Abstract
The piggyBac transposon system provides a non-viral alternative for cost-efficient and simple chimeric antigen receptor (CAR) T cell production. The generation of clinical-grade CAR T cells requires strict adherence to current good manufacturing practice (cGMP) standards. Unfortunately, the high costs of commonly used lentiviral or retroviral vectors limit the manufacturing of clinical-grade CAR T cells in many non-commercial academic institutions. Here, we present a manufacturing platform for highly efficient generation of CD19-specific CAR T cells (CAR19 T cells) based on co-electroporation of linear DNA transposon and mRNA encoding the piggyBac transposase. The transposon is prepared enzymatically in vitro by PCR and contains the CAR transgene flanked by piggyBac 3′ and 5′ arms. The mRNA is similarly prepared via in vitro transcription. CAR19 T cells are expanded in the combination of cytokines interleukin (IL)-4, IL-7, and IL-21 to prevent terminal differentiation of CAR T cells. The accurate control of vector copy number (VCN) is achieved by decreasing the concentration of the transposon DNA, and the procedure yields up to 1 × 108 CAR19 T cells per one electroporation of 1 × 107 peripheral blood mononuclear cells (PBMCs) after 21 days of in vitro culture. Produced cells contain >60% CAR+ cells with VCN < 3. In summary, the described manufacturing platform enables a straightforward cGMP certification, since the transposon and transposase are produced abiotically in vitro via enzymatic synthesis. It is suitable for the cost-effective production of highly experimental, early-phase CAR T cell products.
Keywords: chimeric antigenic receptor, CD19, piggyBac transposon, electroporation, T cells
Graphical abstract

CD19-specific chimeric antigenic receptor-modified T cells were generated by electroporation of linear piggyBac transposon DNA prepared in vitro via PCR and mRNA encoding the piggyBac transposase prepared by in vitro transcription. Generated CAR-T cells contain >60% CAR+ cells with <3 copies of transposon per cell.
Introduction
Chimeric antigen receptors (CARs) are artificial proteins that can redirect the specificity of T cells to any surface antigen. One of these antigens is the B cell antigen CD19, a well-validated target of already commercially available CD19-specific CAR (CAR19) T cells approved against refractory B cell malignancies. The CAR consists of a single-chain antibody segment (scFv) that defines the specificity, a short linker sequence, a transmembrane domain, and an intracellular signaling domain. Upon recognizing the antigen, the CAR initiates a T cell activation and expansion similarly as an endogenous T cell receptor and subsequent elimination of tumor cells.1 The development of novel and experimental CAR-T cell therapies requires a cost-effective and rapid production chain.2 The most complicated step in the manufacture of good manufacturing practice (GMP)-grade CAR T cells is the transfection of T cells, in a majority of cases performed via recombinant lentiviral or retroviral vectors (LV/RV). To meet the current quality standards, these viral vectors must be similarly produced at GMP quality, which is expensive and very slow.3 Several alternative non-viral approaches were thus developed, including transposons such as piggyBac (PB) or Sleeping Beauty (SB) or genome editing via CRISPR-Cas9.3, 4, 5 Transposons are usually used as two plasmids; the first one encodes the CAR19 transgene, and the second one encodes the PB transposase. Both plasmids are electroporated into T cells; the transposase excises the transposon from the first plasmid and randomly integrates it into the genome.6 Notably, the transposase can be introduced into T cells as mRNA to reduce the risk of random integration of the second plasmid.7 Several clinical trials demonstrated the efficiency of both PB and SB transposons for generating CAR T cells and validated this approach as safe and feasible.8 Other non-viral gene engineering techniques such as CRISPR-Cas9-based DNA editing enable transgenesis into a precise DNA locus within the genome. However, this unique feature is impaired by a highly increased risk of chromosomal translocations resulting from the off-target effect of the CRISPR-Cas9 enzyme complex.5 In contrast, PB transposon integrates its cargo randomly within the genome without producing such severe genomic mutations.9
It will be of value to develop a universal and straightforward T cell transfection technology to overcome such barriers. Here, we show that abiogenically (i.e., by in vitro enzymatic synthesis) prepared PB transposon/transposase can be used in this manner to manufacture CD19-specific CAR T cells efficiently. The transposon was prepared by preparative PCR as a linear dsDNA containing the CAR19 transgene flanked with PB 3′ and 5′ arms. The PB transposase is encoded by mRNA and was prepared by in vitro transcription. According to the already established protocol, the transposon/transposase were co-electroporated into T cells, followed by in vitro expansion.10 By titrating down the amount of transposon DNA, it was possible to efficiently produce >60% CAR+ T cells containing less than three copies of CAR19 transgene per transduced cell. Generated CAR19 T cells were of early memory immunophenotype and displayed efficient effector functions. This manufacturing platform reduces the complexity of currently used viral-based gene transfection techniques and supports novel immunotherapeutic product development and rapid clinical testing.
