Abstract
Idiopathic pulmonary fibrosis (IPF) is a fatal disease in which the normal alveolar network is gradually replaced by fibrotic scars. Current evidence suggests that metabolic alterations correlate with myofibroblast activation in IPF. Anlotinib has been proposed to have antifibrotic effects, but the efficacy and mechanisms of anlotinib against lung fibrosis have not been systematically evaluated. The antifibrotic effects of anlotinib were evaluated in bleomycin-induced mouse models and transforming growth factor-beta 1 (TGF-β1)-stimulated lung fibroblasts. We measured lactate levels, 2-NBDG glucose uptake and the extracellular acidification rate (ECAR) to assess glycolysis in fibroblasts. RNA-protein coimmunoprecipitation (RIP) and polysome analyses were performed to investigate novel mechanisms of glycolytic reprogramming in pulmonary fibrosis. We found that anlotinib diminished myofibroblast activation and inhibited the augmentation of glycolysis. Moreover, we show that PCBP3 posttranscriptionally increases PFKFB3 expression by promoting its translation during myofibroblast activation, thus promoting glycolysis in myofibroblasts. Regarding mechanism, anlotinib exerts potent antifibrotic effects by downregulating PCBP3, reducing PFKFB3 translation and inhibiting glycolysis in myofibroblasts. Furthermore, we observed that anlotinib had preventative and therapeutic antifibrotic effects on bleomycin-induced pulmonary fibrosis. Therefore, we identify PCBP3 as a protein involved in the regulation of glycolysis reprogramming and lung fibrogenesis and propose it as a therapeutic target for pulmonary fibrosis. Our data suggest that anlotinib has antifibrotic effects on the lungs, and we provide a novel mechanism for this effect. Anlotinib may constitute a novel and potent candidate for the treatment of pulmonary fibrosis.
Keywords: pulmonary fibrosis, anlotinib, glycolysis, PFKFB3, PCBP3
Introduction
Fibrosis can develop in most organs and cause organ failure. The most common type of lung fibrosis is idiopathic pulmonary fibrosis (IPF), which is highly prevalent and associated with a dramatically increased disease burden worldwide (Wynn and Ramalingam, 2012; Hutchinson et al., 2015). Overall, the development of new therapeutics should be pursued. Currently, only pirfenidone and nintedanib have been approved as therapeutics for IPF (Taniguchi et al., 2010; Sato et al., 2017), and as both drugs have limited efficacy (Spagnolo and Maher, 2017), there is an urgent need to identify new potential therapeutic agents for IPF patients.
Upon chronic microinjury to the alveolar epithelium, fibroblast activation and transdifferentiation into myofibroblasts are among the first responses detectable at the site of damage (Plantier et al., 2018). Myofibroblasts are characterized by de novo expression of α-smooth muscle actin (α-SMA), the formation of stress fibers, and enhanced abilities to proliferate, migrate, and produce extracellular matrix (ECM) (Hinz, 2012; Hinz et al., 2012; Liu et al., 2021). These cells drive a wound-healing response that relies on the deposition of collagen-rich ECM and activates transforming growth factor-beta 1 (TGF-β1) signaling (Kenyon et al., 2003; Sapudom et al., 2015; Nigdelioglu et al., 2016). This transient response must be tightly controlled, otherwise it can become persistent and lead to excessive matrix accumulation and fibrosis. Understanding the molecular bases of fibroblast activation is therefore essential in identifying novel and efficient antifibrotic therapeutic targets to reduce the incidence, morbidity and mortality of people suffering from clinically refractory disorders, including IPF.
Metabolic perturbation is implicated in the pathogenesis of several kinds of tissue fibrosis (DeBerardinis and Thompson, 2012; Chen et al., 2018), including pulmonary fibrosis (Para et al., 2019; Bueno et al., 2020). To cope with the high energy demands of myofibroblasts, including increases in proliferation and matrix production, it is reasonable that activated myofibroblasts exhibit augmented aerobic glycolysis to meet additional bioenergetic and biosynthetic demands, even in oxygen-rich conditions, similar to observations in many cancer cells and other nonmalignant proliferating cells. A major driver of glycolysis is 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3), which produces fructose-2,6-bisphosphate (F2,6BP), the most potent allosteric activator of the glycolytic rate-limiting enzyme phosphofructokinase-1 (PFK1) (Van Schaftingen et al., 1982; Cao et al., 2019). Previous studies have shown that augmentation of aerobic glycolysis is an essential step during myofibroblast activation (Para et al., 2019). Ramping down glycolysis is effective in diminishing myofibroblast activation, thus limiting lung fibrosis. However, metabolism-based therapeutics for treating fibrotic disorders are still lacking.
Anlotinib (AL3818) hydrochloride is a novel multitargeted tyrosine kinase inhibitor (TKI) that targets the receptor tyrosine kinases vascular endothelial growth factor receptor (VEGFR) 1 thru 3, epidermal growth factor receptor (EGFR), fibroblast growth factor receptor (FGFR) 1 thru 4, platelet-derived growth factor receptor (PDGFR) α and β, and stem cell factor receptor (Sun et al., 2016; Xie et al., 2018). Many studies have reported the therapeutic effects of anlotinib in several diseases, such as nonsmall cell lung cancer (Liang et al., 2019), endometrial cancers (Taurin et al., 2018) and osteosarcoma (Liang et al., 2019). Interestingly, the targets of anlotinib are similar to those of nintedanib, a drug that has been approved for the treatment of IPF. A recent report suggested that intraperitoneal administration of anlotinib attenuates bleomycin-induced lung fibrosis in mice by suppressing the TGF-β signaling pathway (Ruan et al., 2020). Nevertheless, the mechanism by which anlotinib resolves lung fibrosis and whether anlotinib may be therapeutically used to improve lung function are not well understood.
In the current study, we analyzed the antifibrotic effect of anlotinib on TGF-β1-induced fibroblast transdifferentiation and bleomycin-induced pulmonary fibrosis. Our data suggest that anlotinib therapy decreases fibrotic markers in vitro and in vivo. The antifibrotic effect of anlotinib is associated with inhibition of PFKFB3-dependent glycolysis, which is posttranscriptionally regulated by the RNA binding protein PCBP3. These findings provide a theoretical basis for the clinical development and application of anlotinib for the treatment of pulmonary fibrosis.
Materials and Methods
Isolation of Primary Mouse Fibroblast Cultures
Normal mouse primary fibroblasts were generated by culturing the lungs of C57BL/6 as previously described (Meng et al., 2014). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, United States) supplemented with 15% fetal bovine serum (FBS, PAN, German). The cells were cultured at 37°C in 5% CO2 and 95% humidity. Unless specifically noted, all experiments were performed with cells at passage 3.
Cell Lines
Human lung fibroblast line IMR90 was purchased from American Type Culture Collection (Manassas, VA). IMR90 were maintained in DMEM supplemented with 10% FBS, 100 units/mL penicillin, and 100 g/ml streptomycin in 5% CO2 and 95% humidity at 37°C.
Western Blotting
Cells or dissected mouse lung tissue samples were lysed in ice-cold RIPA lysis buffer with protease inhibitors. Protein concentrations were determined using a BCA Protein Quantitative Analysis Kit (Fudebio-tech) after which protein samples were separated by 8–12% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were then incubated at room temperature for 1 h in TBST containing 5% BSA. After blocking, the membranes were incubated with primary antibodies for 24 h at 4°C.The following primary antibodies were used: anti-Fibronectin (Abcam, ab268020); anti-Collagen I (affinity, AF7001); anti-alpha smooth muscle (Abcam, ab5694); anti-PFKFB3 (Abcam, ab181861); anti-Beta actin (proteintech, 66009-1-Ig); anti-Hexokinase 2 (proteintech, 22029-1-AP); anti-PKM2 (Proteintech, 15822-1-AP); anti-LDHA (Proteintech, 19987-1-AP); anti-LDHB (Proteintech, 14824-1-AP); and anti-PCBP3 (Abcam, ab154252). Then, the membranes were washed three times with TBST and incubated with donkey anti-rabbit IgG H&L (Abcam, ab175772) for 1 h at room temperature. The membranes were developed using the ECL method according to the manufacturer’s instructions (Millipore) and detected on a GeneGnome XRQ chemiluminescence imaging system (Syngene). ImageJ was used to calculate the relative density of proteins.
Immunofluorescence Staining
The culture medium was washed away with PBS. The cultured cells were fixed with 4% paraformaldehyde for 30 min. Then, the samples were permeabilized with 0.5% Triton X-100 in PBS for 10 min, blocked with 1% BSA in PBS for 1 h at room temperature, and incubated with primary antibodies at 4°C overnight. The primary antibodies included anti- Fibronectin (Abcam, ab268020), anti-alpha smooth muscle (Abcam, ab5694) and anti-PCBP3 (Abcam, ab154252). Then, the cells were washed three times with PBS and incubated with goat anti-rabbit IgG/Alexa Fluor 555-conjugated secondary antibodies (Biosynthesis, bs-0296GA488 and bs-0295G-AF555) for 1 h at room temperature followed by 10 min of DAPI (4’,6-diamidino-2-phenylindole dihydrochloride) staining to visualize cell nuclei visualization as previously described (Chen et al., 2021).
Quantitative RT-PCR (qPCR)
Total RNA was isolated from primary mouse lung fibroblasts using RNA MiniPrep Kits (Zymo Research, R2050). Reverse transcription reactions were performed with a PrimeScriptTM II 1st strand cDNA synthesis Kit (Takara, 6210A/B) according to the manufacturer’s recommendations. qPCR analysis was performed using a HiScript RT- SuperMix for qPCR kit (Vazyme, R223-01) with a CFX96 Touch Real-Time PCR Detection System. The mRNA levels of target genes were normalized to the β-actin mRNA level. Primers used for qPCR are listed in (Table 1).
TABLE 1.
List of primer sequences used in this study.
| Gene | Species | Forward primer | Reverse primer |
|---|---|---|---|
| β-actin | Mus musculus | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
| PFKFB3 | Mus musculus | CCCAGAGCCGGGTACAGAA | GGGGAGTTGGTCAGCTTCG |
Wound-Healing Migration Assay
Cells were seeded in six-well plates and grown until they reach 100% confluence. A “wound” was subsequently created with a sterile 100 μL pipette tip. The cells were pretreated with anlotinib (1 µM) for 3 h and then exposed to TGF-β1 (10 ng/ml) for an additional 24 h. After 24 h, the cells were fixed with 4% paraformaldehyde, and images were obtained using a fluorescence microscope. Wound area can be calculated by manually tracing the cell-free area in captured images using the ImageJ public domain software (NIH, Bethesda, MD).
Cell Proliferation Assay
Cell proliferation was determined by the CCK-8 Kit (Dojindo Laboratories) according to the manufacturer’s instructions. Briefly, 10 μL of CCK-8 solution was added to cultured cells in each well, followed by incubation at 37°C for 1 h. The OD values were measured at 450 nm using a microplate reader. EdU staining was conducted using the BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 594 (Beyotime, Cat. No: C00788L). Cells were washed with PBS. Fresh DMEM was added, and then, 10 µM EdU was added into the medium. The cells were incubated for 2 h at 37°C/5% CO2. After the incubation, the cells were washed with PBS to remove the DMEM and the free EdU probe. The cells were then fixed in 4% paraformaldehyde at room temperature for 30 min before being stained with DAPI for 3 min. After an additional wash in PBS, the cells were observed under Nikon ECLIPSE TS100 (Japan).
Glucose Uptake Assay
Primary mouse lung fibroblasts were pretreated with anlotinib (1 µM) for 3 h and then exposed to TGF-β1 (10 ng/ml) for an additional 24 h. Then, the four types of cells were detached and transferred to a 96-well plate in fresh growth medium at a density of 10,000 cells per well for the direct 2-NBDG glucose uptake assay. The cells were rinsed twice with PBS. Glucose uptake was initiated by the addition of 100 μM 2-NBDG to each well. After 30 min, the medium was removed. The plates were then rinsed with PBS, and the fluorescence was measured at an excitation wavelength of 485 nm and an emission wavelength of 535 nm.
Intracellular and Extracellular Lactate Analysis
To measure lactate production, cells were treated as described for the glucose uptake assay. One hundred thousand cells were then plated into a 12-well plate and incubated in DMEM containing 10% FBS for 10 h. To measure the secretion of lactate, the media were removed, and the cells were incubated in FBS-free DMEM. After incubation for 1 h, the supernatant was collected to measure lactate production (Biovision). The reaction mixture was incubated for 30 min at room temperature in the dark. The lactate levels were measured at 450 nm in a microplate reader and normalized to the protein concentrations. To measure the lactate levels in mouse lung tissue, 10 mg of lung tissues was isolated and homogenized in assay buffer (Biovision). The samples were centrifuged, and the soluble fractions were measured and normalized to the protein concentrations.
Extracellular Acidification Rate
The extracellular acidification rate (ECAR) was measured using the Agilent Seahorse XFp Extracellular Flux Analyzer (Seahorse Bioscience). Experiments were performed according to the manufacturer’s instructions. ECAR were measured using Seahorse XF Glycolysis Stress Test Kit (Agilent Technologies). Briefly, cells were transfected or infected as in glucose uptake assay. The transfected cells were harvested and the cell number was counted. After baseline measurements, glucose, the oxidative phosphorylation inhibitor oligomycin, and the glycolytic inhibitor 2-DG were sequentially injected into each well at the indicated time points. Data were analysed by Seahorse XFp Wave software. ECAR is reported in mpH/minute. The cells in each well were digested by trypsin digestion (Gibco, United States), and count cell numbers by cell counting chamber. The results were normalized to normalized to cell number in each well.
RNA Immunoprecipitation (RIP)
Cells were rinsed twice with ice-cold PBS and lysed with an equal pellet volume of RIPA-2 buffer. Protein-A Dynabeads (Invitrogen) were incubated with either mouse IgG or FLAG antibody (Abcam, ab205606). Beads coated in antibody were resuspended in NT2 buffer. Thawed and clarified lysates were added and the bead/antibody/lysate mixture was incubated at 4°C overnight rotating end-over-end. Beads were washed with cold NT2 buffer five times. Proteinase K treatment released RNAs from bound proteins and input and bound RNA was isolated with TRIzol (Invitrogen) and reverse transcribed as described above.
Polysome Analysis
Cells were transfected with empty vector or Flag-PCBP3 and incubated with 100 g/ml cycloheximide for 10 min and lysed with polysome extraction buffer containing 20 mM Tris–HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2 and 0.5% NP-40 as previously described (Kim et al., 2015). Cytoplasmic lysates were fractionated by ultracentrifugation through 10–50% linear sucrose gradients and divided into 12 fractions. The total RNA in each fraction was extracted and analyzed by quantitative RT-PCR analysis.
Overexpression Experiments and RNA Interference
The Plasmid vector encoding PCBP3 and the empty vector were purchased from Hanbio (Shanghai, China). Primary mouse lung fibroblasts were cultured in six well plates (105 cells/well) and added with 2.5 μg of target plasmid per well. After 12 h, the transfection medium was changed to normal medium. Effects of overexpression on mRNA and protein levels were examined 36 h later. The siRNA targeting mouse PFKFB3 (PFKFB3 siRNA: 5′- CCUCUUGACCCUGAUAAAUTT-3′) were synthesized by Genepharma Co. (Shanghai, China). Primary mouse lung fibroblasts were cultured in six well plates (105 cells/well) and transfected using Lipofectamine 3,000 (Invitrogen, CA) with PFKFB3 siRNA or negative control siRNA (NC siRNA) for 48 h following the manufacturer’s instructions.
Animal Experiments
All experiments were conducted in accordance with protocols approved by the Southern Medical University Institutional Animal Care and Use Committee. Female mice (C57BL/6), 6–8 weeks of age, were purchased from Southern Medical University. The mice were kept on a 12 h light-dark cycle with free access to food and water. For bleomycin administration, the mice were anesthetized with 2, 2, 2-tribromoethanol (Sigma-Aldrich), followed by intratracheal instillation of BLM (5 U/kg, i. t.) in 50 μL phosphate-buffered saline (PBS) or equally volume PBS for 21 days. The mice were administered dimethyl sulfoxide (DMSO) (control group) or anlotinib (1 mg/kg, i. p.) once daily for 21 consecutive days. Further experiments were designed to measure the effects of delayed anlotinib administration. Anlotinib treatment was initiated 1 week after exposure to bleomycin, and the mice were administered with anlotinib (1 or 2 mg/kg/day) for 2 weeks, and the mice were sacrificed at day 21. The lungs were harvested for further analyses.
Pulmonary Function Test
At endpoint, at least 5 mice from each group were anesthetized with 2,2,2-tribromoethanol in saline, tracheotomized below the larynx, and intubated with a tracheal cannula. After the surgery, the mice were placed inside the plethysmographic chamber and the cannula was connected to the machine. Pulmonary function was measured by pulmonary function test system (BUXCO, United States). The system’s software automatically records and displays the pulmonary function parameters.
