Abstract
Accumulating evidence has shown that long noncoding RNAs (lncRNAs) are involved in several biological processes, including immune responses. However, the role of lncRNAs in antiviral innate immune responses remains largely elusive. Here, we identify an uncharacterized human lncRNA AVAN from influenza A virus (IAV) infected patients, that is significantly upregulated following RNA virus infection. During IAV infection, AVAN play an indispensable role in antiviral immune responses. In vivo, we enforced the expression of AVAN in transgenic mice or adeno-associated virus encoding AVAN delivery system and found that AVAN significantly alleviated IAV virulence and virus replication. Mechanistically, nuclear AVAN positively regulates the transcription of forkhead box O3A (FOXO3a) by associating with its promoter and inducing chromatin remodeling to promote neutrophil chemotaxis. Meanwhile, cytoplasmic AVAN binds directly to the E3 ligase TRIM25 and enhances TRIM25-mediated K63-linked ubiquitination of RIG-I, thereby promoting TRIM25- and RIG-I-mediated antiviral innate immune responses, including the induction of type I interferon and ISGs. Moreover, AVAN binds to the B Box/CCD domain of TRIM25 and 1–200nt of AVAN were the functional moieties. Collectively, our findings highlight the potential clinical implications of human lncRNA AVAN as a key positive regulator of the antiviral innate immune response and a promising target for developing broad antiviral therapeutics.
Subject terms: Infectious diseases, Acute inflammation, Signal transduction, Microbiology
Introduction
Long noncoding RNAs (lncRNAs), a large class of noncoding RNAs with no or limited coding potential, are identified as functional RNAs that participate in a wide range of biological processes [1–3], including innate and adaptive immune responses, such as immune cell proliferation and cytokine production [4, 5]. With the discovery and characterization of an increasing number of lncRNAs, several mechanisms of these lncRNAs in virus-host interactions have been elucidated [6–8], however, many others remain uncharacterized.
Influenza A virus (IAV) is a leading cause of respiratory-related morbidity and mortality, posing a substantial threat to global health [9]. However, some mechanisms underlying IAV–host interactions remain unclear. Neutrophils provide one of the early lines of innate immunity and defense against invading microorganisms [10] and can rapidly recruited to infection sites during the innate immune response to IAV [11]. Besides, neutrophils can produce and release abundant cytokines and chemokines, which enable them to influence antiviral defense. However, how lncRNA functions in this process is largely unknown.
The host recognizes the invading IAV mainly through retinoic-acid-inducible gene-I (RIG-I), which is the main sensor involved in recognizing cytoplasmic viral RNA, to induce antiviral innate immunity and eliminate infection [12–14]. The binding of viral RNA to the RIG-I C-terminal regulatory domain results in a conformational change that, in turn, enhances the phosphorylation of IRF3 and eventually leads to the production of type I interferons [6]. RIG-I activation is a complex, multistep process controlled by phosphorylation and ubiquitination, while TRIM25 and RIPLET are two essential E3 ligases important for RIG-I signaling [14, 15]. Several studies reported that RIPLET, not TRIM25, is the obligatory ubiquitin E3 ligase for RIG-I. However, numerous in vivo and in vitro experiments revealed that TRIM25, belonging to the tripartite motif (TRIM) protein family involved in ubiquitination, interacts with the CARD1 of RIG-I and induces Lys63-linked ubiquitination of RIG-I [12, 14, 16]. A stable interaction between TRIM25 and RIG-I is crucial for initiating RIG-I-mediated antivirus responses [17–19]. However, little is known about the roles of lncRNAs in IAV-infected patients or the mechanisms of lncRNAs functioning in TRIM25/RIG-I signaling.
Here, we profile the lncRNAs of IAV-infected patient neutrophils and identify an undescribed human lncRNA, AVAN, which plays a critical role in antiviral innate immunity. We provide evidence that nuclear AVAN positively regulates FOXO3a expression by remodeling the FOXO3a promoter region to promote neutrophil chemotaxis and recruitment. Meanwhile, cytoplasmic AVAN promotes the interaction between TRIM25 and RIG-I by binding to TRIM25 directly, enhancing K63-linked ubiquitination of RIG-I and the activation of RIG-I-mediated antiviral response. Collectively, AVAN enhances neutrophil activation and type I IFN production and defenses against IAV infection in vitro and in vivo. These results reveal AVAN as a therapeutic candidate to maintain innate immune homeostasis after IAV infection.
Results
Human lncRNA AVAN is identified to participate in controlling IAV infection
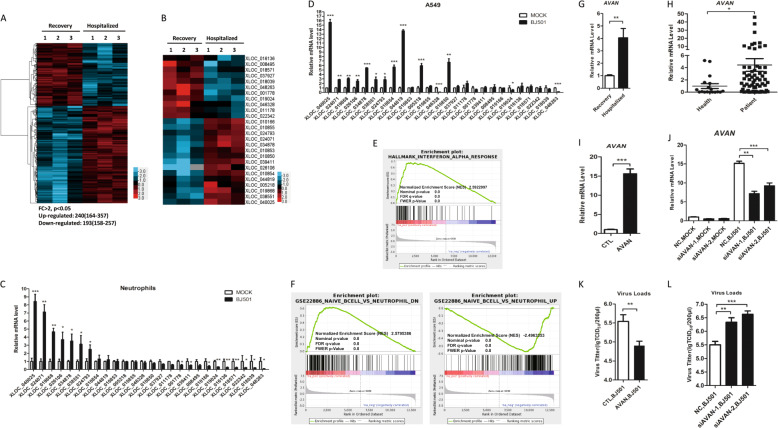
To explore the roles of host lncRNAs during IAV infection, we profiled whole transcriptional alterations using RNA-Seq in neutrophil samples from patients infected with influenza A (H7N9) virus in the acute-stage and their matched recovery-stage samples (GSE108807) (Table S1). We identified an average of 240 upregulated and 193 downregulated novel lncRNAs differentially expressed in acute-stage influenza patients compared with their recovery-stage counterparts (FC > 2, p < 0.05) (Fig. 1A). To investigate the association between lncRNAs and IAV infection, 26 novel lncRNAs with notably aberrant expression were selected as candidates after in silico analysis (Fig. 1B). To confirm the results, we isolated the neutrophils from healthy volunteers and infected with influenza virus A/Beijing/501/2009 (abbreviated as BJ501) for 12 h and found that XLOC_040025 (AVAN), an uncharacterized lncRNA, was the most significantly upregulated after infection (Fig. 1C). In addition, we infected human alveolar epithelial cells (A549) with BJ501 and found that AVAN was also the most strongly upregulated after infection among the 26 lncRNAs (Fig. 1D). Moreover, we also verified the expression of AVAN in peripheral blood mononuclear cell (PBMC), CD14+ monocytes, monocyte-derived macrophages, and BEAS-2B, a human normal lung epithelial cell, after BJ501 infection and found AVAN was also significantly upregulated (Fig. S1A–D). We next analyzed the related gene coexpression networks from the sequencing data and performed gene set enrichment analysis (GSEA) and found that response to influenza viral stimulation as well as chemotaxis and immune cell activation were related to AVAN (Fig. 1E, F; Fig. S1E; Table S2–8), suggesting that AVAN participates in innate antiviral immunity. In addition, the expression of AVAN was quantified in the RNA-seq samples by qRT-PCR (Fig. 1G). Meanwhile, AVAN was significantly upregulated in 63 additional samples from patients confirmed to be infected with the influenza A (H1N1) virus (Fig. 1H; Table S9). We then analyzed the relationship between AVAN abundance and patient’s condition and found that the expression of AVAN is correlated with severity, but not the age or sex (Fig. S1F–H). Functionally, A549 cells overexpressing AVAN displayed a significantly decreased virus titer after BJ501 infection (Fig. 1I–K). In contrast, virus titer was upregulated during AVAN knockdown upon BJ501-infected A549 cells (Fig. 1J–L). The results suggest that AVAN might be involved in the regulation of antiviral responses.
Fig. 1. Human lncRNA AVAN is identified to participated in controlling IAV infection.
