Graphical abstract

Keywords: Candida auris, Transcriptomics, Proteomics, Drug resistance, Stress response
Abstract
Candida auris has emerged as a serious worldwide threat by causing opportunistic infections that are frequently resistant to one or more conventional antifungal medications resulting in high mortality rates. Against this backdrop, health warnings around the world have focused efforts on understanding C. auris fungal biology and effective prevention and treatment approaches to combat this fungus. To date, there is little information about the differentially expressed genes when this fungus is treated with conventional antifungals, and caspofungin is a standard echinocandin deployed in the therapy against C. auris. In this work, we treated two distinct strains of C. auris for 24 h with caspofungin, and the cellular responses were evaluated at the morphological, translational and transcriptional levels. We first observed that the echinocandin caused morphological alterations, aggregation of yeast cells, and modifications in the cell wall composition of C. auris. Transcriptomic analysis revealed an upregulation of genes related to the synthesis of the cell wall, ribosome, and cell cycle after exposure to caspofungin. Supporting these findings, the integrated proteomic analysis showed that caspofungin-treated cells were enriched in ribosome-related proteins and cell wall, especially mannoproteins. Altogether, these results provide further insights into the biology of C. auris and expands our understanding regarding the antifungal activity of caspofungin and reveal cellular targets, as the mannose metabolism, that can be further explored for the development of novel antifungals.
1. Introduction
Invasive fungal infections are responsible for over 1.5 million deaths per year [1]. Among these diverse fungal diseases, bloodstream infections caused by Candida species are the most common cause of invasive disease [2]. Although C. albicans is frequently the most associated species with invasive candidiasis [3], infections by non-albicans species have increased. The non-albicans species commonly display multidrug resistance, representing a major problem of public health globally, particularly in immunocompromised, hospitalized patients [4], [5].
The emerging multidrug resistant fungal pathogen C. auris was responsible for numerous nosocomial outbreaks in healthcare settings [6], [7]. C. auris was first reported in 2009 as the etiological agent of an ear infection in Japan [8], [9], and it remains understudied. The difficulty in identifying the fungus by classic phenotyping and our gaps in understanding its propagation mechanisms in healthcare settings worldwide has impeded our capacity to control its spread [10]. Due to these issues and its remarkable drug resistance, C. auris was the only fungal pathogen classified as a global public health threat [8]. This fungus can be transmitted rapidly and persist on hospital surfaces and medical devices [11], [12], and has a high mortality, particularly among patients who have undergone multiple medical interventions [10], [13], [14], [15].
C. auris has an extremely high frequency of multidrug resistance to polyenes, echinocandins, and azoles, especially fluconazole [16]. Resistant phenotypes to fluconazole and echinocandin are linked to polymorphisms in the erg11 and fks1 genes, respectively [16], [17], but other regulators of resistance are poorly known. Another feature that may contribute to the resistance of C. auris to a variety of antimicrobial agents is biofilm formation [18], [19], [20]. Even though it is not as thick or adherent to medical devices substrates compared to C. albicans biofilms [18], [21], C auris biofilms confer drug resistance to fluconazole [18], [22] and caspofungin [18]. This factor may be associated with an increased activity in efflux pumps [22]. C. auris biofilm transcriptome analysis has revealed that genes coding efflux pumps such as ATP-binding cassette (ABC) and major facilitator superfamily (MFS) transporters are upregulated in the presence of antifungal agents, especially at 24 h post-exposure [22].
The echinocandins are the drugs of choice for the initial treatment of C. auris, as recommended by the CDC (https://www.cdc.gov/fungal/candida-auris/c-auris-treatment.html). On the basis of this recommendation, we analyzed the global cellular response of two isolates of C. auris to caspofungin. Our data showed that caspofungin not only affected cell morphology, but also led to a transcriptional response with upregulation of cell wall associated processes and silencing of pathways that are associated with cellular responses to stress. Our study contributes with new information on C. auris that highlights genetic responses to a standard antifungal.
2. Material and methods
2.1. Fungal growth conditions
C. auris strains MMC1 (isolated in Montefiore Medical Center, Bronx NY) and B11244 (Venezuela Clade) were maintained at −80°C. B11244 was obtained from the CDC (also known as CDC 385). After thawing in Sabouraud broth, suspensions were incubated at 30 °C for 24 h. Yeast cell suspensions were then plated onto Sabouraud agar plates and incubated at 30 °C for 48 h. The plates were then stored at 4 °C (for no longer than 4 weeks) and used in experiments. Yeast cell viability prior to experimental use and during culturing was monitored by propidium iodide (PI) staining and analysis by flow cytometry.
Based on MICs, growth curves of C. auris in the presence of caspofungin were performed and subinhibitory concentrations were determined: 12.5 ng/mL and 10 ng/mL for MMC1 and B11244, respectively. The assays were performed in technical and biological triplicates in a 96-well flat bottom translucent plate to a final volume of 200 μL per well and 5 × 105 cells/mL. The cells were incubated in a microplate reader (Synergy Biotek) for 72 h at 30˚C, with optical density (OD) readings taken every 1 h with a wavelength of 540 nm, with prior shaking of the plate for 30 s before each reading. A non-linear model, Gompertz curve, was used to analyze the growth curves, and the significance was calculated by ANOVA, comparing control versus treatment. The results represent the mean ± standard deviation of three independent experiments. The curves and analyses was performed using the GraphPad Prism 8.0 program.
2.2. Scanning electron microscopy (SEM)
The cells were collected by centrifugation at 5,000 rpm for three minutes and were washed 3 times with PBS buffer and then was fixed with 1 mL of 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2 for 1 h at room temperature and subsequently they were washed 3 times with post-fixation solution (0.1 M cacodylate buffer, 0.2 M sucrose and 2 mM MgCl2). After the fixation step, the cells were adhered in round coverslips with poly-I-lysine for 30 min. The excess was quickly removed, and the samples were gradually dehydrated, by adding sequentially adding ethanol 30%, ethanol 50%, and ethanol 70% for 5 min each and 95% ethanol followed by 100% ethanol, twice, for 10 min each. After dehydration, 100% ethanol was replaced with carbon dioxide (CO2) with a 10 times cycle. At the end of the process, the coverslips were metallized with gold and visualized in a scanning electron microscope (Jeol JSM-6010 Plus-LA) at 5 Kv.
2.3. Cell wall staining
Yeast cells, in Sabouraud broth, were incubated with or without caspofungin at a concentration of 12.5 ng/mL (MMC1 – MIC = 2 μg/mL) and 10 ng/mL (B11244 – MIC = 0.5 μg/mL) for 24 h at 37 °C. Cells were washed with PBS and fixed with paraformaldehyde 4% for 30 min at room temperature (RT). After washing again, cells were blocked with BSA 1% in PBS for 1 h at RT and then incubated with concanavalin A conjugated with Alexa 488 (5 μg/mL), wheat-germ agglutinin conjugated with NHS-rhodamine (WGA – 10 μg/mL) and uvitex 1% for 30 min at RT. After washing with PBS, samples were analyzed under an Observer Z1 (Zeiss) microscope. In parallel, cells that were only stained with concanavalin A were analyzed in a flow cytometer (FacScalibur) for quantification of mannoproteins. First, cells were analyzed by size and intracellular complexity (FSC and SSC) to exclude doublets. The selected population of single yeast cells were then analyzed in FL1 channel to measure the intensity of Alexa 488 fluorescence (relative to the amount of exposed mannoproteins).
2.4. RNA isolation and sequencing
Total RNA was isolated from 1 × 107C. auris cells using the miRCURY RNA isolation kit (Exiqon - Qiagen) with minor adaptations. A 1:1 vol of glass beads was added to the lysis buffer along with the yeast cells, and the mixture was subjected to 10 rounds of 1 min at 4 °C of vortex agitation in order to disrupt the fungal cell wall. After centrifugation, total RNA was isolated according to the manufacturer’s instructions. The DNA cleanup step was performed with all samples using the RNAse-free DNAse protocol (Qiagen). For RNA quantification and integrity analysis we used a Qubit fluorometer (Thermo Fisher) and an Agilent 2100 Bioanalyzer; RNA 6000 pico and RNA small kits (Agilent Technologies).