Hydroxyproline Assay
Lung collagen content was measured with a hydroxyproline (HYP) kit (Nanjing Jian Cheng Institute, Nanjing, China). The lung tissues were prepared for hydrolysis, adjusting the PH value to 6.0–6.8. Subsequently, the developing solution was added to the tissues that were incubated at 37°C for 5 min. Absorbance was read at 550 nm using a microplate reader. Data were expressed as micrograms (µg) of HYP per mg of wet lung tissue.
Materials
TGF-β1 were purchased from R&D Systems, Inc. (Minneapolis, MN, United States). Anlotinib dihydrochloride (AL3818, S8726) were purchased from Selleck (Houston, TX, United States).
Statistical Analysis
The results are expressed as the means ± standard deviation (SD). Multigroup comparisons were performed using one-way ANOVA. Student’s t-test was used for comparisons between two groups. A p value of less than 0.05 was considered significant. Replicates consisted of at least three independent experiments. Analyses were performed on SPSS version 25.0 (IBM) for Windows and GraphPad Prism version 6.0 (GraphPad Software, CA).
Results
Anlotinib Represses Myofibroblast Activation and the Profibrogenic Phenotype in vitro
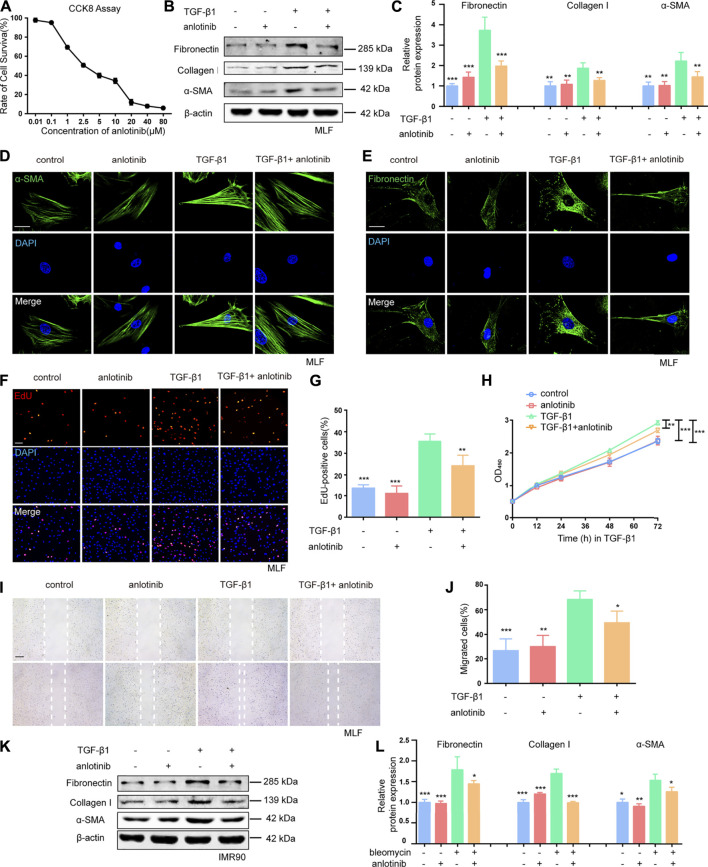
Given that TGF-β1 is the predominant cytokine that stimulates the differentiation of lung fibroblasts into myofibroblasts and induces ECM production (Sapudom et al., 2015; Huang et al., 2020), we examined the effect of anlotinib (the chemical structure is shown in Supplementary Figure S1) on TGF-β1-induced activation of primary mouse lung fibroblasts (MLFs). The CCK-8 assay results showed that anlotinib did not cause significant cytotoxicity at doses of 1 µM (Figure 1A). To mimic the inhibitory effect of anlotinib on the progression of lung fibrosis, primary MLFs were pretreated with anlotinib (1 µM) for 3 h and then exposed to TGF-β1 (10 ng/ml) for an additional 24 h. Our results demonstrated that TGF-β1 induced the expression of fibronectin, collagen I, and α-SMA, but anlotinib reversed the expression of these fibrotic markers (Figures 1B,C). Immunofluorescence analysis of α-SMA and fibronectin showed similar results (Figures 1D,E). We also examined whether anlotinib affected the proliferation and migration of fibroblasts, which have been shown to significantly contribute to many fibrotic pathologies (Jarman et al., 2014; Huang et al., 2020). As shown by the EdU (Figures 1F,G) and CCK-8 results (Figure 1H), anlotinib treatment prevented the TGF-β1-induced proliferation of primary MLFs. Moreover, anlotinib inhibited the TGF-β1-induced migration of fibroblasts (Figures 1I,J). These results were confirmed in the human IMR90 cell line (Figures 1K, L and Supplementary Figures S2A–D). These data indicate that anlotinib can repress myofibroblast activation and the profibrogenic phenotype in vitro.
FIGURE 1.
Anlotinib represses myofibroblast activation and the profibrogenic phenotype in vitro.(A) Dose-dependent cytotoxicity of anlotinib in primary mouse lung fibroblast isolated from healthy mice by CCK-8. (B) Western blots analysis of Fibronectin, Collagen I, α-SMA and β-actin in mouse lung fibroblasts treated with anlotinib (1 µM) for 3 h and then exposed to TGF-β1 (10 ng/ml) for 3 h, followed by TGF-β1 for an additional 24 h. (C) Quantification for the indicated protein (mean ± SD, n = 3). Immunofluorescence for α-SMA (green) (D) and Fibronectin (green) (E). DAPI-stained nuclei (blue). Scale bar, 25 µm. (F) An EdU assay was used to observe the proliferative cells. Scale bar, 100 µm. (G) The number of EdU-positive cells was recorded (mean ± SD, n = 3). (H) A cck8 assay was used to observe the proliferative cells. (I) Representative images to show scratch-wound assay. Scale bars, 100 µm. Experiments were performed as in B. Images were taken 0, and 24 h after assay (white lines indicate wound edge). (J) Quantitative analysis of migration distance (mean ± SD, n = 3). (K) Western blots analysis of Fibronectin, Collagen I, α-SMA and β-actin in IMR90 cells treated with anlotinib (1 µM) for 3 h and then exposed to TGF-β1 (10 ng/ml) for an additional 24 h. (L) Quantification for the indicated proteins in IMR90 cells (mean ± SD, n = 3). *p < 0.05, **p < 0.01, ***p < 0.001 VS TGF-β1-treated group (ANOVA).
Anlotinib Inhibits PFKFB3-Driven Glycolysis in Lung Myofibroblasts
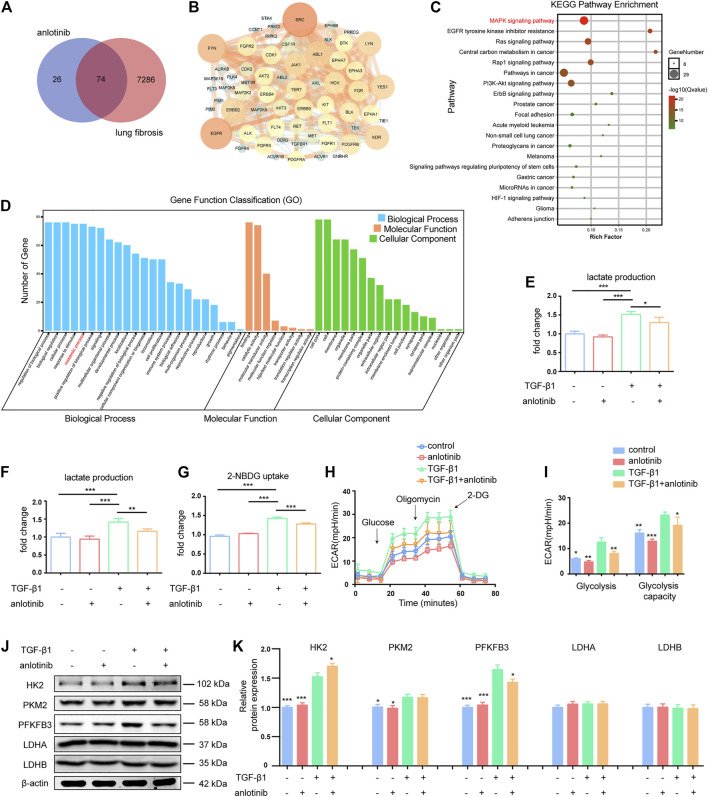
To investigate the potential antifibrotic mechanisms of anlotinib, we estimated the most likely macromolecular targets of anlotinib and obtained 100 potential targets through SwissTargetPrediction (Table 2) (Gfeller et al., 2014). A total of 7,360 lung fibrosis-related targets were obtained from the GeneCards database (Table 3) (Safran et al., 2002). To clarify the interaction between potential anlotinib targets and lung fibrosis-related targets, the intersection of the targets was mapped by drawing a Venn diagram and constructing a target network (Figure 2A). Seventy-four common targets were shared between the potential anlotinib targets and lung fibrosis-related targets (Table 4). STRING (version 11.0) was used for protein-protein interaction (PPI) analysis (Figure 2B) (Szklarczyk et al., 2019). Bioinformatics analysis data identified mitogen-activated protein kinase (MAPK) signaling pathway-related genes as the top hits among the 74 genes (Figure 2C). Given that the MAPK pathway is well recognized as a metabolic regulator and that many of these genes control metabolic processes (Figure 2D) (Ho et al., 2004; Papa et al., 2019; Hu et al., 2020; Wang F. et al., 2020), we first confirmed the presence of glycolytic alterations in lung myofibroblasts. We directly measured the levels of lactate and found that both intracellular and extracellular lactate levels in lung myofibroblasts treated with TGF-β1 were significantly increased (Figures 2E,F). Consistent with the augmented glycolysis in lung myofibroblasts, these cells also demonstrated increased glucose consumption (Figure 2G). However, anlotinib treatment decreased the production and secretion of lactate and reduced the consumption of glucose (Figures 2E–G). Accordingly, extracellular acidification rate (ECAR) analysis indicated that treatment with TGF-β1 increased glycolysis and glycolytic activity in primary MLFs, both of which were also reduced by anlotinib (Figures 2H,I). To delineate the mechanisms by which anlotinib inhibits the augmented glycolysis observed in lung myofibroblasts, we assessed the expression of key glycolytic enzymes in these cells. We found that PFKFB3 was induced by TGF-β1 in lung fibroblasts and that anlotinib significantly decreased its expression at the protein level (Figures 2J,K). PFKFB3 is not a rate-limiting glycolytic enzyme; instead, PFKFB3 catalyzes the conversion of fructose-6-phosphate to fructose-2,6-bisphosphate, which is an allosteric activator of PFK1 and a potent stimulator of glycolysis (Atsumi et al., 2002; De Bock et al., 2013). Taken together, these data suggest that anlotinib can abrogate the PFKFB3-driven increase in glycolysis, participating in myofibroblast activation.
TABLE 2.
Targets of anlotinib obtained through SwissTargetPrediction.
| Target | Common name | Uniprot ID | ChEMBL ID | Target class | Probability* | Known actives (3D/2D) |
|---|---|---|---|---|---|---|
| Tyrosine-protein kinase receptor UFO | AXL | P30530 | CHEMBL4895 | Kinase | 0.106165761464 | 72/8 |
| Tyrosine-protein kinase receptor TYRO3 | TYRO3 | Q06418 | CHEMBL5314 | Kinase | 0.106165761464 | 53/3 |
| Proto-oncogene tyrosine-protein kinase MER | MERTK | Q12866 | CHEMBL5331 | Kinase | 0.106165761464 | 46/3 |
| Receptor protein-tyrosine kinase erbB-2 | ERBB2 | P04626 | CHEMBL1824 | Kinase | 0.106165761464 | 178/5 |
| Tyrosine-protein kinase BRK | PTK6 | Q13882 | CHEMBL4601 | Kinase | 0.106165761464 | 22/2 |
| Serine/threonine-protein kinase Aurora-B | AURKB | Q96GD4 | CHEMBL2185 | Kinase | 0.106165761464 | 143/14 |
| Vascular endothelial growth factor receptor 1 | FLT1 | P17948 | CHEMBL1868 | Kinase | 0.106165761464 | 79/18 |
| Epidermal growth factor receptor erbB1 | EGFR | P00533 | CHEMBL203 | Kinase | 0.106165761464 | 597/31 |
| Vascular endothelial growth factor receptor 2 | KDR | P35968 | CHEMBL279 | Kinase | 0.106165761464 | 485/95 |
| Tyrosine-protein kinase SRC | SRC | P12931 | CHEMBL267 | Kinase | 0.106165761464 | 448/42 |
| Hepatocyte growth factor receptor | MET | P08581 | CHEMBL3717 | Kinase | 0.106165761464 | 314/108 |
| Serine/threonine-protein kinase GAK | GAK | O14976 | CHEMBL4355 | Kinase | 0.106165761464 | 16/2 |
| Kinesin-1 heavy chain/Tyrosine-protein kinase receptor RET | RET | P07949 | CHEMBL2041 | Kinase | 0.106165761464 | 72/11 |
| ALK tyrosine kinase receptor | ALK | Q9UM73 | CHEMBL4247 | Kinase | 0.106165761464 | 207/3 |
| Tyrosine-protein kinase ABL | ABL1 | P00519 | CHEMBL1862 | Kinase | 0.106165761464 | 127/11 |
| Stem cell growth factor receptor | KIT | P10721 | CHEMBL1936 | Kinase | 0.106165761464 | 106/10 |
| Activin receptor type-1 | ACVR1 | Q04771 | CHEMBL5903 | Kinase | 0.106165761464 | 29/1 |
| Vascular endothelial growth factor receptor 3 | FLT4 | P35916 | CHEMBL1955 | Kinase | 0.106165761464 | 37/8 |
| Tyrosine-protein kinase receptor FLT3 | FLT3 | P36888 | CHEMBL1974 | Kinase | 0.106165761464 | 156/14 |
| Platelet-derived growth factor receptor alpha | PDGFRA | P16234 | CHEMBL2007 | Kinase | 0.106165761464 | 57/14 |
| Fibroblast growth factor receptor 1 | FGFR1 | P11362 | CHEMBL3650 | Kinase | 0.106165761464 | 189/9 |
| TGF-beta receptor type I | TGFBR1 | P36897 | CHEMBL4439 | Kinase | 0.106165761464 | 78/4 |
| Tyrosine-protein kinase LCK | LCK | P06239 | CHEMBL258 | Kinase | 0.106165761464 | 167/19 |
| Tyrosine-protein kinase BTK | BTK | Q06187 | CHEMBL5251 | Kinase | 0.106165761464 | 67/5 |
| Tyrosine-protein kinase Lyn | LYN | P07948 | CHEMBL3905 | Kinase | 0.106165761464 | 76/6 |
| Fibroblast growth factor receptor 3 | FGFR3 | P22607 | CHEMBL2742 | Kinase | 0.106165761464 | 37/3 |
| Platelet-derived growth factor receptor beta | PDGFRB | P09619 | CHEMBL1913 | Kinase | 0.106165761464 | 114/8 |
| Tyrosine-protein kinase YES | YES1 | P07947 | CHEMBL2073 | Kinase | 0.106165761464 | 34/3 |
| Fibroblast growth factor receptor 2 | FGFR2 | P21802 | CHEMBL4142 | Kinase | 0.106165761464 | 35/27 |
| Tyrosine-protein kinase FGR | FGR | P09769 | CHEMBL4454 | Kinase | 0.106165761464 | 17/4 |
| Macrophage colony stimulating factor receptor | CSF1R | P07333 | CHEMBL1844 | Kinase | 0.106165761464 | 149/7 |
| Tyrosine-protein kinase BLK | BLK | P51451 | CHEMBL2250 | Kinase | 0.106165761464 | 20/4 |
| Serine/threonine-protein kinase PLK4 | PLK4 | O00444 | CHEMBL3788 | Kinase | 0.106165761464 | 14/2 |
| Ephrin receptor | EPHB4 | P54760 | CHEMBL5147 | Kinase | 0.106165761464 | 30/3 |
| Tyrosine-protein kinase FYN | FYN | P06241 | CHEMBL1841 | Kinase | 0.106165761464 | 36/3 |
| Dual specificity mitogen-activated protein kinase kinase 2 | MAP2K2 | P36507 | CHEMBL2964 | Kinase | 0.106165761464 | 11/3 |