A Cluster heat map showing differentially expressed lncRNAs in IAV-infected patient neutrophils compared with recovery-stage samples based on RNA-seq data (FC > 2; p < 0.05). B Cluster heat map showing 26 lncRNA candidates selected via in silico analysis from the RNA-seq data (FC > 2; p < 0.05). The expression of 26 lncRNA candidates in neutrophils (C) from healthy volunteers and A549 (D) stimulated with BJ501 (MOI = 0.5) or mock for 12 h by qRT-PCR analysis (n = 3; means ± SEM; *p < 0.05; **p < 0.01; ***p < 0.001). E, F GSEA analysis of AVAN related gene coexpression networks from the sequencing data. G AVAN upregulation in IAV-H7N9-infected patient (n = 3; means ± SEM; **p < 0.01). H AVAN expression in IAV-infected patient neutrophils by qRT-PCR analysis (healthy controls = 18; patients = 63; *p < 0.05). The efficiency of AVAN overexpression (I) or knockdown (K) was determined by qRT-PCR in BJ501-uninfected or -infected A549 cells (n = 3; means ± SEM; **p < 0.01; ***p < 0.001). BJ501 replication in AVAN-overexpression (J) and AVAN-knockdown (L) A549 cells examined by the TCID50 assay (MOI = 1). The virus titers in supernatants were measured at 24 h post infection (n = 3; means ± SEM; **p < 0.01; ***p < 0.001).
AVAN is identified as a novel lncRNA and preferentially upregulated following RNA virus infection
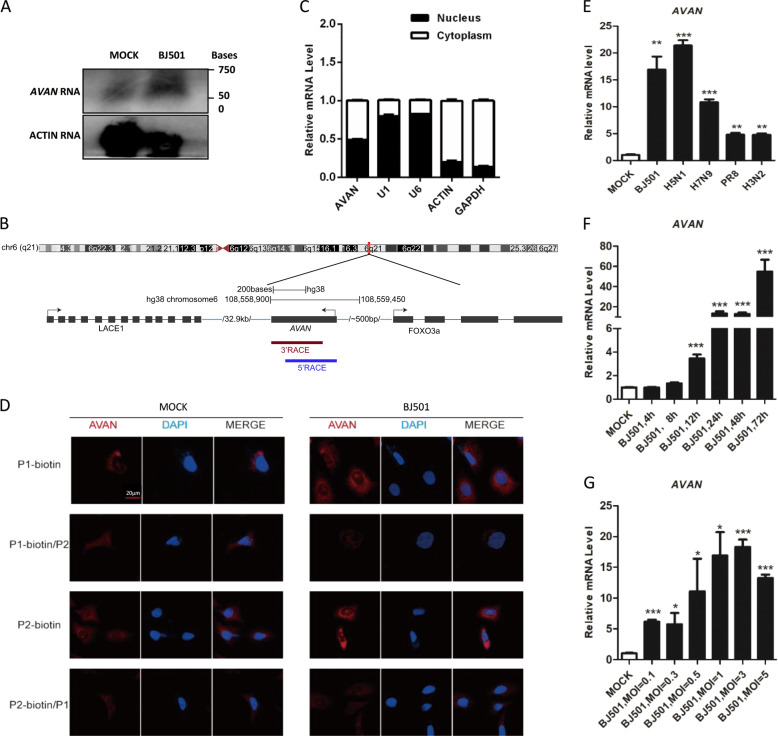
According to ORF finder [20] and the coding potential calculator [21], AVAN exhibited no protein-coding potential (Fig. S2A). Only one transcript of AVAN was found and it was upregulated upon IAV infection using northern blot analysis (Fig. 2A). The exact transcript length (517 nt) of AVAN was confirmed by rapid amplification of cDNA ends (RACE) (Table S10), revealing that AVAN is polyadenylated (Fig. 2B). Subcellular fractionation followed by qRT-PCR in BJ501-infected A549 cells showed that AVAN localized both in the cytoplasm and nucleus (Fig. 2C), which were confirmed by the RNA fluorescence in situ hybridization (RNA-FISH) (Fig. 2D). Furthermore, in competitive association FISH, free labeling-probes weakened the fluorescent dye from biotin labeling-probes, displaying the specific distribution of AVAN (Fig. 2D).
Fig. 2. AVAN is identified as a novel lncRNA and preferentially upregulated following RNA virus infection.
A Northern blotting of AVAN in A549 cells treated with mock or BJ501 (MOI = 1) at 24 h post infection. B Schematic diagram showing full-length AVAN gene locus and RACE products. C Fractionation of BJ501-infected A549 cells followed by qRT-PCR analysis. The U1 and U6 RNAs served as positive controls for nuclear gene expression. The ACTIN and GAPDH RNAs served as positive controls for cytoplasmic gene expression. D AVAN intracellular localization visualized by RNA-FISH in A549 cells stimulated with MOCK (left) or BJ501 (MOI = 1) (right) for 24 h. DAPI, 4′, 6-diamidino-2-phenylindole. Probe 1-biotin and probe 2-biotin, biotinylation AVAN probes; probe 1 and 2, no labeled free AVAN probes. Scale bar, 10 μm. E AVAN expression in A549 cells infected with BJ501 (MOI = 1), H5N1 (MOI = 1), H7N9 (MOI = 1), PR8 (MOI = 1), H3N2 (MOI = 1) for 24 h by qRT-PCR analysis (n = 3; means ± SEM; **p < 0.01; ***p < 0.001). AVAN expression in A549 cells infected with BJ501 at an MOI of 1 for the indicated times (F) or at the indicated MOIs for 24 h (G) by qRT-PCR analysis (n = 3; means ± SEM; *p < 0.05; **p < 0.01; ***p < 0.001).
We confirmed that AVAN could be stimulated by multiple sub-strains of IAV, including H1N1, H7N9, H5N1, H3N2 (Fig. 2E). Interestingly, the expression of AVAN increased more under the stimulation of IAV with stronger pathogenicity, such as H1N1 (BJ501), H7N9, and H5N1. Moreover, ANAN was slightly upregulated by some other respiratory RNA viruses, including Sendai virus (SeV) and respiratory syncytial virus (RSV), but not the DNA virus, adenovirus (ADV) (Fig. S2B). In addition, Poly (I: C), IFN-β treatment could also enhanced the expression of AVAN (Fig. S2B). Furthermore, AVAN expression was upregulated upon BJ501 challenge in a time- and dose-dependent manner in A549 cells (Fig. 2F, G) and the human promyelocytic leukemia cell line (HL60), which is often a substitute for neutrophils (Fig. S2C, D). Together, these results demonstrate that increment of AVAN level is associated with RNA viral infection.
AVAN is essential for antiviral immune responses during IAV infection
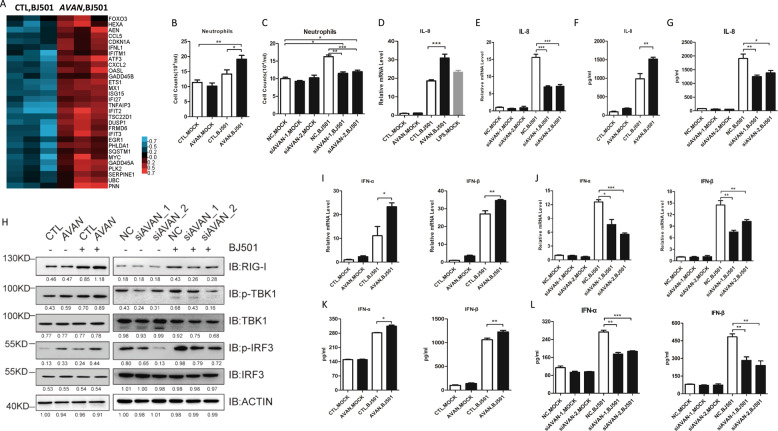
To explore the global influence and detailed mechanism of AVAN during virus infection, we performed transcriptome microarray assay and found that AVAN overexpression altered the expression of 81 genes after 14 h of BJ501 infection (Fig. S3A; Table S11). Most of these differentially expressed genes were associated with antiviral responses and inflammatory diseases according to Reactome pathway analysis (Fig. S3B) and FunDO diseases analysis (Fig. S3C). Notably, genes associated with IFN signaling and cytokine signaling, including chemokines and ISGs (IFN stimulated genes), were markedly upregulated after AVAN overexpression (Fig. 3A). These significantly upregulated genes were confirmed by qRT-PCR (Fig. S4A), and they were abolished after AVAN knockdown upon BJ501 infection (Fig. S4B). These data also reveal that AVAN participates in antiviral innate immune processes.
Fig. 3. AVAN is essential for antiviral immune responses during IAV infection.
