Abstract
CF II, a factor required for cleavage of the 3′ ends of mRNA precursor in Saccharomyces cerevisiae, has been shown to contain four polypeptides. The three largest subunits, Cft1/Yhh1, Cft2/Ydh1, and Brr5/Ysh1, are homologs of the three largest subunits of mammalian cleavage-polyadenylation specificity factor (CPSF), an activity needed for both cleavage and poly(A) addition. In this report, we show by protein sequencing and immunoreactivity that the fourth subunit of CF II is Pta1, an essential 90-kDa protein originally implicated in tRNA splicing. Yth1, the yeast homolog of the CPSF 30-kDa subunit, is not detected in this complex. Extracts prepared from pta1 mutant strains are impaired in the cleavage and the poly(A) addition of both GAL7 and CYC1 substrates and exhibit little processing activity even after prolonged incubation. However, activity is efficiently rescued by the addition of purified CF II to the defective extracts. Extract from a strain with a mutation in the CF IA subunit Rna14 also restored processing, but extract from a brr5-1 strain did not. The amounts of Pta1 and other CF II subunits are reduced in pta1 strains, suggesting that levels of the subunits may be coordinately regulated. Coimmunoprecipitation experiments indicate that the CF II in extract can be found in a stable complex containing Pap1, CF II, and the Fip1 and Yth1 subunits of polyadenylation factor I. While purified CF II does not appear to retain the association with these other factors, this larger complex may be the form recruited onto pre-mRNA in vivo. The involvement of Pta1 in both steps of mRNA 3′-end formation supports the conclusion that CF II is the functional homolog of CPSF.
The posttranscriptional acquisition of a poly(A) tail on the 3′ ends of eukaryotic mRNAs is an essential process which promotes transcription termination (7) and transport of the mRNA from the nucleus (14). The poly(A) tail is also important for optimal translation and for regulating mRNA stability (10, 32, 36, 41). Polyadenylation requires two events—site-specific endonucleolytic cleavage of primary transcripts followed by poly(A) addition to the upstream fragment. While these two steps are closely coupled in vivo, they can be experimentally uncoupled in vitro and assayed separately, allowing biochemical characterization of the protein components required for each individual step. Such studies have revealed a remarkable conservation in the factors needed for polyadenylation in mammals and in the yeast Saccharomyces cerevisiae, in spite of differences in the sequence and organization of RNA signals which specify this processing event (for recent reviews, see references 25, 40, and 42).
However, differences have been found in the composition of the various factors which make up the basic polyadenylation machinery of these organisms. Some proteins appear to be unique to one or the other system. For example, in yeast, three factors (CF IA, CF IB, and CF II) are sufficient for accurate cleavage of precursor. CF IA consists of four polypeptides: Rna14, Rna15, Pcf11, and p50 (19, 33). CF IB is the Hrp1 protein (18). Rna14 and Rna15 are thought to be homologs of the p77 and p64 subunits of the mammalian CstF cleavage factor (39). On the other hand, counterparts to Pcf11 and Hrp1 have not yet been found in the mammalian system.
We recently described the purification of the yeast CF II by use of its ability to reconstitute the cleavage reaction in the presence of CF IA and CF IB (43). It contains four polypeptides, Cft1/Yhh1, Cft2/Ydh1, Brr5/Ysh1, and a 90-kDa protein whose identity was not determined. The three known proteins are homologs to the three largest subunits of the mammalian cleavage-polyadenylation specificity factor (CPSF), an activity required for both cleavage and poly(A) addition. In yeast, the poly(A) addition step needs CF IA, CF IB, polyadenylation factor I (PF I), and the yeast poly(A) polymerase, Pap1 (6, 19). In these studies, CF II activity was not required for poly(A) addition, raising the possibility that it might not be a strict functional analog of CPSF. However, extracts depleted of Brr5/Ysh1 or Cft1/Yhh1 by immunoprecipitation were defective for both cleavage and poly(A) addition (5, 38). Resolution of this apparent difference has come with the recent purification of a multiprotein complex from yeast containing PF I activity (33). This complex contained Pap1, Fip1 (a protein which interacts directly with Pap1), Yth1 (a yeast homolog of the CPSF 30-kDa subunit), Pta1 (an essential gene affecting pre-tRNA splicing), several uncharacterized proteins, and, surprisingly, the Cft1/Yhh1, Cft2/Ydh1, and Brr5/Ysh1 subunits of CF II. A mutation in Pta1 was also shown to affect the poly(A) addition step in vitro (33).
The requirement of specific components of CF II in cleavage and/or poly(A) addition of yeast mRNA precursor can be fully addressed only once an active factor composed of recombinant subunits is available. Until that time, it is important to thoroughly probe the functions of these subunits through the analysis of in vitro systems derived from wild-type and mutant cells. In this report, we use this approach to characterize new properties of Pta1, i.e., its copurification with the CF II cleavage factor, its previously unsuspected contribution to the cleavage step, and the requirement of Pta1 for accumulation of complex containing the other CF II subunits.
MATERIALS AND METHODS
Yeast strains, media, and genetic techniques.
Yeast strains and their relevant genotypes were FY41 (MATa ura3-52 leu2-Δ1 trp1Δ63 his4-917δ; F. Winston, Harvard Medical School, Boston, Mass.), FY1284 (MATa pta1-2 ura3-52 ade8 his4-917δ; F. Winston), POC8-23d (MATa pta1-1 ade2-1 leu2-Δ1 lys2 trp1-Δ101 ura3-52 [31]), LM113 (MATa ran14-1 his3-11,15 ade2-1 ura3-1 [26]), YSN399 (brr5-1 MATα ura3-52 his3-Δ200 ade2-1000 leu2-Δ1 lys2-801a TRP1 [30]), and LM96 (fip1-1 ade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 [34]).
Cell culturing and extract preparation.