The sequencing library was constructed with the TruSeq Stranded mRNA kit (Illumina) prepared according to the manufacturer’s instructions and all the samples were prepared in three independent replicates. RNAseq was performed on a HiSeq 2500 (Illumina, single-end 50-bp SR mid output run) at the Life Sciences Core Facility (LaCTAD), a part of the University of Campinas (UNICAMP).
2.5. Transcriptomic data analysis
The sequences in fastq format were analyzed by CLC Genomics Workbench© v 20.0 (Qiagen), using the corresponding C. auris genome for strain B8441 (GCA_002759435.2V2). The parameters used for the alignments were: mismatch cost (2), insertion cost (3), deletion cost (3), length fraction (0.8), and similarity fraction (0.8). Only uniquely mapped reads were considered in the analysis. The statistical test applied was the DGE (Differential Gene Expression) using the RNA-seq package with CLC Genomics Workbench© v 20.0 (Qiagen). The parameters for the RNA-seq analysis were: strand setting - both, library type setting - bulk, Calculate expression for genes without transcript - Yes. The library size was normalized using the TMM (trimmed mean of M values) method [23]. After TMM normalization calculation for each sample, TMM-adjusted log CPM counts was estimated, similar to the EdgeR tool [23]. For the mapping settings, the software applied the EM (expectation–maximization algorithm) estimation algorithm. The differential expression applied by CLC Genomics Workbench uses multi-factorial statistics based on a negative binomial Generalized Linear Model (GLM). This statistic test considers that the read counts follow a Negative Binomial distribution. The same test is used by other RNA-seq tools, including edgeR and DESeq [24]. The false discovery rate (FDR) adjusted p-value is a multiple-testing correction and follows the same approach as the software DESeq2 [25]. The expression values for the transcripts were registered in TPM (Transcripts per Million), and TMM (trimmed mean of M values) was used as a normalization method. The parameters to select the differentially expressed transcripts were 3-fold change (>3 FC) and False discovery rate (FDR) below or equal to 0.05. For the Heat map clustering the parameters were: Distance measure = Euclidean distance, Linkage criteria = Average linkage, Filter settings = Filter by statistics, Statistical comparison = Caspofungin vs. Control, Minimum absolute fold change = 8.0, Correction = FDR p-value Threshold = 0.001.
The differentially expressed transcripts sequences from MMC1 and B11244 cells were compared with orthologous genes from C. albicans genome strain SC5314 (assembly ASM18296v3) by Reciprocal Best Hit (RBH). For the Reciprocal best hit analysis we used BLASTp and performed protein sequence comparisons between C. albicans strain SC5314 (assembly ASM18296v3) and C. auris strain B8441 (assembly GCA_002759435.2). The options for NCBI BLAST different to the defaults were: a maximum E-value threshold of 1e-6, Number of threads of 4, Mask low complexity regions – yes, word size of 3, match of 2, mismatch of −3, gap cost existence of 5, and gap cost extension of 2. The default scoring matrix for blastp used was BLOSUM62 [26]. The genes that presented >40% of identity were considered for the analysis.
For all the gene ontology enrichment analysis, we used the Database for Annotation, Visualization and Integrated Discovery version 6.8 program (DAVID) [27]. In the Functional Annotation tool, the analysis uses a modified Fisher Exact (EASE), to calculate the gene-enrichment in annotation terms, that all the tables are expressed as FDR (false discovery rate). The C. albicans orthologs Uniprot accession IDs were used to search for the functional annotation with the following parameters: Count −2 and EASE score 0.1. The functional annotation uses a Kappa statistic score to estimate the co-occurrence of any given gene pairs [28]. We applied the Fisher's exact test and considered only the terms with p ≤ 0.05 and we then compared terms for the up- and down-regulated genes to a background of all terms to obtain an overall insight into the effect of caspofungin on C. auris compared to the absence of the antifungal.
The RNA-seq data have been deposited at the Sequence Read Archive (SRA) database under the accession number (SRA: SRP295539 BioProject: PRJNA682185). We had biological triplicates for both strains and conditions tested, except for the third replicate for B11244 with caspofungin, due to sequencing technical problems we obtained biological duplicates. In our RNA seq analysis, we obtained on average 7.3 million reads per sample that mapped >95% of the reference genome (GCA_002759435.2), with a 30x coverage (Table S1).
2.6. Quantitative RT-PCR
For the quantitative real time PCR the experimental design was performed according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines [29]. Total RNA was isolated in duplicate from 1 × 107C. auris cells using the miRCURY RNA isolation kit (Exiqon - Qiagen) with adaptations. A 1:1 vol of glass beads was added to the lysis buffer along with the yeast cells, and the mixture was subjected to 10 rounds of 1 min at 4°C of vortex agitation in order to disrupt the fungal cell wall. After centrifugation, total RNA was isolated according to the manufacturer’s instructions and quantified using Qubit™ fluorometer RNA HS kit (Thermo Fisher) and the RNA integrity was assessed with Bioanalyzer RNA PICO 6000 (Agilent). After isolation, 1 ug (for cellular RNA) was treated with 1 U of DNAse I RNAse-free (#EN0521 PROMEGA) according to manufacturer instructions. After that, the cDNA was synthesized from 1 µg of cellular RNA as template. For the reverse transcriptase reactions 0.3 µM random primer (Invitrogen) and 1 µL of reverse transcriptase (Superscript II, Thermo Scientific), according to the manufacturers’ instructions. PCR was performed with 40 ng of cDNA for the cell and 1.6 ng of cDNA as the template and GoTaqTM master mix according to manufacturer instructions (Promega). The oligonucleotides were designed with PRIMER-Blast using the following parameters: PCR product size maximum of 250 nt. Tm varying from 57 to 63 °C, RefSeq mRNA as a database and Candida auris as the organism. The primer sets used for PCR are described below. The qPCR was performed in four technical replicates for each sample. The following program was used in the Lightcycler 480 (Roche) equipment: initial denaturation at 95 °C for 15 min and 45 cycles of 95 °C for 15 s, 62 or 64°C for 20 s and 72°C for 45 s. The reference gene used was C5 sterol desaturase and the target genes and the primers used are listed in Table 1.
Table 1.
Primers used in this study.
| Gene ID | Direction | Sequence | Start | Stop | Tm | GC% | Product length |
|---|---|---|---|---|---|---|---|
| B9J08_002761 | Forward | GGGCCAAAACTCCTACGGAA | 96 | 115 | 60.0 | 55.0 | 248 |
| Reverse | AGCCGTAAGATCCTGAAGCG | 343 | 324 | 59.9 | 55.0 | ||
| B9J08_003359 | Forward | GCAGAGATCGAGGAAGACGG | 154 | 173 | 60.0 | 60.0 | 150 |
| Reverse | ACGAGACTCGGCCTCTAAGT | 303 | 284 | 60.0 | 55.0 | ||
| B9J08_001469 | Forward | TAGAGCCCATCAGGCTTCCT | 239 | 258 | 60.0 | 55.0 | 166 |
| Reverse | GCGTCTAGCTCGTTCTCCTC | 404 | 385 | 60.0 | 60.0 | ||
| B9J08_005424 | Forward | GTCGAGCGGGGAGTATCAAG | 198 | 217 | 60.0 | 60.0 | 245 |
| Reverse | AACACGCCCAGTCGAAGAAA | 442 | 423 | 60.2 | 50.0 | ||
| B9J08_002817 | Forward | TGGTCTTCTTCCCCATTGGC | 45 | 64 | 60.0 | 55.0 | 171 |
| Reverse | GTCAAAAACGTCCAGGTGCC | 215 | 196 | 60.0 | 55.0 | ||
| B9J08_002202 | Forward | CCCACTCCATCATTGGTGCT | 344 | 363 | 60.0 | 55.0 | 172 |
| Reverse | AAGACACCGAACTTGGCGAT | 515 | 496 | 60.0 | 50.0 | ||
| B9J08_004504 | Forward | GGGCAGCTTGTTCATTGACG | 750 | 769 | 60.1 | 55.0 | 168 |
| Reverse | GTTGCCATATCGGTCAACGC | 917 | 898 | 60.0 | 55.0 | ||
| B9J08_003737 | Forward | ATCTGGCGTGCTACTACTGC | 427 | 446 | 59.9 | 55.0 | 216 |
| Reverse | CTTGTACACGGAAGGCCAGT | 642 | 623 | 60.0 | 55.0 |
2.7. Protein extraction and digestion
Proteins were extracted from 109 cells washed with 1x PBS by suspending the cells in lysis buffer (20 mM HEPES, 50 mM citrate, 10 mM CaCl2, 10% glycerol, 1% Triton, 15 mM MgCl2, 100 mM DTT) with glass beads, and vortexing for 15 cycles of 1 min followed by 1 min on ice. Lysates were cleaned by centrifuging twice, first at 8,000 xg for 5 min at 4 °C, and then at 10,000 xg for 10 min at 4 °C. Protein concentration was determined using the tryptophan fluorescence method [30]. After quantification, 50 µg samples were separated by electrophoresis in SDS-PAGE 10% (v/v) gels and stained with Comassie R250 0,1%. Each lane was destained and dehydrated with ethanol, reduced with 10 mM DTT, alkylated with 50 mM iodacetamide and digested overnight with 12.5 ng/µL trypsin solution in 50 mM ammonium bicarbonate at 37 °C. The peptides were extracted twice with 30% acetonitrile (ACN), 3% trifluoroacetic acid (TFA) and twice with ACN alone, and then dried in a vacuum centrifuge and desalted with C18 Stage Tips prior to nanoLC- ESI-MS/MS.
