Abstract
Sorting nexins (SNXs), the retromer-associated cargo binding proteins, have emerged as critical regulators of the trafficking of proteins involved in the pathogenesis of diverse diseases. However, studies of SNXs in the development of cardiovascular diseases, especially cardiac hypertrophy and heart failure, are lacking. Here, we ask whether SNX3, the simplest structured isoform in the SNXs family, may act as a key inducer of myocardial injury. An increased level of SNX3 was observed in failing hearts from human patients and mice. Cardiac-specific Snx3 knockout (Snx3-cKO) mice and Snx3 transgenic (Snx3-cTg) mice were generated to evaluate the role of Snx3 in myocardial hypertrophy, fibrosis, and heart function by morphology, echocardiography, histological staining, and hypertrophic biomarkers. We report that Snx3-cKO in mice significantly protected against isoproterenol (ISO)-induced cardiac hypertrophy at 12 weeks. Conversely, Snx3-cTg mice were more susceptible to ISO-induced cardiac hypertrophy at 12 weeks and showed aggravated cardiac injury even heart failure at 24 weeks. Immunoprecipitation-based mass spectrometry, immunofluorescent staining, co-immunoprecipitation, localized surface plasmon resonance, and proximity ligation assay were performed to examine the direct interaction of SNX3-retromer with signal transducer and activator of transcription 3 (STAT3). We discovered that STAT3 was a new interacting partner of SNX3-retromer, and SNX3-retromer served as an essential platform for assembling gp130/JAK2/STAT3 complexes and subsequent phosphorylation of STAT3 by direct combination at EE. SNX3-retromer and STAT3 complexes were transiently imported into the nucleus after hypertrophic stimuli. The pharmacological inhibition or knockdown of STAT3 reversed SNX3 overexpression-induced myocardial injury. STAT3 overexpression blunts the beneficial function of SNX3 knockdown on hypertrophic cardiomyocytes. We show that SNX3-retromer promoted importin α3-mediated STAT3 nuclear trafficking and ultimately leading to cardiac injury. Taken together, our study reveals that SNX3 plays a key role in cardiac function and implicates SNX3 as a potential therapeutic target for cardiac hypertrophy and heart failure.
Subject terms: Molecular biology, Cardiovascular diseases
Introduction
By responding to various pathophysiological stresses, the heart undergoes a transition from compensatory hypertrophy to decompensatory phase, in which left ventricular remodeling, systolic and diastolic dysfunction and eventual heart failure (HF) occurs [1–3]. The abnormal activation of multiple cellular signaling pathways, such as protein kinase C (PKC)-mitogen-activated protein kinase (MAPK), a calcineurin-nuclear factor of activated T cells (NFATs), and janus kinase (JAK)-signal transducer and activator of transcription protein (STAT), is the pathological basis for the development of HF [3, 4].
The physiological functions of signaling proteins are closely linked to their intracellular trafficking and subcellular localization, which is determined by sorting nexins (SNXs)–retromer complex and endosomes [5–8]. SNXs are featured by a highly conserved Phox homology (PX)-domain and comprise 33 members in mammals [9]. Structurally, SNXs directly or indirectly bind to vacuolar protein sorting (VPS) family, VPS26 VPS35 and VPS29, to constitute retromer complex; SNXs also target the endosomal membrane through the PX-domain [5, 6]. Functionally, SNXs recruit retromer to early endosomes (EE) or recycling endosomes (RE) and subsequently mediate retrograde transport of cargo proteins via vesicle budding from EE/RE to trans-Golgi network (TGN), plasma membrane (PM), or cell nucleus [5–7]. SNXs have been reported to regulate diverse disease processes, including Alzheimer’s disease, cancer, HF, and arthritis [10–13]. For instance, we previously reported that deficiency of SNX10 prevents inflammation and bone erosion in a mouse model of rheumatoid arthritis through promoting NFATc1 degradation [10]; SNX13 profoundly affects cardiac performance through apoptosis repressor with caspase recruitment domain (ARC)-caspase signaling pathway [11]. Recently, it is found that SNX3 (the simplest structured isoform in the SNXs family)-retromer is required for retrograde recycling of Wntless [14], PC1, and PC2 [15], the cation-independent mannose 6-phosphate receptor (CI-M6PR) [16], and iron transporters [17]. However, the role of SNX3 in the pathogenesis of cardiovascular diseases, especially cardiac hypertrophy and HF, remains unknown.
Signal transducer and activator of transcription 3 (STAT3), a subtype of STAT family, participates in the pathological process of cardiac hypertrophy and HF [4, 18, 19]. Cardiomyocyte-specific overexpressed STAT3 (Stat3-Tg) in mice causes spontaneous concentric cardiac hypertrophy [4]. In response to stimulation with pro-inflammatory cytokines and growth factors, STAT3 is phosphorylated at tyrosine 705 (Y705) by receptor-associated JAK2, then forms homo- or hetero-dimers, and translocate to the cell nucleus where they act as transcription activators [18–20]. Importin α3 (also called Kpna3), a nuclear import factor, is responsible for STAT3 nuclear import independent of tyrosine phosphorylation [21, 22]. Upon axons injury exposure, the newly synthesized STAT3 combines with importin α5 and dynein (a retrograde molecular motor) and transports back to the cell body [23–25]. STAT3 co-localizes with endocytic vesicles in transit from the cell membrane to the perinuclear region, the perinuclear endosomal compartment to sustain phosphorylated STAT3 (Y705) in the nucleus [26–29]. To date, it remains largely unknown whether the retromer-dependent mechanism is involved in the phosphorylation and nuclear trafficking of STAT3.
Here, we demonstrate that the mRNA and protein levels of SNX3 were increased in end-stage failing human hearts and cardiac tissues from isoproterenol (ISO)-induced mouse cardiac injury model. Besides, cardiac-specific Snx3 knockout (Snx3-cKO) in mice efficaciously protected against ISO-induced cardiac injury at 12 weeks. Conversely, cardiac-specific Snx3 transgenic (Snx3-cTg) mice were hypersensitive to ISO-induced cardiac injury at 12 weeks and resulted in cardiac hypertrophy and dysfunction even HF at 24 weeks. Our results reveal the importance of the SNX3–retromer complex for assembling gp130/JAK2/STAT3 complexes and subsequent phosphorylation of STAT3 by direct combination at EE. Hypertrophic stimulation-induced nuclear translocation of STAT3 was facilitated by importin α3-mediated SNX3–retromer importing. The pharmacological inhibition or knockdown of STAT3 reversed SNX3 overexpression-induced myocardial injury. STAT3 overexpression blunts the beneficial function of SNX3 knockdown on hypertrophic cardiomyocytes. SNX3–retromer promoted STAT3 nuclear trafficking and ultimately leading to cardiac injury. Together, our study identifies SNX3 as a potential target for cardiac hypertrophy and HF.
Results
SNX3 expression was up-regulated in human and mouse failing hearts
To explore the potential role of SNX3 in the development of HF, we analyzed heart samples from 9 patients with HF in end-stage and 3 non-failing healthy controls. The SNX3 mRNA level was increased in failing human heart tissues compared with that in the normal control tissues as shown by qPCR (Supplementary Fig. S1). The results from immunofluorescence (IF) and western blot analysis suggest that the protein expression of SNX3 was significantly higher in the failing human heart tissues (Fig. 1A, B). Similarly, the mRNA and protein levels of SNX3 were clearly induced in the myocardium of mouse cardiac hypertrophy and HF model by subcutaneous (s.c.) injection of ISO (3 mg/kg/day, a classic hypertrophic agonist [30–32]) (Supplementary Fig. S2). These results suggest that SNX3 was associated with cardiac hypertrophy and HF in humans and mice.
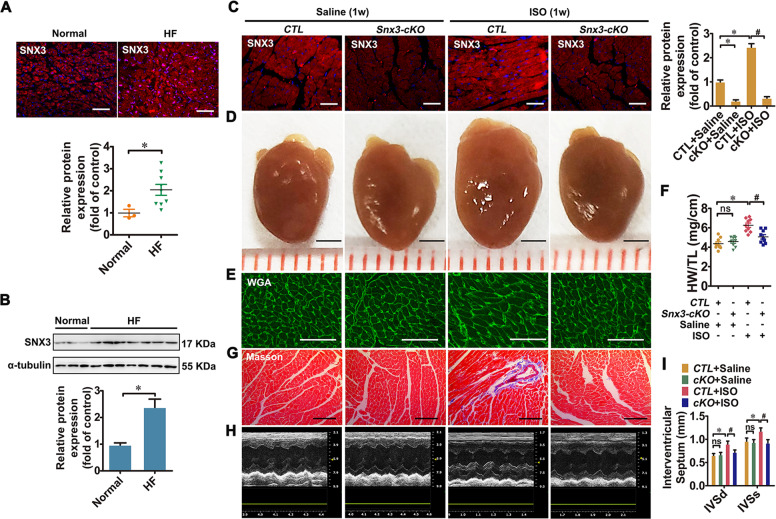
Fig. 1. SNX3 expression was increased in failing human hearts, and cardiac-specific Snx3-cKO mice were protected from ISO-induced cardiac injury.
A and B A significant induction of SNX3 protein expression in end-stage failing human heart tissues (n = 9 per HF, n = 3 per normal control), which was identified by IF assay (Scale bar: 200 μm) and western blot analysis. Male C57BL/6 Snx3-cKO mice and their respective CTL at 11 weeks were injected with ISO (3 mg/kg/day, s.c.) or an equal volume of sterile normal saline for one week. C The protein expression of SNX3 was measured by an IF assay (Scale bar: 100 μm). D and E Gross observation of heart morphology, WGA staining (Scale bar: 50 μm)-stained cross-sections of heart tissues were shown. F The HW/TL ratio was calculated. G Masson staining (Scale bar: 100 μm)-stained cross-sections of heart tissues were shown. H The representative echocardiographic graphs were presented. I The echocardiographic parameters IVS was measured. Representative images of five independent experiments were presented. The data were presented as the means ± SEM. *P < 0.05 vs. Normal or CTL + Saline group, #P < 0.05 vs.. CTL + ISO group, n = 11 per CTL + Saline group, n = 11 per cKO + Saline group, n = 10 per CTL + ISO group, n = 12 per cKO + ISO group. CTL littermate negative control, HF heart failure, HW/TL, the heart weight to the tibia length ratio, IF immunofluorescence, ISO isoproterenol, IVS interventricular septum, ns no statistical difference, s.c. subcutaneously, Snx3-cKO cardiac-specific Snx3 knockout, WGA wheat germ agglutinin, 1w 1 week. See also Supplementary Figs. S1–S4.