S. cerevisiae strains were cultured in YPD (1% yeast extract, 2% peptone, 2% glucose) supplemented with ampicillin (50 mg/liter) at 30°C to reach an optical density at 600 nm of 1.0 to 1.5. Whole-cell extracts were prepared by a modification of the method previously described by Kessler et al. (19). The cultured cells were harvested by centrifugation at 5,000 × g at 25°C for 15 min, weighed, and resuspended at 5 ml/g of cells in buffer A (1 M sorbitol, 50 mM Tris-HCl [pH 7.8], 10 mM MgCl2, 30 mM β-mercaptoethanol). Usually, 3 to 4 g of cells was obtained from 1 liter of culture. The cell suspensions were rotated at 30°C for 30 min and collected by centrifugation at 5,000 × g for 10 min at 4°C. The cell pellets were weighed and resuspended at 2 ml/g of cells in buffer B (10 mM HEPES-KOH [pH 7.0], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, 0.6 μM leupeptin, and 2 μM pepstatin A). The cells were disrupted with an equal volume of glass beads (0.5-mm diameter) by four cycles of 30 s of agitation with 1 min of cooling on ice between cycles. The extracts were then adjusted to 0.2 M KCl, rotated gently for 30 min at 4°C, and cleared by centrifugation at 35,000 × g for 30 min at 4°C. Additional insoluble material was removed by centrifugation at 225,000 × g for 30 min at 4°C. Solid ammonium sulfate was then added to 40% saturation and stirred for 30 min at 4°C. Precipitated proteins were collected by centrifugation at 15,000 × g for 20 min; resuspended in 300 μl of buffer C (20 mM Tris-HCl [pH 7.90], 0.2 mM EDTA, 10% glycerol, 0.5 mM DTT) with 50 mM KCl, 1 mM phenylmethylsulfonyl fluoride, 0.6 μM leupeptin, and 2 μM pepstatin A; and dialyzed against the same buffer (1 liter with one change) for 2 h.
mRNA substrates.
Capped, 32P-labeled mRNAs used as substrates in the processing assays were prepared from the following plasmids by in vitro transcription of linearized DNAs as described by Chen and Moore (6). Full-length GAL7-1 RNA containing the GAL7 poly(A) site and flanking sequences was prepared from pJCGAL7-1 (6). Precleaved substrate GAL7-9 lacking sequences downstream of the poly(A) site was prepared from pJCGAL7-9 (6). CYC1 pre-mRNA was transcribed from pGYC1 (4). All precursor RNAs were purified from 5% acrylamide–8.3 M urea gels (37), precipitated twice with ethanol, and stored frozen at −20°C in 50 mM Tris-HCl, pH 7.0.
Protein isolation and analysis.
CF II was purified from yeast crude whole-cell extracts as described previously (43). The CF II samples used for assays in this study were from the poly(A)-Sepharose step of the published purification protocol (43). The CF I fraction from the heparin-Sepharose step was obtained as described elsewhere (19).
The CF II-containing sample from the poly(A)-Sepharose step was separated on a sodium dodecyl sulfate (SDS)-polyacrylamide gel, and the gel was stained with Coomassie brilliant blue. The 90-kDa protein band was excised from the gel and submitted for microsequencing at the Harvard University Microchemistry Facility (Cambridge, Mass.), by using collisionally activated dissociation on a Finnigan TSO 7000 triple quadruple mass spectrometer.
SDS-polyacrylamide gels were prepared and run according to the method of Laemmli (21). Silver staining was performed according to the method of Gottlieb and Chavko (12) with the Silver Staining Plus kit from Bio-Rad.
Antibodies, Western blotting, and immunoprecipitations.
Polyclonal antibodies against Cft2 were produced from mice immunized with recombinant protein made in Escherichia coli, by a method described by Kessler et al. (19). Rabbit antisera to Cft1 (38), Brr5/Ysh1 (16), and Fip1 (34) were gifts from the respective authors. The monoclonal antibody directed against Pap1 was prepared as tissue culture supernatant (20). For Pta1 antibody, the EcoRI-XhoI fragment of PTA1 was cloned into the same sites in pET23c (Novagen). Recombinant protein was made in the BL21(DE3) strain of E. coli and purified with nickel agarose according to standard protocols from Novagen. Hybridomas secreting monoclonal antibodies directed against Pta1 were produced by immunizing BALB/c mice with this fragment of Pta1. Ascites fluid was made by intraperitoneal injection of hybridoma cell lines into pristane-treated BALB/c mice by standard techniques. The Yth1 polyclonal antibody was produced in mice as described elsewhere (19) with a recombinant glutathione S-transferase–Yth1 fusion protein (Pharmacia), which had been expressed and purified according to the manufacturer’s specifications.
Immunoblotting assays were done according to standard procedures (37). Monoclonal antibodies specific for Pap1 and Pta1 were used at dilutions of 1:50 and 1:1,000, respectively, in phosphate-buffered saline (PBS) with 2% bovine serum albumin. Polyclonal antibodies were diluted 1:1,000 in the above buffer.
For coimmunoprecipitation experiments, 20 μl of protein A-agarose beads (Gibco) was equilibrated in PBS buffer, mixed with 1 to 2 μl of polyclonal antibodies against Fip1 or Cft1 in a final volume of 40 μl or with 50 μl of monoclonal anti-Pap1 antibody, and incubated at room temperature for 2 h to allow antibodies to bind to beads. The beads were then washed once with 1 ml of PBS and twice with 1 ml of 0.2 M sodium borate, pH 9.0. Antibodies were cross-linked to the beads by adding 0.2 ml of 20 mM dimethylsuberimidate (Sigma) in 0.2 M sodium borate, pH 9.0, followed by incubation at room temperature for 30 min (13). The coupling reaction was stopped by washing the beads with 1 ml of 0.2 M ethanolamine (pH 8.0) and incubation of the beads in the same solution for 2 h at room temperature. The beads were then washed once with immunoprecipitation (IP) buffer (20 mM Tris-HCl [pH 7.9], 150 mM KCl, and 0.1% NP-40) followed by washing in sequence with the following solutions to remove uncoupled antibodies (once with 20 mM Tris-HCl [pH 7.9]–150 mM KCl–2 M NaCl and then with 1 ml of 50 mM glycine [pH 3.0]–500 mM NaCl and finally equilibrated with IP buffer). In some experiments, the Pap1 antibody was coupled to protein A-Sepharose beads by using anti-immunoglobulin G antibody as a bridge.
Whole-cell extract (10 μl, ∼50 μg of protein) or fractions containing CF II [30 μl, ∼20 μg of protein from the Q-Sepharose fraction, or 100 μl, ∼3 μg of protein from the poly(A)-Sepharose fraction] were preadsorbed to 200 μl of a 15% protein A–Sepharose slurry in IP buffer to remove proteins that bind to the affinity resin nonspecifically. After incubation on a roller at 4°C for 30 min, the beads were pelleted and the supernatant was recovered. Two hundred microliters of the preadsorbed sample was combined with the antibody beads, and the mixture was incubated for 2 h at 4°C. The supernatant was removed, and the pellet was washed three times with TBS buffer (20 mM Tris-HCl [pH 7.9], 150 mM KCl). Proteins present in the pellet were eluted from the beads by being boiled in 2× sample buffer (21), separated by electrophoresis on 8% polyacrylamide gels containing SDS, and either directly stained with silver or transferred to a polyvinylidene difluoride membrane for immunoblotting.
