Abstract
Homeostasis of oxalic acid appears to be regulated, in part, by the gut-associated bacterium Oxalobacter formigenes. The loss of this bacterium from the gut flora is associated with an increased susceptibility to hyperoxaluria, a condition which can lead to the formation of calcium oxalate crystalluria and kidney stones. In order to identify and quantify the presence of O. formigenes in clinical specimens, a quantitative-PCR-based assay system utilizing a competitive DNA template as an internal standard was developed. This quantitative competitive-template PCR test allows for the rapid, highly specific, and reproducible quantification of O. formigenes in fecal samples and provides a prototype for development of DNA-based quantitative assays for enteric bacteria.
Recent studies are providing compelling evidence that the gut-associated, oxalate-degrading bacterium Oxalobacter formigenes is an important component in regulating oxalate absorption across the intestinal wall and, therefore, oxalate levels in the plasma (2, 14). Thus, it is not surprising that the aberrant homeostasis of oxalate present in several clinical conditions, such as recurrent calcium oxalate urolithiasis (13, 16), inflammatory bowel disease (12), Crohn’s disease and steatorrhea (11), cystic fibrosis (26), and sequelae to jejunoileal bypass surgery (1), is associated with the loss or decreased activity of O. formigenes in the human intestinal tract. Although the absence of O. formigenes within the human gut flora is clinically important in the pathogenesis of oxalate-related diseases, screening for and/or enumerating O. formigenes is not performed as a routine diagnostic test. This is due, in part, to the requirement for special anaerobic culture techniques (6). Recently, we reported the development of a rapid, sensitive, and genus-specific PCR-based assay system capable of detecting O. formigenes in fecal samples (25, 27). Using this system, we have been able to confirm the pathogenic link between the absence of O. formigenes and hyperoxaluria (24, 26).
The testing of fecal samples from healthy males and females has shown that O. formigenes normally is present at between 5 × 106 and 5 × 108 CFU/g (wet weight) of feces (1, 21). In contrast, O. formigenes is not detectable in the feces of cystic fibrosis patients with hyperoxaluria (25) or in most patients with recurrent calcium oxalate kidney stone formation (i.e., patients with more than five recurrent episodes) (16). However, a subpopulation of urolithiasis patients appears to harbor reduced numbers of O. formigenes (102 to 103 CFU/g [wet weight] of feces), and these patients report having fewer kidney stone episodes (fewer than four recurrent episodes) over a 5-year period (13, 16). These findings point to the need for a simple and reliable quantitative assay capable of enumerating O. formigenes in clinical specimens. In this report, we describe a rapid, simple, quantitative-PCR assay that utilizes an internal competitive DNA template for estimating populations of O. formigenes in fecal samples.
MATERIALS AND METHODS
Bacterial strains.
O. formigenes OxB was used as the standard throughout this study. This strain was grown in medium B containing 30 mM oxalate, as described elsewhere (2). Other bacterial strains used in this study were obtained from the American Type Culture Collection (ATCC), Manassas, Va., and are as follows: Alcaligenes sp. ATCC 11883, Bacteroides ovatus ATCC 8483, Citrobacter brakii ATCC 43162, Clostridium perfringens ATCC 13124, Clostridium sordellii ATCC 9714, Enterobacter aerogenes ATCC 13048, Enterobacter cloacae ATCC 13047, Enterococcus faecalis ATCC 29212, Escherichia coli ATCC 25922, Klebsiella oxytoca ATCC 43165, Lactobacillus acidophilus ATCC 314, Moorella thermoacetica ATCC 35608, Moorella thermoautotrophica ATCC 33924, Moraxella osloensis ATCC 10973, Proteus mirabilis ATCC 29245, Proteus vulgaris ATCC 8427, Pseudomonas aeruginosa ATCC 27853, Salmonella typhimurium ATCC 6994, Shigella boydii ATCC 29928, Staphylococcus aureus ATCC 29213, Staphylococcus epidermidis ATCC 49134, Streptococcus bovis ATCC 49147, Streptococcus pneumoniae ATCC 49136, and Veillonella parvula ATCC 10790.
Isolation of genomic DNA from bacterial strains.
Cultures (10 to 15 ml) of O. formigenes were centrifuged at 10,000 × g, the supernatants were discarded, and the bacterial pellets were stored at −80°C. Bacteria from either the frozen O. formigenes samples or the lyophilized ATCC samples were resuspended in 567 μl TE buffer (10 mM Tris-Cl [pH 7.5] and 1 mM EDTA [pH 8.0]), 30 μl of 10% sodium dodecyl sulfate, and 3 μl of proteinase K (20 mg/ml), and each mixture was incubated for 5 h at 37°C to ensure bacterial cell lysis. Nucleic acids were extracted from the lysates with phenol-chloroform-isoamylalcohol (25:24:1). Chromosomal DNA was precipitated from the aqueous phase by adding 1/2 volume of 7.5 M ammonium acetate and 2 volumes of 100% ethanol. DNA was recovered by centrifugation (12,000 × g) and washed once with 70% ethanol. The pellet was resuspended in 15 to 20 μl of H2O.