| Tyrosine-protein kinase HCK | HCK | P08631 | CHEMBL3234 | Kinase | 0.106165761464 | 30/4 |
| Serine/threonine-protein kinase 10 | STK10 | O94804 | CHEMBL3981 | Kinase | 0.106165761464 | 13/4 |
| Tyrosine-protein kinase ABL2 | ABL2 | P42684 | CHEMBL4014 | Kinase | 0.106165761464 | 13/4 |
| Tyrosine-protein kinase TIE-2 | TEK | Q02763 | CHEMBL4128 | Kinase | 0.106165761464 | 24/16 |
| Ephrin type-A receptor 8 | EPHA8 | P29322 | CHEMBL4134 | Kinase | 0.106165761464 | 9/3 |
| Serine/threonine-protein kinase 2 | SLK | Q9H2G2 | CHEMBL4202 | Kinase | 0.106165761464 | 17/4 |
| Tyrosine-protein kinase FRK | FRK | P42685 | CHEMBL4223 | Kinase | 0.106165761464 | 15/3 |
| Ephrin type-A receptor 6 | EPHA6 | Q9UF33 | CHEMBL4526 | Kinase | 0.106165761464 | 10/4 |
| TRAF2- and NCK-interacting kinase | TNIK | Q9UKE5 | CHEMBL4527 | Kinase | 0.106165761464 | 20/4 |
| Serine/threonine-protein kinase MST1 | STK4 | Q13043 | CHEMBL4598 | Kinase | 0.106165761464 | 11/3 |
| Mitogen-activated protein kinase kinase kinase kinase 5 | MAP4K5 | Q9Y4K4 | CHEMBL4852 | Kinase | 0.106165761464 | 12/4 |
| Casein kinase I epsilon | CSNK1E | P49674 | CHEMBL4937 | Kinase | 0.106165761464 | 12/3 |
| Ephrin type-A receptor 3 | EPHA3 | P29320 | CHEMBL4954 | Kinase | 0.106165761464 | 11/3 |
| Tyrosine-protein kinase receptor Tie-1 | TIE1 | P35590 | CHEMBL5274 | Kinase | 0.106165761464 | 11/4 |
| Mitogen-activated protein kinase kinase kinase kinase 3 | MAP4K3 | Q8IVH8 | CHEMBL5432 | Kinase | 0.106165761464 | 14/3 |
| Serine/threonine-protein kinase SIK2 | SIK2 | Q9H0K1 | CHEMBL5699 | Kinase | 0.106165761464 | 12/3 |
| Mitogen-activated protein kinase kinase kinase kinase 1 | MAP4K1 | Q92918 | CHEMBL5749 | Kinase | 0.106165761464 | 14/3 |
| Serine/threonine-protein kinase 33 | STK33 | Q9BYT3 | CHEMBL6005 | Kinase | 0.106165761464 | 16/4 |
| Mitogen-activated protein kinase kinase kinase kinase 4 | MAP4K4 | O95819 | CHEMBL6166 | Kinase | 0.106165761464 | 17/4 |
| Serine/threonine-protein kinase TAO2 | TAOK2 | Q9UL54 | CHEMBL1075195 | Kinase | 0.106165761464 | 7/3 |
| Serine/threonine-protein kinase TAO3 | TAOK3 | Q9H2K8 | CHEMBL5701 | Kinase | 0.106165761464 | 12/2 |
| Voltage-gated calcium channel alpha2/delta subunit 1 | CACNA2D1 | P54289 | CHEMBL1919 | Calcium channel auxiliary subunit alpha2delta family | 0.106165761464 | 15/0 |
| Voltage-gated calcium channel alpha2/delta subunit 2 | CACNA2D2 | Q9NY47 | CHEMBL3896 | Calcium channel auxiliary subunit alpha2delta family | 0.106165761464 | 4/0 |
| Receptor protein-tyrosine kinase erbB-4 | ERBB4 | Q15303 | CHEMBL3009 | Kinase | 0.106165761464 | 29/3 |
| Mitogen-activated protein kinase kinase kinase 8 | MAP3K8 | P41279 | CHEMBL4899 | Kinase | 0.106165761464 | 32/0 |
| Serine/threonine-protein kinase/endoribonuclease IRE1 | ERN1 | O75460 | CHEMBL1163101 | Enzyme | 0.106165761464 | 18/1 |
| Macrophage-stimulating protein receptor | MST1R | Q04912 | CHEMBL2689 | Kinase | 0.106165761464 | 8/11 |
| Serine/threonine-protein kinase Aurora-C | AURKC | Q9UQB9 | CHEMBL3935 | Kinase | 0.106165761464 | 15/2 |
| Fibroblast growth factor receptor 4 | FGFR4 | P22455 | CHEMBL3973 | Kinase | 0.106165761464 | 15/2 |
| Ephrin type-A receptor 7 | EPHA7 | Q15375 | CHEMBL4602 | Kinase | 0.106165761464 | 7/3 |
| Dual specificity mitogen-activated protein kinase kinase 5 | MAP2K5 | Q13163 | CHEMBL4948 | Kinase | 0.106165761464 | 11/4 |
| Serine/threonine-protein kinase RIPK2 | RIPK2 | O43353 | CHEMBL5014 | Kinase | 0.106165761464 | 6/4 |
| Discoidin domain-containing receptor 2 | DDR2 | Q16832 | CHEMBL5122 | Kinase | 0.106165761464 | 13/3 |
| Activin receptor type-1B | ACVR1B | P36896 | CHEMBL5310 | Kinase | 0.106165761464 | 6/1 |
| Epithelial discoidin domain-containing receptor 1 | DDR1 | Q08345 | CHEMBL5319 | Kinase | 0.106165761464 | 14/4 |
| Mitogen-activated protein kinase kinase kinase kinase 2 | MAP4K2 | Q12851 | CHEMBL5330 | Kinase | 0.106165761464 | 15/3 |
| Misshapen-like kinase 1 | MINK1 | Q8N4C8 | CHEMBL5518 | Kinase | 0.106165761464 | 14/4 |
| Leukocyte tyrosine kinase receptor | LTK | P29376 | CHEMBL5627 | Kinase | 0.106165761464 | 10/3 |
| Serine/threonine-protein kinase 35 | STK35 | Q8TDR2 | CHEMBL5651 | Kinase | 0.106165761464 | 9/4 |
| Ephrin type-A receptor 1 | EPHA1 | P21709 | CHEMBL5810 | Kinase | 0.106165761464 | 9/3 |
| Ephrin receptor | EPHB6 | O15197 | CHEMBL5836 | Unclassified protein | 0.106165761464 | 11/4 |
| Receptor tyrosine-protein kinase erbB-3 | ERBB3 | P21860 | CHEMBL5838 | Kinase | 0.106165761464 | 4/3 |
| Serine/threonine-protein kinase MST4 | STK26 | Q9P289 | CHEMBL5941 | Kinase | 0.106165761464 | 12/2 |
| Eukaryotic translation initiation factor 2-alpha kinase 1 | EIF2AK1 | Q9BQI3 | CHEMBL6029 | Kinase | 0.106165761464 | 3/3 |
| SPS1/STE20-related protein kinase YSK4 | MAP3K19 | Q56UN5 | CHEMBL6191 | Kinase | 0.106165761464 | 15/4 |
| Serine/threonine-protein kinase AKT2 | AKT2 | P31751 | CHEMBL2431 | Kinase | 0.106165761464 | 318/0 |
| Protein kinase C gamma | PRKCG | P05129 | CHEMBL2938 | Kinase | 0.106165761464 | 38/0 |
| Serine/threonine-protein kinase AKT | AKT3 | Q9Y243 | CHEMBL4816 | Kinase | 0.106165761464 | 73/0 |
| Serine/threonine-protein kinase PIM1 | PIM1 | P11309 | CHEMBL2147 | Kinase | 0.106165761464 | 638/0 |
| Serine/threonine-protein kinase PIM2 | PIM2 | Q9P1W9 | CHEMBL4523 | Kinase | 0.106165761464 | 448/0 |
| Serine/threonine-protein kinase PIM3 | PIM3 | Q86V86 | CHEMBL5407 | Kinase | 0.106165761464 | 331/0 |
| Cyclin-dependent kinase 2/cyclin E1 | CCNE1 CDK2 | P24864 P24941 | CHEMBL1907605 | Kinase | 0.106165761464 | 74/0 |
| Tyrosine-protein kinase JAK1 | JAK1 | P23458 | CHEMBL2835 | Kinase | 0.106165761464 | 137/0 |
| Dipeptidyl peptidase VIII | DPP8 | Q6V1X1 | CHEMBL4657 | Protease | 0.106165761464 | 346/0 |
| Dipeptidyl peptidase IX | DPP9 | Q86TI2 | CHEMBL4793 | Protease | 0.106165761464 | 239/0 |
| Phosphodiesterase 4B | PDE4B | Q07343 | CHEMBL275 | Phosphodiesterase | 0.106165761464 | 43/0 |
| Protein kinase C iota | PRKCI | P41743 | CHEMBL2598 | Kinase | 0.106165761464 | 287/0 |
| Cyclin-dependent kinase 2 | CDK2 | P24941 | CHEMBL301 | Kinase | 0.106165761464 | 170/0 |
| Cyclin-dependent kinase 1 | CDK1 | P06493 | CHEMBL308 | Kinase | 0.106165761464 | 146/0 |
| Cyclin T1 | CCNT1 | O60563 | CHEMBL2108 | Other cytosolic protein | 0.106165761464 | 111/0 |
| Telomerase reverse transcriptase | TERT | O14746 | CHEMBL2916 | Enzyme | 0.106165761464 | 79/0 |
| Sodium/calcium exchanger 1 | SLC8A1 | P32418 | CHEMBL4076 | Electrochemical transporter | 0.106165761464 | 44/0 |
| Gonadotropin-releasing hormone receptor | GNRHR | P30968 | CHEMBL1855 | Family A G protein-coupled receptor | 0.106165761464 | 431/0 |
| Amine oxidase, copper containing | AOC3 | Q16853 | CHEMBL3437 | Enzyme | 0.106165761464 | 19/0 |
TABLE 3.
Lung fibrosis-related targets obtained from the GeneCards database.
| Number | Gene | Number | Gene | Number | Gene | Number | Gene |
|---|---|---|---|---|---|---|---|
| 1 | CFTR | 335 | PDGFA | 669 | CPLANE1 | 1,003 | HSPH1 |
| 2 | TGFB1 | 336 | MIR197 | 670 | MKI67 | 1,004 | MT-ATP6 |
| 3 | TERT | 337 | MIRLET7B | 671 | TBX1 | 1,005 | MIR212 |
| 4 | TP53 | 338 | MIR195 | 672 | PI3 | 1,006 | CASP10 |
| 5 | TNF | 339 | MIR96 | 673 | BMP4 | 1,007 | PDE5A |
| 6 | SFTPC | 340 | CHI3L1 | 674 | PMS2 | 1,008 | SH2D1A |
| 7 | EGFR | 341 | MIRLET7E | 675 | SLC7A7 | 1,009 | SCN5A |
| 8 | IL6 | 342 | MIR16-1 | 676 | E2F1 | 1,010 | RSPH4A |
| 9 | MUC5B | 343 | MIR199A1 | 677 | S100A1 | 1,011 | PIK3R2 |
| 10 | RTEL1 | 344 | MIR210 | 678 | TKT | 1,012 | DNAH9 |
| 11 | IL10 | 345 | MIR20A | 679 | MYRF | 1,013 | PSMA7 |
| 12 | SFTPB | 346 | MIR378A | 680 | LAMA5 | 1,014 | KIF3A |
| 13 | SFTPA1 | 347 | THBD | 681 | COL2A1 | 1,015 | CYP3A5 |
| 14 | ELN | 348 | DMBT1 | 682 | DEFB1 | 1,016 | LOC111674466 |
| 15 | ABCA3 | 349 | MIR185 | 683 | RPL5 | 1,017 | ADM |
| 16 | CAV1 | 350 | IL9 | 684 | SOD1 | 1,018 | PSMC3 |
| 17 | IFNG | 351 | CFM1 | 685 | PLK1 | 1,019 | WDPCP |
| 18 | MUC1 | 352 | CD79A | 686 | SCGB3A2 | 1,020 | MIR101-1 |
| 19 | SFTPA2 | 353 | DYNC2LI1 | 687 | SLC9A3R1 | 1,021 | PRKCB |
| 20 | SERPINA1 | 354 | NEK1 | 688 | IREB2 | 1,022 | IL13RA2 |
| 21 | STAT3 | 355 | IQCB1 | 689 | LOC113664106 | 1,023 | MYH11 |
| 22 | TERC | 356 | MIR204 | 690 | FADD | 1,024 | PRKG2 |
| 23 | CCN2 | 357 | HLA-A | 691 | CYP3A4 | 1,025 | SUFU |
| 24 | IL13 | 358 | IGF2 | 692 | HSPB1 | 1,026 | PIK3C2A |
| 25 | TLR4 | 359 | FLNA | 693 | MAPK14 | 1,027 | PSMC5 |
| 26 | PARN | 360 | BCL2 | 694 | CTSB | 1,028 | KCNQ1OT1 |
| 27 | CTNNB1 | 361 | GSN | 695 | IGF2R | 1,029 | MIR133A1 |
| 28 | CCR6 | 362 | MAP2K2 | 696 | BAP1 | 1,030 | GOPC |
| 29 | AKT1 | 363 | BPIFA1 | 697 | GATA2 | 1,031 | TRAF2 |
| 30 | MIR21 | 364 | MIR15A | 698 | MMP8 | 1,032 | TNFRSF6B |
| 31 | NKX2-1 | 365 | EPHX1 | 699 | ITGAV | 1,033 | MIR139 |
| 32 | MMP1 | 366 | HPS5 | 700 | PTX3 | 1,034 | LOC111674470 |
| 33 | SMAD4 | 367 | TRAF3IP1 | 701 | MSLN | 1,035 | IGHMBP2 |
| 34 | ACE | 368 | CFTR-AS1 | 702 | AFF4 | 1,036 | PSMA3 |
| 35 | SPP1 | 369 | AP3B1 | 703 | CTCF | 1,037 | PSMC6 |
| 36 | CXCL8 | 370 | GAPDH | 704 | ADA | 1,038 | GADD45B |
| 37 | EGF | 371 | KRT19 | 705 | KITLG | 1,039 | FKRP |
| 38 | FASLG | 372 | MIR107 | 706 | ZNF423 | 1,040 | DNAAF2 |
| 39 | CDH1 | 373 | CP | 707 | IL12A | 1,041 | ERCC5 |
| 40 | KRAS | 374 | HLA-B | 708 | PRKCA | 1,042 | E2F3 |
| 41 | SERPINE1 | 375 | MMP12 | 709 | EGR1 | 1,043 | PIK3R3 |
| 42 | BRAF | 376 | AGER | 710 | DLL4 | 1,044 | PRF1 |
| 43 | SMAD3 | 377 | DDR1 | 711 | IFNA1 | 1,045 | IFNAR1 |
| 44 | PRTN3 | 378 | TNNT2 | 712 | EZR | 1,046 | PF4 |
| 45 | PIK3CA | 379 | CCL18 | 713 | LGALS3 | 1,047 | MIR27B |
| 46 | IL1RN | 380 | MIR148B | 714 | AFP | 1,048 | VTN |
| 47 | HMOX1 | 381 | MIR141 | 715 | TUBB2B | 1,049 | DNAJC5 |
| 48 | IL1B | 382 | INS | 716 | EPCAM | 1,050 | BGLAP |
| 49 | HLA-DRB1 | 383 | GLI3 | 717 | APOB | 1,051 | MIR455 |
| 50 | FAM13A | 384 | JAK2 | 718 | SPPL2C | 1,052 | LOC111674471 |
| 51 | FAS | 385 | EVC | 719 | CDK2 | 1,053 | CSF3R |
| 52 | PTEN | 386 | HPS3 | 720 | GDF2 | 1,054 | VCL |
| 53 | SFTPD | 387 | IL33 | 721 | EPO | 1,055 | NAGLU |
| 54 | ITGAM | 388 | NOS2 | 722 | VCP | 1,056 | GUSB |
| 55 | DSP | 389 | SLPI | 723 | HSPA8 | 1,057 | NEAT1 |
| 56 | STN1 | 390 | MIR133B | 724 | ATP4A | 1,058 | TBX4 |
| 57 | TGFBR1 | 391 | TCTN3 | 725 | HBB | 1,059 | PSMD1 |
| 58 | SRC | 392 | TLR5 | 726 | SELE | 1,060 | PSMD12 |
| 59 | CDKN2A | 393 | MUC4 | 727 | NAT2 | 1,061 | GAST |
| 60 | ERBB2 | 394 | MIR93 | 728 | FGF10 | 1,062 | MIR99A |
| 61 | CTLA4 | 395 | MIR18A | 729 | BMP7 | 1,063 | CREB1 |
| 62 | NFE2L2 | 396 | SLC11A1 | 730 | ETS1 | 1,064 | TRPC6 |
| 63 | MMP9 | 397 | PSMA6 | 731 | DEFB4A | 1,065 | EPAS1 |
| 64 | MTOR | 398 | RNASE3 | 732 | PRDM10 | 1,066 | MIR125B1 |
| 65 | HRAS | 399 | SMAD2 | 733 | GJA1 | 1,067 | IFNGR1 |
| 66 | TINF2 | 400 | ICAM1 | 734 | RPS6KB1 | 1,068 | PGR |
| 67 | FGFR1 | 401 | TGFA | 735 | JAK3 | 1,069 | TLR1 |