3′-end processing assays.
Processing assays were done as described elsewhere (6, 19, 43). Reaction mixtures were assembled on ice in a volume of 12 μl containing 1 mM magnesium acetate, 75 mM potassium acetate, 2% polyethylene glycol 8000 (Fisher), 2 mM ATP, 10 nM radioactive GAL7 RNA or CYC1 (8 ng), 1.5 μM (0.6 μg) tRNA, 1 mM DTT, 0.4 U of RNasin (Promega), and 0.1 mg of bovine serum albumin per ml, 1 to 2 μl of CF I or CF II sample, or whole-cell extracts (∼15 μg of protein). Reaction mixtures were incubated at 30°C for 20 to 30 min, reactions were stopped with proteinase K and SDS as described elsewhere (6), reaction mixtures were diluted to 30 μl with 50 mM Tris-HCl (pH 7.0) and extracted once with phenol-chloroform-isoamyl alcohol (25:24:1), and 1/10 of the reaction mixture was separated by electrophoresis on a 5% acrylamide–8.3 M urea gel and visualized by autoradiography.
RESULTS
Pta1 is a component of CF II.
We have recently purified and characterized CF II from yeast (43). A 5,300-fold purification of CF II indicated that four polypeptides of 150, 105, 100, and 90 kDa copurify with CF II activity. The 150-kDa protein was recognized by antibodies against Cft1/Yhh1, the yeast homolog of the 160-kDa subunit of the mammalian CPSF (38). Protein sequencing showed that the 105-kDa protein, designated Cft2/Ydh1 (cleavage factor 2), exhibits significant homology to the CPSF 100-kDa subunit (15) and that the 100-kDa subunit of CF II was identical to Brr5/Ysh1 (5, 15), a yeast protein with striking similarity to the 73-kDa subunit of CPSF.
To identify the smallest subunit of CF II, the band at 90 kDa was excised from an SDS-polyacrylamide gel stained with Coomassie blue and subjected to microsequencing. The sequence derived from one of the tryptic peptides of the 90-kDa protein was QLSALLSTLGVSTKT. A search of the yeast protein database revealed that this peptide is encoded by the essential gene PTA1. PTA1 was initially defined by a conditional growth mutation, pta1-1, that affects pre-tRNA processing in vivo (31).
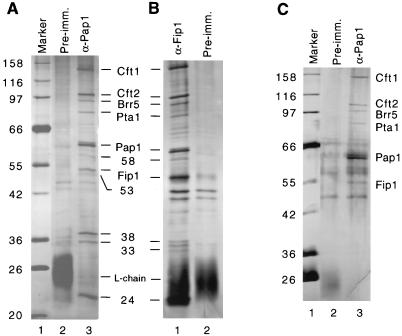
To further confirm the composition of the CF II complex, CF II-containing fractions were subjected to immunoprecipitation with antibodies against Cft1/Yhh1. These samples were obtained from a Q-Sepharose fractionation, an early step during purification (Fig. 1A, lane 3), or poly(A)-Sepharose chromatography, a late step during purification (Fig. 1A, lane 4). Analysis of the immunoprecipitated proteins on an SDS-polyacrylamide gel stained with silver shows four prominent bands which are not found when a control resin coupled to preimmune serum is used (Fig. 1A, lanes 2, 5, and 6). The molecular weights of these proteins exactly match those reported for our previous purification of CF II (43). Two of the immunoprecipitated proteins were recognized by a mixture of anti-Brr5/Ysh1 and anti-Pta1 antibodies, respectively (Fig. 1B, lane 3), and the same two proteins are also present in the purified CF II eluted from a Superose 6 column, the final step of purification (Fig. 1B, lane 2). The anti-Pta1 and anti-Ysh1/Brr5 antibodies do not cross-react with their respective antigens when tested individually against recombinant Brr5/Ysh1 and Pta1 (data not shown). In agreement with the biochemical purification of CF II, the coimmunoprecipitation results show that CF II is a stable complex of four proteins and further establish that the previously uncharacterized 90-kDa subunit is Pta1.
FIG. 1.
Pta1 is tightly associated with the CF II complex. (A) Fractions containing CF II from Q-Sepharose (QS) and poly(A)-Sepharose (PA) fractionations were subjected to immunoprecipitation with anti-Cft1 antibodies. The immunoprecipitated proteins were resolved on an SDS–8% polyacrylamide gel, and the gel was stained with silver. Four polypeptides, with sizes corresponding to those of CF II subunits, were coprecipitated (lanes 5 and 6). Lanes 3 and 4 are the unbound proteins from the supernatant. Lane 1 contains molecular mass standards (in kilodaltons), and lane 2 is an immunoprecipitation of the Q-Sepharose fraction with preimmune serum. The positions of the light and heavy chains of the immunoglobulins in lanes 2, 5, and 6 are indicated. (B) Immunoblot analysis of the CF II fraction from the Superose 6 step of CF II purification (lane 2) and proteins immunoprecipitated from the Q-Sepharose fraction with anti-Cft1 antibodies (lane 3). Blots were stained with a mixture of Brr5/Ysh1 polyclonal antibodies and a monoclonal antibody against Pta1.
CF II directly associates with a Pap1-PF I complex.
Three subunits of CF II are homologous to subunits of the mammalian CPSF. CPSF-160 (the 160-kDa subunit of CPSF) contacts poly(A) polymerase directly (29). To determine whether yeast CF II interacts with Pap1, we examined the proteins in extracts which coimmunoprecipitate with a monoclonal antibody against a C-terminal epitope in Pap1 (20). In addition to Pap1, the precipitate contains all four subunits of CF II, Fip1, and several other proteins (p58, p53, p38, p33, and p24) which are not found when preimmune serum is used (Fig. 2A, lanes 2 and 3). The identities of Pap1, Fip1, and the CF II subunits were confirmed by Western blotting (data not shown). An immunoprecipitation experiment with anti-Fip1 antibodies brought down a set of proteins with a pattern almost identical to that from the immunoprecipitation with anti-Pap1 antibody (Fig. 2B, lane 1).
FIG. 2.