Cardiac-specific Snx3-cKO attenuated ISO-induced cardiac hypertrophy in mice
To examine the function of SNX3 in cardiac hypertrophy, a cardiac-specific Snx3-cKO mice model was generated by breeding Snx3-floxed mice with C57BL/6-Myh6em1(IRES-Cre)Smoc line (Supplementary Fig. S3A). A cDNA encoding internal ribosome entry site (IRES) and Cre recombinase is inserted into the 3′UTR region of α-myosin heavy chain (Myh6) gene to generate this Myh6-IRES-Cre mouse line [33]. As shown in Fig. 1C, Supplementary Fig. S3F and G, endogenous SNX3 protein expression was specifically depleted in heart tissues of Snx3-cKO mice, compared with their littermate negative controls (CTL). Snx3-cKO mice showed no obvious cardiac structural or functional defects at basal conditions.
To assess whether Snx3 deletion influences cardiac dysfunction under stressed conditions, at 11 weeks, both Snx3-cKO mice and CTL mice were randomly injected with ISO (3 mg/kg/day, s.c.) for one week to establish a cardiac hypertrophy model. Compared with the saline group, ISO treatment significantly induced cardiac hypertrophy, fibrosis, and heart dysfunction in CTL mice, evidenced by gross morphologic examination, histological staining, hypertrophic biomarkers (Anf, Bnp, and β-Mhc), heart weight to the tibia length ratio (HW/TL) and echocardiography (Fig. 1D–I, Supplementary Fig. S4). The results of heart morphology, wheat germ agglutinin (WGA) staining, hematoxylin–eosin (HE) staining, HW/TL ratio, as well as the mRNA levels of hypertrophic biomarkers, demonstrated that ISO treatment-induced cardiomyocyte hypertrophy in CTL mice (Fig. 1D–F, Supplementary Fig. S4A, C). Cardiac fibrosis was exhibited by masson staining and picric sirius red (PSR) staining in CTL + ISO group (Fig. 1G, Supplementary Fig. S4B). The increase of cardiomyocyte size and cardiac fibrosis induced by ISO were significantly reduced in Snx3-cKO mice (Fig. 1D–G, Supplementary Fig. S4A–C). Data of echocardiography, such as ejection fraction (EF), fractional shortening (FS), cardiac output (CO), stroke volume (SV), left ventricular diameter (LVID), and left ventricular volume (LVV), were significantly reduced, while left ventricular mass (LVM), interventricular septum (IVS) and left ventricular posterior wall thickness (LVPW) were increased in ISO group, which was partially recovered by Snx3 knockout (Fig. 1I, Supplementary Fig. S4D–L). These results suggest that knockout of Snx3 significantly alleviated ISO-induced cardiac injury and heart dysfunction in mice.
Cardiac-specific Snx3 transgene led to HF in mice
To further determine whether SNX3 contributes to myocardial injury, cardiac-specific Snx3-cTg mice were generated by crossing Snx3 transgenic mice with B6.FVB-Tg(Myh6-Cre)2182Mds/J mice (Fig. 2A, Supplementary Fig. S5). This transgenic strain uses a Cre/loxP approach in which transgenic Cre expression is driven by the mouse Myh6 promoter [34]. Compared with littermate negative controls (N-Tg), the expression of SNX3 protein is robustly expressed in the heart of Snx3-cTg mice, as indicated by western blot, IF assay, and the luciferase activity by live imaging (Fig. 2B, Supplementary Fig. S5E–H).
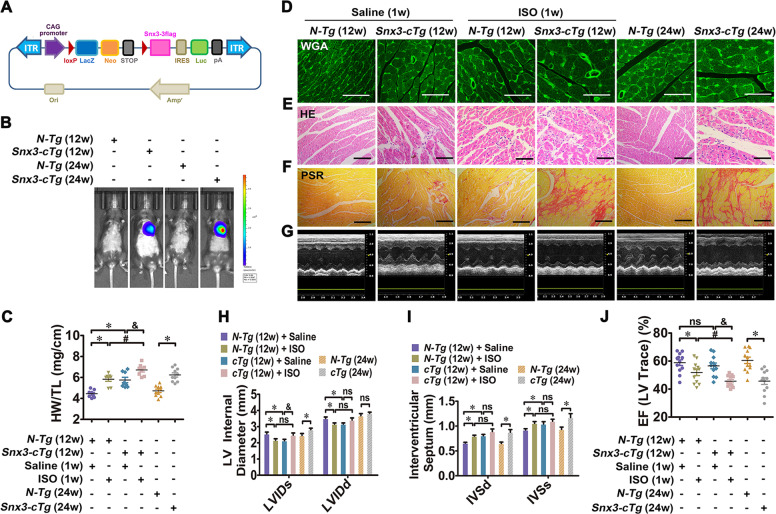
Fig. 2. Cardiac-specific Snx3 transgene led to myocardial injury in mice.
We investigated the changes in cardiac structure and function in Snx3-cTg mice at 12 and 24 weeks. Besides, Snx3-cTg mice (at 11 weeks) were injected with 3 mg/kg/d ISO (s.c.) or an equal volume of sterile normal saline for one week. A The scheme of Snx3-cTg mice construction strategy was shown. B The protein level of SNX3 was detected by live imaging. C The HW/TL ratio was calculated. D–F WGA staining (Scale bar: 50 μm), HE staining (Scale bar: 100 μm) and PSR staining (Scale bar: 100 μm)-stained cross-sections of heart tissues were shown. G The representative echocardiographic graphs were shown. H–J The echocardiographic parameters (LVID, IVS, and EF) were measured. Representative images of five independent experiments were presented. The data were shown as the means ± SEM. *P < 0.05 vs. N-Tg (12w) + Saline or N-Tg (24w) group, #P < 0.05 vs.. N-Tg (12w) + ISO group, &P < 0.05 vs. Snx3-cTg (12w) + Saline group. n = 12 per N-Tg (12w) + Saline group, n = 11 per N-Tg (12w) + ISO group, n = 12 per Snx3-cTg (12w) + Saline group, n = 12 per Snx3-cTg (12w) + ISO group, n = 10 per N-Tg (24w) group, n = 9 per Snx3-cTg (24w) group. EF ejection fraction, HE hematoxylin–eosin, HW/TL the heart weight to the tibia length ratio, ISO isoproterenol, IVS interventricular septum, LVID left ventricular diameter, ns no statistical difference, N-Tg non-transgenic, PSR picric sirius red, s.c. subcutaneously, Snx3-cTg cardiac-specific Snx3 transgenic mice, WGA wheat germ agglutinin, 1w 1 week, 12w 12 weeks, 24w 24 weeks. See also Supplementary Figs. S5–S7.
At 12 weeks of age, Snx3-cTg mice showed a mild degree of cardiac hypertrophy, as indicated by the following observations: (1) the larger heart/cardiac size by gross morphological examination, the increased hypertrophic biomarkers and HW/TL ratio, WGA staining, and HE staining (Fig. 2C–E, Supplementary Fig. S6A, B); (2) deposition of collagen protein by PSR staining (Fig. 2F); (3) echocardiography results (Fig. 2G) showed a decrease in LVID (Fig. 2H), CO (Supplementary Fig. S6C), SV (Supplementary Fig. S6D) and LVV (Supplementary Fig. S6F), as well as an increase in IVS (Fig. 2I), LVM (Supplementary Fig. S6E) and LVPW (Supplementary Fig. S6G). Overexpression of SNX3 did not apparently affect EF, FS, or heart rate (HR) in Snx3-cTg mice at 12 weeks (Fig. 2J, Supplementary Fig. S6H, I).
We also investigated whether overexpression of SNX3 aggravated ISO-induced cardiac hypertrophy in Snx3-cTg mice (12-weeks-old). There were signs of an increase in the cardiomyocyte size in both N-Tg mice and Snx3-cTg mice; however, Snx3-cTg mice have larger cardiomyocyte size than in N-Tg mice (Fig. 2C–E, Supplementary Fig. S6A, B). In addition, ISO-induced cardiac fibrosis was aggravated when SNX3 was overexpressed (Fig. 2F). After ISO infusion, severe heart dysfunction detected by echocardiography was noted in Snx3-cTg mice more than N-Tg mice (Fig. 2G–J, Supplementary Fig. S6C–I). These results suggest that Snx3-cTg mice were more susceptible to ISO-induced cardiac injury.
After 24 weeks, Snx3-cTg mice displayed significant hypertrophic growth of cardiomyocytes, increases in HW/TL ratios and hypertrophic biomarkers, myocardial fibrosis, compared with N-Tg mice (Fig. 2C–F, Supplementary Fig. S7A, B). According to the echocardiography (Fig. 2G), Snx3-cTg mice exhibited increases in LVID (Fig. 2H), IVS (Fig. 2I), LVM (Supplementary Fig. S7C), and LVV (Supplementary Fig. S7D), declines in EF (Fig. 2J), FS (Supplementary Fig. S7E), SV (Supplementary Fig. S7F) and CO (Supplementary Fig. S7G), implying that Snx3-cTg mice spontaneously developed severe heart dysfunction. These results suggest that the cardiac structure was disorganized and systole diastolic performances tend to be worsened with advancing age in Snx3-cTg mice.