A Altered mRNA expression in AVAN-overexpressing A549 cells and empty vector control A549 cells infected with BJ501 (MOI = 1) for 14 h by cDNA microarray (n = 3; FC > 1.3; p < 0.05). The altered expression of mRNAs involved with ISGs mRNAs that were upregulated during AVAN overexpression were clustered. Transwell assay of neutrophil migration in AVAN-overexpressing (B) or AVAN-knockdown (C) A549 culture supernatants (n = 3; means ± SEM; *p < 0.05; **p < 0.01; ***p < 0.001). IL-8 expression in AVAN-overexpressing (D) or AVAN-knockdown (F) A549 cells were measured by qRT-PCR (MOI = 1) at 24 h post infection (n = 3; means ± SEM; ***p < 0.001). LPS stimulation was the positive control. IL-8 expression in AVAN-overexpressing (E) or AVAN-knockdown (G) A549 cells were measured by ELISA (MOI = 1) at 24 h post infection (n = 3; means ± SEM; *p < 0.05; **p < 0.01). H Western blot analysis of RIG-I signaling in A549 cells transfected with AVAN or siRNAs upon BJ501 (MOI = 1) infection for 24 h. All AVAN overexpression data are normalized to the first lane (CTL-) of ACTIN and all AVAN knockdown data are normalized to the first lane (NC-) of ACTIN. IFN-α and IFN-β expression in AVAN-overexpression (I) and AVAN-knockdown (J) A549 cells infected with BJ501 (MOI = 1) measured by qRT-PCR at 24 h post infection (n = 3; means ± SEM; *p < 0.05; **p < 0.01; ***p < 0.001). IFN-α and IFN-β expression in AVAN-overexpression (K) and AVAN-knockdown (L) A549 cells infected with BJ501 (MOI = 1) measured by ELISA at 24 h post infection (n = 3; means ± SEM; *p < 0.05; **p < 0.01; ***p < 0.001).
Neutrophils activation was the main component of innate immune responses against invading microorganisms. The neutrophil transwell assays revealed that AVAN overexpression significantly upregulated neutrophil chemotaxis during BJ501 infection (Fig. 3B). These phenomena were disrupted by knocking down AVAN (Fig. 3C). The chemokines interleukin-8 (CXCL8, IL-8) is a potent neutrophil chemoattractant [22–24]. We found that AVAN overexpression increased IL-8 transcript upon BJ501 infection compared with those of the control vector (Fig. 3D; Fig. S5A) while IL-8 transcript was decreased upon AVAN knockdown in A549 and BEAS-2B cells (Fig. 3E; Fig. S5B). The protein level of IL-8 was also upregulated during AVAN overexpression and decreased during AVAN knockdown upon BJ501 infection in A549 cells (Fig. 3F, G). In addition, we found that IL-8 transcript and protein in BJ501-infected HL60 cells were consistent with those obtained in A549 cells when AVAN overexpressed or knockdown (Figure S5C–H). Meanwhile, IL-8 was also up- or downregulated by AVAN overexpression or knockdown in BJ501-infected primary human neutrophils (Fig. S5I, J).
RIG-I signaling plays a crucial role in antiviral innate immune response. Thus, we explored the impact of AVAN on RIG-I signaling and found that AVAN overexpression increased TBK1 and IRF3 phosphorylation upon BJ501 infection (Fig. 3H; Fig. S6A). In contrast, AVAN knockdown weakened these changes (Fig. 3H; Fig. S6B). Importantly, mRNA levels of IFN-α and IFN-β were significantly upregulated in AVAN-overexpressed A549 and BASE-2B cells upon BJ501 infection (Fig. 3I; Fig. S6C) and reduced when AVAN-knockdown (Fig. 3J; Fig. S6D). And the protein expression trends of IFN-α and IFN-β were consistent with their respective mRNA expression (Fig. 3K, L).
Collectively, it indicates that AVAN plays a vital role in antiviral responses by positively regulating neutrophils chemotaxis and RIG-I signaling activation.
AVAN expression significantly alleviates IAV virulence in transgenic mice
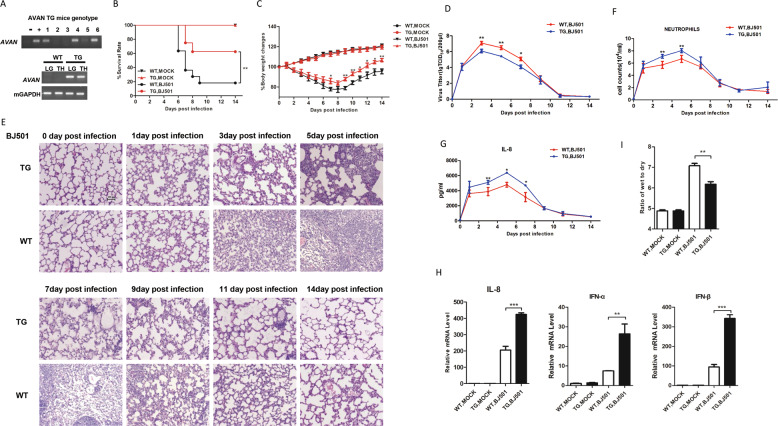
AVAV is not conserved between human and mouse (Fig. S7). To further investigate the in vivo effect of AVAN on BJ501 infection, transgenic mice (TG) mice expressing human AVAN were generated as previously described [7]. TG mice lung with high AVAN expression was identified and selected for this experiment (Fig. 4A). After being intranasal (i.n.) inoculated, BJ501 displayed a considerably lower virulence in TG mice compared with wild-type (WT) littermates. The survival rates were 60% and 20% in TG mice and WT littermates after 14 days post infection, respectively (Fig. 4B). The TG mice exhibited significantly reduced body weight loss at 7 days post infection compared with the WT littermates (Fig. 4C). Meanwhile, the viral titer in TG mice was significantly lower than that in WT littermates from 3 days to 7 days post infection (Fig. 4D). HE staining revealed that histopathologic alterations significantly improved in TG mice from 3 days to 9 days post infection (Fig. 4E). Moreover, neutrophil recruitment in BALF were significantly enhanced in TG cohort from 3 days to 5 days post infection compared with WT upon BJ501 infection (Fig. 4F). The IL-8 concentrations were also increased from 3 days to 7 days post infection in TG mice (Fig. 4G). Subsequently, we found that IL-8 and type I IFN expression in the total lung tissue were promoted in TG mice under BJ501 challenge at 5 days post infection (Fig. 4H). In addition, we examined the lung edema and found that lung wet-to-dry ratio in TG mice was significantly lower than that in WT littermates (Fig. 4I). Taken together, these observations indicate that AVAN expression can ameliorate BJ501-induced acute lung injury in vivo and protect mice from BJ501 infection.
Fig. 4. AVAN expression significantly alleviates IAV virulence in transgenic mice.
A The genotype of C57BL/6J TG mice was identified by PCR of mouse tail DNA and RT-PCR of tissue RNA. LG: lung; TH: Thymus. B, C Four-week-old TG and WT mice were inoculated with 105.125 TCID50 BJ501 virus, respectively. Survival rates and body weight changes of wild-type mice (n = 10 for each group) were monitored for 2 weeks following BJ501 challenge (*p < 0.05; **p < 0.01). D Viral load of lung tissues (n = 3 for each group) at 0, 1, 3, 5, 7, 9, 11, 14 DPI (*p < 0.05; **p < 0.01). E HE-stained images of lung tissues at 0, 1, 3, 5, 7, 9, 11, 14 DPI (magnification = ×200). F Neutrophil recruitment in the bronchoalveolar lavage fluid (BALF) (n = 3 for each group) at 0, 1, 3, 5, 7, 9, 11, 14 DPI (**p < 0.01). G The secretion of IL-8 in the BALF (n = 3 for each group) at 0, 1, 3, 5, 7, 9, 11, 14 DPI (*p < 0.05; **p < 0.01). I Wet-to-dry ratio of lung tissues (n = 5 for each group) at 5 DPI (**p < 0.01). H IL-8, IFN-α, and IFN-β expression in the lung of TG mice measured by qRT-PCR (**p < 0.01; ***p < 0.001).
Adeno-associated virus encoding AVAN delivery significantly alleviates IAV virulence in vivo
Adeno-associated virus (AAV) vector mediated gene in vivo delivery is a great way of drugs administration, among which AAV2/9 can target the lung of mice [25]. To study whether AVAN can be used as a drug, we constructed AVAN-containing AAV2/9 and control vectors, and then delivered them into mice via i.n. administration. We found that AVAN was ectopically expressed in the lung of AAV2/9-AVAN treated mice (Fig. S8A) and the groups pretreated with AAV2/9-AVAN exhibited significantly increased survival rates and reduced body weight loss at 8 days post infection compared with the control group (Fig. S8B, C). Furthermore, ameliorated lung edema, improved lung histopathology, and reduced virus titer were observed in AAV2/9-AVAN-pretreated mice after infection (Fig. S8D–F). In addition, AAV2/9-AVAN promotes IL-8 and type I IFN expression in vivo (Fig. S8G). These results showed that AAV encoding AVAN delivery significantly alleviates IAV virulence in vivo.