CF II interacts with Pap1 and Fip1. Whole-yeast-cell extract was used for coimmunoprecipitation. Samples were analyzed by electrophoresis on an SDS–8% polyacrylamide gel and stained with silver. (A) Immunoprecipitation assay with antibody against Pap1 (lane 3) or with preimmune serum (lane 2). (B) Immunoprecipitation with antibodies against Fip1 brought down the same set of proteins as shown in panel A (lane 1). Treatment with preimmune serum is shown in lane 2. (C) Immunoprecipitation assay with antibody against Pap1, followed by a more stringent wash step. All CF II subunits were retained along with Fip1 and Pap1 (lane 3). Numbers to the left of panels A and C indicate molecular masses in kilodaltons.
The composition of the Pap1 and Fip1 immunoprecipitates is similar to that of a multiprotein complex obtained in a recent purification of PF I activity (33). This complex contains, in addition to Pap1 and Fip1, the entire CF II complex, Yth1 (the yeast homolog of the mammalian CPSF 30-kDa subunit), and two uncharacterized proteins, Pfs1 and Pfs2 (17, 33). The size of the 24-kDa polypeptide in the precipitate from our experiment corresponds to that of Yth1, and the sizes of 58- and 53-kDa polypeptides are close to those of Pfs1 and Pfs2. Two unknown proteins of 35 and 36 kDa also copurify with the PF I complex and may be related to the 33- and 38-kDa polypeptides which coimmunoprecipitate with Pap1 and Fip1 in our study. If the anti-Pap1 immunoprecipitate is washed more stringently with TBS buffer plus 0.1% NP-40, the three smallest peptides are removed (Fig. 2C, lane 3). The retention of Fip1 and the CF II subunits under these conditions indicates that these proteins are more strongly associated with Pap1 than are the smaller ones.
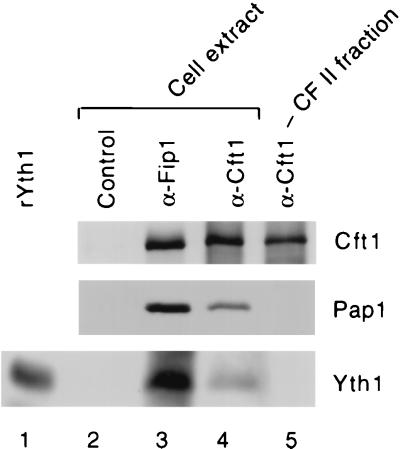
The Yth1 homolog, CPSF-30, is not always found in active preparations of CPSF (11, 28). We have not observed Yth1 in purified CF II (43), a result consistent with the fact that a mutation in Yth1 caused a defect in poly(A) addition but not cleavage (2). However, Yth1 is known to stain poorly with silver (2), and we cannot rule out the possibility that it is present in substoichiometric amounts. To further investigate this issue, we probed various immunoprecipitates with antibody against Yth1. By Western blotting, Yth1 is readily detected as a protein immunoprecipitated from extract with Fip1 antibodies (Fig. 3, lane 3), in agreement with the direct physical interaction of these two proteins (2). As expected given the results shown in Fig. 2, Yth1 and Pap1 are present when extract is treated with Cft1 antibodies (Fig. 3, lane 4). However, neither it nor Pap1 is observed when a CF II fraction is immunoprecipitated with the same antibody (Fig. 3, lane 4). Equivalent amounts of Cft1 are found in the Cft1 precipitates from extract or CF II. These results suggest that Yth1 may not interact directly or strongly with any of the CF II subunits and is likely not to be essential for precursor cleavage in yeast.
FIG. 3.
Yth1 is not detected in Cft1 immunoprecipitates of CF II. Cell extract (lanes 2 to 4) or a CF II phosphocellulose fraction (lane 5) was immunoprecipitated with the indicated antibodies, and blots of the immunoprecipitates were probed with antibody against Cft1, Pap1, or Yth1. Recombinant Yth1 produced in E. coli is shown in lane 1, and an immunoprecipitation with preimmune serum is shown in lane 2.
The levels of Pta1 and other CF II subunits are reduced in strains containing pta1 mutations.
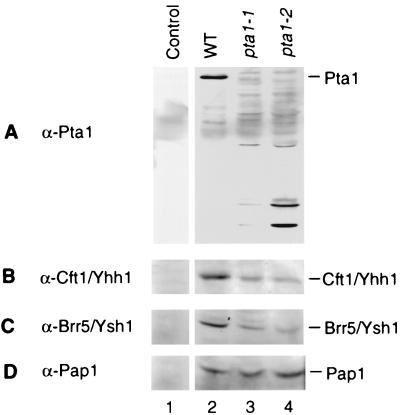
Two pta1 conditional mutants, the pta1-1 and pta1-2 strains, were employed to further characterize Pta1. Both of the mutant strains show slow growth at 30°C and lack of growth at 37°C (22, 31). To determine the level of the Pta1 protein in these mutant cells, a direct immunoblot analysis of cell extracts was carried out with monoclonal antibody against Pta1. The extracts were prepared from cells cultured at 30°C. Pta1 protein of normal size could not be detected in either of the pta1 mutant extracts. Instead, a set of several shorter peptides were stained by the Pta1 antibody in both pta1-1 and pta1-2 mutant extracts (Fig. 4A, lanes 3 and 4). These species are probably degraded fragments or short translation products. O’Connor and Peebles (31) observed that both the temperature-sensitive growth defect and the tRNA splicing defect of the pta1-1 strain could be suppressed by the ochre suppressor tRNA gene SUP11, indicating that a termination codon had been created within the protein reading frame. Because PTA1 is an essential gene, the pta1-1 allele cannot be a null mutation. Thus, a pta1-1 ochre fragment is adequate for Pta1 function, or translational read-through of the ochre stop codon might produce sufficient full-length protein for viability.
FIG. 4.
Immunoblot analysis of cell extract from wild-type and pta1 mutant strains. Equal amounts of protein (15 μg) from wild-type (lanes 1 and 2), pta1-1 (lane 3), or pta1-2 (lane 4) extracts were separated on an SDS–8% polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and immunoblotted with a monoclonal antibody against Pta1 (A), Cft1/Yhh1 (B), Brr5/Ysh1 (C), or Pap1 (D). Lane 1 is a control blot probed with preimmune antiserum from a mouse subsequently immunized with Pap1 antigen.
The immunoblot assay was also used to analyze the presence of other subunits of CF II and Pap1 in the yeast extracts. The amount of Cft1 and Brr5/Ysh1 was reduced in pta1 mutants in comparison with that from wild-type extract (Fig. 4B and C, lanes 2 to 4), while the amount of Pap1 protein remained constant in the wild type and the two pta1 mutants (Fig. 4D, lanes 2 to 4).