Isolation of genomic DNA from human fecal specimens.
Bacterial DNA was isolated directly from fresh stool samples obtained from individuals known to be positive or negative for O. formigenes by using the procedures of Stacey-Phipps et al. (28). Approximately 25 mg of feces was suspended in 1.5 ml of phosphate-buffered saline and centrifuged at low speed to remove debris, and the supernatant was centrifuged at 16,000 × g for 5 min to obtain a bacterial pellet. The pellet was resuspended in 0.6 ml of binding lysis buffer (5.3 M guanidine thiocyanate, 10 mM dithiothreitol, 1% Tween 20, 0.3 M sodium acetate, and 50 mM sodium citrate) and incubated at 65°C for 10 min. Glass matrix (50 μl) (Glass Plac; National Scientific Supply Co., San Rafael, Calif.) was added to absorb DNA. The DNA was eluted from the glass beads with 10 mM TE buffer.
PCR and QC-PCR.
All PCRs and quantitative competitive-template PCR. (QC-PCRs) were performed as described elsewhere (17, 30). In brief, 50-μl reaction mixtures contained 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate, 1.25 U of Taq (GIBCO-BRL, Bethesda, Md.), 1 μg of genomic DNA, and 1 μM concentrations (each) of a 5′ and 3′ primer. The optimal reaction profile proved to be 94°C for 5 min followed by 35 cycles of 94°C for 1 min for denaturation, 60°C for 1 s with a gradual decrease of 0.1°C/S to 55°C for annealing, and 72°C for 1 min for primer extension. For QC-PCR, 1 μl of an appropriate dilution of competitive templates was added to the standard PCR mixture. PCR and QC-PCR products were size separated by gel electrophoresis in 1.2% agarose containing ethidium bromide, illuminated with UV light, and photographed for documentation. Gels were then scanned with an Alpha Inotech Imager ISO1000 (Alpha Inotech Corp., San Leandro, Calif.) to determine the relative band intensities of the PCR products for quantification.
Southern blots.
PCR products were transferred from the agarose gels to positively charged nylon membranes (Boehringer-Mannheim GmBH, Indianapolis, Ind.) by positive-pressure blotting and UV cross-linking. Hybridizations were carried out as described elsewhere (25) by using an internal sequence oligonucleotide probe and the Genius system of Boehringer-Mannheim, following company instructions. The oligonucleotide used as the probe was 5′-GACAATGTAGAGTTGACTGATGGCTTTCATG-3′, synthesized in the University of Florida ICBR Oligonucleotide Synthesis Laboratory (Gainesville, Fla.). This oligonucleotide was end labeled with digoxigenin in a reaction catalyzed by terminal transferase. The digoxigenin-labeled oligonucleotide probe was hybridized to the immobilized DNA fragments, and hybridization was detected colorimetrically by enzyme-linked immunosorbent assay with a digoxigenin-specific antibody linked to alkaline phosphatase according to the protocol provided with the Genius kit.
RESULTS
Specificity of DNA-based detection of O. formigenes.
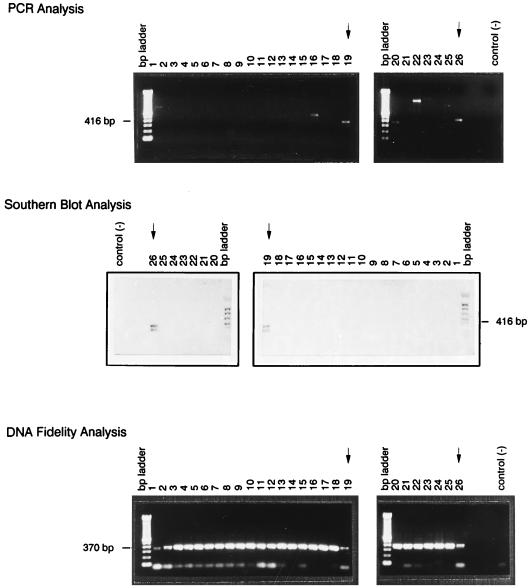
The specificity of this DNA-based assay system was determined by testing its ability to distinguish O. formigenes from other bacterial species. PCRs were carried out with the primer pair OXFfp-OXFrp and with genomic DNA from O. formigenes as well as from each of the 24 bacterial strains listed above. As shown in Fig. 1, only O. formigenes DNA (lanes 19 and 26) gave rise to an amplicon of the correct molecular size (top panel) that hybridized with the genus-specific probe (middle panel). Some PCR-Southern blots of the O. formigenes oxc gene amplicon reveal the presence of a shadow band of approximately 300 bp. This band has been sequenced and found to be homologous to the 337 bp of the 3′ end of the expected 416-bp PCR product. Why a small portion of the 5′ end is absent from these amplicons is unknown.