AVAN triggers FOXO3a expression in the nucleus to enhance neutrophil chemotaxis
Studies show that divergent lncRNAs can positively regulate the transcription of nearby genes through chromatin remodeling [26–31]. The genomic location of AVAN suggested that it was divergently transcribed from upstream of FOXO3a gene (Fig. 2B). Interestingly, microarray data revealed that FOXO3a was upregulated following AVAN overexpression during BJ501 infection (Fig. 3A). The qRT-PCR results showed that the overexpression of AVAN upregulated FOXO3a transcription and protein expression in A549 cells (Fig. 5A; Fig. S9A). Conversely, knocking down AVAN downregulated FOXO3a transcription and protein expression (Fig. 5B; Fig. S9A).
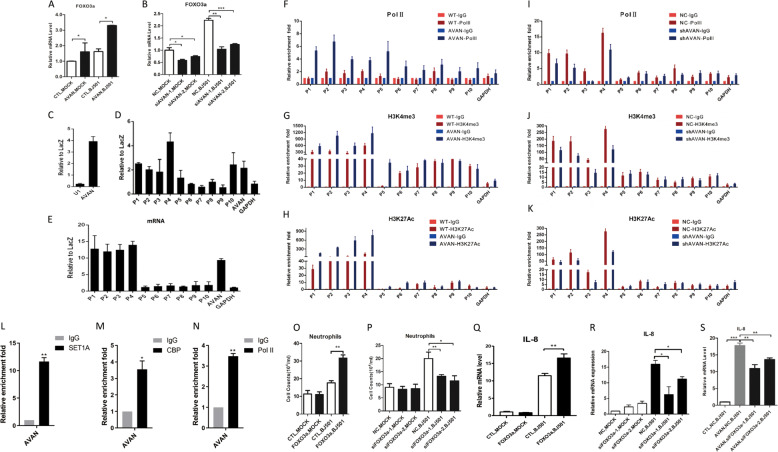
Fig. 5. AVAN enhances FOXO3a expression in nucleus.
A, B FOXO3a expression during virus infection in A549 cells were measured by qRT-PCR. A549 cells were transfected with AVAN plasmids (A) or siRNA (B) (n = 3; means ± SEM; *p < 0.05; **p < 0.01; ***p < 0.001). C Enrichment of AVAN in ChIRP assay analyzed by qRT-PCR, U1 as a negative control. ChIRP assay showing that AVAN binds directly to the FOXO3a promoter DNA in BJ501-uninfected (D) and -infected (E) A549 cells. P1–P10 represent the different regions in FOXO3a promoter or gene body locus. (F–K) H3K4me3 and H3K27ac levels and Pol II binding of the FOXO3a promoter were analyzed through ChIP followed by qRT-PCR in BJ501-infected A549 cells. Endogenous RIP of SET1A (L), CBP (M), and Pol II (N) in BJ501-infected cells using specific or anti-IgG antibodies. The relative enrichment fold of AVAN was calculated by qRT-PCR (n = 3; means ± SEM; *p < 0.05; **p < 0.01). O, P Transwell assay of neutrophil migration in FOXO3a-overexpressing or FOXO3a-knockdown A549 culture supernatant (n = 3; means ± SEM; *p < 0.05; **p < 0.01). (Q, R) IL-8 expression in FOXO3a-overexpressing (N) or FOXO3a-knockdown (O) A549 cells were measured by qRT-PCR (n = 3; means ± SEM; *p < 0.05; **p < 0.01). S IL-8 expression in AVAN-overexpressing or/and FOXO3a-knockdown A549 cells were measured by qRT-PCR (n = 3; means ± SEM; **p < 0.01; ***p < 0.001).
To test whether AVAN interacts with the FOXO3a promoter, we conducted chromatin isolation by RNA purification (ChIRP) using AVAN antisense oligoes followed by qRT-PCR in A549 cells and found that AVAN bound to sequences upstream of FOXO3a (Fig. 5C–E; Fig. S9B). We further verified the potential role of AVAN in modulating chromatin modifications at the FOXO3a promoter region. Chromatin immunoprecipitation (ChIP)-qPCR revealed that ectopic AVAN expression increased Pol II binding, as well as H3K4me3 and H3K27ac levels, at the AVAN-coated FOXO3a promoter in BJ501-infected A549 cells (Fig. 5F-H) and uninfected A549 cells (Fig. S9C–E). In contrast, AVAN knockdown decreased these enrichment (Fig. 5I–K; Fig. S9F–H). Meanwhile, we performed Pol II, SET1A (Histone Lysine methyltransferase) [32, 33] and CBP (Histone acetyltransferase) [34] RNA immunoprecipitation (RIP) to examine if AVAN itself interacts with chromatin modification enzymes and RNA polymerase, and the result showed that AVAN binds to these chromatin modification enzymes and Pol II (Fig. 5L–N; Fig. S9I). This indicated that AVAN may play a direct role in modifying chromatin and recruiting Pol II at the FOXO3a promoter region.
Chemokines were markedly upregulated in cells with ectopic AVAN expression. A previous study reported that FOXO3a could upregulate chemokine to enhance neutrophil accumulation [35]. We also found that ectopic FOXO3a expression significantly promoted IL-8 expression and neutrophil chemotaxis in BJ501-infected A549 cells (Fig. 5O–Q; Fig. S9J). In contrast, FOXO3a knockdown decreased IL-8 expression and neutrophil chemotaxis during BJ501 infection (Fig. 5P–R; Fig. S9K). In addition, knocked down FOXO3a in AVAN-overexpressing A549 cells abrogated AVAN-induced IL-8 expression during BJ501 infection (Fig. 5S). These data indicate that AVAN plays a critical role in regulating FOXO3a expression by remodeling the FOXO3a promoter region, which additionally increased IL-8 and neutrophil chemotaxis.
AVAN binds to TRIM25 directly and enhances the TRIM25-mediated K63-linked ubiquitination of RIG-I
To investigate the molecular mechanism of AVAN, RNA pull-down assays using biotin-labeled AVAN or antisense probe followed by mass spectrometry (MS) analysis were performed. The result of MS revealed that TRIM25 was pulled down by AVAN but not by AVAN antisense control in BJ501-infected A549 cells (Fig. 6A, B). This result was confirmed by RNA pull-down western blot experiments (Fig. 6C). Besides, ChIRP followed by western blot revealed that AVAN-specific probes could pull down TRIM25 while LacZ could not (Fig. 6D). To validate the interaction between AVAN and TRIM25, we immunoprecipitated TRIM25 from BJ501-infected A549 cells and quantified the protein-bound AVAN. AVAN was enriched by exogenous (Fig. 6E) and endogenous (Fig. 6F) TRIM25 relative to the isotype immunoglobulin G (IgG) control. In addition, we found that AVAN also binds to rodent TRIM25 through RNA pull-down using TG mice lung tissue (Figure S10A). However, we failed to detect the interaction of AVAN and RIG-I (data not shown).
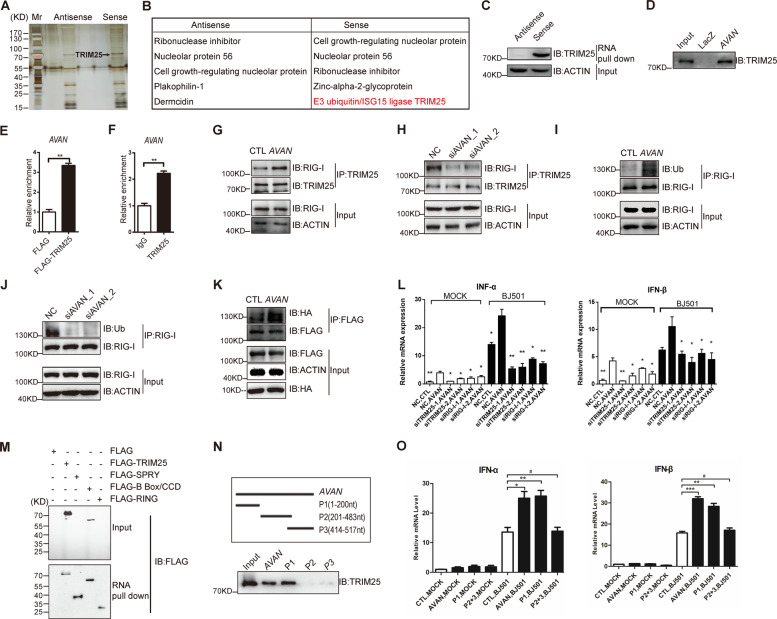
Fig. 6. AVAN binds to TRIM25 directly and enhances the TRIM25-mediated K63-linked ubiquitination of RIG-I.