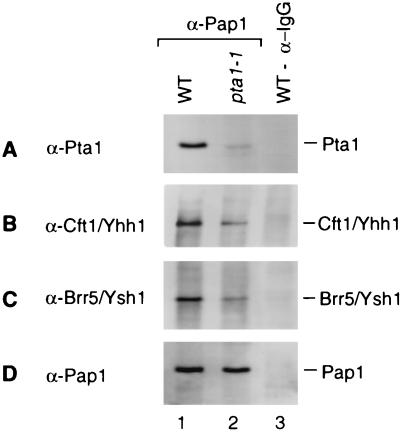
To further investigate whether the pta1 mutation influenced the assembly of CF II complex and the interaction with Pap1, we immunoprecipitated Pap1 from extracts of wild-type and mutant pta1-1 strains. Precipitated proteins were resolved by SDS-polyacrylamide gel electrophoresis and immunoblotted with antibodies against components of CF II. Interestingly, a small amount of full-length Pta1 protein was now detectable in the precipitate from the pta1-1 mutant (Fig. 5A, lane 2). Analysis of the cultures used for extract preparation did not reveal cells which could now grow at the nonpermissive temperature, as might be expected if reversion or suppression of the pta1 alleles had occurred. Instead, the full-length Pta1 was more likely the result of a low-level translational read-through of the mutated pta1 transcripts. The amount of other components of CF II in the Pap1 immunoprecipitate was also reduced in comparison with that from the wild-type strain (Fig. 5B and C, lanes 1 and 2), while the amount of precipitated Pap1 protein from the pta1-1 mutant was similar to that from wild type (Fig. 5D, lanes 1 and 2), consistent with the relative levels of these proteins in the respective extracts. The short fragments of Pta1 were not found in immunoprecipitates (data not shown), suggesting that only full-length Pta1 can be assembled into complex.
FIG. 5.
Full-length Pta1 protein is found in a Pap1-containing complex. An equal amount of extract from wild-type (WT) or pta1-1 mutant cells was subjected to immunoprecipitation with monoclonal antibody against Pap1. The immunoprecipitated proteins were fractionated on an SDS–8% polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and immunostained with antibody against Pta1 (A), Cft1/Yhh1 (B), Brr5/Ysh1 (C), or Pap1 (D). Lane 1, wild-type extract; lane 2, pta1-1 mutant extract; lane 3, wild-type extract immunoprecipitated with anti-immunoglobulin G (IgG) antibody.
Pta1 is required for both cleavage and poly(A) addition.
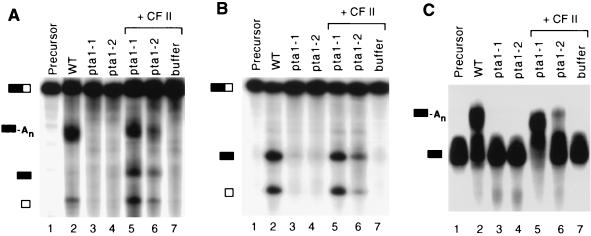
Pta1 has been shown elsewhere to be necessary for efficient poly(A) addition (33). Because of the presence of Pta1 in CF II, we wanted to investigate its role in cleavage. We tested extracts from the two pta1 temperature-sensitive mutant strains, the pta1-1 and pta1-2 strains, for processing of full-length GAL7 pre-mRNA. Extracts from both pta1 mutant strains failed to process the GAL7 substrate (Fig. 6A, lane 2). In comparison, extract from wild-type cells cleaved the full-length RNA substrate and polyadenylated the upstream cleavage product (Fig. 6A, lane 2). These reactions were performed for 20 min, and under these conditions, the maximal amount of processing in wild-type extracts occurred by 40 min. In the pta1-1 mutant extracts, there was no accumulation of product even after 60 min of incubation (data not shown), indicating that the defect was not simply due to a lower rate of processing. Both cleavage and poly(A) addition could be restored to the pta1 extracts by addition of CF II from a poly(A)-Sepharose column, the penultimate step in the published purification protocol (43). The activity of pta1-1 extract was recovered more efficiently than that of pta1-2 extract (Fig. 6A, lanes 5 and 6). CF II alone exhibited no activity (Fig. 6A, lane 7). The rescue by CF II indicates that the non-CF II activities needed for 3′-end processing are not affected by the pta1 mutations. Furthermore, it is consistent with Pta1 being an important subunit of CF II.
FIG. 6.
Pta1 is required for both cleavage and poly(A) addition of GAL7 pre-mRNA substrate. Equal amounts of protein were mixed with 32P-labeled GAL7 substrate and incubated at 30°C for 20 min as described in Materials and Methods. Products were resolved on denaturing polyacrylamide gels and visualized by autoradiography. (A) Processing assay with full-length GAL7-1 RNA substrate and ATP. (B) Cleavage assay in which ATP is replaced with 2′-dATP to block the poly(A) addition step. (C) Poly(A) addition assay with precleaved RNA substrate GAL7-9. Lanes 1, precursor; lanes 2, wild-type (WT) extract; lanes 3, pta1-1 mutant extract; lanes 4, pta1-2 mutant extract; lanes 5, pta1-1 extract supplemented with CF II; lanes 6, pta1-2 extract supplemented with CF II; lanes 7, reaction with CF II only. The positions of the full-length GAL7-1 precursor RNA, poly(A)+ RNA, upstream and downstream cleavage products, and precleaved GAL7-9 precursor (top to bottom, respectively) are indicated by the bars on the left of each panel.
Cleavage and poly(A) addition assays were then carried out separately with the mutant extracts, in order to examine both steps of the reaction independently. For the cleavage assay, the poly(A) addition step was blocked by replacement of ATP with 2′-dATP. Both pta1 mutant extracts were defective for cleavage (Fig. 6B, lanes 3 and 4). The cleavage in pta1-1 extract was almost fully restored by addition of CF II, and in pta1-2 extract, it was partially recovered (Fig. 6B, lanes 5 and 6). The poly(A) addition assay was carried out with a precleaved RNA substrate, GAL7-9. As in the case of cleavage, both mutant extracts failed to polyadenylate the substrate RNA (Fig. 6C, lanes 3 and 4), and the poly(A) addition could be recovered by adding CF II (Fig. 6C, lanes 5 and 6). Again, the poly(A) addition activity with GAL7-9 substrate was recovered more efficiently in pta1-1 extract than in pta1-2 extract. We do not know the nature of the pta1-2 mutation, but the higher level of short Pta1 fragments in the pta1-2 mutant than in the pta1-1 mutant (Fig. 4) may have an inhibitory effect.
