Abstract
The inner membrane-bound protein Ras integrates various extracellular signals that are subsequently communicated from the cytoplasm to the nucleus via the Raf/MEK/MAPK cascade. Here we show that the retinoblastoma protein pRb, previously reported to be a nuclear target of this pathway, can in turn influence the activation state of Ras. Rb-deficient fibroblasts display elevated levels (up to 30-fold) of activated Ras during G1. Expression of wild-type pRb or a number of pRb mutants defective in E2F regulation reverses this effect. We provide evidence that the mid-G1 activation of Ras in Rb-deficient cells, which occurs at the level of guanine nucleotide binding, differs from that of epidermal growth factor-induced stimulation of Ras, being dependent on protein synthesis. The aberrant levels of Ras activity associated with loss of pRb may be responsible for the differentiation defects in Rb-deficient cells, because suppression of Ras activity in Rb−/− fibroblasts restores the transactivation function of MyoD and the expression of a late marker of skeletal muscle differentiation. These data suggest that nuclear-cytoplasmic communication between pRb and Ras is bidirectional.
The three ras proto-oncogenes—encoding H-Ras, K-Ras4A, K-Ras4B, and N-Ras (2)—have each been implicated in the regulation of differentiation, cell growth, and cell proliferation (33, 34). Ras proteins participate in various extracellular signaling cascades initiated from a number of receptor and nonreceptor tyrosine kinases and thus serve to communicate information from the cell surface to the nucleus (50). In this context, they operate as molecular switches activated by guanine nucleotide exchange factor-mediated enrichment of active, GTP-bound forms (45). A number of effector pathways are downstream of Ras, the best characterized of which is the Raf/MEK/MAPK kinase cascade. This mitogenic signaling pathway is perhaps the archetypal example of Ras-mediated communication from the extracellular milieu to the nucleus. Indeed, virtually all studies focused on Ras signaling pertain to the unidirectional flow of information from the cytoplasm to the nucleus.
Like Ras, the retinoblastoma protein pRb is also involved in regulation of cell proliferation during G1 and differentiation processes (7, 17, 60). pRb controls cell cycle progression, at least in part, through regulation of the E2F family of transcription factors, which is in turn mediated by phosphorylation events (14, 60). During mid-G1, the initial phosphorylation events on pRb are controlled by D-type cyclins (cyclins D1, D2, and D3) (55). Consistent with the notion of nuclear proteins being the ultimate target of Ras-mediated signaling, each member of the Ras/Raf/MEK/MAPK pathway has been implicated in the regulation of cyclin D1 and thereby the state of pRb phosphorylation and its cell cycle function (11). Indeed, Rb-deficient fibroblasts are resistant to the G1 arrest induced by either Ras inactivation (29, 37, 44) or cyclin D1 neutralization (32). Thus, pRb appears a legitimate downstream target of Ras action. A connection between Ras and pRb has also recently been demonstrated genetically in Caenorhabditis elegans. However, the LIN-35 pRb/LET-60 Ras communication appears not to be involved in the regulation of proliferation in this system (31).
Both Ras and pRb also control differentiation (5, 7, 28, 34, 62, 63). Unlike cell cycle progression, a link between pRb and Ras during differentiation is not immediately apparent, although available evidence suggests that such a connection may exist. For example, either loss of pRb or ectopic expression of constitutively active Ras impairs the transcriptional functions of MyoD (20, 24, 38, 40, 47, 53). Thus, both the presence of pRb and the regulation of Ras activity appear to be important for proper MyoD function.
It has recently been demonstrated that Ras activation following restimulation of quiescent murine fibroblasts is biphasic, with peaks of activation soon after growth factor addition and then again in mid-G1 (58). In the same body of work, it was shown that the mid-G1 activation of Ras in a human cervical carcinoma line, HeLa, was independent of serum factors, but was dependent on de novo mRNA and protein synthesis (58). Since HeLa cells express the E7 oncoprotein, which binds to and inactivates pRb (15), one interpretation of these results is that pRb might influence Ras activation. We have directly addressed this possibility by using mouse embryo fibroblasts (MEFs) derived from Rb-deficient embryos and show that these cells display elevated levels of activated N- and K-Ras. Additionally, we provide evidence suggesting that the ability of pRb to regulate the activation of Ras may be linked to the influence of pRb on differentiation. These data suggest that signaling from Ras to the nucleus is bidirectional.
MATERIALS AND METHODS
Cell culture.
Rb+/+ and Rb−/− MEFs and their immortalized 3T3 derivatives have been described previously (44). MEFs derived from p107-deficient mice were kindly provided by T. Jacks, N. Dyson, and E. Harlow. 3T3 derivatives of p107−/− MEFs were generated as described previously (59). All MEFs and their 3T3 derivatives were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). NIH 3T3 cells and their cyclin D1 derivatives (46) were cultured in DMEM containing 5% bovine calf serum. Rb−/− 3T3 cells expressing hemagglutinin (HA)-tagged versions of wild type and mutant pRb were created by cotransfecting pSG5L-HA-pRB, pSG5L-HA-pRBΔex22, pSG5L-HA-pRB;661W or pSG5L-HA-pRBΔex4 (53) together with pBABE-puro, with subsequent selection in puromycin (3 μg/ml). Both stable clones (see Fig. 2) and pooled populations (see Fig. 4) were used for analysis. pRb expression was reconstituted in Rb−/− MEFs by retroviral infection. The retroviral vector encoding human pRb, pBPJTR2-pRb, was constructed by subcloning the Rb cDNA from pSG5-Rb into pBPJTR2 (42). The packaging cell line, Bosc23 (43), was transfected with pBPJTR2-pRb, and the resulting retroviral supernatant was used for infections. After 3 days of puromycin selection, pooled populations were used for analysis. Rb−/− 3T3 cells expressing MyoD were similarly constructed by using a retroviral vector encoding MyoD (pBabe-MyoD [38]). Stable lines of Rb+/+ 3T3 cells expressing T antigen and the K1 mutant were created by cotransfecting pSG5-WT T antigen (TAg) or pSG5-K1 TAg (64) together with pBABE-puro, followed by selection in puromycin. For serum starvation, cells were maintained in DMEM containing 0.2% FBS for 72 h.
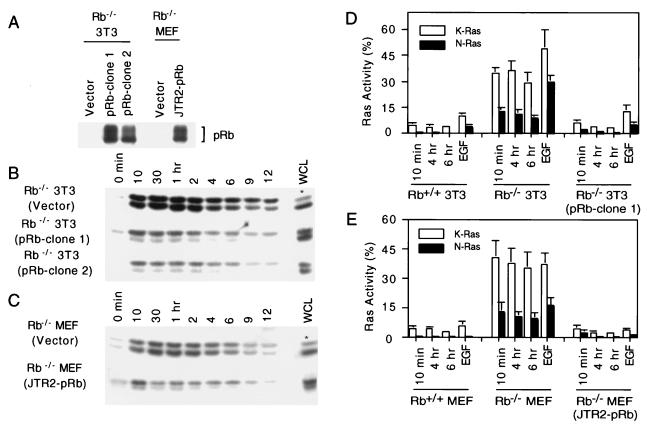
FIG. 2.