A RNA pull-down of AVAN-associated proteins using biotinylated AVAN or antisense probes. Isolated proteins were resolved by SDS-PAGE followed by silver staining. B The list of proteins identified by MS in AVAN sense and antisense group, and particularly TRIM25 was pulled down by AVAN in BJ501-infected A549 cells but not by the AVAN antisense. C Pull-down western blot showing that AVAN can bind directly to TRIM25. D ChIRP followed by western blot shows that AVAN can bind to TRIM25. Exogenous (E) and endogenous (F) RIP of TRIM25 in BJ501-infected cells using anti-TRIM25 or anti-IgG antibodies. The relative enrichment fold of AVAN was calculated by qRT-PCR (**p < 0.01). TRIM25 co-immunoprecipitation with proteins from lysates of BJ501-infected A549 cells transfected with AVAN (G) or AVAN-siRNAs (H), followed by immunoblotting. Anti-TRIM25 and anti-RIG-I antibodies were used for immunoprecipitated. Immunoblot analysis of endogenous RIG-I ubiquitylation in BJ501-infected A549 cells transfected with AVANs (I) or AVAN-siRNAs (J). Anti-RIG-I and anti-Ub antibody were used for immunoprecipitated. K Immunoblot analysis of proteins immunoprecipitated with anti-Flag from lysates of BJ501-infected A549 cells transfected with AVAN, HA-Ub, and Flag-tagged RIG-I. L IFN-α (left) and IFN-β (right) expression upon AVAN transfection in A549 cells that were infected by BJ501 or not (MOI = 1) at 24 h post infection, and then individually knockdown RIG-I or TRIM25, analyzed by qRT-PCR (n = 3; means ± SEM; *p < 0.05; **p < 0.01). M AVAN pull-down western blot with lysates of A549 cells transfected with Flag, Flag-TRIM25, Flag-SPRY, Flag-B Box/CCD, or Flag-Ring. N Truncated AVAN pull-down truncates (upper panel) were obtained via in vitro transcription and incubated with BJ501-infected A549 lysates for RNA pull-down. O IFN-α (left) and IFN-β (right) expression analyzed by qRT_PCR upon AVAN or AVAN truncates transfection in A549 cells that were infected by BJ501 or not (MOI = 1) at 24 h post infection (n = 3; means ± SEM; # p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001).
TRIM25 is crucial for ubiquitination of RIG-I and the stabilization of TRIM25-RIG-I interaction is important for a sustained antiviral IFN response [14]. To explore the effect of AVAN on TRIM25–RIG-I interaction, TRIM25 co-immunoprecipitation from BJ501-infected A549 cells transfected with AVANs or siRNAs were performed. We found that ectopically expressing AVAN markedly enhanced the association between TRIM25 and RIG-I upon BJ501 infection, and AVAN knockdown weakened this association (Fig. 6G-H). Further, the association between TRIM25 and RIG-I was also significantly enhanced in TG mice compared to WT upon infection (Fig. S10B). Moreover, endogenous or exogenous RIG-I ubiquitylation was significantly increased in AVAN-overexpressing cells with BJ501 infection while RIG-I ubiquitylation was decreased in AVAN-knockdown cells (Fig. 6I–K).
To test whether AVAN enhances the IFN-mediated antiviral response through TRIM25-RIG-I signaling, we knocked down endogenous TRIM25 in HEK-293T cells and observed a markedly reduced effect of ectopic AVAN expression on the activation of IFNB1-responsive reporters upon BJ501 infection (Fig. S10C). In addition, the qRT-PCR analysis after knockdown RIG-I or TRIM25 in AVAN-overexpressing cells showed that knocks down RIG-I or TRIM25 abrogated AVAN-induced effects on IFN-α and IFN-β expression during BJ501 infection (Fig. 6L; Fig. S10D, E). Besides, the expression of IFN-α and IFN-β were also weakened after AVAN and TRIM25 double knocks down upon BJ501 infection (Fig. S10F).
Together, these data show that AVAN can directly bind TRIM25 to enhance the interaction between TRIM25 and RIG-I, promoting the ubiquitylation of RIG-I to elicit downstream signaling cascades and initiate the innate antiviral immune response by upregulating type I IFNs.
AVAN binds to the B Box/CCD domain of TRIM25 and 1-200nt of AVAN were the functional moieties
TRIM25 is composed of a RING-finger domain, a B box/coiled-coil domain (B Box/CCD), and a SPRY domain [36, 37]. In order to study the interaction region between AVAN and TRIM25, we constructed Flag-tagged TRIM25 truncated proteins containing the SPRY domain, B Box/CCD domain, or RING domain and performed AVAN RNA pull-down western blot experiments. The results showed that only the B Box/CCD domain was involved in the AVAN-TRIM25 association (Fig. 6M). Furthermore, to determine the functional structures of AVAN, we designed and constructed three truncated probes from AVAN. The result of RNA pull-down assay indicated that the TRIM25-binding activity mapped between nucleotides 1 and 200 of AVAN (Fig. 6N). In addition, different fragments of AVAN were overexpressed in A549. We observed that P1 (1–200 nt) could enhance the association between TRIM25 and RIG-I, and significantly upregulate the expression of IFN-α and IFN-β, as full-length AVAN, while fragment P2 + P3 (201–517 nt) could not (Fig. S10G; Fig. 6O). These results indicate that AVAN physically interacted with B Box/CCD domain of TRIM25 and 1–200 nt of AVAN were crucial for the function of AVAN.
Together, we propose that AVAN physically interacts with TRIM25 to facilitate the interaction between TRIM25 and RIG-I, which is essential for TRIM25-mediated ubiquitination of RIG-I to elicit the antiviral immune response.
Discussion
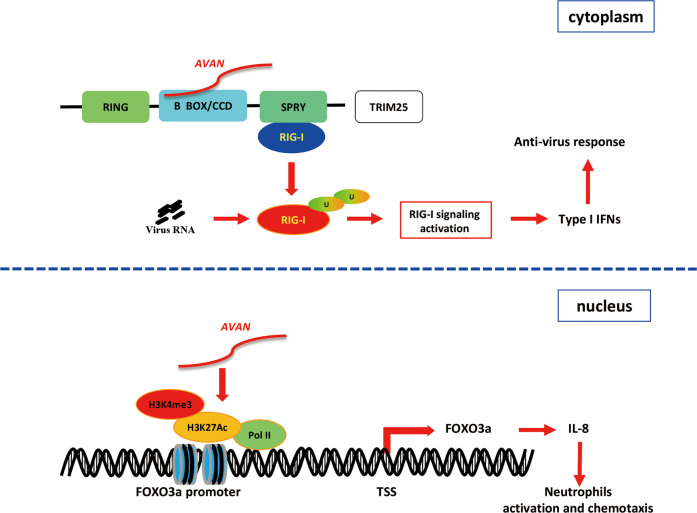
Recently, increasing evidence supports the importance of lncRNAs in host–virus interactions, especially in the regulation of innate immune responses [6]. Here, we presented lncRNA expression profiles from IAV-infected patient neutrophils through RNA-Seq for the first time and characterized a novel lncRNA AVAN, which was stimulated by RNA viruses. Our data show that nuclear AVAN positively regulates FOXO3a expression by chromatin remodeling to promote neutrophil chemotaxis and recruitment. Meanwhile, cytoplasmic AVAN directly binds TRIM25 to enhance TRIM25-mediated ubiquitination of RIG-I, and activate antiviral immune signaling to combat viral infection in vitro and in vivo (Fig. 7).
Fig. 7. Schematic of the mechanisms by which AVAN regulates antivirus innate immune responses.
(Upper) Cytoplasmic AVAN directly binds the B Box/CCD domain of TRIM25, which enhanced TRIM25- and RIG-I-mediated antiviral innate immune responses. (Below) Nuclear AVAN positively regulates the transcription of FOXO3a by associating with its promoter and inducing chromatin remodeling to promote neutrophil chemotaxis.
FOXO3a is a member of the Forkhead box O (FoxO) family of transcription factors, which is a PTEN/PI3K/AKT effector functioning in diverse cellular activities [38–40]. FOXO3a has been shown to directly suppress FASL transcription, thereby limiting the FAS-mediated death receptor pathway of apoptosis and maintained neutrophil survival and activation during inflammation [41]. Another literature showed that circulating extracellular vesicles (vEVs) could activate FOXO3a expression and downstream signaling pathway, leading to the expression of IL-8, CXCL4, and CCL5 to enhance neutrophil accumulation in kidney [35]. However, little is known about FOXO3a function during the immune response in epithelial cells. Previous research reported that FOXO3a negatively regulated cytokine expression in colonic epithelial cells [42]. In our study, FOXO3a plays a different function during IAV infection. FOXO3a promotes IL-8 expression in IAV-BJ501-infected A549 cells, which additionally enhances neutrophil chemotaxis. AVAN is located 419-bp upstream of FOXO3a. We found that AVAN can markedly increase FOXO3a expression through chromatin remodeling, thereby upregulating the expression of IL-8. However, knockdown FOXO3a blocked the upregulation of IL-8 by AVAN. Furthermore, to exclude the possibility that the upregulation of FOXO3a after AVAN overexpression is the result of enhanced IFN response, as opposed to a direct impact by AVAN on FOXO3a locus, we used HL60 cells to evade IFN effects (data not shown). Meanwhile, our data showed that AVAN may have a direct role in modifying the chromatin and interacting with Pol II by enhancing Pol II binding, H3K4me3 and H3K27ac recruiting on FOXO3a promoter (Fig. 5F–N). Thus, we propose that AVAN may enhance neutrophil chemotaxis through the regulation of FOXO3a expression. However, how AVAN is induced in neutrophils and the detailed mechanism by which FOXO3a regulates the expression of chemokines need to be further investigated.