2.8. NanoLC-ESI-MS/MS acquisition
Peptides of each sample were separated by online reversed-phase nanoscale capillary liquid chromatography and analyzed by electrospray mass spectrometry in tandem (ESI-MS/MS). The experiments were performed in the mass spectrometry facility RPT02H of Carlos Chagas Institute (Fiocruz, Parana) with a nanoLC-1D plus (Eksigent) coupled to LTQ Orbitrap XL ETD (Thermo Scientific) mass spectrometer. Chromatographic separation of the peptide mixtures was carried out on an analytical silica column of 15 cm, 75-µm ID and with a 3 µM diameter C18 particles (Dr. Maisch), flow rate of 250 nL/min of mobile phase (ACN, 0.1% formic acid, 5% DMSO) with a linear gradient from 5 to 40% ACN in 180 min. Peptides were ionized by nano-electrospray (voltage 2.7 kV) and injected into the MS. Full-scan MS spectra (at 300.0–1800.0 m/z range) were acquired on an Orbitrap analyzer with a resolution of R = 60,000. The 10 most intense peaks were fragmented by CID and analyzed in the Ion trap. A dynamic exclusion list of 90 sec was applied and the “lock mass” option was enabled (m/z = 401.922718). The MS data was deposited into the Mendeley Data repository with the dataset identifier http://dx.https://doi.org/10.17632/4pbttmb3kx.1.
2.9. Proteomic data analysis
The LC-MS/MS data were matched against the C. auris strain B8441 database from UniProt Knowledgebase (downloaded on November 11, 2020, containing 5,409 sequences) using MaxQuant software version 1.6.17.0 [31]. Among the search parameters were specified a tolerance of 0.5 Da for MS/MS, and 20 ppm for MS first search and 4.5 ppm for MS main search. Quantification was done by the LFQ method. Cysteine carbamidomenthylation were set as fixed modification, methionine oxidation and N-terminal acetylation were set as variable modification. The tables generated by Max Quant were analyzed with Perseus software version 1.6.14.0 [32]. A false discovery rate (FDR) of 1% was applied for both peptide and protein identification. The contaminants and reverse sequences were removed. The LFQ intensity was converted to log2(x) scale and ANOVA multiple-sample test was performed to determine differentially expressed proteins using p-Value ≤ 0.05 and Fold Change > 2 as cut off.
3. Results
3.1. Caspofungin induces drastic alterations in C. auris
We selected for our study two clinical isolates of C. auris with distinct morphological properties and similar caspofungin susceptibility profiles. In standard conditions, C. auris strain B11244 form cellular aggregates, and the MMC1 strain grows homogeneously. We asked if these characteristics would correlate with the morphological alterations in response to a long exposure (24 h) to caspofungin. The caspofungin concentration used was 10 ng/mL for B11244 and 12.5 ng/mL for MMC1, and was set to be high enough to induce a stress response, but also to allow fungal growth (Fig. 1A, B). In untreated MMC1 cells, well-defined, elliptical-shaped yeast morphology, as well as budding cells with typical bud scars, were observed. B11244 cells showed the typical aggregated growth profile (Fig. 1C). In contrast, caspofungin-treated cells exhibited a severely distorted yeast cell topography, with cells fused together and enlarged yeasts forming clumps mixed with cells with normal morphology (Fig. 1C).
Fig. 1.
Caspofungin treatment led to morphological alterations in C. auris. (A) Growth curve of C. auris in the presence or absence of caspofungin. The growth was analyzed, and the assay was performed in a technical and biological triplicate. The significance was calculated by ANOVA, the red arrow and the red dotted line indicate the time that the cells were collected for the study. (B) Effect of sub-optimal concentration of caspofungin on viability of C. auris. Cultures of C. auris were cultivated with or without caspofungin for 24 h and cell viability was addressed by propidium iodide staining and analysis by flow cytometry. Graphs represent average and standard deviation for 4 independent experiments. (C) Scanning electron microscopy images of C. auris at 1400x magnification. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The cell wall of Candida spp. in general consists of an inner layer of chitin, b-1,6-glucan and b-1,3-glucan. The cell wall proteins are in the outer layer and are linked to the b-glucan by glycosylphosphatidylinositol (GPI) anchors [33]. The cell wall is a plastic structure that can dynamically alter the composition to respond to stress conditions, to allow cell survival [34].
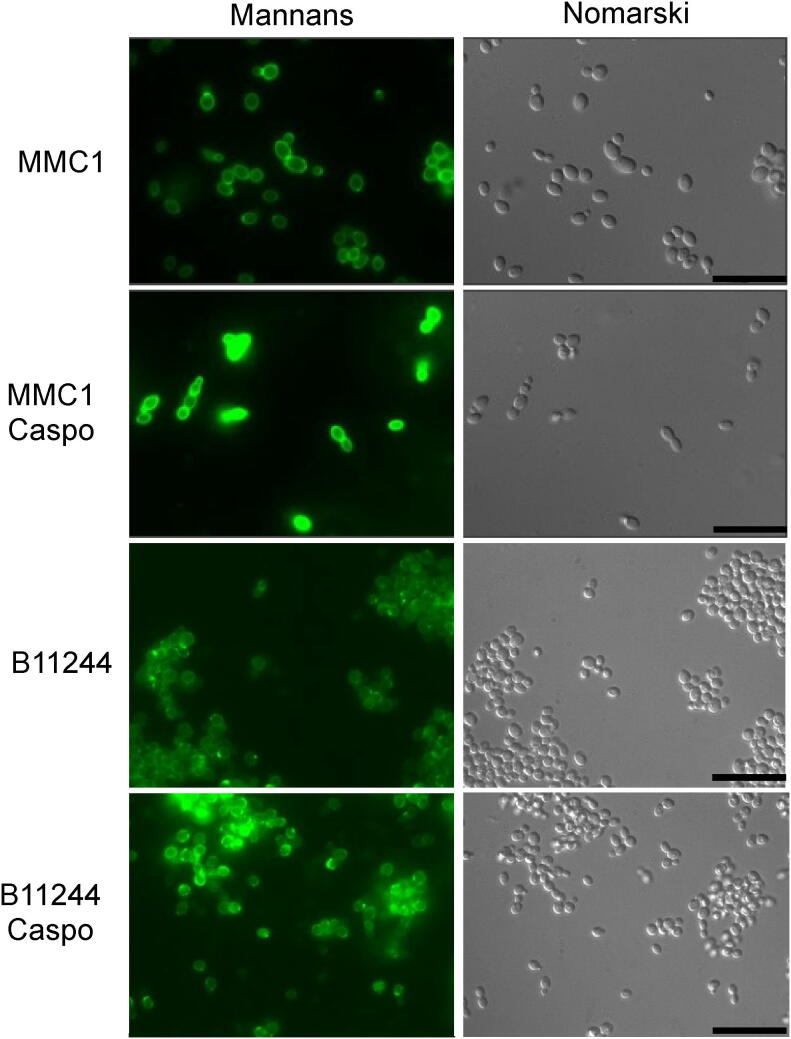
As caspofungin is a drug that interferes with cell wall dynamics, we addressed whether there were alterations of important constituents of the cell wall after the treatment with the antifungal. Indeed, treatment with caspofungin induced cell wall modifications of both strains of C. auris. Yeast cells treated with caspofungin had a thickened mannoprotein layer (Fig. 2). Other cell wall components such as chitin and chitin oligomers were addressed by fluorescence microscopy, however only the levels of mannoproteins were promising and were therefore submitted to validation by flow cytometry. The validation by flow cytometry showed that caspofungin modified the cell wall properties of C. auris by inducing an increase on the exposure of mannoproteins.