SNX3-retromer directly interacted with STAT3 at EE
The prominent effects of SNX3 on cardiac hypertrophy and HF (Figs. 1 and 2, Supplementary Fig. S1–7) prompt us to investigate its underlying mechanisms. Since SNX3 is a key retromer-associated protein [5, 6], we thus examined whether retromer is involved in SNX3-mediated cardiac hypertrophy. By immunoprecipitation-based mass spectrometry (IP-MS), we identified STAT3, as a new interacting partner of SNX3, in addition to known interacting proteins, such as retromer proteins VPS26 and VPS35 (Supplementary Fig. S8A, B). To validate the interaction of SNX3 and STAT3, neonatal rat cardiomyocytes (NRCMs) were infected with Ad-SNX3 (Flag-tagged) or Ad-STAT3 (HA-tagged). Protein interaction was evaluated by IF staining and co-IP assays (Fig. 3A–C). Indeed, IF staining results indicate that HA-labeled STAT3 and retromer proteins (SNX3, VPS26, and VPS35) were located in the same cellular compartments (Fig. 3A). The co-IP results suggest that SNX3–retromer interacted with STAT3 in NRCMs and mouse heart tissues (Fig. 3B, C, Supplementary Fig. S8C–E). According to localized surface plasmon resonance (LSPR) results, recombinant STAT3 protein (117–770aa) directly interacted with SNX3, VPS35, and VPS26 with a binding constant at 7.10e−7, 1.58e−8, 1.30e−8 M (KD values), respectively (Supplementary Fig. S8F–L). The interaction between truncated STAT3 protein (576–678aa) and SNX3–retromer was shown in Fig. 3D with a binding constant at 6.11e–8, 5.06e–8, 1.01e−7 M (KD values) in a cell-free system. Moreover, the proximity ligation assay (PLA) was performed to visually detect the protein–protein interactions using confocal microscopy. As shown in Supplementary Fig. S8M, the binding of SNX3 and STAT3 was located in the cytoplasm of normal cultured cardiomyocytes. However, the SNX3/STAT3 complex was induced by ISO treatment and was substantially localized in the nucleus (Supplementary Fig. S8M).
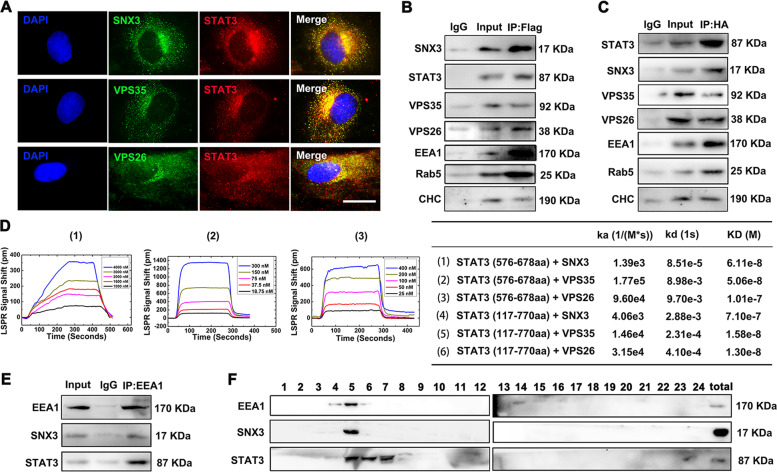
Fig. 3. SNX3–retromer directly interacted with STAT3 at early endosomes in vivo and in vitro.
A NRCMs were infected with Ad-STAT3 (HA-tagged) and were measured by IF staining using confocal microscopy (Scale bar: 25 μm). Representative images of five independent experiments were presented. NRCMs were infected with Ad-SNX3 (Flag-tagged) or Ad-STAT3 (HA-tagged) and were precipitated by anti-Flag (B) or anti-HA (C). D The binding curves of SNX3-retromer and truncated STAT3 protein (576-678aa) were measured by LSPR analysis, and the ka, kd, and KD values for STAT3 (576–678aa, 117–720aa), SNX3, VPS35, and VPS26 were calculated by TraceDrawer™. E NRCMs were precipitated by anti-EEA1 (a marker for early endosome) for SNX3 and STAT3 detection in co-IP assays. F The early endosome fraction was purified from NRCMs using density gradient centrifugation, and detected by western blot. n = 5. CHC clathrin heavy chain; co-IP co-immunoprecipitation, IP–MS immunoprecipitation-based mass spectrometry, LSPR localized surface plasmon resonance, NRCMs neonatal rat cardiomyocytes. See also Supplementary Figs. S8 and S9.
Besides, NRCMs were precipitated by anti-early endosome antigen 1 (EEA1), Rab5, and clathrin heavy chain (CHC) (markers of early endosome), SNX3 and STAT3 were found in the precipitation of EE by co-IP assays (Fig. 3E, Supplementary Fig. S9A and B). However, no significant binding of SNX3 and Rab7, a marker of late endosome (LE), was detected by co-IP assays (Supplementary Fig. S9C). This result is also consistent with the previous report that SNX3–retromer is predominantly localized on EE and has little presence on LE [5, 6]. The early endosome fraction was purified from NRCMs using density gradient centrifugation, and detected by western blot analysis (Fig. 3F). We observed that both STAT3 and SNX3 mainly associated with EE in NRCMs, though some STAT3 positive staining was presented in LE and lysosomes, as suggested by STAT3 combining with the endosomal compartment markers (EEA1, Rab5, Rab7, and CHC) and lysosome marker (Lamp-2) (Fig. 3B, C, E, F, Supplementary Fig. S9D, E). These results imply that STAT3 directly interacted with SNX3–retromer at the early endosome in cardiomyocytes.
SNX3–retromer served as a platform for STAT3 activation
It is well-established that JAK2/STAT3 pathway is involved in the pathogenesis of cardiac hypertrophy and HF [4]. To investigate whether SNX3 cooperates with STAT3 to mediate cardiac hypertrophy, we established three classical cardiac hypertrophic models (induced by ISO, angiotensin II (AngII), and phenylephrine (PE), respectively) in NRCMs (Fig. 4A, B). The phosphorylation levels of JAK2 (Y1007 and Y1008) and STAT3 (Y705), and the protein expression of SNX3 were enhanced in these hypertrophic models, whereas the protein levels of gp130 and STAT3 remained unchanged (Fig. 4C, Supplementary Fig. S10A). The homo-/hetero-dimerization of STAT3 and STAT1 was also increased by treatment with hypertrophy stimuli (Supplementary Fig. S10B).
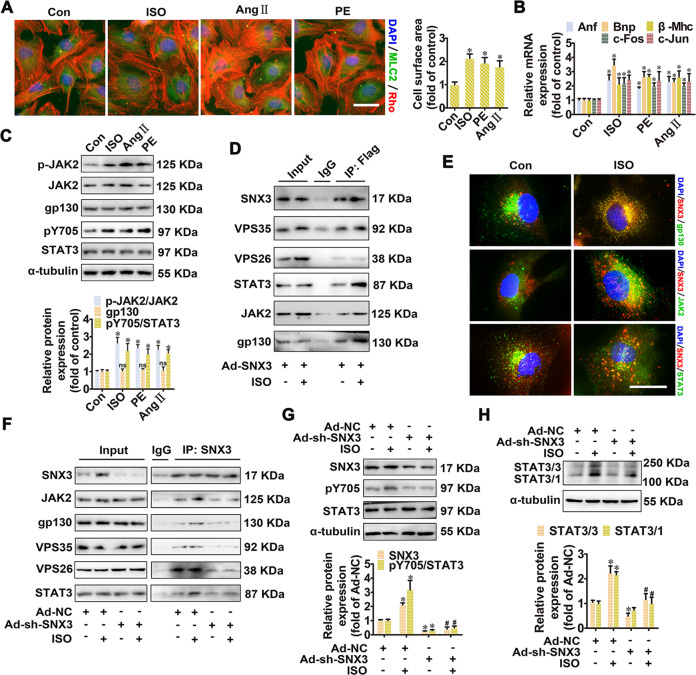
Fig. 4. SNX3–retromer acted as a platform for STAT3 activation in NRCMs.
NRCMs were treated with three hypertrophic stimulants, including ISO (10 μmol/L), Ang II (1 μmol/L), and PE (100 μmol/L), respectively, for the times indicated. A The cell surface area was measured by staining with anti-MLC2 antibody (green) and rhodamine-phalloidin (red) (Scale bar: 25 μm). B The mRNA levels of Anf, Bnp, β-Mhc, c-Fos, and c-Jun were determined by qPCR. C Western blot analysis was performed to detect the phosphorylated JAK2 (at tyrosine 1007 and 1008, p-JAK2) and STAT3 (at tyrosine 705, pY705), as well as the protein expression of JAK2, gp130 and STAT3. D NRCMs infected with Ad-SNX3 (Flag-tagged) were treated with ISO for 1 h and were precipitated by anti-Flag antibody, followed by co-IP assays. E The intracellular co-localization of SNX3 and gp130, JAK2, STAT3 in ISO-treated NRCMs was determined using confocal microscopy (Scale bar: 25 μm). NRCMs were infected with Ad-sh-SNX3 followed by incubation with ISO (10 μmol/L for 1 h), were precipitated by anti-SNX3 in co-IP assays (F), were detected the phosphorylated STAT3 (pY705), the protein expression of SNX3 and STAT3 (G), and were examined the protein expression of STAT3/3 homodimer or STAT3/1 heterodimer (H). Representative images of five independent experiments were presented. The data were shown as the means ± SEM. *P < 0.05 vs. control or Ad-NC group, #P < 0.05 vs.. Ad-NC + ISO group, n = 5. Ang II, angiotensin II; co-IP, co-immunoprecipitation, IF immunofluorescence, ISO isoproterenol, MLC2 myosin light chain 2, NC negative control, NRCMs neonatal rat cardiomyocytes, PE phenylephrine, qPCR quantitative polymerase chain reaction, 1h, 1 h. See also Supplementary Figs. S10 and S11A.
Given that SNX3 is localized in retromer-containing endosomes [6], we investigated whether SNX3–retromer is involved in JAK2/STAT3 signaling. The results of co-IP and IF assays showed that ISO increased the association of STAT3 with SNX3, JAK2, and gp130, whereas knockdown of SNX3 decreased these interactions (Fig. 4D–F, Supplementary Fig. S11A). Consistently, knockdown of SNX3 significantly inhibited ISO-induced phosphorylation and dimerization of STAT3 (Fig. 4G, H), whereas overexpression of SNX3 by infecting with Ad-SNX3 had the opposite effects (Supplementary Fig. S10C–E). Collectively, these results suggest that SNX3–retromer serves as a platform for the assembly of gp130/JAK2/STAT3 complexes and the subsequent phosphorylation of STAT3 in ISO-induced cardiac hypertrophy model in vitro.
