Summary
About 150 modifications have been identified in RNA species. Besides their regulatory roles in the intracellular gene expression, abundant modified RNA nucleosides are catabolized from RNA and released into extracellular fluids, which can impact extracellular signaling as ligands for receptors. Here, we describe a protocol to prepare samples from biological specimens, including cultured cells, extracellular fluid, and tissues, to measure both intracellular and extracellular RNA modifications using mass spectrometry.
For complete details on the use and execution of this protocol, please refer to Ogawa et al. (2021).
Subject areas: Mass Spectrometry, Metabolism, Metabolomics, Molecular Biology, Signal Transduction
Graphical abstract

Highlights
-
•
Isolation of various RNA species from total RNA for analysis of modified nucleosides
-
•
Extraction of modified nucleosides-containing metabolites from biological fluids
-
•
Methods and parameters for measurement of modified nucleosides by LC-MS/MS
About 150 modifications have been identified in RNA species. Besides their regulatory roles in the intracellular gene expression, abundant modified RNA nucleosides are catabolized from RNA and released into extracellular fluids, which can impact extracellular signaling as ligands for receptors. Here, we describe a protocol to prepare samples from biological specimens, including cultured cells, extracellular fluid, and tissues, to measure both intracellular and extracellular RNA modifications using mass spectrometry.
Before you begin
Note: Abbreviations: LMS, L-methionine sulfone; FBS, fetal bovine serum; LC-MS/MS, liquid chromatography-mass spectrometry
-
1.
RNA purification. Fresh biological samples should be processed immediately after collection as described below. Otherwise, samples should be snap-frozen in liquid nitrogen and stored at –80°C until preparation.
Note: To purify RNA from tissues or cells, a phenol-chloroform extraction is recommended to isolate all RNA species. If a silica spin column is used to extract RNA, confirm that the column will retain the desired RNA species. Conventional spin columns will only retain large RNA species (> 200nt).
Note: For cell culture samples, it is highly recommended to use phenol red-free medium to prevent contamination of the HPLC column and mass spectrometer with phenol red.
Note: For tissue samples, record the wet weight after collection or before homogenization. To extract modified nucleosides, ∼100 mg of tissues is sufficient.
-
2.
Prepare nuclease P1 solution and 200 mM filtered-HEPES solution (pH 7.0) to digest RNA. Dissolve 500 U nuclease P1 in 1 mL of 10 mM ammonium acetate (pH 5.3) to make a 0.5 U/μL solution.
-
3.
Prepare standard nucleoside solutions diluted in water (for example, 10 nM, 100 nM, and 1 μM).
Note: If available, stable isotope standard is recommended. Standard solutions should be aliquoted in small volumes and stored at temperatures lower than –20°C. Avoid repeated freeze–thaw cycles.
-
4.Precondition ultrafiltration device before use
-
a.Add 300 μL ultrapure water to the centrifugal filter device.
-
b.Centrifuge at 14,000 × g at room temperature for 30 min.
-
c.Invert the inner filter device and perform a flash centrifugation to completely remove residual water.
-
d.The device is ready for addition of cell or tissue extracts.
-
a.
Note: According to the manufacturer’s datasheet (https://www.merckmillipore.com/JP/en/product/Amicon-Ultra-0.5mL-Centrifugal-Filters-for-DNA-and-Protein-Purification-and-Concentration,MM_NF-C82301#documentation), the Amicon Ultr-0.5 ultrafiltration membrane contains a trace amount of glycerin, which may interfere with the evaporation step and LC-MS/MS analysis. We commend performing this preconditioning step to minimize the unnecessary interference.
Note: Perform preconditioning steps right before extraction process.