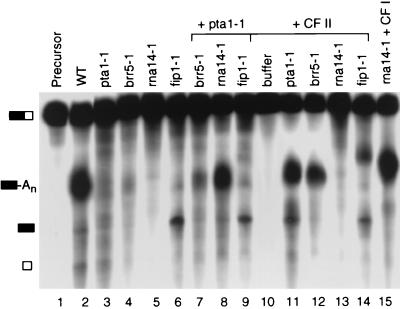
To examine the functional interaction of Pta1 with other proteins involved in 3′-end processing, extracts from strains containing conditional mutations in CF II, CF IA, or PF I subunits were prepared and tested for processing activity and the ability to complement the defect of the pta1-1 extract. Rna14, a subunit of CF IA, is required for both cleavage and poly(A) addition (26). Extract from an rna14-1 strain is defective for processing of CYC1 pre-mRNA (26). Consistent with this finding, extract from the rna14-1 strain was unable to process GAL7-1 pre-mRNA (Fig. 7, lane 5), but processing could be restored by the addition of a CF I-containing fraction (Fig. 7, lane 15). The fip1-1 extract is defective in the poly(A) addition of CYC1 substrate (33). Similarly, we found that this extract could cleave the GAL7 precursor but failed to polyadenylate the upstream cleavage product (Fig. 7, lane 6), a result expected for a subunit of PF I. Although Brr5/Ysh1 was first identified as a subunit of PF I (16), it has also been found to be a component of CF II (43). The brr5-1 extract shows a very low level of activity, as indicated by a small amount of the polyadenylated product, but with no accumulation of cleavage product (Fig. 7, lane 4), indicating that Brr5 is also involved in cleavage.
FIG. 7.
Complementation of polyadenylation activity in the pta1-1 mutant extract. Reactions were performed as described in the legend to Fig. 6, and reaction mixtures were incubated at 30°C for 30 min. 32P-labeled GAL7-1 RNA was incubated with extracts of wild-type (WT) (lane 2), pta1-1 (lane 3), brr5-1 (lane 4), rna14-1 (lane 5), and fip1-1 (lane 6) strains. Lanes 7 to 9 are reaction mixtures containing pta1-1 extracts combined with other mutant extracts as indicated. Lanes 10 to 14 are reaction mixtures containing different mutant extracts or buffer as indicated and supplemented with CF II. The reaction mixture of rna14-1 extract in the presence of a CF I fraction (from the heparin-Sepharose step) is shown in lane 15. See the Fig. 6 legend for symbol definitions.
The defective processing of the pta1-1 extract could be enhanced significantly by addition of rna14-1 mutant extract but not by addition of brr5-1 and fip1-1 extracts (Fig. 7, lanes 3 and 7 to 9), supporting the idea that Pta1 is associated with Brr5/Ysh1 and Fip1 in complexes required for processing of mRNA precursor. Addition of CF II efficiently restored the processing activities of pta1-1 and brr5-1 mutant extracts (Fig. 7, lanes 11 and 12), confirming that these two proteins are in the CF II complex. A small amount of product with a longer poly(A) tail is visible in the reaction with the fip1-1 mutant extract which has been supplemented with CF II (Fig. 7, lane 14). This could be due to a low level of PF I activity in this CF II fraction or could reflect the possibility that the levels of CF II are limiting in the fip1-1 extract and that increasing the amount of CF II can partially suppress the fip1-1 defect.
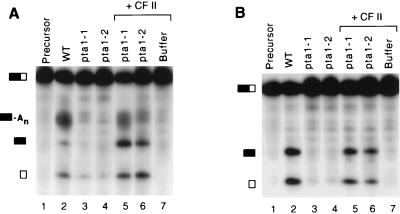
In a previous study, extract from the pta1-1 mutant was found to be normal in cleavage and defective in poly(A) addition with CYC1 pre-mRNA (33). To determine if the defects that we were observing were substrate specific, we also tested pta1 mutants with full-length CYC1 RNA as substrate. Similar to the results found with the GAL7 pre-mRNA, extracts from pta1 mutants failed to process CYC1 RNA in complete reactions containing ATP (Fig. 8A, lanes 3 and 4) or cleavage-only reactions in which ATP was replaced by 2′-dATP (Fig. 8B, lanes 3 and 4). For CYC1, a low level of cleavage in mutant extracts may be evident from the appearance of a small amount of RNA migrating at the position of the downstream cleavage product (Fig. 8, lanes 3 and 4). The processing activities can be restored to pta1 mutant extracts when they are supplemented with CF II (Fig. 8, lanes 5 and 6). In contrast to the GAL7 substrate, the processing of CYC1 precursor in both mutant extracts was recovered almost equally efficiently. In coupled cleavage-poly(A) addition assays with either substrate, there is often more accumulation of cleaved products and less polyadenylated RNA in pta1 mutant extracts complemented with CF II than in wild-type extract (Fig. 6A and 8A, lanes 2, 5, and 6), suggesting that cleavage activity is recovered more efficiently than is poly(A) addition. Taken together, these results strongly argue that Pta1 is required for both cleavage and poly(A) addition in yeast mRNA 3′-end formation.
FIG. 8.
Pta1 is required for both cleavage and poly(A) addition of CYC1 pre-mRNA substrate. Reactions were carried out as described in the legend to Fig. 6. (A) Coupled cleavage-polyadenylation assays in the presence of ATP. (B) Cleavage assays with 2′-dATP. 32P-labeled CYC1 substrates were combined with extracts of wild-type (WT) (lane 2), pta1-1 (lane 3), and pta1-2 (lane 4) strains and pta1-1 and pta1-2 strains plus CF II (lanes 5 and 6, respectively). In lane 6, a reaction was performed with CF II only. See the Fig. 6 legend for symbol definitions.
DISCUSSION
In this report, we demonstrate that Pta1 is the previously uncharacterized 90-kDa subunit of the CF II cleavage factor needed for mRNA 3′-end formation in the yeast S. cerevisiae. This protein had been found in the most highly purified preparations of CF II, in which only four polypeptide bands were visible when the preparation was assayed on an SDS-polyacrylamide gel stained with silver (43). Consistent with this chromatographic behavior, we also show that Pta1 and the other three CF II subunits of CF II are immunoprecipitated from CF II-containing fractions with antibodies against Cft1/Yhh1. This result confirms that CF II is a tightly associated complex of Cft1/Yhh1, Cft2/Ydh1, Brr5/Ysh1, and Pta1.