| 68 | COL1A1 | 402 | RPGR | 736 | TP63 | 1,070 | CD40 |
| 69 | NPHP3 | 403 | ANXA5 | 737 | HNF1B | 1,071 | DLK1 |
| 70 | F2 | 404 | CXCR1 | 738 | BBS10 | 1,072 | FOS |
| 71 | DPP9 | 405 | IL2 | 739 | ITGA2 | 1,073 | DNAL1 |
| 72 | TGFB2 | 406 | GREM1 | 740 | HOTAIR | 1,074 | MAD1L1 |
| 73 | ATP11A | 407 | CD274 | 741 | SGK1 | 1,075 | PSMD11 |
| 74 | HFE | 408 | MIR142 | 742 | DNAAF1 | 1,076 | HBA2 |
| 75 | PDGFRB | 409 | GSTP1 | 743 | DNMT1 | 1,077 | RAB11B |
| 76 | ITGA3 | 410 | PHOX2A | 744 | DAB2 | 1,078 | CCKBR |
| 77 | MIRLET7D | 411 | SOS2 | 745 | CDH2 | 1,079 | CLCN3 |
| 78 | MBL2 | 412 | LMNA | 746 | HJV | 1,080 | PPARA |
| 79 | NOTCH1 | 413 | ENPP2 | 747 | PRKAA1 | 1,081 | MB |
| 80 | MET | 414 | TOP1 | 748 | HSPA5 | 1,082 | GAS5-AS1 |
| 81 | TIMP1 | 415 | APEX1 | 749 | CHUK | 1,083 | LOC111674474 |
| 82 | SERPINH1 | 416 | CXCL9 | 750 | SKP2 | 1,084 | KCNJ1 |
| 83 | SCNN1A | 417 | PLAU | 751 | ILK | 1,085 | KIAA0319L |
| 84 | MMP2 | 418 | MIRLET7C | 752 | ADORA2B | 1,086 | PSMA2 |
| 85 | ELANE | 419 | MIR30E | 753 | SST | 1,087 | PSMD3 |
| 86 | VEGFA | 420 | MIR214 | 754 | CFLAR | 1,088 | LOC111674473 |
| 87 | PLG | 421 | MIR486-1 | 755 | CCNB1 | 1,089 | ABCF2 |
| 88 | MIR130A | 422 | KRT7 | 756 | ADCY10 | 1,090 | PSMD8 |
| 89 | ADRB2 | 423 | TNFRSF13B | 757 | HDAC9 | 1,091 | MIR590 |
| 90 | KIT | 424 | MIR106B | 758 | ARG1 | 1,092 | PSMD7 |
| 91 | CC2D2A | 425 | MIR181A1 | 759 | BAK1 | 1,093 | RNF5 |
| 92 | TGFBR2 | 426 | MTHFR | 760 | DICER1 | 1,094 | GLIS3 |
| 93 | PDGFRA | 427 | BTNL2 | 761 | IKBKG | 1,095 | CHAT |
| 94 | IL4 | 428 | PTPN11 | 762 | MYH6 | 1,096 | LOC111674476 |
| 95 | NHP2 | 429 | KDR | 763 | COL4A3 | 1,097 | MIR23B |
| 96 | STK11 | 430 | ELMOD2 | 764 | FGA | 1,098 | MIR130B |
| 97 | NOP10 | 431 | STING1 | 765 | ABL1 | 1,099 | MIR151A |
| 98 | ALOX5 | 432 | THY1 | 766 | GJA5 | 1,100 | LOC111674478 |
| 99 | TMEM67 | 433 | MIR140 | 767 | ARHGAP31 | 1,101 | CD82 |
| 100 | CCL2 | 434 | VCAM1 | 768 | FCGR3B | 1,102 | E2F2 |
| 101 | MIR34C | 435 | CCR2 | 769 | SLC26A9 | 1,103 | RNF185 |
| 102 | STAT1 | 436 | CDKN1A | 770 | LRBA | 1,104 | MIR423 |
| 103 | ALB | 437 | STAT4 | 771 | PMM2 | 1,105 | DEFB103B |
| 104 | HGF | 438 | CEP164 | 772 | ACD | 1,106 | NFKB2 |
| 105 | RPGRIP1L | 439 | MIR128-2 | 773 | CHIT1 | 1,107 | MIR186 |
| 106 | NPHP1 | 440 | S100A9 | 774 | CDK6 | 1,108 | AFAP1-AS1 |
| 107 | TSC2 | 441 | TLR3 | 775 | CSF1 | 1,109 | PSMC2 |
| 108 | MDM2 | 442 | SERPINA3 | 776 | ABCG2 | 1,110 | ERLIN2 |
| 109 | TTC21B | 443 | MIR25 | 777 | CASP1 | 1,111 | GADD45G |
| 110 | EDN1 | 444 | CD44 | 778 | P2RX7 | 1,112 | CS |
| 111 | WDR19 | 445 | MALAT1 | 779 | ASCL1 | 1,113 | CD22 |
| 112 | CRP | 446 | COL3A1 | 780 | VWF | 1,114 | PSMB1 |
| 113 | CCL11 | 447 | CD28 | 781 | PROM1 | 1,115 | LOC111674479 |
| 114 | AGT | 448 | MIR23A | 782 | PDPN | 1,116 | LOC111674467 |
| 115 | CCND1 | 449 | MIR19A | 783 | PRODH | 1,117 | SDHB |
| 116 | COL1A2 | 450 | HAMP | 784 | ALG9 | 1,118 | LOC111674465 |
| 117 | COPA | 451 | MIR429 | 785 | HOXD13 | 1,119 | PSMD14 |
| 118 | CASP8 | 452 | MIR335 | 786 | TNFRSF10A | 1,120 | IFNB1 |
| 119 | DKC1 | 453 | TTR | 787 | IL12B | 1,121 | PLAT |
| 120 | ACTC1 | 454 | C2CD3 | 788 | BBS9 | 1,122 | PSMA5 |
| 121 | FGFR2 | 455 | NR1H4 | 789 | NR3C1 | 1,123 | NEB |
| 122 | IL17A | 456 | G6PD | 790 | MIR122 | 1,124 | CHGA |
| 123 | SCNN1B | 457 | MIR127 | 791 | ITGB3 | 1,125 | MIR187 |
| 124 | ATM | 458 | TNNI3 | 792 | ERF | 1,126 | LOC113523647 |
| 125 | MIR155 | 459 | CXCL1 | 793 | CCR5 | 1,127 | HSD3B7 |
| 126 | SHH | 460 | MIR424 | 794 | ADIPOQ | 1,128 | LOC108491823 |
| 127 | RB1 | 461 | IFT52 | 795 | TF | 1,129 | IL4R |
| 128 | CSF2 | 462 | ZCCHC8 | 796 | RXRA | 1,130 | PSMB4 |
| 129 | CEP290 | 463 | AGTR2 | 797 | FAT4 | 1,131 | DRC1 |
| 130 | FCGR2A | 464 | NSMCE3 | 798 | PSMD4 | 1,132 | MIR361 |
| 131 | PKHD1 | 465 | TYR | 799 | UMOD | 1,133 | COL11A2 |
| 132 | NPHP4 | 466 | ABCB4 | 800 | TMEM107 | 1,134 | MIR497 |
| 133 | MIR200B | 467 | NOS3 | 801 | BLOC1S6 | 1,135 | SBDS |
| 134 | MKS1 | 468 | LPAR1 | 802 | MMP14 | 1,136 | TLR6 |
| 135 | SCNN1G | 469 | CXCR2 | 803 | PTK2B | 1,137 | MIR339 |
| 136 | FAM111B | 470 | ZIC3 | 804 | PIEZO2 | 1,138 | RECQL4 |
| 137 | FN1 | 471 | LOC111674475 | 805 | CDK1 | 1,139 | DERL2 |
| 138 | INVS | 472 | CCR7 | 806 | SIRT3 | 1,140 | SH2B3 |
| 139 | STAT6 | 473 | ANKS6 | 807 | IL15 | 1,141 | GUCA2A |
| 140 | WRAP53 | 474 | CALR | 808 | TAP1 | 1,142 | CCDC103 |
| 141 | FGFR3 | 475 | TCTN1 | 809 | GATA6 | 1,143 | PCNA |
| 142 | RET | 476 | MIR224 | 810 | EPOR | 1,144 | PSME2 |
| 143 | NRAS | 477 | IGF1R | 811 | PVT1 | 1,145 | NCOR1 |
| 144 | DDR2 | 478 | XRCC1 | 812 | MUSK | 1,146 | TTC37 |
| 145 | FBN1 | 479 | CD34 | 813 | ESR2 | 1,147 | PSMB5 |
| 146 | OFD1 | 480 | EVC2 | 814 | MAP1B | 1,148 | PRKG1 |
| 147 | MAP2K1 | 481 | CEACAM5 | 815 | MT-CYB | 1,149 | LEPR |
| 148 | ENG | 482 | MIR338 | 816 | TRAF6 | 1,150 | TNC |
| 149 | NEK8 | 483 | GATA4 | 817 | PKD1L1 | 1,151 | PTGS1 |
| 150 | FGF2 | 484 | MIRLET7G | 818 | RBPJ | 1,152 | IFNA2 |
| 151 | MUC5AC | 485 | SOD2 | 819 | DMD | 1,153 | DZIP1L |
| 152 | NFKB1 | 486 | MIR191 | 820 | FABP4 | 1,154 | MIR296 |
| 153 | RARB | 487 | CCR3 | 821 | CD80 | 1,155 | CD86 |
| 154 | TNFRSF1A | 488 | CXCL5 | 822 | PSMA4 | 1,156 | MYCL |
| 155 | IL5 | 489 | MYLK | 823 | IGHE | 1,157 | VPS45 |
| 156 | APC | 490 | H2AC18 | 824 | EGFR-AS1 | 1,158 | GBE1 |
| 157 | SMPD1 | 491 | GPC3 | 825 | DNMT3B | 1,159 | IL7R |
| 158 | MPO | 492 | TMEM138 | 826 | MIR26A1 | 1,160 | CYP27A1 |
| 159 | ACTA2 | 493 | MIR377 | 827 | FLT4 | 1,161 | PLA2G7 |
| 160 | MIR126 | 494 | CCL22 | 828 | CCNA2 | 1,162 | NOTCH2 |
| 161 | NOD2 | 495 | SLC34A2 | 829 | DNAI2 | 1,163 | CEACAM1 |
| 162 | MIR17 | 496 | PTPN22 | 830 | TUG1 | 1,164 | MIR485 |
| 163 | FGF7 | 497 | PKD2 | 831 | IFIH1 | 1,165 | LOC113633876 |
| 164 | IFT140 | 498 | DNAH5 | 832 | KATNIP | 1,166 | REG3A |
| 165 | KRT18 | 499 | S100A4 | 833 | MMP3 | 1,167 | EPRS1 |
| 166 | REN | 500 | BAX | 834 | KLF6 | 1,168 | LOC113633875 |
| 167 | PDCD1 | 501 | THPO | 835 | BLOC1S3 | 1,169 | LOC113604967 |
| 168 | VEGFC | 502 | MIR137 | 836 | SERPINB1 | 1,170 | ZEB1 |
| 169 | IFT80 | 503 | FOXJ1 | 837 | GLA | 1,171 | FOXM1 |
| 170 | SCGB1A1 | 504 | BCL2L1 | 838 | LPO | 1,172 | CASR |
| 171 | BRCA2 | 505 | TRPV4 | 839 | F2R | 1,173 | TALDO1 |
| 172 | AHI1 | 506 | CCR4 | 840 | BTK | 1,174 | MIR219A1 |
| 173 | TCTN2 | 507 | CADM1 | 841 | SOCS3 | 1,175 | CCL26 |
| 174 | BMPR2 | 508 | TMEM237 | 842 | COL4A2 | 1,176 | LOC113664107 |
| 175 | RAF1 | 509 | LOC111674463 | 843 | NBAS | 1,177 | CLCN4 |
| 176 | CXCR4 | 510 | GBA | 844 | CEP104 | 1,178 | SNHG1 |
| 177 | KIAA0586 | 511 | EZH2 | 845 | MAGEA4 | 1,179 | LAMA2 |
| 178 | HIF1A | 512 | SOX9 | 846 | DERL1 | 1,180 | TCF7 |
| 179 | FARSB | 513 | MIR24-2 | 847 | AKT3 | 1,181 | TFRC |
| 180 | TLR2 | 514 | SPARC | 848 | NPPA | 1,182 | PLCZ1 |
| 181 | AGTR1 | 515 | S100A8 | 849 | BCL2L11 | 1,183 | POMC |
| 182 | BMP6 | 516 | LTA | 850 | LAMA1 | 1,184 | ABCC3 |
| 183 | CDK4 | 517 | MKKS | 851 | IL3 | 1,185 | CRYAB |
| 184 | TMEM216 | 518 | HYDIN | 852 | ITGA5 | 1,186 | LOC110806263 |
| 185 | RELA | 519 | GLI1 | 853 | CYP2D6 | 1,187 | MCM4 |
| 186 | CSF3 | 520 | BIRC5 | 854 | DOCK6 | 1,188 | TNFSF13B |
| 187 | CCL5 | 521 | MAPK3 | 855 | CANX | 1,189 | MIR149 |
| 188 | ESR1 | 522 | PSMB8 | 856 | AXIN2 | 1,190 | PLA2G6 |
| 189 | MIR223 | 523 | AP3D1 | 857 | SCARB2 | 1,191 | KNG1 |
| 190 | FOXP3 | 524 | MIRLET7A3 | 858 | CDX2 | 1,192 | HOTTIP |
| 191 | TTN | 525 | IDH1 | 859 | SLC9A3 | 1,193 | AR |
| 192 | MARS1 | 526 | CYP2E1 | 860 | ICOS | 1,194 | PSMB2 |
| 193 | MAPK8 | 527 | TNFSF10 | 861 | PTGER4 | 1,195 | LSM1 |
| 194 | MEG3 | 528 | PRKCD | 862 | SLC17A5 | 1,196 | NIPBL |
| 195 | CCL3 | 529 | MIR181C | 863 | CXCR5 | 1,197 | PSMA8 |
| 196 | HPS4 | 530 | CYCS | 864 | TCTEX1D2 | 1,198 | IL16 |
| 197 | BRCA1 | 531 | IFT27 | 865 | MXRA5 | 1,199 | CCAT1 |
| 198 | IL2RA | 532 | TUBB3 | 866 | FOXP1 | 1,200 | SPRY2 |
| 199 | HPS1 | 533 | HP | 867 | MIR138-1 | 1,201 | RIPK1 |
| 200 | MIR34A | 534 | ALMS1 | 868 | CLDN1 | 1,202 | PHKG2 |
| 201 | MIR200C | 535 | NOX4 | 869 | MIR154 | 1,203 | MIR503 |
| 202 | B9D1 | 536 | RYR1 | 870 | INSR | 1,204 | GSTT1 |
| 203 | Dnase1 | 537 | TSLP | 871 | UCHL1 | 1,205 | PRKCI |
| 204 | TSC1 | 538 | VDAC1 | 872 | DCN | 1,206 | AGL |
| 205 | WDR35 | 539 | U2AF1 | 873 | XPNPEP3 | 1,207 | RAG2 |
| 206 | WT1 | 540 | CAT | 874 | KRT13 | 1,208 | HNF1A-AS1 |
| 207 | INPP5E | 541 | CALCA | 875 | COMT | 1,209 | CCAT2 |
| 208 | MPL | 542 | DNAH11 | 876 | EDNRA | 1,210 | ERLIN1 |
| 209 | MIR125A | 543 | NF1 | 877 | CA4 | 1,211 | MIR125B2 |
| 210 | CSPP1 | 544 | PTK2 | 878 | HLA-DQA1 | 1,212 | MIR15B |
| 211 | GRP | 545 | SP110 | 879 | BBS12 | 1,213 | CREBBP |
| 212 | MIR144 | 546 | IL12RB1 | 880 | KRT8 | 1,214 | CTSL |
| 213 | RTEL1-TNFRSF6B | 547 | NQO1 | 881 | CMA1 | 1,215 | NR5A1 |
| 214 | SERPINC1 | 548 | GAA | 882 | AKR1B10 | 1,216 | MSR1 |
| 215 | CD4 | 549 | MIR409 | 883 | LOC113633877 | 1,217 | UTP4 |
| 216 | DYNC2H1 | 550 | VIP | 884 | CEP55 | 1,218 | MIR301A |
| 217 | MIR145 | 551 | GPT | 885 | MAP2K4 | 1,219 | MUC7 |
| 218 | FLT1 | 552 | MIR708 | 886 | CDK5 | 1,220 | CASC2 |
| 219 | TNFRSF1B | 553 | NFKBIA | 887 | P2RY2 | 1,221 | ZNRD1ASP |
| 220 | MIR29A | 554 | KEAP1 | 888 | NLRP3 | 1,222 | PLCG2 |
| 221 | HLA-DQB1 | 555 | LOC111674477 | 889 | PSMC4 | 1,223 | MIR124-1 |
| 222 | FHIT | 556 | CD36 | 890 | HSPA4 | 1,224 | MIR382 |
| 223 | H19 | 557 | IGFBP3 | 891 | AVPR2 | 1,225 | LAMP1 |
| 224 | PKD1 | 558 | NKX2-5 | 892 | RPS27A | 1,226 | CD69 |
| 225 | SOS1 | 559 | CEP83 | 893 | MIR375 | 1,227 | DANCR |
| 226 | SPINK1 | 560 | BDNF | 894 | MIR33A | 1,228 | TP53COR1 |
| 227 | MIR31 | 561 | ABCB11 | 895 | ATF6 | 1,229 | MYL3 |
| 228 | IFT172 | 562 | FGFR4 | 896 | RXRB | 1,230 | CYSLTR2 |
| 229 | MMP7 | 563 | CTSG | 897 | UBC | 1,231 | SELL |
| 230 | CD8A | 564 | ITGB1 | 898 | NPC2 | 1,232 | LINC-ROR |
| 231 | PDGFB | 565 | CSF2RA | 899 | CLCN2 | 1,233 | NOS1 |
| 232 | SLC2A1 | 566 | DNAI1 | 900 | FOXE1 | 1,234 | IFT20 |
| 233 | JUN | 567 | ARL3 | 901 | MIR132 | 1,235 | MIR10B |
| 234 | BMP2 | 568 | MECP2 | 902 | HDGF | 1,236 | CD81 |
| 235 | HMGB1 | 569 | CDKN1B | 903 | WNT7B | 1,237 | ANTXR1 |
| 236 | ERBB3 | 570 | DNMT3A | 904 | SLC9A3R2 | 1,238 | PRSS2 |
| 237 | MIR150 | 571 | VHL | 905 | H2AX | 1,239 | SNHG20 |
| 238 | PRSS1 | 572 | THBS1 | 906 | PRKAG2 | 1,240 | MIR95 |
| 239 | MIR29C | 573 | YAP1 | 907 | SERPINB3 | 1,241 | GALC |
| 240 | MIR146B | 574 | TIMP2 | 908 | LRP1B | 1,242 | DGCR5 |
| 241 | ABCC1 | 575 | TOLLIP | 909 | DNAAF3 | 1,243 | HNMT |
| 242 | BBS2 | 576 | KCNK3 | 910 | BCL10 | 1,244 | SLCO2A1 |
| 243 | ABCB1 | 577 | NOTCH3 | 911 | RAG1 | 1,245 | MLH1 |
| 244 | CXCL10 | 578 | RASSF1 | 912 | LAMA4 | 1,246 | PLA2G2A |
| 245 | XIAP | 579 | SP1 | 913 | HLA-G | 1,247 | MME |
| 246 | KIF21A | 580 | RHOA | 914 | PSMB3 | 1,248 | TYMS |
| 247 | PIK3R1 | 581 | ABCC2 | 915 | SIRT1 | 1,249 | MIR198 |
| 248 | BBS1 | 582 | IKBKB | 916 | MIRLET7A1 | 1,250 | JAK1 |