Optional: Microman E (Gilson) is recommended for accurately dispensing volatile liquids such as methanol.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Ultrapure Water (LC/MS grade) | Wako | Cat#214-01301 |
| Methanol (LC/MS grade) | Wako | Cat#134-14523 |
| Chloroform (HPLC grade) | Wako | Cat#033-08631 |
| Acetonitrile (LC/MS grade) | Wako | Cat#018-19853 |
| 2-Propanol (LC/MS grade) | Wako | Cat#168-25531 |
| 10× Loading dye | Takara | Cat#9157 |
| Agarose S | Nippon Gene | Cat#316-01191 |
| Alkaline phosphatase (E. coli C75) | Takara | Cat#2120A |
| Nuclease P1 | Wako | Cat#145-08221 |
| L-Methionine sulfone | Alfa Aesar | Cat# A17027 |
| Pseudouridine | Tokyo Chemical Industry | Cat#P2396; CAS:1445-07-4 |
| 5-Methylcytidine | Tokyo Chemical Industry | Cat#M1931; CAS:2140-61-6 |
| 2′-O-Methylcytidine | Tokyo Chemical Industry | Cat#M2317; CAS:2140-72-9 |
| 2′-O-Methyluridine | Tokyo Chemical Industry | Cat#M2290; CAS:2140-76-3 |
| 2′-O-Methylguanosine | Tokyo Chemical Industry | Cat#M2318; CAS:2140-71-8 |
| 2′-O-Methyladenosine | Tokyo Chemical Industry | Cat#M2291; CAS:2140-79-6 |
| 5,6-Dihydrouridine | Toronto Research Chemicals | Cat#D449668; CAS:5627-05-4 |
| 5-Formylcytidine | Toronto Research Chemicals | Cat#F698965; CAS:148608-53-1 |
| N6, 2′-O-Dimethyladenosine | Toronto Research Chemicals | Cat#D447415; CAS:57817-83-1 |
| 3-Methyluridine | Sigma-Aldrich | Cat#M4129; CAS:2140-69-4 |
| 7-Methylguanosine | Sigma-Aldrich | Cat#M0627; CAS:20244-86-4 |
| 1-Methylguanosine | Santa Cruz Biotechnology | Cat#sc-500889; CAS:2140-65-0 |
| 1-Methylinosine | Santa Cruz Biotechnology | Cat#sc-483758; CAS:2140-73-0 |
| 2′-O-Methylinosine | Santa Cruz Biotechnology | Cat#sc-283498; CAS:3881-21-8 |
| N2-Methylguanosine | Biosynth Carbosynth | Cat#NM35522; CAS:2140-77-4 |
| 1-Methyladenosine | Cayman Chemical | Cat#16937; CAS:15763-06-1 |
| N6-Methyladenosine | Abcam | Cat#ab145715; CAS:1867-73-8 |
| N4-Acetylcytidine | Combi-Blocks | Cat#QB-9019; CAS:3768-18-1 |
| N2,N2-Dimethylguanosine | MedChem Express | Cat#HY-113137; CAS:2140-67-2 |
| N6,N6-dimethyladenosine | MedChem Express | Cat#HY-101984; CAS:2620-62-4 |
| RNA Clean & Concentrator-5 | Zymo Research | Cat#R1015 |
| Oligotex-dT30<Super> mRNA Purification Kit (From Total RNA) | Takara | Cat#9086 |
| Zymoclean Gel RNA Recovery Kit | Zymo Research | Cat#R1011 |
| Other | ||
| Submerge-Mini (electrophoresis device) | ATTO | Cat#WSE-1710 |
| UV transilluminator | Bio Craft | Cat#CI-110B |
| TissueRuptor Disposable Probes | QIAGEN | Cat#990890 |
| TissueRuptor II | QIAGEN | Cat#9002754 |
| Amicon Ultra-0.5 Centrifugal Filter Devices | Millipore | Cat#UFC500396 |
| Rotating incubator | Taitec | Cat#RT-50 |
| SpeedVac vacuum concentrators | Thermo Fisher Scientific | Cat#SPD1010 |
| High Performance Liquid Chromatograph Triple Quadrupole Mass Spectrometer | Shimadzu | Cat#LCMS-8050 |
| Inertsil ODS-3 column | GL Sciences | Cat#5020-84655 |
Materials and equipment
[Nuclease P1 solution]
| Reagent | Final concentration | Amount |
|---|---|---|
| Nuclease P1 | 0.5 U/μL | 500 U |
| Ammonium acetate (pH 5.3) | 10 mM | 1 mL |
| Total | n/a | 1mL |
Storage: Dispense the solution into aliquots after the initial dissolution and store for up to 1 year at –20°C. Avoid repeated freeze and thaw.
[Extraction buffer 1]
| Reagent | Final concentration | Amount |
|---|---|---|
| LMS (1mM) | 1 μM | 50 μL |
| 99.7% methanol | 99.7% | 50 mL |
| Total | n/a | 50 mL |
Storage: Store for up to 6 months at –80°C.
[Extraction buffer 2]
| Reagent | Final concentration | Amount |
|---|---|---|
| LMS (1mM) | 1 μM | 45 μL |
| 99.7% methanol | n/a | 25 mL |
| Ultrapure water | n/a | 10 mL |
| Chloroform | n/a | 10 mL |
| Total | n/a | 50 mL |
Storage: Store for up to 6 months at –80°C.
[Extraction buffer 3]
| Reagent | Final concentration | Amount |
|---|---|---|
| LMS (1mM) | 1 μM | 50 μL |
| 99.7% methanol | n/a | 25 mL |
| Ultrapure water | n/a | 25 mL |
| Total | n/a | 50 mL |
Storage: Store for up to 6 months at –80°C.