SNX3–retromer promoted the nuclear localization of STAT3
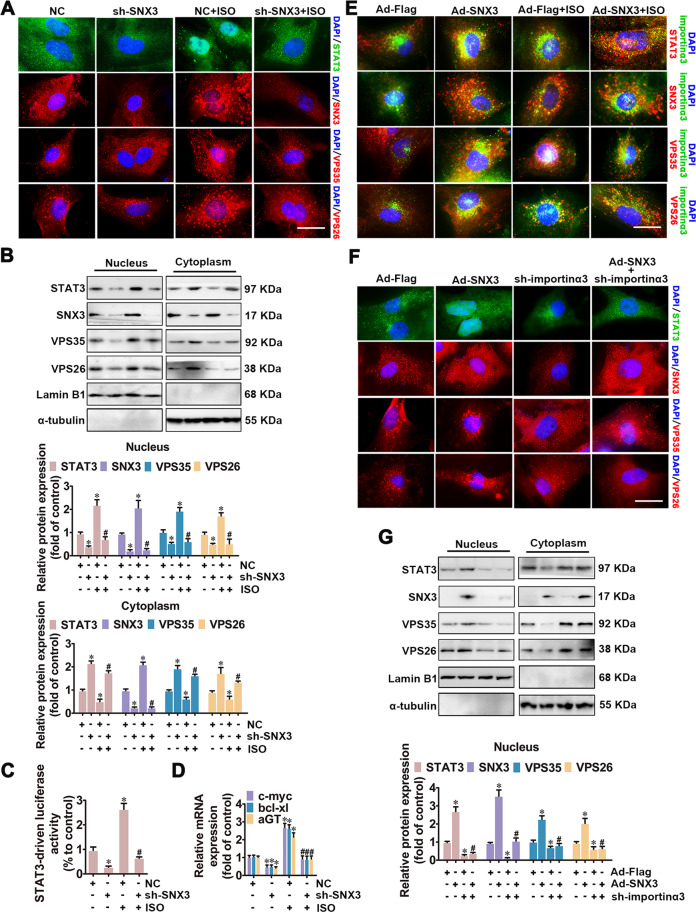
After treatment with stimuli of cardiac hypertrophy, cytoplasmic STAT3 shuttles to the nucleus (Supplementary Figs. S12A and S13), where STAT3 orchastreates hypertrophy-related target gene expression (such as Anf, c-fos, c-myc, aGT) [4, 35, 36]. Given that the SNX3–retromer complex plays a vital role in sorting cargoes from endosomes to the intracellular trafficking [6], we examined the effects of the SNX3–retromer on the subcellular distribution of STAT3 in the process of cardiac hypertrophy. NRCMs were treated with ISO, AngII, or PE, or were infected with Ad-SNX3 or Ad-sh-SNX3 before ISO treatment. The nuclear and cytoplasmic proteins were extracted from NRCMs, and were detected by western blot analysis. The results showed that transient nuclear import of STAT3, SNX3, VPS35, and VPS26, after treatment with hypertrophic stimuli for 1 h (Supplementary Fig. S12A). By contrast, SNX3 silencing reversed ISO-induced nuclear import of STAT3 using IF and western blot analysis (Fig. 5A, B). Similarly, the ISO-induced increase of transcriptional activity and target gene expression of STAT3 was inhibited by SNX3 knockdown in vitro (Fig. 5C, D) or by SNX3 knockout in vivo (Supplementary Fig. S14). JAK2 knockdown reversed overexpressed SNX3-induced nuclear import of STAT3 (Supplementary Fig. S11C). Knockdown of retromer components VPS35 attenuated the effects of SNX3 overexpression on STAT3 nuclear localization (Supplementary Fig. S15B). These results suggest that JAK2 was involved in SNX3–retromer-mediated STAT3 nuclear localization.
Fig. 5. SNX3–retromer promoted the nuclear localization of STAT3 in NRCMs.
NRCMs were infected with Ad-sh-SNX3 or Ad-SNX3 before ISO treatment for 1 h. Besides, another group of NRCMs was infected with Ad-SNX3 followed by transfection with shRNAs of importin α3. A, E, and F IF assay was performed to detect the subcellular distribution of STAT3, SNX3, VPS35, and VPS26. Representative images of five independent experiments were presented. B and G The nuclear and cytoplasmic proteins were extracted from NRCMs, and were detected by western blot analysis. The results were normalized to those of α-tubulin/Lamin B1. C Luciferase reporter gene assays showed the transcriptional activity of STAT3. D The mRNA levels of the target genes of STAT3 (c-myc, bcl-xl, and aGT) were confirmed by qPCR. The data were shown as the means ± SEM. *P < 0.05 vs. NC or Ad-Flag group, #P < 0.05 vs.. Ad-sh-SNX3 or Ad-SNX3 group, n = 5. IF immunofluorescence, ISO isoproterenol, NC negative control, NRCMs neonatal rat cardiomyocytes, qPCR quantitative polymerase chain reaction. See also Supplementary Figs. S11–S16.
Previous studies have shown that importin α3, a nuclear importing factor, is responsible for STAT3 nuclear import induced by hormones or intracellular tyrosine kinases [21, 22]. The endogenous importin α3 was knocked down using the appropriate shRNA (Supplementary Fig. S12B, C). By IF staining, we observed that Ad-SNX3 treatment significantly increased ISO-induced interaction of importin α3 and STAT3–SNX3-retromer complexes (Fig. 5E). However, importin α3 knockdown attenuated Ad-SNX3-triggered the nuclear importing of STAT3 and SNX3-retromer (Fig. 5F, G, Supplementary Fig. S12D), suggesting that SNX3-retromer promoted the importin α3-mediated nuclear localization of STAT3 in cardiomyocytes.
Involvement of STAT3 in SNX3-mediated cardiomyocyte hypertrophy
We then asked whether SNX3 induces cardiomyocyte hypertrophy through STAT3 activation. Knockdown of SNX3 significantly suppressed the hypertrophic growth of cardiomyocytes induced by ISO, which was inhibited by STAT3 overexpression (Fig. 6A, B, Supplementary Fig. S17C and D). Conversely, Ad-SNX3 led to increased hypertrophic responses, which were partly inhibited by stattic (an inhibitor of STAT3) or STAT3 knockdown, implying that STAT3 was involved in SNX3-mediated cardiomyocyte hypertrophy (Fig. 6A, B, Supplementary Fig. S17C, D). Knockdown of retromer components VPS35 significantly suppressed the hypertrophic responses induced by Ad-SNX3 (Supplementary Fig. S15C, D). Similarly, SNX3 overexpression led to increased hypertrophic responses, which were obviously inhibited by JAK2 knockdown (Supplementary Fig. S11D, E). These data pinpoint that JAK2/STAT3 was involved in SNX3-retromer-mediated cardiac hypertrophy.
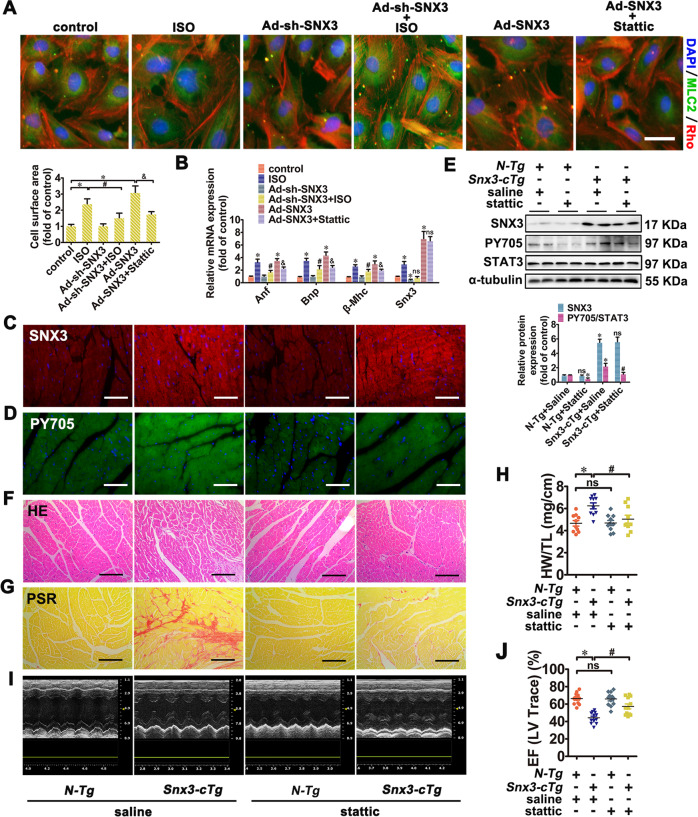
Fig. 6. STAT3 was involved in SNX3-mediated cardiomyocyte hypertrophy in NRCMs and in Snx3-cTg mice.
NRCMs were divided into four groups: (1) infected with Ad-sh-SNX3 before ISO treatment; (2) infected with Ad-SNX3 with or without stattic (5 μmol/L, 24 h); (3) transfected with si-STAT3 before infection of Ad-SNX3; (4) infected with Ad-STAT3 before transfection of Ad-sh-SNX3 in ISO-treated NRCMs. A The cell surface area was measured by staining with anti-MLC2 (green) and rhodamine-phalloidin (red) (Scale bar: 25 μm). B The mRNA expression of Anf, Bnp, β-Mhc, and Snx3 were measured by qPCR. Additionally, male Snx3-cTg mice (10-weeks-old) were administrated with stattic (40 mg/kg/d, i.p.) or an equal volume of vehicle for 2 weeks. C–E The protein expression of SNX3 and the phosphorylated STAT3 (pY705) were measured by IF (Scale bar: 100 μm) and western blot analysis. F and G HE staining (Scale bar: 100 μm) and PSR staining (Scale bar: 100 μm)-stained cross-sections of heart tissues were shown. H The HW/TL ratio was calculated. I The representative echocardiographic graphs was presented. J The echocardiographic parameter EF was measured. Representative images of five independent experiments were presented. The data were presented as the means ± SEM. *P < 0.05 vs. control, Ad-Flag, Ad-NC or N-Tg + Saline, group, #P < 0.05 vs.. ISO, Ad-SNX3 or Snx3-cTg + Saline group, &P < 0.05 vs. Ad-sh-SNX3 or Ad-SNX3 group, n = 5 for cell test. n = 12 per N-Tg (12w) + Saline group, n = 11 per Snx3-cTg (12w) + Saline group, n = 12 per N-Tg (12w) + Stattic group, n = 12 per Snx3-cTg (12w) + Stattic group. EF ejection fraction, HE hematoxylin–eosin, HW/TL the heart weight to the tibia length ratio, IF immunofluorescence, i.p. intraperitoneally, ISO isoproterenol, MLC2 myosin light chain 2, NC negative control, NRCMs neonatal rat cardiomyocytes, ns no statistical difference, N-Tg non-transgenic, PSR picric sirius red, qPCR quantitative polymerase chain reaction, Snx3-cTg cardiac-specific Snx3 transgenic, 1h 1 h. See also Supplementary Figs. S11, S15, S17–S19.