| 249 | IRF1 | 583 | GZMB | 917 | PLA2G1B | 1,251 | PDE4D |
| 250 | IL1A | 584 | LEP | 918 | KDM4C | 1,252 | LAMC2 |
| 251 | PTPRC | 585 | BBS5 | 919 | MUC6 | 1,253 | AHR |
| 252 | HLA-DPB1 | 586 | APOE | 920 | TUBB1 | 1,254 | TPM1 |
| 253 | ERCC6 | 587 | NHLRC2 | 921 | SKIV2L | 1,255 | MT-CO2 |
| 254 | STX1A | 588 | NPPB | 922 | TPM2 | 1,256 | ASCC1 |
| 255 | SETD2 | 589 | SYP | 923 | CCDC40 | 1,257 | EOGT |
| 256 | CCL17 | 590 | ACTB | 924 | TET2 | 1,258 | CCL7 |
| 257 | IDH2 | 591 | PSMA1 | 925 | TRIP11 | 1,259 | TRPM4 |
| 258 | CXCL12 | 592 | PTCH1 | 926 | ITGA2B | 1,260 | GPSM2 |
| 259 | MIR27A | 593 | LOXL2 | 927 | ENO2 | 1,261 | TARS1 |
| 260 | GSTM1 | 594 | CEP41 | 928 | EIF2AK3 | 1,262 | TPM3 |
| 261 | DCTN4 | 595 | MYPN | 929 | RSPH9 | 1,263 | CD14 |
| 262 | PRKN | 596 | CR1 | 930 | MIR193A | 1,264 | MIR216A |
| 263 | CXCR3 | 597 | ERCC1 | 931 | PXN | 1,265 | CTAG1B |
| 264 | MIR200A | 598 | IGFBP5 | 932 | DNAAF5 | 1,266 | MIR22HG |
| 265 | ARL13B | 599 | MGMT | 933 | LAT | 1,267 | MIR10A |
| 266 | PTGS2 | 600 | VIM | 934 | RSPH1 | 1,268 | PCAT1 |
| 267 | MIR148A | 601 | ENO1 | 935 | CDKN2B-AS1 | 1,269 | SNHG15 |
| 268 | MYC | 602 | NLRC4 | 936 | ADORA1 | 1,270 | BANCR |
| 269 | IGF1 | 603 | PIK3CG | 937 | RAD51 | 1,271 | PSAP |
| 270 | IL18 | 604 | MYH7 | 938 | DIABLO | 1,272 | IL11 |
| 271 | CYP1A1 | 605 | F13A1 | 939 | TFR2 | 1,273 | MIR362 |
| 272 | NEK9 | 606 | WNT4 | 940 | RSPO2 | 1,274 | UCA1 |
| 273 | LOX | 607 | NME1 | 941 | C4A | 1,275 | UBE2L3 |
| 274 | MIR30D | 608 | CD63 | 942 | IFI27 | 1,276 | FBLN5 |
| 275 | MIR146A | 609 | RPGRIP1 | 943 | MIR29B1 | 1,277 | GC |
| 276 | IFT122 | 610 | ATP8B1 | 944 | HSPD1 | 1,278 | TP73-AS1 |
| 277 | B9D2 | 611 | GNAS | 945 | CCDC39 | 1,279 | PPBP |
| 278 | KCNQ1 | 612 | TUBB | 946 | PDE4A | 1,280 | LINC00473 |
| 279 | CYP2A6 | 613 | IFRD1 | 947 | TRIM21 | 1,281 | SOX2-OT |
| 280 | MIR483 | 614 | CASP9 | 948 | PLCG1 | 1,282 | MIR181B1 |
| 281 | MIR183 | 615 | LAMP2 | 949 | IL1RL1 | 1,283 | XIST |
| 282 | IL1R1 | 616 | HDAC2 | 950 | BBIP1 | 1,284 | MIR129-1 |
| 283 | SDCCAG8 | 617 | ANXA1 | 951 | MIR196A1 | 1,285 | PRL |
| 284 | PTRH2 | 618 | IFT74 | 952 | BAD | 1,286 | MIR193B |
| 285 | MIR182 | 619 | ROS1 | 953 | MYBPC3 | 1,287 | NR3C2 |
| 286 | CDKN3 | 620 | CLEC7A | 954 | ARAF | 1,288 | MYL1 |
| 287 | GUCY2C | 621 | AKT2 | 955 | VDR | 1,289 | SDC1 |
| 288 | MIR222 | 622 | WRN | 956 | MAGEA1 | 1,290 | MIR24-1 |
| 289 | LOC111674472 | 623 | DTNBP1 | 957 | TAC1 | 1,291 | PHB |
| 290 | KIF7 | 624 | CD19 | 958 | IDUA | 1,292 | MYL2 |
| 291 | FLNC | 625 | SMARCA4 | 959 | EPX | 1,293 | APOA1 |
| 292 | PPARG | 626 | ANGPT2 | 960 | ASXL1 | 1,294 | SNHG12 |
| 293 | XRCC3 | 627 | PARP1 | 961 | MAGEA3 | 1,295 | CEP57 |
| 294 | SNAI1 | 628 | BBS7 | 962 | RAC1 | 1,296 | IL6R |
| 295 | POSTN | 629 | MIF | 963 | GATA1 | 1,297 | SERPINF2 |
| 296 | MIR192 | 630 | SELP | 964 | GAS5 | 1,298 | CALB2 |
| 297 | JAG1 | 631 | HSPG2 | 965 | MIR9-1 | 1,299 | MIR152 |
| 298 | CD40LG | 632 | ATP12A | 966 | RYR2 | 1,300 | ADK |
| 299 | DYNC2I1 | 633 | HSP90AA1 | 967 | JPH2 | 1,301 | NRG1 |
| 300 | ACVRL1 | 634 | CHRM3 | 968 | RIOX2 | 1,302 | MIR501 |
| 301 | MIR221 | 635 | MVP | 969 | LOC111674464 | 1,303 | GPRC5A |
| 302 | IFT43 | 636 | DCDC2 | 970 | TNFAIP3 | 1,304 | LZTR1 |
| 303 | DES | 637 | ERCC2 | 971 | PIK3CB | 1,305 | TLR7 |
| 304 | MIR22 | 638 | PPP2R1B | 972 | DHCR7 | 1,306 | POT1 |
| 305 | TNFRSF10B | 639 | TYMP | 973 | C1S | 1,307 | EP300 |
| 306 | IRF5 | 640 | CLCA1 | 974 | SNAI2 | 1,308 | FBL |
| 307 | ALK | 641 | OGG1 | 975 | ERBB4 | 1,309 | PLOD2 |
| 308 | CEP120 | 642 | MCL1 | 976 | ACVR1 | 1,310 | DLL1 |
| 309 | MIR451A | 643 | CCN4 | 977 | NPC1 | 1,311 | TOP2A |
| 310 | DYNC2I2 | 644 | F3 | 978 | LNX1 | 1,312 | MAGEC2 |
| 311 | CASP3 | 645 | STAT5B | 979 | GRB2 | 1,313 | MIR499A |
| 312 | CCL4 | 646 | FUZ | 980 | TFAP2B | 1,314 | ENSG00000266919 |
| 313 | HPS6 | 647 | LBR | 981 | SLC6A4 | 1,315 | TNFRSF11B |
| 314 | MIR143 | 648 | CXCL2 | 982 | IFT88 | 1,316 | HOXA11-AS |
| 315 | ICOSLG | 649 | PLAUR | 983 | NPHS1 | 1,317 | RASGRP1 |
| 316 | CLCA4 | 650 | ASAH1 | 984 | MAP2K7 | 1,318 | SPRY4-IT1 |
| 317 | HLA-DPA1 | 651 | BPI | 985 | PSMD2 | 1,319 | FIP1L1 |
| 318 | FOXF1 | 652 | NPM1 | 986 | DNAH8 | 1,320 | GSR |
| 319 | BIRC3 | 653 | ACTA1 | 987 | B2M | 1,321 | CST3 |
| 320 | SMAD7 | 654 | ANGPT1 | 988 | INTU | 1,322 | LTBP4 |
| 321 | MIR203A | 655 | GGT1 | 989 | ENTPD1 | 1,323 | ZFAS1 |
| 322 | MIR30A | 656 | CAMP | 990 | PRSS8 | 1,324 | F5 |
| 323 | MIR324 | 657 | SOD3 | 991 | CCNE1 | 1,325 | STMN1 |
| 324 | MIR199B | 658 | F2RL3 | 992 | LRRC56 | 1,326 | AIRE |
| 325 | ACP5 | 659 | TEK | 993 | MAP3K8 | 1,327 | RETN |
| 326 | MIR205 | 660 | LTF | 994 | SHC1 | 1,328 | NTS |
| 327 | TP73 | 661 | MT-CO1 | 995 | CCDC114 | 1,329 | KRT5 |
| 328 | TLR9 | 662 | LIPA | 996 | ACHE | 1,330 | F2RL1 |
| 329 | MAPK1 | 663 | WNT3 | 997 | TBX20 | 1,331 | TNFSF11 |
| 330 | CLCN5 | 664 | GDF1 | 998 | AURKB | 1,332 | COL4A5 |
| 331 | TMEM231 | 665 | SLC40A1 | 999 | ASL | 1,333 | PIK3CD |
| 332 | BBS4 | 666 | GLIS2 | 1,000 | MIR30C1 | 1,334 | FGF9 |
| 333 | RMRP | 667 | AREG | 1,001 | IL2RB | 1,335 | KRT20 |
| 334 | TGFB3 | 668 | CDKN2B | 1,002 | NCF2 |
FIGURE 2.
Anlotinib inhibits PFKFB3-driven glycolysis in lung myofibroblasts. (A) Venn diagram to show the overlaps between anlotinib targets and lung fibrosis-related targets. (B) Protein-protein interaction (PPI) network of common targets between anlotinib and IPF. (C) The KEGG enrichment analysis of 74 targets of common targets. (D)The GO enrichment for each section listed. The mouse lung fibroblasts were pretreated with anlotinib (1 µM) for 3 h and then exposed to TGF-β1 (10 ng/ml) for an additional 24 h, and then the cells were lysed and lactate contents in the cellular lysates (E) and culture media (F) were determined. The data are presented as fold change relative to the levels of the untreated control group (mean ± SD, n = 3). (G) Glucose uptake detected with 2-NBDG were determined. The data are presented as fold change relative to the levels of the untreated control group (mean ± SD, n = 3). (H) Extracellular acidification rate (ECAR) was assessed. (I) Glycolysis and glycolysis capacity were quantified and shown as histograms (mean ± SD, n = 3). (J) Western blot analysis of HK2、PKM2、PFKFB3、LDHA and LDHB, β-actin was used as a loading control. (K) Quantification of HK2、PKM2、PFKFB3、LDHA and LDHB protein levels relative to β-Actin is shown (mean ± SD, n = 3). I, K, *p < 0.05, **p < 0.01, ***p < 0.001 VS TGF-β1-treated group by ANOVA.
TABLE 4.
Common targets shared between the potential anlotinib targets and lung fibrosis-related targets.
| Number | Gene | Number | Gene |
|---|---|---|---|
| 1 | AXL | 38 | EPHA3 |
| 2 | MERTK | 39 | TIE1 |
| 3 | ERBB2 | 40 | CACNA2D1 |
| 4 | AURKB | 41 | ERBB4 |
| 5 | FLT1 | 42 | MAP3K8 |
| 6 | EGFR | 43 | ERN1 |
| 7 | KDR | 44 | MST1R |
| 8 | SRC | 45 | FGFR4 |
| 9 | MET | 46 | EPHA7 |
| 10 | RET | 47 | MAP2K5 |
| 11 | ALK | 48 | RIPK2 |
| 12 | ABL1 | 49 | DDR2 |
| 13 | KIT | 50 | ACVR1B |
| 14 | ACVR1 | 51 | DDR1 |
| 15 | FLT4 | 52 | MAP4K2 |
| 16 | FLT3 | 53 | EPHA1 |
| 17 | PDGFRA | 54 | EPHB6 |
| 18 | FGFR1 | 55 | ERBB3 |
| 19 | TGFBR1 | 56 | EIF2AK1 |
| 20 | BTK | 57 | MAP3K19 |
| 21 | LYN | 58 | AKT2 |
| 22 | FGFR3 | 59 | PRKCG |
| 23 | PDGFRB | 60 | AKT3 |
| 24 | YES1 | 61 | PIM1 |
| 25 | FGFR2 | 62 | PIM2 |
| 26 | FGR | 63 | JAK1 |
| 27 | CSF1R | 64 | DPP8 |
| 28 | BLK | 65 | DPP9 |
| 29 | PLK4 | 66 | PDE4B |
| 30 | FYN | 67 | PRKCI |
| 31 | MAP2K2 | 68 | CDK2 |
| 32 | HCK | 69 | CDK1 |
| 33 | STK10 | 70 | CCNT1 |
| 34 | ABL2 | 71 | TERT |
| 35 | TEK | 72 | SLC8A1 |
| 36 | SLK | 73 | GNRHR |
| 37 | STK4 | 74 | AOC3 |
PCBP3 Posttranscriptionally Increases PFKFB3 Expression by Promoting Its Translation During Myofibroblast Activation
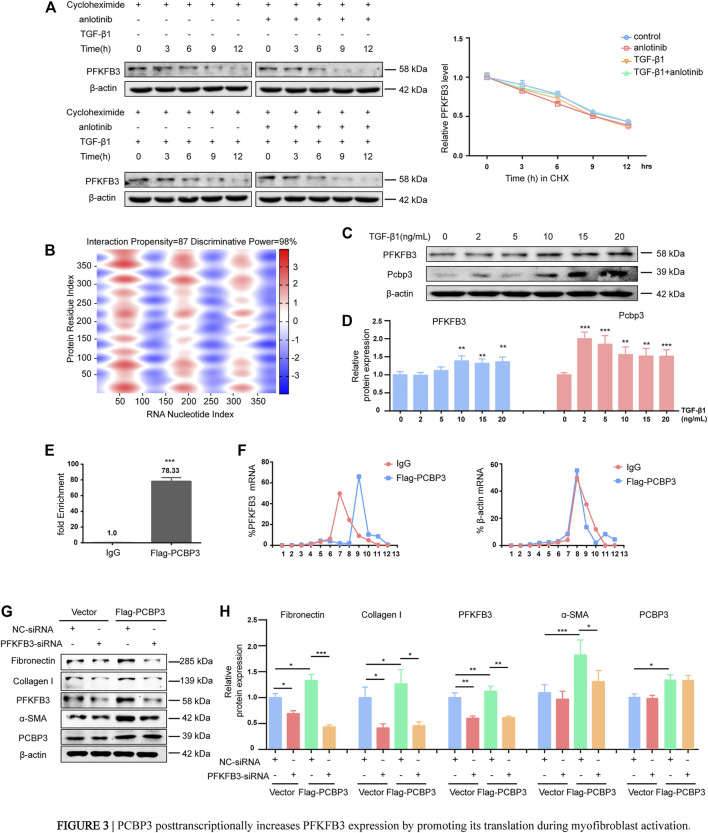
Interestingly, the progressive upregulation of PFKFB3 during myofibroblast activation induced by TGF-β1 that was observed at the protein level was not confirmed at the mRNA level, as measured by RT-PCR (Supplementary Figures S3A, B). These results indicate that TGF-β1-induced overexpression does not require de novo transcription of PFKFB3. To further verify these findings, primary MLFs were incubated with cycloheximide to block new protein synthesis, and immunoblotting was used to measure PFKFB3 levels (Figure 3A). The half-life of PFKFB3 was not significantly altered, indicating that TGF-β1 does not influence PFKFB3 protein stability. Therefore, we postulated that PFKFB3 upregulation is modulated through posttranscriptional mechanisms in this context. To verify this hypothesis, we used the online tool catRAPID to screen for potential proteins that may interact with PFKFB3 mRNA and identified that PCBP3 (Table 5) (Agostini et al., 2013; Livi et al., 2016), a member of the PCBP family, has a high probability of directly interacting with PFKFB3 mRNA (Figure 3B) (Choi et al., 2007; Kang et al., 2012; Leidgens et al., 2013; Wang J. et al., 2020). We comparatively analyzed the expression of PCBP3 after treatment with different doses of TGF-β1 by immunoblot analysis and found that PCBP3 protein expression was increased in primary MLFs after TGF-β1 treatment (Figures 3C,D), which correlated with PFKFB3 overexpression. To better define the connection between PCBP3 function and PFKFB3, we performed RNA-protein coimmunoprecipitation (RIP) studies in primary MLFs transfected with FLAG-tagged PCBP3 (FLAG-PCBP3). An antibody targeting the FLAG protein was used to immunoprecipitate FLAG-PCBP3 and any interacting molecules from the cell lysates. Reverse transcription followed by PCR was then used to identify individual PFKFB3 mRNAs isolated with FLAG-PCBP3. We found that PFKFB3 transcripts were enriched by PCBP3 coimmunoprecipitation compared to control IgG coimmunoprecipitation (Figure 3E), demonstrating that PFKFB3 mRNA is indeed a direct target of PCBP3 in MLFs. To test the possibility that PCBP3 may influence PFKFB3 translation, we performed polysome analysis in cells transfected with FLAG-PCBP3. Cytoplasmic lysates were fractionated through sucrose gradients to separate ribosomal subunits (40S and 60S), monosomes (80S) and progressively larger polysomes. RNA was extracted from each of the 12 fractions, and the levels of PFKFB3 and β-actin mRNA were quantified by quantitative RT-PCR. While PFKFB3 mRNA levels peaked in fraction 7 in control cells, the distribution of PFKFB3 mRNA shifted rightward when PCBP3 was overexpressed, peaking in fraction 9, indicating that PFKFB3 mRNA formed, on average, larger polysomes after PCBP3 overexpression (Figure 3F). The distribution of β-actin mRNA was not affected by PCBP3 overexpression. These results indicated that overexpression of PCBP3 increases the translation of PFKFB3. Overall, these results suggest that PCBP3 improves PFKFB3 expression levels by increasing its translation rather than by influencing its protein stability.