FIG. 1.
Specificity of the PCR-based identification of O. formigenes. PCRs were carried out with 1 ng of genomic DNA isolated from each of the following: Bacteroides ovatus, Proteus vulgaris, Enterobacter cloacae, Escherichia coli, Enterobacter aerogenes, Clostridium perfringens, Clostridium sordellii, Veillonella parvula, Streptococcus pneumoniae, Staphylococcus aureus, Pseudomonas aeruginosa, Streptococcus bovis, Enterococcus faecalis, Staphylococcus epidermidis, Moorella thermoautotrophica, Shigella boydii, Proteus mirabilis, Salmonella typhimurium, Oxalobacter formigenes, Citrobacter brakii, Lactobacillus acidophilus, Moraxella osloensis, Moorella thermoacetica, Alcaligenes sp., Klebsiella oxytoca, and Oxalobacter formigenes (top panel, lanes 1 to 26, respectively). Southern blots of the gels were carried out with an Oxalobacter genus-specific DNA oligonucleotide probe (middle panel). To test the ability of each genomic-DNA preparation to support a PCR, PCRs were carried out with 1 ng of genomic DNA isolated from O. formigenes or from one of the 24 other bacterial strains and a set of universal primers for bacterial 16S rRNA (bottom panel). Arrows indicate lanes containing O. formigenes DNA.
A problem inherent in all DNA-based detection systems, especially those using PCR, is the possibility of false-negative reactions resulting from inadequate DNA preparations. To determine if this may have played a role in the specificity experiment, each DNA preparation was simultaneously tested for suitability for PCR amplification by using the universal primer pair, 5′ primer (5′-AACTGGAGGAAGGTGGGGAT-3′) and 3′ primer (5′-AGGAGGTGATCCAACCGCA-3′), to amplify the bacterial gene encoding 16S rRNA (15). As shown in Fig. 1 (bottom panel), all DNA preparations permitted the amplification of a proper 16S rRNA PCR product, indicating that each DNA preparation contained appropriate DNA and that the specificity obtained for O. formigenes detection was not due to false-negative reactions.
QC-PCR.
O. formigenes expresses a unique gene required for the catabolism of oxalate: oxc (encoding oxalyl-coenzyme A decarboxylase). This gene has been cloned and sequenced (17). Sequencing of the 5′ ends of the oxc genes from numerous isolates of O. formigenes identified unique, highly conserved regions which permitted the synthesis of a genus-specific PCR primer pair (forward primer, 5′-AATGTAGAGTTGACTGA-3′ [OXFfp]; reverse primer, 5′-TTGATGCTGTTGATACG-3′ [OXFrp]) (25, 27). This PCR primer pair amplifies a 416-bp product (in group II strains) or a 413-bp product (in group I strains) of the 5′ end of the oxc gene which is unique to O. formigenes.
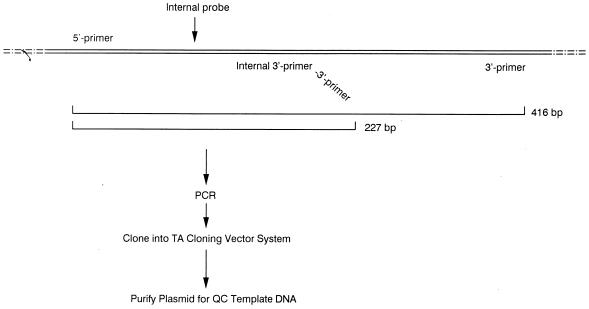
QC-PCR is based on the assumptions that a genomic DNA template and a competitive DNA template containing homologous primer sites will compete equally for PCR primers and that both experimental- and competitive-template PCR products will subsequently be amplified colinearly. To construct a suitable competitive-DNA template for use as the internal control, a 227-bp fragment of the oxc gene flanked by sequences homologous for the OXFfp-OXFrp primer pair and containing a genus-specific probe was generated (Fig. 2). To accomplish this, a PCR was performed with the OXFfp 5′ primer plus a modified OXFrp 3′ primer. The modified 3′ primer (5′-TTGATGCTGTTGATACGGTCAAGCAAACGCC-3′) consisted of two portions: a 5′ end which contained the 3′ primer sequence (underlined) within the oxc gene and a 3′ end which annealed at a site located approximately 200 bp downstream of the 5′ primer site. The PCR that used the primer pair OXFfp-modified OXFrp amplified the 210-bp segment and synthesized the 17-bp OXFrp primer site at the 3′ end. This PCR fragment was purified and ligated into pCR-2.1 (Invitrogen, Inc., San Diego, Calif.). A recombinant pCR-2.1 plasmid with the proper insert (confirmed by sequencing) was selected for use as the internal competitive template.
