MS/MS) and heavy to light tryptic probe-modified peptide ratios, representing control versus treated IA-alkyne labeled sites, were quantified. Harrpernoid D treated wild-type, CYP27A1high HCT116 cells had an enrichment in a number of mitochondrial and endoplasmic reticulum proteins compared to the comparably treated CYP27A1−/− HCT116 cells (Figure 4B and Data S2). Interestingly, teucrin A and furan, which are activated by CYP3A have been shown to covalently target mitochondrial proteins and enzymes responsible for lipid, amino acid, and drug metabolism (Druckova, et al., 2007; Moro, et al., 2012; Lekehal, et al., 1996).
Genome-wide siRNA–compound epistasis screening reveals mitochondrial pathway-specific mechanism of action for harrpernoid D
In an effort to add clarity to the chemoproteomics data, we performed a genome-wide siRNA-compound epistasis screen. Three independent viability screens were conducted in the presence of harrpernoid D, BRAF inhibitor (dabrafenib), or control (DMSO) to identify the genes that alter the cellular viability in response to harrpernoid D but not to BRAFi or control treatment. Both RSA and GESS off-target analysis identified CYP27A1 as a hit (target gene of depleted siRNA) that suppresses sensitivity to harrpernoid D (but not to BRAFi) (Figure 4C, SI Figure 3B, and Data S3). In addition, GESS analysis revealed that knockdown of ferredoxin 1 (FDX1), which assists in transferring electrons from NADPH to CYP27A1 (Pikuleva, et al., 1999), also suppressed compound activity (SI Figure 3C). Notably, knocking down genes that are involved with oxidative phosphorylation (OxPhos)/mitochondrial electron transport chain (ETC) complex sensitized cells to harrpernoid D (but not to BRAFi). Those genes include mitochondrial chaperones (HSPA9 and HSPD1), a mitochondrial tRNA synthetase (TARS2), a mitochondrial protein transporter (TIMM23), and ETC complex subunit/assembly factors (NDUFB2, ATP5B, FOXRED1, and ECSIT) (Figure 4D). Based on the epistasis between harrpernoid D and silencing mitochondrial pathway genes, we hypothesized that regulating mitochondrial pathway components is functionally relevant for the mechanism of action for harrpernoid D. To validate a mitochondrial pathway-specific mechanism of action, we used a cell-based assay where cells were grown under conditions that shift the metabolic balance toward oxidative phosphorylation (Lai, et al., 2013). Cells that are metabolically adapted for growth under hypoxic and anaerobic conditions under high glucose media can derive most of their energy from glycolysis rather than mitochondrial oxidative phosphorylation. When galactose is substituted for glucose as source of energy, however, cells rely nearly exclusively on oxidative phosphorylation for ATP synthesis and are more sensitive to compounds that block mitochondrial function than cells grown in media with glucose (Marroquin, et al., 2007). We observed a mild increased sensitivity to haperforin G, harrpernoid D, and strong sensitivity to an established mitochondrial ETC inhibitor (ETCi) antimycin A, but not to BRAFi (dabrafenib), of cells cultured in galactose-containing media when compared to those cultured in glucose-containing media (Figure 4E). These observations together with the pathway epistasis from the siRNA screen suggest that harrpernoid D impairs mitochondrial function as a primary effect rather than secondary to other cytotoxic mechanisms. RNA-seq analysis of cell lines treated with harrpernoid D corroborated these observations. Functional gene set enrichment analysis (GSEA) revealed that a significant fraction of genes associated with mitochondrial homeostasis and mitochondrial stress response pathway is down-regulated at an early time point (48 hours after treatment) and up-regulated at a later time point (72 hours after treatment) in cells treated with harrpernoid D (-Log10 P < 5; SI Figure 5). This finding indicates that harrpernoid D affects mitochondrial homeostasis and leads to mitochondrial stress response pathway activation at the later time point (72 hours after treatment). We did not find any evidence of regulation of mitochondrial pathway genes after harrpernoid D treatment in CYP27A1−/− cells (SI Figure 5).
Harrpernoid D inhibits mitochondrial biogenesis and maintenance of mitochondrial integrity
To evaluate harrpernoid D induced effects on mitochondrial biogenesis, we compared the expression level of two proteins (SDH-A and MT-COX1) which are each subunits of a different oxidative phosphorylation enzyme complex. Complex IV includes several proteins that are encoded in the mitochondrial genome, while the proteins of Complex II are entirely encoded in the nucleus. MT-COX1 is the subunit I of Complex IV (COX-I) encoded by mitochondrial DNA (mtDNA), and SDH-A is a subunit of Complex II encoded by nuclear DNA (nDNA). The expression level of MT-COX1 is markedly decreased by harrpernoid D treatment in two cell lines that are sensitive to harrpernoid D (MELHO and UACC257) but not in an insensitive cell line (MM127) (Figure 5A), while the expression level of SDH-A was not changed by harrpernoid D treatment. Of note, the apparent molecular weight of SDH-A from harrpernoid D treated cells was slightly larger compared to control cells, which might be due to the mitochondrial signal peptides that are not efficiently processed. This was also observed in other subunits of oxidative phosphorylation complex (ATP5A of complex V, UQCRC2 of complex III, and SDH-B of complex II) (SI Figure 6A).
Figure 5. Harrpernoid D inhibits mitochondrial biogenesis.
(a) Immunoblot analysis of mitobiogenesis associated protein (SDH-A and MT-COX1) in cells treated with harrpernoid D 2.5μM (IC50) for 72 hours. ACTIN was used as a loading control. (b) Volcano plot of genes from RNA-seq analysis on cells treated with 4.1μM of compound (IC70) for indicated time. X-axis: log2 fold change relative to DMSO control at each time point. Genes encoded by mitochondrial DNA are marked. (c) Representative high-content images of cells treated with compound (IC50) for 72 hours and stained with an antibody for TFAM (green), mitochondria (red), and nucleus (blue). (d) Subcellular fractionation and immunoblot analysis on TFAM in indicated cell lines treated with compound (IC50) or DMSO control. TUBULIN: cytoplasmic loading control. ALAS1: mitochondrial loading control. Data is representative of at least three independent experiments. Amount of protein loaded: 5μg (Cyto; cytoplasmic fraction), 5μg (mito; mitochondrial fraction), and 25μg (WCL; whole cell lysate).
RNA-seq analysis showed that downregulation of MT-COX1 expression occurs at the transcriptional level. Mitochondrial DNA encodes 13 mRNAs (MT-ND1, MT-ND2, MT-ND3, MT-ND4, MT-ND4L, MT-ND5, MT-ND6, MT-CYB, MT-CO1, MT-CO2, MT-CO3, MT-ATP, and MT-ATP8), each of which encodes a polypeptide component of the mitochondrial oxidative phosphorylation system (Anderson, et al., 1981; Mishra & Chan, 2014; Chinnery & Hudson, 2013). Notably, the levels of all 13 mRNAs expression were down regulated over the course of harrpernoid D treatment, which was consistent with progressive proliferative arrest (Figure 5B and SI Figure 6B). We next assessed the effects of harrpernoid D on mitochondria abundance using two types of mitochondria-specific dyes (MitoTracker Green; stains total mitochondria and MitoTracker Red; stains respiring mitochondria). Harrpernoid D treatment reduced the total level of mitochondria (SI Figure 6C). To provide an independent confirmation of the decrease in the number of mitochondrion in cells treated with harrpernoid D, we quantified the amount of mitochondrial DNA present in the cells in relation to genomic DNA by quantitative PCR (Wen, et al., 2013). While harrpernoid D treatment in CYP27A1−/− cells did not alter the relative mtDNA copy number, it significantly reduced the mtDNA copy number in wild-type cells (SI Figure 6D). To rule out the possibility of direct inhibitory effect on mitochondrial membrane potential by harrpernoid D, we measured mitochondrial membrane potential at two different time points. Harrpernoid D did not decrease mitochondrial membrane potential at early time point (2 hours after treatment), whereas an established uncoupler CCCP (2-[2-(3-chlorophenyl)hydrazinylyidene]propanedinitrile) significantly reduced membrane potential, suggesting that harrpernoid D does not directly inhibit mitochondrial membrane potential as a primary effect (SI Figure 6E). The modest reduction of membrane potential at the later time point (24 hours after treatment) is likely due to a secondary inhibitory effect of harrpernoid D on the mitochondrial pathway.