CF II subunits also copurify with PF I (33), an activity necessary for poly(A) addition but not for cleavage of yeast precursor (6). In support of this finding, our coimmunoprecipitation experiments indicate that the CF II in processing extract can be found in a stable complex containing Pap1, CF II, and the Fip1 and Yth1 subunits of PF I. Purified CF II does not appear to retain the association with these other factors and is sufficient for cleavage activity. The fact that unpolyadenylated product often accumulates in reactions containing ATP suggests that two forms of CF II (free and associated with PF I-specific subunits) may also exist in extracts, and it is not yet clear whether these factors work together in vivo only as a preassembled complex. In any event, the interactions which recruit CF II to a CF II-PF I-Pap1 complex are not known. CPSF-160, the mammalian homolog of Cft1/Yhh1, has been shown elsewhere to bind poly(A) polymerase (29). However, in yeast, this function may have been assumed by Fip1, a protein without a mammalian homolog but with some similarity to CPSF-160 in sequence (29). Like CPSF-160, Fip1 interacts directly with the poly(A) polymerase (34, 44) and inhibits the enzyme’s activity (29, 44). A physical interaction has also been demonstrated between Fip1 and the CF IA subunit Rna14 (34), as has been shown elsewhere for CPSF-160 and the Rna14 homolog CstF-77 (29).
When the PTA1 gene is mutated and the production of full-length Pta1 protein is highly reduced, the amount of other CF II subunits recruited into a complex with Pap1 is decreased. Thus, it is possible that Pta1 facilitates the assembly of this complex. However, the total amounts of other CF II components in a pta1-1 mutant extract are also reduced, whereas the amount of Pap1 is not affected. These observations suggest that free CF II subunits or ones assembled into a partial complex without Pta1 may not be stable. Alternatively, the reduced Pta1 production or the short Pta1 fragments may inhibit the synthesis of other CF II components. In either case, Pta1 seems necessary for the accumulation of CF II. A previous study showed that the amount of the CF IA subunit Rna15 was severely reduced not only in an rna15 mutant strain but also in rna14 and pap1 mutant cells (1), a finding in agreement with the idea that levels of some of the proteins involved in 3′-end processing may be coordinately regulated in yeast.
The interactions described above indicate a role for CF II subunits in both cleavage and polyadenylation. Support for Pta1 function in 3′-end processing is provided by the in vitro analysis of cell extracts from pta1 conditional mutants. In our experiments, extracts from the pta1-1 and pta1-2 strains are defective in the cleavage and the poly(A) addition of at least two pre-mRNA substrates, GAL7 and CYC1. This defect can be restored by addition of CF II. Our data differs from that of Preker et al. (33), which showed that a pta1-1 mutant extract failed to polyadenylate CYC1 substrate but still gave normal cleavage. We observed no processing in pta1 mutant extracts, and cleavage product did not accumulate even after prolonged incubation. If poly(A) addition is more sensitive to limiting amounts of CF II than is the cleavage step, the discrepancy in these two studies could be accounted for by differences in extract preparation or culture conditions which might affect the concentration of full-length Pta1 in the mutant extract and thus the quantity of assembled CF II.
We have also found that the extract from the brr5-1 mutant is impaired in both cleavage and poly(A) addition, similar to what we found for the pta1-1 mutant. Consistent with Brr5/Ysh1 and Pta1 being part of the same complex, the brr5-1 and pta1-1 extracts could not complement each other, but their processing activities could be restored by addition of rna14-1 extract or the CF II fraction. Chanfreau et al. (5) have shown that extracts from this brr5-1 mutant strain were defective in poly(A) addition but not in cleavage, while wild-type extracts immunodepleted of Brr5/Ysh1 exhibited loss of cleavage activity as well. In a different study, extracts depleted of this protein by transcriptional repression of the gene were impaired in both cleavage and poly(A) addition, though more severely affected for poly(A) addition (16). Depletion of extract with antibodies to Cft1/Yhh1 abolished both cleavage and poly(A) addition (38). Restoration of cleavage required only the addition of the CF II-containing fraction, while poly(A) addition needed the further addition of Pap1 and a fraction containing PF I (38). While a functional analysis of Cft2/Ydh2 has not been performed, its presence in highly purified CF II strongly supports the idea that it participates with the other three CF II subunits in 3′-end formation.
The three largest subunits of yeast CF II are homologous to ones in mammalian CPSF, a complex needed for both the cleavage and the poly(A) addition phases of the reaction. One of the functions of CPSF is to recognize the AAUAAA signal element in the pre-mRNA, and an analogous role for CF II is suggested by the finding that the interaction of CF II with RNA substrate depends on the UAUAUA type of polyadenylation signal (43). The yeast CF I factor was originally proposed as the functional homolog of mammalian CPSF in that, like CPSF, it was required for cleavage and polyadenylation (19), even though some of its subunits bear a resemblance to subunits of the mammalian cleavage factor CstF. It has now been shown that even CstF can behave as a poly(A) addition factor, if the pre-mRNA has a CstF binding site upstream of the AAUAAA sequence (27). However, our current work and the studies described above indicate that CF II is the genuine yeast counterpart of mammalian CPSF, with homology in both function and sequence. Like CPSF, it plays a central role in yeast pre-mRNA 3′-end processing, recognizing critical cis-acting signals and interacting with other factors to facilitate cleavage and poly(A) addition.
A mammalian homolog of the fourth subunit of CF II, Pta1, has not been found in the known cleavage-polyadenylation factors. A mammalian homolog may have evolved to have a greater role in other cell processes and less of a role in mRNA 3′-end formation, as perhaps suggested by the observations that mutations in Pta1 also lead to defects in the splicing of pre-tRNA or cause synthetic lethality in combination with a disruption of the gene encoding Spt3 (22), a polymerase II (Pol II) transcription factor which interacts with the TATA-binding protein (TBP) (9). The cause of the tRNA splicing defect in yeast is not known. However, in this regard, it is intriguing that a genetic interaction has been discovered between PAP1 and RET1, which encodes the second largest subunit of Pol III (3), and that a mutation in a tRNA synthetase gene has been shown to affect transcription termination downstream of yeast poly(A) sites (23).