FIG. 2.
Synthesis pathway of a competitive DNA template for quantifying the PCR. A truncated form of the 416-bp fragment of the oxc gene containing both the 5′ and 3′ primer sites was synthesized by PCR by using a modified 3′ primer. This truncated and modified sequence was ligated into the pCR2.1 vector system in order to produce high copy numbers.
Quantification CFU with QC-PCR.
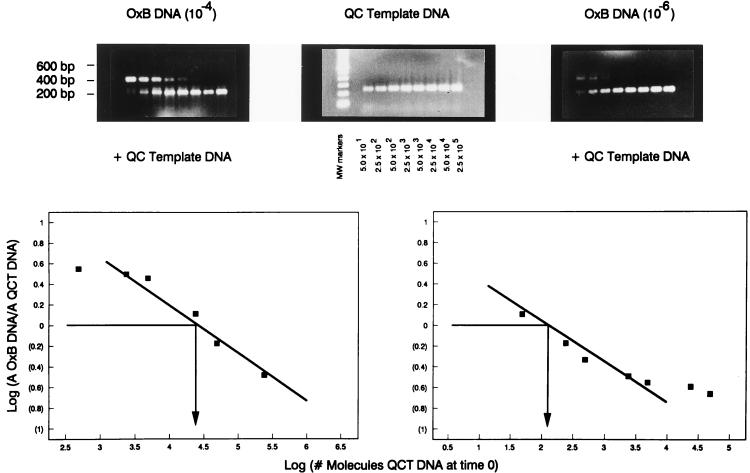
To determine the accuracy of the QC-PCR in quantifying the number of CFU in of O. formigenes samples, QC-PCRs were established with two dilutions of an O. formigenes DNA preparation which had a starting spectrophotometric reading of 1.126 μg of DNA/μl. Assuming that the genome of O. formigenes is similar in size to that of E. coli (4.7 × 103 kb), 1 μg of O. formigenes genomic DNA contains 1.8 × 108 molecules (or gene copies). Thus, the original genomic DNA preparation of O. formigenes OxB contained approximately 2 × 108 molecules/μl. Two dilutions, fourfold (20,000 genomes) and sixfold (200 genomes), of this DNA were used as templates in the QC-PCR with dilutions of competitive templates ranging from 50 to 250,000 molecules. The PCR products were size separated by electrophoresis through 1.5% agarose gels and visualized with UV light (Fig. 3, top panels). PCR bands were scanned for intensities and normalized for differences in molecular mass, and the log ratios of O. formigenes to template band intensities were plotted against the log of the copy number of synthetic template added per reaction to determine log equivalence. Quantitation of oxc genes, and thereby bacteria, revealed the accuracy of this QC-PCR detection system: the log equivalence revealed that the number of molecules of O. formigenes OxB in the reaction was estimated to be 19,900 to 25,100 and 126 to 158, respectively (Fig. 3, bottom panels).
FIG. 3.
Quantification of the number of O. formigenes genomes in a sample by using the QC-PCR. Dilutions, ranging from 50 to 250,000 molecules of purified plasmid containing the competitive template, were used either as DNA templates in PCR to establish standard curves (top, center panel) or as competitive DNA by mixing with 2 × 104 (left panel) or 2 × 102 (right panel) copies of purified O. formigenes OxB genomes. PCR band intensities were scanned and normalized for molecular mass, and the log ratios of O. formigenes to template band intensities were graphed to determine log equivalence (bottom panels). QCT, quantitative competitive template.
Correlation between CFU detected by culture versus by QC-PCR.