LncRNAs are reported to function as important regulators in various critical steps of the antiviral response. They exert roles in regulating chromatin accessibility, mRNA stability, and protein activity [6, 26, 43]. A recent study shows that lnczc3h7a binds to both TRIM25 and RIG-I, facilitates the RIG-I–TRIM25 interaction and thus promotes downstream signaling transduction [6]. In addition, previous study showed that a stabilization of TRIM25-RIG-I interaction is important for a sustained antiviral IFN response [14, 17]. However, several recent studies have shown that RIPLET, but not TRIM25 is essential for RIG-I activation [18, 19], and the RIG-I-independent antiviral activities of TRIM25 was recently described [44, 45]. Despite these recent contradictory results, many studies have identified regulatory factors that regulate TRIM25-medaited RIG-I polyubiquitination, and many viral antagonists of TRIM25 that function to inhibit the polyubiquitination and signaling of RIG-I have been discovered. Herein, our data displayed that AVAN bind B Box/CCD domain of TRIM25 directly to enhance the association between TRIM25 and RIG-I, but we failed to observe a direct correlation between AVAN and RIG-I. Through truncating AVAN, we found that the P1 fragment of AVAN (1–200 nt) is necessary to bind TRIM25, and overexpression of P1 could enhance the interaction between TRIM25 and RIG-I while other fragments could not, these results showed that the physical interaction between TRIM25 and AVAN was important for TRIM25-RIG-I axis.
The N-terminal RING domain dimerization is required for E3 ubiquitin ligase activity of TRIM25 and TRIM25-mediated RIG-I ubiquitination [14, 17]. The coiled-coil domain of TRIM25 is an essential domain that mediates higher-order oligomerization of TRIM25, and is necessary for TRIM25 RING domain dimerization to productively engage Ub-conjugated E2 enzymes and become catalytically active, which directly affects the interaction between TRIM25 and RIG-I and the activation of RIG-I signaling pathway [37]. On the other hand, IAV nonstructural protein 1 (NS1) was shown to directly target the TRIM25 CCD domain, thus blocking TRIM25 multimerization and TRIM25-mediated RIG-I CARD ubiquitination [12, 13], which inhibited the antiviral response. Given that AVAN can also be integrated into the CCD domain, we speculate that there may be two possible mechanisms about AVAN promoting TRIM25-mediated RIG-I activation, the first is to facilitate the CCD scaffold to enhance the multimerization of TRIM25; the second is competitively binding to CCD domain to inhibit the interaction between TRIM25 and NS1 to prevent viral immune escape. Therefore, additional experiments were needed to solidify the TRIM25–RNA interaction and verify the hypothesis.
AVAN are upregulated by multiple RNA viruses, Poly (I:C) and interferon, but not DNA virus, suggesting that AVAN is RNA virus derived and the expression is not entirely IFN-I-dependent. Interestingly, our results show that the more virulent the virus is, the more likely it is to stimulate the production of AVAN, and the expression of AVAN strongly correlates with the severity of the influenza patients. However, the specific mechanism of how AVAN is stimulated needs to be further explored.
AVAN is not conserved between human and mouse. However, we find that AVAN also binds to rodent TRIM25 in mouse pull-down and enhances the interaction between TRIM25 and RIG-I in vivo. Our data show that overexpression of AVAN can effectively stimulate the production of IL-8 and recruit more neutrophils in the early stage of IAV infection, which is consistent with the trend of pathological changes in lung tissues. AVAN overexpression also reduce IAV infection and alleviate lung injury in TG and AAV2/9-AVAN delivered mice. Innate immune response is the first line of defense against pathogens, and neutrophils are the most important effector cells in innate immunity. Increasingly evidences show that neutrophils play a key role in the antiviral effect. Although immune response plays a double-edged role in infectious diseases, our results suggest that effective immune response in the early stage of infection may contribute to pathogen clearance and disease recovery.
In conclusion, we provide the first lncRNA landscape of IAV-infected patient neutrophils and identify the function of novel lncRNA AVAN in the innate antiviral immune response. Understanding the powerful roles of versatile lncRNAs and identifying these IAV-related lncRNAs, especially AVAN, may provide insights into the pathogenesis of viral infection, with AVAN potentially representing an essential target for intervention.
Materials and methods
Cells and viruses
A549 and BEAS-2B cells were obtained from ATCC and cultured in DMEM/F12 (1:1) supplemented with 10% (v/v) FBS and penicillin–streptomycin (100 U/ml); 293T cells were cultured in DMEM/High Glucose supplemented with 10% (v/v) FBS and penicillin–streptomycin (100 U/ml). The IAV A/Beijing/501/2009 (BJ501, H1N1), A/Puerto Rico/8/1934 (PR8, H1N1), A/Hebei/01/2013 (H7N9), A/Anhui/01/2005 (H5N1), A/Wisconsin/67/2005(H3N2) used in this study were propagated by inoculation into 9 to 11-day-old specific pathogen-free (SPF) embryonated chicken eggs via the allantoic route as described previously [46]. SeV, RSV, and ADV were used in this study. Virus stocks were aliquoted and stored at −80 °C until use.
Mice
Pathogen-free 4–6-week-old female C57BL/6 mice were purchased from the Laboratory Animal Center, AMMS, Beijing, China. The AVAN transgenic C57BL/6 mice expressing AVAN under the control of the CAG promoter were generated by the Institute of Laboratory Animal Science of Peking Union Medical College as previously described [47]. The transgenic mouse line (TG-AVAN) was bred by mating hemizygous males to WT C57BL/6 females. Half of the offspring was hemizygous for the transgene while the other half was WT and served as a control. All experiments were performed in 4–6 week-old females and their WT female littermates. All procedures, including animal studies, were conducted following the National Guidelines for the Care of Laboratory Animals (2006-398) and performed in accordance with institutional regulations after protocol review and approval by the Institutional Animal Care and Use Committee of the Academy of Military Medical Sciences (project no. 2012-005). All animals are randomly grouped. Mice were lightly anesthetized and subjected to i.n. inoculation with a tissue culture infective dose (TCID) of 105.125 TCID50 of BJ501 influenza virus in a volume of 20 μl per mouse. Control mice were inoculated with 20 μl of allantonic fluid. Inoculated mice were maintained under SPF conditions and monitored daily for weight loss and mortality or infection for 14 days post infection. The survival rate, body weight changes, viral loads, histological examination, and acute lung edema (wet-to-dry ratio) were determined as described previously [46, 48]. The number of infiltrating inflammatory cells was counted and presented as the number of cells per 200× field.
Constructs and viral packaging
The AAV2/9-AVAN and AAV2/9 empty vectors were constructed by OBiO Technology Corp., Ltd (Shanghai, China) with standard methods. In brief, AVAN was sub-cloned into pAAV-EF1a-EGFP-3FLAG plasmids to produce pAAV-EF1a-EGFP-3FLAG-CMV-AVAN. For pAAV-EF1a-EGFP-3FLAG-CMV-AVAN plasmid, CMV promote and AVAN with hGH-polyA sequences was inserted into pAAV-EF1a-EGFP-3FLAG backbone with Hind III and Blp I. The identification primer of CMV fragment in pAAV-EF1a-EGFP-3FLAG-CMV-AVAN is 5′-CGCAAATGGGCGGTAGGCGTG-3′, the primer for AVAN is 5′-GATTCCGAACAACTGAGAAGG-3’ and the primer for hGH-PolyA is 5′-ACTGGAGTGGCAACTTCCAGGGC-3′. pAAV-EF1a-EGFP-3FLAG-CMV-AVAN was produced by transfection of AAV-293 cells with AAV helper plasmid (pHelper) and AAV Rep/Cap expression plasmid (pAAV-RC). Viral particles were purified by an iodixanol step-gradient ultracentrifugation method.
RNA interference
A549 cells were transfected with siRNA targeting AVAN, TRIM25, and RIG-I using jetPRIME (Polyplus) according to the manufacturer’s instructions. The siRNA sequences used in the experiments are listed in Supplementary Table S12.