[1% TAE-Agarose gel for the extraction of rRNA]
| Reagent | Final concentration | Amount |
|---|---|---|
| Agarose | 1% | 1 g |
| 50× TAE Buffer | 1× | 2 mL |
| ddH2O | n/a | 98 mL |
| Ethidium bromide solution (10 mg/mL) | 300 ng/mL | 3 μL |
| Total | n/a | 100mL |
CRITICAL: Before use, treat all equipment to inactivate RNases, such as by using RNaseZAP.
Storage: Do not store the gel. Use the gel immediately after solifification.
Alternatives: Many popular ethidium bromide alternatives, such as SYBR Gold (Thermo Fischer Scientific) can be used instead.
Step-by-step method details
Timing: 3–4 h for step a
Timing: 1–2 h for step b
Timing: 3–4 h for step c
Timing: 3–24 h for step d
Timing: 2–3 h for step e
Timing: 2–3 h for step f
Timing: 2–3 h for step g
Timing: 3–4 h for step h
Timing: 33 min/sample for step 2
-
1.Sample preparation
-
a.Isolation of small RNAsThis section describes procedures to isolate small RNAs from total RNA for enzymatic digestion and LC-MS/MS analysis.
-
i.Purify total RNA from cells or tissues by using a phenol-containing reagent, such as TRIzol (Thermo Fisher), according to the manufacturer's instruction (https://tools.thermofisher.com/content/sfs/manuals/trizol_reagent.pdf). Use of a reagent similar to TRIzol will isolate cell debris and protein isolated to the acidic phenol-containing layer whereas the aqueous layer will contain the desired total RNA.
-
ii.Resuspend RNA in ultrapure water to a concentration of 1 μg/μL. More than 100 μg RNA is needed to perform all downstream procedures.
-
iii.Use an RNA clean & concentrator kit to separate small (17–200 nt) and large (> 200 nt) RNA species from total RNA, following the manufacturer’s instruction (https://files.zymoresearch.com/protocols/_r1013_r1014_r1015_r1016_rna_clean_concentrator-5.pdf). Briefly, add the appropriate volume of RNA Binding Buffer and ethanol to each sample. Transfer the mixture to a silica-based Zymo-Spin IC Column and centrifuge. Large RNAs are retained in the column and small RNAs are in the flow-through. Mix the flow-through with ethanol and load the mixture to a new Zymo-Spin IC Column and centrifuge. After several cleanup procedures with wash buffer, large and small RNAs can be eluted from the column, respectively.Note: The yield of small RNAs is usually less than one-fourth the yield of large RNAs.Note: Alternatively, other commercially available RNA extraction kit which can isolate small RNAs can be used such as miRNeasy Mini Kit (QIAGEN).
-
iv.Measure the concentration of both small RNAs and large RNAs. If the concentration of small RNAs is more than one-fourth that of large RNAs, it is likely that the small RNAs are contaminated by large RNAs. Proceed to step d i. for digestion. RNA can be stored at −80°C if it is not immediately subjected to digestion.
-
i.
-
b.Isolation of mRNAsThis section describes procedures to isolate mRNA from total RNA for enzymatic digestion and LC-MS/MS analysis.
-
i.Purify polyA+ mRNA from either large RNA or total RNA by using the Oligotex-dT30<Super> mRNA Purification Kit according to the manufacturer’s instruction (https://www.qiagen.com/us/products/discovery-and-translational-research/dna-rna-purification/rna-purification/mrna/oligotex-mrna-kits/). Briefly, mix RNA with Binding Buffer and Oligotex-dT30 magnetic beads. Heat-denature the RNA at 70°C for 3 min and then let stand at room temperature for 10 min to hybridize magnetic beads and mRNA. After centrifugation, remove the supernatant and suspend the magnetic particles with wash buffer, then transfer the mixture to a spin column and centrifuge the column. Repeat the washing step and polyA+ mRNA can be eluted from the column with RNase free dH2O.
-
ii.Measure the concentration of mRNA, which should be approximately 1%–2% of the input concentration. Proceed to step d i. for digestion. RNA can be stored at −80°C if it is not directly subjected to digestion.
 CRITICAL: Do not exceed the maximum capacities of either spin columns or Oligo-dT beads. If excessive RNA is applied, mRNA fraction will be easily contaminated with rRNA and tRNA.
CRITICAL: Do not exceed the maximum capacities of either spin columns or Oligo-dT beads. If excessive RNA is applied, mRNA fraction will be easily contaminated with rRNA and tRNA. CRITICAL: If contamination is suspected, it is recommended to perform agarose gel electrophoresis to check the purity. 1% TAE-agarose gel containing ethidium bromide and RNA denature with formamide can be used (as described below). Purified mRNA fraction can be subject to a second round of purification if contamination is confirmed.