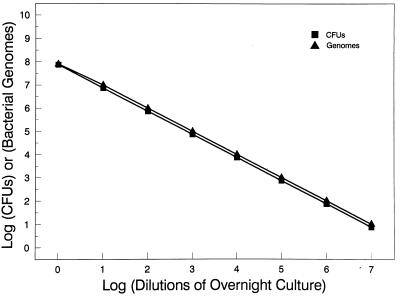
To correlate the number of CFU of O. formigenes detected via culture plating versus QC-PCR, an overnight culture of O. formigenes was prepared and determined to have a titer of approximately 0.7 × 108 CFU/ml. This culture was serially diluted 10-fold to 0.7 × 101 CFU/ml. DNA isolated from 1 ml of each dilution was mixed with competitive template diluted from 1010 to 102 copies/reaction. The PCR products were size separated, visualized with UV light, and photographed. Photographs were scanned for relative band intensities and normalized for differences in molecular weight, and the log ratios of O. formigenes to template band intensities were plotted against the log of the copy number of synthetic template added per reaction. As shown in Fig. 4, the number of CFU for each dilution, as estimated by optical density and determined by QC-PCR, were nearly identical. This experiment was performed in triplicate, and the coefficients of variation proved to be less than 6% at all dilutions of the O. formigenes cultures.
FIG. 4.
Comparison of the numbers of O. formigenes CFU in a bacterial culture detected by culture and QC-PCR. An overnight culture of O. formigenes OxB containing approximately 0.7 × 108 CFU/ml, as determined by optical density at 600 nm, was serially diluted 10-fold. DNA isolated from each dilution was used in QC-PCR. Log equivalences of O. formigenes to template band intensities were determined and compared to the estimated number of CFU for each dilution.
Similar results were obtained when the numbers of CFU in clinical fecal specimens were obtained by culture and compared to the results of QC-PCR (Table 1). Fecal samples were collected from individuals known to be positive or negative for O. formigenes. The number of CFU was determined for each sample either by standard culture methods or by the QC-PCR detection system. Again, whether the sample was analyzed by culture or by QC-PCR, the number of CFU detected proved to be quite similar for all specimens.
TABLE 1.
Correlation of CFU quantification by standard culturing and QC-PCR
| Clinical sample no.a | CFU/g of feces (wet weight) quantified by:
|
|
|---|---|---|
| Culture method | QC-PCR | |
| 1 | 0 | 0 |
| 2 | 0 | 0 |
| 3 | 0 | 0 |
| 4 | 2.3 × 106 | 2.5 × 106 |
| 5 | 5.5 × 106 | 3.0 × 106 |
| 6 | 2.1 × 107 | 3.5 × 107 |
| 7 | 3.1 × 107 | 3.1 × 107 |
| 8 | 1.0 × 107 | 1.6 × 107 |
Specimens were collected in North Carolina, where CFU were quantified by culture; samples were mailed to Florida, where CFU were quantified by QC-PCR.
Clinical sample stability and reproduction.
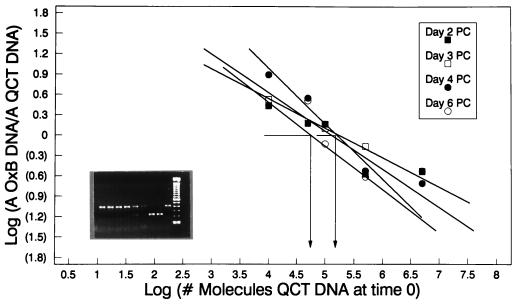
By using the Cary-Blair culture swab transport system (Difco Laboratories, Detroit, Mich.), four fecal swabs (ca. 20 mg of stool sample/swab) were obtained from an individual living in North Carolina who was known to be positive for O. formigenes, and these swabs were shipped through the mail to Florida. The swabs were transported and maintained at room temperature. Upon arrival in Florida (day 2 postcollection), one swab was vortexed in 500 μl of sterile H2O, and the genomic DNA was extracted. The extract was tested for the presence of O. formigenes DNA with QC-PCR, which detected a robust colonization of 8.0 × 108 bacteria/g (wet weight) (Fig. 5). The remaining three swabs were treated identically, except that DNA was extracted on days 3, 4, and 6 postcollection. Results consistent with the day 2 determinations were obtained with specimens prepared on days 3 and 4 postcollection; however, by day 6, the number of detectable bacteria began to decline, probably due to the death of the microorganisms. Quantification of bacteria in the clinical samples tested on days 2, 3, and 4 showed a coefficient of variation of less than 10%.
FIG. 5.
Testing clinical samples for stability and reproducibility in detecting O. formigenes. Four fecal swabs collected and transported in the Cary-Blair culture transport system were processed on days 2, 3, 4, and 6 following collection to analyze the sample stability and the reproducibility of the QC-PCR assay system. DNA was isolated from approximately 20 mg of fecal specimen eluted from each swab, and the presence of O. formigenes genomes was determined by QC-PCR (insert). Log equivalences of O. formigenes to template band intensities were graphed to quantify the number of genomes present in each sample. The number of CFU detected on days 2, 3, and 4 was 8 × 108/g (wet weight), while the number of CFU detected on day 6 was 2.8 × 108/g (wet weight). QCT, quantitative competitive template.
Detecting temporal changes in O. formigenes colonization.