Reconstitution of pRb in Rb-deficient fibroblasts and the effect on Ras activation. (A) Individual clones of Rb−/− 3T3 cells transfected with or without a pRb-encoding plasmid were analyzed for pRb expression by immunoprecipitation followed by Western blot analysis (pRb-clone 1 and -2 and vector). Rb−/− MEFs were infected with a pRb retrovirus (JTR2-pRb) or control retrovirus (vector), and pooled populations were analyzed for pRb expression. (B) Rb−/− 3T3 clones described for panel A were analyzed for the presence of activated N- and K-Ras as described in the legend to Fig. 1. Fifteen micrograms of whole-cell lysate (WCL) was analyzed for total Ras proteins; an asterisk indicates that 30 μg instead of 15 μg of WCL was loaded. (C) Same as panel B, except Rb−/− MEFs were analyzed. (D) N- and K-Ras activation during reentry to the cell cycle from G0. Rb−/− 3T3, Rb+/+ 3T3, and an Rb−/− 3T3 clone in which pRb expression was reconstituted (clone 1) were used for the analysis. Ras activation was expressed as the ratio of activated N- or K-Ras to total N- or K-Ras expressed as a percentage following densitometric scanning of autoradiograms represented in Fig. 1 and 2B. Serum-starved and EGF-treated cells (10 ng/ml for 10 min) were analyzed in parallel. Bars represent the averages plus standard deviations for at least three independent experiments. (E) Same as panel D, except primary MEFs were used.
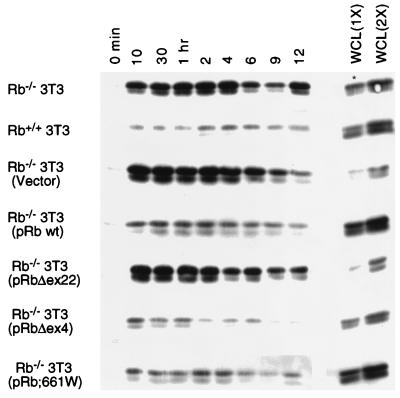
FIG. 4.
Effect of protein products encoded by partially penetrant mutants of Rb on Ras activation. Individual lines of Rb−/− 3T3 cells expressing pRb;661W, pRbΔex4, or pRbΔex22 and control (empty vector) lines were analyzed for the presence of activated N- and K-Ras as described in the legend to Fig. 1. Whole-cell lysates (WCL) at 15 μg (1X) or 30 μg (2X) were analyzed for total Ras protein; an asterisk indicates that 30 and 60 μg were loaded. wt, wild type. Results are representative of five independent experiments.
Ras activation assays.
Cells were washed twice with ice-cold HBS (25 mM HEPES [pH 7.5], 150 mM NaCl) and lysed in Mg2+-containing lysis buffer (25 mM HEPES [pH 7.5], 150 mM NaCl, 1% NP-40, 0.25% Na deoxycholate, 10% glycerol, 25 mM NaF, 10 mM MgCl2, 1 mM EDTA, 1 mM Na3VO4, 250 μM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, and 10 μg of aprotinin per ml). Lysates were clarified by centrifugation, and protein concentrations were determined (Bio-Rad protein assay). In all assays, 300 μg of the supernatants was incubated with the Ras binding domain (RBD) of cRaf-1 fused to glutathione S-transferase (GST-RBD) to isolate GTP-bound Ras (58). Glutathione-Sepharose beads (15 μl of packed beads [Pharmacia]) were preloaded with GST-RBD (10 μg). After incubation for 40 min at 4°C, beads were washed four times in lysis buffer. Bound proteins, separated on sodium dodecyl sulfate-polyacrylamide gels (12%), were transferred to polyvinylidene difluoride membranes, probed with pan Ras antibody (Ab-3; Oncogene Science), and visualized by enhanced chemiluminescence (ECL; Amersham).
Cell permeabilization and Ras guanine nucleotide binding.
Rb−/− and Rb+/+ 3T3 cells were serum starved and stimulated with DMEM containing 10% FBS for 5 min or 4 h before being washed with warm phosphate-buffered saline. To each dish, 0.36 ml of permeabilization buffer (Trans-port transient cell permeabilization kit; GIBCO/BRL) was added and diluted with Trans-port reagent, immediately followed by the addition of 10 μCi of [α-32P]GTP (3,000 Ci/mmol; NEN) (time zero). At various times thereafter, the supernatant was removed, and cells were lysed in 1% Triton X-100 buffer (50 mM HEPES [pH 7.4], 1% Triton X-100, 100 mM NaCl, 5 mM MgCl2, 1 mg of bovine serum albumin per ml, 250 μM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml) containing 0.1 mM unlabeled GTP. Ras was immunoprecipitated from equal amounts of total cellular protein by using Y13-259 (American Type Culture Collection) as described previously (12). Total specific radioactivity was determined by Cerenkov counting for 32P. Duplicate samples were analyzed, and values were averaged.
Transcriptional transactivation assays.
For MyoD transactivation assays, Rb+/+ and Rb−/− 3T3 fibroblasts were plated onto 60-mm-diameter dishes at 1.5 × 105 and 1 × 105 cells per plate, respectively. The cells were transfected (44) as indicated with 1 μg of pCSA-MyoD (38), 2 μg of pMCK-Luc (gift from A. Lassar), 1 μg of pCMV-βGal, 2 to 4 μg of pSG5L-HA-RB (53), 0.25 to 0.75 μg of pMT-RasN17 (18), or empty vector plasmid. Forty-eight hours after transfection, the cell culture medium was changed to differentiation medium (DMEM containing 2% horse serum). Luciferase and β-galactosidase activities were assayed 48 h later. For glucocorticoid receptor alpha (GRα) transactivation assays, Rb+/+ and Rb−/− 3T3 fibroblasts were plated onto 60-mm-diameter dishes at 2 × 105 and 1.5 × 105 cells per plate, respectively. The cells were transfected as indicated with 0.5 μg of pRS-hGRα (19), 2 μg of pMMTV-GRE-Luc (gift from W. Chin), 1 μg of pCMV-βGal, 1 to 2 μg of pSG5L-HA-RB, 0.25 to 0.75 μg of pMT-RasN17, or empty vector plasmid. Twenty-four hours after transfection, dexamethasone (1 μM) was added. Luciferase and β-galactosidase activities were determined 24 h later. For E2F transactivation assays, 2 × 105 Rb−/− 3T3 cells were transfected as indicated with 150 to 450 ng of pRc/CMV-E2F-1 (gift from E. Flemington), 1 μg of 3X(E2F)DHFR-Luc (gift from E. Flemington), 1 μg of pCMV-βGal, 0.25 to 0.75 μg of pMT-RasN17, or empty vector plasmid. Luciferase and β-galactosidase activities were determined 48 h later.
MHC induction.
Rb−/− 3T3 cells stably expressing MyoD were transfected with plasmids encoding RasN17, pRb, or the vector control and subsequently placed in differentiation medium. Forty-eight hours later, cell lysates were prepared, and 10 μg was resolved on a denaturing gel. After transfer to a polyvinylidene difluoride membrane, the blot was probed with a monoclonal antibody to myosin heavy chain (MHC) (MF20 [1]; Developmental Studies Hybridoma Bank) followed by enhanced chemiluminescence detection (Amersham).