Results
CAR19 T cells can be generated with enzymatically in vitro prepared linear DNA transposon and mRNA transposase
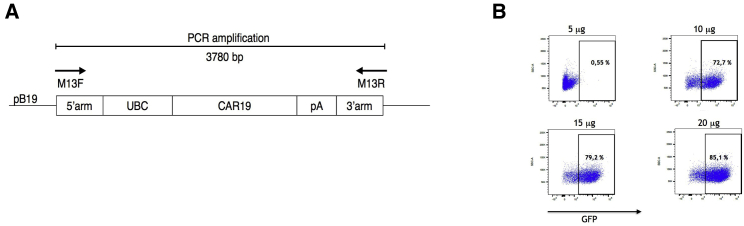
A second-generation CAR19 construct previously generated by us11þ was used as a template for the amplification of the transposon by preparative PCR (Figure 1A). The CAR19 construct contains anti-CD19 scFv derived from hybridoma B-D3 and has the following structure: CD8 leader sequence, anti-CD19 scFv, CD8 hinge domain, CD8 transmembrane domain, 4-1BB intracellular domain, TCR zeta intracellular domain; its full sequence is provided in Materials and methods. The manufacturing of the transposon DNA was performed commercially as described in Materials and methods, the purity of DNA was >99%, and there were no mutations within the coding sequence. Next, to manufacture the transposase mRNA, a plasmid pST containing a hyperactive PB transposase (hyPBase) under the T7 promoter was used as a template for the mRNA production by in vitro transcription. As a control reagent for initial experiments, an mRNA encoding GFP was similarly produced. First, we determined the electroporation efficiency of mRNA in T cells under conditions (voltage and pulse length) optimized for the electroporation of DNA by electroporating increasing amounts of GFP mRNA. The results in Figure 1B show that an amount between 10 and 15 μg of mRNA per electroporation should transduce a sufficient number of cells. Based on this estimate, we decided always to use 12 μg of transposase mRNA per one electroporation of 1 × 107 peripheral blood mononuclear cells (PBMCs) in a volume of 100 μL, since our goal was to transduce a sufficient number of cells with a lesser amount of transposase to ensure that hyPBase remains active intracellularly only for a short time.
Figure 1.
The design of linear DNA transposon encoding the CAR19 and optimization of mRNA electroporation
(A) The image shows the scheme of the DNA transposon. M13F and M13R primers were used to generate amplicon 3,780 bp in length by PCR as described in Materials and methods. (B) To determine the efficiency of electroporation with various amounts of mRNA, the PBMCs were electroporated with the increasing amounts of mRNA encoding EGFP. The number in each dot plot indicates the percentage of transduced (GFP+) PBMCs after 3 days. One representative experiment is shown.
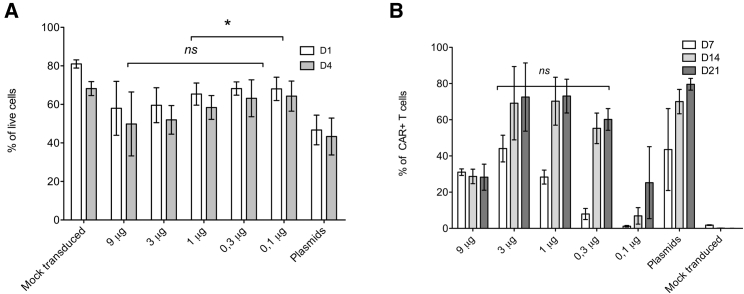
To determine the optimal concentration of the transposon, PBMCs were electroporated with decreasing amounts of DNA (from 9 to 0.1 μg/100 μL). The transfection efficiency was compared to the procedure based on the electroporation of plasmids encoding the transposon/transposase. The data in Figure 2A show that the viability at day 1 and day 4 post electroporation was slightly higher upon electroporation of PCR CAR19 + mRNA hyPBase than the control PBMCs electroporated with plasmids (4 μg transposon plasmid + 2 μg transposase plasmid). As expected, the transfection efficiency depended on the DNA concentration (Figure 2B). The percentage of CAR+ T cells was initially low at day 7 post electroporation, but at later time points (day 14, day 21), we observed an increase in CAR+ cells reaching ∼60%–70%. The efficient generation of CAR19 T cells was possible in the range of 3–0.3 μg of DNA per one (100 μL) electroporation. A further decrease below 0.3 μg/100 μL resulted in a substantial reduction of the percentage of transfected cells.
Figure 2.
The transduction efficiency of PBMCs with PCR DNA/mRNA
(A) Viability after electroporation with decreasing amounts of linear transposon or after electroporation with transposon/transposase plasmids. (B) Percentage of CAR19 T cells 7, 14, and 21 days post-electroporation, n = 4. ∗p < 0.005. ns, Not significant.
Low VCN can be achieved by transfection with minimal amount of transposon DNA using both PCR DNA/mRNA and plasmid/plasmid approach
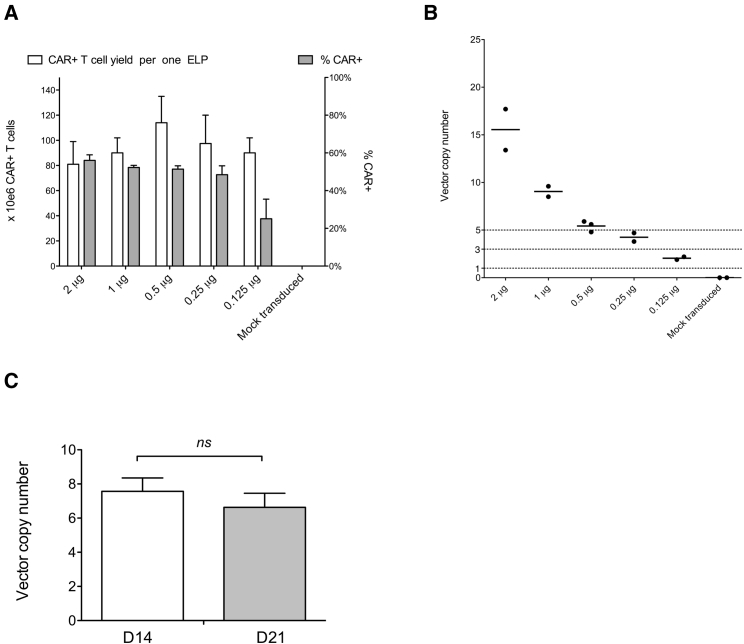
The vector copy number (VCN) is a critical parameter for the production of clinical-grade CAR T cells. Thus, we quantified the number of integrated CAR19 transposons (i.e., VCN) per cell by digital PCR (ddPCR) and corrected the value to the percentage of CAR+ cells within the sample, which was determined by fluorescence-activated cell sorting (FACS). The data in Figure 3A show that the VCN of CAR19 transposon correlated with the decreasing amount of electroporated PCR CAR19 (the amount of mRNA was constant for all samples). Importantly, the generation of CAR19 T cells with as low as 1–3 copies of transgene was possible by reducing the PCR CAR19 concentration to 0.3 μg/100 μL. In contrast, control electroporation with a high amount of transposon/transposase plasmids (4 μg CAR19 + 2 μg PBASE) produced CAR19 T cells with approximately VCN = 23.