CRITICAL: If contamination is suspected, it is recommended to perform agarose gel electrophoresis to check the purity. 1% TAE-agarose gel containing ethidium bromide and RNA denature with formamide can be used (as described below). Purified mRNA fraction can be subject to a second round of purification if contamination is confirmed.
-
i.
-
c.Isolation of 18s rRNA and 28s rRNAThis section describes procedures to isolate 28S rRNA and 18S rRNA from large RNA or total RNA by agarose gel extraction.
-
i.Prepare 1% TAE-agarose gel. Measure 1 g agarose. Mix agarose powder with 100 mL 1× TAE (2 mL RNAse-free 50× TAE + 98 mL ultrapure water) in a microwavable flask. Microwave for 1–3 min until the agarose is completely dissolved. After the complete melt of agarose, let agarose solution cool down to about 50°C. Add 3 μL ethidium bromide solution (10 mg/mL) and swirl. Pour the agarose solution into an assembled gel tray and cover the tray with saran wrap. Let the agarose gel solidify for about 60 min at room temperature.
-
ii.Mix 10 μL RNase-free formamide with 10 μL total RNA or large RNA (≤5 μg RNA), and denature samples by heating at 60°C for 15 min followed by quick cooling on ice.
-
iii.Add 3 μL of 10× loading buffer composed of 50% glycerol, 0.9% SDS, and 0.05% Bromophenol Blue.
-
iv.Electrophorese the entire sample (23 μL) with voltage 100V for 20 min (until the dye has migrated two-thirds of the length of the gel). RNA molecular size marker such as DynaMarker (BioDynamics Laboratory) can be used to confirm the size of rRNAs.
-
v.Visualize the gel on a UV transilluminator and excise the 28S or 18S rRNA bands using razor blade. Usually RNA shows a 28S rRNA band at 4.5 kb and an 18S rRNA band at 1.9 kb, and small RNA species that possibly contained tRNA and other small RNA species. For a typical photograph of the gel, please refer to the Figure 4G of Ogawa et al. (2021).
-
vi.Extract 28S or 18S rRNAs from the excised gel slices by using the Zymoclean Gel RNA Recovery Kit. When using 5 μg RNA as starting materials, the yield of 28S rRNA should be >900 ng; the yield of 18S rRNA >450 ng. The ratio of 28S rRNA to 18S rRNA should be approximately 2:1. Proceed to step d i. for digestion. RNA can be stored at −80°C if it is not directly subject to digestion.
-
i.
-
d.RNA digestion
-
i.Mix up to 2.5 μg of each RNA sample with 2 μL nuclease P1 solution (0.5 U/μL), 0.5 μL bacteria alkaline phosphatase (BAP), and 2.5 μL of 200 mM HEPES (pH 7.0), and bring the total volume to 25 μL with ultrapure water.
-
ii.Digest RNA for 3 h at 37°C. For 2′-O-methylated nucleosides, which are resistant to RNase (Sproat et al., 1989), prolonged digestion (up to 24 h) can increase the yield. For a prolonged digestion, use a PCR instrument to prevent evaporation.
-
iii.After digestion, the digested samples should be immediately subjected to LC-MS/MS analysis.
-
i.
-
e.Extracellular nucleosides from cultuerd cellsThis section describes extraction of metabolites containing modified nucleosides from supernatant of cell culture.
-
i.Thaw cell culture medium (if frozen) and gently vortex.
-
ii.Transfer 50 μL to a 1.5 mL tube.
-
iii.Add 250 μL of pre-cooled extraction buffer 1 and gently vortex.
-
iv.Rotate the mixture using a rotator for 15 min at 4°C.
-
v.Centrifuge at 15,000 × g for 10 min at 4°C.
-
vi.Transfer 250 μL supernatant to a new 1.5 mL tube.
-
vii.Add 100 μL ultrapure water to the sample and gently vortex.
-
viii.Transfer the mixture into a preconditioned ultrafiltration tube.
-
ix.Centrifuge at 16,000 × g for 1 h at 4°C.
-
x.Transfer the entire ultrafiltrate volume into a new 1.5 mL tube and puncture the lid by using a 20-gauge needle.
-
xi.Evaporate samples by using a SpeedVac Vacuum Concentrator.
-
xii.Dissolve each dried sample in 25 μL ultrapure water.
-
xiii.Centrifuge at 16,000 × g for 10 min at 10°C.