RESULTS
Rb-deficient fibroblasts display elevated levels of activated N- and K-Ras.
To determine whether loss of pRb influences Ras activity, we analyzed the activation state of Ras during the G1 interval in MEFs derived from genotyped Rb−/− and Rb+/+ littermate embryos, and their derivatives were immortalized according to a defined 3T3 protocol. The activation state of Ras was determined with an assay that is based on the fact that active, GTP-bound forms of Ras bind to Raf-1, while inactive, GDP-bound Ras does not (58).
In Rb+/+ MEFs and 3T3 cells released from quiescence by serum stimulation, K-Ras was activated throughout G1 with the highest levels of activation in mid-G1 (Fig. 1A and B). Rb−/− MEFs and their immortalized derivatives showed a similar K-Ras activation profile, although the extent of activation was higher than with their Rb-positive counterparts. During the same time course, little activated N-Ras was detected in Rb-positive fibroblasts. In striking contrast, the GTP-bound form of N-Ras was readily detectable in Rb-deficient fibroblasts (Fig. 1A and B). Quantitative analysis revealed a significantly higher level (at least 30-fold) of N-Ras activation in Rb-deficient cells than that in their wild-type counterparts (Fig. 2D and E). The identity and migration pattern of N- and K-Ras were confirmed by Western blot analysis with antibodies specific to the various Ras isoforms. H-Ras was barely detectable in either cell line (data not shown).
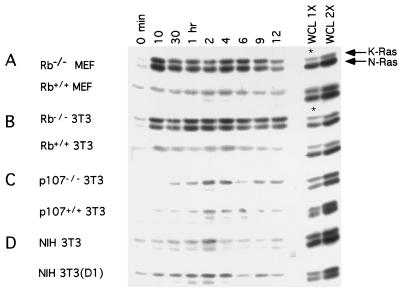
FIG. 1.
Ras activation in Rb−/− and Rb+/+ fibroblasts and other cell strains. (A to D) The indicated cell types were serum starved for 72 h and restimulated by the addition of serum. At the indicated times, lysates were prepared, and the presence of activated N- and K-Ras in equal amounts of total protein was assayed. Whole-cell lysates (WCL) at 15 μg (1X) or 30 μg (2X) were analyzed for total Ras protein; asterisks indicate that 30 and 60 μg were loaded. Each panel is representative of at least five independent experiments.
Ras activation during the G1 interval in p107−/− 3T3 cells was similar to that found in Rb-positive cells, with very low levels of N-Ras activation (Fig. 1C). Similar results were obtained with p130−/− fibroblasts (data not shown). Thus, of the pRb family members, the effects observed on N- and K-Ras activation appear to be a specific function of pRb. Together, these results suggest that the absence of pRb, one of the ultimate targets of Ras-mediated signaling, can itself affect Ras activity.
We determined whether upstream regulators of pRb that modulate its function by effecting phosphorylation would mimic the alteration in N- and K-Ras activation observed in Rb−/− fibroblasts. To this end, we analyzed Ras activation in NIH 3T3 cells in which cyclin D1 is ectopically expressed. Little difference was found in the Ras activation profile during G1 when comparing parental NIH 3T3 cells and their cyclin D1 derivatives (Fig. 1D), suggesting that premature activation of CDK4 does not influence Ras activity.
Reconstitution of pRb in Rb-deficient fibroblasts decreases levels of activated N- and K-Ras.
To rule out the possibility that the effect of pRb on the levels of GTP-bound N- and K-Ras might be due to a genetic event other than loss of Rb, we reintroduced Rb into Rb-deficient fibroblasts and analyzed Ras activity. The expression of pRb in two such representative clones is shown in Fig. 2A. These clones showed a pattern of Ras activation during G1 that was similar to that found with Rb-positive fibroblasts (Fig. 2B). Similar results were obtained when a pooled population of Rb−/− primary MEFs, in which pRb expression had been reconstituted by retroviral infection, was analyzed (Fig. 2A and C). In both the primary and immortalized Rb−/− cells, reintroduction of pRb decreased the level of N- and K-Ras activation in mid-G1 by approximately 13- and 10-fold, respectively (Fig. 2D and E). Thus, at this level of analysis, the effect of pRb loss on N- and K-Ras activation appears to be reversible and not attributable to an adaptive mutation.
We consistently observed lower levels of total Ras protein in Rb-deficient fibroblasts than in their wild-type counterparts, with the effect being more pronounced for K-Ras (Fig. 1). Reintroduction of Rb into Rb−/− fibroblasts appears to restore the total levels of Ras (Fig. 2). The fold reduction in total Ras (approximately twofold for N-Ras and three- to fourfold for K-Ras) cannot account for the differences noted in the levels of activated Ras (the ratio of GTP-bound Ras to total Ras). In addition, we have created stable lines of Rb−/− and Rb+/+ 3T3 cells expressing matched amounts of epitope-tagged Ras. Analysis of these lines reveals higher levels of activated (endogenous and exogenous) Ras in the Rb-deficient cells than in the Rb+/+ cells (data not shown). Furthermore, the levels of endogenous activated Ras in these lines are unaltered. The total amount of Ras protein does not appear to have a direct bearing upon the proportion of active to total Ras. Thus, we feel that the effects on the total levels of Ras observed do not explain how loss of pRb leads to elevated levels of activated Ras.
Viral oncoproteins induce activation of Ras.
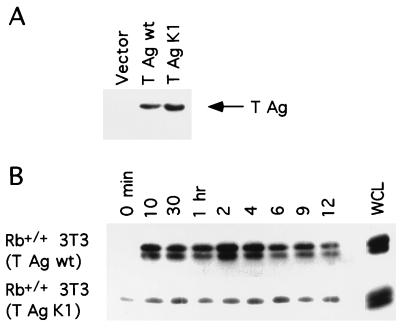
To further exclude the possibility that a pRb-independent mechanism might be responsible for the observed aberrant levels of active Ras in Rb−/− cells, we inactivated pRb function in Rb+/+ cells and determined the effect on Ras. To this end, Rb+/+ 3T3 cells expressing simian virus 40 large TAg which can bind to and inactivate pRb, were generated (Fig. 3A). The profile of Ras activation during the G1 interval in Rb-positive cells expressing T antigen was similar to that observed in Rb−/− 3T3 cells (Fig. 3B). Analogous results have been reported for murine C3H10T1/2 cells expressing TAg (48). In contrast, Rb+/+ 3T3 cells expressing a mutant of TAg (K1) that fails to bind to pRb (10) did not exhibit elevated levels of activated N- and K-Ras (Fig. 3). Similar observations were made with Rb+/+ and NIH 3T3 cells expressing human papillomavirus E7 (data not shown). Thus, functional inactivation of pRb through the expression of viral oncoproteins results in increased levels of Ras activity.
FIG. 3.
Ras activation in Rb+/+ 3T3 cells expressing simian virus 40 TAg. (A) Rb+/+ 3T3 lines expressing wild-type (wt) TAg mutant TAg (K1), or the vector control were analyzed for expression of TAg. (B) Rb+/+ 3T3 lines described for panel A were analyzed for the presence of activated N- and K-Ras as described in the legend to Fig. 1. WCL, whole-cell lysate. Results are representative of at least five independent experiments.