Fig. 2.
Effect of caspofungin on C. auris yeast cells. Yeast cells were treated with sub-lethal concentrations of caspofungin for 24 h until the evaluation of cell wall components by microscopy. Bars = 20 µm. CAS – caspofungin.
3.2. Caspofungin treatment induces dynamic changes of gene expression related to the cell wall and other cellular processes
We investigated C. auris gene expression changes in response to caspofungin treatment using RNA seq. In three independent experiments, C. auris strains B11244 and MMC1 were cultivated for 24 h in the absence or presence of sub-inhibitory concentrations of caspofungin. We used principal-component analysis (PCA) and a heat map for the hierarchical clustering to analyze the similarities between the replicates and the differences among the treated and untreated samples. The samples clustered together, indicating a high level of correlation, whereas the treated and untreated samples clustered separately, consistent with a specific and global transcriptome response (Fig. S1).
To assess whether the transcripts were differentially expressed, we set the statistical significance of false-discovery rate (FDR) smaller than 5% and a fold change of at least 2x as requirements for differentially expressed transcripts. We found that 1088 transcripts were differentially expressed in the B11244 strain and 1589 in the MMC1 strain (Table S2). When we compared the transcripts common to both strains, we observed 657 mRNAs, which corresponds to 60% and 41% of the differentially expressed transcripts in B11244 and MMC1 strains, respectively (Table S2). The Table 2, Table 3 present the most expressed transcripts identified in both strains and also those exclusive to B11244 and MMC1.
Table 2.
The top upregulated transcripts upon caspofungin treatment. Max group mean – the average TPM values.
 |
Table 3.
The top upregulated transcripts under control conditions. Max group mean – the average TPM values.
 |
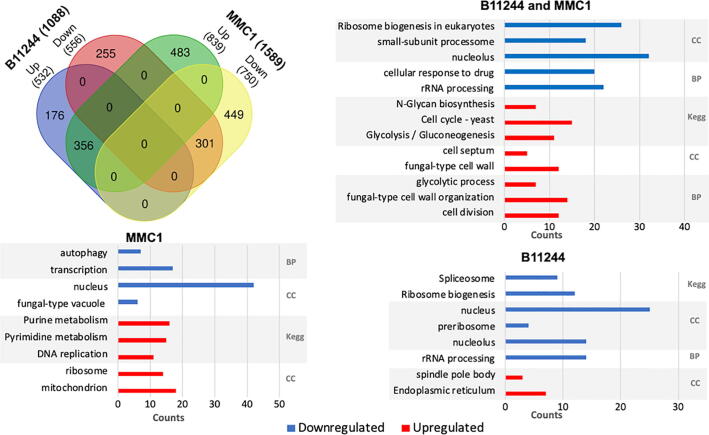
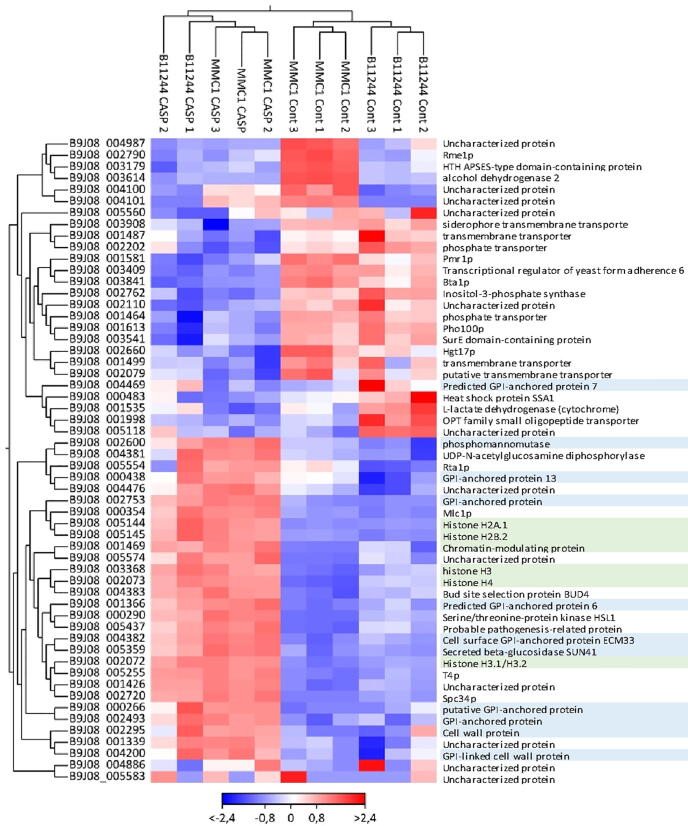
To evaluate the impact of caspofungin treatment in the gene expression of C. auris, we performed a functional enrichment analysis of the differentially expressed transcripts regulated in the presence of caspofungin in both MMC1 and B11244 strains. Firstly, we explored the 356 transcripts upregulated in the presence of the antifungal. Caspofungin induced changes in many important pathways such as cell cycle, glycolysis/gluconeogenesis (fold change 4.56, FDR 0%), cell wall formation (fold change 4.16, FDR 0%) and various types of N-glycan biosynthesis (fold change 4.71, FDR 3%), which are also linked to the synthesis of the cell wall (Fig. 3). The most expressed transcripts in response to the antifungal were histones and glycosylphosphatidylinositol (GPI)-anchored proteins in both MMC1 and B11244 strains (Table 2, Fig. 3 and Fig. 4). Among the 301 downregulated transcripts, we identified ribosome biogenesis in eukaryotes (fold change 9.93, FDR 0%) as the more prevalent pathway (Table 3 Fig. 3 and Fig. 4).
Fig. 3.
Venn diagram of the mRNAs identified in the B11244 (n = 1088) and MMC1 (n = 1588) strains after the statistical filters were applied. The transcripts equally expressed in both strains are the intersection of the diagram (n = 657). The bar charts represent the enriched functional categorization of the transcripts based on gene ontology (GO) annotations, in blue the terms associated to the downregulated and in red the upregulated transcripts. The x-axis represents the counts for each term and for the GO analysis the terms were statistically filtered by FDR ≤ 5%. BP – biological process, CC – cellular component, Kegg – Kyoto encyclopedia of genes and genomes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Heat map of the top most differentially expressed transcripts common to both B11244 and MMC1 strains (n = 11). The color scheme refers to log2 of the Fold change. Each column refers to one of the experimental replicates. The genes names were colored: In blue represent the transcripts related to cell wall and in green with nucleosome. The parameters for this analysis were: FDR < 0.01 and absolute fold change > 8. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We then analyzed the transcripts exclusively changed in the MMC1 strain. With these parameters, 483 transcripts were upregulated in the presence of caspofungin. The enriched pathways associated with these mRNAs were related to the functionality of ribosome (fold change 2.91, FDR 5%), mitochondrial (fold change 2.74, FDR 3%) and DNA replication (fold change 4.0, FDR 2%) (Fig. 3 and Table S3). The 449 downregulated transcripts upon caspofungin treatment in the MMC1 strain included numerous mRNAs coding for transcription factors (fold change 3.36, FDR 5%), nuclear functionality (fold change 1.67, FDR 3%), and autophagy pathways (fold change 6.45, FDR 2%) (Table S3).
A similar analysis with the B11244 strain revealed 176 transcripts that were upregulated during caspofungin treatment. The enriched terms reflected changes in protein export (fold change 6.71, FDR 1%), endoplasmic reticulum functionality (fold change 5.39, FDR 2%), and spindle body formation (fold change 22.31, FDR 4%) (Fig. 3 and Table S3). As for downregulated mRNAs (n = 255), the associated pathways were ribosome biogenesis (fold change 5.61, FDR 0%), spliceosome functionality (fold change 4.71, FDR 1%), and rRNA processing (fold change 6.51, FDR 0%) (Fig. 3 and Table S3).