Because the transcription of all 13 mRNAs was down regulated, we hypothesized that the transcriptional regulator of mtDNA, TFAM (transcriptional factor A, mitochondrial) might be associated with the inhibitory action of harrpernoid D on mitochondrial biogenesis. To interrogate this possibility, we performed high-content imaging analysis to visualize co-localization of TFAM with mitochondria. In control cells, TFAM is co-localized to the mitochondria, which are composed of a reticulum of distinct tubular and round mitochondrion. However, cells treated with harrpernoid D display more dispersed localization of TFAM throughout the cell periphery (Figure 5C). To further analyze the localization of TFAM upon harrpernoid D treatment, we performed subcellular fractionation in harrpernoid D sensitive (MELHO and UACC257) and insensitive cell lines (MELHO CYP27A1−/−). While the level of TFAM in whole-cell lysates remained relatively constant after harrpernoid D treatment, the level of TFAM in mitochondrial fraction was markedly decreased in harrpernoid D sensitive cell lines (Figure 5D). Reduction of TFAM localization to mitochondria was also observed by treatment with MKT-077 a HSPA9 inhibitor (SI Figure 6F). HSPA9 serves as a key regulatory component for the import of mitochondrial proteins (Schneider, et al., 1994), and inhibition of HSPA9 by MKT-077 perturbed the mitochondrial import of reporter protein, MTS-EGFP (mitochondrial targeting sequence of COX8 fused to EGFP). However, harrpernoid D did not disrupt the mitochondrial import of MTS-EGFP, suggesting harrpernoid D may not directly inhibit mitochondrial protein targeting (SI Figure 6G). Taken together, these results effectively demonstrate inhibition of mitochondrial biogenesis by harrpernoid D.
Targeting mitobiogenesis by harrpernoid D can overcome drug resistance to BRAFi/MAPKi
Several studies have reported that inhibition of BRAF by small molecules induces an oxidative phosphorylation gene program and increases mitochondrial biogenesis. Especially, treatment of BRAF-mutated melanomas with BRAFi renders them addicted to oxidative phosphorylation and such adaptive metabolic program limits the efficacy of BRAFi (Haq, et al., 2013; Baenke, et al., 2016; Corazao-Rozas, et al., 2013; Canor & Sabatini, 2012).
To assess the opportunity to target mitobiogenesis and overcome resistance to BRAF/MAPK inhibition by harrpernoid D, we generated melanoma cell lines (MELHO and UACC257) that have acquired resistance to BRAFi (dabrafenib) or combination of BRAFi (dabrafenib) and MEKi (trametinib) (SI Figure 6H). Consistent with a previous report (Baenke, et al., 2016), cells with acquired resistance displayed increased level of maximal respiratory capacity compared to the parental cells (SI Figure 6I). To assess the efficacy of harrpernoid D in limiting proliferation of cells with acquired resistance to BRAFi, we treated the cells with a combination of BRAFi (dabrafenib) and harrpernoid D or haperforin G. Combination of BRAFi and harrpernoid D treatment resulted in synergistic anti-proliferative activity as determined by the Bliss independence model (Keith, et al., 2005). Notably, combination of harrpernoid D and BRAFi effectively inhibited proliferation of cells with acquired resistance to BRAFi (Figure 6A). Similar results were obtained with a triple combination of harrpernoid D, BRAFi, and MEKi. Harrpernoid D and haperforin G could drive profound anti-proliferative response in cells that have acquired resistance to combination of BRAFi and MEKi (Figure 6B). Collectively, this data supports that harrpernoid D can enhance therapeutic benefits of BRAF/MAPK inhibition and combined treatment of harrpernoid D and BRAFi/MAPKi may overcome acquired resistance to BRAFi/MAPKi.
Figure 6. Combinatory treatment of harrpernoid D with BRAFi/MAPKi can overcome acquired resistance to BRAFi/MAPKi.
Percent viability relative to vehicle (DMSO) control for the indicated melanoma cell lines (parental cells and cells with acquired resistance to BRAFi (a) or combination of BRAFi and MEKi (b) after treatment for 120 hours with indicated concentrations of compound. (a) Cells were treated with the indicated concentrations of BRAFi in combination with harrpernoid D. (b) Cells were treated with indicated concentrations of BRAFi with fixed dose (IC50) of MEKi in combination with harrpernoid D. Error bars indicate mean ± s.d. of at least 3 replicates.
Discussion
Here we report the anti-melanoma activity and mechanism of action of three limonoid natural products. Limonoids are highly oxygenated nortriterpenoids found in the Meliaceae, Rutaceae, Cneoraceae, and the Harrisonia of Simaroubaceae (Yan, et al., 2011). Limonoids exhibit a large structural diversity and a range of biological activities such as insecticidal, antibacterial, antimalarial, antiviral, and anticancer (MacKinnon, et al., 1997; Roy & Saraf, 2006; Manners, 2007; Yan, et al., 2016). A large-scale cell line profiling on 498 cell lines across 25 tissue lineages allowed us to identify unique anti-melanoma activity of limonoid natural products isolated from Harrisonia perforata. Global transcriptome analysis on unique gene signatures of sensitive versus insensitive cells revealed that MITFhigh melanoma cells are enriched for sensitivity. MITF is considered a 'lineage survival' or 'lineage addiction' oncogene required for both tissue-specific cancer development and tumor progression (Garraway, et al., 2005; Shtivelman, et al., 2014) and hence is an important target to limit proliferation of melanoma. Of note, ChIP-seq (chromatin immunoprecipitation coupled to high throughput sequencing) identified putative binding site of MITF within the promoter region of CYP27A1 (Webster, et al., 2014) and modulation of MITF expression resulted in concomitant regulation of CYP27A1 expression, consistent with a previous report (Hoek, et al., 2008), suggesting that CYP27A1 is a transcriptional target of MITF. Our data with genetic manipulation of CYP27A1 and chemical perturbation of CYP27A1 activity established that harrpernoid D activity is dependent on CYP27A1 expression and its enzyme activity, which contributes to the enrichment of MITFhigh cells among group of cells that are sensitive to harrpernoid D. This is the initial case of an anti-melanoma compound, to our knowledge, where a transcriptional target gene of MITF is required for anti-proliferative activity of a compound. At least 15% of human melanomas have MITF gene amplification (Garraway, et al., 2005; Hodis, et al., 2012) and this correlates with poor prognosis of malignant melanoma. This might indicate innovative opportunities for melanoma-specific targeting by compound(s) that are highly dependent on expression of MITF or transcriptional target gene(s) of MITF such as CYP27A1.
CYP27A1 is a mitochondrial cytochrome P450 enzyme which can hydroxylate vitamin D3 and cholesterol at carbons 25 and 26, respectively (Tieu, et al., 2012; Gupta, et al., 2007). Because of the structural similarity of haperforin G, haperforin C2, and harrpernoid D to vitamin D3 and cholesterol, it seemed possible that CYP27A1 metabolized the parent precursors to a biologically active form. Similar prodrug activation of furanoid natural products such as teucrin A are activated by CYP3A enzymes to their active enedial which can covalently modify cysteine and lysine residues of proteins (Druckova, et al., 2007). Biotransformation, synthetic manipulation, as well as isoTOP-ABPP experiments confirmed the conversion of harrpernoid D to an active enedial metabolite that covalently modifies cysteine residues of mitochondrial and endoplasmic reticulum proteins. It will be of future interest to investigate the mechanism of toxicity at the level of metabolite-target specificity. For example, whether a combination of targets identified by chemoproteomic approaches acts in concert and if there are other modifier proteins that contribute to the observed cytotoxic effects.