Protein products encoded by partially penetrant alleles of Rb retain the ability to regulate Ras activity.
Classical familial retinoblastoma is attributable to germ line mutations in the Rb gene. These mutations result in bilateral tumors in 90% of carriers. However, Rb mutations have been identified in which the carriers are either absent of disease, develop unilateral retinoblastoma, or suffer benign retinomas (13, 25, 30). The protein products encoded by such “partially penetrant Rb alleles” have been shown to be defective for a subset of known pRb functions (25, 53, 61), including the ability to bind E2F. In an effort to gain further insight into which functions of pRb might be involved in the regulation of Ras activity, we determined whether the protein products encoded by two partially penetrant Rb alleles, 661W (amino acid substitution) and Δex4 (deletion of exon 4), retain the ability to affect Ras activation. Expression of either of these pRb mutants in Rb−/− 3T3 cells resulted in a marked downmodulation of Ras activation, similar to that observed upon reintroduction of wild-type pRb (Fig. 2 and 4). In contrast, ectopic expression of a pRb mutant defective in all known functions of pRb, Δex22 (deletion exon 22), had no effect on Ras activation (Fig. 4). At this level of analysis, these results indicate that the protein products of two partially penetrant alleles of Rb retain the ability to regulate the activation state of Ras and suggest that this function of pRb is separable from its role in E2F regulation.
Inhibition of Ras activity restores pRb-dependent transcription and the expression of a late marker of skeletal muscle differentiation.
In an effort to demonstrate a biological consequence of elevated Ras activation resulting from loss of pRb, we considered the known properties of Rb-deficient fibroblasts. If aberrant Ras activation was indeed responsible for the characteristic cell cycle or differentiation defects associated with pRb loss, we would predict that inhibition of Ras activity in a pRb-negative background should restore these functions. Although not ruling out the possibility that the effect of pRb loss on Ras activation does, in some way, impinge upon cell cycle and/or growth control, it has already been established that Ras inactivation in Rb-deficient fibroblasts is without significant impact on G1 cell cycle progression (29, 37, 44). The mechanisms by which pRb controls differentiation are less well characterized than those of pRb-mediated regulation of E2F, although pRb appears to regulate a number of transcription factors involved in promoting these processes (5, 6, 20, 57). Importantly, the pRb mutants 661W and Δex4, which have lost the capacity to regulate E2F while retaining the ability to potentiate certain differentiation processes (53), behave essentially like the wild-type protein in relation to Ras activation (Fig. 4). We considered the possibility that loss of pRb leading to aberrant levels of N- and K-Ras activation might be linked to the inability of Rb-deficient cells to support certain aspects of differentiation.
Loss of pRb has been shown to lead to defects in skeletal muscle cell differentiation (20, 51, 63). Furthermore, the transcriptional activity of MyoD, a key regulator of muscle differentiation, is impaired in Rb-deficient cells, and this defect can be corrected by reintroduction of pRb (38, 53). Likewise, constitutively activated Ras has been shown to inhibit skeletal myoblast differentiation and the transcriptional transactivation function of MyoD in a cell cycle-independent fashion (24, 40, 47). Thus, we tested the possibility that the aberrant levels of Ras activity resulting from loss of pRb might contribute to the defect in MyoD function in Rb-deficient cells.
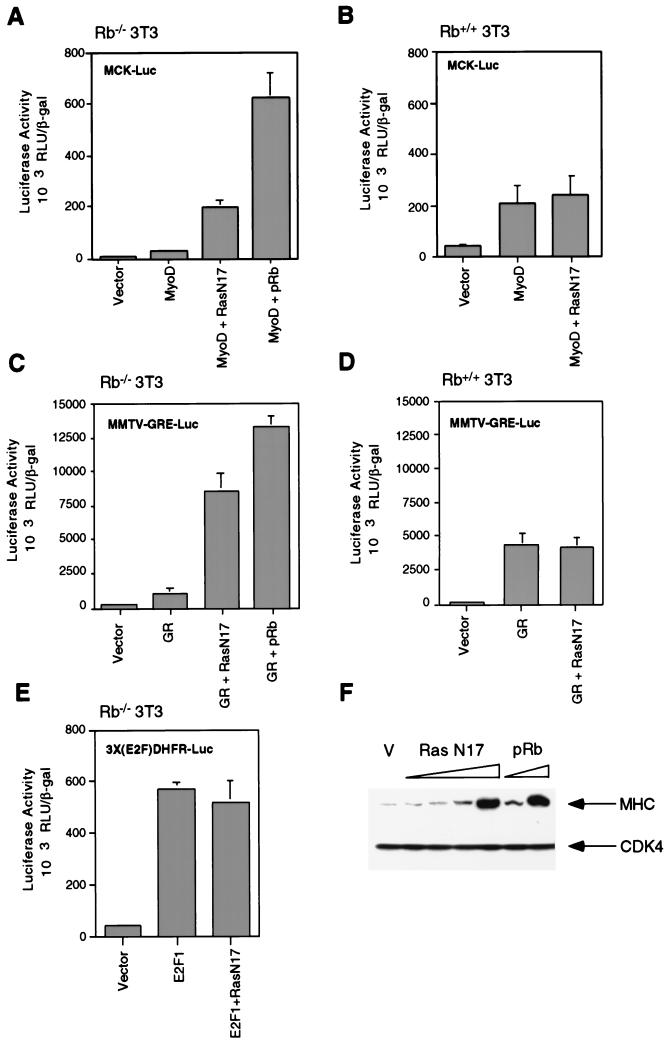
Rb−/− 3T3 cells were transfected with plasmids encoding MyoD and a muscle creatine kinase reporter, together with a plasmid encoding a dominant-negative Ras protein, RasN17. We did not want to completely abolish Ras activity, since Rb-positive cells do contain appreciable levels of activated Ras. We therefore titrated the RasN17-encoding plasmid, in an attempt to match this level of Ras activation, and thereby found a concentration optimal for activation of MyoD in Rb-deficient cells. As shown in Fig. 5A, expression of this chosen level of RasN17 plasmid in Rb-deficient fibroblasts led to an 8- to 10-fold increase in MyoD activation, compared to the level in cells transfected with a MyoD-encoding plasmid alone. Indeed, the absolute level of transcriptional activation achieved in Rb-deficient cells was comparable to that seen with Rb+/+ 3T3 cells transfected with only MyoD and the reporter construct (Fig. 5B). Expression of the RasN17-encoding plasmid was without effect on MyoD transactivation in Rb+/+ 3T3 cells (Fig. 5B).
FIG. 5.