PBMC, CD14+ monocytes, neutrophils isolation
This study was approved by the Ethics Committees of the PLA General Hospital. PBMC was isolated from fresh peripheral blood using Ficoll Pague PLUS (GE Healthcare) according to the instructions supplied by the manufacturer. The CD14+ Monocytes were isolated from PBMC with anti-human-CD14-PE using a FACS Calibur flow cytometer (BD Biosciences). Neutrophils were isolated from fresh peripheral blood using Percoll PLUS (GE Healthcare) according to the instructions supplied by the manufacturer. For neutrophils collection, briefly, fresh peripheral blood was taken from IAV patients and healthy human donors and layered on a two-step Percoll PLUS gradient (75% and 60%), then centrifuged at 500 × g for 25 min. The cells at the interface between 75% and 60% were collected and washed twice with PBS and re-suspended in RPMI-1640 medium (Gibco) supplemented with 10% FBS and 100 U/ml penicillin–streptomycin at 37 °C in a humidified atmosphere of 5% (v/v) CO2.
Monocyte-derived macrophages management
Fresh human monocytes were isolated from PBMC and co-cultured with 50 ng/ml M-CSF in RPMI-1640 medium (Gibco) supplemented with 10% FBS at 37 °C in a humidified atmosphere of 5% (v/v) CO2 for 7 days. The culture medium was changed every 2 days.
ELISA
Cytokines levels were measured using an ELISA kit (Dakewe, Beijing).
Antibodies and reagents
The primary antibodies anti-RIG-I(D14G6), anti-TBK1(D1B4), anti-phospho-TBK1 (Ser172, D52C2), anti-IRF3(D6I4C), anti-phospho-IRF3 (Ser396,4D4G), anti-β-Actin(13E5), anti-FOXO3a, anti-H3K4me3, anti-rabbit IgG, anti-SET1A, and anti-CBP were purchased from Cell Signaling Technology. Anti-human-CD14-PE were purchased from BD Biosciences. Anti-Flag, anti-HA, anti-H3K27ac, and anti-TRIM25 were purchased from Sigma. Anti-Pol II was purchased from Abcam. Streptavidin C1 Beads were purchased from Invitrogen. Protein A/G PLUS-Agarose was purchased from Santa Cruz. Protein A/G Beads were purchased from Thermo Scientific. The western chemiluminescent HRP substrate was purchased from Millipore Corporation.
Western blotting
All cells were lysed in RIPA (Solarbio) supplemented with protease and phosphatase inhibitor cocktail (100×, Thermo Fisher) and lysed for 10 min on ice. The supernatant was mixed with 1/4 volume of 5× loading dye. The mixtures were then heated at 95 °C and stored at –80 °C. The samples were separated by SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were then blocked with 5% nonfat milk (BD) in 1× Tris-buffered saline and 0.1% Triton 100 for 1 h while shaking at room temperature. Next, the membranes were incubated with primary antibodies and horseradish peroxidase-conjugated secondary antibodies. Bands were visualized using the Kodak film exposure detection system. The film was scanned, and the band intensity was analyzed using Quantity One software.
Quantitative real-time PCR
Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen). cDNA was generated by reverse transcription with commercial PrimeScript RT Master Mix (Takara). Primer pairs (Table S12) were designed using Primer Premier Software 5.0 (Premier Biosoft International, Palo Alto, CA) and synthesized by Invitrogen. Quantitative real-time PCR was performed in triplicate wells of a 96-well reaction plate on an ABI 7500 PCR System (Applied Biosystems). GAPDH was used as the endogenous control. The 2−∆∆Ct method was used to calculate expression relative to the internal control. The data were analyzed using ABI 7500 SDS software v.1.3.
Luciferase assays
For IFNB1 transcriptional activity assays, 100 ng of pGL3-IFNB1 luciferase plasmid was cotransfected with 20 ng of pRL-TK vector into the cells using jetPRIME Transfection reagent (Polyplus). At 24 h after transfection, the cells were harvested according to the manufacturer’s protocol (Promega), and firefly and Renilla luciferase signals were measured using a dual-luciferase reporter assay system (Promega) on a Promega GloMax 96 machine (Promega) according to the protocol provided by the manufacturer.
Northern blotting
For northern blotting, total RNA was isolated from A549 cells using TRIzol reagent. Probes (Custom LNA mRNA Detection) were designed and synthesized by Exiqon and were also used for FISH. Northern blotting was performed as described previously [31].
5′ and 3′ RACE
The 5′ and 3′ RACE analyses were performed using the SMARTer RACE 5′/3′ Kit (Clontech) according to the manufacturer’s instructions. The RACE PCR products were cloned into pMD-19Tvector (Takara) and sequenced.
RNA pull-down assays
RNA pull-downs were performed as described [31]. In vitro biotin-labeled RNAs (lncRNA AVAN and its antisense RNA) were transcribed with biotin RNA labeling mix (Roche) and T7 RNA polymerase (Roche), treated with RNase-free DNase I (Promega) and purified using the RNeasy Mini Kit (QIAGEN). Biotinylated RNA was incubated with cell lysate, and precipitated proteins were separated via SDS-PAGE. Then the gel was stained using SilverQuest™ Silver Staining Kit (Invitrogen™) according to the manufacturer’s instructions. Following silver staining, specific bands in the sense group and control bands with equal molecular weight in the antisense group were cut and fed into nanoLC-LTQ-Orbitrap XL for MS analysis.
RNA fluorescence in situ hybridization (RNA-FISH)
RNA-FISH was performed as described previously [49]. Hybridization was carried out using DNA probe sets (Biosearch Technologies) according to the protocol provided by Biosearch Technologies. Cells were observed on a FV1000 confocal laser microscope (Olympus).
RNA immunoprecipitation (RIP) and ChIRP assays
RIP assays were performed as described [49] with minor modification. Briefly, after incubation, the magnetic beads were washed with high salt lysis buffer (containing 500 mM NaCl) five times. RIP products were analyzed by qRT-PCR using the primer pairs listed in Table S12. ChIRP was performed according to Chu et al. [50]. The ChIP-enriched FOXO3a promoter was quantified by qRT-PCR using the primer pairs listed in Table S12.
Chemokine measurement
Bronchoalveolar lavage fluid (BALF) samples were collected from the infected mice at indicated time points. For chemokine measurements, BALF samples were processed using ELISA following the manufacturer’s instructions.
Neutrophil recruitment measurement
BALF samples were collected from the infected mice at indicated time points. After centrifugation of BALF, the precipitated cells were collected and re-suspended with 400 μl pre-cooled PBS. The neutrophils were measured by an automatic blood cell analyzer (SYSMEX, Japan) following the manufacturer’s instructions.
Transwell assays
A549 cells overexpressed or knocked down AVAN or FOXO3a, and then infected with BJ501 (MOI = 1). After 24 h infection, the supernatant was collected and added into the lower chamber of chemotactic chamber. Neutrophils were isolated from peripheral blood of healthy people and added into the upper chamber of chemotactic chamber. The cells were cultured at 37 °C in a humidified atmosphere of 5% (v/v) CO2 for 1 h, and then the number of neutrophils in the lower chamber was calculated.
Bronchoalveolar epithelial cells isolation
Mice were killed at indicated time points. Agar containing 0.25% trypsin was poured into the trachea of mice and fixed for a moment. After the agar was coagulated, it was placed in the medium containing 0.25% trypsin, 0.01 g L−1 DNase I, and 5% fetal bovine serum, and incubated at 37 °C for 2–4 h. The lung tissue was cut up with scissors, the cells were suspended and centrifuged. The cell precipitates were resuspended in DMEM/F12 medium supplemented with 10% FBS and cultured in mouse IgG pre-coated culture dish for 3 h. The non-adherent cells were aspirated and centrifuged for precipitation.
Statistical analyses
All data are presented as the means ± SEM. Data were analyzed using Student’s t test. Survival data were analyzed by Kaplan–Meier survival analysis, and single time points were analyzed using ANOVA. All analyses are performed using Instat software (Version 5.0, GraphPad prism). p < 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
We thank Dr. Pin Wang (Navy Medical University) and Dr. Qiang Wang (Harvard Medical School) for providing plasmids for this research. We are grateful to Prof. Bing Ni (Army Medical University) for insightful discussions and helpful comments on the article. We thank Dr. Hongjing Gu (Beijing Institute of Microbiology and Epidemiology), Dr. Zhongpeng Zhao (Beijing Institute of Microbiology and Epidemiology), Fang Sun (Fifth Medical Center of Chinese PLA General Hospital), and Jinxia Cheng (Fifth Medical Center of Chinese PLA General Hospital) for their help in this research.
Funding
This work was supported in part by funding from the National Programs for High Technology Research and Development of China (SS2015AA020924), the National Major Research and Development Program of China (No. 2017YFC1200800), and the Natural Science Foundation of China (81771700).