To investigate proteins that direct anti-melanoma activity we ran a genome-wide siRNA–compound epistasis screen that confirmed mitobiogenesis as a target pathway of harrpernoid D. It is unknown at this time why liver cancer cell lines in our CCLE profiling showed little sensitivity to haperforin G compared to melanoma lines, given the high expression of CYP27A1 in normal liver for its cholesterol metabolism function (SI Figure 1E). We speculate that other cytochrome p450 metabolic enzymes in liver cell lineages process the limonoids to an inactive form, ameliorating toxicity by the CYP27A1-induced metabolite. This proposal is supported by the fast clearance of harrpernoid D by human and rat liver microsomes (human: half life (t1/2) 3.9 min, intrinsic clearance (CLint) 356uL/min/mg protein & rat: half life t1/2 <1.98 min, CLint >700 uL/min/mg protein).
Targeting mitobiogenesis might be effective in improving the efficacy of BRAFi/MAPKi and preventing the acquisition of drug resistance to BRAFi/MAPKi (Zhang, et al., 2016; Baenke, et al., 2016). A previous study on human melanoma xenograft models showed that tumors with acquired resistance to vemurafenib showed continued dependency on BRAF(V600E)/MAPK signaling owing to elevated BRAF(V600E) expression (Das Thakur, et al., 2013). Increased BRAF(V600E) expression is in part due to BRAF copy number amplification as shown in pre-clinical and clinical case of melanoma (Shi, et al., 2012; Wagle, et al., 2014) and one of the most significant regions of copy gain was located on chromosome 7q34 which contains both BRAF and NDUFB2 (Lin, et al., 2008; Miller, et al., 2010). NDUFB2 encodes a polypeptide which is a subunit of the NADH:ubiquinone oxidoreductase (complex I of electron transport chain complex) catalyzing the first step of mitochondrial oxidative phosphorylation. Therefore, increased level of mitochondrial respiration in melanoma with acquired resistance to BRAFi as shown previously by others (Baenke, et al., 2016; Corazao-Rozas, et al., 2013) and by our study might be attributed to collateral co-amplification of a neighboring OxPhos gene with BRAF.
Approximately 60% of malignant melanomas harbor the BRAFV600 missense mutation, resulting in constitutively activated MAPK signaling (Davies, et al., 2002). BRAF inhibitors, such as dabrafenib and vemurafenib elicit rapid antitumor responses however, intrinsic and acquired resistance and clonal evolution of melanoma during BRAFi/MAPKi therapy often leads to a rapid tumor relapse by reactivation of MAPK or PI3K pathway and limits the overall response and therapeutic benefit of BRAFi/MAPKi therapy (Shi, et al., 2014). Metabolic reprogramming is a hallmark of cancer modulated in part through deregulating cellular energetics to support effective neoplastic proliferation (Hanahan & Weinberg, 2011). The Warburg effect illustrates that aerobic glycolysis is the predominant metabolic pathway for cancer cells to support continuous cell growth and proliferation (Warburg, 1956). However, slow-cycling melanoma cells that are characterized by high expression levels of the histone demethylase JARID1B predominantly utilize mitochondrial oxidative phosphorylation (OxPhos) to generate ATP and are intrinsically resistant to multiple drugs such as cisplatin and vemurafenib (Roesch, et al., 2013). BRAF-mutated melanoma cells treated with vemurafenib, caused upregulation of the MITF/PGC1A signaling axis, resulting in metabolic reprogramming toward OxPhos and conferring intrinsic resistance to BRAF inhibitors (Haq, et al., 2013). A subset of tumor biopsies from patients with disease progression despite BRAFi/MAPKi treatment showed increased mitochondrial DNA (mtDNA) content and upregulated mitochondrial biogenesis and tumor bioenergetics (Zhang, et al., 2016). Efforts have been made to address the clinical and biological relevance of targeting mitochondrial biogenesis (mitobiogenesis) and tumor bioenergetics in the context of intrinsic and acquired drug resistance (Zhang, et al., 2016; Baenke, et al., 2016; Schockel, et al., 2015; Corazao-Rozas, et al., 2013).
We therefore suggest that inhibition of mitobiogenesis might indicate emerging opportunities for combination therapies with BRAFi/MAPKi to maximize patient benefit and to deploy rational design of therapeutic combination for durable disease control. Our study characterizes harrpernoid D as a CYP27A1 activated prodrug that regulates proliferation and survival of melanoma cells by targeting mitobiogenesis.
Significance
Melanoma cells are able to overcome BRAF inhibitors, such as vemurafenib, by upregulating oxidative phosphorylation (OxPhos), inducing resistance. Combination therapy targeting OxPhos dependency with BRAF inhibition has previously been demonstrated. In this study, we identified a set of compounds that disrupt mitochondrial biogenesis in melanoma lineage specific cells. Melanoma lines sensitive to compound treatment had high expression of MITF, a melanocytic transcriptional master regulator. Expression of CYP27A1, a transcriptional target of MITF, was essential for cytotoxic activity of these compounds. These compounds, members of the limonoid natural product family, were determined to be prodrugs activated by CYP27A1 to generate a broadly reactive enedial metabolite. This transient enedial metabolite covalently modify ER and mitochondrial proteins and disrupts mitochondrial biogenesis. These finding suggest the possibility to exploit tissue-selective inhibition of mitochondrial biogenesis through a lineage-specific prodrug mechanism. Such an approach could expand the therapeutic window for the suppression of OxPhos dependency in BRAF mutant melanomas.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and request for resources and reagents will be fulfilled by the lead contact, William Forrester, william.forrester@novartis.com.
Materials availability
Plasmids and compounds generated in this study will be made available upon reasonable request. Material supply of haperforin G, haperforin C2, and harrpernoid D are limited and subject to handling under Nagoya protocol.
Data and code availability
The RNA-Seq data are available in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov) with accession number SRP108654. Data generated in this study will be made available upon reasonable request. No code was developed for this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines used in this study are part of the Novartis Cancer Cell Line Encyclopedia collection (CCLE). Detailed information about these cell lines (including gender, primary site (tumor type), source) are described in (Barretina, et al., 2012) and are also included in Data S1.
METHOD DETAILS
Cell line profiling
Cell line profiling experiment was performed as described (Carson, et al., 2015). All assays were automated and performed with an ultra-high throughput screening system built by the Genomics Institute of the Novartis Research Foundation (Melnick, et al., 2006) (http://www.gnfsystems.com). Briefly, 498 cell lines across 25 tissue lineages (Data S1) were plated into 1,536-micro plates (Greiner) at a density of 250 cells per well with a final volume of 5μL. 12 to 24 hours after plating, 15 nL of each compound dilution series was transferred to the cells using an Echo acoustic liquid dispenser (Labcyte) to yield final compound concentration ranges of 30 μM to 0.3μM (11 point dose response) in 3.16-fold dilutions. Cell viability was determined by CellTiter-Glo (Promega) 72 hours after compound treatment and data was captured using ViewLux plate reader (Perkin Elmer) according to the manufacturer’s instructions. Absolute maximum cytotoxicity for each cell line was benchmarked against the impact on viability by exposure to 10μM of MG132 (proteasome inhibitor that shows pan-toxicity).
Cell lines and Cell culture
Melanoma cell lines were obtained from the Novartis cell line encyclopedia collection (Barretina, et al., 2012) and were cultured in RPMI 1640 medium containing 10% Fetal Bovine Serum (FBS), 1% penicillin/streptomycin (P/S), and 2mM Glutamine.
Cell line manipulation
CYP27A1−/− cell line clones were generated using CRISPR-Cas9 (pU6g-cas9-puromycin; gift from Dr. Yi Yang at Novartis) mediated knockout followed by single-cell sorting. Loss of CYP27A1 protein expression and homogenous biallelic knockout of CYP27A1 in each cell clones was confirmed by Western blot analysis and sequencing. For CYP27A1-overexpressing cell lines, CYP27A1 cDNA was cloned into pCDH lentiviral vector (System Biosciences), cells then were transduced with packaged lentivirus and selected for stable expression of CYP27A1 using antibiotics treatment (1000μg/mL neomycin or 2μg/mL puromycin as indicated).
Generation of BRAFi/MAPKi resistant cell lines
Resistant cell lines (MEL-HO and UACC257) were generated as described previously (Corcoran, et al., 2010). Briefly, dabrafenib or combination of dabrafenib and trametinib was added at a starting concentration of 10 nM (dabrafenib) or 0.316 nM (trametinib), and cells were maintained in fresh drug-containing medium changed every 72 hours. Cells were passaged when they reached ~70% confluence. After every two or three passages at a given concentration of drug, the concentration of drug was increased in half-log intervals until a final concentration was achieved.