Effect of inhibition of Ras activity on transcriptional activation in Rb−/− and Rb+/+ 3T3 cells. (A) Rb−/− 3T3 cells were transfected with plasmids encoding MyoD, RasN17, and pRb. A MyoD-responsive reporter, pMCK-Luc, and a β-galactosidase-encoding plasmid were included in each case. After transfection, cells were placed in differentiation medium, and luciferase and β-galactosidase activities were determined 48 h later. Relative luciferase activities, normalized for β-galactosidase activity, were calculated. Bars represent the averages plus standard deviations for four independent experiments. (B) Same as panel A, except Rb+/+ 3T3 cells were used. (C) Rb−/− 3T3 cells were transfected with plasmids encoding GRα, RasN17, and pRb. A GR-responsive reporter, MMTV-GRE-Luc, and a β-galactosidase-encoding plasmid were included. After transfection, cells were treated with dexamethasone for 24 h, at which time, relative luciferase activities, normalized for β-galactosidase activity, were determined. Bars represent the averages plus standard deviations for five independent experiments. (D) Same as panel C, except Rb+/+ 3T3 cells were used. (E) Rb−/− 3T3 cells were transfected with plasmids encoding E2F-1 and RasN17. An E2F-responsive reporter, 3X(E2F)DHFR-Luc, and a β-galactosidase-encoding plasmid were included. Twenty-four hours later, relative luciferase activities, normalized for β-galactosidase activity, were determined. Bars represent the averages plus standard deviations for three independent experiments. (F) Rb−/− 3T3 cells infected with a retrovirus encoding MyoD were transfected with plasmids encoding RasN17 (0.5, 1, 2.5, or 5 μg), pRb (0.5 or 1 μg), or a vector control (V). After transfection, cells were placed in differentiation medium for 48 h. At this time, the expression of MHC and CDK4 (as a loading control) was monitored by Western blot analysis. The results are representative of at least four independent experiments.
Like MyoD, the transcriptional activity of the GR is impaired in Rb-deficient cells (57), and oncogenic Ras can inhibit the activity of the GR (21, 52). As shown in Fig. 5C, GR transcriptional activity was potentiated 8- to 10-fold in Rb−/− 3T3 cells by cotransfection of a plasmid encoding RasN17. These findings again suggest that downregulation of Ras activity in Rb-deficient fibroblasts can, at this level of analysis, mimic reintroduction of pRb. Inhibition of Ras activity was without effect on GR transactivation in Rb+/+ 3T3 cells (Fig. 5D) or on E2F-1-mediated transcription in Rb−/− 3T3 cells (Fig. 5E). Thus, the aberrant levels of Ras activity in Rb-deficient fibroblasts appear to contribute to the transcriptional defect associated with MyoD and the GR in these cells.
In addition to the activation of MyoD, the expression of late markers of differentiation is attenuated in Rb−/− myoblasts (20, 38, 39, 51, 63). To determine whether modulation of Ras activity in Rb-deficient cells can restore certain aspects of the myogenic differentiation program, we analyzed the expression of MHC, a late marker of muscle differentiation. To this end, Rb−/− 3T3 cells expressing ectopic MyoD were generated by using a MyoD retrovirus. These cells were transfected with expression plasmids for either RasN17, pRb, or a vector control, placed in differentiation medium, and assayed for MHC expression 48 h later. As shown in Fig. 5F, expression of dominant-negative Ras led to a significant induction of MHC compared to that in vector-transfected cells. Likewise, as a positive control, expression of pRb led to a significant induction of MHC. The induction of MHC by ectopic expression of pRb or RasN17 was approximately 50 to 75% of that seen during differentiation of C2C12 myoblasts (pRb positive) (data not shown). These results suggest that inhibition of the aberrant levels of Ras activity in Rb-deficient fibroblasts can, at least in part, substitute for pRb in the induction of MHC. This result is consistent with an earlier demonstration by others that the protein products encoded by a partially penetrant allele of Rb, pRb;661W, retain the ability to restore the expression of MHC during myogenic differentiation (53) and our result that these pRb mutants retain the ability to regulate Ras activity. Together, these results support the notion that the high levels of Ras activity in Rb-deficient fibroblasts might be responsible, in part, for the failure of these cells to execute a differentiation program.
Increased Ras activation in Rb-deficient fibroblasts occurs at the level of guanine nucleotide exchange.
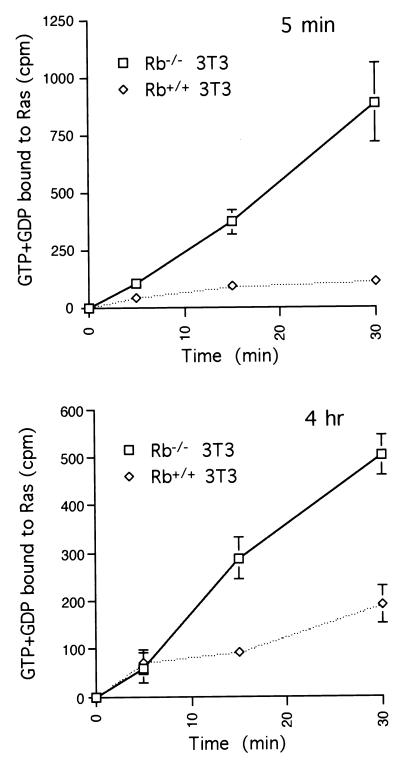
To explore the mechanism by which loss of pRb leads to increased levels of Ras activation, we considered processes known to be involved in the regulation of Ras. There are two complementary ways in which wild-type Ras activation can be upregulated: (i) increased guanine nucleotide exchange, mediated by guanine nucleotide exchange factors (45); and (ii) decreased GTPase activity through downmodulation of GTPase activating proteins (3). We attempted to discriminate between these two alternative mechanisms by measuring the rate of guanine nucleotide binding to Ras, an event thought to be regulated by exchange factors.
Rb−/− 3T3 and Rb+/+ 3T3 cells, in either early or mid-G1, were permeabilized to allow added [α-32P]GTP to enter the cells, and at various times thereafter, the levels of radioactivity bound to Ras were determined. In this comparison, the rate of nucleotide binding to Ras was significantly higher in an Rb-null background during both early and mid-G1 (Fig. 6). This result suggests that elevated levels of activated Ras associated with loss of pRb occur via stimulation of guanine nucleotide exchange, which presumably occurs at the level of Ras nucleotide exchange factors. These data do not, however, allow us to rule out the involvement of GAPs.
FIG. 6.
Ras guanine nucleotide binding in Rb−/− and Rb+/+ 3T3 cells. Rb−/− and Rb+/+ 3T3 cells were serum starved for 72 h before restimulation. Five minutes (top panel) or 4 h (bottom panel) later, cells were permeabilized and incubated with [α-32P]GTP. At the indicated times thereafter, cell lysates were prepared and subjected to immunoprecipitation with anti-Ras antibody. The recovered radioactivity was quantified and plotted as described in Materials and Methods. Points represent the averages plus standard deviations for three independent experiments.
Ras activation in Rb-deficient fibroblasts is cycloheximide sensitive.
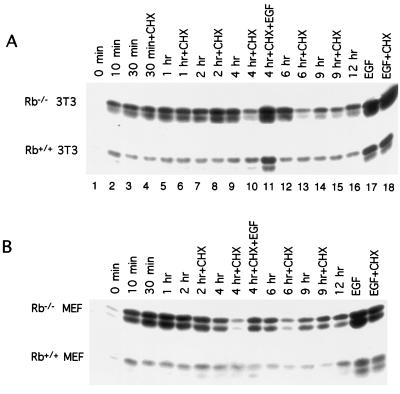
The mid-G1 activation of Ras in HeLa cells, unlike growth factor-induced activation of Ras, has been reported to be sensitive to cycloheximide-induced inhibition of protein synthesis (58). This information was used to further characterize the pathway leading to aberrant levels of Ras activity induced by loss of pRb. Specifically, we sought to determine the effect of cycloheximide treatment on Ras activation in our experimental system induced by either loss of pRb or growth factor stimulation. However, even a moderate inhibition of protein synthesis can have a profound impact upon fibroblast cell cycle progression (4, 41). So, in an attempt to minimize possible effects secondary to cell cycle perturbation and to map the periods of cycloheximide sensitivity with high resolution, cells were treated for 30 min at various times after release from quiescence to determine the effect of cycloheximide on Ras activation during G1.