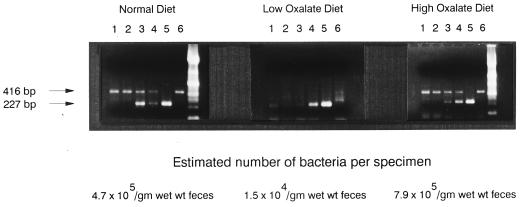
Changes in the amount of dietary oxalate consumed by humans is known to affect the robustness of O. formigenes colonization; that is, increases in oxalate consumption increase colonization, whereas decreases in oxalate consumption result in a decrease of colonization (10). As presented in Fig. 6, by using the QC-PCR detection system, it was possible to quantitate temporal changes in numbers of bacteria induced in a human through changes in diet.
FIG. 6.
Quantifying temporal changes in O. formigenes colonization induced by changes in diet. Detection of O. formigenes in fecal samples obtained from a subject on a normal diet who was then placed on a diet low in oxalate-containing foods followed by a diet high in oxalate-containing foods. Amplifications were carried out with 1 μl of sample DNA alone (lanes 6) and in the presence of 42 (lanes 1), 83 (lanes 2), 415 (lanes 3), 830 (lanes 4), or 8,300 (lanes 5) copies of the competitive template.
DISCUSSION
Application of modern biotechnology to microbiological testing has resulted in the development of “rapid methods” that no longer rely solely on culturing (18). These assays include direct detection of cell components (e.g., use of bioluminescence or fluorescence), antibody-based detection of cellular antigens (e.g., use of enzyme-linked immunosorbent assay), biochemical tests which identify endogenous enzyme activities (e.g., trypsin-like protease activity of Porphyromonas gingivalis, Bacteroides forsythus, and Treponema denticola), and DNA-based detection (e.g., DNA probe or PCR-based tests). DNA-based tests identify specific sequences of genomic nucleic acid unique to individual microorganisms. PCR-based assays have already been developed for viral pathogens (5), sexually transmitted pathogens like Chlamydia trachomatis (20), air-borne pathogens like Mycobacterium tuberculosis (22), and a variety of food-borne pathogens like Listeria monocytogenes, E. coli, Salmonella spp., Campylobacter spp., Yersinia enterocolitica, and Vibrio vulnificus (3, 15, 19, 29, 31). Interestingly, many of these techniques remain deficient in sensitivity because they require a preenrichment step. Selective enrichment prior to PCR amplification has been used as a way to concentrate the bacteria and/or dilute potential inhibitory factors. Unfortunately, these steps tend to negate the sensitivity and speed of PCR-based tests. Obviously, any procedure needed to either concentrate or enrich the bacteria sacrifices both time and the ability to accurately enumerate the contaminating bacteria (18, 29).
Quantitative PCR has been used primarily for the measurement of viral loads within cell populations (7, 9). Currently, there are three general strategies for the quantification of amplified target DNA. One approach involves coamplification of an internal standard and the target of interest followed by the size separation of the specific amplicons by electrophoresis (30). Electrophoretic size separation schemes have several potential problems, including the dependency of amplification ratios for amplicons on DNA quality and length. A second approach uses hybridization with capture probes based on hybridization of the entire amplified sequence (8). Capture probe methods are inappropriate for the quantification of amplicons that contain homologous regions (i.e., competitive templates), but this technique can be suitably modified for automation (8, 23). The latest approach is real-time detection using quantitative-PCR thermocyclers, like the ABI PRISM 7700 (4). Real-time detection, while rapid and accurate, cannot be easily adapted to large multiplex assays due to the limited availability of fluorochromes with nonoverlapping spectra.
In this report, we have described a simple method for the direct quantification of O. formigenes in clinically derived specimens by using QC-PCR. This method utilizes a known quantity of a synthesized DNA template as an internal control that competes for PCR primers with the experimental DNA, in this case the bacterial genome. Theoretically, at log template equivalence, the internal controls and the bacterial genomes should amplify colinearly.
Our results indicate that the QC-PCR test is considerably faster than, and as sensitive as, current culture techniques, is highly reproducible in quantifying the number of O. formigenes genomes present in culture or clinical samples, and is uniquely specific for the target bacterium. This assay can quantify as few as 10 CFU of O. formigenes/ml, and this quantitation is linear up to 108 CFU/ml (Fig. 4). Furthermore, the specificity of the QC-PCR is not affected by the presence of other microorganisms or factors in either culture or fecal matter, and the number of O. formigenes identified by QC-PCR has proven to be consistent with the levels of oxalate degradation observed in media containing dilutions of this oxalate-degrading bacterium. In the present study, the potential clinical utility of QC-PCR was illustrated by quantifying O. formigenes in swabs of fecal specimens transported through routine mail service. Using common, inexpensive, and user-friendly transport systems for enteric bacteria (e.g., the Cary-Blair or Stuart’s culture swab), we have shown that detection of O. formigenes was consistent for up to 4 days before the number of detectable genomes began to decline. Lastly, but perhaps just as importantly, the entire QC-PCR procedure, as described here, can be accomplished within an 8-h work shift, and automation could reduce this time even further. Thus, QC-PCR appears to have broad application in the clinical setting for the quantification of bacteria in clinical samples.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grants DK-20586 (R.P.H.) and DK-53556 (A.B.P.) from the National Institutes of Health and NAG5-3968 (R.P.H.) from NASA.