Cell viability/cytotoxicity assay
Cells were plated into each well of Greiner 384-well plates at a density of 1,500 cells/well. Cells were incubated with increasing concentration of compounds (0.1% vol/vol DMSO) for 72 to 96 hours and cell viability/cytotoxicity was measured by EnVison plate reader (Perkin Elmer) with CellTiter-Glo (Promega) or CellTox Green (Promega) according to the manufacturer’s protocol.
Glucose-Galactose assay
Mitochondrial toxicity by compound was assessed by Glucose-Galactose assay as described previously (Lai, et al., 2013).
Cell cycle analysis/cell proliferation assay
For S phase analysis, cells were stained using Click-iT® EdU Alexa Fluor® 488 Flow Cytometry Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. To distinguish cells in G0/G1 phase versus S phase, or G2, cells were stained using FxCycle™ PI/RNase Staining Solution (Thermo Fisher Scientific) according to the manufacture’s protocol. Cells were analyzed using BD FACSCANTO II (BD Biosciences) and FlowJo software.
ChIP-qPCR
ChIP was performed as described previously (Salma, et al., 2017). Briefly, protein:chromatin-crosslinked complexes were immunoprecipitated with either MITF-specific antibody (Cell Signaling) or isotype control antibody. qRT-PCR was performed on samples to amplify a fragment occupied by MITF using primers spanning promoter region of CYP27A1, TYR, or RPL11 gene body. Primers used were: 5’-GGAAGGCCATTGGGAGAAG-3’ and 5’-GATTCTTTCGAAATCGGGAAGC-3’ for CYP27A1 promoter, 5’-GTGGGATACGAGCCAATTCGAAAG-3’ and 5-TCCCACCTCCAGCATCAAACACTT-3’ for TYR, and 5-ACACAGATCATGGGTCTTGCTCCA-3’ and 5-AGCACCTGTCCCAGGAATAACCAA-3’ for RPL11 gene body.
Western blot analysis
Whole cell lysates were isolated with RIPA Lysis and Extraction buffer (Thermo Fisher Scientific) supplemented with Halt™ protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific) according to the manufacturer’s protocol. All antibodies used were from Cell Signaling except for: CYP27A1 (Abcam), Mitobiogenesis cocktail (Abcam), Alas1 (abcam), MITF (Thermo Fisher Scientific)
Subcellular fractionation
Subcellular fractionation and mitochondria isolation was performed by using differential centrifugation with Mitochondria Isolation Kit (Thermo Scientific) according to the manufacturer’s protocol.
High-content imaging
High-content staining for mitochondria, TFAM, SDHA was performed using an IN Cell Analyzer 6000 Cell Imaging System (GE Healthcare Life Sciences). Antibodies used were: MitoTracker® Red CMXRos (Thermo Fisher Scientific), TFAM (D5C8) (Cell Signaling), SDHA (D6J9M) (Cell Signaling)
mtDNA copy number analysis
Total genomic DNA was isolated using QIAamp DNA Kit (QIAGEN) and a quantitative PCR method was used to determine the mtDNA copy number as described (Wen, et al., 2013). The primers of β-actin (product length, 138 bp) were forward, 5′-CGGGAAATCGTGCGTGACAT-3′ and reverse, 5′-GAA GGAAGGCTGGAAGAGTG-3′. The primers of mtDNA (product length, 487 bp) were forward, 5′-TACTCACCAGACGCCTCAACCG-3′ and reverse, 5′-TTA TCGGAATGGGAGGTGATTC-3′.
Quantitation of mitochondrial membrane potential
Mitochondrial membrane potential was measured using mitochondrial membrane potential indicator (MMPI, Codex) according to the manufacturer’s instructions.
Metabolic assays
Oxygen consumption rate (OCR) was measured with the optical fluorescent oxygen/hydrogen sensor XFe96 Seahorse analyzer according the manufacturer’s protocol. During the continuous OCR measure, the following compounds were sequentially administered: oligomycin 1mM, FCCP 2mM, and rotenone/antimycin A 1mM. The data was normalized to total cell number as measured using the CellTag700 stain and Odyssey imaging system (Li-COR, Lincoln, NE).
Genome-wide RNAi screening
60nL of 2uM siRNAs were stamped using an Echo acoustic liquid dispenser (Labcyte) on each well of 1,536-micro plates (Greiner) for final concentration of 20nM. Cells were then reverse-transfected using Dharmafect 4 (Dharmacon) according to the manufacturer’s protocol. 24 hours after transfection, cells were incubated for 96 hours with DMSO (vehicle), IC30 of harrpernoid D, or dabrafenib until lysed with CellTiter-Glo (Promega) for viability assay. Luminescence signal was measured with ViewLux microplate reader (Perkin Elmer) according to the manufacturer’s instructions. Screen data were processed using RSA (redundant siRNA activity) analysis and GESS (genome-wide enrichment of seed sequence matches) off-target analysis as described previously (Sigoillot, et al., 2012; Birmingham, et al., 2009).
RNA-sequencing (RNA-seq)
Three biological replicates of MEL-HO wild-type and MEL-HO cyp27a1−/− were treated with DMSO, IC20, and IC70 of harrpernoid D (0.1% DMSO vol/vol) for 24 hours, 48 hours, and 72 hours. Total RNA was isolated according to the manufacturer’s protocol with an RNeasy kit with on-column DNA digestion (Qiagen). RNA concentration and integrity was measured by fragment analyzer with Standard Sensitivity RNA kit according to the manufacturer’s protocol (Advanced Analytical Technologies). RNA-seq libraries were prepared using the TruSeq Stranded mRNA Sample Preparation kit (Illumina) and sequenced in strand specific paired-end mode, 2x76bp, using the HiSeq2500 platform. Alignment against Human Ensembl genome reference GRCh38 (release 78) utilized the exon quantification pipeline (EQP) (Schuierer & Roma, 2016) with the Bowtie2 (Langmead & Salzberg, 2012) alignment module for exon- and gene-level quantitation. Alignment quality was assessed using Picard (http://broadinstitute.github.io/picard/) and ArrayQualityMetrics (Kauffmann, et al., 2009), passing internal criteria. Differential expression comparisons were calculated using limma linear modeling (M.E., et al., 2015) with voom precision weighting (Law, et al., 2014).
Isolation of harrpernoid D
30 kg leaves of Harrisona perforate were milled and extracted with ethyl acetate. The resulting extract was purified using a Daisogel SP 200-15-C8 column using water and methanol as solvents (gradient 30% to 100% methanol). Fractions containing the target molecule were further purified using a Sunfire RP18 column followed by final purification using a Synergi Polar-RP column (0.1% formic acid / acetonitrile containing 0.1% formic acid as solvents). 115 mg pure (>95%) harrpernoid D could be isolated. 1H NMR: (d6-DMSO) δ 7.90 (1H, d, 5.6Hz), 7.73 (1H, br), 7.72 (1H, m), 6.63 (1H, m), 6.14 (1H, d, 5.6Hz), 5.69 (1H, d, 2.1Hz), 5.23 (1H, s), 4.19 (1H, m), 3.89 (1H, br), 2.68 (1H, dd, 13.5 & 6.1Hz), 1.95 (2H, m), 1.73 (2H, m), 1.64 (1H, m), 1.48 (3H, s), 1.34 (3H, s), 1.34 (1H, m), 1.11 (3H, s), 0.81 (3H, s), The structure was confirmed by x-ray analysis. The measured crystal is a di hydrate. The furanyl ring C27 to C31 is disordered and refined on two positions (A/B): ratio 55:45.