The genetic linkage of PTA1 and SPT3 is also very interesting, in light of much recent evidence supporting the coupling of Pol II transcription and mRNA 3′-end formation (for a review, see references 25, 40, and 42). Two findings are particularly relevant to the Pta1-Spt3 interaction. First, CPSF, the mammalian homolog of yeast CF II, is recruited to the mammalian preinitiation complex by TFIID, which is composed of TBP and TBP-associated factors (8). Second, Pta1 has been shown to associate specifically with the phosphorylated carboxyl-terminal domain of Pol II, and mutations in Kin28, the TFIIH-associated C-terminal domain kinase, result in reduced levels of Pta1 in the cell (35). Further work is needed to define the exact role of Pta1 in yeast mRNA 3′-end processing and explore other cellular functions which might be provided by this protein.
ACKNOWLEDGMENTS
We thank J. Madson and F. Winston for the pta1-2 strain; L. Minvielle-Sebastia, P. J. Preker, and W. Keller for the rna14-1 and fip1-1 strains; and S. Noble and C. Guthrie for the brr5-1 strain. We also are grateful to A. Zhelkovsky for critically reading the manuscript.
This work was supported by U.S. Public Health Service grant RO1 GM41752 from the National Institutes of Health to C. Moore.
REFERENCES
- 1.Amrani N, Dufour M E, Bonneaud N, Lacroute F. Mutations in STS1 suppress the defect in 3′ mRNA processing caused by the rna15-2 mutation in Saccharomyces cerevisiae. Mol Gen Genet. 1996;252:552–562. doi: 10.1007/BF02172401. [DOI] [PubMed] [Google Scholar]
- 2.Barabino S M, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 3.Briggs M W, Butler J S. RNA polymerase III defects suppress a conditional-lethal poly(A) polymerase mutation in Saccharomyces cerevisiae. Genetics. 1996;143:1149–1161. doi: 10.1093/genetics/143.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler J S, Platt T. RNA processing generates the mature 3′ ends of yeast CYC1 mRNA in vitro. Science. 1988;242:1270–1274. doi: 10.1126/science.2848317. [DOI] [PubMed] [Google Scholar]
- 5.Chanfreau G, Noble S M, Guthrie C. Essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF) Science. 1996;274:1511–1514. doi: 10.1126/science.274.5292.1511. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Moore C L. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol Cell Biol. 1992;12:3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colgan D, Manley J. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 8.Dantonel J C, Murthy K G, Manley J L, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 9.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 10.Ford L P, Bagga P S, Wilusz J. The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol Cell Biol. 1997;17:398–406. doi: 10.1128/mcb.17.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmartin G M, Fleming E S, Oetjen J, Graveley B R. CPSF recognition of an HIV-1 mRNA 3′-end processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 12.Gottlieb M, Chavko M. Silver staining of native and denatured eucaryotic DNA in agarose gels. Anal Biochem. 1981;165:33–37. doi: 10.1016/0003-2697(87)90197-7. [DOI] [PubMed] [Google Scholar]
- 13.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 14.Huang Y, Carmichael G. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenny A, Hauri H-P, Keller W. Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol Cell Biol. 1994;14:8183–8190. doi: 10.1128/mcb.14.12.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenny A, Minvielle-Sebastia L, Preker P, Keller W. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science. 1996;274:1514–1517. doi: 10.1126/science.274.5292.1514. [DOI] [PubMed] [Google Scholar]
- 17.Keller W, Minvielle-Sebastia L. A comparison of mammalian and yeast pre-mRNA 3′ end processing. Curr Opin Cell Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- 18.Kessler M, Henry M, Gross S, Shen E, Zhao J, Silver P, Moore C. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kessler M M, Zhao J, Moore C L. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. J Biol Chem. 1996;271:27167–27175. doi: 10.1074/jbc.271.43.27167. [DOI] [PubMed] [Google Scholar]
- 20.Kessler M M, Zhelkovsky A M, Skvorak A, Moore C L. Monoclonal antibodies to yeast poly(A) polymerase (PAP) provide evidence for association of PAP with cleavage factor I. Biochemistry. 1995;34:1750–1759. doi: 10.1021/bi00005a032. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Madison J, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magrath C, Hyman L. A mutation in Grs1, a glycine tRNA synthetase, affects mRNA 3′ end formation in S. cerevisiae. Genetics. 1999;152:129–141. doi: 10.1093/genetics/152.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minvielle-Sebastia L, Beyer K, Krecic A, Hector R E, Swanson M S, Keller W. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 1998;17:7454–7468. doi: 10.1093/emboj/17.24.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minvielle-Sebastia L, Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- 26.Minvielle-Sebastia L, Preker P J, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 27.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley J L, Proudfoot N J. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murthy K G, Manley J L. Characterization of the multisubunit cleavage-polyadenylation specificity factor. J Biol Chem. 1992;267:14804–14811. [PubMed] [Google Scholar]
- 29.Murthy K G K, Manley J L. The 160 kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′ end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 30.Noble S M, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Connor J, Peebles C. PTA1, an essential gene of Saccharomyces cerevisiae affecting pre-tRNA processing. Mol Cell Biol. 1992;12:3843–3856. doi: 10.1128/mcb.12.9.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preiss T, Hentze M W. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 33.Preker P, Ohnacker M, Minvielle-Sebastia L, Keller W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preker P J, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez, C. R., E.-J. Cho, M. C. Keogh, C. L. Moore, A. L. Greenleaf, and S. Buratowski. Kin28, the TFIIH-associated CTD kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 36.Sachs A, Sarnow P, Hentze M. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Stumpf G, Domdey H. Dependence of yeast pre-mRNA 3′-end processing on Cft1: a sequence homolog of the mammalian AAUAAA binding factor. Science. 1996;274:1517–1520. doi: 10.1126/science.274.5292.1517. [DOI] [PubMed] [Google Scholar]
- 39.Takagaki Y, Manley J L. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature. 1994;372:471–474. doi: 10.1038/372471a0. [DOI] [PubMed] [Google Scholar]
- 40.Wahle E, Ruegsegger U. 3′-end processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 41.Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 42.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao J, Kessler M, Moore C. Cleavage factor II of S. cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J Biol Chem. 1997;272:10831–10838. doi: 10.1074/jbc.272.16.10831. [DOI] [PubMed] [Google Scholar]
- 44.Zhelkovsky A, Helmling S, Moore C. Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol Cell Biol. 1998;18:5942–5951. doi: 10.1128/mcb.18.10.5942. [DOI] [PMC free article] [PubMed] [Google Scholar]