-
xiv.Transfer 20 μL of the supernatant into a new tube suitable for LC-MS/MS measurement.Note: Samples should preferably be subjected to LC-MS/MS analysis immediately after preparation, but samples can be kept frozen below –20°C after evaporation (step xi.). In this case, cover the punctured lid with parafilm to avoid contamination.
-
i.
-
f.Intracellular nucleosides from cultured cellsThis section describes procedures to extract intracellular metabolites containing modified nucleosides from HEK 293A cells plated in 6-well plates at a density of 400,000–500,000 cells per well.
-
i.Aspirate supernatant from cell culture dish.
-
ii.Wash cells with ice-cold 5% mannitol or PBS, and completely aspirate the fluid.
-
iii.Add 500 μL pre-chilled extraction buffer 1 and incubate for 10 min on ice.
-
iv.Transfer the liquid to a 1.5 mL tube and gently vortex.
-
v.Add 400 μL pre-chilled chloroform and vortex.
-
vi.Add 200 μL pre-chilled ultrapure water and vortex.
-
vii.Further mix the tube contents on a rotating mixer for 15 min at 4°C.
-
viii.Centrifuge at 10,000 × g for 5 min at 4°C.
-
ix.Transfer 350 μL of the top layer into a preconditioned ultrafiltration tube.
-
x.Centrifuge at 16,000 × g for 1 h at 4°C.
-
xi.Transfer the entire ultrafiltrate volume into a new 1.5 mL tube and puncture the lid by using a 20-gauge needle.
-
xii.Evaporate samples by using a SpeedVac Vacuum Concentrator.
-
xiii.Dissolve each dried sample in 25 μL ultrapure water.
-
xiv.Centrifuge at 16,000 × g for 10 min at 10°C.
-
xv.Transfer 20 μL of the supernatant into a new tube suitable for LC-MS/MS measurement.Note: See section 4 in “before you begin” for the procedures of preconditioning ultrafiltration device.Note: Samples should preferably be subjected to LC-MS/MS analysis immediately after preparation, but samples can be kept frozen below –20°C after evaporation (step f xii.). In this case, cover the punctured lid with parafilm to avoid contamination.
-
i.
-
g.Nucleosides in mammalian extracellular fluidThis section describes extraction of metabolites containing modified nucleosides from extracellular fluids such as plasma, urine, and aqueous humor.
-
i.Thaw biological fluids (if frozen) and gently vortex.
-
ii.Transfer 25 μL of each sample into a 1.5 mL tube.
-
iii.Add 450 μL pre-chilled extraction buffer 2 (water: methanol: chloroform = 1 : 2.5 : 1) and gently vortex.
-
iv.Mix the tube contents on a rotator for 15 min at 4°C.
-
v.Centrifuge at 16,000 × g for 3 min at 4°C.
-
vi.Transfer 315 μL of the top layer into a new 1.5 mL tube.
-
vii.Add 140 μL ultrapure water to the sample and gently vortex.
-
viii.Centrifuge at 16,000 × g for 3 min at 4°C.
-
ix.Transfer 300 μL supernatant into a preconditioned ultrafiltration tube.
-
x.Centrifuge at 16,000 × g for 1 h at 4°C.
-
xi.Transfer the entire ultrafiltrate volume into a new 1.5 mL tube and puncture the lid by using a 20-gauge needle.
-
xii.Evaporate samples by using a SpeedVac Vacuum Concentrator.
-
xiii.Dissolve each dried sample in 25 μL ultrapure water.
-
xiv.Centrifuge at 16,000 × g for 10 min at 10°C.
-
xv.Transfer 20 μL of the supernatant into a new tube suitable for LC-MS/MS measurement.Note: Samples should preferably be subjected to LC-MS/MS analysis immediately after preparation, but samples can be kept frozen below –20°C after evaporation (step xii.). In this case, cover the punctured lid with parafilm to avoid contamination.
-
i.
-
h.Nucleosides in mammalian tissuesThis section describes extraction of metabolites containing modified nucleosides from tissues.
-
i.Add 1.2 mL pre-chilled extraction buffer 3 (water: methanol = 1 : 1) to each tissue sample (∼ 100 mg) and homogenize in an ice bucket by using a TissueRuptor II with a disposable probe.
-
ii.Transfer entire homogenate into a new 1.5 mL tube and centrifuge at 16,000 × g for 15 min at 4°C.
-
iii.Transfer 500 μL supernatant into a preconditioned ultrafiltration tube.
-
iv.Centrifuge at 16,000 × g for 1 h at 4°C.
-
v.Transfer the entire ultrafiltrate volume into a new 1.5 mL tube and puncture the lid by using a 20-gauge needle.