Chemical oxidation of harrpernoid D
Harrpernoid D (1.0 mg, 2.3 μmol, 1.0 eq.) was dissolved in 0.3 mL of acetone-d6 and placed in an NMR tube under nitrogen atmosphere. To this solution was added at once DMDO-d6 (23 mM in acetone-d6, 0.10 mL, 1.0 eq.) and the resulting mixture was let it stand at rt. The conversion was monitored by 1H NMR. After 24 h, most of the harrpenoid D was converted to the enedial which was unstable towards purification: 1H NMR: (600 MHz, acetone-d6) δ 10.77 – 10.72 (m, 1H), 10.65 – 10.63 (m, 1H), 7.81 – 7.77 (m, 1H), 6.64 – 6.55 (m, 1H), 6.06 – 6.03 (m, 1H), 5.66 – 5.63 (m, 1H), 5.28 – 5.25 (m, 1H), 4.30 – 4.24 (m, 1H), 3.81 – 3.76 (m, 1H), 2.76 – 2.70 (m, 1H), 1.99 – 1.96 (m, 1H), 1.97 – 1.92 (m, 1H), 1.91 – 1.85 (m, 1H), 1.85 – 1.77 (m, 2H), 1.57 (s, 3H), 1.50 – 1.44 (m, 1H), 1.43 (s, 3H), 1.21 – 1.17 (m, 3H), 1.01 – 0.98 (m, 3H). HRMS: (ESI-TOF, m/z) calcd. For C25H29O8 [M+H] + calc.: 457.1857; found: 457.1861.
CYP27A1 biotransformation assay
Human CYP27A1 was expressed in Bacillus megaterium as described (Ehrhardt, et al., 2016). 7.5 uL of substrate solution (harrpernoid D (4 mg/mL) with 10 mg/mL cyclodextrin in EtOH) was added to 5 mL of 100 mM pH 7.0 phosphate buffered saline and 2 mL of suspension buffer (100 mM phosphate, 2 g/L xylose, 0.1 g/L δ-aminolevulinic acid hydrochloride adjusted to a pH of 7.5 with NaOH solution). The mixture was incubated at 30°C and 200 rpm. Each reaction was mixed with 5g of lysozyme and sonicated 6 x 30 seconds, with a small tip 2 sec cycle at 85% power. Then the reactions were incubated for 1h at 30°C and 200 rpm before 2 uL of benzonase (Merck) was added. Additional incubation for 1h at 30°C and 200 rpm before the reactions were frozen in dry ice for storage before analysis. Lysate was extracted with acetonitrile, ethyl acetate, or methanol and analyzed by LC/MS.
IsoTOP-ABPP
IsoTOP-ABPP studies were done as previously reported (Spradlin, et al., 2019). Cells were lysed by probe sonication in PBS and protein concentrations were measured by BCA assay35. For in situ experiments, cells were treated for 90 min with either DMSO vehicle or covalently acting small molecule (from 1,000× DMSO stock) before cell collection and lysis. Proteomes were subsequently labeled with N-5-Hexyn-1-yl-2-iodoacetamide (IA-alkyne) labeling (100 μM) for 1 h at room temperature. CuAAC was used by sequential addition of tris(2-carboxyethyl) phosphine (1 mM, Sigma), tris[(1-benzyl-1H-1,2,3-triazol-4-yl) methyl]amine (34 μM, Sigma), copper(II) sulfate (1 mM, Sigma) and biotin-linker-azide—the linker functionalized with a tobacco etch virus (TEV) protease recognition sequence as well as an isotopically light or heavy valine for treatment of control or treated proteome, respectively. After CuAAC, proteomes were precipitated by centrifugation at 6,500g, washed in ice-cold methanol, combined in a 1:1 control:treated ratio, washed again, then denatured and resolubilized by heating in 1.2% SDS–PBS to 80C for 5 min. Insoluble components were precipitated by centrifugation at 6,500g and soluble proteome was diluted in 5 mL 0.2% SDS–PBS. Labeled proteins were bound to avidin-agarose beads (170 mL resuspended beads per sample, Thermo Pierce) while rotating overnight at 4C. Bead-linked proteins were enriched by washing three times each in PBS and water, then resuspended in 6 M urea and PBS (Sigma), and reduced in TCEP (1 mM, Sigma), alkylated with iodoacetamide (18 mM, Sigma), before being washed and resuspended in 2 M urea and trypsinized overnight with 0.5 μg ml−1 sequencing grade trypsin (Promega). Tryptic peptides were eluted off. Beads were washed three times each in PBS and water, washed in TEV buffer solution (water, TEV buffer, 100 μM dithiothreitol) and resuspended in buffer with Ac-TEV protease and incubated overnight. Peptides were diluted in water and acidified with formic acid (1.2 M, Spectrum) and prepared for analysis.
Mass spectrometry analysis
Total peptides from TEV protease digestion for isoTOP-ABPP were pressure loaded onto 250 mm tubing packed with Aqua C18 reverse phase resin (Phenomenex no. 04A4299), which was previously equilibrated on an Agilent 600 series high-performance liquid chromatograph using the gradient from 100% buffer A to 100% buffer B over 10 min, followed by a 5 min wash with 100% buffer B and a 5 min wash with 100% buffer A. The samples were then attached using a MicroTee PEEK 360 mm fitting (Thermo Fisher Scientific no. p-888) to a 13 cm laser pulled column packed with 10 cm Aqua C18 reverse-phase resin and 3 cm of strong-cation exchange resin for isoTOP-ABPP studies. Samples were analyzed using an Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) using a five-step Multidimensional Protein Identification Technology (MudPIT) program, using 0, 25, 50, 80 and 100% salt bumps of 500 mM aqueous ammonium acetate and using a gradient of 5–55% buffer B in buffer A (buffer A: 95:5 water:acetonitrile, 0.1% formic acid; buffer B 80:20 acetonitrile:water, 0.1% formic acid). Data were collected in data dependent acquisition mode with dynamic exclusion enabled (60 s). One full mass spectrometry (MS1) scan (400–1,800 mass-to-charge ratio (m/z)) was followed by 15 MS2 scans of the nth most abundant ions. Heated capillary temperature was set to 200C and the nanospray voltage was set to 2.75 kV, as previously described.
Data were extracted in the form of MS1 and MS2 files using Raw Extractor v.1.9.9.2 (Scripps Research Institute) and searched against the Uniprot human database using ProLuCID search methodology in IP2 v.3 (Integrated Proteomics Applications, Inc.) (Xu, et al., 2015) Probe-modified cysteine residues were searched with a static modification for carboxyaminomethylation (+57.02146) and up to two differential modifications for methionine oxidation and either the light or heavy TEV tags (+464.28596 or +470.29977, respectively). Peptides were required to be fully tryptic peptides and to contain the TEV modification. ProLUCID data was filtered through DTASelect to achieve a peptide false-positive rate below 5%. Only those probe-modified peptides that were evident across two out of three biological replicates were interpreted for their isotopic light to heavy ratios. Light versus heavy isotopic probe-modified peptide ratios are calculated by taking the mean of the ratios of each replicate paired light vs. heavy precursor abundance for all peptide spectral matches (PSM) associated with a peptide. The paired abundances were also used to calculate a paired sample t-test p-value in an effort to estimate constancy within paired abundances and significance in change between treatment and control. P-values were corrected using the Benjamini/Hochberg method.