FIGURE 3.
PCBP3 posttranscriptionally increases PFKFB3 expression by promoting its translation during myofibroblast activation. (A) PFKFB3 degradation in mouse lung fibroblasts when protein synthesis was inhibited by 50 μM cycloheximide (mean ± SD, n = 3). (B) Prediction of RNA–protein interaction of PFKFB3 mRNA with PCBP3 protein using the catRAPID algorithm. Red represents interaction strength. (C) Western blot analysis of PFKFB3 and PCBP3 protein levels in mouse lung fibroblasts stimulated with TGF-β1 for the indicated concentrations. (D) Quantification of PFKFB3 and PCBP3 protein levels relative to β-actin is shown (mean ± SD, n = 3, **p < 0.01, ***p < 0.001 compared with 0 by one-way ANOVA). (E) Quantitative RT-PCR (qPCR) to show the effect of PCBP3 overexpressing on PFKFB3 RNA immunoprecipitation (RIP) in mouse lung fibroblasts. Values were plotted as mean ± SD from three independent experiments. p value was calculated by Student t test. ***p < 0.001. (F) Mouse lung fibroblasts expressing Flag-PCBP3 were fractionated into cytoplasmic extracts through sucrose gradients. The arrow indicates the direction of sedimentation. The distribution of PFKFB3 and β-actin mRNAs was quantified by RT-PCR analysis of RNA isolated from 12 gradient fractions. Statistical analyses were performed using Student t test. ***p < 0.001. (G) Mouse lung fibroblasts were transfected with Flag-PCBP3, and then transfected with PFKFB3-siRNAs or NC-siRNA. The levels of Fibronectin、Collagen I、PFKFB3、α-SMA、PCBP3 and β-actin assessed by western blot. (H) Graphical representation of the relative levels of indicated proteins (mean ± SD, n = 3, *p < 0.05, **p < 0.01, ***p < 0.001 by ANOVA).
TABLE 5.
Potential proteins may interact with PFKFB3 mRNA through the catRAPID algorithm.
| # | Protein D | RNA ID | Z score? | Discriminative power (%)? | Interaction strength (%)? | Domain? | Motif? | Ranking? |
|---|---|---|---|---|---|---|---|---|
| 1 | ELAV1_MOUSE_247-308 | NC_000068.7:c11_1_5924-6036 | −0.13 | 50 | 98 | yes | yes | |
| 2 | ELAV1_MOUSE_247-308 | NC_000068.7:c11_1_26816-27008 | −0.08 | 67 | 99 | yes | yes | |
| 3 | ELAV1_MOUSE_247-308 | NC_000068.7:c11_1_15806-15983 | −0.23 | 40 | 90 | yes | yes | |
| 4 | PCBP3_MOUSE_301-351 | NC_000068.7:c11_1_41382-41564 | −0.69 | 14 | 13 | yes | yes | |
| 5 | ELAV1_MOUSE_247-308 | NC_000068.7:c11_1_77223-77340 | −0.47 | 22 | 74 | yes | yes | |
| 6 | ELAV1_MOUSE_247-308 | NC_000068.7:c11_1_10399-10532 | −0.50 | 20 | 64 | yes | yes | |
| 7 | ELAV1_MOUSE_109-176 | NC_000068.7:c11_1_5924-6036 | −0.14 | 50 | 98 | yes | yes | |
| 8 | ELAV1_MOUSE_109-176 | NC_000068.7:c11_1_45706-45815 | −0.39 | 26 | 85 | yes | yes | |
| 9 | ELAV1_MOUSE_109-176 | NC_000068.7:c11_1_26816-27008 | −0.04 | 63 | 99 | yes | yes | |
| 10 | ELAV1_MOUSE_109-176 | NC_000068.7:c11_1_15806-15983 | −0.24 | 40 | 90 | yes | yes | |
| 11 | ELAV1_MOUSE_109-172 | NC_000068.7:c11_1_5924-6036 | −0.15 | 47 | 97 | yes | yes | |
| 12 | ELAV1_MOUSE_109-172 | NC_000068.7:c11_1_45706-45815 | −0.40 | 26 | 85 | yes | yes | |
| 13 | ELAV1_MOUSE_109-172 | NC_000068.7:c11_1_15806-15983 | −0.26 | 37 | 87 | yes | yes | |
| 14 | PCBP3_MOUSE_301-351 | NC_000068.7:c11_1_77461-77636 | −0.71 | 14 | 9 | yes | yes | |
| 15 | PCBP3_MOUSE_301-351 | NC_000068.7:c11_1_66050-66214 | −0.66 | 14 | 17 | yes | yes |
To determine the functional impact of PCBP3-mediated regulation of PFKFB3 expression in lung fibrosis, we transfected lung fibroblasts with FLAG-PCBP3. Expression of PFKFB3 was significantly increased by PCBP3 overexpression compared to that of the empty vector control. Reliable markers of the phenotypic transformation of fibroblasts into myofibroblasts, fibronectin, collagen I and α-SMA, were markedly increased in FLAG-PCBP3-treated cells at the protein level (Figures 3G,H) compared with vector-treated cells. In turn, using small interfering RNA (siRNA) to silence PFKFB3, the FLAG-PCBP3-induced overexpression of fibronectin, collagen I and α-SMA was abolished (Figures 3G,H). These findings suggest that PCBP3 protein upregulation is an early and sustained event during fibroblast activation and that the profibrogenic effects of PCBP3 are mediated by PFKFB3 expression. Taken together, these data suggest that PCBP3 posttranscriptionally increases PFKFB3 expression by promoting its translation during myofibroblast activation.
Anlotinib Represses PCBP3 Expression Levels During Myofibroblast Activation
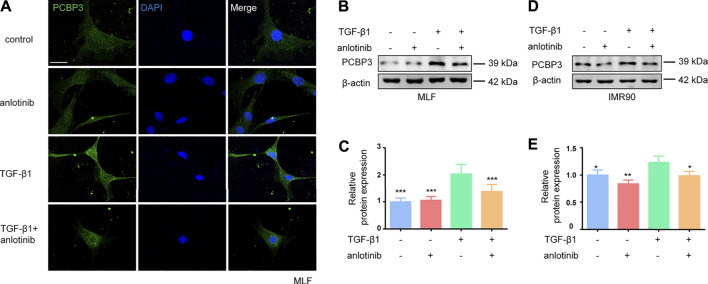
To confirm the regulation of PCBP3 by anlotinib in vitro, we evaluated the protein expression of PCBP3 in MLFs and IMR90 cells. We found that TGF-β1 induced the expression of PCBP3 in MLFs and that anlotinib prevented PCBP3 expression by immunofluorescence analysis (Figure 4A). Western blot analysis of PCBP3 showed a similar result (Figures 4B,C) in MLFs, and these results were confirmed in the human IMR90 cell line (Figures 4D,E). Taken together, these data suggest that anlotinib can repress PCBP3 expression levels during myofibroblast activation in vitro.
FIGURE 4.
Anlotinib represses PCBP3 expression levels during myofibroblast activation. (A) Immunofluorescence for PCBP3 (green) in mouse lung fibroblasts treated with anlotinib for 3 h, followed by TGF-β1 for an additional 24 h. DAPI-stained nuclei (blue). Scale bar, 25 µm. (B) Western blots analysis of PCBP3 and β-actin in primary mouse lung fibroblasts. (C) Quantification for the indicated proteins (mean ± SD, n = 3). (D) The Western blots analysis of PCBP3 and β-actin in IMR90 cells treated with anlotinib for 3 h, followed by TGF-β1 for an additional 24 h. (E) Quantification for the indicated proteins (mean ± SD, n = 3). *p < 0.05, **p < 0.01, ***p < 0.001 VS TGF-β1-treated group by ANOVA.
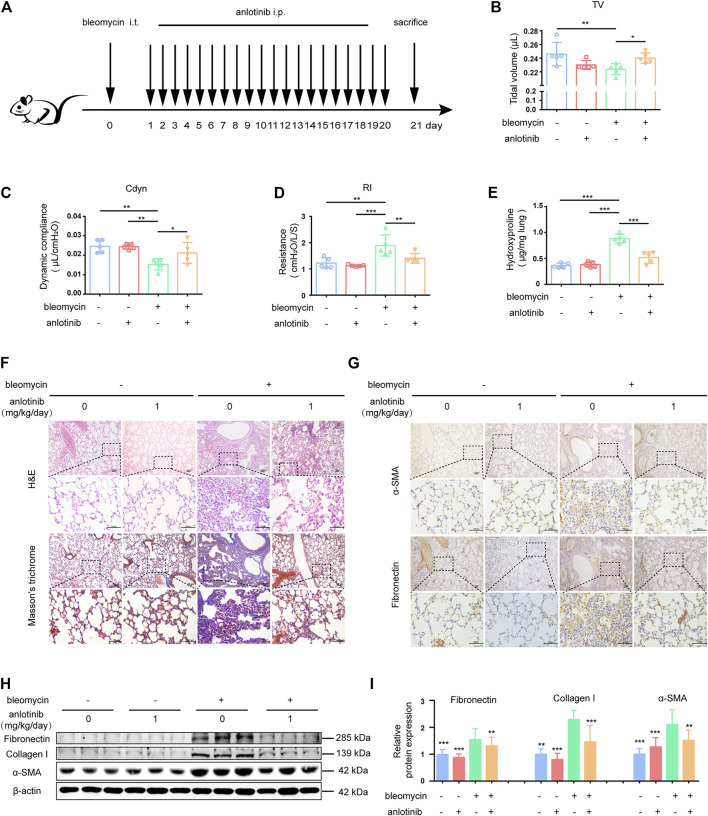
Anlotinib Attenuates Bleomycin-Induced Pulmonary Fibrosis
To investigate the biological effects of anlotinib on pulmonary fibrosis in vivo, we established a bleomycin (BLM)-induced mouse model of pulmonary fibrosis. The mice were intraperitoneally injected with 1 mg/kg anlotinib daily after BLM administration (Figure 5A). From the first week after bleomycin instillation, the bleomycin-treated mice showed a certain reduction in activity, accompanied by slight shortness of breath. 21 days after bleomycin administration, bleomycin-treated mice showed obvious hyperventilation, accompanied by reduced activity and weight loss, but no similar symptoms were observed in the control group. A single dose of BLM (5 mg/kg) administered by intratracheal instillation successfully induced pulmonary fibrosis in C57BL/6 mice, as evidenced by a decline in pulmonary function, decreased tidal volume (TV, Figure 5B) and dynamic compliance (Cdyn, Figure 5C), and increased lung resistance (RI, Figure 5D). However, treatment with anlotinib significantly reversed bleomycin-induced pulmonary dysfunction. Moreover, we evaluated collagen deposition in the lung tissues by analyzing the hydroxyproline (HYP) content and found that anlotinib treatment reduced the amount of collagen in the lungs of bleomycin-treated mice (Figure 5E). Hematoxylin and eosin (H&E) staining indicated that anlotinib-treated mice had decreased lung inflammation and reduced lung architectural damage (Figure 5F). Accordingly, Masson’s trichrome staining showed decreased collagen deposition in anlotinib-treated mice compared with vehicle-treated mice (Figure 5F). Furthermore, attenuated fibrosis was supported by decreased protein levels of fibronectin and α-SMA by immunohistochemical (IHC) staining (Figure 5G). We also found that anlotinib treatment reduced fibronectin, collagen I and α-SMA expression by western blotting (Figures 5H,I). Taken together, these data show that anlotinib attenuates bleomycin-induced pulmonary fibrosis in vivo.
FIGURE 5.
Anlotinib attenuates bleomycin-induced pulmonary fibrosis. (A) Intervention dosing regimen of anlotinib in experimental mouse model of fibrosis. C57BL/6 mice were intraperitonealy injuected with 1 mg/kg of anlotinib or vehicle (n = 5-6 per group) daily after bleomycin instillation. Lungs were harvested at 21 days for the following analyses. Analysis of tidal volume (TV) (B), dynamic compliance (Cdyn) (C), and lung resistance (RI) (D) (mean ± SD, n = 5). (E) Hydroxyproline (HYP) contents in lung tissues from mice (mean ± SD, n = 5). Representative images show haematoxylin and eosin (H&E), Masson’s trichrome (F), α-SMA and Fibronectin staining (G) of lung sections from the indicated groups of mice. Scale bars, 100 µm. (H) Western blot analysis of Fibronectin、Collagen I and α-SMA, β-actin was used as a loading control. (I) Quantification of Fibronectin、Collagen I and α-SMA protein levels relative to β-actin is shown (mean ± SD, n = 6). *p < 0.05, **p < 0.01, ***p < 0.001 VS BLM-treated group by one-way ANOVA.