To further confirm the role of STAT3 in SNX3-induced cardiac injury in vivo, Snx3-cTg mice were treated with stattic (40 mg/kg/d, i.p.) or an equal volume of vehicle for 2 weeks. The phosphorylation level of STAT3 at the tyrosine 705 (pY705) was significantly inhibited by stattic, suggesting that the activation STAT3 was inhibited by stattic (Fig. 6D, E). After treatment with stattic, N-Tg mice showed no noticeable changes in cardiomyocytes (Fig. 6F–J, Supplementary Fig. S18A–M). However, the degree of cardiac hypertrophy (determined by gross observation of heart morphology, hypertrophic biomarkers, HW/TL ratio, WGA staining, and HE staining) of Snx3-cTg mice was significantly alleviated in stattic treatment group compared with that in the vehicle-treated group (Fig. 6F, H, Supplementary Fig. S18A, B, D). Besides, Snx3-cTg mice showed increased cardiomyocyte interstitial fibrosis (determined by PSR staining and masson staining), while that effect was widely suppressed upon stattic treatment (Fig. 6G, Supplementary Fig. S18C). The echocardiography data showed that Snx3-cTg reduced EF, FS, CO, LVV, LVID, and SV, increased LVPW, LVM, and IVS, which could be antagonized by stattic in different degrees (Fig. 6J, Supplementary Fig. S18E–L).
These results suggest that STAT3 was involved in SNX3 overexpression-induced cardiomyocyte hypertrophy in vivo and in vitro.
Discussion
SNX3 plays a crucial role in the pathogenesis of diverse diseases, including Parkinson’s disease [17], Alzheimer’s disease [37], and autosomal dominant polycystic kidney disease [15]. However, it remains unknown whether SNX3 plays a role in cardiac diseases. In this study, we observed that SNX3 expression was increased in end-stage failing human hearts and cardiac tissues from a mouse models of ISO-induced cardiac injury. In addition to ISO, two other classical neurohumoral stimuli (AngII and PE) can also increase the mRNA and protein levels of SNX3 in vitro. These findings indicate that SNX3 is a common mediator of neurohumoral stimulation-induced pro-hypertrophic signaling.
By using gain-of-function and loss-of-function experiments in vivo, we revealed a novel role of SNX3 in the development of cardiac hypertrophy and HF. This was evidenced by the fact that cardiac-specific Snx3-cKO protected mice against ISO-induced cardiac hypertrophy, conversely, Snx3-cTg mice were hypersensitive to ISO-induced cardiac hypertrophy and aggravated cardiac injury with advancing age.
It has been reported that the SNXs family affects a wide range of biological processes by regulating the intracellular trafficking of diverse signaling proteins [10–13, 38]. For instance, deficiency of SNX10 prevents inflammation and bone erosion in rheumatoid mouse arthritis through promoting NFATc1 degradation; [10] SNX13 profoundly affects cardiac performance through the SNX13–PXA–ARC–caspase signaling pathway; [11] Deletion of SNX27 reverses epithelial–mesenchymal transition in highly aggressive breast cancer cells [13]; SNX27 serves as an essential adaptor protein, mediates beta 2ARs to the retromer tubule and endosome-to-PM trafficking [38]. However, the effect of SNX3 on the intracellular transportation of cargo proteins, which is tightly associated with cardiovascular diseases, has not been evaluated yet. Considering that SNX3-retromer was found in the nucleus of hypertrophic cardiomyocytes in this study, IP–MS was performed to identify the possible nuclear proteins that might bind to SNX3. According to our results, we mainly focus on the specific role of SNX3–STAT3 interaction in cardiomyocytes. SNX3 was required for STAT3 activation, inhibition of STAT3 could reduce the detrimental role of overexpressed SNX3 in cardiomyocytes and Snx3-cTg mice. Of course, in addition to STAT3, there must be other target proteins of SNX3-retromer, which is also the focus of our future work.
It was previously believed that retromer (VPS26–VPS35–VPS29 heterotrimers) act as the cargo-sorting complex [39, 40]. Recent studies have shown that SNX proteins are indispensable for cargo recognition, select, and binding [6, 41]. A T-shaped architecture of SNX3-retromer was identified, and this complex comprised SNX3, VPS26, and VPS35 [6]. SNX3–retromer complex play the central role in recycling various proteins (such as Wntless, transferrin receptor, and Fet3-Ftr1) from the endosomes to TGN or PM [14, 17, 37, 42–46]. SNX3–retromer mediate Wntless sorting and Wnt secretion [14, 42–44], which mediates cellular crosstalk to regulate glucose metabolism and cardiac homeostasis [47, 48]. SNX3 regulates the recycling of transferrin receptors and iron assimilation [37], which regulates cellular iron homeostasis in cardiomyocytes [49]. SNX3–retromer mediate retrograde recycling of iron transporters in S. cerevisiae and C. elegans models of Parkinson’s disease [17, 37]. In iron-starved cells, Fet3-Ftr1 is sorted by SNX3/Grd19 and retromer into a recycling pathway that delivers it back to the PM [45, 46]. In this study, we identified STAT3 as a new interacting partner of the SNX3–retromer complex mainly in EE in cardiomyocytes.
STAT3 activity is tightly regulated for proper physiological processes, and its aberrant and persistent activation will result in pathological cardiac hypertrophy [4, 18]. Over-activation of STAT3 could be mainly attributed to either the stimulation of STAT3 activators (such as ISO and IL-6) or the aberrantly activated upstream tyrosine kinases (such as JAK2 and EGFR) [18–20]. The present work suggests that SNX3–retromer acts as an essential platform for the assembly of gp130/JAK2/STAT3 complexes induced by ISO, and subsequent phosphorylation of STAT3 by direct combining at EE.
To date, the main research of SNXs-mediated intracellular transportation is focused on the subcellular trafficking from the endosomes to TGN or PM [6]. It is rarely reported that the nuclear transport of intracellular proteins mediated by SNXs, except that SNX11 is required for the translocation of factor II receptor-like 1 (F2rl1) from the PM to the cell nucleus in retinal ganglion cells [7]. We report that the SNX3–retromer complex was transiently imported into the nucleus after hypertrophic stimuli, and promoted importin α3-dependent nuclear translocation of STAT3 in the cardiac hypertrophy model.
Considering that isolated cardiomyocytes in vitro could mainly simulate compensated cardiac hypertrophy, but could not simulate the stage of decompensated cardiac hypertrophy or HF, this may be the reason why the change of subcellular localization of SNX3 was not very consistent in vivo and in vitro. We speculate that SNX3 protein is increased in both cytoplasm and nucleus with the continuous stimulation of cardiac hypertrophy stimulant, the nuclear and cytoplasmic SNX3 proteins collaborate to play a role in the process of HF.
According to our work, it is plausible that SNX3–retromer promotes both STAT3 activation and nuclear translocation. The detailed mechanism whereby the complex is recruited to membrane receptor to be activated and subsequently directed to nuclear after activation is not clear yet. There may be another protein or posttranslational modification that determines the fate of the complex for intracellular trafficking.
In conclusion, the present study demonstrates that SNX3–retromer plays a critical role in cardiac function and suggests that SNX3 could be exploited as a potential new therapeutic target for cardiac hypertrophy and HF.
Materials and methods
Human heart samples
The study conforms to the principles that govern the use of human tissues outlined in the Declaration of Helsinki. Approval was obtained from the human ethics committee of First Affiliated Hospital of Sun Yat-sen University. All patients or the family of prospective heart donors gave written informed consent prior to participation. Failing human heart samples were collected from 9 patients undergoing heart transplantation because of end-stage HF (Table S1). Three non-failing control heart samples were obtained from prospective multi-organ donors, which did not exhibit cardiovascular pathology but were unable to be transplanted due to technical reasons. Tissue samples were collected at the time of explantation and rapidly frozen in liquid nitrogen or fixed with 4% paraformaldehyde.
Animal studies
Snx3-floxed mice were constructed in the Shanghai Model Organisms Center by CRISPR/Cas9 technology. Briefly, the donor vector containing four guide RNAs and Cas9 mRNA targeting Snx3 introns 2 and 3 (Table S2) was microinjected into C57BL/6 mouse fertilized eggs. The positive founder mice were backcrossed with wild-type C57BL/6 mice to obtain heterozygous Snx3flox/+ mice with germline transmission. Snx3flox/+ mice were self-crossed to generate homozygous Snx3flox/flox mice. Snx3flox/flox mice were crossed with C57BL/6J-Myh6em1(IRES-Cre)Smoc mice (Shanghai Model Organisms Center, Stock No. NM-KI-00083. MGI ID: 97255) to generate Myh6-Cre+; Snx3flox/flox mice, i.e. cardiac-specific Snx3-cKO mice.
Snx3 transgenic mice were constructed in the Shanghai Model Organisms Center using standard methods. Briefly, the Piggybac vector harboring mouse SNX3 cDNA was microinjected into the fertilized egg of the C57BL/6 mouse, and the transgenic founder mice were obtained. Three generations of a backcross between each Snx3 transgenic mice and wild-type (C57BL/6) mice were adopted to breed two independent Snx3 transgenic lines. The two transgenic lines were, respectively, crossed with B6.FVB-Tg(Myh6-Cre)2182Mds/J mice (Jackson Laboratory (Bar Harbor, ME), Stock No. 011038. MGI ID: 2386742) to generate cardiac-specific overexpressed Snx3 (Snx3-cTg) mice.
The mice were genotyped by PCR and further confirmed by western blot analysis (Tables S3–S5). All animal protocols were conducted under the institutional guidelines of the Animal Care and Use Committee and were approved by the Research Ethics Committee, Sun Yat-sen University. Experimental animals were housed, bred, and maintained in the specific pathogen-free (SPF) facility of the Experimental Animal Center of Sun Yat-sen University. As we previously described [30], the transthoracic 2D-guided M-mode echocardiography (such as heart function and global cardiac volumes) was assessed using a Technos MPX ultrasound system (ESAOTE, SpAESAOTE SpA, Italy) equipped with a 40-MHz scan probe by an investigator who was blinded to the specific group assignment. The Vevo 2100 imaging software was used for measurements and calculations. Then, the treated mice were sacrificed, and hearts were rapidly sampled for further experiments.