To validate the consistency of our analysis of the response to caspofungin in C. auris, we performed qPCR (Fig. S2). We selected transcripts with distinct levels of expression in order to validate the RNA seq data (Fig. S2). As a reference for the relative expression analysis, we selected the C5 sterol desaturase because this transcript expression level did not vary comparing the control and treatment conditions. Indeed, the relative quantification of the transcripts were in accordance with the expression levels detected by the RNA-seq comparing the experimental conditions (Fig. S2).
3.3. Correlation between transcriptome and cellular metabolism
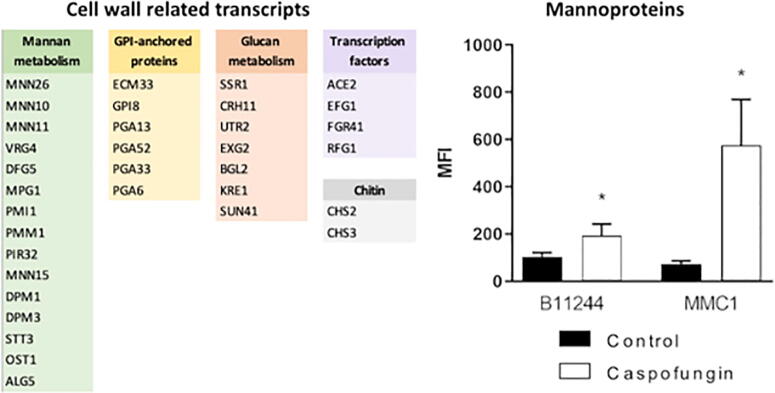
We asked whether the transcriptomic responses correlated with cellular alterations. For example, there were modifications in the cell wall of C. auris upon caspofungin treatment (Fig. 2), in addition to a high number of upregulated mRNAs related to cell wall biogenesis and organization, including mannan metabolism. Mannosylation levels of the cell surface were measured by flow cytometry and, indeed, mannosyl detection was greater when the cells were submitted to the caspofungin treatment (Fig. 5).
Fig. 5.
Cell wall alterations and correlation with the transcriptomic data. The transcripts associated to cell wall and mannoproteins measurement by flow cytometry. * indicates p < 0.05 by one-way Anova followed by Bonferroni’s multicomparisons test for four independent experiments. MMC1: Control vs caspo p-value = 0.0459. B11244: Control vs caspo p-value = 0.0253.
3.4. Proteomic analysis in response to caspofungin
Proteomic analyses of C. auris B11244 and MMC1 strains were performed in biological triplicates and technical duplicates. In total, 2004 proteins from the control and caspofungin groups were identified. The data was filtered based only in peptides identified in at least 2 replicates, and a minimum value of log fold change of 2 was established. Next, the proteins were filtered based on the value of p ≤ 0.05, in the ANOVA tests with multiple samples, totaling 632 proteins for the B11244 strain and 628 for the MMC1 strain that presented a fold change of at least 2 when control conditions and caspofungin treatment was considered (Table 4). The values of zero were kept in the matrices, without choosing to substitute the value of the normal distribution or the value of a constant. The control and treatment groups shared 328 proteins in common, but also have an expressive number of exclusive proteins for the studied strains.
Table 4.
Enriched pathways or gene ontology terms associated to the proteomic data in response to caspofungin treatment and the control conditions.
| Category | Term | P-Value | Fold Enrichment | Fisher Exact | ||
|---|---|---|---|---|---|---|
| B11244 MMC1 | Caspofungin | GOTERM_BP | Fungal-type cell wall organization | 1.10E−03 | 16.7 | 6.30E−05 |
| GOTERM_MF | 1,3-beta-glucanosyltransferase activity | 1.30E−02 | 138.3 | 8.30E−05 | ||
| GOTERM_BP | Translation | 3.10E−02 | 9.6 | 3.10E−03 | ||
| GOTERM_CC | Yeast-form cell wall | 8.70E−02 | 19.9 | 4.30E−03 | ||
| KEGG_PATHWAY | Ribosome | 5.00E−02 | 7.2 | 6.80E−03 | ||
| Control | KEGG_PATHWAY | Biosynthesis of secondary metabolites | 8.40E−05 | 4.9 | 1.70E−05 | |
| KEGG_PATHWAY | Biosynthesis of amino acids | 1.20E−04 | 9 | 1.30E−05 | ||
| KEGG_PATHWAY | Histidine metabolism | 1.60E−03 | 43.2 | 3.40E−05 | ||
| B11244 | Caspofungin | GOTERM_CC | Yeast-form cell wall | 3.50E−04 | 14.2 | 2.30E−05 |
| KEGG_PATHWAY | Protein processing in endoplasmic reticulum | 7.60E−03 | 4.6 | 1.60E−03 | ||
| GOTERM_BP | GDP-mannose biosynthetic process | 2.50E−02 | 75.7 | 2.70E−04 | ||
| GOTERM_BP | thiamine biosynthetic process | 3.50E−02 | 54.1 | 5.60E−04 | ||
| Control | KEGG_PATHWAY | Biosynthesis of secondary metabolites | 6.20E−09 | 3.3 | 1.80E−09 | |
| KEGG_PATHWAY | Biosynthesis of amino acids | 1.90E−08 | 5.3 | 3.30E−09 | ||
| KEGG_PATHWAY | Biosynthesis of antibiotics | 7.00E−05 | 2.9 | 2.30E−05 | ||
| KEGG_PATHWAY | 2-Oxocarboxylic acid metabolism | 2.50E−04 | 7.5 | 2.90E−05 | ||
| KEGG_PATHWAY | Histidine metabolism | 2.50E−04 | 15 | 1.40E−05 | ||
| KEGG_PATHWAY | Valine, leucine and isoleucine biosynthesis | 2.90E−03 | 13.3 | 1.80E−04 | ||
| MMC1 | Caspofungin | GOTERM_MF | structural constituent of ribosome | 2.00E−03 | 13.6 | 1.40E−04 |
| KEGG_PATHWAY | Ribosome | 3.00E−03 | 10.7 | 2.70E−04 | ||
| GOTERM_BP | Translation | 4.10E−03 | 10.9 | 3.60E−04 | ||
| Control | KEGG_PATHWAY | Biosynthesis of amino acids | 7.20E−07 | 6.9 | 9.90E−08 | |
| KEGG_PATHWAY | Histidine metabolism | 4.70E−04 | 24 | 1.70E−05 | ||
| KEGG_PATHWAY | Biosynthesis of secondary metabolites | 2.00E−03 | 2.8 | 6.90E−04 | ||
| KEGG_PATHWAY | 2-Oxocarboxylic acid metabolism | 9.50E−03 | 8.5 | 1.00E−03 | ||
| KEGG_PATHWAY | Lysine biosynthesis | 1.40E−02 | 15.8 | 8.00E−04 | ||
| KEGG_PATHWAY | Biosynthesis of antibiotics | 1.60E−02 | 2.7 | 5.60E−03 |
Using a fold change cutoff for protein detection equal to or >2, between the control group and caspofungin-treated cells, 63 proteins were positively regulated and 85 negatively regulated in strain B11244 after antifungal treatment. In strain MMC1, 40 proteins were positively regulated, while 44 proteins were down regulated (Table S4).
The biological processes predicted by the ontology confirms the stress state of the cell. It was also possible to establish a relationship between the observed response and the previously identified antifungal mechanisms of caspofungin. There were a number of proteins enriched in caspofungin-treated cells related to cell wall synthesis/integrity and ribosomes, which were common to both strains (Fig. 6). Caspofungin treatment also induced a common enrichment of proteins related to the cell-wall organization (FDR 0.35%), in addition to translation (FDR 4.90%) and enzymes related to b-glucan metabolism (FDR 1.20%) (Table 4). Among the most differentially expressed proteins, we highlight the observation that Mkc1 and Phr2 have been previously implicated in stress response, cell wall biogenesis, and virulence (Fig. 4) [35]. In strain B11244, a response associated with protein processing in endoplasmic reticulum (FDR 4%) was exclusively observed. More specifically, mitochondrial heat shock protein 60, pleiotropic ABC efflux transporter of multiple drugs CDR1, hsp70 family ATPase, and mitochondrial heat shock protein 78 were upregulated (Fig. S3 and Table S4). Hog1, which interacts with Mkc1 and plays a role in the integrity of the cell wall in addition to presenting different responses to stress, was also enriched in caspofungin-treated cells. In the MMC1 strain, the most abundant proteins in caspofungin-treated cells were ribosomal (FDR 0.21%, 50% of the total proteins identified as exclusive to MMC1), and cell wall-related (FDR 4.7%) (Fig. S4 and Table S4). As for the proteins identified in the control condition, the most observed terms were amino acid metabolism and secondary metabolites in both strains (FDR 0.15%) (Table 4).