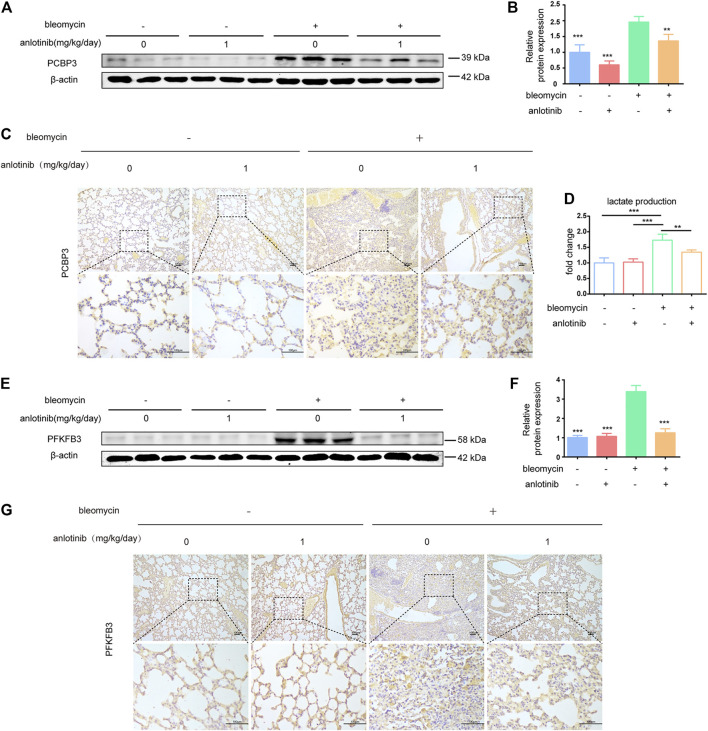
Anlotinib Decreases PCBP3 Expression and Inhibits PFKFB3-Driven Glycolysis in Fibrotic Rodent Lungs
We next examined whether the levels of PCBP3 were regulated by anlotinib in vivo. We evaluated the expression of PCBP3 in lung tissues and found that the protein levels of PCBP3 were markedly increased after bleomycin instillation, while anlotinib treatment decreased PCBP3 expression (Figures 6A,B). Accordingly, IHC staining showed decreased PCBP3 protein levels in anlotinib-treated mice compared with vehicle-treated mice (Figure 6C). In addition, to confirm the regulation of PFKFB3-driven glycolysis by anlotinib in vivo, we measured the levels of lactate and the expression of PFKFB3 in the lungs of mice. We found that there were significantly higher levels of lactate in the lungs of bleomycin-treated mice than in the lungs of control mice, and anlotinib decreased lactate levels (Figure 6D). Western blot and IHC staining studies revealed that bleomycin-induced PFKFB3 expression in the lungs of mice was prevented by anlotinib (Figures 6E–G). Overall, these results suggest that anlotinib decreases PCBP3 expression and inhibits PFKFB3-driven glycolysis in fibrotic rodent lungs.
FIGURE 6.
Anlotinib decreases PCBP3 expression and inhibit the PFKFB3-driven glycolysis in fibrotic rodent lungs. (A) Intervention dosing regimen of anlotinib in experimental mouse model of fibrosis. C57BL/6 mice were intraperitonealy injuected with 1 mg/kg of anlotinib or vehicle (n = 5-6 per group) daily after bleomycin instillation. Lungs were harvested at 21 days. Western blot analysis of PCBP3, β-actin was used as a loading control (n = 6). (B) Quantification of PCBP3 protein levels relative to β-actin is shown (mean ± SD, n = 6). (C) Representative images show PCBP3 staining of lung sections from the indicated groups of mice. Scale bars, 100 µm. (D) lactate contents in lung tissues from mice (mean ± SD, n = 5). (E) Western blot analysis of PFKFB3, β-actin was used as a loading control (n = 6). (F) Quantification of PFKFB3 protein levels relative to β-actin is shown (mean ± SD, n = 6). (G) Representative images show PFKFB3 staining of lung sections from the indicated groups of mice. Scale bars, 100 µm. **p < 0.01, ***p < 0.001 VS BLM-treated group by one-way ANOVA.
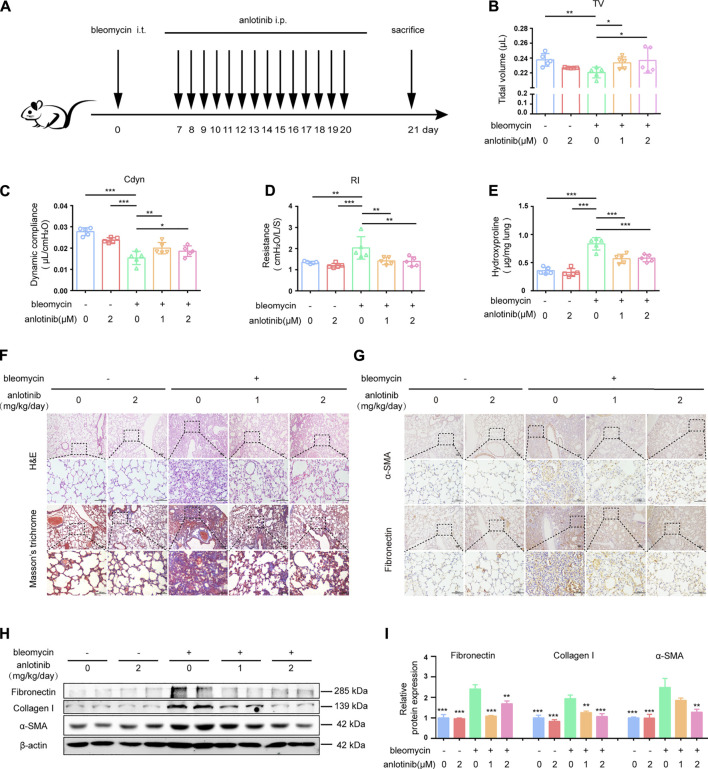
Anlotinib Accelerates the Resolution of Bleomycin-Induced Lung Fibrosis
We demonstrated that anlotinib treatment could attenuate bleomycin-induced pulmonary fibrosis. In that in vivo experiment, anlotinib was administered at approximately the same time as bleomycin instillation. We further examined whether anlotinib could postpone the progression of established fibrosis. Therefore, we performed another in vivo experiment in which anlotinib was intraperitoneally injected 7 days after bleomycin instillation (Figure 7A). As interventions beginning 7 days post bleomycin were classified as therapeutic (Izbicki et al., 2002; Moeller et al., 2008), we initially treated mice with anlotinib (1 mg/kg/day or 2 mg/kg/day) beginning on day 7 after bleomycin instillation. Pulmonary function tests showed that anlotinib treatment reversed the bleomycin-induced decline in pulmonary function, with increases in TV (Figure 7B) and Cdyn (Figure 7C) and a decrease in RI (Figure 7D). HYP measurements showed that the collagen content was significantly decreased in anlotinib-treated mice compared with vehicle-treated mice (Figure 7E). H&E staining and Masson’s trichrome staining of lungs collected at day 21 showed enhanced recovery from fibrosis upon anlotinib treatment (Figure 7F). Correspondingly, IHC staining showed that anlotinib treatment reduced fibronectin and α-SMA expression in the lungs (Figure 7G). Western blot analysis also showed that anlotinib decreased the protein levels of fibronectin, collagen I and α-SMA in the lungs (Figures 7H,I). Collectively, these data clearly demonstrate that anlotinib accelerates fibrosis resolution in vivo even after the establishment of fibrosis.
FIGURE 7.
Anlotinib accelerates resolution of bleomycin-induced pulmonary fibrosis. (A) Intervention dosing regimen of anlotinib in established pulmonary fibrosis. Bleomycin instillation was used to induce fibrosis and no treatment was given during the first 7 d. Then, mice were intraperitonealy injuected with 1 or 2 mg/kg of anlotinib or vehicle (n = 5-6 per group) daily. Lungs were harvested at 21 days. Tidal volume (TV) (B), dynamic compliance (Cdyn) (C), and lung resistance (RI) (D) of mice were measured (mean ± SD, n = 5). (E) HYP contents in lung tissues from mice (mean ± SD, n = 5). Representative images show H&E, Masson’s trichrome (F), α-SMA and Fibronectin staining (G) of lung sections from the indicated groups of mice. Scale bars, 100 µm. (H) Western blot analysis of Fibronectin、Collagen I and α-SMA, β-actin was used as a loading control (n = 4). (I) Quantification of Fibronectin、Collagen I and α-SMA protein levels relative to β-actin is shown (mean ± SD, n = 5). *p < 0.05, **p < 0.01, ***p < 0.001 VS BLM-treated group by one-way ANOVA.
Discussion
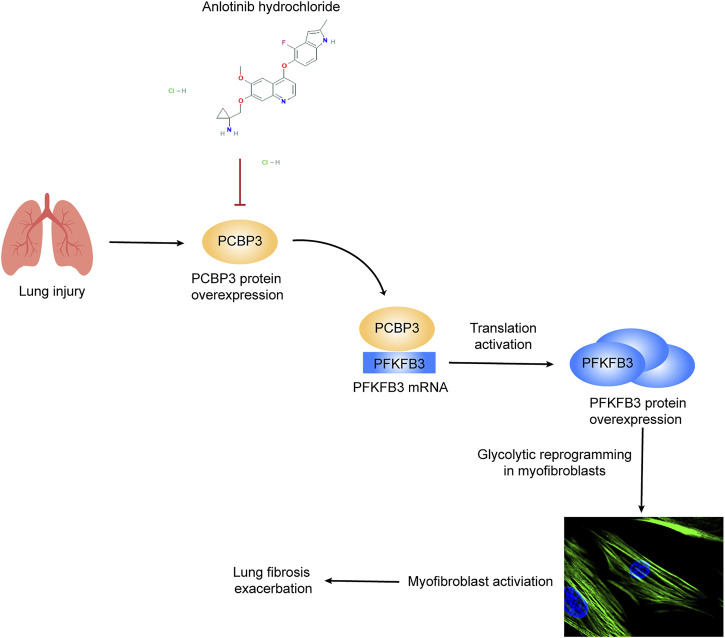
Despite recent advances in our understanding of IPF pathology, there is still no curative treatment for this disease; indeed, the currently available antifibrotic treatment modalities slow but do not completely stop the progression of the disease (Spagnolo and Maher, 2017). In this study, we demonstrate that anlotinib strongly inhibits fibroblast-to-myofibroblast transdifferentiation and reduces extracellular matrix production in primary MLFs and in the human IMR90 cell line. Accordingly, preventative and therapeutic administration of anlotinib to bleomycin-administered mice resulted in accelerated resolution of fibrosis. No adverse, systemic side effects were observed. Here, we demonstrate a novel mechanism by which anlotinib exerts antifibrotic effects by downregulating PCBP3, reducing PFKFB3 translation and inhibiting glycolysis in myofibroblasts (Figure 8).
FIGURE 8.
Schematic representation of PCBP3-PFKFB3-dependent glycolysis and the inhibitory effect of anlotinib on this pathway. Lung injury induces PCBP3 expression, which results in an increase in PFKFB3 expression by promoting its translation, resulting in the augmentation of glycolysis in lung fibroblasts. Glycolytic reprogramming participates in myofibroblast activation and furthers lung fibrosis. The tyrosine kinase inhibitor anlotinib inhibits PFKFB3-driven glycolysis by decreasing the expression of PCBP3, thereby suppressing myofibroblast activation and inhibiting the exacerbation of lung fibrosis.
A previous study revealed that male and female C57BL/6 mice did not differ in terms of their lung fibrotic responses, including cellular infiltration, collagen deposition, and quantifiable morphological changes in the lung architecture, but that the bleomycin-induced decrease in static compliance was significantly greater in males than in females (Voltz et al., 2008). This adverse effect on lung function was found to be due to male sex hormones. So sex differences should be carefully considered when interpreting experimental models of pulmonary fibrosis in mice (Blaauboer et al., 2014). In our study, we used only female mice to avoid the sex differences. We found that anlotinib exerted the preventative effects on bleomycin model of pulmonary fibrosis. Furthermore, anlotinib can also accelerate fibrosis resolution after the establishment of fibrosis.
A recent report showed that anlotinib inhibits the profibrotic effect of TGF-β1 in lung fibroblasts by attenuating inflammation and oxidative stress (Ruan et al., 2020). Our data are in line with that report and reveal an additional mechanism by which anlotinib acts on lung fibroblasts to attenuate fibrosis. There is emerging evidence about the association between metabolic disorders and IPF (Yin et al., 2019; Cho et al., 2020; Hu et al., 2020). Similar to highly proliferative cancer cells, myofibroblasts are highly dependent on glycolysis in vitro (Bueno et al., 2020). Furthermore, glycolysis is necessary not only for fibroblast growth and migration but also for the acquisition and maintenance of a myofibroblastic phenotype (Xu et al., 2017; Para et al., 2019). A previous study revealed that inhibition of glycolysis by the PFKFB3 inhibitor 3PO or by genomic disruption of the PFKFB3 gene blunted the differentiation of lung fibroblasts into myofibroblasts and attenuated profibrotic phenotypes in myofibroblasts (Xie et al., 2015). Another study revealed that lung fibroblasts displayed augmented aerobic glycolysis through activation of the PI3K-Akt-mTOR/PFKFB3 pathway in LPS-induced pulmonary fibrosis (Hu et al., 2020). Our data, along with previous studies, demonstrated that glycolytic reprogramming was critical to lung myofibroblast activation and pulmonary fibrosis. Furthermore, we demonstrated that anlotinib could strongly inhibit glycolytic reprogramming in vitro and in vivo.
The results presented herein provide new insights into the molecular mechanisms of lung fibrogenesis. This work unveils a previously unrecognized posttranscriptional regulation in activated lung fibroblasts composed of the RNA binding protein PCBP3 and the critical glycolytic enzyme PFKFB3, which maintains fibroblasts with higher glycolytic activity in fibrotic lungs compared to normal lung fibroblasts in healthy lungs. PCBP family members perform multiple functions by binding to the poly(C) sequence in both DNA and RNA to modulate mRNA stabilization, translation silencing, or translation enhancement (Blyn et al., 1997; Andino et al., 1999; Ostareck et al., 2001). Our present findings showing that PCBP3 plays an important role in myofibroblast activation and fibrogenesis and significantly extends our previous understanding by identifying an additional node of interaction between PCBP3-mediated posttranscriptional dysregulation and lung disease. We found that PFKFB3 protein overexpression was not accompanied by PFKFB3 mRNA upregulation, indicating that this increase was not transcriptionally derived. Instead, we observed that high PFKFB3 protein levels were maintained during fibroblast transdifferentiation, owing to PCBP3-mediated translational activation. Thus, the PCBP3 protein is upregulated during myofibroblast activation and binds directly to PFKFB3 during transcription. This binding activates PFKFB3 mRNA translation and generates high levels of the glycolysis activator PFKFB3. This mechanism does not exclude additional pathways of regulating PFKFB3 expression. Hence, it is not unusual for key proteins to be regulated at multiple levels, including through transcription, translation, and posttranslational modifications.
Our study is the first to report that anlotinib inhibits PFKFB3-mediated glycolysis in myofibroblasts. Moreover, anlotinib attenuates glycolysis in myofibroblasts by repressing PCBP3 expression levels rather than directly regulating the expression of PFKFB3, as anlotinib treatment does not decrease the mRNA levels of PFKFB3. Our work contributes novel mechanistic insight into the action of anlotinib. However, one of the limitations of this study is that we didn’t knock out PCBP3/PFKFB3 in mice to verify their effects in lung fibrosis, which may be explored in the further research. This future direction may be important to better understand how PCBP3 regulates PFKFB3-mediated glycolysis in pulmonary fibrosis. The other one is that this study only used bleomycin mice model for the research. Although the bleomycin model is the most widely used and best-characterized mouse model, the fibrosis of the bleomycin model is self-resolving, which contrasts with the progressive chronic fibrosis typical of human IPF (Liu et al., 2017). Therefore, whether anlotinib could attenuate fibrosis in human IPF still requires ex vivo models of pulmonary fibrosis.
In conclusion, our study demonstrated a clear antifibrotic role for anlotinib in the lungs. Its antifibrotic activity is mediated by its ability to decrease PCBP3 expression and attenuate PFKFB3-driven glycolysis, thereby inhibiting myofibroblast activation. Anlotinib might be considered as a potential therapeutic option for IPF patients.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by Southern Medical University Institutional Animal Care and Use Committee.
Author Contributions
WC, JZ, and WZ performed study concept and design; YL, YL, and ZZ performed development of methodology and writing; HH and XW provided acquisition, analysis and interpretation of data, and statistical analysis; XM and FZ provided technical and material support; SC and HD performed review and revision of the paper.
Funding
This work was supported by National Natural Science Foundation of China (No. 81870058, 81600648, 81970032, and 81870058), National Natural Science Foundation of Guangdong Province (NO. 2017A030313849).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.744826/full#supplementary-material
Abbreviations
IPF, idiopathic pulmonary fibrosis; BLM, bleomycin; RBP, RNA binding protein; α-SMA, α-smooth muscle actin; ECM, extracellular matrix; PFKFB3, 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase-3; F2, 6BP, fructose-2, 6-bisphosphate; PFK1, phosphofructokinase-1; TKI, tyrosine kinase inhibitor; VEGFR, vascular endothelial growth factor receptor; EGFR, epidermal growth factor receptor; FGFR, fibroblast growth factor receptor; PDGFR, platelet-derived growth factor receptor; TGF-β1, transforming growth factor-beta 1; TV, tidal volume; Cdyn, dynamic compliance; RI, resistance; HYP, hydroxyproline; IHC, immunohistochemical; H&E, Hematoxylin and eosin; MLF, mouse lung fibroblasts; PPI, protein-protein interaction; MAPK, mitogen-activated protein kinase; ECAR, extracellular acidification rate; siRNA, small interfering RNA; DMSO, dimethylsulfoxide; PBS, phosphate buffered saline; DMEM, Dulbecco’s Modified Eagle Medium; FBS, fetal bovine serum; DAPI, 4′,6-diamidino-2-phenylindole.