Figure 3.
PCR DNA/mRNA approach enables efficient generation of CAR19 T cells with low VCN
(A) The integration efficiency of the transposon was determined by quantifying the number of integrated CAR19 transgenes per transduced T cell by digital PCR. The graph shows the correlation of VCN with the amount of electroporated transposon. The control group included CAR19 T cells prepared via a plasmid approach and mock-transduced T cells. The bar indicates the mean value. (B) The efficiency of generation of CAR19 T cells with decreasing amounts of transposon was determined by calculating the yield of produced CAR19 T cells 7, 14, and 21 days post-electroporation and was compared to the CAR19 T cells generated via plasmids. n = 4. ns, Not significant.
Next, we determined the maximal number of CAR19 T cells generated by this protocol by large-scale cultivations in G-Rex bottles (Figure 3B). Electroporation of DNA in the range 3–0.3 μg, which yields CAR T cells with an acceptable VCN (<10), produced equal numbers of CAR T cells compared to electroporation of plasmids. At day 14 the CAR19 T cell yield was ∼1 × 107 CAR+ cells per one electroporation, and at day 21 the CAR-T yield was ∼1 × 108 CAR+ cells per one electroporation. Control PBMCs electroporated with the transposon DNA without transposase mRNA did not expand in vitro and therefore were not further analyzed (data not shown). These data demonstrate that such a transposon dilution technique enables accurate titration of transposon DNA and reliably generates high numbers of CD19-specific CAR T cells with VCN < 5.
Similarly to the manufacturing of CAR19 T cells via PCR-made transposon DNA, we also used this transposon dilution technique to generate CAR19 T cells via electroporation of transposon/transposase plasmids. PBMCs were electroporated with a constant amount of transposase plasmid (1 μg) plus decreasing amounts of CAR19 transposon plasmids. The transfection and CAR19 T cell yield efficiency decreased with the reduced concentration of transposon plasmid, similarly to the PCR DNA/mRNA approach (Figure 4A). The optimal amount of transposon plasmid that enabled an efficient generation of CAR19 T cells with approximately VCN = 2-5 was in the range of 0.125–0.25 μg DNA per electroporation (Figure 4B). Interestingly, the comparison of the plasmid/plasmid approach with the PCR DNA/mRNA approach (Figure 3A versus Figure 4B) showed that the transfection with 1 μg of plasmid transposon produced CAR19 T cells with ∼9 copies of transgene per cell and the transfection with PCR DNA/mRNA produced CAR19 T cells with ∼3 copies of transgene per cell, while the overall yield of cultivation was similar under both conditions. This finding suggests that the transposition via the plasmid/plasmid approach is more efficient. Next, to determine any possible changes of VCN during the in vitro expansion, CAR-T cells were generated using 1 μg of transposon/transposase plasmids per electroporation and VCN was measured on day 14 and at the end (day 21) of in vitro expansion. The results presented in (Figure 4C) show that VCN did not significantly change between day 14 and day 21, since ∼7 versus 6 copies per cell were detected.
Figure 4.
Generation of CAR19 T cells with low copies of the transgene using plasmid DNA
(A) PBMCs were electroporated with decreasing amounts of PB19 plasmids, shown as μg per one electroporation, while the amount of the transposase plasmid was the same in all groups (1 μg). The values on the left y-axis indicate the number of generated CAR+T cells per one electroporation after 21 days of in vitro expansion. The values on the right y-axis indicate the percentage of transduced T cells at the same time. n = 2. (B) The graphs show the CAR transgene copy number in the same cell cultures. ns, Not significant. (C) To determine the changes of CAR transgene copies over time, the quantification was performed 14 and 21 days post-electroporation. n = 3. ns, Not significant.