Fig. 6.
Comparison between differentially expressed proteins (FC > 2) in caspofungin treated and control cells common in B11244 and MMC1 strains (n = 12). The B11244 or MMC1 strains (columns) and protein groups (lines) were hierarchically clustered. The protein intensity was represented by color scale (blue, lower intensity; red, higher intensity). Highlighted in grey are the proteins more abundant in the caspofungin treatment. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Comparison between transcriptomic and proteomic responses in response to caspofungin.
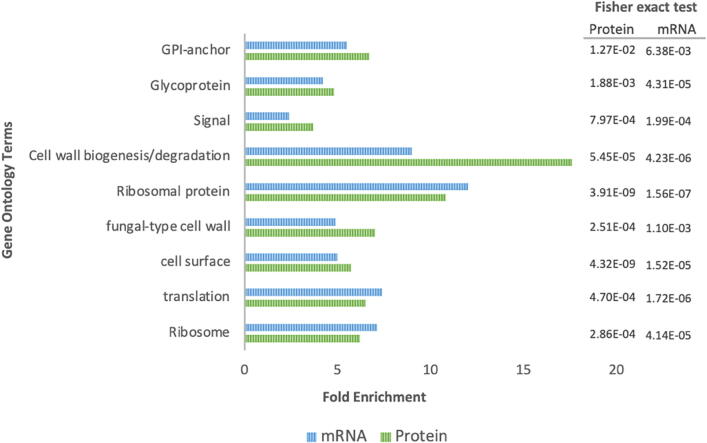
The combined results of RNA and protein analyses indicated an integrated response of the cells after exposure to caspofungin. Specifically, cell wall proteins, ribosomes and those involved with the protein processing in the endoplasmic reticulum were identified as participants of the response of C. auris to caspofungin (Fig. 7), as concluded from the increased detection of these classes after drug exposure in both RNA and protein analyses. Proteins associated with virulence were upregulated in both mRNA and protein analyses, including Hog1, Phr2, in addition to those related to cell wall synthesis (mannose-6-phosphate isomerase, mannose-1-phosphate guanylyltransferase, mannan endo-1,6-alpha-mannosidase DFG5, glucan 1,3-beta-glucosidase BGL2, glucan 1,3-beta-glucosidase, and 1,3-beta-glucanosyltransferase PGA4).
Fig. 7.
Comparison of the gene ontology terms enriched in the transcriptome and proteome upon caspofungin treatment. The bar charts represent the enriched functional categorization of the transcripts based on gene ontology (GO) annotations. In blue the terms associated to mRNAs and in green the terms associated to the proteomic data from both strains. The x-axis represents the fold enrichment for each term and the numbers on the right express the statisctic values - Fisher exact test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
5. Discussion
C. auris is a major threat to public health, as extensively discussed in this study and many others [6], [36], [37], [38], [39], [40], [41]. To understand how C. auris responds to major antifungals is essential for the design of therapeutic strategies, and for the identification of cellular targets for novel antifungals. The mechanism of antifungal activity of caspofungin, the first-line treatment against C. auris, is already known. However, how the inhibition of cell wall synthesis is orchestrated with other cellular responses remains unknown. The caspofungin antifungal drug acts by inhibiting the β-1,3-glucan synthase. It targets the catalytic glucan synthase FKS subunits, therefore suppressing the synthesis of b-1,3-glucan in the fungal cell wall [42], [43], [44].
We identified multiple pathways that were significantly modified by the treatment of C. auris with caspofungin. Molecules involved in N-glycan biosynthesis were enriched in C. auris treated with caspofungin, which is consistent with cell wall rearrangement. In C. neoformans, N-linked glycosylation is an important modulator of host cell death, and therefore has a critical role in pathogenicity [45]. Cryptococcal mutants harboring truncated N-glycans were not pathogenic in mice, despite being able to attach to lung epithelial cells and enter them through phagocytosis. Also, C. neoformans capacity for cell wall remodeling was maintained, but they induced less cell death in macrophages, a mechanism used for pulmonary escape and dissemination in vivo [45]. These results indicate that, if a similar process occurs in C. auris, caspofungin treatment could impact fungal virulence.
Cell wall related transcripts, such as GPI-anchored associated genes, were the most expressed genes in the presence of caspofungin for both strains in our study. Interestingly, cell wall genes were previously shown to be upregulated during the initial hours of biofilm formation in C. auris [22]. This high expression levels of cell wall related transcripts could be a compensatory mechanism to circumvent the absence of glucans with other components, such as chitin and mannans, which may reduce C. auris susceptibility to antifungal drugs. In fact, our results support such compensatory mechanisms, as evidenced by altered levels of the transcripts, as well as the altered detection of mannans and chitin at the cell wall. This outcome has been described for several Candida species that rapidly responded to caspofungin by increasing the cell wall chitin content [46], [47]. In addition, a similar result was observed in Aspergillus fumigatus where caspofungin treatment led to an increased chitin content, which reduced susceptibility to the antifungal, and altered cell morphology [48]. As for the mannan synthesis, the mRNA coding the GPI-anchored protein ECM33 was one of the most upregulated transcripts in both C. auris strains under caspofungin treatment. ECM33 helps the assembling of the mannoprotein outer layer of the cell wall [49]. Supporting our mRNA data, C. auris yeast cells from both strains presented a dramatic increase in their mannoprotein layer under caspofungin treatment.
Our results demonstrated a high expression of mRNAs coding for histones. In C. albicans, the availability of specific chromatin modifiers can affect drug resistance [50]. The transcription factor Cas5 has been implicated in stress responses, drug resistance and cell cycle regulation in C. albicans (52).
We also observed a higher expression of mRNAs coding for ribosomal proteins in C. auris treated with caspofungin, which was more evident in the MMC1 strainThese results are in agreement with a previous study characterizing the C. auris transcriptome during amphotericin B and voriconazole treatment [39]. Upon amphotericin B treatment, the upregulated genes were related to translation and ribosomal proteins [39]. Compared to our data, we found 78 common transcripts also upregulated in the presence of caspofungin. Most of the mRNAs responding to amphotericin B treatment (46) were also identified in our study (Table S5). We also found 30 transcripts upregulated during both caspofungin (this study) and voriconazole treatments [39]. In these common transcripts, the majority of the mRNAs code for ribosomal proteins (Table S5). In addition, during the biofilm formation in C. auris, the most consistent mRNAs upregulated were those coding ribosomal proteins [22] (Table S5). In a study that combined proteomics and microarray analyses of A. fumigatus treated for 24 h with caspofungin, 81% of the overexpressed proteins and 86.4% of the mRNAs were ribosomal [52]. This change was suggested to be associated with a ribosomal reshuffling response, which reflects a requirement for more protein synthesis to overcome the inhibition caused by the antifungal drug [53]. In a recent work, a C. auris strain was adapted to high concentrations of caspofungin and subjected to RNA-seq. Similar to our observations, most of the differentially expressed transcripts enriched in response to caspofungin coded for cell wall proteins [51]. We then compared our data with the results obtained of C. auris after 3 h in the presence of high caspofungin concentration and it was possible to observe a partial overlap in the transcripts differentially expressed. The common enriched pathways were fungal cell wall, amino sugar metabolic process, chitin synthase and mannan endo-1,6-alpha-mannosidase activity (Table S5). These similarities confirm our data and reinforces the important role not only of chitin, but also other cell wall proteins as a compensatory mechanism to allow cell survival.