Treatment of Rb+/+ MEFs and their 3T3 derivatives with cycloheximide during early and mid-G1 had no significant impact on the level of K-Ras activation (Fig. 7). A different picture emerged in the analysis of Rb-deficient fibroblasts, in which cycloheximide markedly reduced the activation of both N- and K-Ras during mid-G1 (4 to 6 h) (Fig. 7). Cycloheximide had no significant effect on the level of either K- or N-Ras activation in early G1 (0 to 2 h). Ras activation in Rb+/+ 3T3 cells expressing wild-type T antigen also revealed a sensitivity to an inhibition of protein synthesis in mid-G1 (data not shown).
FIG. 7.
Effect of cycloheximide treatment of Ras activation. (A) Quiescent Rb−/− and Rb+/+ 3T3 cells were restimulated with serum. At the indicated times, the activation state of Ras was measured. Cycloheximide (CHX) was added (25 μg/ml) to cultures 30 min before the indicated time point when levels of GTP-bound Ras were determined. Also shown are the levels of GTP-bound Ras determined 10 min after the addition of EGF alone (lane 17) or EGF with cycloheximide (lane 18) at time zero. In lane 11, cycloheximide was added at 3.5 h after serum stimulation and EGF was added 20 min later. Analysis of the activation state of Ras was performed at 4 h. The results shown are representative of at least 10 independent experiments. (B) Same as panel A, except primary MEFs were used, and the cycloheximide treatments at 30 min and 1 h were not included.
For comparison, we also determined the effect of cycloheximide treatment on Ras activation induced by epidermal growth factor (EGF). Although EGF treatment led to a significant activation of both K- and N-Ras in early (10 min) and mid-G1 (4 h) in Rb−/− MEFs, 3T3 cells, and their wild-type counterparts, cycloheximide was without effect on Ras activation at either time point (Fig. 7). Together, these data suggest that the mechanism by which Ras activity is increased following loss of pRb differs from that associated with EGF treatment. Specifically, while EGF-induced stimulation of Ras is insensitive to cycloheximide treatment, Ras activation in Rb−/− fibroblasts is sensitive to inhibition of protein synthesis during mid-G1. The pathway leading from pRb to Ras and the critical protein product(s) whose synthesis is required for Ras activation following pRb loss or inactivation remain to be identified.
DISCUSSION
Both Ras and pRb regulate proliferation and differentiation. It would seem appropriate then, that in order to bring about orderly changes in cell behavior, each regulated these cellular processes in a coordinate manner. One means of achieving this is through communication. Indeed, in the context of cell cycle progression, the G1 arrest induced by Ras inactivation is pRb dependent, placing pRb downstream of Ras (29, 37, 44). Here we provide evidence that this communication can also operate in the other direction because pRb has been shown to regulate N- and K-Ras activation.
pRb, Ras activation, and cell cycle control.
A current model of pRb function known as the pRb pathway indicates that the upstream regulators of pRb function (G1 cyclins, cyclin-dependent kinases [CDKs], and CDK inhibitors) influence the phosphorylation status of pRb and thereby its ability to regulate its best characterized downstream target, the E2F family of transcription factors. This model explains how pRb regulates proliferation and describes how deregulation or inactivation of each component of this pathway can confer a selective proliferative advantage and thus predispose to cancer (54). The results presented here suggest that the effect of pRb on N- and K-Ras activation may not be a function of the status of pRb phosphorylation, but rather the absence of pRb or its functional inactivation by viral oncoproteins (Fig. 1 and 3). Ectopic expression of cyclin D1 in fibroblasts has been shown to lead to premature activation of CDK4 and phosphorylation of pRb (26, 36, 49). However, enforced expression of cyclin D1 does not appear to significantly alter the activation state of Ras (Fig. 1D). Furthermore, in these cells as well as in parental NIH 3T3 cells, Ras activation peaks at approximately 2 h after growth factor stimulation (Fig. 1), several hours before the activation of cyclin D1-CDK4 in these cells (26).
The protein products of two partially penetrant mutant Rb alleles used in this study, 661W and Δex4, have previously been shown to lack the ability to bind to E2F and repress E2F-dependent transcription, while retaining the ability to promote certain differentiation processes (25, 53, 61). Together with the data presented here showing these mutants behave essentially like the wild-type protein in relation to Ras activity (Fig. 4), these findings suggest that the ability of pRb to regulate Ras activation is not linked to its ability to regulate E2F function. It is noteworthy, however, that the character of N- and K-Ras activation following pRb loss does appear to change as a function of cell cycle position. This was revealed in experiments analyzing the cycloheximide sensitivity of Ras activation, in which elevated levels of N- and K-Ras activation seen in Rb−/− fibroblasts in early G1 were insensitive to a block in protein synthesis, while in contrast, the mid-G1 activation of Ras was sensitive to cycloheximide treatment (Fig. 7). At present, it is difficult to put these observations into mechanistic terms, but they do suggest that the means by which pRb imposes itself on regulation of Ras activity is different in early versus mid-G1.
pRb, Ras activation, and the control of differentiation-specific transcription.
The role of pRb in differentiation has been studied mostly in the context of skeletal muscle. Levels of pRb increase during myoblast differentiation (9, 16, 35), and although Rb−/− embryos die at approximately day 13 of gestation with what appears to be normal skeletal muscle (8, 22, 27), partial restoration of pRb function does reveal critical defects in muscle differentiation later in development (63). MyoD is comparatively inactive in a pRb-negative background, and in such cells, wild-type pRb and a partially penetrant mutant of pRb have been shown to restore MyoD-dependent transactivation and the expression of late markers of differentiation (20, 38, 53). Given these observations, we considered the possibility that the ability of pRb to regulate the activation state of Ras may be part of the mechanism by which pRb operates as a regulator of differentiation. To this end, we tested whether the effect of pRb loss on the activation state of Ras was causally related to the inability of MyoD to promote differentiation of Rb-deficient cells. Our data suggest that reintroduction of pRb and inhibition of Ras activity in Rb-deficient fibroblasts are, to a certain degree, functionally equivalent with respect to MyoD function (Fig. 5). Additionally, we show that the expression of MyoD together with dominant-negative Ras in Rb−/− fibroblasts can induce the expression of MHC, a late marker of muscle differentiation (Fig. 5).
There are some striking parallels between the results presented here and the temporal location of endogenous MyoD action. MyoD expression is absent in G0 cells, but increases to maximum levels in mid-G1. It has been suggested that myoblasts, correspondingly, have the capacity to differentiate in mid-G1, but not G0 (23). We found that the mid-G1 activation of Ras in Rb-deficient cells was characteristically dependent upon ongoing protein synthesis (Fig. 7). Taken together, these observations suggest the possibility that the link between pRb and Ras activation in mid-G1 might, in some way, be related to the ability of pRb to cooperate with MyoD. It is conceivable that the communication between pRb and Ras in mid-G1 allows MyoD to bring about differentiation during this window.