QUANTIFICATION AND STATISTICAL ANALYSIS
Cell line growth curve data were graphed using a nonlinear regression fitting model ([inhibitor] vs. response - four parameters) calculated by GraphPad Prism® software, Version 6.07 (GraphPad Software, Inc. [La Jolla, CA, USA]). Error bars indicate mean ± s.d. of (at least) 3 replicates. qRT-PCR/ChIP-qPCR data were graphed using GraphPad Prism® software, Version 6.07 (GraphPad Software, Inc. [La Jolla, CA, USA]) and error bars indicate mean ± s.d. of (at least) 3 replicates. Cell line confluency was measured IncuCyte™ (Essen BioScience) proliferation assay for live-cell analysis software and error bars indicate mean ± s.d. of 3 replicates. Flow cytometry data and cell cycle analysis was performed using the Univariate Models with Watson Pragmatic algorithm in FlowJo® software. Statistical analysis for cell line transcriptome data was performed by Wilcox test using effect size calculated by mean difference between two arrays of cell lines divided by standard deviation. Box-and-whisker plots indicate the values of median, 25th percentiles, and 75th percentiles. siRNA analysis was performed using the redundant siRNA activity (RSA) method for eight individual siRNAs per gene and GESS (genome-wide enrichment of seed sequence matches) off-target analysis. Detailed statistical methods were described previously (Sigoillot, et al., 2012; Birmingham, et al., 2009). RNA-seq alignment against Human Ensembl genome reference GRCh38 (release 78) utilized the exon quantification pipeline (EQP) (Schuierer & Roma, 2016) with the Bowtie2 (Langmead & Salzberg, 2012) alignment module for exon- and gene-level quantitation. Alignment quality was assessed using Picard (http://broadinstitute.github.io/picard/) and ArrayQualityMetrics (Kauffmann, et al., 2009), passing internal criteria. Differential expression comparisons were calculated using limma linear modeling (M.E., et al., 2015) with voom precision weighting (Law, et al., 2014). For quantification of Western blots, bands were quantified using Image J and normalized to protein loading controls. Statistical analysis was performed using a Student’s unpaired two-tailed t-test. For isoTOP-ABPP data analysis, data were extracted in the form of MS1 and MS2 files using Raw Extractor v.1.9.9.2 (Scripps Research Institute) and searched against the Uniprot human database using ProLuCID search methodology in IP2 v.3 (Integrated Proteomics Applications, Inc.) (Xu, et al., 2015). Cysteine residues were searched with a static modification for carboxyaminomethylation (+57.02146) and up to two differential modifications for methionine oxidation and either the light or heavy TEV tags (+464.28596 or +470.29977, respectively). Peptides were required to be fully tryptic peptides and to contain the TEV modification. ProLUCID data were filtered through DTASelect to achieve a peptide false-positive rate below 5%. Only those probe-modified peptides that were evident across two out of three biological replicates were interpreted for their isotopic light to heavy ratios. For those probe-modified peptides that showed ratios greater than two, we only interpreted those targets that were present across all three biological replicates, were statistically significant and showed good quality MS1 peak shapes across all biological replicates. Light versus heavy isotopic probe-modified peptide ratios are calculated by taking the mean of the ratios of each replicate paired light versus heavy precursor abundance for all peptide-spectral matches associated with a peptide. The paired abundances were also used to calculate a paired sample t-test P value in an effort to estimate constancy in paired abundances and significance in change between treatment and control. P values were corrected using the Benjamini–Hochberg method.
Supplementary Material
Cell lines and MITF/CYP27A1 microarray data, relating to Figure 1 & SI Figure 1
isoTOP-ABPP data, related to Figure 4
siRNA genome-wide screen, related to Figure 4
Microarray data, relating to Figure 2
Structural characterization of harrpernoid D, relating to Figure 1
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PhosphoPlus® p38 MAPK (Thr180/Tyr182) Antibody Duet | Cell Signaling Technology | Cat#8203 |
| PhosphoPlus® MEK1/2 (Ser217/Ser221) Antibody Duet | Cell Signaling Technology | Cat#8211 |
| PhosphoPlus® p44/42 MAPK (Erk1/2) (Thr202/Tyr204) Antibody Duet | Cell Signaling Technology | Cat#8201 |
| Anti-alpha Tubulin antibody [DM1A] | Abcam | ab7291 |
| Phospho-B-Raf (Ser445) Antibody | Cell Signaling Technology | Cat#2696 |
| Microphthalmia (Transcription Factor) (MiTF) Ab-1 (C5) | Thermo Scientific | MS-771-P1 |
| MitoBiogenesis™ Western Blot Cocktail | Abcam | ab123545 |
| Total OXPHOS Human WB Antibody Cocktail | Abcam | ab110411 |
| TFAM (D5C8) Rabbit mAb | Cell signaling | Cat#8076 |
| Recombinant Anti-Alas1 antibody [EPR10247] | Abcam | ab154860 |
| Anti-CYP27A1 antibody | Sigma-Aldrich | ab82926 |
| Monoclonal Anti-β-Actin antibody produced in mouse | Sigma-Aldrich | A5441 |
| Bacterial and Virus Strains | ||
| Bacillus megaterium | Ehrhardt et. al. (2016) | N/A |
| Chemicals, Peptides, and Recombinant proteins | ||
| MG-132 | Selleck Chemicals | S2619-5MG (CAS 1211877-36-9) |
| MitoTracker™ Green FM | Invitrogen™ | M7514 |
| Vybrant® DyeCycle™ Green Stain | Invitrogen™ | V35004 |
| FxCycle™ PI/RNAse Staining Solution | Invitrogen™ | F10797 |
| Glucose Solution | Invitrogen | A2494001 |
| 20% Galactose Solution | Teknova | G0505 |
| Fetal Bovine Serum | Gibco™ | Cat#10082147 |
| Penicillin-Streptomycin (5,000 U/mL) | Gibco™ | Cat#15070063 |
| L-Glutamine (200 mM) | Gibco™ | Cat#25030149 (CAS 56-85-9) |
| RIPA lysis and extraction buffer | Invitrogen | Cat#89900 |
| DMSO (Hybri-Max™, sterile-filtered, BioReagent, suitable for hybridoma, ≥99.7%) | Sigma-Aldrich | D2650-5X5ML (CAS 67-68-5) |
| Rotenone | Enzo lifesciences | ALX-350-360-G001 (CAS 83-79-4) |
| Antimycin A | Alfa aeser | J63522 (CAS 1397-94-0) |
| Tetramethylrhodamine, Methyl Ester, Perchlorate (TMRM) | Invitrogen™ | T668 (CAS 115532-50-8) |
| Hoechst 33342 Solution | Thermo Scientific | Cat#62249 (CAS 875756-97-1) |
| Iodoacetamide, 98% | ACROS Organics | Cat#31985062 |
| Urea | Fisher Chemical | U15-500 (CAS 57-13-6) |
| ProteaseMAX Surfactant, lyopholized | Promega | REF#V2072 |
| Ammonium Bicarbonate | Fisher Chemical | A643-500 (CAS 1066-33-7) |
| TCEP 99% | Strem Chemicals | Cat#15-7400 (CAS 51805-45-9) |
| Formic Acid, 99.0+%, Optima LC/MS Grade | Fisher Chemical | A117-50 (CAS 64-18-6) |
| Sequencing Grade Modified Trypsin, Porcine | Promega | V511A |
| Laemmli SDS sample buffer, reducing (4x) | Alfa Aesar | J60015 |
| Tris(benzyltriaxole methylamine) (TBTA) | Sigma-Aldrich | Cat#678937 (CAS 510758-28-8) |
| Biotin-TEV-azide | Weerapana et. al. (2010) | N/A |
| Copper (II) sulfate | Sigma-Aldrich | Cat#451657 |
| AcTEV Protease | Invitrogen™ | Cat#12575-015 |
| N-Hex-5-ynyl-2-iodo-acetamide (IA-alkyne) | Chess Gmbh | Cat#3187 |
| Pierce protease inhibitor mini tablets, EDTA-free | Thermo Scientific | A32955 |
| High Capacity Streptavidin Agarose Resin | Thermo Scientific | Cat#20357 |
| Streptavidin Agarose Resin | Thermo Scientific | Cat#20349 |
| MKT-077 | Sigma-Aldrich | M5449-5MG (CAS 147366-41-4) |
| Fadrozole | Novartis compound library | N/A (CAS 102676-31-3) |
| Dabrafenib | Novartis compound library | N/A (CAS 1195765-45-7) |
| Trametinib | Novartis compound library | N/A (CAS 871700-17-3) |
| CCCP (Carbonyl cyanide m-chlorophenyl hydrazone) | Novartis compound library | N/A (CAS 555-60-2) |
| Critical Commercial Assays | ||
| CellTiter-Glo® Luminescent Cell Viability Assay | Promega | G7571 |
| CellTox™ Green Cytotoxicity Assay | Promega | G8741 |
| RealTime ready DNA Probes Master, 5 x 1 ml | Roche | Cat#5502381001 |
| Mitochondria Isolation Kit for Cultured Cells | Thermo Scientific | 89874 |
| MitoTracker™ Red CMXRos | Invitrogen™ | M7512 |
| MITF TaqMan™ Gene Expression Assay | Applied Biosystems™ | 4453320 (Hs01117294_m1) |
| Click-iT® Plus EdU Alexa Fluor® 488 Flow Cytometry Assay Kit | Invitrogen™ | C10632 |
| Mitochondrial Membrane Potential Indicator | Codex BioSolutions | CB80600010 |
| Agilent Seahorse XF Cell Mito Stress Test Kits | Agilent Technologies | Cat#103015-100 |
| Deposited Data | ||
| Small molecule x-ray structure of harrpernoid D | Cambridge Crystallographic Data Center (CCDC); ccdc.cam.ac.uk | Cat#2061432 |