Functional assays of CAR T cells and immunophenotype of CAR19 T cells
To examine the effector functions of CAR19 T cells engineered with PCR DNA/mRNA, their cytotoxic activity and interferon-gamma (IFN-γ) production was measured after antigenic challenge with CD19+ B cell line Ramos and compared to CAR19 T cells generated via DNA plasmids. The amount of transposon used for transfection was the lowest concentration that effectively produced CAR19 T cells with VCN < 5, i.e., 0.3 μg PCR DNA/12 μg mRNA and 0.25 μg transposon plasmid/1 μg transposase plasmid. Both methods produced CAR19 T cells with similar effector functions, as the cytotoxicity (Figure 5A) and the production of IFN-γ (Figure 5B) were not significantly different. Next, we determined the immunophenotype of generated CAR19 T cells via flow cytometry. The representative dot plots in (Figure 5D) demonstrate that both methods produced CAR T cells with very similar immunophenotype, characterized by the high level of expression of early memory antigens CD62L, CD27, and CD28 and absence of expression of the exhaustion marker PD-1. The graph in (Figure 5C) shows the immunophenotype of all donors analyzed. In summary, we found no significant differences in immunophenotype or the cytotoxic activity of CAR19 T cells prepared by these two approaches.
Figure 5.
Generated CAR19 T cells are functional and express early memory surface antigens
The graph shows the cytotoxicity of CAR19 T cells generated with PCR DNA/mRNA or with plasmids against target Ramos cells at ratios of 1:1 and 3:1; black bars show the cytotoxicity of control non-transduced T cells. n = 4. (B) The production of IFN-γ was measured after restimulation of CAR19 T cells with Ramos cells at 1:1 ratio after 24 h. n = 4. ns, Not significant. (C) The immunophenotype of generated CAR19 T cells was determined by multiparameter flow cytometry. The graphs show the percentages of individual T cell memory subsets among CAR+ T cells. n = 5. ns, Not significant. (D) The dotplots show the expression of antigens CD4, CD8, CD45RA, CD62L, CD27, CD28, and PD-1 on either CD4+ or CD8+ CAR19 T cells. One representative matched sample out of five donors is shown.
Discussion
The manufacturing of clinical-grade CAR T cells via transfection by LV/RV is a complicated multi-step process with high costs. Additionally, difficulties in obtaining commercially produced GMP-grade viral vectors within a reasonable time window create challenging obstacles, especially for academic centers not equipped with virus production facilities.12,13
The presented work demonstrates as proof of principle that CAR19 cells can be efficiently manufactured via transfection with abiogenically prepared transposon/transposase vectors. The described procedure enables accurate control of VCN per cell while providing sufficient yield of CAR19 T cells for therapeutic purposes. The control of VCN is achieved by a “transposon dilution technique” that enables CAR19 T cell generation with VCN below 3, not only after modification via PCR DNA/mRNA but also after traditional modification with plasmids encoding the transposon and transposase. The in vitro expansion of CAR19 T cells is performed in cytokines interleukin (IL)-4, IL-7, and IL-21, similarly, as we have already described11,14 to preserve an early memory phenotype to enable cultivation for up to 21 days. The use of PCR DNA/mRNA has no significant adverse effects on the CAR T cell memory phenotype or effector activity compared to the transfection with plasmids. Notably, the generation of transposon DNA by preparative PCR and the generation of transposase mRNA are both rapid and straightforward procedures with no relevant biohazard issues. The simplicity and the speed of vector preparation via this method might enable faster clinical testing of novel types of CAR T cells.
A similar method of T cell transfection was tested by Bishop et al.15 The authors attempted to generate CAR T cells by electroporation of in vitro-amplified “doggy bone” DNA (dbDNA) combined with dbDNA encoding the PB transposase. Our data similarly demonstrate that enzymatically prepared genetic vectors represent a feasible alternative. The transposon copy number can be accurately controlled by a simple dilution of the PCR DNA without adverse effects on the cultivation yield.
Another experimental non-viral technique used to produce CAR T cells is based on CRISPR-Cas9 genome editing. CRISPR-Cas9 enables the integration of the transgene into a precise locus within the genome. It has been shown that the insertion of CAR transgene into the TCR locus disrupts the expression of endogenous TCR, which leads to improved function of CAR T cells by reducing the tonic TCR signaling.16 However, CRISPR-Cas9 has significant off-target effects and produces chromosomal translocations.6 Additionally, the procedure for T cell editing via CRISPR-Cas9 is far more complex than a modification via transposons and cannot be as quickly GMP certified.17
Lentiviral vectors are currently the gold standard for genetic engineering of T cells, and their safety was reliably proven in thousands of patients. This type of vector is especially suited for large-scale pharmaceutical processes where costs, biohazard, and time are not an issue. However, their use for small-scale generation of experimental phase 1 products is problematic. The presented technique might fill this gap and provide a rapid methodology for clinical testing of novel types of CAR constructs with unproven medical efficiency. Undeniably, the clinical experience with transposon vectors is insufficient, and thus the likelihood of oncogenesis induced by transposons cannot be reliably determined at this moment.
Moreover, animal studies can hardly answer such complex questions. On the other hand, mature T cells are inherently resistant to malignant transformation, compared to, for example, undifferentiated hematopoietic precursors, or B cells. In our experience, pharmaceutical regulatory agencies require additional quality control (QC) tests such as mapping of integration sites and monitoring of T cell clonality for approval of clinical trials with transposon-engineered T cells.
In summary, the manufacture of clinical-grade CAR T cells via lentiviral or retroviral transfection is a complicated and expensive task primarily resulting from GMP rules regulating the preparation of viral vectors. Currently, the major bottleneck is the difficulty in obtaining the GMP-grade viral vectors within a reasonable time. Enzymatically prepared transposon vectors are surprisingly highly efficient for small-scale manufacturing of this type of therapeutical product. In our opinion, the development of an easily certifiable GMP-grade method of T cell transfection is critical for the rapid development of T cell-based therapies in the future.