Our proteomic analysis supported the transcriptomic data. The cell wall biogenesis and degradation were enriched in our proteomic and transcriptomic analyses after exposure to caspofungin. The enrichment of transcripts and proteins related to biofilm formation, translation, GPI-anchored proteins, and mannose related proteins during caspofungin treatment was also consistently verified in both analyses. It is already known that the establishment of biofilms contributes to the success of the infection [54], just as they confer a greater resistance of the organism to antifungals [55]. C. auris expresses proteins related to biofilm formation and maintenance in higher levels compared to C. albicans [56]. It is worth mentioning the presence of two important proteins that play roles in various metabolic pathways Mkc1 and Hog1. Mkc1 is an important kinase in pathogenic fungi, is part of a MAPK pathways, associated to many metabolic pathways in the cell. Mkc1 is involved in cell cycle and also in maintaining cell wall integrity [57]. It is also involved in the tolerance of C. albicans to caspofungin [35], [58]. Hog1 is also part of the MAPK response and was identified more expressed in the B11244 strain. The HOG pathway plays an essential role in C. albicans, including infection, virulence, and stress response [59]. In C. auris, Hog1 is also associated to virulence and stress response [60]. The deletion of hog1 in C. auris lead to a reduced resistance to caspofungin, amphotericin B and cell wall composition [61]. This result is in accordance with our observation that in the presence of caspofungin HOG1 was more expressed, and this could allow adaptation in the presence of the antifungal and cell wall remodeling.
We showed that an extensive cell wall remodeling, transcriptional and translational alterations occur in C. auris upon caspofungin treatment. In distinct Candida species, it has been described that in response to caspofungin, the cells increased the chitin content and b-1,3-glucan become more exposed on the cell surface [46], [47], [62]. We did not observe a clear enrichment in chitin; however, we observed an increased detection of mannans. In accordance with our current observations in C. auris, in Candida glabrata caspofungin treatment led to higher amounts of mannans on the cell wall [63]. Therefore, our results consolidate the notion that not only chitin but also mannan synthesis, is an important of C. auris to a long exposure to caspofungin. Interestingly, C. auris mannoproteins are unique compared to other pathogenic Candida species [64]. In fact, the presence of these unique mannans in C. auris affects the recognition by and modulation of the host immune cells. When cell wall components derived from C. albicans and C. auris were used to stimulate PBMCs, it was observed that after 4 h, the PBMC transcriptomic alteration was due to the β-glucan in both species. However, in a late response (24 h), C. auris mannans were the main component eliciting the response in PBMC gene expression, and this was not observed in C. albicans, maybe due to the specific mannan composition in C. auris cell wall [65]. Our study demonstrates, by transcriptomics, proteomics and cellular measurement, an important role for mannan in the adaptation of C. auris during growth with caspofungin, in addition to chitin, which has been extensively explored. This biosynthesis pathway can be a drug target and should be better explored, as already described for host-pathogen interaction.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the staff of the Genomics section of the Life Sciences Core Facility (LaCTAD), part of the University of Campinas (UNICAMP), for their contributions to RNA-sequencing. J.D.N., D.Z-M. and E.S.N. were partially supported by NIH R21 AI124797. M.L.R. was supported by grants from the Brazilian Ministry of Health (grant number 440015/2018-9), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grants 405520/2018-2, and 301304/2017-3) and Fiocruz (grants VPPCB-007-FIO-18 and VPPIS-001-FIO18). The authors also acknowledge support from the Instituto Nacional de Ciência e Tecnologia de Inovação em Doenças de Populações Negligenciadas (INCT-IDPN). M.L.R. is currently on leave from the position of Associate Professor at the Microbiology Institute of the Federal University of Rio de Janeiro, Brazil. LRA received financial support from Inova Fiocruz/Fundação Oswaldo Cruz [Grant number VPPCB-07-FIO-18-2-52] and CNPq [Grant number 442317/2019-0]. L.R.A is a research fellow awardee from CNPq.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.09.007.
Contributor Information
Joshua D. Nosanchuk, Email: josh.nosanchuk@einsteinmed.org.
Lysangela R. Alves, Email: lysangela.alves@fiocruz.br.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bongomin F., Gago S., Oladele R., Denning D. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi. 2017;3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortegiani A., Misseri G., Chowdhary A. What’s new on emerging resistant Candida species. Intensive Care Med. 2019;45(4):512–515. doi: 10.1007/s00134-018-5363-x. [DOI] [PubMed] [Google Scholar]
- 3.Guinea J. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect. 2014;20:5–10. doi: 10.1111/1469-0691.12539. [DOI] [PubMed] [Google Scholar]
- 4.de Jong A.W., Hagen F. Attack, defend and persist: how the fungal pathogen Candida auris was able to emerge globally in healthcare environments. Mycopathologia. 2019;184(3):353–365. doi: 10.1007/s11046-019-00351-w. [DOI] [PubMed] [Google Scholar]
- 5.Schelenz S., Hagen F., Rhodes J.L., Abdolrasouli A., Chowdhary A., Hall A. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 2016;5(1) doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lockhart S.R., Etienne K.A., Vallabhaneni S., Farooqi J., Chowdhary A., Govender N.P. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhodes J., Abdolrasouli A., Farrer R.A., Cuomo C.A., Aanensen D.M., Armstrong-James D. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect. 2018;7(1):1–12. doi: 10.1038/s41426-018-0045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nett JE. Candida auris : An emerging pathogen “ incognito ”? 2019:6–11. [DOI] [PMC free article] [PubMed]
- 9.Satoh K., Makimura K., Hasumi Y., Nishiyama Y., Uchida K., Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 10.Jeffery-Smith A., Taori S.K., Schelenz S., Jeffery K., Johnson E.M., Borman A. Candida auris: a review of the literature. Clin Microbiol Rev. 2018;31(1) doi: 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eyre D.W., Sheppard A.E., Madder H., Moir I., Moroney R., Quan T.P. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018;379(14):1322–1331. doi: 10.1056/NEJMoa1714373. [DOI] [PubMed] [Google Scholar]
- 12.Welsh R.M., Bentz M.L., Shams A., Houston H., Lyons A., Rose L.J. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol. 2017;55(10):2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams E., Quinn M., Tsay S., Poirot E., Chaturvedi S., Southwick K. Candida auris in healthcare facilities, New York, USA, 2013–2017. Emerg Infect Dis. 2018;24(10):1816–1824. doi: 10.3201/eid2410.180649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamoth F., Kontoyiannis D.P. The Candida auris Alert: facts and perspectives. J Infect Dis. 2018;217:516–520. doi: 10.1093/infdis/jix597. [DOI] [PubMed] [Google Scholar]
- 15.Rudramurthy S.M., Chakrabarti A., Paul R.A., Sood P., Kaur H., Capoor M.R. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J Antimicrob Chemother. 2017;72:1794–1801. doi: 10.1093/jac/dkx034. [DOI] [PubMed] [Google Scholar]
- 16.Chowdhary A., Prakash A., Sharma C., Kordalewska M., Kumar A., Sarma S. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother. 2018;73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 17.Kordalewska M., Lee A., Park S., Berrio I., Chowdhary A., Zhao Y. Understanding Echinocandin resistance in the emerging pathogen Candida auris. Antimicrob Agents Chemother. 2018;62:e00238–e318. doi: 10.1128/AAC.00238-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherry L., Ramage G., Kean R., Borman A., Johnson E.M., Richardson M.D. Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis. 2017;23(2):328–331. doi: 10.3201/eid2302.161320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton M.V., Nett J.E. Candida auris infection and biofilm formation: Going beyond the surface. Curr Clin Microbiol Rep. 2020;7(3):51–56. doi: 10.1007/s40588-020-00143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez E.G., Zarnowski R., Choy H.L., Zhao M., Sanchez H., Nett J.E. Conserved role for biofilm matrix polysaccharides in Candida auris drug resistance. MSphere. 2019;4(1) doi: 10.1128/mSphereDirect.00680-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang M.Y., Woolford C.A., May G., McManus C.J., Mitchell A.P., Lin X. Circuit diversification in a biofilm regulatory network. PLoS Pathog. 2019;15(5):e1007787. doi: 10.1371/journal.ppat.1007787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kean R, Delaney C, Sherry L, Borman A, Johnson EM, Richardson MD, et al. Transcriptome Assembly and Profiling of Candida auris Reveals Novel Insights into Biofilm-Mediated Resistance. MSphere 2018;3. 10.1128/mSphere.00334-18. [DOI] [PMC free article] [PubMed]