Bioluminescence imaging
XenoLightTM d-luciferin potassium salt (PerkinElmer, P/N 122799) was diluted in sterile PBS to 15 mg/mL and was, respectively, injected into the luciferase-labeled Snx3-cTg mice or N-Tg mice (150 mg/kg). A few minutes after injection, mice were anesthetized using 2% isoflurane inhalation (with a 2 L/min oxygen flow rate), and were placed inside the camera box of the IVIS Lumina XR small animal optical imaging system (PerkinElmer). The sequential images of the mice were run every 2 min. Lumazone Version 2.0 software was used to analyze the intensity of the fluorescence (intensity/s) in mice [50].
Histological analysis
Myocardial tissue samples were fixed in 10% paraformaldehyde and embedded in paraffin for sectioning. Sections were stained with WGA, HE, Masson’s, and PSR for histopathological examination under a light microscope.
For immunofluorescent (IF), paraffin sections of myocardial samples were treated with primary anti-SNX3 (Proteintech, #10772-1-AP) or anti-p-STAT3 (Y705, Cell Signaling Technology, CST, Beverly, MA, USA, #9145) overnight at 4 °C. Then, the samples were incubated with CoraLite488/594-conjugated anti-rabbit IgG (diluted 1:200, Proteintech, #SA00013-6 or #SA00013-8), were counter-stained with 4′,6-diamidino-2-phenylindole (DAPI, CST, #4083). Fluorescence was captured by EVOS FL Auto (Life Technologies, Bothell, WA, USA).
Plasmid, recombinant adenoviral vectors, and recombinant protein
Vps26 (NM_001007740.1), Vps35 (NM_001105718.2), Snx3 (NM_001044283.1), and Stat3 (NM_012747.2) were constructed by ligating respective full-length cDNA into pcDNA3.1 (+) with Flag or HA-tag. Similarly, short hairpin (sh) RNA targeting the rat’s importin α3 gene (also called as Kpna3, NM_001014792) (sh-importin α3), the rat’s Snx3 gene (sh-SNX3), and non-targeting control shRNA (sh-NC) sequences (Tables S6 and S7) were, respectively, subcloned into pcDNA3.1 (+) with Flag-tag.
Recombinant adenoviral vectors expressing rats Snx3 cDNA (Ad-SNX3, with Flag-tag), rats STAT3 cDNA (Ad-STAT3, with HA-tag), sh-SNX3 sequence (Ad-sh-SNX3, with Flag-tag), and control vectors were generated by standard procedures. Briefly, the pAdTrack plasmids containing corresponding genes were linearized by using restriction endonuclease PmeI, and were transformed into Escherichia coli strain BJ5183 cells carrying the Ad-Easy-1 plasmid. The successful recombinant plasmids were digested with restriction endonuclease PacI, and were transfected into HEK293A cells to generate Ad-SNX3, Ad-STAT3, or Ad-sh-SNX3. The vectors were purified by plaque, cultured on a large scale, and purified by CsCl step-gradient and isopycnic-gradient centrifugation.
GST-STAT3, GST-SNX3, GST-VPS35, and GST-VPS26 protein (pGEX-4T-1 vector) were, respectively, expressed in E. coli BL21 (DE3) maintained in Luria-Bertani (LB) medium (including 50 μg/mL ampicillin) at 37 °C. By the addition of isopropyl-β-d-thiogalactopyranoside (IPTG, 0.5 mmol/L), protein expression was induced at 16 °C overnight to an OD600 value of 0.8. Cells were re-suspended in 20 mmol/L Tris + 200 mmol/L NaCl + 1 mmol/L PMSF, were broken using ultrasonic instrument in ice, and were collected supernatant after centrifugation (12,000 rpm for 3 h at 4 °C). These recombinant protein were purified by GST-Sefinose Gravity Column, were enzyme digested at room temperature overnight. All purified protein were dissolved in sterile phosphate-buffered saline (PBS), their concentration were measured and stored at −80 °C.
LSPR assays
To examine the direct interaction between STAT3 and SNX3-retromer, LSPR assays were conducted on an OpenSPR system (Nicoya Lifesciences, Waterloo, Canada). Recombinant proteins STAT3 (117-770aa, 576-678aa), SNX3, and VPS35 served as the ligand and were, respectively, immobilized on a gold nanoparticle sensor chip via capture-coupling. Subsequently, the recombinant protein SNX3, VPS35, and VPS26 at different concentrations were sequentially injected into the chamber in running buffer (filtered PBS) with a constant flow rate of 20 μL/min, and were passed over the sensor (about 5 min) for the association of two protein. Following each recombinant protein injection (all concentrations were performed in triplicate), the chip was completely dissociated with the complex and regenerated by injecting hydrochloric acid (pH 2.0). As recommended by the manufacturer, the results were analyzed by Trace Drawer software (Ridgeview Instruments AB). The kinetic parameters, including the association constant (ka), dissociation constant (kd) and affinity (KD, KD = kd/ka), were calculated by a simple 1:1 dilution corrected model, which adjusted to the wavelength shifts consisting with the varied concentration of protein [51].
Proximity ligation assay
NRCMs cultured in coverslips were infected with Ad-SNX3 (Flag-tagged) and Ad-STAT3 (HA-tagged) before ISO (10 μM) treatment for 1 h. After treatment, cells were washed with filtered PBS for three times, fixed with 4% paraformaldehyde for 10 min, and permeabilized with 0.3% Triton X-100 for 5 min. After washing with filtered PBS, add blocking solution to each sample for 1 h at room temperature. NRCMs were stained with anti-Flag rabbit antibody and anti-HA mouse antibody in a humidity chamber overnight at 4 °C. Then, the PLA was performed according to the manufacturer instructions (Duolink™ In Situ Red Starter Kit Mouse/Rabbit (red), catalog #DUO92101, Sigma-Aldrich). The cells were incubated with the PLA probe solution (1 h, 37 °C), and were incubated with ligation–ligase solution (30 min, 37 °C) in a pre-heated humidity chamber. After washing, coverslips were incubated with the amplification reaction mixture (100 min, 37 °C) in a pre-heated humidity chamber, washed, and counterstained with DAPI (blue). The images were observed by a confocal microscope (Zeiss, Germany) and analyzed by the Axiovision software (Zeiss). Each red dot represents the detection of protein–protein interactions (the distance between the two proteins is <40 nm) [52].
Primary culture of NRCMs
As reported before [30], NRCMs were isolated from the hearts of one to 3-day-old SD rats. Cardiomyocytes were plated into six-well microplates (Corning, USA) comprising Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) supplemented with 10% newborn calf serum (NBCS) and 5-bromodeoxyuridine (Sigma, #B5002, 0.1 mmol/L), at a density of 1 × 106 cells/well.
Plasmid transfection and virus infections
NRCMs were transfected with sh-SNX3 plasmid (Table S6), sh-importin α3 (Table S7), si-STAT3 (Table S8), si-VPS35 (Table S9), or si-JAK2 (Table S10) together with lipofectamine 3000 reagent (Invitrogen, USA) in OptiMEM medium as per the manufacturer’s instructions. The medium was changed to DMEM complete medium after 8–72 h of transfection. NRCMs were infected with recombinant adenoviruses (including Flag-tagged SNX3, Flag-tagged sh-SNX3, or HA-tagged STAT3) at a multiplicity of infection (MOI) of 20 particles per 5 cells. Western blot and/or Quantitative RT-PCR were performed to confirm the efficiency of overexpression or depletion [53].
Measurement of the cell surface area
NRCMs were seeded in 24-well microplates, fixed with paraformaldehyde (4%, Beyotime, #P0099) for 10 min at room temperature. After permeabilizing with Triton X-100 (0.3%, Beyotime, #P0096) and blocking with goat serum (Beyotime, #C0265), the cells were incubated with primary antibody myosin light chain 2 (MLC2, Proteintech, #10906-1-AP) overnight at 4 °C, treated with secondary antibody anti-alexa fluor 488 (Proteintech, #SA00013-6, for 2 h) and rhodamine-phalloidin (0.1%, Invitrogen #R415, for 30 min) at room temperature. After washing with filtered PBS, NRCMs were mounted by using DAPI (CST, #4083). The images were taken using the High Content Screening System (Thermo Fisher Scientific, USA), and the cell surface area from randomly selected fields (50 for each group) was analyzed by the built-in image analysis software [30, 53, 54].
Isolation of EE by continuous density gradient centrifugation
The nuclear and cytoplasmic protein was successively extracted from NRCMs using a kit (SC-003, Inventbiotech, MN, USA). The postnuclear fraction was suspended in buffer, and was used as a continuous sucrose density gradient. After centrifugation (210,000×g for 3 h at 4 °C), a milky band should be visible at each interface. 24 consecutive fractions were collected from each interface into tubes, and were subjected to western blot analysis for detection of protein EEA1, which is an early endosomal marker [55, 56].
Low temperature SDS–PAGE, Western blot and co-immunoprecipitation (co-IP) analysis
Low-temperature SDS–PAGE was conducted to investigate the homodimers (STAT3/STAT3) or heterodimer (STAT3/STAT1). In short, total protein from NRCMs were incubated in loading buffer (without 2-mercaptoethanol) for 5 min at 37 °C, were separated by 8% SDS–PAGE for 4–5 h at a constant current of 40 mA, and were transferred to a PVDF membranes (Millipore). The whole process of the experiment must be made under the low temperature. After blocking with 10% blocking buffer (Beyotime, #P0023B), the membranes were incubated with individual antibodies at 4 °C overnight [57].