Materials and methods
Preparation of DNA and mRNA amplicons and plasmid cloning
The mRNA encoding the hyperactive PB transposase (hyPbase) was prepared by in vitro transcription with the HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs). To increase the stability of mRNA, we used 3′-O-Me-m7G (5′) PPP(5′) G RNA cap structure analog at a concentration of 8 mM (New England Biolabs). Before an in vitro transcription, the pSTI plasmid template encoding the hyPBase, or GFP, was linearized via BspQI (New England Biolabs) and gel-purified. The reaction was carried out for 3 h at 37°C using 1 μg of linearized plasmid template per 50-μL reaction. Prepared mRNA was purified with the MEGAclear Transcription Clean-Up Kit (Thermo Fisher Scientific) and analyzed with the Experion Automated Electrophoresis System (Bio-Rad Laboratories). The yield was ∼100 μg per one 50-μL reaction.
The transposon containing the CAR19 transgene was prepared commercially by preparative PCR (LinearX, USA) using M13 fwd and M13 rev primers and a Taq DNA polymerase, followed by a purification step on a chromatographic column. The declared purity of the PCR product was >99%. Furthermore, no mutations in the coding sequence were detected by Sanger sequencing. Plasmids were amplified in E. coli and purified by QIAGEN plasmid kits.
T cell cultivation and electroporation
PBMCs were separated from buffy coats obtained from healthy donors with Ficoll density gradient centrifugation (Ficoll-Paque, GE Healthcare). All donors gave written informed consent in accordance with Helsinki ethical guidelines. Cells were cultivated at 37°C in 5% CO2 in CellGenix GMP DC medium (CellGenix) supplemented with heat-inactivated 10% fetal bovine serum (Thermo Fisher Scientific), penicillin/streptomycin 100 U/mL (Thermo Fisher Scientific), and cytokines IL-4 (20 ng/mL), IL-7 (10 ng/mL), and IL-21 (40 ng/ml), all from Miltenyi Biotec. For large-scale expansion, we used G-Rex10 flasks (Wilson Wolf). For electroporations, 1 × 107 PBMCs were resuspended in 100 μL of buffer T containing plasmid DNA or linear transposon+ mRNA. Cells were then electroporated with the Neon Electroporation System (Thermo Fisher Scientific, USA) by a single 20-ms/2,300-V pulse and then were transferred to complete cell media. Next day, cells were polyclonally stimulated with TransAct reagent (anti-CD3/CD28, Miltenyi Biotec, Germany) and expanded for 21 days in the presence of IL-4, IL-7, and IL-21, identically as already described.13
Functional assays and flow cytometry
The functionality of CAR19 T cells was tested by FACS-based cytotoxic assay against Ramos B cellsþ. Briefly, Ramos cells were labeled with 0.5 μM CFSE (Thermo Fisher Scientific) in PBS for 10 min at room temperature, washed in CellGenix medium, and mixed with target cells at a ratio of 3:1 and 1:1. After 24-h incubation, cells were harvested, labeled with DAPI, and analyzed by FACS to determine the percentage of dead Ramos cells.
To determine the production of IFN-γ, we similarly co-cultivated CAR19 T cells with Ramos B cells at a 1:1 ratio overnight. The production of IFN-γ was determined in supernatants with the ELISA MAX Deluxe Set Kit (BioLegend). The absorbance at 450 nm was measured on an ELISA reader (Tecan Infinite 200 Microplate Reader). The IFN-γ concentration was calculated according to the diluted IFN-γ standard calibration curve in the GraphPad Prism software.
To determine the immunophenotype of CAR19 T cells, the cells were washed in 1× PBS, stained with fluorochrome-conjugated monoclonal antibodies (mAbs) in FACS buffer (1% BSA, 0.1% sodium azide, 1× PBS) for 30 min on ice, washed with PBS, and resuspended in FACS buffer containing DAPI 100 ng/mL (Merck). In the case of indirect labeling, the primary antibody was washed out with PBS, and cells were then stained with fluorochrome-conjugated secondary antibody for 30 min on ice. The expression of CAR19 transgene was determined with Alexa Fluor 647-conjugated F(ab′)2 fragment of Goat Anti-Mouse immunoglobulin G (IgG) (Jackson ImmunoResearch). This polyclonal antibody reacts with mouse anti-CD19 scFv. The immunophenotype of CAR T cell was determined similarly as already described.13 First, cells were stained either with Alexa Fluor 647- or Alexa Fluor 488-labeled F(ab′)2 fragment of Goat Anti-Mouse IgG (Jackson ImmunoResearch, UK) to detect the CAR transgene. Cells were then washed twice and blocked with 10% mouse serum, followed by staining with fluorescently labeled mouse mAbs specific to human antigens: CD45RA-BUV737 (clone HI100), CD62L-BV650 (clone DREG-56), and CD3-BV786 (clone UCHT1) were purchased from BD Biosciences (USA). CD4-qDOT605 (clone S3.5) and CD28-PE-Cy7 (clone CD28.2) were from BioLegend (USA). CD8-AF700 (clone MEM-31) and CD27-PE-Dy590 (clone LT27) were from EXBIO. Live cells were identified with the Fixable Blue Dead Cell Stain Kit (Thermo Fischer Scientific, USA). Staining specificity was extensively tested to rule out possible binding of mouse antibodies to cells via goat anti-mouse Ab used to detect CAR. Cells were analyzed with BD LSRFortessa (BD Biosciences), and the data were processed with FlowJo software.