-
vi.Evaporate samples by using a SpeedVac Vacuum Concentrator.
-
vii.Dissolve each dried sample in 25 μL ultrapure water.
-
viii.Centrifuge at 16,000 × g for 10 min at 10°C.
-
ix.Transfer 20 μL of the supernatant into a new tube suitable for LC-MS/MS measurement.Note: If a sample is too big or too hard to homogenize, chop the sample to reduce the size before adding extraction buffer 3.Note: Samples should preferably be subjected to LC-MS/MS analysis immediately after preparation, but samples can be kept frozen below –20°C after evaporation (step h vi.). In this case, cover the punctured lid with parafilm to avoid contamination.
-
i.
-
a.
-
2.
Measurement of nucleosides by LC-MS/MS
This section describes measurement of modified/unmodified nucleosides after preparation of the samples using LCMS-8050 (Shimadzu) with Inertsil ODS-3 Column (GL science). Obtained data were analyzed using LabSolutions software (Shimadzu).
These parameters have been developed and optimized for the Shimadzu LCMS-8050 mass spectrometer equipped with binary pump, autosampler and electrospray ionization (ESI) using Inertsil ODS-3 Column (reversed phase C18 Column, particle size, 2 μm, I.D., 2.1 mm, length, 150mm, GL science), but these parameters can be adapted for other instruments.-
a.Set up parameters for liquid chromatography.
-
b.Flow rate = 0.4 mL/min
-
c.Mobile phase: Buffer A: 5 mM ammonium acetate (pH 5.3) in water; Buffer B: 60% acetonitrile in water
-
d.The LC time gradient was created using two Nexera LC-30AD solvent delivery unit (Shimadzu) as follows:
Time (min) Buffer B (%) 0 0 10 22.1 15 63.1 17 100 22 100 23 0.6 33 0.6 -
e.Set up the temperature parameters for mass spectrometer:
-
i.Interface temperature = 300°C
-
ii.Desolvation line temperature = 250°C
-
iii.Heat block temperature = 400°C
-
i.
-
f.Nitrogen gas was supplied by an N2 supplier Model T24FD (System Instruments) for nebulization and drying, and argon gas was used for collision-induced dissociation. Nebulizing gas flow is 3.0 L/min, heating gas flow is 10.0 L/min, and drying gas flow is 10.0 L/min.
-
g.Set capillary needle in the interface and needle protrusion to 1.0 mm.
-
h.The MS/MS conditions for each target is optimized using the automated MRM optimization procedure in LabSolutions software (Shimadzu). The MRM transitions as shown in the following table:
Nucleosides Precursor ion (m/z) Product ion (m/z) Retention time Q1 pre-Bias (V) CE Q3 pre-Bias (V) Polarity LMS 182 56 1.078 16 21 24 + D 247 115 1.771 12 9 27 + Y 245 209 1.78 14 10 23 + τm5U 380 124 1.802 19 25 18 - C 244 112 2.168 12 15 22 + m3C 258 126 2.23 11 14 21 + acp3U 346 214 2.371 20 18 24 + m1A 282 150 2.481 10 20 30 + U 245 113 2.716 12 11 20 + m7G 298 166 2.956 12 16 30 + τm5s2U 396 124 3.436 19 25 18 - m3U 259 127 3.445 14 11 26 + m5C 258 126 3.454 11 14 21 + I 269 137 3.654 14 21 28 + Um 259 113 3.706 13 11 21 + Cm 258 112 3.745 13 13 21 + G 284 152 3.835 14 15 30 + f5C 272 140 4.288 12 15 22 + m1I 283 151 4.946 20 18 29 + m1G 298 166 5.001 12 16 30 + Im 283 137 5.027 15 12 25 + Gm 298 152 5.069 13 12 30 + m2G 298 166 5.353 12 16 30 + ac4C 286 154 5.476 15 13 28 + A 268 136 5.512 12 17 24 + m22G 312 180 6.552 11 18 11 + Am 282 136 6.89 15 16 26 + t6A 413 281 6.916 21 15 20 + m6A 282 150 7.855 10 20 30 + ms2t6A 459 327 9.157 25 14 25 + m6Am 296 150 9.73 12 21 29 + m62A 296 164 11.176 12 21 29 + i6A 336 204 14.548 10 15 13 + ms2i6A 382 182 16.939 17 29 12 + -
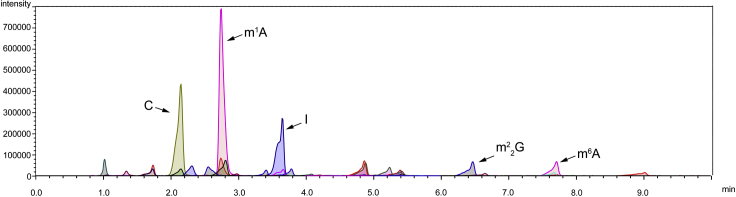
i.Set samples into the rack in autosampler (SIL-30AC, Shimadzu), and measure the nucleoside levels. We recommend running standard nucleoside solution before actual samples for quantification of results and validation of the sensitivity of the mass spectrometer. The representative mass chromatographs of extracellular fluids are shown below. The representative peak area of individual standard modified or unmodified nucleoside is also listed below.