We acknowledge the advice of, and useful discussions with, Saeed Khan.
REFERENCES
- 1.Allison M J, Cook H M, Milne D B, Gallaher S, Clayman R V. Oxalate degradation by gastrointestinal bacteria from humans. J Nutr. 1986;116:455–460. doi: 10.1093/jn/116.3.455. [DOI] [PubMed] [Google Scholar]
- 2.Allison M J, Dawson K A, Mayberry W R, Foss J G. Oxalobacter formigenes gen. nov., sp. nov.: oxalate degrading bacteria that inhabit the gastrointestinal tract. Arch Microbiol. 1985;141:1–7. doi: 10.1007/BF00446731. [DOI] [PubMed] [Google Scholar]
- 3.Barrette T J, Feng P, Swaminathan B. Amplification methods for detection of food-borne pathogens. In: Lee H, Morse S, Olsvik O, editors. Nucleic acid amplification technologies. Natick, Mass: Eaton Publishing Co.; 1997. pp. 171–181. [Google Scholar]
- 4.Batt C A. Molecular diagnostics for dairy-borne pathogens. J Dairy Sci. 1997;80:220–229. doi: 10.3168/jds.S0022-0302(97)75931-9. [DOI] [PubMed] [Google Scholar]
- 5.Clewley J P, editor. The polymerase chain reaction (PCR) for human viral diagnosis. Boca Raton, Fla: CRC Press; 1994. [Google Scholar]
- 6.Daniel S L, Cook H M, Hartman P A, Allison M J. Enumeration of anaerobic oxalate-degrading bacteria in the ruminal contents of sheep. FEMS Microbiol Ecol. 1989;62:329. [Google Scholar]
- 7.Davis G L, Lau J Y, Urdea M S, Neuwald P D, Wilber J C, Lindsay K, Perrillo R P, Albrecht J. Quantitative detection of hepatitis C virus RNA with a solid-phase signal amplification method: definition of optimal conditions for specimen collection and clinical application in interferon-treated patients. Hepatology. 1994;19:1337–1341. [PubMed] [Google Scholar]
- 8.Denis M, Soumet C, Legeay O, et al. Development of a semiquantitative PCR assay using internal standard and colorimetric detection on microwell plates for pseudorabies virus. Mol Cell Probes. 1997;11:439–448. doi: 10.1006/mcpr.1997.0139. [DOI] [PubMed] [Google Scholar]
- 9.Detmer J, Lagier R, Flynn J, Zayati C, Kolberg J, Collins M, Urdea M, Sánchez-Pescador R. Accurate quantification of hepatitis C virus (HCV) RNA from all HCV genotypes by using branched-DNA technology. J Clin Microbiol. 1996;34:901–907. doi: 10.1128/jcm.34.4.901-907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doane L T, Liebman M, Caldwell D R. Microbial oxalate degradation: effects on oxalate and calcium balance in humans. Nutr Res. 1989;9:957–964. [Google Scholar]
- 11.Goldkin L, Cave D R, Jaffin B, Bliss C M, Allison M J. Bacterial oxalate metabolism in the human colon: a possible factor in enteric hyperoxaluria. Gastroenterology. 1986;90:1431–1432. [Google Scholar]
- 12.Goldkin L, Cave D R, Jaffin B, Robinson W, Bliss C M. A new factor in enteric hyperoxaluria: Oxalobacter formigenes. Am J Gastroenterol. 1985;80:860. [Google Scholar]
- 13.Han J Z, Zhang X, Li J G, Zhang Y S. The relationship of Oxalobacter formigenes and calcium oxalate calculi. J Tongji Med Univ. 1995;15:249–252. doi: 10.1007/BF02887957. [DOI] [PubMed] [Google Scholar]
- 14.Hatch M, Freel R W. Oxalate transport across intestinal and renal epithelia. In: Khan S, editor. Calcium oxalate in biological systems. Boca Raton, Fla: CRC Press; 1996. pp. 217–238. [Google Scholar]
- 15.Hill W E. The polymerase chain reaction: applications for the detection of food-borne pathogens. Crit Rev Food Sci Nutr. 1996;36:123–173. doi: 10.1080/10408399609527721. [DOI] [PubMed] [Google Scholar]
- 16.Kleinschmidt K, Mahlmann A, Hautmann R. Anaerobic oxalate-degrading bacteria in the gut decrease faecal and urinary oxalate concentrations in stone formers. In: Ryall R, Bais R, Marshall V R, Rofe A M, Smith L H, Walker V R, editors. Urolithiasis 2. New York, N.Y: Plenum Press; 1993. pp. 439–441. [Google Scholar]