- 23.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research 2009. 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed]
- 28.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 2009. 10.1373/clinchem.2008.112797. [DOI] [PubMed]
- 30.Wiśniewski J.R., Gaugaz F.Z. Fast and sensitive total protein and peptide assays for proteomic analysis. Anal Chem. 2015;87(8):4110–4116. doi: 10.1021/ac504689z. [DOI] [PubMed] [Google Scholar]
- 31.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 32.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 33.Gow N.A.R., Latge J.-P., Munro C.A., Heitman J. The fungal cell wall: structure, biosynthesis, and function. Microbiol Spectr. 2017;5(3) doi: 10.1128/microbiolspec.FUNK-0035-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heilmann C.J., Sorgo A.G., Mohammadi S., Sosinska G.J., de Koster C.G., Brul S. Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans. Eukaryot Cell. 2013;12(2):254–264. doi: 10.1128/EC.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaFayette S.L., Collins C., Zaas A.K., Schell W.A., Betancourt-Quiroz M., Gunatilaka A.A.L. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, Calcineurin, and Hsp90. PLoS Pathog. 2010;6(8):e1001069. doi: 10.1371/journal.ppat.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casadevall A., Kontoyiannis D.P., Robert V. Environmental Candida auris and the global warming emergence hypothesis. MBio. 2021;12(2) doi: 10.1128/mBio.00360-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du H., Bing J., Hu T., Ennis C.L., Nobile C.J., Huang G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16(10):e1008921. doi: 10.1371/journal.ppat.1008921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodrigues M.L., Nosanchuk J.D., Reynolds T.B. Fungal diseases as neglected pathogens: A wake-up call to public health officials. PLoS Negl Trop Dis. 2020;14(2):e0007964. doi: 10.1371/journal.pntd.0007964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muñoz J.F., Gade L., Chow N.A., Loparev V.N., Juieng P., Berkow E.L. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun. 2018 doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lockhart S.R., Etienne K.A., Vallabhaneni S., Farooqi J., Chowdhary A., Govender N.P. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chowdhary A., Sharma C., Meis J.F., Hogan D.A. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13(5):e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurtz M.B., Douglas C.M. Lipopeptide inhibitors of fungal glucan synthase. Med Mycol. 1997;35(2):79–86. doi: 10.1080/02681219780000961. [DOI] [PubMed] [Google Scholar]
- 43.Saravolatz L.D., Deresinski S.C., Stevens D.A. Caspofungin. Clin Infect Dis. 2003;36:1445–1457. doi: 10.1086/375080. [DOI] [PubMed] [Google Scholar]
- 44.Song J.C., Stevens D.A. Caspofungin: Pharmacodynamics, pharmacokinetics, clinical uses and treatment outcomes. Crit Rev Microbiol. 2016;42(5):813–846. doi: 10.3109/1040841X.2015.1068271. [DOI] [PubMed] [Google Scholar]
- 45.Thak EJ, Lee S Bin, Xu-Vanpala S, Lee DJ, Chung SY, Bahn YS, et al. Core N-glycan structures are critical for the pathogenicity of cryptococcus neoformans by modulating host cell death. MBio 2020;11. 10.1128/mBio.00711-20. [DOI] [PMC free article] [PubMed]
- 46.Walker L.A., Munro C.A., de Bruijn I., Lenardon M.D., McKinnon A., Gow N.A.R. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008;4(4):e1000040. doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker L.A., Gow N.A.R., Munro C.A. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob Agents Chemother. 2013;57(1):146–154. doi: 10.1128/AAC.01486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker L.A., Lee K.K., Munro C.A., Gow N.A.R. Caspofungin treatment of Aspergillus fumigatus results in ChsG-dependent upregulation of chitin synthesis and the formation of chitin-rich microcolonies. Antimicrob Agents Chemother. 2015;59(10):5932–5941. doi: 10.1128/AAC.00862-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pardo M., Monteoliva L., Vázquez P., Martínez R., Molero G., Nombela C. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology. 2004 doi: 10.1099/mic.0.26924-0. [DOI] [PubMed] [Google Scholar]
- 50.O’Kane C.J., Weild R., M. Hyland E. Chromatin structure and drug resistance in candida spp. MDPI AG. 2020;6(3):121. doi: 10.3390/jof6030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lara-Aguilar V, Rueda C, García-Barbazán I, Varona S, Monzón S, Jiménez P, et al. Adaptation of the emerging pathogenic yeast Candida auris to high caspofungin concentrations correlates with cell wall changes. Virulence n.d.;12:1400–17. 10.1080/21505594.2021.1927609. [DOI] [PMC free article] [PubMed]
- 52.Cagas S.E., Jain M.R., Li H., Perlin D.S. Profiling the Aspergillus fumigatus proteome in response to caspofungin. Antimicrob Agents Chemother. 2011;55:146–154. doi: 10.1128/AAC.00884-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shishodia S.K., Tiwari S., Shankar J. Resistance mechanism and proteins in Aspergillus species against antifungal agents. Mycology. 2019;10:151–165. doi: 10.1080/21501203.2019.1574927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nailis H., Kucharíková S., Řičicová M., Van Dijck P., Deforce D., Nelis H. Real-time PCR expression profiling of genes encoding potential virulence factors in Candida albicans biofilms: identification of model-dependent and -independent gene expression. BMC Microbiol. 2010;10:114. doi: 10.1186/1471-2180-10-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seneviratne C.J., Wang Y., Jin L., Abiko Y., Samaranayake L.P. Proteomics of drug resistance in Candida glabrata biofilms. Proteomics. 2010;10:1444–1454. doi: 10.1002/pmic.200900611. [DOI] [PubMed] [Google Scholar]
- 56.Zamith-Miranda D., Heyman H.M., Cleare L.G., Couvillion S.P., Clair G.C., Bredeweg E.L. Multi-omics Signature of Candida auris, an Emerging and Multidrug-Resistant Pathogen. MSystems. 2019 doi: 10.1128/msystems.00257-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Navarro-García F., Alonso-Monge R., Rico H., Pla J., Sentandreu R., Nombela C. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology. 1998;144:411–424. doi: 10.1099/00221287-144-2-411. [DOI] [PubMed] [Google Scholar]
- 58.Caplan T., Polvi E.J., Xie J.L., Buckhalter S., Leach M.D., Robbins N. Functional Genomic Screening Reveals Core Modulators of Echinocandin Stress Responses in Candida albicans. Cell Reports. 2018;23:2292–2298. doi: 10.1016/j.celrep.2018.04.084. [DOI] [PubMed] [Google Scholar]
- 59.Alonso-Monge R., Guirao-Abad J.P., Sánchez-Fresneda R., Pla J., Yagüe G., Argüelles J.C. The Fungicidal Action of Micafungin is Independent on Both Oxidative Stress Generation and HOG Pathway Signaling in Candida albicans. Microorganisms. 2020;8:1867. doi: 10.3390/microorganisms8121867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Day A.M., McNiff M.M., da Silva D.A., Gow N.A.R., Quinn J. Hog1 Regulates Stress Tolerance and Virulence in the Emerging Fungal Pathogen Candida auris. MSphere. 2018 doi: 10.1128/msphere.00506-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shivarathri R, Jenull S, Stoiber A, Chauhan M, Mazumdar R, Singh A, et al. The Two-Component Response Regulator Ssk1 and the Mitogen-Activated Protein Kinase Hog1 Control Antifungal Drug Resistance and Cell Wall Architecture of Candida auris. MSphere 2020;5. 10.1128/mSphere.00973-20. [DOI] [PMC free article] [PubMed]
- 62.Lee K.K., MacCallum D.M., Jacobsen M.D., Walker L.A., Odds F.C., Gow N.A.R. Elevated Cell Wall Chitin in Candida albicans Confers Echinocandin Resistance In Vivo. Antimicrob Agents Chemother. 2012;56:208–217. doi: 10.1128/AAC.00683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodrigues C.F., Rodrigues M.E., Henriques M. Susceptibility of Candida glabrata biofilms to echinocandins: alterations in the matrix composition. Biofouling. 2018;34:569–578. doi: 10.1080/08927014.2018.1472244. [DOI] [PubMed] [Google Scholar]
- 64.Yan L., Xia K., Yu Y., Miliakos A., Chaturvedi S., Zhang F. Unique Cell Surface Mannan of Yeast Pathogen Candida auris with Selective Binding to IgG. ACS Infect Dis. 2020;6:1018–1031. doi: 10.1021/acsinfecdis.9b00450. [DOI] [PubMed] [Google Scholar]
- 65.Bruno M., Kersten S., Bain J.M., Jaeger M., Rosati D., Kruppa M.D. Transcriptional and functional insights into the host immune response against the emerging fungal pathogen Candida auris. Nat Microbiol. 2020 doi: 10.1038/s41564-020-0780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.