Clues to the mechanism of Ras activation following pRb loss.
Our data indicate that the rate of guanine nucleotide binding to Ras is higher in Rb−/− fibroblasts than in Rb+/+ fibroblasts (Fig. 6). This suggests that the observed elevation of activated N- and K-Ras resulting from loss of Rb is due to an enhanced rate of GTP binding to these proteins. However, we have not ruled out the possibility that the intrinsic GTPase activity of N- and K-Ras is modulated as a function of Rb status. In preliminary studies, we have compared the levels of mSos1, mSos2, EGF receptor, Shc, GRB2, and p120RasGAP in Rb−/− and Rb+/+ 3T3 cells and found no significant difference in the abundance of these proteins (data not shown). Thus, at this level of analysis, the abundance of the well-characterized upstream regulators of Ras activation does not appear to explain why Ras activation is elevated in Rb-deficient fibroblasts.
Our results suggest that Ras activation (during mid-G1) following pRb loss is dependent on de novo protein synthesis. Since pRb acts principally as a transcriptional regulator, the simplest model of how loss of pRb might influence Ras activation is that pRb regulates the expression of a gene(s) whose protein product impinges on GTP loading of Ras. We would predict that the levels of such a protein would be significantly reduced by short treatment with cycloheximide. In this scenario, pRb would negatively regulate the synthesis of a positive regulator of guanine nucleotide exchange on Ras. It is noteworthy that pRb does not appear to be essential for Ras activation but is rather a modifier of this process. In this regard, modifiers of Ras-dependent signaling have already been identified in C. elegans, some of which have mammalian homologues that directly interact with Ras (see reference 56 and references therein). Future studies will be directed at determining the level at which pRb impinges on the regulation of guanine nucleotide exchange on Ras.
Concluding remarks.
We have demonstrated that loss of pRb leads to a significant elevation in the levels of GTP-bound, active N- and K-Ras in murine fibroblasts, suggesting communication from the nucleus to the inner plasma membrane. The mid-G1 activation of Ras in Rb-deficient fibroblasts requires de novo protein synthesis, indicating that immediate-early activation and mid-G1 activation of Ras occur via distinct mechanisms. These observations likely have direct bearing on the roles of pRb and Ras in differentiation and possibly restriction point control. We have provided evidence suggesting that the aberrant levels of activated Ras in Rb-deficient cells are causally linked to the failure of these cells to execute a differentiation program. These findings reveal an additional component of pRb function in tumor suppression.
ACKNOWLEDGMENTS
We thank D. Shalloway and S. Taylor for the GST-RBD construct; S. Reeves for the retroviral vector; W. Pear and D. Baltimore for Bosc23 cells; G. Cooper for the dominant-negative Ras plasmid; A. Lassar for the MCK reporter, MyoD plasmid, and retrovirus; W. Chin for the GR plasmid and GRE reporter; W. Sellers, F. Kaye, and W. Kaelin for pRb plasmids; J. DeCaprio for TAg plasmids; C. Sherr and M. Roussel for the NIH 3T3 cells and their cyclin D1 derivative; T. Jacks, N. Dyson, and E. Harlow for p107-deficient MEFs; T. Upton for p107−/− 3T3 cells; E. Flemington for the E2F-1 and reporter plasmid; and J. Lamb, R. Weinberg, O. Iliopoulos, W. Sellers, M. Weber, J. Griffin, D. Livingston, T. Roberts, and the members of the Ewen Laboratory for critical review of the manuscript.
This work was supported by National Cancer Institute grant CA65842 to M.E.E. M.E.E. is a Scholar for the Leukemia Society of America.
REFERENCES
- 1.Bader D, Masaki T, Fischman D A. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J Cell Biol. 1982;95:763–770. doi: 10.1083/jcb.95.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 3.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: a conserved switch for diverse functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 4.Brooks R F. Continuous protein synthesis is required to maintain the probability of entry into S phase. Cell. 1977;12:311–317. doi: 10.1016/0092-8674(77)90209-4. [DOI] [PubMed] [Google Scholar]
- 5.Chen P-L, Riley D J, Chen Y, Lee W-H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 6.Chen P-L, Riley D J, Chen-Kiang S, Lee W-H. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P-L, Riley D J, Lee W-H. The retinoblastoma protein as a fundamental mediator of growth and differentiation signals. Crit Rev Eukaryot Gene Expr. 1995;5:79–95. [PubMed] [Google Scholar]
- 8.Clarke A R, Maandag E R, van Roon M, van der Lugt N M T, van der Valk M, Hooper M L, Berns A, Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 9.Coppola J A, Lewis B A, Cole M D. Increased retinoblastoma gene expression is associated with late stages of differentiation in many different cell types. Oncogene. 1990;5:1731–1733. [PubMed] [Google Scholar]
- 10.DeCaprio J A, Ludlow J W, Figge J, Shew J-Y, Huang C-M, Lee W-H, Marsilio E, Paucha E, Livingston D M. SV40 large T antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 11.Downward J. Cell cycle: routine role for Ras. Curr Biol. 1997;7:R258–R260. doi: 10.1016/s0960-9822(06)00116-3. [DOI] [PubMed] [Google Scholar]
- 12.Downward J, Graves J D, Warne P H, Rayter S, Cantrell D A. Stimulation of p21ras upon T-cell activation. Nature. 1990;346:719–723. doi: 10.1038/346719a0. [DOI] [PubMed] [Google Scholar]
- 13.Dryja T P, Rapaport J, McGee T L, Nork T M, Schwartz T L. Molecular etiology of low-penetrance retinoblastoma in two pedigrees. Am J Genet. 1993;52:1122–1128. [PMC free article] [PubMed] [Google Scholar]
- 14.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 15.Dyson N, Guida K, Munger K, Harlow E. The human papillomavirus 16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 16.Endo T, Goto S. Retinoblastoma gene product Rb accumulates during myogenic differentiation and is deinduced by the expression of SV40 large T antigen. J Biochem. 1992;112:427–430. doi: 10.1093/oxfordjournals.jbchem.a123916. [DOI] [PubMed] [Google Scholar]
- 17.Ewen M E. Regulation of the cell cycle by the Rb tumor suppressor family. In: Pagano M, editor. Cell cycle control. Vol. 22. Heidelberg, Germany: Springer-Verlag; 1998. pp. 149–179. [DOI] [PubMed] [Google Scholar]
- 18.Feig L A, Cooper G M. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gigure V, Hollenberg S, Rosenfeld M, Evans R. Functional domains of the human glucocorticoid receptor. Cell. 1986;46:645–652. doi: 10.1016/0092-8674(86)90339-9. [DOI] [PubMed] [Google Scholar]
- 20.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 21.Hock J R, Ziemiecki A, Klemenz R, Friis R, Groner B. Oncogene mediated repression of glucocorticoid hormone response elements and glucocorticoid receptor levels. Cancer Res. 1989;49:2266s–2274s. [PubMed] [Google Scholar]