Duplex digital droplet PCR
To determine the copy number of CAR19 transgene, the genomic DNA was isolated with the QIAamp DNA Mini Kit (QIAGEN). The PrimerQuest Tool online software (ITD, USA) was used to design primers and probes listed in Table 1. Sequences of primers and double-quenched probes for the human albumin gene that served as a reference gene were previously described.18 Duplex PCR reactions contained ddPCR Supermix for Probes (no dUTP) (cat. no. 186-3024, Bio-Rad Laboratories, USA), 900 nM of each primer pair, 250 nM of each FAM- and HEX-labeled probe, and 30–40 ng of DNA template. The reaction mix was split into ∼20,000 droplets with a QX200 Droplet Generator (Bio-Rad Laboratories). The PCR was performed on a C1000 Touch Thermal Cycler (Bio-Rad) using the following amplification conditions: 10 min at 95°C, 45 cycles of a two-step of 30 s at 94°C and 60 s at 54°C, ending with a final hold of 10 min at 98°C for droplet stabilization and cooling to 4°C. Droplets were counted on/divided by a QX200 droplet reader based on their fluorescence amplitude into positive or negative. Data were analyzed with QuantaSoft Analysis Pro version 1.0.596 software (Bio-Rad), including automatic Poisson distribution. The VCN was determined as the ratio of CAR copies/albumin copies × 2 and divided by the fraction of CAR+ T cells in the sample.
Table 1.
The sequences of primers and probes
| CAR19 | forward | 5′ atcggatatcgtgatgacac 3′ |
| reverse | 5′ gactagacctgcaagagatg 3′ | |
| probe | 5′ FAM-ctctccctgcctgtcagtcttgga-BHQ1 3′ | |
| PBASE | forward | 5′ CCCTGAACATCGTGAGAA 3′ |
| reverse | 5′ ATCTCGTCGGTGAAGAAC 3′ | |
| probe | 5′ FAM-CCCACCAGGATG TGCAGGAACATCT-BHQ1 3′ |
|
| Albumin | forward | 5′ TGAAACATACGTTCCCAAAGAGTTT 3′ |
| reverse | 5′ CTCTCCTTCTCAGAAAGTGTGCATAT 3′ | |
| probe | 5′ HEX-TGCTGAAAC-ZEN-ATTCACCTTCCATGCAGAT-IABkFQ 3′ |
DNA sequence of the CAR19 transposon
(M13F)GTAAAACGACGGCCAGTGAGCGCGCGTAATACGACTCACTATAGGGCGAATTGGGGCGCGCCATTCTAGATTAACCCTAGAAAGATAGTCTGCGTAAAATTGACGCATGCATTCTTGAAATATTGCTCTCTCTTTCTAAATAGCGCGAATCCGTCGCTGTGCATTTAGGACATCTCAGTCGCCGCTTGGAGCTCCCGTGAGGCGTGCTTGTCAATGCGGTAAGTGTCACTGATTTTGAACTATAACGACCGCGTGAGTCAAAATGACGCATGATTATCTTTTACGTGACTTTTAAGATTTAACTCATACGATAATTATATTGTTATTTCATGTTCTACTTACGTGATAACTTATTATATATATATTTTCTTGTTATAGATATCAACTAGAATGCTAGCCTCGAGGGCCTCCGCGCCGGGTTTTGGCGCCTCCCGCGGGCGCCCCCCTCCTCACGGCGAGCGCTGCCACGTCAGACGAAGGGCGCAGCGAGCGTCCTGATCCTTCCGCCCGGACGCTCAGGACAGCGGCCCGCTGCTCATAAGACTCGGCCTTAGAACCCCAGTATCAGCAGAAGGACATTTTAGGACGGGACTTGGGTGACTCTAGGGCACTGGTTTTCTTTCCAGAGAGCGGAACAGGCGAGGAAAAGTAGTCCCTTCTCGGCGATTCTGCGGAGGGATCTCCGTGGGGCGGTGAACGCCGATGATTATATAAGGACGCGCCGGGTGTGGCACAGCTAGTTCCGTCGCAGCCGGGATTTGGGTCGCAGTTCTTGTTTGTGGATCGCTGTGATCGTCACTTGGTGAGTAGCGGGCTGCTGGGCTGGCCGGGGCTTTCGTGGCCGCCGGGCCGCTCGGTGGGACGGAGGCGTGTGGAGAGACCGCCAAGGGCTGTAGTCTGGGTCCGCGAGCAAGGTTGCCCTGAACTGGGGGTTGGGGGGAGCGCAGCAAAATGGCGGCTGTTCCCGAGTCTTGAATGGAAGACGCTTGTGAGGCGGGCTGTGAGGTCGTTGAAACAAGGTGGGGGGCATGGTGGGCGGCAAGAACCCAAGGTCTTGAGGCCTTCGCTAATGCGGGAAAGCTCTTATTCGGGTGAGATGGGCTGGGGCACCATCTGGGGACCCTGACGTGAAGTTTGTCACTGACTGGAGAACTCGGTTTGTCGTCTGTTGCGGGGGCGGCAGTTATGGCGGTGCCGTTGGGCAGTGCACCCGTACCTTTGGGAGCGCGCGCCCTCGTCGTGTCGTGACGTCACCCGTTCTGTTGGCTTATAATGCAGGGTGGGGCCACCTGCCGGTAGGTGTGCGGTAGGCTTTTCTCCGTCGCAGGACGCAGGGTTCGGGCCTAGGGTAGGCTCTCCTGAATCGACAGGCGCCGGACCTCTGGTGAGGGGAGGGATAAGTGAGGCGTCAGTTTCTCTGGTCGGTTTTATGTACCTATCTTCTTAAGTAGCTGAAGCTCCGGTTTTGAACTATGCGCTCGGGGTTGGCGAGTGTGTTTTGTGAAGTTTTTTAGGCACCTTTTGAAATGTAATCATTTGGGTCAATATGTAATTTTCAGTGTTAGACTAGTAAATTGTCCGCTAAATTCTGGCCGTTTTTGGCTTTTTTGTTAGACGGATCCATGAATTCGCCAGCatggccctgcctgtgacagccctgctgctgcctctggctctgctgctgcatgccgctagacccacgcgtgaagtgcagctgcagcagtctggacctgagctggtaaagcctggggcttcagtgaagatgtcctgcaaggcttctggatacacattcactagctatgttatgcactgggtgaagcagaagcctgggcagggccttgagtggattggatatgttaatccttacaatgatggtactaagtacaatgagaagttcaaaggcaaggccacactgacttcagacaaatcctccagcacagcctacatggagctcagcagcctgacctctgaggactctgcggtctattactgtgcaagagggccttattactacggtagtagcccctttgactactggggccaagggaccacggtcaccgtctccggaggtggcggttcaggcggtggcggatccggcggtggcggatccggcggtggcggatcggatatcgtgatgacacagtctccactctccctgcctgtcagtcttggagatcaagcctccatctcttgcaggtctagtcagagccttgaaaacagtaatggaaacacctatttgaactggtacctccagaaaccaggccagtctccacagctcctgatctacagggtttccaaccgattttctggggtcctagacaggttcagtggtagtggatcagggacagatttcacactgaaaatcagcagagtggaggctgaggatttgggagtttatttctgcctccaagttacacatgtccctcccacgttcggtgctgggaccaagctcgagatcaaacgtactagtcccacaaccacccctgcccctagacctccaacacccgcccctacaatcgccagccagcctctgtctctgaggcccgaggcttgtagacctgctgcaggcggagccgtgcacaccagaggactggatttcgcctgcgacatctacatctgggcccctctggccggcacatgcggagtgctgctgctgagcctcgtgatcaccctgtactgcaaccaccggaaccggcggagagtgaagcggggcagaaagaagctgctgtacatcttcaagcagcccttcatgcggcccgtgcagaccacccaggaagaggacggctgctcctgcagattccccgaggaagaagaaggcggctgcgagctgagagtgaaattcagcagatccgccgacgcccctgcctaccagcagggacagaaccagctgtacaacgagctgaacctgggcagacgggaagagtacgacgtgctggacaagcggagaggcagggaccctgagatgggcggaaagccccagcggagaaagaacccccaggaaggcctgtataacgaactgcagaaagacaagatggccgaggcctacagcgagatcggaatgaagggcgagcggagaagaggcaagggccacgatggactgtatcagggcctgagcaccgccaccaaggacacctatgacgccctgcacatgcaggccctgccccccagatgaTGAGAATTCCGACTGTGCCTTCTAGTTGCCAGCCATCTGTTGTTTGCCCCTCCCCCGTGCCTTCCTTGACCCTGGAAGGTGCCACTCCCACTGTCCTTTCCTAATAAAATGAGGAAATTGCATCGCATTGTCTGAGTAGGTGTCATTCTATTCTGGGGGGTGGGGTGGGGCAGGACAGCAAGGGGGAGGATTGGGAAGACAATAGCAGGCATGCTGGGGATGCGGTGGGCTCTATGGGTCGACATACTAGTTAAAAGTTTTGTTACTTTATAGAAGAAATTTTGAGTTTTTGTTTTTTTTTAATAAATAAATAAACATAAATAAATTGTTTGTTGAATTTATTATTAGTATGTAAGTGTAAATATAATAAAACTTAATATCTATTCAAATTAATAAATAAACCTCGATATACAGACCGATAAAACACATGCGTCAATTTTACGCATGATTATCTTTAACGTACGTCACAATATGATTATCTTTCTAGGGTTAATCTAGTATACGCGTATGCGGCCGCTTAATTAATCCAGCTTTTGTTCCCTTTAGTGAGGGTTAATTGCGCGCTTGGCGTAATCATGGTCATAGCTGTTTCCTG (M13R)
Amino acid sequence of CAR19 construct
MALPVTALLLPLALLLHAARPTREVQLQQSGPELVKPGASVKMSCKASGYTFTSYVMHWVKQKPGQGLEWIGYVNPYNDGTKYNEKFKGKATLTSDKSSSTAYMELSSLTSEDSAVYYCARGPYYYGSSPFDYWGQGTTVTVSGGGGSGGGGSGGGGSGGGGSDIVMTQSPLSLPVSLGDQASISCRSSQSLENSNGNTYLNWYLQKPGQSPQLLIYRVSNRFSGVLDRFSGSGSGTDFTLKISRVEAEDLGVYFCLQVTHVPPTFGAGTKLEIKRTSPTTTPAPRPPTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFACDIYIWAPLAGTCGVLLLSLVITLYCNHRNRRRVKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPQRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR∗
Acknowledgments
This work was supported by grants OPVVV CZ.02.1.01/0.0/0.0/16_025/0007428 and AZV 19-08-00147.