| Experimental Models: Cell Lines | ||
| Cell lines used in this study | Novartis cell line encyclopedia collection | N/A (Barretina, J. et al. Nature 483, 603-7 (2012)); Data S1 |
| Oligonucleotides | ||
| See Data S1 Oligonucleotides Reagents tab | ||
| Recombinant DNA | ||
| pCDH-CMV-MCS-EF1α-Puro lentiviral vector | System Biosciences | CD510B-1 |
| pTurboGFP-mito vector | Clontech | 632432 |
| Sigma CRISPR dCas9-p300 Activator Expression Plasmid | Sigma-Aldrich | DCAS9P300-1EA |
| Software and Algorithms | ||
| ImageJ | (Schneider, et al., 2012) | https://imagej.nih.gov/ij/ |
| IP2 proteomics pipeline 5.0.1 | Integrated Proteomics Applications | N/A |
| Pipeline pilot | Biovia - Dassault Systèmes® | N/A |
| PLATON | (Spek, 2003) | https://www.platonsoft.nl/platon/ |
Acknowledgements
D.E.F. gratefully acknowledges grant support from NIH (5P01 CA163222-07 and 5R01 AR043369-24) and the Dr. Mirian and Sheldon G. Adelson Medical Research Foundation. We thank the teams of the NIBR postdoc program, CBT High Throughput Biology, CBT Data Science, GDC Natural Product Unit, and CBT Genomics Sciences (Novartis, Cambridge, USA and Basel, Switzerland) and Shu Li (Analytical Sciences, Novartis, Cambridge, USA) for collaboration and support. We thank Yi Yang for pU6g-cas9-puromycin (Novartis, Cambridge, USA). We appreciate crystallography support from Ina Dix and Trixie Wagner (Novartis, Basel, Switzerland). We thank Markus Schirle and Jason R. Thomas for helpful discussion. We also thank the members of the David Fisher laboratory (Massachusetts General Hospital, Boston, USA) and Stephen Helliwell (Novartis, Basel, Switzerland) for critical reading of the manuscript and discussions.
Footnotes
Declaration of interests
All authors (except otherwise noted) are or were at the time of their involvement with the research employees of Novartis Institutes for BioMedical Research and may hold stock in Novartis. This study was funded by the Novartis Institutes for BioMedical Research and the Novartis-Berkeley Center for Proteomics and Chemistry Technologies. D.K.N. is a co-founder, shareholder, and adviser for Frontier Medicines. D.E.F. has a financial interest in Soltego, a company developing salt inducible kinase inhibitors for topical skin-darkening treatments that might be used for a broad set of human applications. The interests of D.E.F. were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
References
- Anderson S et al. , 1981. Sequence and organization of the human mitochondrial genome. Nature, Volume 290, pp. 457–465. [DOI] [PubMed] [Google Scholar]
- Backus KM et al. , 2016. Proteome-wide covalent ligand discovery in native biological systems. Nature, Volume 534, pp. 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baenke F et al. , 2016. Resistance to BRAF inhibitors induces glutamine dependency in melanoma cells. Mol. Oncol, Volume 10, pp. 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J et al. , 2012. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, Volume 483, pp. 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A et al. , 2006. "Bayes affinity fingerprints" improve retrieval rates in virtual screening and define orthogonal bioactivity space: when are multitarget drugs a feasible concept?. J. Chem. Inf. Model, Volume 46, pp. 2445–2456. [DOI] [PubMed] [Google Scholar]
- Birmingham A et al. , 2009. Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods, 6(8), pp. 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canor J & Sabatini D, 2012. Cancer cell metabolism: one hallmark, many faces. Cancer Discov., Volume 2, pp. 881–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson C et al. , 2015. Englerin A agonizes the TRPC4/C5 cation channels to inhibit tumor cell line proliferation. PLoS One, 10(6), p. e0127498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery P & Hudson G, 2013. Mitochondrial genetics. Br. Med. Bull, Volume 106, pp. 135–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corazao-Rozas P et al. , 2013. Mitochondrial oxidative stress is the Achille's heel of melanoma cells resistant to Braf-mutant inhibitor. Oncotarget, Volume 4, pp. 1986–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran R et al. , 2010. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci. Signal, 3(149), p. ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Richa A, Lopes R & Schwartsmann G, 2001. Natural products in anticancer therapy. Curr. Opin. Pharmacol, Volume 1, pp. 364–369. [DOI] [PubMed] [Google Scholar]
- Das Thakur M et al. , 2013. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature, Volume 494, pp. 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H et al. , 2002. Mutations of the BRAF gene in human cancer. Nature, Volume 417, pp. 949–954. [DOI] [PubMed] [Google Scholar]
- Dias D, Urban S & Roessner U, 2012. A historic overview of natural products in drug discovery. Metabolites, Volume 2, pp. 303–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckova A, Mernaugh R, Ham A-J & Marnett L, 2007. Identification of the protein targets of the reactive metabolite of teucrin A in vivo in the rat. Chem. Res. Toxicol, Volume 20, pp. 1393–1408. [DOI] [PubMed] [Google Scholar]
- Druckovia A & Marnett L, 2006. Characterization of the amino acid adducts of the enedial derivative of teucrin A. Chem. Res. Toxicol, Volume 19, pp. 1330–1340. [DOI] [PubMed] [Google Scholar]
- Du J et al. , 2003. MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. Am. J. Pathol, Volume 163, pp. 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt M, Gerber A, Hannemann F & Bernhardt R, 2016. Expression of human CYP27A1 in B. megaterium for the efficient hydroxylation of cholesterol, vitamin D3 and 7-dehydrocholesterol. Journal of Biotechnology, Volume 218, pp. 34–40. [DOI] [PubMed] [Google Scholar]
- Garraway L et al. , 2005. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature, Volume 436, pp. 117–122. [DOI] [PubMed] [Google Scholar]
- Gupta R, Patrick K & Bell N, 2007. Mutational analysis of CYP27A1: assessment of 27-hydoxylation of cholesterol and 25-hydroxylation of vitamin D. Metabolism, Volume 56, pp. 1248–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D & Weinberg R, 2011. Hallmarks of cancer: the next generation. Cell, Volume 144, pp. 646–674. [DOI] [PubMed] [Google Scholar]
- Haq R et al. , 2013. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell, Volume 23, pp. 302–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey A, 2008. Natural products in drug discovery. Drug Discov. Today, Volume 13, pp. 894–901. [DOI] [PubMed] [Google Scholar]
- Harvey A, Edrada-Ebel R & Quinn R, 2015. The re-emergence of natural products for durg discovery in the genomics era. Nat. Rev. Drug Discov, Volume 14, pp. 111–129. [DOI] [PubMed] [Google Scholar]
- Hodis E et al. , 2012. A landscape of driver mutations in melanoma. Cell, Volume 150, pp. 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek K et al. , 2008. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res., Volume 21, pp. 665–676. [DOI] [PubMed] [Google Scholar]
- Hou L, Panthier J & Arnheiter H, 2000. Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF. Development, Volume 127, pp. 5379–5389. [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R & Huber W, 2009. arrayQualityMetrics--a bioconductor package for quality assessment of microarray data. Bioinformatics, 25(3), pp. 415–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith C, Borisy A & Stockwell B, 2005. Multicomponent therapeutics for networked systems. Nat. Rev. Drug Discov, Volume 4, pp. 71–78. [DOI] [PubMed] [Google Scholar]
- Khuong-Huu Q et al. , 2000. New rearranged lmonoids from Harrisonia perforata. J. Nat. Prod, Volume 63, pp. 1015–1018. [DOI] [PubMed] [Google Scholar]
- Khuong-Huu Q et al. , 2001. New rearranged limonoids from Harrisonia perforata. III. J. Nat. Prod, Volume 64, pp. 634–637. [DOI] [PubMed] [Google Scholar]
- Kornienko A & La Clair J, 2017. Covalent modification of biological targets with natural products through Paal-Knorr pyrrole formation. Nat. Prod. Rep, Volume 34, pp. 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K et al. , 2013. Integrated compound profiling screens identify the mitochondrial electron transport chain as the molecular target of the natural products manassantin, sesquicillin, and arctigenin. ACS Chem. Bio, Volume 8, pp. 257–267. [DOI] [PubMed] [Google Scholar]