Western blot analysis was performed as previously reported [53, 54]. Immunoblots were labeled with the following primary antibodies: primary antibodies against p-JAK2 (Y1007 and Y1008, rabbit, diluted 1:500, #3776), JAK2 (rabbit, diluted 1:1000, #3230), gp130 (rabbit, diluted 1:1000, #3732), p-STAT3 (Y705, rabbit, diluted 1:1000, #9145), STAT3 (rabbit, diluted 1:2000, #9139) were bought form CST. Primary antibodies against CHC (rabbit, diluted 1:1000), EEA1 (rabbit, diluted 1:1,000) and Rab5 (rabbit, diluted 1:1000) were purchased from CST (endosomal marker antibody sampler kit, #12666). SNX3 (rabbit, diluted 1:1000, #10772-1-AP), VPS26 (rabbit, diluted 1:800, #15915-1-AP) and VPS35 (rabbit, diluted 1:800, #10236-1-AP) were products of Proteintech. Primary antibodies against importin α3 (rabbit, diluted 1:1000, #I9783), HA (rabbit, diluted 1:5000, #H6908), and α-tubulin (mouse, diluted 1:5000, #T8203) were purchased from Sigma-Aldrich. Anti-Flag (mouse, diluted 1:5000, #PM185), anti-Lamin B1 (rabbit, diluted 1:1000, # PM064) were purchased from MBL. The enzyme horseradish peroxidase (HRP)-conjugated secondary antibodies (CST, #7074 and #7076) were applied to chemiluminescence detection and the protein band intensities were quantified by LabWorks software (Bio-Rad, USA).
For co-IP, anti-EEA1 (rabbit, diluted 1:50), anti-CHC (rabbit, diluted 1:50), anti-Rab5 (rabbit, diluted 1:20) were purchased from CST (endosomal marker antibody sampler kit, #12666), anti-VPS26 (rabbit, diluted 1:20, #15915-1-AP), and anti-VPS35 (rabbit, diluted 1:20, #10236-1-AP) were products of Proteintech. Anti-Flag (rabbit, diluted 1:50, #PM185) was purchased from MBL. Anti-HA (rabbit, diluted 1:50, #H6908) was purchased from Sigma-Aldrich. The rabbit normal IgG (#3900) and mouse normal IgG (#53484) were purchased from CST. NRCMs were harvested with IP lysis buffer (Beyotime, # P0013) supplemented with protease and phosphatase inhibitor cocktails (Bimake, #B14012 and #B15002). After clarification by centrifugation, 400–600 μg of total protein cell lysate were incubated with the indicated primary antibodies overnight at 4 °C, and were incubated with protein G-agarose beads (Pierce, Rockford, IL, USA) at 4 °C for 4 h. Normal IgG was served as an control. The immunoprecipitated proteins were detected by western blot.
IF assay
NRCMs were cultured in chamber slides (ThermoFisher Scientific). After treatment, cells were washed with filtered PBS for three times, fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.3% Triton X-100 for 5 min and followed by blocking with goat serum for 1 h at room temperature. The cells were further treated with the following primary antibodies overnight at 4 °C: STAT3 (CST, #9139), SNX3 (Proteintech, #10772-1-AP), VPS35 (Proteintech, #10236-1-AP), VPS26 (Proteintech, #15915-1-AP), EEA1 (CST, #12666), Rab5 (CST, #12666), Rab7 (CST, #12666), CHC (CST, #12666), Lamp-2 (Proteintech, #66301-1-Ig), gp130 (CST, #3732), JAK2 (CST, #3230), and importin α3 (Sigma-Aldrich, #I9783). Fluorescence emitted by fluorescence-conjugated secondary antibodies (Proteintech, #SA00013-6 and #SA00013-8) at room temperature for 2 h. The slides were mounted with DAPI (CST, #4083) and were observed by a confocal microscope (Zeiss, Germany) or EVOS FL Auto (Life Technologies).
Total RNA isolation, cDNA synthesis, and real-time polymerase chain reaction (qPCR)
Total RNA was extracted from snap-frozen cardiac tissues or NRCMs by using Trizol reagent (Invitrogen, #15596026), and its concentration was measured with a Nanodrop 2000 (Thermo Fisher Scientific, USA). The RNA extract (1000 ng) was reversely transcribed to first strand cDNA using the One-step Reverse Transcription (RT) Kit (Thermo Fisher Scientific, USA). Quantitative SYBR Green-based PCR (TOYOBO, Janpan) was conducted on the iCycler iQ system (Bio-Rad, USA). GAPDH was used as reference gene. All PCR assays were performed in triplicate. Data were analyzed using the 2−ΔΔCT method. The oligonucleotide sequences are synthesized by Sangon (Shanghai, China), and listed in Table S11.
Dual-luciferase reporter gene assay
The conserved DNA-binding sequence of rats STAT3 (TTCCGGGAA) were subcloned into pGL3 Basic plasmid (Promega, #E1751) [58]. NRCMs were seeded at 5 × 104 cells per well into 96-well microplates, and were transiently co-transfected with the luciferase reporter (100 ng/well) and pRL-TK reporter constructs (Promega, E2241) at 20 ng/well. The total content of transfected DNA was normalized by empty vector. After the indicated treatments, the luciferase activity was determined by the dual-luciferase reporter assay system (Promega, #E1980) on a microplate reader (TECAN Infinite M1000).
Statistical analysis
Graph Pad Prism 6.0 (Graph Pad software) or SPSS Version 21 was used for statistical analysis. Normality of the obtained data was assessed using a Shapiro–Wilk test. When normality was confirmed, statistical differences among groups were analyzed using either Student’s t test (for two groups) or one (or two)-way analysis of variance (ANOVA, for more than two groups). Otherwise, the non-parametric test Kruskal–Wallis test followed by the Dunn’s post-hot test was used to correct for multiple comparisons. The Levene or Brown–Forsynth test was used to compare the variance between the two groups. In all cases, differences were considered statistically significant at a P value (two-sided) < 0.05.
Supplementary information
Acknowledgements
We thank all patients who participated in this study for their cooperation. We also thank Jiantao Ye, Min Li, and Zhiping Liu for their excellent technical assistance.
Author contributions
JL and PQL conceived the project. JL, ZKW, YHH, JJW, and XLZ designed and performed majority of the experiments and data analyses. SWX, YQH, and PQL provided scientific advice. DPS, PXW, ZML, and MYL performed several in vitro experiments. JL, ZKW, and PQL wrote the manuscript. JL, SWX, YQH, and DPS. critically revised this paper. All authors critically evaluated the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81803521, 81872860, 82003710, 82070464), Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2017BT01Y093), National Major Special Projects for the Creation and Manufacture of New Drugs (2019ZX09301104), National Engineering and Technology Research Center for New drug Druggability Evaluation (Seed Program of Guangdong Province, 2017B090903004), Special Program for Applied Science and Technology of Guangdong Province (2015B020232009), Natural Science Foundation of Guangdong Province (2019A1515010273, 2021B1515020100), and Fundamental Research Funds for the Central Universities (19ykpy131).
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Ethics approval
All the sample collection and experimental protocols were approved by the Research Ethics Committee of Sun Yat-sen University.
Footnotes
Edited by R. Kitsis
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhongkai Wu, Email: wuzhk@mail.sysu.edu.cn.
Peiqing Liu, Email: liupq@mail.sysu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00789-w.
References
- 1.Brown DA, Perry JB, Allen ME, Sabbah HN, Stauffer BL, Shaikh SR, et al. Mitochondrial function as a therapeutic target in heart failure. Nat Rev Cardiol. 2017;14:238–50. doi: 10.1038/nrcardio.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, et al. Amer Heart Assoc Stat, S. Stroke Stat, Heart disease and stroke statistics-2012 update a report from the American Heart Association. Circulation. 2012;125:E2–E220. doi: 10.1161/CIR.0b013e318245fac5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 4.Kunisada K, Negoro S, Tone E, Funamoto M, Osugi T, Yamada S, et al. Signal transducer and activator of transcription 3 in the heart transduces not only a hypertrophic signal but a protective signal against doxorubicin-induced cardiomyopathy. Proc Natl Acad Sci USA. 2000;97:315–9. doi: 10.1073/pnas.97.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovtun O, Leneva N, Bykov YS, Ariotti N, Teasdale RD, Schaffer M, et al. Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature. 2018;561:561–4. doi: 10.1038/s41586-018-0526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas M, Gershlick DC, Vidaurrazaga A, Rojas AL, Bonifacino JS, Hierro A. Structural mechanism for cargo recognition by the retromer complex. Cell. 2016;167:1623–35.e14. doi: 10.1016/j.cell.2016.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joyal JS, Nim S, Zhu T, Sitaras N, Rivera JC, Shao Z, et al. Subcellular localization of coagulation factor II receptor-like 1 in neurons governs angiogenesis. Nat Med. 2014;20:1165–73. doi: 10.1038/nm.3669. [DOI] [PubMed] [Google Scholar]
- 8.Cullen PJ, Korswagen HC. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol. 2011;14:29–37. doi: 10.1038/ncb2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen PJ. Endosomal sorting and signalling: an emerging role for sorting nexins. Nat Rev Mol Cell Biol. 2008;9:574–82. doi: 10.1038/nrm2427. [DOI] [PubMed] [Google Scholar]
- 10.Zhou C, You Y, Shen W, Zhu YZ, Peng J, Feng HT, et al. Deficiency of sorting nexin 10 prevents bone erosion in collagen-induced mouse arthritis through promoting NFATc1 degradation. Ann Rheum Dis. 2016;75:1211–8. doi: 10.1136/annrheumdis-2014-207134. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Li CM, Zhang DS, Shi D, Qi M, Feng J, et al. SNX13 reduction mediates heart failure through degradative sorting of apoptosis repressor with caspase recruitment domain. Nat Commun. 2014;5:5177. doi: 10.1038/ncomms6177. [DOI] [PubMed] [Google Scholar]