Author contributions
I.K., P.P., and M.Š. designed and performed experiments; H.Ž., V.Š., and K.Š. analyzed data; P.O. conceptualized the study and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Shi H., Sun M., Liu L., Wang Z. Chimeric antigen receptor for adoptive immunotherapy of cancer: latest research and future prospects. Mol. Cancer. 2014;13:219. doi: 10.1186/1476-4598-13-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y., Huo Y., Yu L., Wang J. Quality Control and Nonclinical Research on CAR-T Cell Products: General Principles and Key Issues. Engineering (Beijing) 2019;5:122–131. [Google Scholar]
- 3.Ausubel L.J., Hall C., Sharma A., Shakeley R., Lopez P., Quezada V., Couture S., Laderman K., McMahon R., Huang P. Production of CGMP-grade lentiviral vectors. Bioprocess Int. 2012;10:32–43. [PMC free article] [PubMed] [Google Scholar]
- 4.Singh H., Manuri P.R., Olivares S., Dara N., Dawson M.J., Huls H., Hackett P.B., Kohn D.B., Shpall E.J., Champlin R.E., Cooper L.J. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manuri P.V.R., Wilson M.H., Maiti S.N., Mi T., Singh H., Olivares S., Dawson M.J., Huls H., Lee D.A., Rao P.H. piggyBac transposon/transposase system to generate CD19-specific T cells for the treatment of B-lineage malignancies. Hum. Gene Ther. 2010;21:427–437. doi: 10.1089/hum.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stadtmauer E.A., Fraietta J.A., Davis M.M., Cohen A.D., Weber K.L., Lancaster E., Mangan P.A., Kulikovskaya I., Gupta M., Chen F. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367:eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang X., Guo H., Tammana S., Jung Y.C., Mellgren E., Bassi P., Cao Q., Tu Z.J., Kim Y.C., Ekker S.C. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells. Mol. Ther. 2010;18:1803–1813. doi: 10.1038/mt.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Z., Maiti S., Huls H., Singh H., Olivares S., Mátés L., Izsvák Z., Ivics Z., Lee D.A., Champlin R.E., Cooper L.J. The hyperactive Sleeping Beauty transposase SB100X improves the genetic modification of T cells to express a chimeric antigen receptor. Gene Ther. 2011;18:849–856. doi: 10.1038/gt.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deniger D.C., Yu J., Huls M.H., Figliola M.J., Mi T., Maiti S.N., Widhopf G.F., 2nd, Hurton L.V., Thokala R., Singh H. Sleeping Beauty transposition of chimeric antigen receptors targeting receptor tyrosine kinase-like orphan receptor-1 (ROR1) into diverse memory T-cell populations. PLoS ONE. 2015;10:e0128151. doi: 10.1371/journal.pone.0128151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yusa K., Zhou L., Li M.A., Bradley A., Craig N.L. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. USA. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ptáčková P., Musil J., Štach M., Lesný P., Němečková Š., Král V., Fábry M., Otáhal P. A new approach to CAR T-cell gene engineering and cultivation using piggyBac transposon in the presence of IL-4, IL-7 and IL-21. Cytotherapy. 2018;20:507–520. doi: 10.1016/j.jcyt.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Poorebrahim M., Sadeghi S., Fakhr E., Abazari M.F., Poortahmasebi V., Kheirollahi A., Askari H., Rajabzadeh A., Rastegarpanah M., Linē A., Cid-Arregui A. Production of CAR T-cells by GMP-grade lentiviral vectors: latest advances and future prospects. Crit. Rev. Clin. Lab. Sci. 2019;56:393–419. doi: 10.1080/10408363.2019.1633512. [DOI] [PubMed] [Google Scholar]
- 13.Wiesinger M., März J., Kummer M., Schuler G., Dörrie J., Schuler-Thurner B., Schaft N. Clinical-Scale Production of CAR-T Cells for the Treatment of Melanoma Patients by mRNA Transfection of a CSPG4-Specific CAR under Full GMP Compliance. Cancers (Basel) 2019;11:1198. doi: 10.3390/cancers11081198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Štach M., Ptáčková P., Mucha M., Musil J., Klener P., Otáhal P. Inducible secretion of IL-21 augments anti-tumor activity of piggyBac-manufactured chimeric antigen receptor T cells. Cytotherapy. 2020;22:744–754. doi: 10.1016/j.jcyt.2020.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Bishop D.C., Caproni L., Gowrishankar K., Legiewicz M., Karbowniczek K., Tite J., Gottlieb D.J., Micklethwaite K.P. CAR T Cell Generation by piggyBac Transposition from Linear Doggybone DNA Vectors Requires Transposon DNA-Flanking Regions. Mol. Ther. Methods Clin. Dev. 2020;17:359–368. doi: 10.1016/j.omtm.2019.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J.C., Hamieh M., Cunanan K.M., Odak A., Gönen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basar R., Daher M., Uprety N., Gokdemir E., Alsuliman A., Ensley E., Ozcan G., Mendt M., Hernandez Sanabria M., Kerbauy L.N. Large-scale GMP-compliant CRISPR-Cas9-mediated deletion of the glucocorticoid receptor in multivirus-specific T cells. Blood Adv. 2020;4:3357–3367. doi: 10.1182/bloodadvances.2020001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Velden V.H.J., Cazzaniga G., Schrauder A., Hancock J., Bader P., Panzer-Grumayer E.R., Flohr T., Sutton R., Cave H., Madsen H.O., European Study Group on MRD detection in ALL (ESG-MRD-ALL) Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007;21:604–611. doi: 10.1038/sj.leu.2404586. [DOI] [PubMed] [Google Scholar]