Author contributions
Authors LL, PY, RC, and XW conceived and designed the experiments; CL, LL, QL, SC, KW, LNZ, and MX performed the experiments; YD, CW, LLZ, ZL, and JL analyzed the data; and CL, LL, KW, and QL wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Ethical approval
All experimental procedures in mice were approved by the Institutional 609 Animal Care and Use Committee of the Academy of Military Medical Sciences. This study 610 was approved by the Ethics Committee of the Chinese PLA General Hospital
Footnotes
Edited by K. Newton
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chengcai Lai, Lihui Liu, Qinghua Liu, Keyu Wang, Sijie Cheng
Contributor Information
Lihui Liu, Email: lihui.mm@163.com.
Xiliang Wang, Email: xiliangw@126.com.
Runsheng Chen, Email: rschen@ibp.ac.cn.
Penghui Yang, Email: ypenghuiamms@hotmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00791-2.
References
- 1.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 2.Nair L, Chung H, Basu U. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat Rev Mol Cell Biol. 2020;21:123–36. doi: 10.1038/s41580-019-0209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fitzgerald KA, Caffrey DR. Long noncoding RNAs in innate and adaptive immunity. Curr Opin Immunol. 2014;26:140–6. doi: 10.1016/j.coi.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42:792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Lin H, Jiang M, Liu L, Yang Z, Ma Z, Liu S, et al. The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat Immunol. 2019;20:812–23. doi: 10.1038/s41590-019-0379-0. [DOI] [PubMed] [Google Scholar]
- 7.Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16:616–26. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang M, Zhang S, Yang Z, Lin H, Zhu J, Liu L, et al. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell. 2018;173:906–19. doi: 10.1016/j.cell.2018.03.064. [DOI] [PubMed] [Google Scholar]
- 9.Livingston E, Bucher K, Rekito A. Coronavirus disease 2019 and influenza. JAMA. 2020;323:1122. [DOI] [PubMed]
- 10.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13:159–75. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 11.Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM, Tumpey TM. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4:e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–49. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koliopoulos MG, Lethier M, van der Veen AG, Haubrich K, Hennig J, Kowalinski E, et al. Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition. Nat Commun. 2018;9:1820. doi: 10.1038/s41467-018-04214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–20. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 15.Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010;8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Wu C, Pan Y, Liu H, Wang X, Yang Y, et al. NDR2 promotes the antiviral immune response via facilitating TRIM25-mediated RIG-I activation in macrophages. Sci Adv. 2019;5:eaav0163. doi: 10.1126/sciadv.aav0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauli EK, Chan YK, Davis ME, Gableske S, Wang MK, Feister KF, et al. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal. 2014;7:ra3. doi: 10.1126/scisignal.2004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadena C, Ahmad S, Xavier A, Willemsen J, Park S, Park J, et al. Ubiquitin-dependent and -independent roles of E3 ligase RIPLET in innate immunity. Cell. 2019;177:1187–e1116. doi: 10.1016/j.cell.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayman T, Hsu A, Kolesnik T, Dagley L, Willemsen J, Tate M, et al. RIPLET, and not TRIM25, is required for endogenous RIG-I-dependent antiviral responses. Immunol Cell Biol. 2019;97:840–52. doi: 10.1111/imcb.12284. [DOI] [PubMed] [Google Scholar]
- 20.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Boer WI, Sont JK, van Schadewijk A, Stolk J, van Krieken JH, Hiemstra PS. Monocyte chemoattractant protein 1, interleukin 8, and chronic airways inflammation in COPD. J Pathol. 2000;190:619–26. doi: 10.1002/(SICI)1096-9896(200004)190:5<619::AID-PATH555>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 23.Chu CQ, Lu XJ, Li CH, Chen J. Molecular characterization of a CXCL8-like protein from ayu and its effect on chemotaxis of neutrophils and monocytes/macrophages. Gene. 2014;548:48–55. doi: 10.1016/j.gene.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Scapini P, Lapinet-Vera JA, Gasperini S, Calzetti F, Bazzoni F, Cassatella MA. The neutrophil as a cellular source of chemokines. Immunological Rev. 2000;177:195–203. doi: 10.1034/j.1600-065X.2000.17706.x. [DOI] [PubMed] [Google Scholar]
- 25.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–55. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CK, Blanco M, Jackson C, Aznauryan E, Ollikainen N, Surka C, et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354:468–72. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- 27.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–6. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–5. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paralkar VR, Taborda CC, Huang P, Yao Y, Kossenkov AV, Prasad R, et al. Unlinking an lncRNA from Its Associated cis Element. Mol Cell. 2016;62:104–10. doi: 10.1016/j.molcel.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao T, Liu L, Li H, Sun Y, Luo H, Li T, et al. Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPalpha. Stem cell Rep. 2015;5:856–65. doi: 10.1016/j.stemcr.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu M, Wang P, Lee J, Martin-Brown S, Florens L, Washburn M, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–44. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26:1714–28. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth S, Denu J, Allis C. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 35.Yuan H, Chen C, Li Y, Ning D, Li Y, Chen Y, et al. Circulating extracellular vesicles from patients with valvular heart disease induce neutrophil chemotaxis via FOXO3a and the inhibiting role of dexmedetomidine. Am J Physiol Endocrinol Metab. 2020;319:E217–E231. doi: 10.1152/ajpendo.00062.2020. [DOI] [PubMed] [Google Scholar]
- 36.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350:217–21. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez JG, Chiang JJ, Sparrer KMJ, Alam SL, Chi M, Roganowicz MD, et al. Mechanism of TRIM25 Catalytic Activation in the Antiviral RIG-I Pathway. Cell Rep. 2016;16:1315–25. doi: 10.1016/j.celrep.2016.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 39.Furukawa-Hibi Y, Yoshida-Araki K, Ohta T, Ikeda K, Motoyama N. FOXO forkhead transcription factors induce G(2)-M checkpoint in response to oxidative stress. J Biol Chem. 2002;277:26729–32. doi: 10.1074/jbc.C200256200. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto K, Miyamoto T, Kato R, Yoshimura A, Motoyama N, Suda T. FoxO3a regulates hematopoietic homeostasis through a negative feedback pathway in conditions of stress or aging. Blood. 2008;112:4485–93. doi: 10.1182/blood-2008-05-159848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jonsson H, Allen P, Peng SL. Inflammatory arthritis requires Foxo3a to prevent Fas ligand-induced neutrophil apoptosis. Nat Med. 2005;11:666–71. doi: 10.1038/nm1248. [DOI] [PubMed] [Google Scholar]
- 42.Snoeks L, Weber CR, Turner JR, Bhattacharyya M, Wasland K, Savkovic SD. Tumor suppressor Foxo3a is involved in the regulation of lipopolysaccharide-induced interleukin-8 in intestinal HT-29 cells. Infect Immun. 2008;76:4677–85. doi: 10.1128/IAI.00227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of Long Noncoding RNAs. Annu Rev Immunol. 2017;35:177–98. doi: 10.1146/annurev-immunol-041015-055459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M, Lau Z, Cheung P, Aguilar E, Schneider W, Bozzacco L, et al. TRIM25 enhances the antiviral action of zinc-finger antiviral protein (ZAP) PLoS Pathog. 2017;13:e1006145. doi: 10.1371/journal.ppat.1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyerson N, Zhou L, Guo Y, Zhao C, Tao Y, Krug R, et al. Nuclear TRIM25 specifically targets influenza virus ribonucleoproteins to block the onset of RNA chain elongation. Cell Host Microbe. 2017;22:627–e627. doi: 10.1016/j.chom.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai C, Wang K, Zhao Z, Zhang L, Gu H, Yang P, et al. C-C motif chemokine ligand 2 (CCL2) mediates acute lung injury induced by lethal influenza H7N9 virus. Front Microbiol. 2017;8:587. doi: 10.3389/fmicb.2017.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Lu D, Zhang W, Quan X, Dong W, Xu Y, et al. Cardioprotection by Hepc1 in cTnT(R141W) transgenic mice. Transgenic Res. 2012;21:867–78. doi: 10.1007/s11248-011-9582-y. [DOI] [PubMed] [Google Scholar]
- 48.Wang W, Yang P, Zhong Y, Zhao Z, Xing L, Zhao Y, et al. Monoclonal antibody against CXCL-10/IP-10 ameliorates influenza A (H1N1) virus induced acute lung injury. Cell Res. 2013;23:577–80. doi: 10.1038/cr.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu L, Yue H, Liu Q, Yuan J, Li J, Wei G, et al. LncRNA MT1JP functions as a tumor suppressor by interacting with TIAR to modulate the p53 pathway. Oncotarget. 2016;7:15787–15800. doi: 10.18632/oncotarget.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP). J Vis Exp. 2012;:3912. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.