References
- Agostini F., Zanzoni A., Klus P., Marchese D., Cirillo D., Tartaglia G. G. (2013). catRAPID Omics: a Web Server for Large-Scale Prediction of Protein-RNA Interactions. Bioinformatics 29 (22), 2928–2930. 10.1093/bioinformatics/btt495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R., Böddeker N., Silvera D., Gamarnik A. V. (1999). Intracellular Determinants of Picornavirus Replication. Trends Microbiol. 7 (2), 76–82. 10.1016/s0966-842x(98)01446-2 [DOI] [PubMed] [Google Scholar]
- Atsumi T., Chesney J., Metz C., Leng L., Donnelly S., Makita Z., et al. (2002). High Expression of Inducible 6-Phosphofructo-2-Kinase/fructose-2,6-Bisphosphatase (iPFK-2; PFKFB3) in Human Cancers. Cancer Res. 62 (20), 5881–5887. 10.2337/diabetes.54.12.3349 [DOI] [PubMed] [Google Scholar]
- Blaauboer M. E., Boeijen F. R., Emson C. L., Turner S. M., Zandieh-Doulabi B., Hanemaaijer R., et al. (2014). Extracellular Matrix Proteins: a Positive Feedback Loop in Lung Fibrosis? Matrix Biol. 34, 170–178. 10.1016/j.matbio.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Blyn L. B., Towner J. S., Semler B. L., Ehrenfeld E. (1997). Requirement of poly(rC) Binding Protein 2 for Translation of Poliovirus RNA. J. Virol. 71 (8), 6243–6246. 10.1128/jvi.71.8.6243-6246.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno M., Calyeca J., Rojas M., Mora A. L. (2020). Mitochondria Dysfunction and Metabolic Reprogramming as Drivers of Idiopathic Pulmonary Fibrosis. Redox Biol. 33, 101509. 10.1016/j.redox.2020.101509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Zhang X., Wang L., Yang Q., Ma Q., Xu J., et al. (2019). PFKFB3-mediated Endothelial Glycolysis Promotes Pulmonary Hypertension. Proc. Natl. Acad. Sci. U S A. 116 (27), 13394–13403. 10.1073/pnas.1821401116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Xu T., Li M., Li C., Ma Y., Chen G., et al. (2021). Inhibition of SENP2-Mediated Akt deSUMOylation Promotes Cardiac Regeneration via Activating Akt Pathway. Clin. Sci. (Lond) 135 (6), 811–828. 10.1042/cs20201408 [DOI] [PubMed] [Google Scholar]
- Chen Z., Liu M., Li L., Chen L. (2018). Involvement of the Warburg Effect in Non-tumor Diseases Processes. J. Cel Physiol. 233 (4), 2839–2849. 10.1002/jcp.25998 [DOI] [PubMed] [Google Scholar]
- Cho S. J., Moon J. S., Nikahira K., Yun H. S., Harris R., Hong K. S., et al. (2020). GLUT1-dependent Glycolysis Regulates Exacerbation of Fibrosis via AIM2 Inflammasome Activation. Thorax 75 (3), 227–236. 10.1136/thoraxjnl-2019-213571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. S., Kim C. S., Hwang C. K., Song K. Y., Law P. Y., Wei L. N., et al. (2007). Novel Function of the Poly(C)-binding Protein Alpha CP3 as a Transcriptional Repressor of the Mu Opioid Receptor Gene. FASEB J. 21 (14), 3963–3973. 10.1096/fj.07-8561com [DOI] [PubMed] [Google Scholar]
- De Bock K., Georgiadou M., Schoors S., Kuchnio A., Wong B. W., Cantelmo A. R., et al. (2013). Role of PFKFB3-Driven Glycolysis in Vessel Sprouting. Cell 154 (3), 651–663. 10.1016/j.cell.2013.06.037 [DOI] [PubMed] [Google Scholar]
- DeBerardinis R. J., Thompson C. B. (2012). Cellular Metabolism and Disease: what Do Metabolic Outliers Teach Us? Cell 148 (6), 1132–1144. 10.1016/j.cell.2012.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller D., Grosdidier A., Wirth M., Daina A., Michielin O., Zoete V. (2014). SwissTargetPrediction: a Web Server for Target Prediction of Bioactive Small Molecules. Nucleic Acids Res. 42, W32–W38. 10.1093/nar/gku293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B. (2012). Mechanical Aspects of Lung Fibrosis: a Spotlight on the Myofibroblast. Proc. Am. Thorac. Soc. 9 (3), 137–147. 10.1513/pats.201202-017AW [DOI] [PubMed] [Google Scholar]
- Hinz B., Phan S. H., Thannickal V. J., Prunotto M., Desmoulière A., Varga J., et al. (2012). Recent Developments in Myofibroblast Biology: Paradigms for Connective Tissue Remodeling. Am. J. Pathol. 180 (4), 1340–1355. 10.1016/j.ajpath.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho R. C., Alcazar O., Fujii N., Hirshman M. F., Goodyear L. J. (2004). p38gamma MAPK Regulation of Glucose Transporter Expression and Glucose Uptake in L6 Myotubes and Mouse Skeletal Muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 286 (2), R342–R349. 10.1152/ajpregu.00563.2003 [DOI] [PubMed] [Google Scholar]
- Hu X., Xu Q., Wan H., Hu Y., Xing S., Yang H., et al. (2020). PI3K-Akt-mTOR/PFKFB3 Pathway Mediated Lung Fibroblast Aerobic Glycolysis and Collagen Synthesis in Lipopolysaccharide-Induced Pulmonary Fibrosis. Lab. Invest. 100 (6), 801–811. 10.1038/s41374-020-0404-9 [DOI] [PubMed] [Google Scholar]
- Huang Y., Wang Y., Wang X., Lin L., Wang P., Sun J., et al. (2020). The Effects of the Transforming Growth Factor-Β1 (TGF-Β1) Signaling Pathway on Cell Proliferation and Cell Migration Are Mediated by Ubiquitin Specific Protease 4 (USP4) in Hypertrophic Scar Tissue and Primary Fibroblast Cultures. Med. Sci. Monit. 26, e920736. 10.12659/msm.920736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J., Fogarty A., Hubbard R., McKeever T. (2015). Global Incidence and Mortality of Idiopathic Pulmonary Fibrosis: a Systematic Review. Eur. Respir. J. 46 (3), 795–806. 10.1183/09031936.00185114 [DOI] [PubMed] [Google Scholar]
- Izbicki G., Segel M. J., Christensen T. G., Conner M. W., Breuer R. (2002). Time Course of Bleomycin-Induced Lung Fibrosis. Int. J. Exp. Pathol. 83 (3), 111–119. 10.1046/j.1365-2613.2002.00220.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman E. R., Khambata V. S., Cope C., Jones P., Roger J., Ye L. Y., et al. (2014). An Inhibitor of NADPH Oxidase-4 Attenuates Established Pulmonary Fibrosis in a Rodent Disease Model. Am. J. Respir. Cel. Mol. Biol. 50 (1), 158–169. 10.1165/rcmb.2013-0174OC [DOI] [PubMed] [Google Scholar]
- Kang D. H., Song K. Y., Choi H. S., Law P. Y., Wei L. N., Loh H. H. (2012). Novel Dual-Binding Function of a Poly (C)-binding Protein 3, Transcriptional Factor Which Binds the Double-Strand and Single-Stranded DNA Sequence. Gene 501 (1), 33–38. 10.1016/j.gene.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Kenyon N. J., Ward R. W., McGrew G., Last J. A. (2003). TGF-beta1 Causes Airway Fibrosis and Increased Collagen I and III mRNA in Mice. Thorax 58 (9), 772–777. 10.1136/thorax.58.9.772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Noren Hooten N., Dluzen D. F., Martindale J. L., Gorospe M., Evans M. K. (2015). Posttranscriptional Regulation of the Inflammatory Marker C-Reactive Protein by the RNA-Binding Protein HuR and MicroRNA 637. Mol. Cel. Biol. 35 (24), 4212–4221. 10.1128/mcb.00645-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidgens S., Bullough K. Z., Shi H., Li F., Shakoury-Elizeh M., Yabe T., et al. (2013). Each Member of the poly-R(C)-binding Protein 1 (PCBP) Family Exhibits Iron Chaperone Activity toward Ferritin. J. Biol. Chem. 288 (24), 17791–17802. 10.1074/jbc.M113.460253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Hui K., Hu C., Wen Y., Yang S., Zhu P., et al. (2019). Autophagy Inhibition Potentiates the Anti-angiogenic Property of Multikinase Inhibitor Anlotinib through JAK2/STAT3/VEGFA Signaling in Non-small Cell Lung Cancer Cells. J. Exp. Clin. Cancer Res. 38 (1), 71. 10.1186/s13046-019-1093-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Philp A. M., Corte T., Travis M. A., Schilter H., Hansbro N. G., et al. (2021). Therapeutic Targets in Lung Tissue Remodelling and Fibrosis. Pharmacol. Ther. 225, 107839. 10.1016/j.pharmthera.2021.107839 [DOI] [PubMed] [Google Scholar]
- Liu T., De Los Santos F. G., Phan S. H. (2017). The Bleomycin Model of Pulmonary Fibrosis. Methods Mol. Biol. 1627, 27–42. 10.1007/978-1-4939-7113-8_2 [DOI] [PubMed] [Google Scholar]
- Livi C. M., Klus P., Delli Ponti R., Tartaglia G. G. (2016). catRAPID Signature: Identification of Ribonucleoproteins and RNA-Binding Regions. Bioinformatics 32 (5), 773–775. 10.1093/bioinformatics/btv629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Yu C. H., Li W., Li T., Luo W., Huang S., et al. (2014). Angiotensin-converting Enzyme 2/angiotensin-(1-7)/Mas axis Protects against Lung Fibrosis by Inhibiting the MAPK/NF-κB Pathway. Am. J. Respir. Cel. Mol. Biol. 50 (4), 723–736. 10.1165/rcmb.2012-0451OC [DOI] [PubMed] [Google Scholar]
- Moeller A., Ask K., Warburton D., Gauldie J., Kolb M. (2008). The Bleomycin Animal Model: a Useful Tool to Investigate Treatment Options for Idiopathic Pulmonary Fibrosis? Int. J. Biochem. Cel. Biol. 40 (3), 362–382. 10.1016/j.biocel.2007.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigdelioglu R., Hamanaka R. B., Meliton A. Y., O'Leary E., Witt L. J., Cho T., et al. (2016). Transforming Growth Factor (TGF)-β Promotes De Novo Serine Synthesis for Collagen Production. J. Biol. Chem. 291 (53), 27239–27251. 10.1074/jbc.M116.756247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck D. H., Ostareck-Lederer A., Shatsky I. N., Hentze M. W. (2001). Lipoxygenase mRNA Silencing in Erythroid Differentiation: The 3'UTR Regulatory Complex Controls 60S Ribosomal Subunit Joining. Cell 104 (2), 281–290. 10.1016/s0092-8674(01)00212-4 [DOI] [PubMed] [Google Scholar]
- Papa S., Choy P. M., Bubici C. (2019). The ERK and JNK Pathways in the Regulation of Metabolic Reprogramming. Oncogene 38 (13), 2223–2240. 10.1038/s41388-018-0582-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para R., Romero F., George G., Summer R. (2019). Metabolic Reprogramming as a Driver of Fibroblast Activation in PulmonaryFibrosis. Am. J. Med. Sci. 357 (5), 394–398. 10.1016/j.amjms.2019.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantier L., Cazes A., Dinh-Xuan A. T., Bancal C., Marchand-Adam S., Crestani B. (2018). Physiology of the Lung in Idiopathic Pulmonary Fibrosis. Eur. Respir. Rev. 27 (147), 170062. 10.1183/16000617.0062-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H., Lv Z., Liu S., Zhang L., Huang K., Gao S., et al. (2020). Anlotinib Attenuated Bleomycin-Induced Pulmonary Fibrosis via the TGF-Β1 Signalling Pathway. J. Pharm. Pharmacol. 72 (1), 44–55. 10.1111/jphp.13183 [DOI] [PubMed] [Google Scholar]
- Safran M., Solomon I., Shmueli O., Lapidot M., Shen-Orr S., Adato A., et al. (2002). GeneCards 2002: towards a Complete, Object-Oriented, Human Gene Compendium. Bioinformatics 18 (11), 1542–1543. 10.1093/bioinformatics/18.11.1542 [DOI] [PubMed] [Google Scholar]
- Sapudom J., Rubner S., Martin S., Thoenes S., Anderegg U., Pompe T. (2015). The Interplay of Fibronectin Functionalization and TGF-Β1 Presence on Fibroblast Proliferation, Differentiation and Migration in 3D Matrices. Biomater. Sci. 3 (9), 1291–1301. 10.1039/c5bm00140d [DOI] [PubMed] [Google Scholar]
- Sato S., Shinohara S., Hayashi S., Morizumi S., Abe S., Okazaki H., et al. (2017). Anti-fibrotic Efficacy of Nintedanib in Pulmonary Fibrosis via the Inhibition of Fibrocyte Activity. Respir. Res. 18 (1), 172. 10.1186/s12931-017-0654-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo P., Maher T. M. (2017). Clinical Trial Research in Focus: Why Do So many Clinical Trials Fail in IPF? Lancet Respir. Med. 5 (5), 372–374. 10.1016/s2213-2600(17)30122-4 [DOI] [PubMed] [Google Scholar]
- Sun Y., Niu W., Du F., Du C., Li S., Wang J., et al. (2016). Safety, Pharmacokinetics, and Antitumor Properties of Anlotinib, an Oral Multi-Target Tyrosine Kinase Inhibitor, in Patients with Advanced Refractory Solid Tumors. J. Hematol. Oncol. 9 (1), 105. 10.1186/s13045-016-0332-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. (2019). STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-wide Experimental Datasets. Nucleic Acids Res. 47, D607–D613. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H., Ebina M., Kondoh Y., Ogura T., Azuma A., Suga M., et al. (2010). Pirfenidone in Idiopathic Pulmonary Fibrosis. Eur. Respir. J. 35 (4), 821–829. 10.1183/09031936.00005209 [DOI] [PubMed] [Google Scholar]
- Taurin S., Yang C. H., Reyes M., Cho S., Coombs D. M., Jarboe E. A., et al. (2018). Endometrial Cancers Harboring Mutated Fibroblast Growth Factor Receptor 2 Protein Are Successfully Treated with a New Small Tyrosine Kinase Inhibitor in an Orthotopic Mouse Model. Int. J. Gynecol. Cancer 28 (1), 152–160. 10.1097/igc.0000000000001129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schaftingen E., Lederer B., Bartrons R., Hers H. G. (1982). A Kinetic Study of Pyrophosphate: Fructose-6-Phosphate Phosphotransferase from Potato Tubers. Application to a Microassay of Fructose 2, 6-bisphosphate. Eur. J. Biochem. 129 (1), 191–195. 10.1111/j.1432-1033.1982.tb07039.x [DOI] [PubMed] [Google Scholar]
- Voltz J. W., Card J. W., Carey M. A., Degraff L. M., Ferguson C. D., Flake G. P., et al. (2008). Male Sex Hormones Exacerbate Lung Function Impairment after Bleomycin-Induced Pulmonary Fibrosis. Am. J. Respir. Cel. Mol. Biol. 39 (1), 45–52. 10.1165/rcmb.2007-0340OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Qi X. M., Wertz R., Mortensen M., Hagen C., Evans J., et al. (2020a). p38γ MAPK Is Essential for Aerobic Glycolysis and Pancreatic Tumorigenesis. Cancer Res. 80 (16), 3251–3264. 10.1158/0008-5472.CAN-19-3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Guo X., Wang D., Yang S. (2020b). Expression Patterns of Pcbp Gene Family Members during Zebrafish Embryogenesis. Gene Expr. Patterns 35, 119097. 10.1016/j.gep.2020.119097 [DOI] [PubMed] [Google Scholar]
- Wynn T. A., Ramalingam T. R. (2012). Mechanisms of Fibrosis: Therapeutic Translation for Fibrotic Disease. Nat. Med. 18 (7), 1028–1040. 10.1038/nm.2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Wan X., Quan H., Zheng M., Fu L., Li Y., et al. (2018). Preclinical Characterization of Anlotinib, a Highly Potent and Selective Vascular Endothelial Growth Factor Receptor-2 Inhibitor. Cancer Sci. 109 (4), 1207–1219. 10.1111/cas.13536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie N., Tan Z., Banerjee S., Cui H., Ge J., Liu R. M., et al. (2015). Glycolytic Reprogramming in Myofibroblast Differentiation and Lung Fibrosis. Am. J. Respir. Crit. Care Med. 192 (12), 1462–1474. 10.1164/rccm.201504-0780OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li J., Yu Z., Rao H., Wang S., Lan H. (2017). HMGB1 Promotes HLF-1 Proliferation and ECM Production through Activating HIF1-α-Regulated Aerobic Glycolysis. Pulm. Pharmacol. Ther. 45, 136–141. 10.1016/j.pupt.2017.05.015 [DOI] [PubMed] [Google Scholar]
- Yin X., Choudhury M., Kang J. H., Schaefbauer K. J., Jung M. Y., Andrianifahanana M., et al. (2019). Hexokinase 2 Couples Glycolysis with the Profibrotic Actions of TGF-β. Sci. Signal. 12 (612), eaax4067. 10.1126/scisignal.aax4067 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.