- 22.Jacks T, Fazeli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 23.Kitzmann M, Carnac G, Vandromme M, Primig M, Lamb N J C, Fernandez A. The muscle regulatory factors MyoD and Myf-5 undergo distinct cell cycle-specific expression in muscle cells. J Cell Biol. 1998;142:1447–1459. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong Y, Johnson S E, Taparowsky E J, Konieczny S F. Ras p21Val inhibits myogenesis without altering the DNA binding or transcriptional activities of the myogenic basic helix-loop-helix factors. Mol Cell Biol. 1995;15:5205–5213. doi: 10.1128/mcb.15.10.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kratzke R A, Otterson G A, Hogg A, Coxon A B, Geradts J, Cowell J K, Kaye F J. Partial inactivation of RB product in a family with incomplete penetrance of familial retinoblastoma and benign retinal tumors. Oncogene. 1994;9:1321–1326. [PubMed] [Google Scholar]
- 26.Ladha M H, Lee K Y, Upton T M, Reed M F, Ewen M E. Regulation of exit from quiescence by p27 and cyclin D1-CDK4. Mol Cell Biol. 1998;18:6605–6615. doi: 10.1128/mcb.18.11.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee E Y-H P, Chang C-Y, Hu N, Wang Y-C J, Lai C-C, Herrup K, Lee W-H, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 28.Lee E Y-H P, Hu N, Yuan S-S F, Cox L A, Bradley A, Lee W-H, Herrup K. Dual roles of the retinoblastoma protein in cell cycle regulation and neuron differentiation. Genes Dev. 1994;8:2008–2021. doi: 10.1101/gad.8.17.2008. [DOI] [PubMed] [Google Scholar]
- 29.Leone G, DeGregori J, Sears R, Jakoi L, Nevins J R. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997;387:422–426. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- 30.Lohmann D R, Brandt B, Hopping W, Passarge E, Horsthemke B. Distinct RB1 gene mutations with low penetrance in hereditary retinoblastoma. Hum Genet. 1994;94:349–354. doi: 10.1007/BF00201591. [DOI] [PubMed] [Google Scholar]
- 31.Lu X, Horvitz H R. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–991. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- 32.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malumbres M, Pellicer A. Ras pathways to cell cycle control and cell transformation. Front Biosci. 1998;3:d887–d912. doi: 10.2741/a331. [DOI] [PubMed] [Google Scholar]
- 34.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 35.Martelli F, Cenciarelli C, Santarelli G, Polikar B, Felsani A, Caruso M. MyoD induces retinoblastoma gene expression during myogenic differentiation. Oncogene. 1994;1994:3579–3590. [PubMed] [Google Scholar]
- 36.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J-Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mittnacht S, Paterson H, Olson M F, Marshall C J. Ras signalling is required for inactivation of tumour suppressor pRb cell-cycle control protein. Curr Biol. 1997;7:219–221. doi: 10.1016/s0960-9822(97)70094-0. [DOI] [PubMed] [Google Scholar]
- 38.Novitch B G, Mulligan G J, Jacks T, Lassar A B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novitch B G, Spicer D B, Kim P S, Cheung W L, Lassar A B. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr Biol. 1999;9:449–459. doi: 10.1016/s0960-9822(99)80210-3. [DOI] [PubMed] [Google Scholar]
- 40.Olson E N, Spizz G, Tainsky M A. The oncogenic forms of N-ras or H-ras prevent skeletal myoblast differentiation. Mol Cell Biol. 1987;7:2104–2111. doi: 10.1128/mcb.7.6.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pardee A B. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 42.Paulus W, Baur I, Boyce F M, Breakefield X O, Reeves S A. Self-contained, tetracycline-regulated retroviral vector system for gene delivery to mammalian cells. J Virol. 1996;70:62–67. doi: 10.1128/jvi.70.1.62-67.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peeper D S, Upton T M, Ladha M H, Neuman E, Zalvide J, Bernards R, DeCaprio J A, Ewen M E. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature. 1997;386:177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- 45.Pronk G J, Bos J L. The role of p21ras in receptor tyrosine kinase signalling. Biochim Biophys Acta. 1994;1198:131–147. doi: 10.1016/0304-419x(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 46.Quelle D E, Ashmun R A, Shurtleff S A, Kato J, Bar-Sagi D, Roussel M F, Sherr C J. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- 47.Ramocki M B, Johnson S E, White M A, Ashendel C L, Konieczny S F, Taparowsky E J. Signaling through mitogen-activated protein kinase and Rac/Rho does not duplicate the effects of activated Ras on skeletal myogenesis. Mol Cell Biol. 1997;17:3547–3555. doi: 10.1128/mcb.17.7.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raptis L, Brownell H L, Corbley M J, Wood K W, Wang D, Haliotis T. Cellular ras gene activity is required for full neoplastic transformation by the large tumor antigen of SV40. Cell Growth Differ. 1997;8:891–901. [PubMed] [Google Scholar]
- 49.Resnitzky D, Reed S I. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol Cell Biol. 1995;15:3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlessinger J, Bar-Sagi D. Activation of Ras and other signaling pathways by receptor tyrosine kinases. Cold Spring Harbor Symp Quant Biol. 1994;59:173–179. doi: 10.1101/sqb.1994.059.01.021. [DOI] [PubMed] [Google Scholar]
- 51.Schneider J W, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- 52.Screpanti V A, Maroder M, Petrangeli E, Frati L, Gulino A. Tumor-promoting phorbol ester and ras oncogene expression inhibit the glucocorticoid-dependent transcription from the mouse mammary tumor virus long terminal repeat. Mol Endocrinol. 1989;3:1659–1665. doi: 10.1210/mend-3-10-1659. [DOI] [PubMed] [Google Scholar]
- 53.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G J. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor growth. Genes Dev. 1998;12:96–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 55.Sherr C J. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 56.Sieburth D S, Sun Q, Han M. SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell. 1998;94:119–130. doi: 10.1016/s0092-8674(00)81227-1. [DOI] [PubMed] [Google Scholar]
- 57.Singh P, Coe J, Hong W. A role for the retinoblastoma protein in potentiating transcriptional activation of the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 58.Taylor S J, Shalloway D. Cell cycle-dependent activation of Ras. Curr Biol. 1996;6:1621–1627. doi: 10.1016/s0960-9822(02)70785-9. [DOI] [PubMed] [Google Scholar]
- 59.Todaro G J, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 61.Whitaker L L, Su H, Baskaran R, Knudsen E S, Wang J Y J. Growth suppression by an E2F-binding-defective retinoblastoma protein (RB): contribution from the RB C pocket. Mol Cell Biol. 1998;18:4032–4042. doi: 10.1128/mcb.18.7.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wood K W, Sarnecki C, Roberts T M, Blenis J. ras mediates nerve growth factor receptor modulation of three signal-transducing protein kinases: MAP kinase, Raf-1, and RSK. Cell. 1992;68:1041–1050. doi: 10.1016/0092-8674(92)90076-o. [DOI] [PubMed] [Google Scholar]
- 63.Zacksenhaus E, Jiang Z, Chung D, Marth J D, Phillips R A, Gallie B L. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- 64.Zalvide J, DeCaprio J A. Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol Cell Biol. 1995;15:5800–5810. doi: 10.1128/mcb.15.10.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]