- 12.Okada H, Zhang W, Peterhoff C, Hwang JC, Nixon RA, Ryu SH, et al. Proteomic identification of sorting nexin 6 as a negative regulator of BACE1-mediated APP processing. FASEB J. 2010;24:2783–94. doi: 10.1096/fj.09-146357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang JL, Li K, Zhang YG, Lu R, Wu SP, Tang JR, et al. Deletion of sorting nexin 27 suppresses proliferation in highly aggressive breast cancer MDA-MB-231 cells in vitro and in vivo. BMC Cancer. 2019;19:555. doi: 10.1186/s12885-019-5769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGough IJ, de Groot REA, Jellett AP, Betist MC, Varandas KC, Danson CM, et al. SNX3-retromer requires an evolutionary conserved MON2:DOPEY2:ATP9A complex to mediate Wntless sorting and Wnt secretion. Nat Commun. 2018;9:3737. doi: 10.1038/s41467-018-06114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng S, Streets AJ, Nesin V, Tran U, Nie H, Onopiuk M, et al. The sorting nexin 3 retromer pathway regulates the cell surface localization and activity of a Wnt-activated polycystin channel complex. J Am Soc Nephrol. 2017;28:2973–84. doi: 10.1681/ASN.2016121349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Y, Carosi JM, Yang Z, Ariotti N, Kerr MC, Parton RG, et al. Retromer has a selective function in cargo sorting via endosome transport carriers. J Cell Biol. 2019;218:615–31. doi: 10.1083/jcb.201806153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel D, Xu C, Nagarajan S, Liu Z, Hemphill WO, Shi R, et al. Alpha-synuclein inhibits Snx3-retromer-mediated retrograde recycling of iron transporters in S. cerevisiae and C. elegans models of Parkinson’s disease. Hum Mol Genet. 2018;27:1514–32. doi: 10.1093/hmg/ddy059. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Li Z, Tan Y, Li Q, Yang H, Wang P, et al. PARP1 interacts with STAT3 and retains active phosphorylated-STAT3 in nucleus during pathological myocardial hypertrophy. Mol Cell Endocrinol. 2018;474:137–50. doi: 10.1016/j.mce.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W, Qu X, Chen B, Snyder M, Wang M, Li B, et al. Critical roles of STAT3 in beta-adrenergic functions in the heart. Circulation. 2016;133:48–61. doi: 10.1161/CIRCULATIONAHA.115.017472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang HX, Xu ZS, Lin H, Li M, Xia T, Cui K, et al. TRIM27 mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis. Nat Commun. 2018;9:3441. doi: 10.1038/s41467-018-05796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha 3. Proc Natl Acad Sci USA. 2005;102:8150–5. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reich NC, Liu L. Tracking STAT nuclear traffic. Nat Rev Immunol. 2006;6:602–12. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 23.Ohara R, Fujita Y, Hata K, Nakagawa M, Yamashita T. Axotomy induces axonogenesis in hippocampal neurons through STAT3. Cell Death Dis. 2011;2:e175. doi: 10.1038/cddis.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Yaakov K, Dagan SY, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, et al. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 2012;31:1350–63. doi: 10.1038/emboj.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Liberto V, Cavalli V. Ready, STAT, go: transcription factors on the move. EMBO J. 2012;31:1331–3. doi: 10.1038/emboj.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukhopadhyay S, Shah M, Xu F, Patel K, Tuder RM, Sehgal PB. Cytoplasmic provenance of STAT3 and PY-STAT3 in the endolysosomal compartments in pulmonary arterial endothelial and smooth muscle cells: implications in pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol. 2008;294:L449–68. doi: 10.1152/ajplung.00377.2007. [DOI] [PubMed] [Google Scholar]
- 27.Bild AH, Turkson J, Jove R. Cytoplasmic transport of Stat3 by receptor-mediated endocytosis. EMBO J. 2002;21:3255–63. doi: 10.1093/emboj/cdf351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.German CL, Sauer BM, Howe CL. The STAT3 beacon: IL-6 recurrently activates STAT 3 from endosomal structures. Exp Cell Res. 2011;317:1955–69. doi: 10.1016/j.yexcr.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kermorgant S, Parker PJ. Receptor trafficking controls weak signal delivery: a strategy used by c-Met for STAT3 nuclear accumulation. J Cell Biol. 2008;182:855–63. doi: 10.1083/jcb.200806076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Huang J, Lu J, Guo Z, Li Z, Gao H, et al. Sirtuin 1 represses PKC-zeta activity through regulating interplay of acetylation and phosphorylation in cardiac hypertrophy. Br J Pharmacol. 2019;176:416–35. doi: 10.1111/bph.14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashimoto T, Kim GE, Tunin RS, Adesiyun T, Hsu S, Nakagawa R, et al. Acute enhancement of cardiac function by phosphodiesterase type 1 inhibition: translational study in the dog and rabbit. Circulation. 2018;138:1974–87. doi: 10.1161/CIRCULATIONAHA.117.030490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang N, Zhang Y, Qian H, Wu S, Cao L, Sun Y. Selective targeting of ubiquitination and degradation of PARP1 by E3 ubiquitin ligase WWP2 regulates isoproterenol-induced cardiac remodeling. Cell Death Differ. 2020;27:2605–19. doi: 10.1038/s41418-020-0523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Lv Z, He L, Huang X, Zhang S, Zhao H, et al. Genetic tracing identifies early segregation of the cardiomyocyte and nonmyocyte lineages. Circ Res. 2019;125:343–55. doi: 10.1161/CIRCRESAHA.119.315280. [DOI] [PubMed] [Google Scholar]
- 34.Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Investig. 1997;100:169–79. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunisada K, Tone E, Fujio Y, Matsui H, Yamauchi-Takihara K, Kishimoto T. Activation of gp130 transduces hypertrophic signals via STAT3 in cardiac myocytes. Circulation. 1998;98:346–52. doi: 10.1161/01.CIR.98.4.346. [DOI] [PubMed] [Google Scholar]
- 36.Frias MA, Rebsamen MC, Gerber-Wicht C, Lang U. Prostaglandin E2 activates Stat3 in neonatal rat ventricular cardiomyocytes: a role in cardiac hypertrophy. Cardiovasc Res. 2007;73:57–65. doi: 10.1016/j.cardiores.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 37.Chen C, Garcia-Santos D, Ishikawa Y, Seguin A, Li L, Fegan KH, et al. Snx3 regulates recycling of the transferrin receptor and iron assimilation. Cell Metab. 2013;17:343–52. doi: 10.1016/j.cmet.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Temkin P, Lauffer B, Jager S, Cimermancic P, Krogan NJ, von Zastrow M. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat Cell Biol. 2011;13:715–21. doi: 10.1038/ncb2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nothwehr SF, Bruinsma P, Strawn LA. Distinct domains within Vps35p mediate the retrieval of two different cargo proteins from the yeast prevacuolar/endosomal compartment. Mol Biol Cell. 1999;10:875–90. doi: 10.1091/mbc.10.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–81. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallon M, Clairfeuille T, Steinberg F, Mas C, Ghai R, Sessions RB, et al. A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc Natl Acad Sci USA. 2014;111:E3604–13. doi: 10.1073/pnas.1410552111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P, Wu YH, Belenkaya TY, Lin XH. SNX3 controls Wingless/Wnt secretion through regulating retromer-dependent recycling of Wntless. Cell Res. 2011;21:1677–90. doi: 10.1038/cr.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorenowicz MJ, Macurkova M, Middelkoop TC, de Groot R, Betist MC, Korswagen HC. Inhibition of late endosomal maturation restores Wnt secretion in Caenorhabditis elegans vps-29 retromer mutants. Cell Signal. 2014;26:19–31. doi: 10.1016/j.cellsig.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 44.Harterink M, Port F, Lorenowicz MJ, McGough IJ, Silhankova M, Betist MC, et al. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat Cell Biol. 2011;13:914–23. doi: 10.1038/ncb2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strochlic TI, Schmiedekamp BC, Lee J, Katzmann DJ, Burd CG. Opposing activities of the Snx3-retromer complex and ESCRT proteins mediate regulated cargo sorting at a common endosome. Mol Biol Cell. 2008;19:4694–706. doi: 10.1091/mbc.e08-03-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strochlic TI, Setty TG, Sitaram A, Burd CG. Grd19/Snx3p functions as a cargo-specic adapter for retromer-dependent endocytic recycling. J Cell Biol. 2007;177:115–25. doi: 10.1083/jcb.200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu WJ, Barrientos T, Mao L, Rockman HA, Sauve AA, Andrews NC. Lethal cardiomyopathy in mice lacking transferrin receptor in the heart. Cell Rep. 2015;13:533–45. doi: 10.1016/j.celrep.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gross JC, Zelarayán LC. The mingle-mangle of Wnt signaling and extracellular vesicles: functional implications for heart research. Front Cardiovasc Med. 2018;5:10. doi: 10.3389/fcvm.2018.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lakhal-Littleton S. Mechanisms of cardiac iron homeostasis and their importance to heart function. Free Radic Biol Med. 2019;133:234–37. doi: 10.1016/j.freeradbiomed.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin CR, Zhang T, Qu XY, Zhang YG, Putatunda R, Xiao X, et al. In vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol Ther. 2017;25:1168–86. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tao Z, Yang H, Shi Q, Fan Q, Wan L, Lu X. Targeted delivery to tumor-associated pericytes via an affibody with high affinity for PDGFRbeta enhances the in vivo antitumor effects of human TRAIL. Theranostics. 2017;7:2261–76. doi: 10.7150/thno.19091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cottrell GS, Soubrane CH, Hounshell JA, Lin H, Owenson V, Rigby M, et al. CACHD1 is an α2δ-Like protein that modulates Ca(V)3 voltage-gated calcium channel activity. J Neurosci. 2018;38:9186–201. doi: 10.1523/JNEUROSCI.3572-15.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu J, Zhang RW, Hong HQ, Yang ZL, Sun DP, Sun SY, et al. The poly(ADP-ribosyl)ation of FoxO3 mediated by PARP1 participates in isoproterenol-induced cardiac hypertrophy. Biochim Biophys Acta. 2016;1863:3027–39. doi: 10.1016/j.bbamcr.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 54.Lu J, Sun DP, Liu ZP, Li M, Hong HQ, Liu C, et al. SIRT6 suppresses isoproterenol-induced cardiac hypertrophy through activation of autophagy. Transl Res. 2016;172:96–112. doi: 10.1016/j.trsl.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 55.de Araujo ME, Lamberti G, Huber LA. Isolation of early and late endosomes by density gradient centrifugation. Cold Spring Harb Protoc. 2015;2015:1013–6. doi: 10.1101/pdb.prot083444. [DOI] [PubMed] [Google Scholar]
- 56.Nakatsuka S, Hayashi M, Muroyama A, Otsuka M, Kozaki S, Yamada H, et al. D-aspartate is stored in secretory granules and released through a Ca2+-dependent pathway in a subset of rat pheochromocytoma PC12 cells. J Biol Chem. 2001;276:26589–96. doi: 10.1074/jbc.M011754200. [DOI] [PubMed] [Google Scholar]
- 57.Luo WW, Wang Y, Yang HW, Dai CM, Hong HL, Li JY, et al. Heme oxygenase-1 ameliorates oxidative stress-induced endothelial senescence via regulating endothelial nitric oxide synthase activation and coupling. Aging. 2018;10:1722–44. doi: 10.18632/aging.101506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horvath CM, Wen Z, Darnell JEA. STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–94. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.