- 17.Lung H-Y, Baetz A L, Peck A B. Molecular cloning, DNA sequence, and gene expression of the oxalyl-coenzyme A decarboxylase gene, oxc, from the bacterium Oxalobacter formigenes. J Bacteriol. 1994;176:2468–2472. doi: 10.1128/jb.176.8.2468-2472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. [Google Scholar]
- 19.Pillai S D, Ricke S C. Strategies to accelerate the applicability of gene amplification protocols for pathogen detection in meat and meat products. Crit Rev Microbiol. 1995;21:239–261. doi: 10.3109/10408419509113542. [DOI] [PubMed] [Google Scholar]
- 20.Quinn T C. Recent advances in diagnosis of sexually transmitted diseases. Sex Transm Dis. 1994;21:S19–S27. [PubMed] [Google Scholar]
- 21.Robinson W R, Cave D R, Bliss C M, Daniel S, Allison M J. Bacterial degradation of oxalate in feces. Gastroenterology. 1985;88:1557. [Google Scholar]
- 22.Roth A, Schaberg T, Mauch H. Molecular diagnosis of tuberculosis: current clinical validity and future perspectives. Eur Respir J. 1997;10:1877–1891. doi: 10.1183/09031936.97.10081877. [DOI] [PubMed] [Google Scholar]
- 23.Rudi K, Skulberg O M, Larsen F, Jakobsen K S. Quantification of toxic cyanobacteria in water by use of competitive PCR followed by sequence-specific labelling of oligonucleotide probes. Appl Environ Microbiol. 1998;64:2639–2643. doi: 10.1128/aem.64.7.2639-2643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sidhu H, Allison M J, Peck A B. Role of the gut inhabiting bacterium, Oxalobacter formigenes, in oxalate-related disorders. Biosci Microflora. 1997;16:21. [Google Scholar]
- 25.Sidhu H, Allison M, Peck A B. Identification and classification of Oxalobacter formigenes strains by using oligonucleotide probes and primers. J Clin Microbiol. 1997;35:350–353. doi: 10.1128/jcm.35.2.350-353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sidhu H, Hoppe B, Hesse A, Tenbrock K, Bromme S, Rietschel E, Peck A B. Antibiotic induced loss of the gut associated bacterium, O. formigenes: a risk factor for hyperoxaluria in cystic fibrosis patients. Lancet. 1998;352:1026–1029. doi: 10.1016/S0140-6736(98)03038-4. [DOI] [PubMed] [Google Scholar]
- 27.Sidhu H, Yenatska L, Ogden S D, Allison M J, Peck A B. Natural colonization of children in the Ukraine with the intestinal bacterium, O. formigenes, using a PCR-based detection system. Mol Diagn. 1997;2:89–97. doi: 10.1054/MODI00200089. [DOI] [PubMed] [Google Scholar]
- 28.Stacy-Phipps S, Mecca J J, Weiss J B. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J Clin Microbiol. 1995;33:1054–1059. doi: 10.1128/jcm.33.5.1054-1059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swaminathan B, Feng P. Rapid detection of food-borne pathogenic bacteria. Annu Rev Microbiol. 1994;48:401–426. doi: 10.1146/annurev.mi.48.100194.002153. [DOI] [PubMed] [Google Scholar]
- 30.Tarnuzzer R W, Macauley S P, Farmerie W, Cabellaro S, Ghassemifar M R, Robinson C, Grant M B, Humphreys-Beher M G, Franzen L, Peck A B, Schultz G S. Competitive RNA templates for detection and quantitation of growth factors, cytokines, extracellular matrix components and matrix metallo-proteinases by RT-PCR. Bio/Technology. 1996;20:670–674. doi: 10.2144/19962004670. [DOI] [PubMed] [Google Scholar]
- 31.Wang R F, Cao W W, Cerniglia C E. A universal protocol for PCR detection of 13 species of food-borne pathogens in food. J Appl Microbiol. 1997;83:727–736. doi: 10.1046/j.1365-2672.1997.00300.x. [DOI] [PubMed] [Google Scholar]