- Langmead B & Salzberg S, 2012. Fast gapped-read alignment with Bowtie2. Nat. Methods, 9(4), pp. 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C, Chen Y, Shi W & Smyth G, 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol., 15(2), p. R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekehal M et al. , 1996. Hepatotoxicity of the herbal medicine germander: Metabolic activation of its furano diterpenoids by cytochrome P450 3A depletes cytoskeleton-associated protein thiols and forms plasma membrane blebs in rat hepatocytes. Hepatology, Volume 24, pp. 212–218. [DOI] [PubMed] [Google Scholar]
- Lin D et al. , 2014. Cytochrome P450-mediated metabolic activation of diosbulbin B. Drug Metab. Dispos, Volume 42, pp. 1727–1736. [DOI] [PubMed] [Google Scholar]
- Lin D et al. , 2016. Lysine- and cysteine-based protein adductions derived from toxic metabolites of 8-epidiosbulbin E acetate. Toxicology Lett., Volume 264, pp. 20–28. [DOI] [PubMed] [Google Scholar]
- Lin W et al. , 2008. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res., Volume 68, pp. 664–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M.E. R et al. , 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res., 43(7), p. e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon S et al. , 1997. Antimalarial activity of tropical Meliaceae extracts and gedunin derivatives. J. Nat. Prod, Volume 60, pp. 336–341. [DOI] [PubMed] [Google Scholar]
- Manners G, 2007. Citrus limonoids: analysis, biactivity, and biomedical prospects. J. Agric. Food Chem, Volume 55, pp. 8285–8294. [DOI] [PubMed] [Google Scholar]
- Marroquin L et al. , 2007. Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to motochondrial toxicants. Toxicol. Sci, Volume 97, pp. 539–547. [DOI] [PubMed] [Google Scholar]
- Mast N, Lin J & Pikuleva I, 2015. Marketed drugs can inhibit cytochrome p450 27A1, a potential new target for breast cancer adjuvant therapy. Mol. Pharmacol, Volume 88, pp. 428–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick J et al. , 2006. An efficient rapid system for profiling the celluar activities of molecular libraries. Proc. Natl. Acad. Sci USA, 103(9), pp. 3153–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D et al. , 2010. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet, Volume 86, pp. 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P & Chan D, 2014. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell. Biol, Volume 15, pp. 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro S et al. , 2012. Identification and pathway mapping of furan target proteins reveal mitochondrial energy production and redox regulation as critical targets of furan toxicity. Toxicol. Sci, Volume 126, pp. 336–352. [DOI] [PubMed] [Google Scholar]
- Newman D & Cragg G, 2016. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod, Volume 79, pp. 629–661. [DOI] [PubMed] [Google Scholar]
- Pikuleva I, Cao C & Waterman M, 1999. An additional electrostatic interaction between adrenodoxin and p450c27 (CYP27A1) results in tighter binding than between adrenodoxin and p450scc (CYP11A1). J. Biol. Chem, Volume 274, pp. 2045–2052. [DOI] [PubMed] [Google Scholar]
- Roesch A et al. , 2013. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell, Volume 23, pp. 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman D et al. , 2015. Metabolic Enzyme Sulfotransferase 1A1 is the trigger for N-benzyl indole carbinol tumor growth supression. Chem. Biol, Volume 22, pp. 1228–1237. [DOI] [PubMed] [Google Scholar]
- Roy A & Saraf S, 2006. Limonoids: Overview of significant biactive triterpenes distributed in plant kingdom. Biol. Pharm. Bull, Volume 29, pp. 191–201. [DOI] [PubMed] [Google Scholar]
- Salma N et al. , 2017. Tfe3 and Tfeb transcriptionally regulate Ppargamma2 expression in adipocytes and mediate adiponectin and glucose levels in mice. Mol Cell. Biol, 37(15), pp. e00608–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Rasband W & Eliceiri K, 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, Volume 9, pp. 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H et al. , 1994. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature, Volume 371, pp. 768–774. [DOI] [PubMed] [Google Scholar]
- Schockel L et al. , 2015. Targeting mitochondrial complex I using BAY 87-2243 reduces melanoma tumor growth. Cancer Metab., Volume 3, p. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuierer S & Roma G, 2016. The exon quantification pipeline (EQP): a comprehensive approach to the quantification of gene, exon and junction expression from RNA-seq data. Nucleic Acids Res., 44(16), p. e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, 2015. A new golden age of natural product drug discovery. Cell, Volume 163, pp. 1297–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H et al. , 2014. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov., Volume 4, pp. 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H et al. , 2012. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun, Volume 3, p. 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R, 2006. The NCI60 human tumor cell line anticancer drug screen. Nat. Rev. Cancer, Volume 6, pp. 813–823. [DOI] [PubMed] [Google Scholar]
- Shtivelman E et al. , 2014. Pathways and therapeutic targets in melanoma. Oncotarget, Volume 5, pp. 1701–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigoillot F et al. , 2012. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat. Methods, 9(4), pp. 363–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spek A, 2003. Single-crystal structure validation with the program PLATON. J. Appl. Cryst, Volume 36, pp. 7–13. [Google Scholar]
- Spradlin J et al. , 2019. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat. Chem. Bio, Volume 15, pp. 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu E et al. , 2012. Metabolism of cholesterol, vitamin D3, and 20-hydroxyvitamin D3 incorporated into phospholipid vesicles by human CYP27A1. J. Steroid Biochem. Mol. Biol, Volume 129, pp. 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N et al. , 2014. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov., Volume 4, pp. 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Weerapana E, Blewett MM & Cravatt BF, 2014. A chemoproteomic platform to quantitatively map targes of lipid-derived electrophiles. Nat Methods, Volume 11, pp. 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O, 1956. On respiratory inpairment in cancer cells. Science, Volume 124, pp. 269–270. [PubMed] [Google Scholar]
- Webster D et al. , 2014. Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition. Genome Res., Volume 24, pp. 751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E et al. , 2010. Quantatative reactivity profiling predicts functional cysteines in proteomes. Nature, Volume 468, pp. 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S, Zhang F & Feng S, 2013. Decreased copy number of mitochondrial DNA: A potential diagnostic criterion for gastric cancer. Oncol Lett, Volume 6, pp. 1098–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T et al. , 2015. ProLuCID: an improved SEQUEST-like algorithm with enhanced snesitivity and specificity. J. Proteomics, Volume 129, pp. 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X-H et al. , 2011. Chemical constituents from fruits of harrisonia perforata. Phytochemistry, Volume 72, pp. 508–513. [DOI] [PubMed] [Google Scholar]
- Yan X-H et al. , 2016. 16-nor limonoids from Harrisonia perforata as promising selective 11B-HSD1 inhibitors. Sci. Rep, Volume 6, p. 36927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G et al. , 2016. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J. Clin. Invest, Volume 126, pp. 1834–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W et al. , 2020. Total synthesis of (+)-haperforin G. J. Am. Chem. Soc, 142(46), pp. 19487–19492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell lines and MITF/CYP27A1 microarray data, relating to Figure 1 & SI Figure 1
isoTOP-ABPP data, related to Figure 4
siRNA genome-wide screen, related to Figure 4
Microarray data, relating to Figure 2
Structural characterization of harrpernoid D, relating to Figure 1
Data Availability Statement
The RNA-Seq data are available in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov) with accession number SRP108654. Data generated in this study will be made available upon reasonable request. No code was developed for this study.