-
a.
Expected outcomes
Modified nucleosides and unmodified nucleosides in the prepared sample are trapped in C18 reverse phase column and eluted from the column at different time points in response to the increase in acetonitrile concentration. Each nucleoside is introduced into the mass spectrometer and detected using the specific MRM parameter. The mass chromatograph of each nucleoside is generated by the software and the area of the peak can be used to calculate the concentration of the −ucleoside.
Here we show the representative peak areas of standard nucleosides (Figures 1, 2, and 3).
| A | m1A | m6A | t6A | ms2t6A | m62A | i6A | ms2i6A | |
|---|---|---|---|---|---|---|---|---|
| 10 nM | 295,277 | 213,584 | 123,372 | 43,402 | 10,679 | 1,026,826 | 170,483 | 94,871 |
| 100 nM | 2,683,129 | 1,908,278 | 1,340,535 | 360,191 | 130,869 | 9,599,842 | 1,685,403 | 1,272,560 |
| m6Am | Am | I | m1I | Im | U | Y | D | |
|---|---|---|---|---|---|---|---|---|
| 10 nM | 835,757 | 277,998 | 70,039 | 270,216 | 65,669 | 1,501 | 6,568 | 24,642 |
| 100 nM | 8,545,363 | 2,651,182 | 765,996 | 2,710,965 | 756,950 | 18,044 | 48,492 | 150,425 |
| m3U | acp3U | Um | G | m1G | m2G | m7G | m22G | |
|---|---|---|---|---|---|---|---|---|
| 10nM | 5,613 | 2,142 | 1,708 | 190,829 | 407,747 | 304,146 | 479,999 | 423,290 |
| 100 nM | 59,429 | 33,564 | 17,355 | 1,275,513 | 3,684,919 | 2,690,115 | 4,507,921 | 3,805,035 |
| Gm | C | m3C | m5C | ac4C | f5C | Cm | |
|---|---|---|---|---|---|---|---|
| 10 nM | 173,000 | 89,806 | 556,717 | 199,492 | 68,001 | 27,231 | 102,212 |
| 100 nM | 1,531,681 | 750,689 | 5,879,203 | 1,599,454 | 577,325 | 233,768 | 905,150 |
Note: These values depend on LC-MS/MS type and manufacture, and on column type and usage status. Optimization is needed for each LC-MS/MS machine.
Figure 1.
Representative LC-MS/MS peaks of human plasma
Figure 2.
Representative LC-MS/MS peaks of human urine
Figure 3.
Representative LC-MS/MS peaks of human aqueous humor
Quantification and statistical analysis
Quantification of modified nucleosides in biological samples can be performed using standard nucleosides with known concentration. Prepare a serial dilution of a standard nucleoside, followed by mass spectrometry analysis. Use the peak areas of the standard nucleosides to generate calibration curve. For any given sample, use the peak area to fit this calibration curve to obtain the concentration. For modified nucleosides in RNA, the unit can be nmol / ng RNA; for modified nucleosides in biological samples, the unit can be nmol / μL. Representative calibration curve of i6A is shown in Figure 4.
Figure 4.
Example of calibration curve for quantitative analysis of i6A
Limitations
This protocol only covers a subset of the RNA modifications that have been identified so far. Matrix effect is one of the main concerns in quantitative LC-MS/MS analysis, especially when using biological fluid samples (such as serum and urine). It is strongly recommended to use stable isotope-labeled nucleosides as internal standards for reliable measurement and for comparing samples.
This protocol was developed to screen all of the modified nucleosides that are listed above. If only a few of these nucleosides are being studied, then optimize the extraction and detection conditions (which are determined by each nucleoside’s polarity) accordingly.
Troubleshooting
Problem 1
RNA yield is low (steps 1-a, 1-b, and 1-c).
Potential solution
Low yield is caused either by overloading the column (which can clog the column or prevent optimal RNA binding) or by RNase contamination. Confirm the binding capacity of the column before use. Also use nuclease-free grade tubes and tips. Wear masks while handling RNA and use an RNase-cleaning agent. RNA contents vary between tissues and cell types. If you cannot improve the yield by addressing these concerns, then increase the amount of starting material to obtain more RNA.
Problem 2
Evaporation process takes too much time (steps 1-e-xi, 1-f-xii, 1-g-xii, and 1-h-vi).
Potential solution
The primary cause of this problem is the residual glycerin of ultrafiltration membrane. Confirm the preconditioning steps are done right before the extraction process. Extend preconditioning centrifugation time from 30 min to 60 min.
Problem 3
Results are not stable when the sample is repeatedly measured (step 2-i).
Potential solution
This problem can be caused by HPLC malfunction, such as the abnormal sample intake or leakage. Prepare a vial containing 50 μL ultrapure water and weigh the vial using an analytic balance. Perform a test run by injecting 5 μL water and weigh the vial again. If HPLC works properly, a weight reduction of 5 mg is expected. If not, then contact the technical service of the manufacturer. Alternatively, check the pressure endurance of the HPLC following the instruction. When the leakage is suspected, then contact the technical service and replace the appropriate parts.
Problem 4
Intensities of peaks are lower than expected (step 2-i).
Potential solution
This is the most frequent problem. Check the peak areas or height of the standard nucleosides. If the results of standard nucleosides are lower than expected, then check mass spectrometer. Usually, accumulation of sample remaining in the mass spectrometer can cause the decrease in peak intensity. From our experience working with Shimazu LCMS-8050, replacing Orifice (Part # S225-15479) or ESI Capillary (Part # S225-14948-91) will often restore the intensity.
Problem 5
Retention time of nucleosides is different (step 2-i).
Potential solution
The primary cause of this problem is the inappropriate pH of buffer A. Check and adjust the pH to 5.3. Alternatively, a shift of retention time can be caused by the composition (or matrix) of a sample. For example, samples prepared from serum or urine contain thousands of molecules in addition to nucleosides, whereas samples prepared from purified RNA basically only contain nucleosides. The difference of the sample composition can potentially cause the change of retention time. Nucleoside with an early retention time, such as uridine and m1A, tends to be easily affected when compared to nucleosides with a late retention time. If samples with different origins are subject to measurement, it is highly recommended not to mix the samples. For example, if an experimenter has 3 blood samples and 3 digested RNA samples, run blood samples and RNA samples separately. It is also recommended to run 2–3 dummy blood samples or RNA samples before the sample of interest.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Akiko Ogawa (accogawa@tohoku.ac.jp).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We are grateful for suggestions from all members of the Department of Molecular Physiology, Kumamoto University, and the Department of Modomics Biology & Medicine, Institute of Development, Aging and Cancer (IDAC), Tohoku University. We thank Y. Takahata and H. Miyamoto for technical assistance. This work was supported by JSPS KAKENHI grants 18H02599 (F.-Y.W.), 18K19521 (F.-Y.W.), 20H05309 (F.-Y.W.), and 20K18371 (A.O.), and by grants from the Japan Science and Technology Agency (JST), SAKIGAKE JPMJPR1532 (F.-Y.W.), Exploratory Research for Advanced Technology (ERATO) JPMJER2002 (F.-Y.W.), Fusion Oriented Research for disruptive Science and Technology (FOREST) JPMJFR205Y (F.-Y.W.), the Takeda Science Foundation (F.-Y.W.), the Uehara Memorial Foundation (F.-Y.W.), the Astellas Foundation for Research on Metabolic Disorders (A.O.), and the Japan Medical Women’s Association Foundation (A.O.).
Author contributions
A.O. performed experiments and analysis used in this study. F.-Y.W. developed the protocol. A.O. and F.-Y.W. wrote and revised this manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Akiko Ogawa, Email: accogawa@tohoku.ac.jp.
Fan-Yan Wei, Email: fanyan.wei.d3@tohoku.ac.jp.
Data and code availability
The published article includes all generated or analyzed during this study.
References
- Ogawa A., Nagiri C., Shihoya W., Inoue A., Kawakami K., Hiratsuka S., Aoki J., Ito Y., Suzuki T., Suzuki T. N6-methyladnenosine (m6A) is an endogenous A3 adenosine receptor ligand. Mol. Cell. 2021;81:1–16. doi: 10.1016/j.molcel.2020.12.038. [DOI] [PubMed] [Google Scholar]
- Sproat B.S., Lamond A.I., Beijer B., Neuner P., Ryder U. Highly efficient chemical synthesis of 2'-O-methyloligoribonucleotides and tetrabiotinylated derivatives; novel probes that are resistant to degradation by RNA or DNA specific nucleases. Nucleic Acids Res. 1989;17:3373–3386. doi: 10.1093/nar/17.9.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article includes all generated or analyzed during this study.